Abstract
Several studies have shown that direct brain stimulation can enhance memory in humans and animal models. Investigating the neurophysiological changes induced by brain stimulation is an important step towards understanding the neural processes underlying memory function. Furthermore, it paves the way for developing more efficient neuromodulation approaches for memory enhancement. In this study, we utilized a combination of unsupervised and supervised machine learning approaches to investigate how amygdala stimulation modulated hippocampal network activities during the encoding phase. Using a sliding window in time, we estimated the hippocampal dynamic functional network connectivity (dFNC) after stimulation and during sham trials, based on the covariance of local field potential recordings in 4 subregions of the hippocampus. We extracted different network states by combining the dFNC samples from 5 subjects and applying k-means clustering. Next, we used the between-state transition numbers as the latent features to classify between amygdala stimulation and sham trials across all subjects. By training a logistic regression model, we could differentiate stimulated from sham trials with 67% accuracy across all subjects. Using elastic net regularization as a feature selection method, we identified specific patterns of hippocampal network state transition in response to amygdala stimulation. These results offer a new approach to better understanding of the causal relationship between hippocampal network dynamics and memory-enhancing amygdala stimulation.
I. Introduction
The hippocampus has a long-established role in declarative memory, with its various subregions thought to mediate different processes related to memory formation [1]. The hippocampal network receives input from a variety of brain areas, including the basolateral amygdala [2]. This connection is thought to be particularly important in the context of memory, since amygdala regulates emotional processing, which can affect how memories are consolidated [3], [4].
A recent study explored how amygdala affects memory consolidation in humans, by performing direct electrical stimulation using depth electrodes in epilepsy patients [5]. When the amygdala was stimulated during presentation of novel images, the patients subsequently showed stronger memory of these images, compared to sham trials. Furthermore, amygdala stimulation led to increased spectral coherence between the hippocampus and perirhinal cortex during retention, indicating that hippocampal connectivity with other important brain regions was affected. However, it is still unclear how stimulating the amygdala affects hippocampal network connectivity at the time of stimulation, and why that leads to better memory. In this study, we are leveraging this dataset of intracranial recordings of hippocampal subregions to explore the effects of the amygdala stimulation on hippocampal networks.
To study the interactions between different hippocampal subregions and how they evolve over time, we employed the use of dynamic functional connectivity (dFNC), which is a metric for time-varying changes in interregional dynamics, as measured by functional connectivity [6]. dFNC has been commonly used in fMRI recordings during cognitive tasks, but recent studies have used it in on EEG signals [7].
To shed light on hippocampal dynamics over time, we are measuring the dFNC of all recorded hippocampal subregions - CA1, CA3, dentate gyrus (DG) and, subiculum (SUB) – and how it is affected by amygdala stimulation. Furthermore, we are assessing the differences of stimulated and sham trials by comparing the transitions observed between different hippocampal dFNC states in both cases. Through this novel approach, we can start exploring how different network states relate to memory formation and how use this knowledge to potentially improve direct brain stimulation.
II. Method
A. Experiment Procedure
Five patients with drug-resistant epilepsy that were implanted with intracranial EEG electrodes in the hippocampus participated in an image recognition memory task, as described in the original study [5]. During memory encoding, 160 images were presented to the patients for three seconds each. Half of the images followed by amygdala stimulation were labeled as Stimulation trials and the other half were considered as Sham trials. The local field potential (LFP) signals of the hippocampus were recorded using depth electrodes (Ad-Tech; 0.86 mm diameter, 2 mm length platinum-coated contacts, typically spaced along 5-mm intervals) and recording systems (XLTEK EMU 128FS; Natus Medical) at each clinical site. LFPs were recorded at a sampling rate of either 500 Hz (3 subjects) or 1000 Hz (2 subjects). A research neurostimulator (CereStim M96; Blackrock Microsystems) was used to deliver the current regulated, charge-balanced, biphasic rectangular pulses at 0.5 mA for 1s in eight trains of four pulses at 50 Hz to the amygdala precisely at the offset of image presentation for a randomized half of the studied images. No seizure activity or after discharges to stimulation were detected during testing or in a thorough post-test review of all recorded LFP channels by a clinical epileptologist [5]. All procedures were by the Emory University Institutional Review Board.
B. Preprocessing
In this study, 2800 ms of the LFP signal, recorded from CA1, CA3, dentate gyrus (DG), and subiculum (SUB), at the offset of electrical (shown in red) and sham (shown in blue) stimulation has been used as shown in Fig. 1. To eliminate the residual effect of the stimulation, 200 ms after the stimulation offset was discarded. The data were first digitally filtered with a low pass cutoff of 1 Hz to attenuate low frequency artifacts and a high pass cutoff of 249 Hz. The median LFP across all available recording electrodes was then subtracted from each channel to remove non-local (global) artifacts.
Fig.1. Schematic of the analysis pipeline.
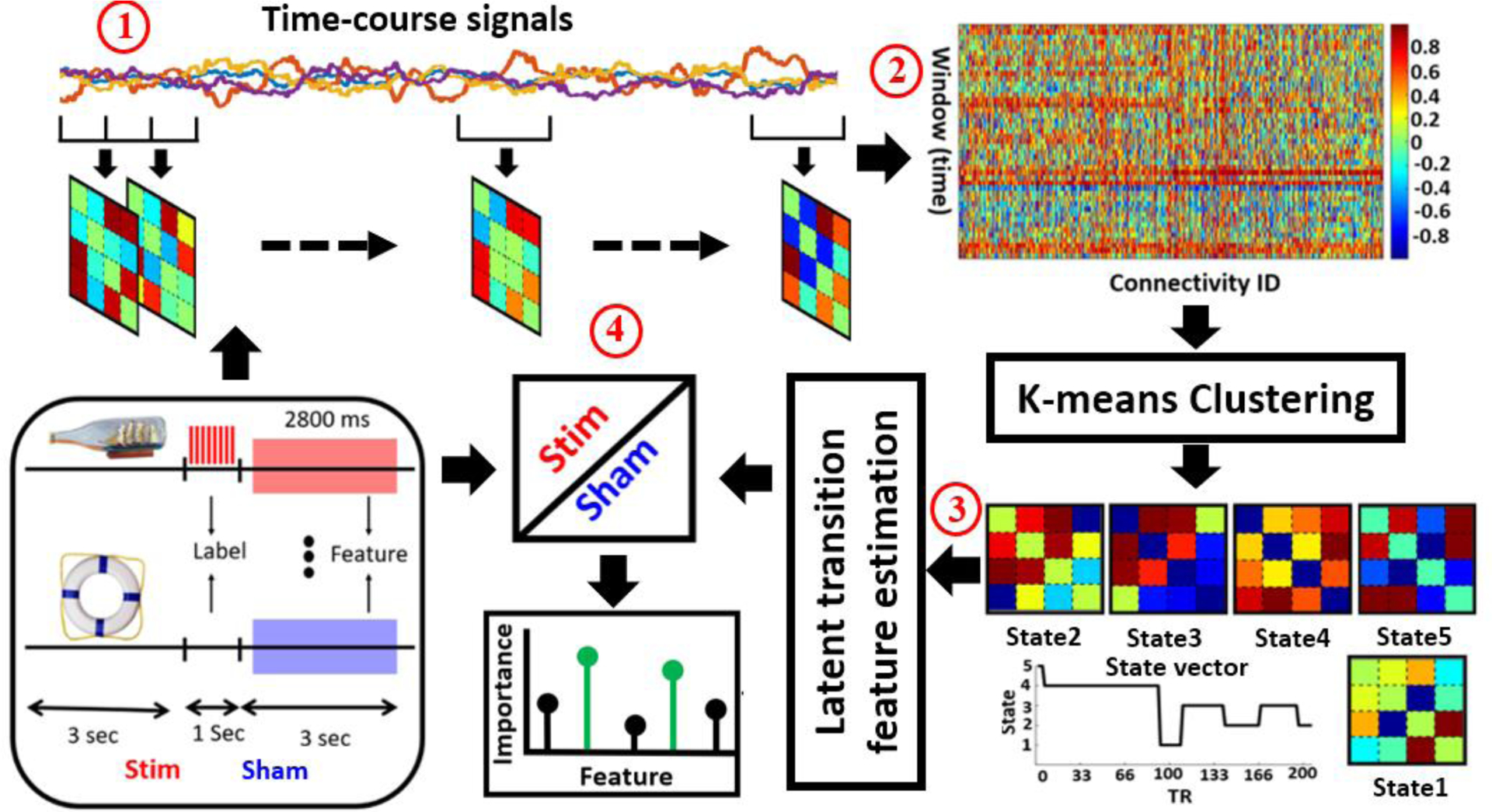
A sliding window over the time course signal of independent components was used to calculate the dFNC of hippocampus in each trial (Step 1). Then, the dynamic states matrix and state transition vectors were computed by applying a k-means clustering method across all windows of all subjects (Step 2). The dFNC between-state transition numbers were calculated as a latent feature to model the temporal pattern of the state vector of each trial (Step 3). These latent features were used as an input to fit logistic regression to classify Stimulated (Stim) from Non-stimulated (Sham). We assessed classifier performance using the area under the receiver-operating characteristics curve (AUC). Feature selection used to model the difference between Stim and Sham trials and find the feature was the most predictive in discriminating between the two classes (Step 4).
C. Dynamic Functional Network Connectivity (dFNC)
The dFNC of each trial (N=160 for each subject), was estimated using a sliding window with a window size of 14 ms in steps of 3.5 ms. First, we segmented the time-course signal to 270 windows, Then, we calculated the covariance matrix of each window to measure the dFNC between channels. We concatenated dFNC estimates of each window for each subject to form a C × C × T array (where C=4 denoted the number of channels and T=270 denoted the number of windows in time), which represented the changes in brain connectivity between channels as a function of time. This process is shown in Step 1 of Fig. 1 [8].
D. Clustering and finding the latent features
After calculating the dFNC for each trial, we concatenated all trials across all subjects as shown in Step 2 of Fig. 1 and applied a k-means clustering algorithm to these dFNC windows to partition the data into a set of separated clusters [8]. The optimal number of centroid states was estimated using the elbow criterion based on the ratio of within to between cluster distance [9]. In a search window of k from 3 to 8, we found that the optimal number of clusters was 5. In addition, the correlation between samples was used as a distance metric in this k-means clustering algorithm with 1000 iterations. After applying k-means clustering, we will have a state vector for each trial as shown in Fig. 1. A state vector showed how the hippocampus changes between any pair of states over the time. Next, using the state vector, we calculated the number of times that the hippocampus network transitions from one state to another one within a trial. This state transition modeled by the equations below:
| (1) |
where Si denotes the state and aij, used as a latent feature showed the number (N) of transition from state j at time t to state i at time step t+1. Overall, this led to 25 latent features for each trial (Step 3 of Fig.1).
E. Classification and feature learning
A logistic regression (LR) was used to perform classification between stimulated and sham trials, by using the latent features of each trial across all five subjects. We utilized elastic net regularization (ENR), which used both L1-and L2-penalization to find the most important features in this classification [10], [11]. We used leave-one subject-out nested cross-validation (CV), in which one subject was used for testing and the remaining four subjects were used for training and validation [12]. In the nested CV, we divided data into training and test sets in any outer-fold. Then, we divided the training data into another training and validation dataset in any inner-fold. The best set of parameters were chosen by training different models using inner-loop training data and validating using a validation dataset. In this approach, the hyperparameters of each model were tuned to minimize the inner-fold CV error of the generalization performance. By sweeping the penalty parameter logarithmically from 10−5 to 105, we found the best value of the parameter that minimizes the CV error. The receiver operating characteristic (ROC) of the CV was computed, and the area under the curve (AUC) was calculated as a measure of the accuracy of the classifier (Step 4 of Fig. 1).
III. Results
A. Hippocampal dFNC states
Five reoccurring dFNC states, named States 1–5, were identified by k-means clustering as shown in Fig. 2a. In this figure, each state represents the centroid of the associated cluster. As shown in this figure, there appeared to be some differences in the connectivity patterns of the different states. In State 1, we observed high connectivity between DG, CA1 and SUB, with CA3 mostly unengaged. On the other hand, in State 2, a high serial connectivity was observed. In State 3, a strong connectivity among CA1, CA3, and SUB was observed. In State 4, there was a negative connectivity between DG and the rest of the nodes. Finally, in State 5 there was high connectivity between SUB and CA3.
Fig.2: k-means clustering and the feature selection results.
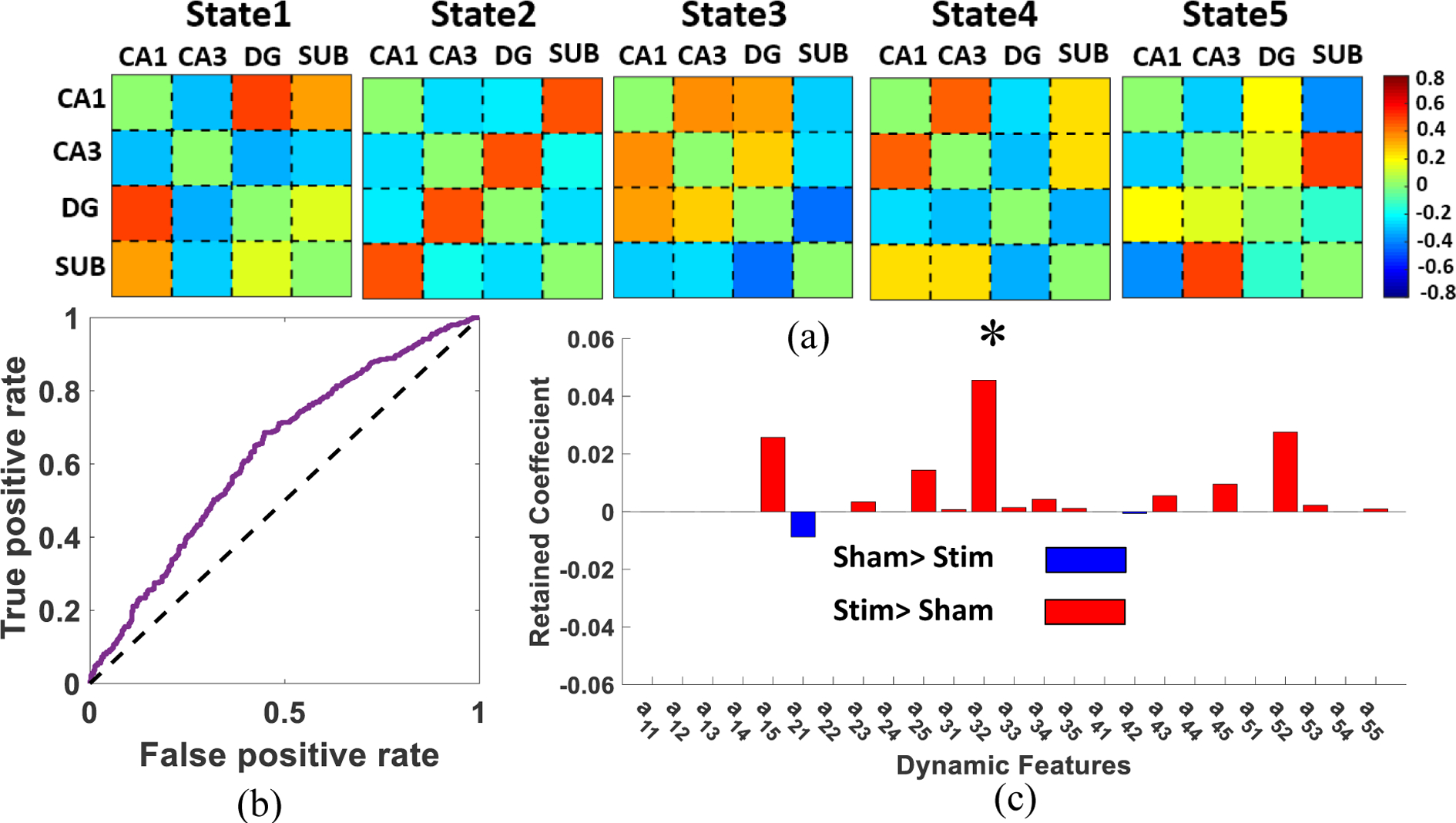
a) The five dFNC states identified by the k-means clustering method. b) ROC of LR with 5-fold ENR prediction of stimulated (Stim) from Sham trials across all subjects using latent feature obtained by k-means clustering method. (AUC: 0.67±0.08). the dashed line shows the change level. c) The distribution of biomarkers identified by ENR. The most important feature is a32 after adjusting p value using Benjamini-Flochberg approach (corrected p<0.05).
B. Classification result between stimulated and non-stimulated trials
Using the number of between-state transitions as a latent feature and 5-fold nested CV, we trained a LR to classify between stimulated and sham trials. In each fold, we left one subject out for testing and trained a LR model using the other four subjects. The overall AUC of the model was 0.67 ± 0.08, that is shown in Fig. 2b. Changes in the dFNC state transitions may be used as a potential biomarker of the effect of amygdala stimulation on the hippocampal networks. Fig. 2c shows the important sparse features in the ENR process that minimized classification error. In this graph, the red color shows higher transition probability in stimulated trial and blue color shows a lower transition probability in stimulated trials. Based on the ENR results, only one transition probability (i.e. transition from state 1 to state 2) was higher in the sham trials. All other transition probabilities were higher in the stimulated trials. Specifically, we observed a relatively higher transition from state 5 to state 1, from state 2 to state 3, and from state 2 to state 5 in the stimulated trials, in which only the transition from state 2 to state 3, i.e. a32, was still statistically significant after a multiple comparison using the Benjamini-Hochberg approach (corrected p<0.05). Therefore, amygdala stimulation increased transition from State 2, which shows the regular serial connectivity expected from hippocampal networks, to State 3 with higher intrinsic connectivity between these subregions.
IV. Discussion
In this study, we developed a novel approach to analyze LFP recordings to investigate the dynamics of the hippocampal functional connectivity that are induced by amygdala stimulation in human. We used recordings from four hippocampal subregions including CA1, CA3, SUB, and DG. Using a sliding window, we estimated the dFNC of each trial. We partitioned the dFNC state space into five different states, using the k-means clustering method. One of the network states (State 2) showed high serial connectivity agreeing with the information flow in the trisynaptic hippocampal circuit [13], as well as recent findings from our group [14]. State 3 showed strong interactions between DG, CA3 and CA1, while SUB, largely considered to be regulating hippocampal output [15], was showing weak connectivity. In addition, a segregated CA1 and DG was observed in State 1 and State 4, respectively. These results suggest that the functional connectivity in the hippocampal networks was highly dynamic, possibly representing different states of coordination between hippocampal subregions.
Next, to model the temporal changes of hippocampal FNC, we calculated the between-state transition number as a latent feature. Following the calculation of this latent feature for all the trials in all subjects, we classified stimulated and sham trials using an LR classifier with leave-one subject-out nested cross-validation. A 67 % AUC suggest between-state transitions may represent a potential biomarker of the effect of stimulation that differentiates stimulated trials from sham.
Finally, using an ENR as a feature learning method embedded in the LR classifier, we found that the between-state transition was statistically higher in the stimulated trials. This potentially suggests the effect of amygdala stimulation in increasing the dynamic of the hippocampal network. In addition, stimulation caused a transition from a state with less connectivity between CA1 and other subregion such as CA3 and DG (State 2) to a state with relatively higher connectivity between CA1/CA3/DG (State 3). This result was consistent with previous findings regarding the role of CA1/CA3/DG connectivity in both encoding and retention, and its association with successful memory [16].
Identifying and comparing dynamic network connectivity between stimulated and sham trials can help better understanding the underlying mechanisms of hippocampal network dynamics effected by amygdala stimulation and inform more effective stimulation paradigms. In sum, this work represented a novel approach to quantify the effect of amygdala stimulation on functional connectivity dynamics of the hippocampal networks in human.
V. Conclusion
In this study, we presented a novel approach for quantifying functional network dynamics in the hippocampus Using ENR feature learning method embedded in a LR classifier, we characterized functional connectivity state transitions as potential biomarkers of the effect of amygdala stimulation on the hippocampal network. The results of this study suggest that amygdala stimulation may enhance functional connectivity in the hippocampal networks.
Acknowledgments
Research supported by NIH grants R01EB028350 and UG3NS100559
References
- [1].Eichenbaum H, “Hippocampus: Cognitive processes and neural representations that underlie declarative memory,” Neuron, vol. 44, no. 1, pp. 109–120, 2004. [DOI] [PubMed] [Google Scholar]
- [2].Pitkanen A, Pikkarainen M, Nurminen N, and Ylinen A, “Reciprocal Connections between the Amygdala and the Hippocampal Formation, Perirhinal Cortex, and Postrhinal Cortex in Rat: A Review,” Annals of the New York Academy of Sciences, vol. 911, no. 1, pp. 369–391, 2006. [DOI] [PubMed] [Google Scholar]
- [3].McGaugh JL, “the Amygdala Modulates the Consolidation of Memories of Emotionally Arousing Experiences,” Annual Review of Neuroscience, vol. 27, no. 1, pp. 1–28, 2004. [DOI] [PubMed] [Google Scholar]
- [4].LeDoux J, “The emotional brain, fear, and the amygdala,” Cellular and Molecular Neurobiology, vol. 23, no. 4–5, pp. 727–738, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Inman CS et al. , “Direct electrical stimulation of the amygdala enhances declarative memory in humans,” Proceedings of the National Academy of Sciences of the United States of America, vol. 115, no. 1, pp. 98–103, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hutchison RM et al. , “Dynamic functional connectivity: Promise, issues, and interpretations,” NeuroImage, vol. 80, pp. 360–378, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Allen EA, Damaraju E, Eichele T, Wu L, and Calhoun VD, “EEG Signatures of Dynamic Functional Network Connectivity States,” Brain Topography, vol. 31, no. 1, pp. 101–116, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Damaraju E et al. , “Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia,” Neuroimage: Clinical, vol. 5, no. July, pp. 298–308, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li M and Zoltàn Z, “The Application Of Cluster Analysis In Ecnomics Science,” vol. 17, no. October 1995, pp. 441–458, 2017. [Google Scholar]
- [10].Tibshiran R, “Regression shrinkage and selection via the lasso : a retrospective,” Journal of the Royal Statistical Society. Series B: Statistical Methodology, vol. 73, no. 3, pp. 273–282, 2016. [Google Scholar]
- [11].Zou H and Hastie T, “Regularization and variable selection via the elastic net,” Journal of the Royal Statistical Society. Series B: Statistical Methodology, vol. 67, no. 2, pp. 301–320, 2005. [Google Scholar]
- [12].Wainer J and Cawley G, “Nested cross-validation when selecting classifiers is overzealous for most practical applications,” pp. 1–9, 2018. [Google Scholar]
- [13].Andersen J, Morris P, Amaral R, Bliss, T. D, & O’Keefe, “The Hippocampus Book,” Oxford university press, 2006. [Google Scholar]
- [14].Trimper JB, Galloway CR, Jones AC, Mandi K, and Manns JR, “Gamma Oscillations in Rat Hippocampal Subregions Dentate Gyrus, CA3, CA1, and Subiculum Underlie Associative Memory Encoding,” Cell Reports, vol. 21, no. 9, pp. 2419–2432, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].O’Mara S, “Controlling hippocampal output: The central role of subiculum in hippocampal information processing,” Behavioural Brain Research, vol. 174, no. 2, pp. 304–312, 2006. [DOI] [PubMed] [Google Scholar]
- [16].Duncan K, Tompary A, and Davachi L, “Associative encoding and retrieval are predicted by functional connectivity in distinct hippocampal area CA1 pathways,” Journal of Neuroscience, vol. 34, no. 34, pp. 11188–11198, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]


