Abstract
Owing to the urgent need for therapeutic interventions against the SARS-coronavirus 2 (SARS-CoV-2) pandemic, we employed an in silico approach to evaluate the SARS-CoV-2 inhibitory potential of newly synthesized imidazoles. The inhibitory potentials of the compounds against SARS-CoV-2 drug targets - main protease (Mpro), spike protein (Spro) and RNA-dependent RNA polymerase (RdRp) were investigated through molecular docking analysis. The binding free energy of the protein-ligand complexes were estimated, pharmacophore models were generated and the absorption, distribution, metabolism, excretion and toxicity (ADMET) properties of the compounds were determined. The compounds displayed various levels of binding affinities for the SARS-CoV-2 drug targets. Bisimidazole C2 scored highest against all the targets, with its aromatic rings including the two imidazole groups contributing to the binding. Among the phenyl-substituted 1H-imidazoles, C9 scored highest against all targets. C11 scored highest against Spro and C12 against Mpro and RdRp among the thiophene-imidazoles. The compounds interacted with HIS 41 - CYS 145 and GLU 288 – ASP 289 – GLU 290 of Mpro, ASN 501 of Spro receptor binding motif and some active site amino acids of RdRp. These novel imidazole compounds could be further developed as drug candidates against SARS-CoV-2 following lead optimization and experimental studies.
Keywords: Drug discovery, Drug target, Molecular docking, Coronavirus
1. Introduction
SARS-CoV-2 (severe acute respiratory syndrome - coronavirus 2) has since been declared a pandemic by the World Health Organization (WHO) and it requires immediate health intervention.1 , 2 Presently, there are limited effective treatment options and targeted therapeutics approved for the clinical management of SARS-CoV-2.3 , 4 SARS-CoV-2 is a member of Beta-coronaviruses which is similar to Severe Acute Respiratory Syndrome Human coronavirus (SARS-CoV-1) and the Middle-East Respiratory Syndrome Human coronavirus (MERS CoV).5 Coronavirus is a positive-sense, single-stranded RNA virus comprising of over 30,000 nucleotides. The replication and transcription of the viral genome require the replicase gene responsible for encoding overlapping polyproteins, pp1a, and pp1ab.6 SARS-CoV-2 attacks the respiratory tract and has become a lethal viral respiratory disease with symptoms ranging from breathing difficulties, sore throat, high fever, diarrhea, cough to multiple organ failure and ultimately death.7
The infection occurs by the binding of SARS-CoV-2 spike protein (Spro) to the Angiotensin-Converting Enzyme-2 (ACE-2) in the alveoli of the host.8 Upon entry, viral RNA translation results in the synthesis of polyproteins responsible for the production of new virions from single-stranded RNA with the aid of RNA-dependent RNA polymerase (RdRp). These proteins are believed to represent primary druggable targets to contrast SARS-CoV-2 growth and replication.9, 10, 11 Analysis of several viruses have also shown that the viral protease is a common target for antiviral drug development,12, 13, 14 hence, the main protease (Mpro) of coronavirus could also be targeted due to its role in viral replication.15 , 16 Development of small molecule inhibitors of these protein target could therefore offer an effective treatment options against SARS-CoV-2.
Traditional methods of developing and discovering new therapeutic agents require rigorous scientific procedures, which consume time and cost. Computational tools, however, offer considerable promise for determining the affinities of small drug-like molecules for protein targets.17 This approach is a viable option for the designing and development of new drugs of biomedical interest.
Imidazoles are heterocyclic compounds with biological potency and proven pharmacological properties that include antiparasitic,18, 19, 20 anti-inflammatory,21 anticancer,22 antifungal,23 antibacterial,24 antimalarial,25 antitubercular,26 and antiviral.27 They are also reported to be carboxypeptidase inhibitors, β-lactamase inhibitors, heme oxygenase inhibitors and 20-HETE synthase inhibitors.18 , 19 , 28 , 29 The Imidazole ring is a nitrogen-containing heterocyclic aromatic molecule with different synthetic strategies and pharmaceutical importance.30 , 31 Thus, the incorporation of imidazole nucleus and electron-rich imidazole ring have become essential in the design, formulation, and development of imidazole-based drugs by pharmaceutical industries. The antiproliferative properties of imidazole derivatives have encouraged their use and synthesis for antiviral research.22 Their ability to attack enzymes and/or proteins critical to the viral life cycle is also a major contributing factor to their antiviral properties.32 , 33
In a previous study, we reported the synthesis and in vitro anti-parasitic activities of a series of imidazole derivatives.19 Owing to the urgency for therapeutic intervention against the coronavirus, we employed the computational approach for evaluating the therapeutic potential of these imidazole compounds against SARS-CoV-2.
2. Materials and methods
2.1. Imidazole derivatives
The test compounds which are mainly imidazole derivatives (Fig. 1 ) were synthesized and characterized as previously described.19 , 34 , 35 Compounds C1 to C5 are bisimidazoles, C6 to C10 are phenyl-substituted 1H-imidazoles and C11 to C14 are thiophene-imidazoles.
Fig. 1.
Structures of imidazole derivatives.
2.2. Ligand preparation
The canonical SMILES of compounds C1 to C14 were converted to PDB format using Chimera 1.14 while the structure data file (SDF) format of standard ligands: Benzyl (Z, 4S)-4-[[(2S)-4-methyl-2-[[(2S)-3-methyl-2-[[(2S)-2-[(5-methyl-1,2-oxazole-3-carbonyl)amino]propanoyl]amino]butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate (inhibitor N3), Pravastatin and remdesivir were obtained from PubChem database. The SDF format of compounds and standard ligands were uploaded to PyRx software and converted to PDBQT format using the OpenBabel plugin. The output files were minimized to obtain the minimum energy for the ligand docking.
2.3. Protein preparation
The crystal structures of the SARS-CoV-2 target proteins were obtained from the RCSB protein data bank (PDB). Main protease (Mpro: 6LU7) was in complex with inhibitor N3, obtained through X-RAY diffraction method, with a resolution 2.16 Å, R-Value free 0.235, R-Value work 0.202 and R-Value observed 0.204.36 Spike receptor-binding domain in complex with its receptor ACE2 (Spro: 6LZG) was through X-RAY diffraction, resolution 2.50 Å, R-Value free 0.216, R-Value work 0.188 and R-Value observed 0.190.37 RNA-dependent RNA polymerase in complex with cofactors (RdRp: 6M71) was obtained through electron microscopy with a Resolution of 2.90 Å.38
The PDB format of the structures were uploaded to Chimera 1.14 workspace and the non-standard residues including ions, water and bounded ligands were first removed. The proteins were structurally minimized at 100 steepest descent steps, 0.02 steepest descent steps size (Ᾰ), 10 conjugate gradient steps, 0.02 conjugate gradient steps size (Ᾰ), and 10 update intervals, using the structure editing wizard Chimera 1.14. Furthermore, solvents were removed, hydrogen bonds were added, charges were assigned using Gasteiger force field and histidine was set for the protonation state. Every available selenomethione (MSE) were changed to methionine (MET), bromo-UMP (5BU) to UMP (U), methylselenyl-dUMP (UMS) to UMP (U) and methylselenyl-dCMP (CSL) to CMP (C). The prepared proteins were uploaded to the PyRx software for molecular docking analysis.
2.4. Molecular docking
Molecular docking of the prepared ligands and proteins were performed using AutoDock vina in the PyRx workspace. Grid space was set by targeting important amino acid residues selected through literature39 and from UniProtKB. Grid box size x = 52.07 Å, y = 65.24 Å and z = 58.07 Å and grid centre dimensions x = −22.94, y = 14.30, z = 58.65 were set for Mpro: 6LU7; grid box size x = 43.86 Å, y = 46.19 Å and z = 58.59 Å and grid center dimensions x = −32.42, y = 30.30, z = 22.14 for Spro: 6LZG; and x = 78.79 Å, y = 83.87 Å, z = 84.28 Å and x = 121.71, y = 122.39, z = 113.69 respectively for RdRp: 6M71. The output files were uploaded to Chimera 1.14 workspace for post docking analysis and preparation of the 3D views of the protein-ligand complex. The 2D views of the molecular interactions were generated using UCSF Chimera 1.14 and Discovery Studio 2020.
2.5. Binding free energy calculation
The binding free energy of the protein-ligand complexes was employed to determine the stability of their complexes via Prime MM-GBSA program (Schrödinger suite version 20,018–4). Before-hand, the imidiazole derivatives were prepared by ligprep, while the respective proteins were prepared using the protein preparation wizard, methods as previously described.40 The active sites of the proteins were predicted by sitemap. Subsequently, the compounds were docked with proteins using glide extra precision (XP) docking. The Prime MM-GBSA panel was used to calculate binding free energy for ligand–protein complexes using the MM-GBSA technology available with Prime.41 OPLS3 force field was selected and VSGB was used as the continuum solvent model. Other options were set as default.
2.6. Receptor-ligand complex pharmacophore modelling
The highest-ranking compound based on binding affinity against the target proteins was selected to develop a receptor-ligand complex pharmacophore model using the PHASE module of Schrödinger suite. The auto (E-pharmacophore) method was used to generate ligand-based pharmacophore hypotheses. The maximum number of features to be generated was set at 7, minimum feature–feature distance was at 2.00, minimum feature–feature distance for feature of the same type at 4.00 and donors as vectors.
2.7. ADMET predictions
The absorption, distribution, metabolism, excretion and toxicity (ADMET) properties (lipophilicity (log Po/w), water solubility (log S), druglikeness, bioavailability score, pharmacokinetics and toxicity profile) of the test compounds were determined using in silico integrative model predictions at the SwissADME, ADMETsar and ProTox-II online servers. Lipophilicity was measured using the partition coefficient between n-octanol and water (log Po/w) according to XLOGP, WLOGP, MLOGP, iLOGP and SILICOS-IT predictive models. The arithmetic mean of the values predicted by the five models is the consensus log P.42 Water solubility was estimated as the logarithm of the molar solubility in water (log S) using the SILICOS-IT predictive model.42 Druglikeness was according to the rule-based filters namely – Lipinski and Verber.43 , 44 Pharmacokinetic properties predicted include: skin permeation, gastrointestinal (GI) absorption permeation, blood–brain (BBB) permeation, substrate and inhibitor of permeability glycoprotein (P-gp) and cytochrome p450 (CYP) respectively. Toxicity properties considered are carcinogenicity, eye corrosion, eye irritation, Ames mutagenesis and hepatotoxicity.
3. Results
3.1. Binding affinities and stability of test compounds with SARS-CoV-2 drug targets
The compounds exhibited various levels of binding affinities with Gibbs free energy (ΔG kcal/mol) ranging from −10.8 to −6.5 for Mpro (6LU7), −10.2 to −6.6 for Spro (6LZG) and −11.4 to −6.3 for RdRp (6M71). Among the three classes of imidazole derivatives, the bisimidazole compound C2 exhibits the highest binding affinity for all the drug targets (−10.8, −10.2, −11.4 kcal/mol for Mpro, Spro and RdRp respectively). Among the phenyl-substituted 1H-imidazoles, C9 has the highest binding affinity for Mpro (−8.0 kcal/mol), Spro (−7.4 kcal/mol) and RdRp (−7.6 kcal/mol). For the thiophene-imidazoles, C11 exhibits the highest binding affinity for Spro (−7.1 kcal/mol) and C12 for Mpro (−7.7 kcal/mol) and RdRp (−7.8 kcal/mol). Many of the test compounds including the 5 bisimidazoles gave binding affinities higher than those of the standard inhibitors (Table 1 ). The estimated binding free energy of the imidiazole derivatives with the protein targets are shown in Table 2 . The major energy contributors to the compounds binding were non-polar salvation terms (ΔGlipo), van der Waals (ΔGvdW), and covalent energy (ΔGcovalent).
Table 1.
Binding affinities (ΔG in kcal/mol) of test compounds for SARS-CoV-2 drug targets.
|
Compounds |
ΔG Energy (Kcal/mol) |
||
|---|---|---|---|
| Mpro (6LU7) |
Spro (6LZG) |
RdRp (6M71) |
|
| Standard ligands | |||
| N3 | −7.4 | ||
| Pravastatin | 6.3 | ||
| Remdesivir | −7.4 | ||
| Bisimidazoles | |||
| C1 | −8.6 | −8.6 | −9.1 |
| C2 | −10.8 | −10.2 | −11.4 |
| C3 | −7.6 | −7.9 | −9 |
| C4 | −10.3 | −10 | −8.4 |
| C5 | −7.9 | −6.7 | −8.2 |
| Phenyl-substituted 1H-imidazoles | |||
| C6 | −7.4 | −7.2 | −7.2 |
| C7 | −7.1 | −6.9 | −6.8 |
| C8 | −6.9 | −6.6 | −6.9 |
| C9 | −8 | −7.4 | −7.6 |
| C10 | −7.8 | −7.2 | −7.3 |
| Thiophene-imidazoles | |||
| C11 | −6.8 | −7.1 | −6.8 |
| C12 | −7.7 | −7.0 | −7.8 |
| C13 | −6.5 | −6.8 | −7 |
| C14 | −6.9 | −6.9 | −6.3 |
Table 2.
Binding free energy of the imidazole derivatives bound to proteins as calculated by MM-GBSA.
|
Compounds |
ΔGbind Energy (Kcal/mol) |
||
|---|---|---|---|
| Mpro (6LU7) |
Spro (6LZG) |
RdRp (6M71) |
|
| C1 | −53.26 | −31.18 | −76.23 |
| C2 | −64.23 | −35.30 | −45.43 |
| C3 | −41.05 | −37.42 | −27.45 |
| C4 | −33.56 | −29.43 | −14.30 |
| C5 | −26.56 | −37.36 | −45.76 |
| C6 | −42.42 | −17.04 | −33.47 |
| C7 | −36.17 | −44.98 | −21.54 |
| C8 | −53.29 | −29.70 | −17.25 |
| C9 | −41.07 | −28.97 | −57.22 |
| C10 | −36.17 | −58.63 | −56.55 |
| C11 | −44.88 | −50.772 | −25.32 |
| C12 | −38.07 | −29.270 | −67.25 |
| C13 | −39.78 | −28.097 | −53.56 |
| C14 | −36.58 | −32.47 | −38.63 |
3.2. Pharmacophore modelling
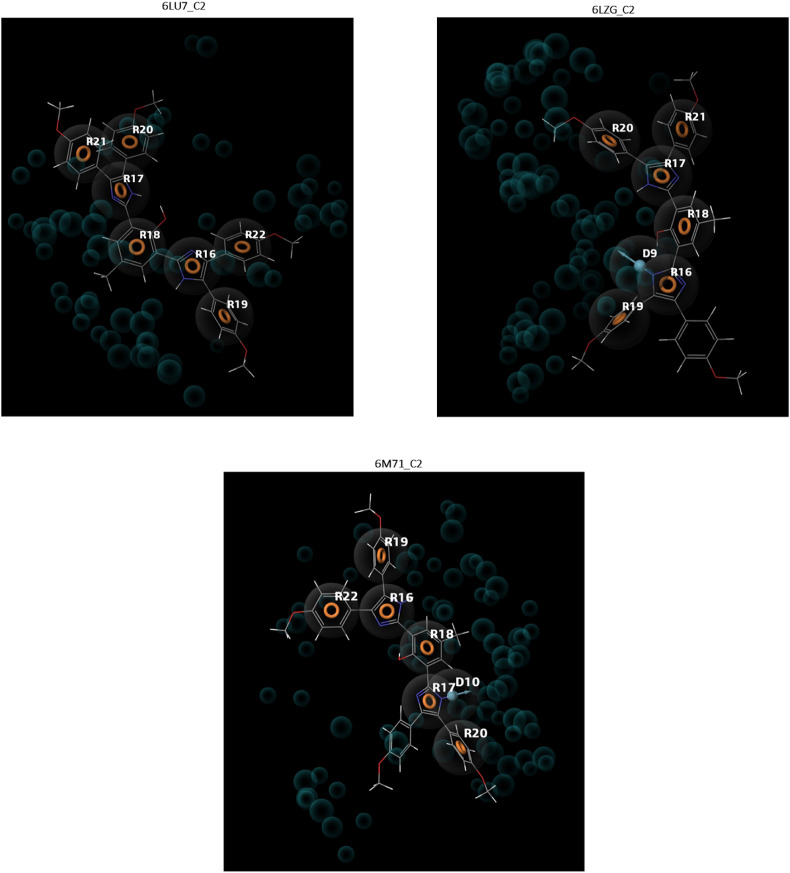
The pharmacophore models of the highest affinity compound (bisimidazole C2) are shown on Fig. 2 . The models showed that the two imidazole rings of the compound contributed to its binding affinity for the protein targets. Apart from forming two aromatic interactions with the three target proteins, one of the imidazole rings of the compound acts as a hydrogen bond donor to Spro and RdRp. The remaining five aromatic rings of Mpro and four of Spro and RdRp are all involved in the receptor-ligand binding.
Fig. 2.
Pharmacophore models of Bisimidazole (C2) on SARS-COV-2 main protease (6LU7), Spike protein (6LZG) and RNA-dependent RNA polymerase (6M71). R = Aromatic ring, D = Hydrogen bond donor.
3.3. ADMET profile
Table 3 shows the SwissADME predicted lipophilicity, water solubility, druglikeness and bioavailability scores of the compounds. The bisimidazole compounds have Log P values as high as 9.03 (C3) and 8.19 (C5), the least value being 4.37 (C2). The Log P values of the phenyl-substituted 1H-imidazoles ranged between 4.3 and 5.87 while those of the thiophene-imidazoles ranged between 4.02 and 6.15. The bisimidazoles C2 and C4 are moderately soluble while the remaining three are poorly soluble. The other two classes of imidazole compounds are either moderately soluble or soluble. For the druglikeness prediction, apart from C2 which violates only one rule, each of the bisimidazoles violates two Lipinski rules, while the remaining two classes of compounds violate only one rule. Besides, all the compounds pass the Veber's rule. For the bioavailability prediction, the bisimidazoles apart from C2 have a score of 0.17 while the remaining compounds score 0.55.
Table 3.
Predicted Lipophilicity (Log P), Water solubility (Log Sw), Druglikeness and Bioavailability scores of test compounds.
| Compounds | Molecular Weight | Consensus Log P | Log Sw (Silicos-IT) | Solubility Class | Lipinski #violations | Veber #violations | Bioavailability Score |
|---|---|---|---|---|---|---|---|
| C1 | 582.95 | 6.23 | −6.39 | Poorly soluble | 2 | 0 | 0.17 |
| C2 | 703.05 | 4.37 | −5.81 | Moderately soluble | 1 | 0 | 0.55 |
| C3 | 747.23 | 9.06 | −7.59 | Poorly soluble | 2 | 0 | 0.17 |
| C4 | 578.91 | 5.5 | −5.46 | Moderately soluble | 2 | 0 | 0.17 |
| C5 | 763.23 | 8.19 | −6.76 | Poorly soluble | 2 | 0 | 0.17 |
| C6 | 416.68 | 5.34 | −4.25 | Moderately soluble | 1 | 0 | 0.55 |
| C7 | 446.71 | 5.01 | −4.12 | Moderately soluble | 1 | 0 | 0.55 |
| C8 | 506.76 | 4.3 | −3.85 | Soluble | 1 | 0 | 0.55 |
| C9 | 389.61 | 4.8 | −3.76 | Soluble | 1 | 0 | 0.55 |
| C10 | 445.72 | 5.87 | −4.68 | Moderately soluble | 1 | 0 | 0.55 |
| C11 | 336.58 | 4.43 | −4.11 | Moderately soluble | 1 | 0 | 0.55 |
| C12 | 483.59 | 6.15 | −5.17 | Moderately soluble | 1 | 0 | 0.55 |
| C13 | 462.73 | 4.51 | −3.89 | Soluble | 1 | 0 | 0.55 |
| C14 | 321.52 | 4.02 | −3.18 | Soluble | 1 | 0 | 0.55 |
Table 4 shows the result of pharmacokinetics prediction of the test compounds. As highlighted in the table, skin permeation values (log Kp in cm/s) of the test compounds ranged from −6.8 (least permeant) to −1.02 (most permeant). Bisimidazoles C3 and C5 are the most skin permeant of all the compounds. All the test compounds apart from C3 and C5 possess high GI absorption potential and 8 of the compounds (1 bisimidazole, 4 phenyl-substituted 1H-imidazoles and 3 thiophene-imidazoles) displayed the ability to penetrate the blood–brain barrier. Compounds C3, C5 and C13 are substrates of Pgp. C3 and C5 were predicted to be inhibitors of CYP3A4, C11 an inhibitor of CYP2C9 and C14 an inhibitor of CYP1A2, CYP2C19, and CYP2C9. As shown on Table 5 , none of the test compounds has the tendency for carcinogenicity, mutagenesis, hepatotoxicity, eye corrosion, eye irritation and human either-a-go-go (hERG) inhibition.
Table 4.
Pharmacokinetics prediction output of test compounds.
| Compounds | GI absorption | Blood–brain permeant | Pgp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Skin permeation log Kp (cm/s) |
|---|---|---|---|---|---|---|---|---|---|
| C1 | High | No | No | No | No | No | No | No | −3.25 |
| C2 | High | No | No | No | No | No | No | No | −6.8 |
| C3 | Low | No | Yes | No | No | No | No | Yes | −1.02 |
| C4 | High | Yes | No | No | No | No | No | No | −3.83 |
| C5 | Low | No | Yes | No | No | No | No | Yes | −1.8 |
| C6 | High | Yes | No | No | No | No | No | No | −3.53 |
| C7 | High | Yes | No | No | No | No | No | No | −4.15 |
| C8 | High | Yes | No | No | No | No | No | No | −5.27 |
| C9 | High | Yes | No | No | No | No | No | No | −3.93 |
| C10 | High | No | No | No | No | No | No | No | −3.25 |
| C11 | High | Yes | No | No | No | Yes | No | No | −4.23 |
| C12 | High | No | No | No | No | No | No | No | −3.16 |
| C13 | High | Yes | Yes | No | No | No | No | No | −4.74 |
| C14 | High | Yes | No | Yes | Yes | Yes | No | No | −4.54 |
Table 5.
Toxicity profiles of imidazole molecules.
| Compounds | Carcinogenicity | Eye corrosion | Eye irritation | Ames mutagenesis | human either-a-go-go inhibition | Hepatotoxicity |
|---|---|---|---|---|---|---|
| C1 | – | – | – | – | – | – |
| C2 | – | – | – | – | – | – |
| C3 | – | – | – | – | – | – |
| C4 | – | – | – | – | – | – |
| C5 | – | – | – | – | – | – |
| C6 | – | – | – | – | – | – |
| C7 | – | – | – | – | – | – |
| C8 | – | – | – | – | – | – |
| C9 | – | – | – | – | – | – |
| C10 | – | – | – | – | – | – |
| C11 | – | – | – | – | – | – |
| C12 | – | – | – | – | – | – |
| C13 | – | – | – | – | – | – |
| C14 | – | – | – | – | – | – |
(−) = Inactive.
3.4. Post docking analysis of selected compounds
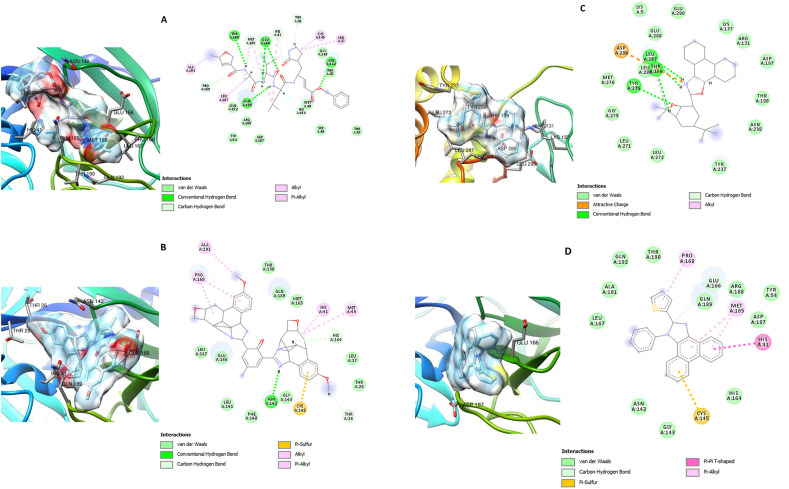
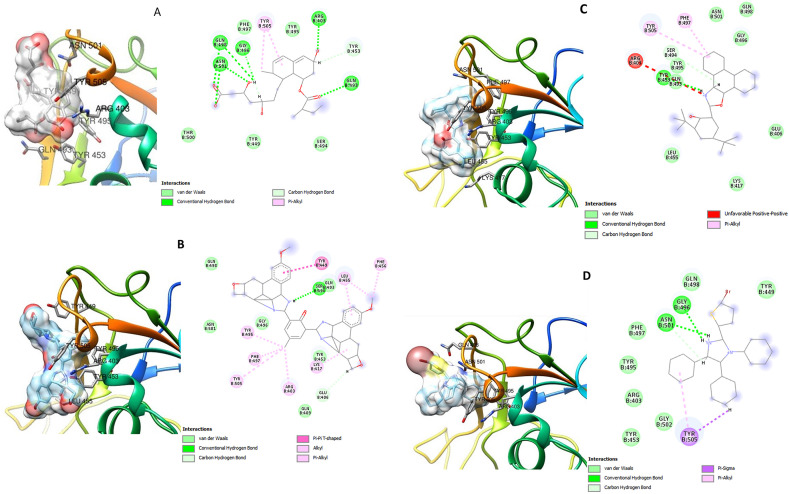
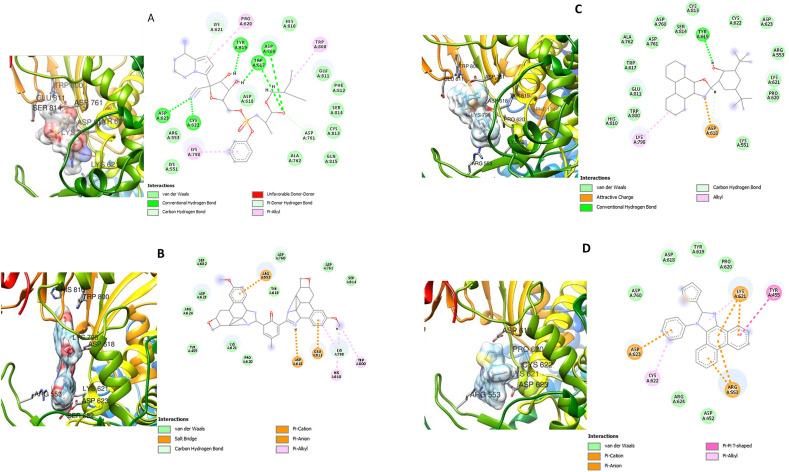
Fig. 3, Fig. 4, Fig. 5 show the three-dimensional (3D) and two-dimensional (2D) structures of the SARS-CoV-2 target proteins in complex with the standard inhibitors and the highest affinity compounds from each of the classes (bisimidazole C2 and phenyl-substituted 1H-imidazole C9 for Mpro, Spro and RdRp; thiophene-imidazole C11 for Spro and C12 with Mpro and RdRp).
Fig. 3.
3D (left) and 2D (right) views of the molecular interactions of amino-acid residues of Mpro (6LU7) with (A) N3 (B) C2 (C) C9 (D) C12.
Fig. 4.
3D (left) and 2D (right) views of the molecular interactions of amino-acid residues of Spro (6LZG) with (A) Pravastatin (B) C2 (C) C9 (D) C11.
Fig. 5.
3D (left) and 2D (right) views of the molecular interactions of amino-acid residues of RdRp (6M71) with (A) Remdesivir (B) C2 (C) C19 (D) C12.
C2 and C12 interacted with ASN 142, GLU 166, CYS 145, HIS 41 and other amino acid residues at the inhibitor (N3) binding site of SARS-CoV-2 Mpro. C9 interacted with residues like GLU 288, ASP 289 and GLU 290 at a different binding pocket on Mpro (Fig. 3). The SARS-CoV-2 Spro formed a complex with C2, C9, C11 and Pravastatin through various types of interactions with some amino acids residues of its receptor binding domain (RBD) (Fig. 4). ASN 501 of Spro formed van der Waals interaction with C2 and C9 (Fig. 3 B and C) and hydrogen bond interaction with Pravastatin and C11 (Fig. 3 A and D). Furthermore, C2, C9, C12 and remdesivir interacted with some amino acid residues at the binding pocket of SARS-CoV-2 RdRp including: ASP 618, TYR 619, ASP 760, ASP 761, TRP 800, GLU 811, CYS 813, and SER 814 in (Fig. 5).
4. Discussion
The novel coronavirus is a global health challenge that calls for emergency and urgent antiviral strategies for the treatment and prevention of the infection. Some antiviral agents including remdesivir were currently repurposed against COVID-19 to allow rapid availability of treatments options3 , 4 as the search for new anti-SARS-CoV-2 drugs continues. The use of heterocyclic compounds as templates for antiviral drug development has become very beneficial as they have been reported to be potent against several viral infections including hepatitis C (HCV), hepatitis B (HBV), human immunodeficiency virus (HIV), herpes simplex (HSV), rotavirus, adenoviruses and coxsackie.27 , 32 , 33 Antiviral imidazole compounds are said to exhibit their anti-viral activity by targeting viral proteases and other critical enzymes/proteins essential to the viral life cycle.32 , 33
The high binding affinities exhibited by the imidazole compounds for SARS-CoV-2 target proteins in this study is an indication of the inhibitory potential of these compounds against these biomolecules and their possible roles as therapeutic agents against SARS-CoV-2. Due to structural similarity with the amino acid histidine, an imidazole can easily interact with important protein molecules thereby modifying their functions.32 , 33 In this study, it was observed that the bisimidazoles possess higher binding affinity for all the target proteins than the other two classes of imidazoles. This could be linked to the two imidazole rings and several other aromatic rings contained in the structure of these compounds. From the result of the pharmacophore modelling of C2 (the highest affinity compound), it was observed that the two imidazole rings together with the other aromatic groups contributed to the binding of this compound to the targets. Interactions involving aromatic rings are very essential to biological recognition including protein-ligand interaction as about 20% of amino acids are aromatic in nature. Aromatic interactions are known to be of paramount importance in drug design for improved efficacy and lead optimization.45
The binding free energy (Table 2) of the complexes agrees with the docking score shown on Table 1. Determination of protein-ligand stability is regarded as a very reliable method for validating docking score9 , 41 and the free energy of favorable reaction is always negative. The results obtained in this study showed that the imidazole derivatives formed a stable complex with the drug targets upon binding, affirming the robustness of the docking results.
In addition to the inhibitory potential demonstrated by the imidazole compounds, quite a number of them possessed moderate ADMET properties. However, some of the compounds may require lead optimization to improve these properties and still maintain the binding affinities. ADMET analysis is a procedure used for defining whether the compounds can be easily absorbed, transported to their target site of action, metabolized in a way that does not instantly remove the activity and easily eliminated from the body while preventing toxic effects. These properties are collectively referred to as ADMET (absorption, distribution, metabolism, elimination and toxicity).46 In silico prediction of ADMET properties is a cheap and time saving alternative to standard experimental approaches47 , 48 and its inclusion at earlier stages of drug discovery programs49 , 50 has become essential51, 52, 53 for reducing the rate of pharmacokinetics-related failure of drugs in the clinical phases.
Lipophilicity and water solubility are critical physicochemical properties that determines the ADMET behaviors of a drug. An orally administered drug should be sufficiently lipophilic to pass through the intestinal lining, penetrate the membrane of target cells and sufficiently hydrophilic to travel in the aqueous blood. The higher the log P value of a compound, the higher its lipophilicity and the lower its water solubility.41 Hence, the compounds with Log P values between 4.02 and 6.15 (all the phenyl substituted imidazoles, the thiophene imidazoles and bisimidazoles C2 and C4) came out to be either soluble or moderately soluble while the bisimidazoles C1, C3 and C5 with higher log P values were predicted to be poorly soluble (Table 3). The presence of more aromatic rings and the higher molecular weights of the bisimidazoles may have contributed to the poor solubility of these compounds, however the ones possessing additional polar side chains (C2 and C4) are moderately soluble.
Drug-likeness analysis is a qualitative assessment of oral bioavailability and it is developed based on structural or physicochemical evaluation of compounds in the advanced stages of drug development.42 The Lipinski's Rule states that an orally active drug should have: not more than 5 hydrogen bond donors, not more than 10 hydrogen bond acceptors, a molecular weight under 500 g/mol and a log P less than 5.43 A molecule would not be orally active if it violates two or more of the rules. Based on these criteria, only C2 meet the requirements for oral bioavailability among the bisimidazoles as it violates only one of the rules, but all phenyl substituted and thiophene imidazoles meet the requirements. Nonetheless, all the test compounds pass the Veber's rule comprising of only two criteria of: 10 or fewer rotatable bonds and polar surface (TPSA) area not greater than 140 Å2.44 Furthermore, the bioavailability score which uses total charge, TPSA and the Lipinski filter54 gave a semi-quantitative estimate of the probability of the compounds to be good oral drugs. The 0.17 bioavailability score of the bisimidazoles shows that these compounds have approximately 17% probability of at least 10% oral bioavailability in rat or measurable human colon carcinoma (Caco-2) permeability, hence these compounds apart from C2 will not be easily bioavailable orally. However, 0.55% bioavailability score of C2 and the remaining 2 classes of test compounds represents a 55% probability, hence, these compounds are likely to be good as oral drugs.
According to the pharmacokinetic predictions of the test compounds, the bisimidazoles C3 and C5 and the thiophene imidazoles C11 and C14 could possibly cause drug–drug interactions as CYP inhibitors. Cytochrome P450 (CYP) is a superfamily of isoenzymes that catalyzes many reactions in the phase I of drug metabolism.55 It has been estimated that 50–90% of drugs are substrates of five major isoforms (CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4)42 and their inhibition is a major cause of pharmacokinetics-related drug–drug interactions.56 , 57 Likewise, as substrates of Pgp, compounds C3, C5 and C13 are likely to be prevented from entering their target site of action. Pgp is an important member of the ATP-binding cassette transporters and it is responsible for the active efflux of xenobiotics through biological membranes for the purpose of protecting the body from foreign chemicals. This efflux pump also contributes to drug resistance by limiting the entry of some drugs into the sensitive areas. Nevertheless, the results of the toxicity prediction showed that none of the test compounds has the tendency for any of the toxicity parameters tested, hence the compounds could be both effective and safe as potential therapeutic agents against SARS-CoV-2.
Analysis of the 3D and 2D structures of the docked SARS-CoV-2 target–imidazole complex showed that these compounds possess inhibitory potentials against the proteins. C2, C12 and the standard inhibitor N3 interacted with CYS 145 and HIS 41 at the binding pocket of the SARS-CoV-2 Mpro while C9 interacted with GLU 288, ASP 289 and Glu 290 (Fig. 3). CYS 145 and HIS 41 are very important amino acids at the catalytic site of the main protease of SARS coronavirus. The protease is a homodimer with the two subunits arranged perpendicularly to each other and each protomer have three discrete structural domains. The substrate binding site of each subunit comprises of a HIS 41–CYS 145 catalytic dyad situated at the interface between two of the structural domains. While HIS 41 acts as a general base, CYS 145 mediates an electrostatic trigger that initiates the first step of the catalytic mechanism of action of the enzyme.58 Mutations at these catalytic structure (HIS 41 - CYS 145) have been reported to almost totally eliminate Mpro enzyme activity.59 , 60 Residues around GLU 288 – ASP 289 – GLU 290 are reported to be associated with Mpro dimerization and mutations in GLU 288 and ASP 289 gave rise to enzymes with reduced activities.61 Hence, interaction of the compounds with these amino acids suggests that these compounds could possibly interfere with the catalytic activity of SARS-CoV-2 main protease and consequently inhibit the replication of the virus.
The SARS-CoV-2 Spro interacted with C2, C9, C11 and Pravastatin through some important amino acids residues including ASN 501 of its receptor binding domain (RBD) (Fig. 4). ASN 501 has been listed as one of the key residues responsible for binding of Spro to its human host receptor (ACE2). ASN 501 of the spike receptor binding motif is reported to form hydrogen bond with Tyr 41 of ACE2. ASN 501 also interacts with ACE2 Lys353, Gly354 and Asp355.36 , 62 Binding of the test compounds to this critical amino acid residue could therefore limit the binding of SARS-CoV-2 Spro to its human host receptor thereby limiting viral entry and disease progression.
Furthermore, C2, C9 and C12 demonstrated inhibitory potentials against SARS-CoV-2 RdRp. These compounds interacted with the same amino acid residues as Remdesivir (a known RdRp inhibitor) in the binding pocket of SARS-CoV-2 RdRp (Fig. 5). The four compounds interacted with residues ASP 618, TYR 619, ASP 760 and other amino acids that have been previously identified at the enzyme's active site.63 RdRp as a crucial enzyme in viral replication, catalyzes the synthesis of the RNA genome by producing a complementary RNA strand from an RNA template,64 hence its inhibition by these imidazole compounds could offer therapeutic benefits against SARS-CoV-2.
5. Conclusions
Three classes of newly synthesized imidazole derivatives were evaluated for therapeutic potentials against SARS-CoV-2 Mpro, Spro and RdRp. The compounds showed exciting binding affinities and stability with the target proteins. Bisimidazole C2 gave the highest binding affinity for all the drug targets among the three classes of compounds. Within the phenyl-substituted 1H-imidazole class, C9 gave the highest docking score against the three targets. For the thiophene-imidazoles, C11 scored highest against Spro and C12 against Mpro and RdRp. The compounds interacted with HIS 41 - CYS 145 and GLU 288 – ASP 289 – GLU 290 of Mpro, ASN 501 of Spro receptor binding motif and some active site amino acids of RdRp. These novel imidazoles could offer therapeutic benefits against SARS-CoV-2, however, lead optimization may be required to modify the ADMET properties of some of the compounds while maintaining their biological activities and wet laboratory experiments will be necessary to further validate the results of this in silico study.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to acknowledge the healthcare frontline, social workers and World Health Organization for their enormous contributions in combating COVID-19 outbreak around the globe. Also, authors appreciate the support of Taif University Researchers Supporting Program (project number: TURSP-2020/151), Taif University, Saudi Arabia.
Footnotes
Peer review under responsibility of Japanese Pharmacological Society.
References
- 1.Hsu L.Y., Chia P.Y., Lim J.F. The novel coronavirus (SARS-CoV-2) pandemic. Ann Acad Med Singapore. 2020;49(105):105–107. [PubMed] [Google Scholar]
- 2.Jebril N. 2020. World Health Organization declared a pandemic public health menace: a systematic review of the coronavirus disease 2019 “COVID-19”, up to 26th March 2020. Available at: SSRN 3566298. [DOI] [Google Scholar]
- 3.Esakandari H., Nabi-Afjadi M., Fakkari-Afjadi J., Farahmandian N., Miresmaeili S.M., Bahreini E. A comprehensive review of COVID-19 characteristics. Biol Proced Online. 2020;22:1–10. doi: 10.1186/s12575-020-00128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parlakpinar H., Polat S., Acet H.A. Pharmacological agents under investigation in the treatment of coronavirus disease 2019 and the importance of melatonin. Fundamental & cli. Pharm. 2020 doi: 10.1111/fcp.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liya G., Yuguang W., Jian L. Studies on viral pneumonia related to novel coronavirus SARS-CoV-2, SARS-CoV, and MERS-CoV: a literature review. APMIS. 2020;128:423–432. doi: 10.1111/apm.13047. [DOI] [PubMed] [Google Scholar]
- 6.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. In silico screening of FDA approved drugs reveals ergotamine and dihydroergotamine as potential coronavirus main protease enzyme inhibitors. Saudi J Biol Sci. 2020;27(10):2674–2682. doi: 10.1016/j.sjbs.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han W., Quan B., Guo Y. The course of clinical diagnosis and treatment of a case infected with coronavirus disease 2019. J. of med. virology. 2020;92(5):461–463. doi: 10.1002/jmv.25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C., Liu Y., Yang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elekofehinti O.O., Iwaloye O., Josiah S.S., Lawal A.O., Akinjiyan M.O., Ariyo E.O. Molecular docking studies, molecular dynamics and ADME/tox reveal therapeutic potentials of STOCK1N-69160 against papain-like protease of SARS-CoV-2. Mol Divers. 2020:1–13. doi: 10.1007/s11030-020-10151-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang K.O., Kim Y., Lovell S., Rathnayake A.D., Groutas W.C. Antiviral drug discovery: norovirus proteases and development of inhibitors. Viruses. 2019;11(2):197. doi: 10.3390/v11020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzon M., Marsh M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Research. 2019;8:1–10. doi: 10.12688/f1000research.19694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Shabat S., Yarmolinsky L., Porat D., Dahan A. Antiviral effect of phytochemicals from medicinal plants: applications and drug delivery strategies. Drug Del. and Trans. Research. 2020:1–14. doi: 10.1007/s13346-019-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elekofehinti O.O., Iwaloye O., Famusiwa C.D., Akinseye O., Rocha J.B. Identification of main protease of coronavirus SARS-CoV-2 (Mpro) inhibitors from melissa officinalis”. Curr Drug Discov Technol. 2020;17:1. doi: 10.2174/157016381799920091810370. [DOI] [PubMed] [Google Scholar]
- 17.Hiramoto T., Nonaka Y., Inoue K. Identification of endogenous surrogate ligands for human P2Y receptors through an in silico search. J Pharmacol Sci. 2004;95(1):81–93. doi: 10.1254/jphs.95.81. [DOI] [PubMed] [Google Scholar]
- 18.Adeyemi O.S., Molina M.T., Eseola A.O., Fonseca-Berzal C., Gómez-Barrio A. New imidazole-based compounds active against Trypanosoma cruzi. Comb Chem High Throughput Screen. 2017;20(1):20–24. doi: 10.2174/1386207320666170110141907. [DOI] [PubMed] [Google Scholar]
- 19.Adeyemi O.S., Eseola A.O., Plass W. Imidazole derivatives as antiparasitic agents and use of molecular modeling to investigate the structure–activity relationship. Parasitol Res. 2020;119:1925–1941. doi: 10.1007/s00436-020-06668-6. [DOI] [PubMed] [Google Scholar]
- 20.de Araújo J.S., Garcia-Rubia A., Sebastian-Perez V. Imidazole derivatives as promising agents for the treatment of Chagas disease. Antimicrob Agents Chemother. 2019;63(4) doi: 10.1128/AAC.02156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raman N., Mitu L., Sakthivel A., Pandi M.S.S. Studies on DNA cleavage and antimicrobial screening of transition metal complexes of 4-aminoantipyrine derivatives of N 2 O 2 type. J Iran Chem Soc. 2009;6(4):738–748. doi: 10.1007/BF03246164. [DOI] [Google Scholar]
- 22.Sharma D., Narasimhan B., Kumar P. Synthesis, antimicrobial and antiviral evaluation of substituted imidazole derivatives. European j. of medicinal chem. 2009;44(6):2347–2353. doi: 10.1016/j.ejmech.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Emami S., Foroumadi A., Falahati M. 2-Hydroxyphenacyl azoles and related azolium derivatives as antifungal agents. Bioorganic & med. chem. letters. 2008;18(1):141–146. doi: 10.1016/j.bmcl.2007.10.111. [DOI] [PubMed] [Google Scholar]
- 24.Bhandari K., Srinivas N., Keshava G.S., Shukla P.K. Tetrahydronaphthyl azole oxime ethers: the conformationally rigid analogues of oxiconazole as antibacterials. European j. of medicinal chem. 2009;44(1):437–447. doi: 10.1016/j.ejmech.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Iman M., Davood A., Khamesipour A. Design of antimalarial agents based on pyrimidine derivatives as methionine aminopeptidase 1b inhibitor: molecular docking, quantitative structure activity relationships, and molecular dynamics simulation studies. J Chin Chem Soc. 2020:1–11. doi: 10.1002/jccs.201900165. [DOI] [Google Scholar]
- 26.Papadopoulou M.V., Bloomer W.D., Rosenzweig H.S. The antitubercular activity of various nitro (triazole/imidazole)-based compounds. Bioorg Med Chem. 2017;25(21):6039–6048. doi: 10.1016/j.bmc.2017.09.037. [DOI] [PubMed] [Google Scholar]
- 27.Liu L., Hu Y., Shen Y.F., Wang G.X., Zhu B. Evaluation on antiviral activity of coumarin derivatives against spring viraemia of carp virus in epithelioma papulosum cyprini cells. Anti. Research. 2017;144:173–185. doi: 10.1016/j.antiviral.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Eseola A.O., Sun W.H., Li W., Woods J.A. Syntheses of new imidazole ligand series and evaluation of 1-, 2-and 4, 5-imidazole substituent electronic and steric effects on N-donor strengths. J Mol Struct. 2010;984(1–3):117–124. doi: 10.1016/j.molstruc.2010.09.015. [DOI] [Google Scholar]
- 29.Kumari A., Singh R.K. Medicinal chemistry of indole derivatives: current to future therapeutic prospectives. Bioorg Chem. 2019;89:103021. doi: 10.1016/j.bioorg.2019.103021. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X., Ma Z., Zhang D. Synthesis of imidazole-based medicinal molecules utilizing the van leusen imidazole synthesis. Pharmaceuticals. 2020;13(3):37. doi: 10.3390/ph13030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerru N., Gummidi L., Maddila S., Gangu K.K., Jonnalagadda S.B. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules. 2020;25(8):1909. doi: 10.3390/molecules25081909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Clercq E. Antiviral drugs in current clinical use. J. of clinical virology. 2004;30(2):115–133. doi: 10.1016/j.jcv.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Kanwal A., Ahmad M., Aslam S., Naqvi S.A.R., Saif M.J. Recent advances in antiviral benzimidazole derivatives: a mini review. Pharmaceut Chem J. 2019;53(3):179–187. doi: 10.1007/s11094-019-01976-3. [DOI] [Google Scholar]
- 34.Adeyemi O.S., Eseola A.O., Plass W., Otuechere C.A., Elebiyo T.C. New imidazoles cause cellular toxicity by impairing redox balance, mitochondrial membrane potential, and modulation of HIF-1α expression. Biochem Biophys Res Commun. 2020;529(1):23–27. doi: 10.1016/j.bbrc.2020.05.059. [DOI] [PubMed] [Google Scholar]
- 35.Eseola A.O., Görls H., Woods J.A.O., Plass W. Single monodentate N-donor ligands versus multi-ligand analogues in Pd (II)-catalysed CC coupling at reduced temperatures. Polyhedron. 2020:114507. doi: 10.1016/j.poly.2020.114507. [DOI] [Google Scholar]
- 36.Jin Z., Du X., Xu Y. Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Zhang Y., Wu L. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y., Yan L., Huang Y. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang G.G. Molecular biology of the SARS-coronavirus. Springer; Berlin, Heidelberg: 2010. Quaternary structure of the SARS coronavirus main protease; pp. 115–128. [DOI] [Google Scholar]
- 40.Iwaloye O., Elekofehinti O.O., Oluwarotimi E.A., Babatomiwa K., Momoh I. In silico molecular studies of natural compounds as possible anti-Alzheimer’s agents: ligand-based design. Network Modeling Analysis in Health Informatics and Bioinfo. 2020;9:54. doi: 10.1007/s13721-020-00262-77. [DOI] [Google Scholar]
- 41.Schrödinger release 2020-2: Prime. Schrödinger, LLC; New York, NY: 2020. [Google Scholar]
- 42.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 44.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. of medicinal chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 45.Singh V.P. Aromatic interactions in biological systems. Sci Technol Jpn. 2015;3:42–48. [Google Scholar]
- 46.Sun H., Li Y., Shen M. Assessing the performance of the MM/PBSA and MM/GBSA methods. 5, improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys Chem Chem Phys. 2014;16:22035–22045. doi: 10.1039/C4CP03179B. [DOI] [PubMed] [Google Scholar]
- 47.Ntie-Kang F. An in silico evaluation of the ADMET profile of the StreptomeDB database. SpringerPlus. 2013;2(1):353. doi: 10.1186/2193-1801-2-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiMasi J.A., Hansen R.W., Grabowski H.G. The price of innovation: new estimates of drug development costs. J. of health economics. 2003;22(2):151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 49.Darvas F., Keseru G., Papp A., Dorman G., Urge L., Krajcsi P. In silico and ex silico ADME approaches for drug discovery. Current topics in med. chem. 2002;2(12):1287–1304. doi: 10.2174/1568026023392841. [DOI] [PubMed] [Google Scholar]
- 50.Hodgson J. ADMET—turning chemicals into drugs. Nat Biotechnol. 2001;19(8):722–726. doi: 10.1038/90761. [DOI] [PubMed] [Google Scholar]
- 51.Navia M.A., Chaturvedi P.R. Design principles for orally bioavailable drugs. Drug Discov Today. 1996;1(5):179–189. doi: 10.1016/1359-6446(96)10020-9. [DOI] [Google Scholar]
- 52.Lombardo F., Gifford E., Shalaeva M.Y. In silico ADME prediction: data, models, facts and myths. Mini Rev Med Chem. 2003;3(8):861–875. doi: 10.2174/1389557033487629. [DOI] [PubMed] [Google Scholar]
- 53.Gleeson P.M., Hersey A., Hannongbua S. In-silico ADME models: a general assessment of their utility in drug discovery applications. Current topics in medicinal chem. 2011;11(4):358–381. doi: 10.2174/156802611794480927. [DOI] [PubMed] [Google Scholar]
- 54.Testa B., Kramer S. The biochemistry of drug metabolism-an introduction. Chem Biodivers. 2009;6(5):591. doi: 10.1002/cbdv.200900022. [DOI] [PubMed] [Google Scholar]
- 55.Hollenberg P.F. Characteristics and common properties of inhibitors, inducers, and activators of CYP enzymes. Drug Metab Rev. 2002;34(1–2):17–35. doi: 10.1081/DMR-120001387. [DOI] [PubMed] [Google Scholar]
- 56.Huang S.M., Strong J.M., Zhang L. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. The J. of cli. Pharm. 2008;48(6):662–670. doi: 10.1177/0091270007312153. [DOI] [PubMed] [Google Scholar]
- 57.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43(15):4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 58.Chang H.P., Chou C.Y., Chang G.G. Reversible unfolding of the severe acute respiratory syndrome coronavirus main protease in guanidinium chloride. Biophys J. 2007;92(4):1374–1383. doi: 10.1529/biophysj.106.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lan J., Ge J., Yu J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 60.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Novak J., Rimac H., Kandagalla S., Pathak P., Grishina M., Potemkin V. 2020. Proposition of a new allosteric binding site for potential SARS-CoV-2 3CL protease inhibitors by utilizing molecular dynamics simulations and ensemble docking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aftab S.O., Ghouri M.Z., Masood M.U. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J translational med. 2020;18(1):1–15. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J Med Virol. 2020:1–40. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 64.Martin Y.A. Bioavailability score. J Med Chem. 2005;48:3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]