Abstract
Background & Aims
The circadian clock is crucial for physiological homeostasis including gut homeostasis. Disorder of the circadian clock may contribute to many diseases including inflammatory bowel disease (IBD). However, the role and the mechanisms of circadian clock involvement in IBD still are unclear.
Methods
Disorder of the circadian clock including chronic social jet lag and circadian clock gene deficiency mice (Bmal1-/-, and Per1-/-Per2-/-) were established. Dextran sulfate sodium (DSS) and/or azoxymethane were used to induce mouse models of colitis and its associated colorectal cancer. Flow cytometry, immunohistochemistry, immunofluorescence, Western blot, and reverse-transcription quantitative polymerase chain reaction were used to analyze the characteristics of immune cells and their related molecules.
Results
Mice with disorders of the circadian clock including chronic social jet lag and circadian clock gene deficiency were susceptible to colitis. Functionally, regulatory B (Breg) cells highly expressing Programmed cell death 1 ligand 1 (PDL1) in intestinal intraepithelial lymphocytes (IELs) helped to alleviate the severity of colitis after DSS treatment and was dysregulated in DSS-treated Bmal1-/- mice. Notably, interleukin 33 in the intestinal microenvironment was key for Bmal1-regulated PDL1+ Breg cells and interleukin 33 was a target of Bmal1 transcriptionally. Dysregulated PDL1+ B cells induced cell death of activated CD4+ T cells in DSS-treated Bmal1-/- mice. Consequently, circadian clock disorder was characterized as decreased numbers of Breg+ PDL1+ cells in IELs and dysfunction of CD4+ T cells promoted colitis-associated colorectal cancer (CRC) in mice. In clinical samples from CRC patients, low expression of Bmal1 gene in paracancerous tissues and center area of tumor was associated closely with a poorer prognosis of CRC patients.
Conclusions
Our study uncovers the importance of the circadian clock regulating PDL1+ Breg+ cells of IELs in IBD and IBD-associated CRC.
Keywords: Bmal1, B Cells, Colitis, Colitis-Associated Colorectal Cancer, PDL1
Abbreviations used in this paper: AOM, azoxymethane; Breg, regulatory B; CD, Crohn’s disease; ChIP, chromatin immunoprecipitation; CJ, chronic jet lag; CRC, colorectal cancer; CT, tumor center; DMEM, Dulbecco’s modified Eagle medium; DSS, dextran sulfate sodium; FBS, fetal bovine serum; GranzB, _; IEL, intraepithelial lymphocyte; IFNγ, interferon γ; IL, interleukin; IM, invasive margin; MCP-1, monocyte chemoattractant protein-1; MFI, mean fluorescence intensity; mRNA, messenger RNA; PBL, peripheral blood lymphocyte; PBS, phosphate-buffered saline; PDL1, Programmed cell death 1 ligand 1; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; SPL, spleen lymphocyte; TGFβ, transforming growth factor β; Th, T helper; UC, ulcerative colitis; WT, wild-type
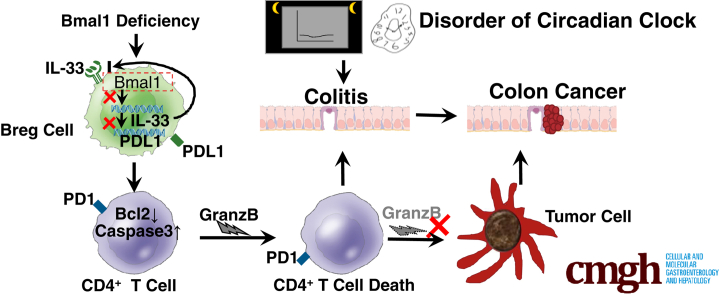
Graphical abstract
Summary.
Deficiency or disorder of circadian rhythms leads to dysfunctions of Programmed cell death 1 ligand 1+ regulatory B cells in intestinal intraepithelial lymphocytes, which results in apoptosis of CD4+ T cells, and ultimately promotes the development of enteritis and even enteritis-associated colorectal cancer.
The maintenance of intestinal homeostasis depends on 3 key factors including the intestinal flora, the intestinal lymphocytes, and the host's immune system. Once any aspect of these factors breaks down, it may cause an inflammatory bowel disease (IBD).
The Toll-like receptor pathway and T helper (Th)1/Th2 paradigm are well known to participate in the pathogenesis of IBD.1 In addition to these, dendritic cells, macrophages, and B cells can specifically present antigens to T cells, and then form multiple T-cell immune responses to participate in intestinal immunity that will contribute to IBD pathogenesis.2 Among these cells, B cells not only produce antibodies but also present antigens or secrete cytokines.3 B cells are involved in many inflammatory diseases,3,4 including intestinal inflammation.5 It is well known that B cells can specifically differentiate into plasma cells and most intestinal plasma cells secrete IgA, acting as a barrier to protect the epithelium against pathogenic microorganisms.6,7 Nevertheless, the role of B cells in IBD still is unclear.
Circadian homeostasis in mammals is maintained by a central clock located in the suprachiasmatic nucleus of the brain and operated by circadian genes such as Bmal1 and Clock.8,9 Circadian rhythms are important drivers for most physiological processes8 and the development of colitis in animal models10; preliminary human studies have shown that patients with IBD are at increased risk for altered sleep patterns,11 and the rs2797685 variant of the PER3 gene was increased significantly in both 1082 CD and 972 UC patients.12 However, the functional link between the circadian clock and B lymphocytes in IBD is unclear.
In this study, we characterized Programmed cell death 1 ligand 1 (PDL1)+ regulatory B (Breg) cells in intestinal intraepithelial lymphocytes (IELs), which played a key role in colitis after dextran sulfate sodium (DSS) treatment and were dysregulated in mice with circadian clock disorders. Our study established a kind of new subset of Breg cells with circadian clock disorders in IBD pathogenesis, suggesting the circadian clock regulates PDL1+ Breg cells as a candidate for the prevention and treatment of IBD, and even IBD-associated colorectal cancer (CRC) in immunopathology.
Results
Circadian Clock Disorders in Mice Are Susceptible to Colitis
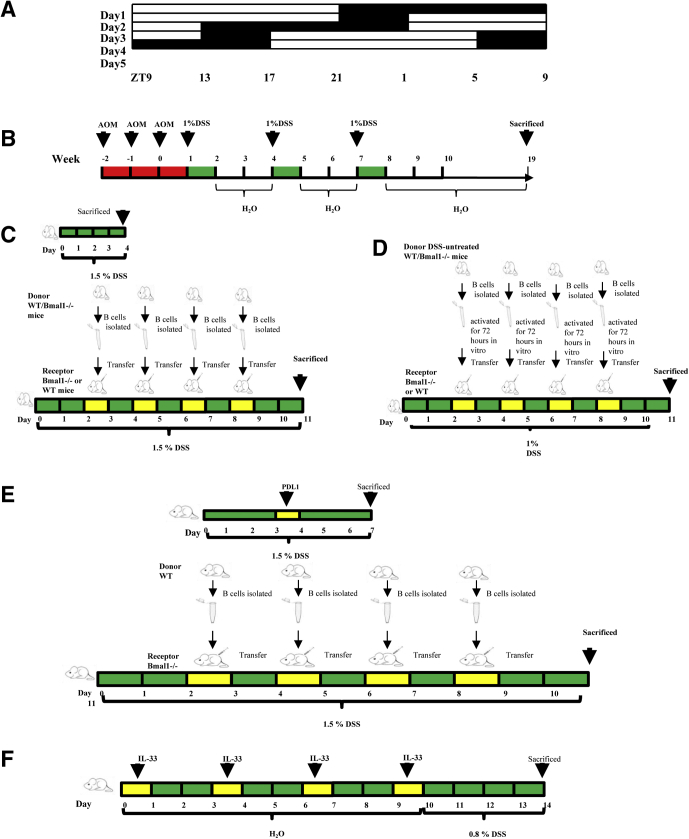
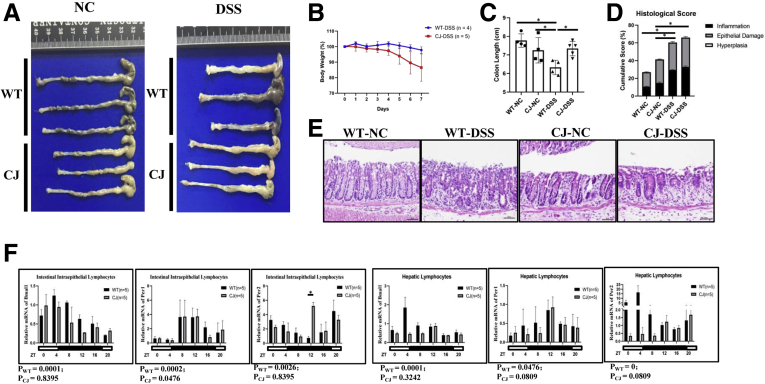
To study the effects of circadian clock disorders on IBD, chronic social jet lag (CJ) (Figure 1A) and circadian clock gene Bmal1 deficiency mouse models were established. We found that mice with CJ are susceptible to DSS-induced colitis (Figure 2A–E). Interestingly, Jonckheere-Terpstra-Kendall (JTK) analysis showed that the rhythm of most clock genes in either intestinal IELs or in hepatic lymphocytes of CJ mice were impaired significantly (Figure2F).
Figure 1.
Schematic diagram of experimental animal induction method. (A) Schematic diagram of CJ mouse model. (B) Mouse model of colitis-associated CRC, (C) Bmal1-/- or WT mice receiving B cells donated from DSS-treated WT or Bmal1-/- mice, (D) Bmal1-/- or WT mice receiving B cells donated from DSS-untreated WT or Bmal1-/- mice, (E) Bmal1-/- mice receiving B cells from WT mice with antagonist antibody to PDL1, (F) Bmal1-/- or WT mice with antagonist antibody to IL33. ZT, Zeitgeber time.
Figure 2.
DSS-induced colitis in mice with CJ. (A) Gross appearance of colorectum, (B) body weight, (C) colorectal length, (D) histologic score, (E) H&E of DSS-treated CJ and WT mice. (F) Transcription levels of core clock genes Bmal1, Per1, and Per2 in intestinal IELs and hepatic lymphocytes of CJ and WT mice from ZT 0 to 20. The P value is estimated by JTK cycle analysis. Zeitgeber time (ZT).
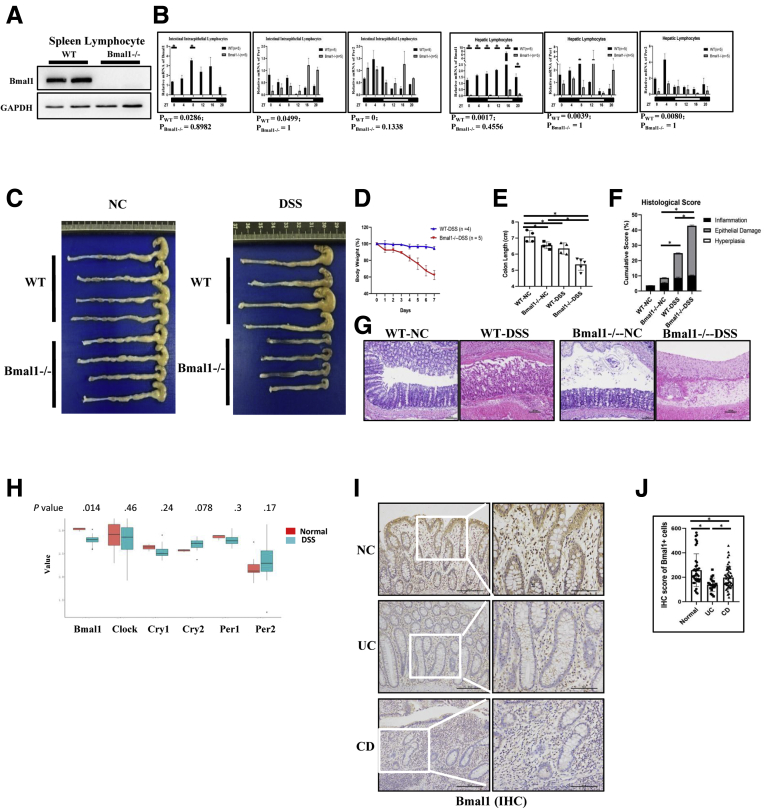
Because Bmal1 is one of the key components of core clock genes, we further studied the role of biological rhythm regulated by clock genes in colitis using Bmal1 knockout (Bmal1-/-) mice. Western blot showed that there was no expression of the Bmal1 gene in spleen lymphocytes (Figure 3A) in Bmal1-/- mice. We further analyzed the rhythm of Bmal1, Per1, and Per2 genes in IELs and hepatic lymphocytes of mice by JTK analysis.13,14 Five mice were selected every 4 hours to detect the transcription levels of the 3 genes within 24 hours by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) for analysis of the rhythm by JTK cycle analysis. JTK results showed that deficiency of Bmal1 impaired the rhythm of the 3 genes in the hepatic lymphocytes and IELs (Figure 3B).
Figure 3.
DSS-induced colitis in Bmal1-/-mice. (A) Bmal1 expression in spleen lymphocytes of Bmal1-/- and WT mice by Western blot. (B) Transcription levels of core clock genes Bmal1, Per1, and Per2 in the intestinal IELs and hepatic lymphocytes of Bmal1-/- and WT mice among 24 hours. The P value is estimated by JTK cycle analysis. (C) Gross appearance of colorectum, (D) body weight, (E) colorectal length, (F) histologic score, (G) H&E of DSS-treated Bmal1-/- and WT mice. (H) The mRNA expression levels of Bmal1, Clock, Cry1, Cry2, Per1, and Per2 in colons of DSS-treated mice by GEO2R of NCBI. A box plot with P value (calculated by the Wilcox test) was used. (I) Immunohistochemical staining and (J) semiquantitative analysis of Bmal1+ lymphocytes in IELs from UC and CD patients. ∗P < .05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NC, non-specific control.
After DSS treatment, the weight of Bmal1-/- mice were decreased significantly on the fourth day compared with that of control wild-type (WT) mice. On the seventh day, the length of colon in DSS-treated Bmal1-/- mice was shortened significantly compared with that of untreated Bmal1-/- mice (Figure 3C–E).
H&E staining showed that the height of intestinal villus in untreated Bmal1-/- mice was significantly shorter than that in untreated WT mice. Moreover, in the DSS-induced enteritis model, approximately 15% of the intestinal epithelium was damaged in WT mice, while more than 30% of the intestinal epithelium was damaged in Bmal1-/- mice (Figure 3F and G).
Using published data,15 we found that the expression of the Bmal1 gene in mice with ulcerative colitis (UC) was significantly lower than that in normal controls (Figure 3H). We further detected the expression of Bmal1 in 4 UC samples, 29 Crohn's disease (CD) samples (Table 1), and 10 normal counterparts by immunohistochemistry. The expression of Bmal1 in CD and UC groups was decreased significantly compared with that in the normal group (Figure 3I and J).
Table 1.
Patients With UC and CD
| Classification | Pathogenic site | Patients, n | % |
|---|---|---|---|
| CD | Small intestine | 16 | 84 |
| Large intestine | 13 | 68 | |
| Other (ileocecal junction) | 0 | 0 | |
| UC | Small intestine | 0 | 0 |
| Large intestine | 3 | 75 | |
| Other (ileocecal junction) | 1 | 25 |
Bmal1 Deficiency Leads to Decreased Proportions of Breg Cells in IELs and Results in Loss of Response and Circadian Rhythm in the Number of Breg Cells in IELs After DSS Treatment
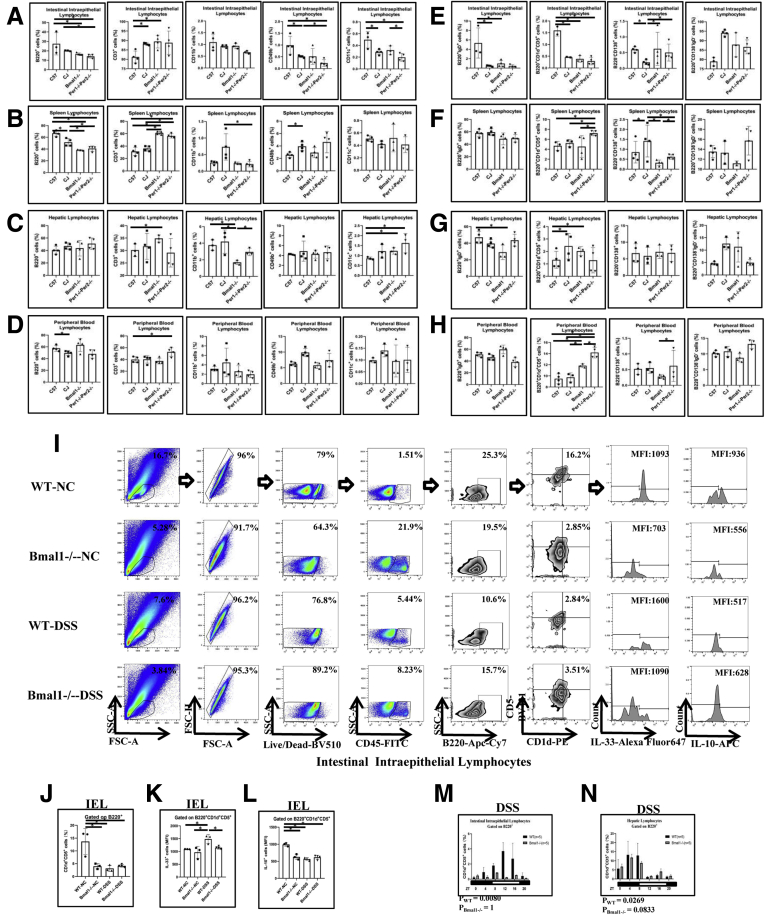
Proportions of main immune cells in peripheral organs of CJ mice, Bmal1-/- mice, and Per1-/-Per2-/- mice were analyzed by flow cytometry. B cells in IELs and spleen lymphocytes (SPLs) either from CJ mice or core clock gene deletion mice were significantly lower than that in WT mice. In addition, T cells in IELs, SPLs, and hepatic lymphocytes either from CJ mice or core clock gene deletion mice were higher than that in WT mice. In addition, monocytes in hepatic lymphocytes from core clock gene deletion mice were lower than that in WT mice. Natural killer cells in IELs from either CJ mice or core clock gene deletion mice were lower than that in WT mice, while natural killer cells in spleen from CJ mice were higher than that in WT mice. Dendritic cells in hepatic lymphocytes either from CJ mice or core clock gene deletion mice were higher than that in WT mice, while dendritic cells in IELs from Per1-/-Per2-/- mice were lower than that in WT mice (Figure 4A–D).
Figure 4.
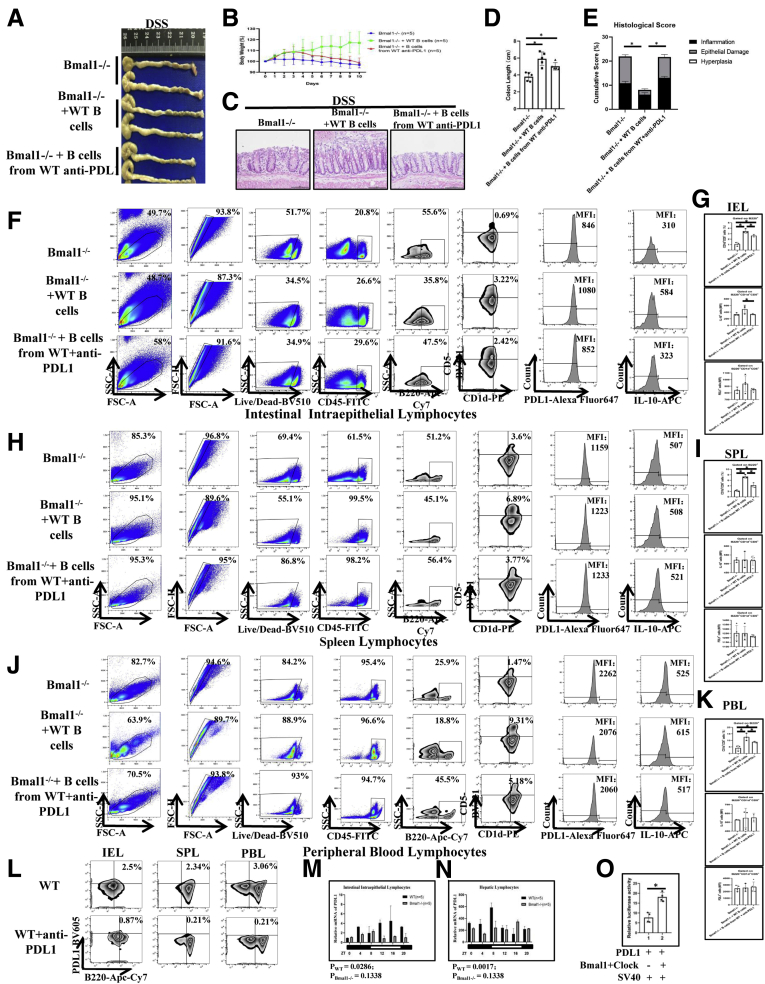
Proportion of main immune cells in peripheral organs of CJ mice, Bmal1-/-mice, and Per1-/-Per2-/-mice. (A–D) Proportion of B220+ cells, CD3+ cells, CD11c+ cells, CD49b+ cells, and CD11b+ cells in peripheral organs of CJ mice, Bmal1-/- mice, and Per1-/-Per2-/- mice. (E–H) Proportion of subpopulations of B cells in peripheral organs of CJ mice, Bmal1-/- mice, and Per1-/-Per2-/- mice. (I) Fluorescence-activated cell sorter (FACS) graphs showing B220+CD1d+CD5+ cells, MFI of IL10 and IL33 on B220+CD1d+CD5+ cells in IELs from DSS-treated or untreated Bmal1-/- and WT mice. (J) Proportion of CD1d+CD5+cells of B220+ cells in IELs. (K) MFI of IL33 and (L) IL10 on B220+CD1d+CD5+ in IELs. (M) Rhythmic oscillation of proportion of B220+CD1d+CD5+ cells in the IELs and (N) hepatic lymphocytes from DSS-treated Bmal1-/- and WT mice within 24 hours. The P value is estimated by JTK cycle analysis. FSC, forward scatter; NC, non-specific control; PE, Phycoerythrin; SSC, side scatter.
We further examined the proportions of subgroups of B cells in the peripheral organs. Naive B cells in IELs and hepatic lymphocytes either from CJ mice or core clock gene deletion mice were lower than that in WT mice. Notably, Breg cells in IELs from CJ mice or core clock gene deletion mice were lower than that in WT mice, while Breg cells in SPLs, hepatic lymphocytes, and peripheral blood lymphocytes (PBLs) from CJ mice or core clock gene deletion mice were higher than that in WT controls. Moreover, plasma cells in IELs from CJ mice were lower than that in WT mice, while plasma cells in liver from CJ mice were higher than that in WT mice. There was no significant difference about memory B cells in spleen, liver, peripheral blood, and intestinal epithelium from CJ mice and core clock gene deletion mice compared with that in controls (Figure 4E–H).
Interestingly, we found that the proportion of classic Breg cells (B220+CD5+CD1d+) in IELs and hepatic lymphocytes of WT mice was decreased significantly and fluctuated in a clock rhythm after DSS treatment (Figure 4I and J). Notably, Bmal1 deficiency abolished the response and circadian rhythm of the number of Breg cells in IELs and hepatic lymphocytes after DSS treatment (Figure 4M and N). Meanwhile, we found that there were no significant changes in the number of Breg cells of spleen and peripheral blood between WT and Bmal1-/- mice with or without DSS treatment (Figure 5C and D).
Figure 5.
The proportion of Breg cells expressing PDL1 in IELs, SPLs, and PBLs in DSS-treated and untreated Bmal1-/-mice. (A) Fluorescence-activated cell sorter (FACS) graphs showing B220+CD1d+CD5+ cells and MFI of PDL1 on B220+CD1d+CD5+ cells in IELs, (C) SPLs, and (E) PBLs of DSS-treated and untreated Bmal1-/- and WT mice. (B) MFI of PDL1 on B220+CD1d+CD5+ cells and proportion of B220+PDL1+ cells, CD5+PDL1+ cells, and CD1d+PDL1+ cells from CD45+ cells in IELs of DSS-treated and untreated Bmal1-/- and WT mice. MFI of PDL1 on B220+CD1d+CD5+ cells in (D) SPLs and (F) PBLs of DSS-treated and untreated Bmal1-/- and WT mice. APC, Allophycocyanin; FITC, Fluoresceine Isothiocyanate; FSC, forward scatter; NC, non-specific control; PE, Phycoerythrin; SSC, side scatter.
Interestingly, in WT mice, the levels of interleukin (IL)33 secreted by Breg cells in IELs from DSS-treated mice were increased by approximately 50% of that in DSS-untreated mice, while Bmal1 knockout attenuated the increased levels of IL33 in DSS-treated mice (Figure 4I and K). Meanwhile, Bmal1 knockout attenuated the decreased levels of IL10 secreted by Breg cells of IELs in DSS-treated mice (Figure 4I and L).
Adoptive Cell Transfer Treatment Using B Cells Isolated From WT Mice Successfully Alleviates Colitis in Bmal1-/- Mice While Adoptive Cell Transfer Treatment Using B Cells From Bmal1-/- Mice Greatly Accelerates the Progress of Colitis in WT Mice
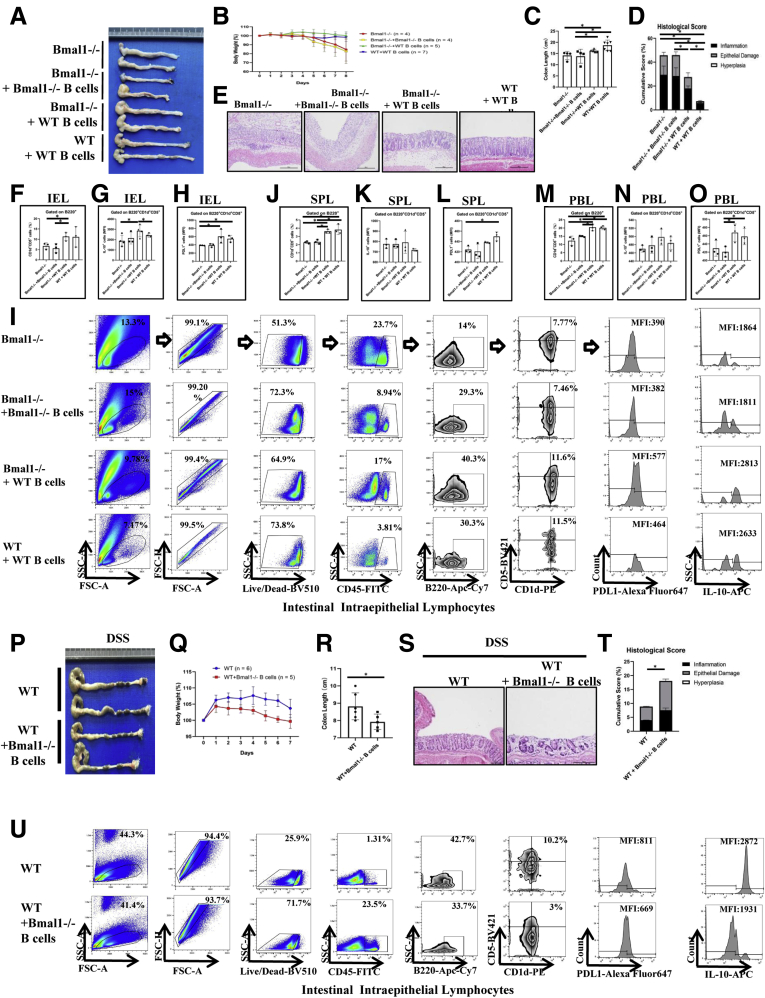
To further identify the role of Breg cells regulated by the clock gene Bmal1 in colitis, we used an adoptive cell transfer strategy by transferring B cells isolated from Bmal1-/- mice or WT mice to Bmal1-/- mice or WT mice (Figure 6A). We found that the body weight of Bmal1-/- mice receiving B cells from WT mice was almost not affected, while the body weight of Bmal1-/- mice without adoptive cell transfer treatment was decreased greatly beginning the fourth day after DSS treatment (Figure 6B). In addition, the colorectal length of the Bmal1-/- mice receiving B cells from WT mice was significantly longer than that of untreated Bmal1-/- mice (Figure 6C). Histologic severities of inflammatory responses indexed by cumulative scores showed that inflammatory responses of colon in Bmal1-/- mice receiving B cells from WT mice was decreased significantly compared with that in untreated Bmal1-/- mice and Bmal1-/- mice receiving B cells from Bmal1-/- mice (Figure 6D and E). Notably, Bmal1-/- mice receiving B cells from WT mice have more than a 2 times higher proportion of Breg cells of IELs, in which the mean fluorescence intensities (MFIs) of IL10 and PDL1 gated on Breg cells were increased significantly, compared with that of untreated Bmal1-/- mice and Bmal1-/- mice receiving B cells from Bmal1-/- mice (Figure 6F–I). In addition, we observed that Bmal1-/- mice receiving B cells from WT mice have increasing proportions of Breg cells in SPLs and PBLs, in which MFIs of IL10 and PDL1 gated on Breg cells were not changed significantly (Figure 6J–O and 7A and B).
Figure 6.
Effects of adoptive cell transfer treatment using B cells isolated from WT mice in DSS-treated Bmal1-/-mice or using B cells from Bmal1-/-mice in DSS-treated WT mice. (A) Gross appearance of colorectum, (B) body weight, (C) colon length, (D) histologic score of colon, and (E) H&E of colon from DSS-treated Bmal1-/- mice, Bmal1-/- mice receiving B cells isolated from Bmal1-/- mice, and from WT mice, and WT mice receiving B cells isolated from WT mice. (F) Proportion of CD1d+CD5+ cells gated from B220+ cells in IELs. (G) MFI of IL10 and (H) PDL1 on B220+CD1d+CD5+ in IELs. (I) Fluorescence-activated cell sorter (FACS) graphs of B220+CD1d+CD5+ cells, and MFI of PDL1 and IL10 on B220+CD1d+CD5+ cells in IELs. (J–L) Proportion of CD1d+CD5+ cells gated from B220+ cells, MFI of IL10 and PDL1 on B220+CD1d+CD5+ cells in SPLs. (M–O) Proportion of CD1d+CD5+ cells from B220+ cells, MFI of IL10 and PDL1 on B220+CD1d+CD5+ cells in PBLs. (P) Gross appearance of colorectum, (Q) body weight, (R) colon length, (S) H&E of colon, and (T) histologic score of colon from WT mice and WT mice receiving B cells from Bmal1-/- mice. (U) FACS graphs showing B220+CD1d+CD5+ cells, and MFI of IL10 and PDL1 on B220+CD1d+CD5+ cells in IELs. ∗P < .05. APC, Allophycocyanin; FITC, Fluoresceine Isothiocyanate; FSC, forward scatter; PE, Phycoerythrin; SSC, side scatter.
Figure 7.
Fluorescence-activated cell sorter (FACS) graphs showing B220+CD1d+CD5+cells, and the MFI of PDL1 and IL10 on (A) B220+CD1d+CD5+cells in SPLs and (B) PBLs from DSS-treating Bmal1-/-mice, Bmal1-/-receiving B cells from Bmal1-/-mice, Bmal1-/-receiving B cells from WT mice, and WT receiving B cells from WT mice. FACS graphs showing B220+CD1d+CD5+ cells, and MFI of PDL1 and IL10 on B220+CD1d+CD5+ cells in (C) SPLs and (D) PBLs from DSS-treating WT mice and WT receiving B cells from Bmal1-/- mice. APC, Allophycocyanin; FITC, Fluoresceine Isothiocyanate; FSC, forward scatter; PE, Phycoerythrin; SSC, side scatter.
Furthermore, we isolated the B cells from DSS-treated Bmal1-/- mice and transferred them into DSS-treated WT mice (Figure 6P). We found that the weight of WT mice transferred with B cells from DSS-treated Bmal1-/- mice (WT receivers) decreased more rapidly than control WT mice after DSS treatment (Figure 6Q). The colorectal length of WT receivers was significantly shorter than that of control WT mice (Figure 6R). Accordingly, histologic severities of inflammatory responses indexed by cumulative scores showed that inflammatory responses of the colon in WT mice receiving B cells from Bmal1-/- mice was increased significantly compared with that in control WT mice (Figure 6S and T).
Notably, the proportion of Breg cells of IELs in receiver WT mice were approximately one third of that in WT controls. WT mice receiving B cells from Bmal1-/- mice have decreased proportions of Breg cells of IELs, in which MFIs of IL10 and PDL1 were decreased, compared with that of untreated WT mice (Figure 6U). In addition, we observed that WT mice receiving B cells from Bmal1-/- mice have no significant changes in the proportion of Breg cells and MFIs of IL10 or PDL1 gated on Breg cells in SPLs and PBLs (Figure 7C and D).
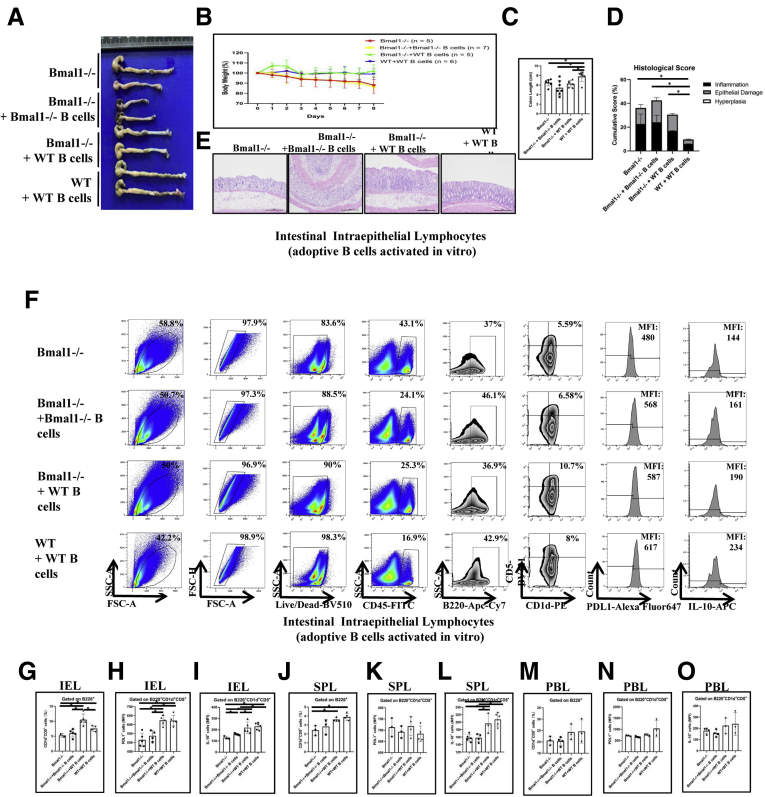
Moreover, we isolated the spleen B cells from DSS-untreated WT mice or Bmal1-/- mice and activated them in vitro, these B cells then were used for adoptive cell transfer treatment to the recipient mice (Figure 8A). Body weight (Figure 8B), length (Figure 8C), and inflammatory responses of colon in Bmal1-/- mice receiving in vitro activating B cells from WT mice were partially restored compared with that in DSS-treated Bmal1-/- mice (Figure 8D and E). The proportion of Breg cells in IELs and SPLs of Bmal1-/- mice receiving in vitro activating B cells from WT mice was increased significantly (Figure 8F, G, and J). MFIs of IL10 gated on Breg cells of IELs (Figure 8F, G, and I) and SPLs (Figure 8J and L), but not MFIs of PDL1 on Breg cells in SPLs (Figure 8K) or MFIs of PDL1 and IL10 gated on Breg cells in PBLs (Figure 8M–O), was increased significantly, in Bmal1-/- mice receiving in vitro activating B cells from WT mice compared with that in DSS-treated Bmal1-/- mice.
Figure 8.
Effects of adoptive cell transfer treatment using adoptive B cells activated in vitro and isolated from WT mice in DSS-untreated Bmal1-/-mice or using B cells from Bmal1-/-mice in DSS-untreated WT mice. (A) Gross appearance of colorectum, (B) body weight, (C) colon length, (D) histologic score of colon, (E) H&E of colon from DSS-treated Bmal1-/- mice, Bmal1-/- mice receiving B cells isolated from Bmal1-/- mice and from WT mice, and WT mice receiving B cells isolated from WT mice. (F) Fluorescence-activated cell sorter (FACS) graphs showing B220+CD1d+CD5+ cells, and MFI of IL10 and PDL1 on B220+CD1d+CD5+ in IELs from models. (G) Proportion of CD1d+CD5+ cells from B220+ cells, and MFI of (H) PDL1 and (I) IL10 on B220+CD1d+CD5+ cells in IELs from models. (J) Proportion of CD1d+CD5+ cells from B220+ in SPLs, and (K) MFI of PDL1 and (L) IL10 on B220+CD1d+CD5+ cells in SPLs from models. (M) Proportion of CD1d+CD5+ cells from B220+ in PBLs, and (N) MFI of PDL1 and (O) IL10 on B220+CD1d+CD5+ cells in SPLs from models. ∗P < .05.
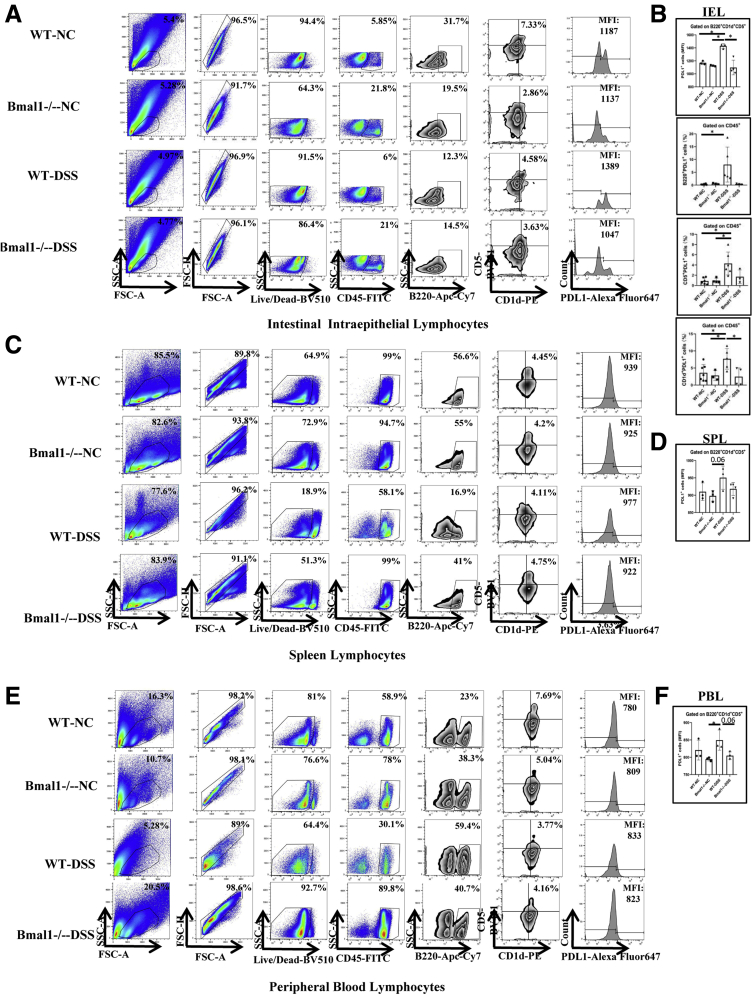
Breg Cells Expressing PDL1 in IELs Are Activated to Alleviate DSS-Induced Severity of Colitis and Dysregulated in DSS-Treated Bmal1-/- Mice
Our results indicate that Breg cells regulated by Bmal1 are very important for IBD development. The characteristics of Breg cells regulated by Bmal1 still are unclear. PDL1, a key immune checkpoint molecule, is important to protect normal, healthy tissue from an immune response. Importantly, we found that the proportion of Breg cells expressing PDL1 of IELs in DSS-treated WT mice was increased significantly by approximately 17% compared with that in untreated WT mice, while the MFI of PDL1 on Breg cells of IELs in DSS-treated Bmal1-/- mice had no significant changes compared with that in untreated Bmal1-/- mice (Figure 5A and B). In addition, MFI values of PDL1 on Breg cells in PBLs and SPLs had no significant changes in DSS-untreated WT mice, DSS-treated WT mice, DSS-untreated Bmal1-/- mice, and DSS-treated Bmal1-/- mice (Figure 5C–F).
To study the functions of PDL1+B220+CD5+CD1d+ cells in colitis, we isolated B cells from WT mice with or without early immunization against PDL1 (Figure 9L and 12E), and then transferred these B cells into Bmal1-/- mice (Figure 9A). We found that, on the fourth day of DSS induction, the body weight of Bmal1-/- mice receiving B cells from WT mice were significantly restored compared with that in Bmal1-/- mice (Figure 9B). On the 12th day, the length of the colon in Bmal1-/- mice receiving B cells from WT mice was significantly longer than that in Bmal1-/- mice (Figure 9D). Histologically, the inflammation and epithelial damage of the colon in receiver Bmal1-/- mice with WT B-cell transfer treatment were alleviated significantly. However, antagonizing PDL1 greatly abolished the recovery effects in Bmal1-/- mice receiving B cells from WT mice (Figure 9C and E).
Figure 9.
The effects of Breg cells expressing PDL1 in IELs in DSS-treated Bmal1-/-mice. (A) Gross appearance of colorectum, (B) body weight of mice, (C) H&E of colon, (D) colon length, (E) histologic score of colon from DSS-treated Bmal1-/- mice, Bmal1-/- mice receiving B cells donated from WT mice, and Bmal1-/- mice receiving B cells donated from WT mice and PDL1 antagonist antibody. (F) Fluorescence-activated cell sorter (FACS) graphs showing B220+CD1d+CD5+ cells, and MFI of PDL1 and IL10 on B220+CD1d+CD5+ in IELs, (H) SPLs, and (J) PBLs. (G) Proportion of CD1d+CD5+ gated on B220+ cells, and MFI of IL10 and PDL1 on B220+CD1d+CD5+ cells in IELs, (I) SPLs, and (K) PBLs. (L) FACS graphs showing PDL1+B220+ cells in IELs, SPLs, and PBLs of donor mice that antagonize PDL1. (M) Transcription levels of PDL1 in the IELs and (N) hepatic lymphocytes of Bmal1-/- and WT mice among 24 hours. The P value is estimated by JTK cycle analysis. (O) The effects of Bmal1 and clock complexes on luciferase activities of PDL1 promoter reporter. ∗P < .05. APC, Allophycocyanin; FITC, Fluoresceine Isothiocyanate; FSC, forward scatter; PE, Phycoerythrin; SSC, side scatter.
Figure 12.
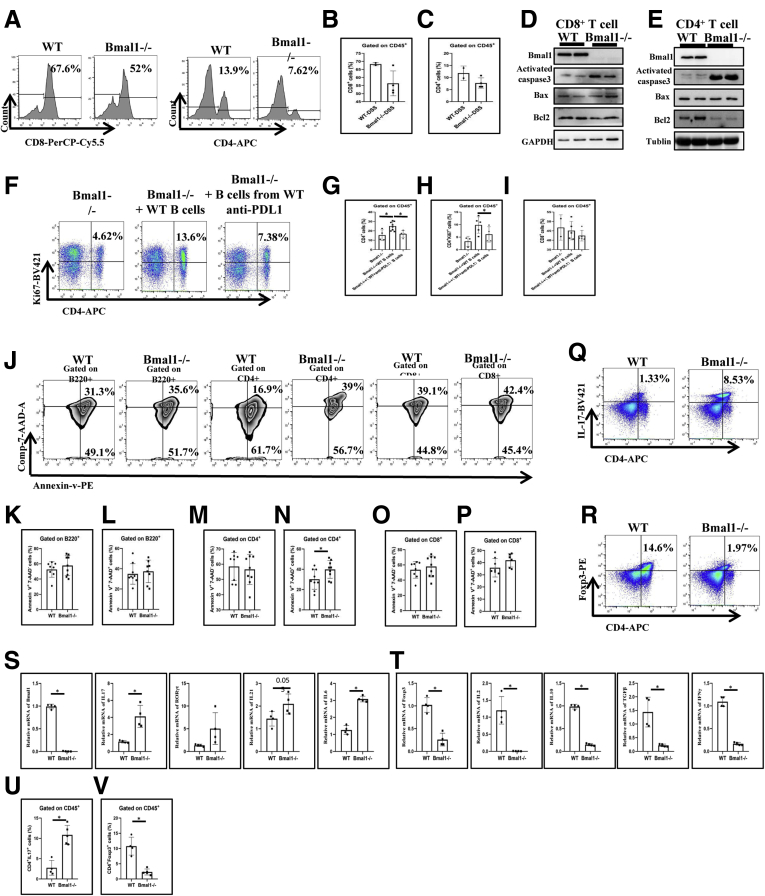
Cell death of activated CD4+T cells in DSS-treated Bmal1-/-mice and the effects of PDL1 antagonist antibody. (A) Fluorescence-activated cell sorter (FACS) graphs and (B) proportions of CD8+ and (C) CD4+ T cells in the colon of DSS-treated Bmal1-/- mice and WT mice. (D) Protein expression of Bmal1, activated caspase 3, Bcl-2 Associated X Protein, and Bcl2 in CD8+ T cells and (E) CD4+ T cells in the colons of DSS-treated Bmal1-/- mice and WT mice. (F) FACS graphs of CD4+Ki67+ cells, (G) proportions of CD4+ T cells, (H) CD4+Ki67+ cells, and (I) CD8+ T cells in the IELs of Bmal1-/- mice, Bmal1-/- mice receiving B cells from WT mice, Bmal1-/- mice receiving B cells from WT mice and PDL1 antagonist antibodies. (J) FACS graphs and proportion of (K) Annexin-v+Comp-7-AAD-B220+ or (L) Annexin-v+Comp-7-AAD+ B220+ cells, (M) Annexin-v+Comp-7-AAD-CD4+ or (N) Annexin-v+Comp-7-AAD+ CD4+ cells, and (O) Annexin-v+Comp-7-AAD-CD8+ or (P) Annexin-v+Comp-7-AAD+ CD8+ cells in IELs from DSS-treated WT mice and Bmal1-/- mice. (Q) FACS graphs and (U) proportions of CD4+IL17+ cells in IELs. (R) FACS graphs and (V) proportions of CD4+Foxp3+ cells in IELs. (S) Transcription expression of Bmal1, IL17, RORγt, IL21, and IL6 in IELs of DSS-treated WT and Bmal1-/- mice. (T) Transcription expression of Forkhead Box P3, IL2, IL10, TGFβ, and IFNγ in IELs of DSS-treated WT and Bmal1-/- mice. ∗P < .05. APC, Allophycocyanin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The Breg cells of IELs in receiver Bmal1-/- mice with WT B-cell transfer treatment (Bmal1-/- receivers) were nearly 4 times those in Bmal1-/- control mice, and PDL1 antagonism successfully suppressed the increase of the number of Breg cells of IELs in receiver Bmal1-/- mice with WT B-cell transfer treatment (<2 times that in Bmal1-/- control mice) (Figure 9F and G). Meanwhile, we found that the MFI of PDL1 on Breg cells of IELs in receiver Bmal1-/- mice with WT B-cell transfer treatment was nearly 25% of that in Bmal1-/- control mice (Figure 9F and G). In addition, there were no differences in the MFIs of PDL1 or IL10 gated on Breg cells in PBLs and SPLs in DSS-treated Bmal1-/- mice, DSS-treated Bmal1-/- mice with WT B-cell transfer treatment, DSS-treated Bmal1-/- mice with WT B-cell transfer treatment, and PDL1 antagonism (Figure 9H–K).
Functionally, PDL1 reporter luciferase activity assays showed that the Bmal1/clock complex promoted the transcription of PDL1 (Figure 9O). Moreover, we found that PDL1 showed rhythmic fluctuations in the hepatic lymphocytes and IELs of WT mice, but no rhythm in Bmal1-/- mice (Figure 9M and N).
IL33 in the Intestinal Microenvironment Is Key for Bmal1-Regulated PDL1+ Breg Cells
To find the key factors involved in Bmal1-regulated PDL1+ Breg cells, we examined the messenger RNA (mRNA) expression of inflammatory factors including IL4, IL6, IL10, IL21, IL33, transforming growth factor β (TGFβ), and monocyte chemoattractant protein-1 (MCP-1) in IELs from DSS-treated or -untreated Bmal1 knockout and WT mice.
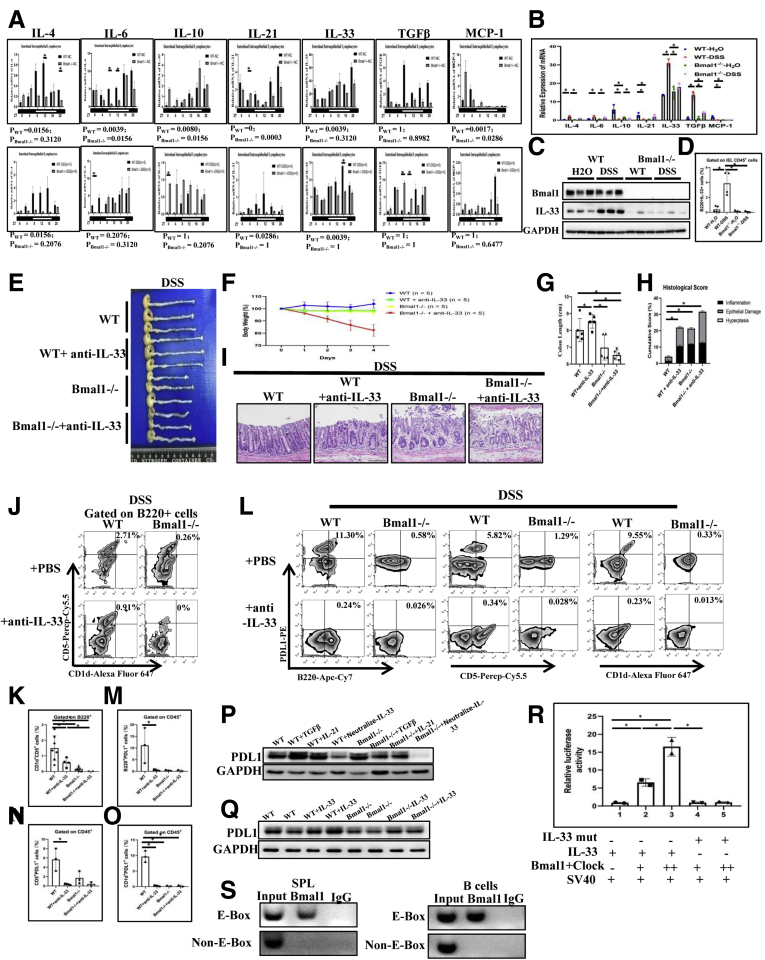
Interestingly, we detected circadian oscillation for mRNA levels of IL4, IL21, and IL33 among IL4, IL6, IL10, IL21, IL33, TGFβ, and MCP-1 in the IELs of DSS-treated WT mice, while mRNA levels of these 7 cytokines had no rhythm at all in the IELs of DSS-treated Bmal1-/- mice within 24 hours (Figure 10A). Notably, the oscillation of IL33 transcripts in IELs of DSS-treated WT was more than twice that in DSS-treated Bmal1-/- mice at Zeitgeber time 16 (Figure 10A). Expression levels of IL4, IL6, IL10, IL21, IL33, and MCP-1, except for TGFβ, in the IELs of DSS-untreated WT mice, showed physiological circadian oscillation. Meanwhile, oscillation of IL6 and IL21 transcripts showed decreased amplitude, while oscillation of IL10 and MCP-1 transcripts showed increased amplitude in DSS-untreated Bmal1-/- mice than oscillation of IL10 and MCP-1 transcripts in DSS-untreated WT mice (Figure 10A).
Figure 10.
The effects of IL33 in intestinal microenvironment on PDL1+Breg cells in DSS-treated Bmal1-/-mice. (A) Transcription levels of IL4, IL6, IL10, IL21, IL33, TGFβ, and MCP-1 in the IELs of DSS-treated and untreated Bmal1-/- and WT mice among 24 hours. The P value was estimated by JTK cycle analysis. (B) Transcription levels of inflammatory cytokines IL4, IL6, IL10, IL21, IL33, TGFβ, and MCP-1 in the IELs of DSS-treated Bmal1-/- and WT mice at the same time. (C) Protein expressions of Bmal1 and IL33, (D) proportion of B220+IL33+ cells in IELs of DSS-treated Bmal1-/- and WT mice. (E) Gross appearance of colorectum, (F) body weight during DSS induction, (G) colorectal length, (H) histologic score of colon, (I) H&E of colon from DSS-treated WT mice, WT mice with IL33 antagonist antibody, Bmal1-/- mice, and Bmal1-/- mice with IL33 antagonist antibody. (J) FACS graphs and (K) proportion of CD5+CD1d+ cells from B220+ cells in IELs. (L) Fluorescence-activated cell sorter (FACS) graphs and (M) proportion of B220+PDL1+, (N) CD1d+PDL1+, and (O) CD5+PDL1+ cells in IELs. (P) PDL1 expressions in B cells isolated from WT and Bmal1-/- spleen treated with IL21, TGFβ, or IL33 neutralizing antibodies by Western blot. (Q) PDL1 expression in B cells isolated from WT and Bmal1-/- spleen treated with IL33 by Western blot. (R) The effects of Bmal1 and clock complexes on luciferase activities of IL33 promoter reporter with or without E-box site mutation. (S) ChIP assay to detect the binding of IL33 gene promoter with Bmal1 in SPLs and B cells. ∗P < .05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
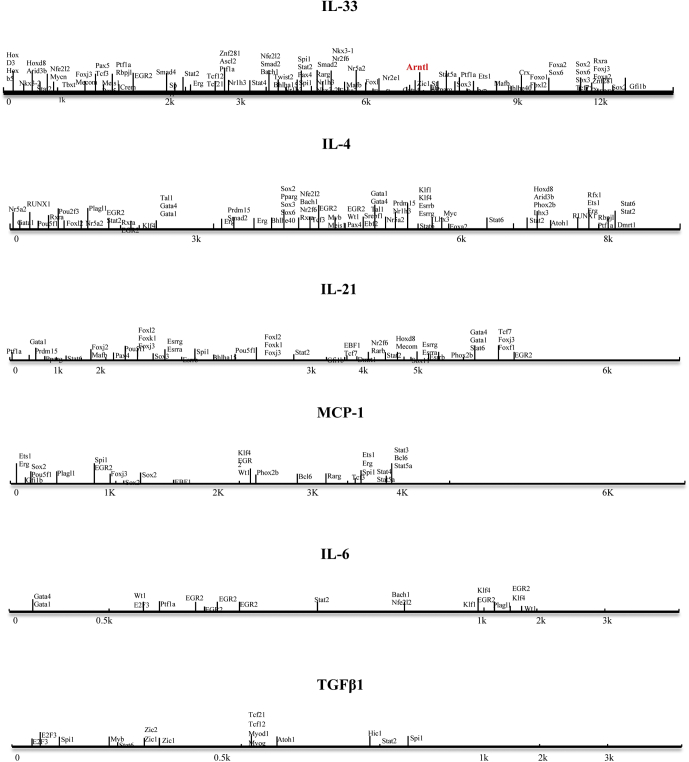
In addition, we also detected IL4, IL6, IL10, IL21, IL33, TGFβ, and MCP-1 of IELs in DSS-untreated WT and Bmal1-/- mice, DSS-treated WT, and Bmal1-/- mice at the same specific time. We found that DSS treatment significantly affected mRNA expression of inflammatory factors in IELs (among them, IL4, IL6, IL33, and TGFβ were up-regulated while IL10, IL21, and MCP-1 were down-regulated) of WT mice (Figure 10B). However, in IELs of Bmal1-/- mice, DSS treatment only significantly up-regulated mRNA expression of IL4, IL6, IL10, IL33, and TGFβ (Figure 10B). Furthermore, Bmal1 defects (Bmal1-/- mice) significantly impaired DSS-induced mRNA increases of intestinal IL33 and TGFβ when compared with that in WT mice (Figure 10B). Among these inflammatory factors, the IL33 expression level in IELs was the highest. At the same time, Western blot indicated that DSS treatment greatly increased intestinal IL33 expression in WT mice and Bmal1 deficiency (Bmal1-/- mice) significantly alleviated the DSS-treated increased expression of intestinal IL33 (Figure 10C). Flow cytometry analysis showed that the number of IL33+B220+ in IELs of DSS-treated WT mice was increased significantly and Bmal1 deficiency greatly eliminated the DSS-induced increasing number of IL33+B220+ cells (Figure 10D). In addition, we tested whether there were some binding sites for Bmal1 in promoter regions among IL4, IL6, IL10, IL21, IL33, TGFβ, and MCP-1 in the JASPAR database (http://jaspar.genereg.net). We found that there was an E-box site (CACGTG) with high confidence (13.2571) and an E’-box site (CACGTT) with confidence (6.11163) in the promoter of IL33, while among other cytokines, only the IL4 promoter region has an E'-box binding site, with a confidence value of 6.11163 (Figure 11).
Figure 11.
Binding sites for Bmal1 in promoter regions among 7 cytokine genes were predicted in the JASPAR database.
To further determine the role of IL33 in Bmal1-regulated colitis in WT and Bmal1-/- mice, IL33 were antagonized with specific antibodies in the following experiments (Figures 10E and F). Ten days after IL33 antagonism, we analyzed the degree of colorectal inflammation and the proportion of PDL1+B220+CD5+CD1d+ cells among WT and IL33 neutralized WT mice, Bmal1-/- and IL33 neutralized Bmal1-/- mice, respectively. In DSS-treated Bmal1-/- mice, antagonizing IL33 worsened the weight loss (Figure 10F) and inflammatory damage of the colon in Bmal1-/- mice (Figure 10H and I). In DSS-treated WT mice, antagonizing IL33 aggravated weight loss, inflammatory damage of the colon, and prolonged the length of colon (Figure 10F–I).
Functionally, we studied the effects of IL33 antagonism on PDL1+Breg+ cells in DSS-treated WT mice and Bmal1-/- mice. We found that IL33 antagonism significantly reduced the number of Breg cells in IELs from DSS-treated WT mice (approximately one third of that in untreated WT mice). However, IL33 neutralization only slightly cut off the number of Breg cells in IELs from DSS-treated Bmal1-/- mice (Figure 10J and K). Furthermore, we found that IL33 neutralization greatly decreased the numbers of PDL1+B220+ cells including PDL1+CD1d+ cells and PDL1+CD5+ cells - in IELs from DSS-treated WT mice. Similarly, IL33 neutralization had little effect on the number of PDL1+B220+ cells, including PDL1+CD1d+ cells and PDL1+CD5+ cells (PDL1+ Breg cells) in IELs from DSS-treated Bmal1-/- mice (Figure 10L–O).
Western blot showed that IL33 neutralizing antibody decreased expressions of PDL1 in B cells from both WT mice and Bmal1-/- mice, while exogenous TGFβ or IL21 significantly increased the expression of PDL1 in B cells from WT mice, but had no effect on expression of PDL1 in B cells from Bmal1-/- mice (Figure 10P). Furthermore, exogenous IL33 increased the expression of PDL1 in B cells both from WT mice and Bmal1-/- mice, however, the effects of exogenous IL33-induced increased expression of PDL1 were impaired significantly in B cells from Bmal1-/- mice (Figure 10Q). These results suggest that IL33 is key for PDL1 expression in B cells.
Using a luciferase reporter system of the IL33 promoter, we found that the Bmal1/clock complex significantly promotes luciferase activity of the IL33 promoter while the Bmal1/clock complex had no effect on luciferase activity of the IL33 promoter if the E-box binding site for Bmal1/clock complex was mutated (Figure 10R). Moreover, in mouse IELs and even B cells, we found that Bmal1 could bind directly to the IL33 promoter through the E-box binding site by chromatin immunoprecipitation (ChIP) (Figure 10S), further supporting IL33 as a target of Bmal1 transcriptionally.
PDL1+ B Cells Induce Cell Death of Activated CD4+ T Cells in DSS-Treated Bmal1-/- Mice
To further study the effects of Bmal1-regulated PDL1+ B cells, especially Breg cells in IBD and its related diseases, we analyzed the number of CD4+ T cells and CD8+ T cells in IELs of DSS-treated WT and Bmal1-/- mice. Surprisingly, the number of CD4+ T cells in Bmal1-/- mice was approximately half of that in WT mice, while the number of CD8+ T cells in Bmal1-/- mice was approximately 18% of that in WT mice (Figure 12A–C). Then, CD8+ T cells and CD4+ T cells from spleens of DSS-treated WT and Bmal1-/- mice were isolated to detect protein levels of activated caspase 3, Bcl-2 Associated X Protein, and Bcl2, respectively. The results showed that the expression of activated caspase 3 in CD4+ T cells and CD8+ T cells from Bmal1-/- mice was higher than that in WT mice. The expression of Bcl2 in CD4+ T cells from Bmal1-/- mice was lower than that in WT mice obviously, while Bcl2 expression in CD8+ T cells between Bmal1-/- and WT mice had no significant difference (Figure 12D and E).
To further identify that the B cells expressing PDL1 are key for proliferation CD4+ T cells or CD8+ T cells of the colon in the DSS-induced mouse colitis model, we transferred the B cells of WT mice back to Bmal1-/- mice or transferred the B cells from WT mice antagonized with PDL1 antibody to Bmal1-/- mice. We found the proportion of CD4+ T cells or Ki67+ CD4+ T cells in Bmal1-/- mice receiving B cells from WT mice was increased significantly and PDL1 antagonism partly reduced the effects of adoptive cell transfer therapy using B cells isolated from WT mice in Bmal1-/- mice (Figure 12F–H). However, the proportion of CD8+ T cells in both recipients was not significantly changed compared with control DSS-treated Bmal1-/- mice (Figure 12I).
Furthermore, we analyzed early and late apoptosis of B220+ cells, CD4+ T cells, and CD8+ T cells of IELs in Bmal1-/- and WT mice treated by DSS for 3 days. We found that only the late apoptosis index of CD4+ T cells not B220+ cells, CD8+ T cells of IELs in Bmal1-/- mice were significantly higher than that in WT mice (Figure 12J–P). Functionally, the proportion of Th17 cells in Bmal1-/- mice was approximately 3–7 times that of WT mice (Figure 12Q and U), and the proportion of Treg cells in Bmal1-/- mice was approximately one seventh to one fifth of the control group (Figure 12R and V). The transcription levels of cytokines IL17, IL21, and IL6 closely related to Th17 function in Bmal1-/- IELs were significantly higher than those in the control group (Figure 12S). The transcription levels of cytokines Forkhead Box P3, IL6, TGFβ, and interferon γ (IFNγ), which are closely related to Treg cell function, were significantly lower than those of the control group (Figure 12T).
Circadian Clock Disorders Characterized as Decreased Numbers of Breg+PDL1+ Cells and Dysfunction of CD4+ T Cells Promotes Colitis-Associated CRC
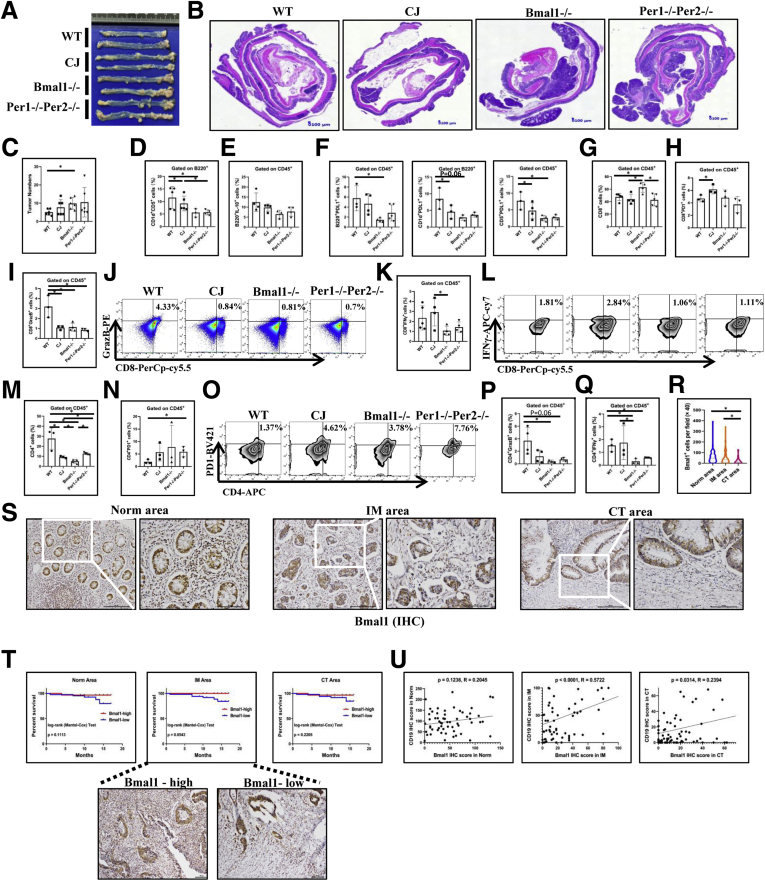
Azoxymethane (AOM) and DSS were used to produce colitis-associated CRC in CJ, Bmal1-/-, and Per1-/-Per2-/- mice (Figure 1B). The results showed that circadian clock disorders in mice were susceptible to pathogenesis of colon cancer. The number of colorectal tumors in CJ, Bmal1-/-, and Per1-/-Per2-/- mice was increased significantly compared with that in control WT mice. For example, the number of colorectal tumors in Bmal1-/- mice was more than twice that in WT mice (Figure 13A–C).
Figure 13.
Effects of disorders of biological rhythm on colitis-associated CRC and expression and prognosis of Bmal1 in clinical samples of CRC. (A) Gross appearance of colorectum, (B) H&E of colon, and (C) tumor numbers from CJ, Bmal1-/-, Per1-/-Per2-/-, and WT mice of enteritis-related bowel cancer model. (D) Proportion of CD1d+CD5+ cells from B220+ cells, (E) IL10+B220+ cells, (F) B220+/CD1d+/CD5+PDL1+ cells in IELs of models. (G) Proportion of CD8+ cells, (H) PD1+ CD8+ cells, (I) CD8+GrazB+ cells and (J) Fluorescence-activated cell sorter (FACS) graphs of CD8+GrazB+ in IELs of models. (K) Proportions and (L) FACS graphs of CD8+IFNγ+ cells in IELs of models. (M) Proportion of CD4+ T cells in IELs of models. (N) Proportion and (O) FACS graphs of CD4+PD1+ cells in IELs of models. (P) Proportion of GranzB+CD4+ cells, (Q) IFNγ+CD4+ cells in the IELs of models. (R) Semiquantitative analysis and (S) immunochemical staining of Bmal1+ immune cells from CT, the front of the IM, and paracancerous area (Norm) in 91 CRC patients. (T) Survival curves of CRC patients with low or high expression of Bmal1 in the 3 regions of tumor samples. (U) Correlation of CD19+ cells and Bmal1+ immune cells in the 3 regions of tumor samples. ∗P < .05.
First, we analyzed the Breg cells in IELs containing infiltrating lymphocytes in these tumors among CJ, Bmal1-/-, and Per1-/-Per2-/- and WT mice of enteritis-related bowel cancer model. The proportion of Breg cells in AOM-/DSS-induced mice with circadian clock disorders (CJ, Bmal1-/-, and Per1-/-Per2-/-mice) was significantly lower than that in control WT mice (Figure 13D). The number of B220+IL10+ cells also was lower in Bmal1-/- and Per1-/-Per2-/- mice than that in WT mice (Figure 13E). Furthermore, we detected the cell proportions of PDL1+B220+, PDL1+CD1d+, and PDL1+CD5+. We found that the proportion of PDL1+Breg+ cells in IELs of Bmal1-/- mice was significantly lower than that of controls (approximately one third of the control) (Figure 13F).
Then, we examined the number of CD8+ T cells and only the number of CD8+ T cells in Bmal1-/- mice was slightly higher than those in control mice (Figure 13G). Furthermore, only the number of CD8+ T cells expressing PD1 in the CJ group was significantly higher than that in the control group (Figure 13H). We also examined the effective molecules killing tumor cells, including IFNγ and Granzyme B (GranzB) in CD8+ T cells. The proportion of GranzB+CD8+ cells, not IFNγ+CD8+ cells, was significantly lower in mice with circadian clock disorders than that in WT mice (Figure 13I–K).
At the same time, we observed the proportion of CD4+ T cells in IELs including infiltrating lymphocytes in intestinal adenoma tissue in mice. The results showed that the number of CD4+ cells in Bmal1-/- and Per1-/-Per2-/- mice with internal circadian rhythm disorder or CJ mice with external circadian rhythm disorder were significantly lower than that in WT mice (Figure 13M). Notably, among these CD4+ cells in mice with abnormal biological clocks, especially in Per1-/-Per2-/- mice, the number of immunosuppressive CD4+ cells, such as PD1+CD4+ cells, was higher than that in WT mice (Figure 13N and O). The number of effective CD4 cells such as IFNγ+CD4+ and GranzB+CD4+ in mice with internal rhythm disorder were both significantly lower than those in WT (Figure 13P and Q).
To further identify the potential role of Bmal1-regulated lymphocytes in the pathogenesis of CRC, we examined the expression of the Bmal1 gene in the lymphocytes among tumor centers (CT), the front of the tumor invasion (invasive margin [IM]), and the paracancerous area in 91 cases of colorectal specimens of CRC patients without metastasis (Table 2). Semiquantitative analyses showed that the number of Bmal1+ lymphocytes in the paracancerous area was significantly higher than that in CT, and the number of Bmal1+ lymphocytes in the IM area was significantly higher than that in the CT (Figure 13R and S). In addition, we found that patients with high expression of Bmal1 (more Bmal1+ lymphocytes) in the IM area had a better prognosis than those with low expression of Bmal1 (few Bmal1+ lymphocytes) (Figure 13T). The correlation analysis showed that the expression of Bmal1 in lymphocytes correlated positively with the expression of CD19 in the IM area (Figure 13U).
Table 2.
Patients With CRC Without Distant Metastasis (n = 91)
| Characteristic | Patients, n | % |
|---|---|---|
| Age, y | ||
| ≤60 | 46 | 50.55 |
| >60 | 45 | 49.45 |
| Tumor | ||
| T1 | 9 | 9.89 |
| T2 | 19 | 20.88 |
| T3 | 28 | 30.77 |
| T4 | 35 | 38.46 |
| Regional lymph node | ||
| N0 | 90 | 100 |
| N1 | 0 | 0.00 |
| N2 | 0 | 0.00 |
| N3 | 0 | 0.00 |
| Molecular subtypes | ||
| MLH1(+) | 85 | 93.41 |
| MSH2(+) | 86 | 94.51 |
| MSH6(+) | 85 | 93.41 |
| PMS2(+) | 84 | 92.31 |
| ERCC1(+) | 64 | 70.33 |
| Ki67(+) | 86 | 94.51 |
| NSE(+) | 5 | 5.49 |
| Syn(+) | 7 | 7.69 |
| CgA(+) | 2 | 2.20 |
| CDX2(+) | 3 | 3.30 |
| Villin | 2 | 2.20 |
| Ck | 2 | 2.20 |
| CK7 | 0 | 0.00 |
| CK8/18 | 0 | 0.00 |
| CK19 | 1 | 1.10 |
| P53 | 0 | 0.00 |
| CK20 | 6 | 6.59 |
| CD56 | 1 | 1.10 |
Discussion
B cells are important components of acquired immune systems in the gut, which exert an effect in a microenvironment where they are exposed to commensal microflora and food antigens, but at the same time they fight against harmful pathogens. Clinical evidence has shown that shift work and CJ can cause sleep disorders and increased risk for pathogenesis of IBD.14,16, 17, 18, 19, 20 However, the role of the circadian clock, especially the mechanism by which the circadian clock regulates IBD, still largely is unclear.
Here, we found that mice with circadian clock disorders are susceptible to colitis. We used different mouse models for circadian clock disorders including CJ mice, or mice lacking the core clock gene Bmal1, to establish the relationship between regulation of the circadian clock and IBD. It was observed that shifting the 12:12-hour light–dark cycle in mice led to more severe enteritis than a constant 12:12-hour light–dark cycle in mice.21 In addition, several studies have suggested that the core clock gene Bmal1 is involved in regulating colonic permeability in a circadian manner22 intestinal regeneration.23
Moreover, we found that circadian clock disorders promote colitis in mice by regulating Breg cells in IELs. Breg cells have been suggested in the inhibition of excessive inflammation24 and experimental enteritis in mice.3, 4, 5 In addition, Breg cells also are important for antitumor immune responses as suppressors.25 Notably, we found that Bmal1 deficiency lead to loss of rhythmic oscillation for the number of Breg cells in colons from DSS-treated mice. Our study reports on the role of circadian clock–regulated B cells in colitis mouse models. Several studies have reported that the circadian protein Nuclear Receptor Subfamily 1 Group D Member 1 directly regulates Th17 differentiation by directly binding and inhibiting Nuclear Factor, Interleukin 3 Regulated.26, 27, 28, 29 Notably, we characterized a type of Breg cells highly expressing PDL1 in IELs, which are key for circadian clock disorder–mediated colitis and colitis-associated CRC. Interestingly, a recent study found that recombinant PDL1 protein significantly ameliorated DSS-induced acute and CD4 CD45RBhigh T-cell–induced chronic colitis in mice by pathogenic Th17 responses and the production of cytokines form PD-1–expressing dendritic cells in diseased colons.30 Here, our study not only characterized a potentially important source for PDL1 from B cells, but also uncovered a new and key subset of Breg cells, PDL1+ Breg cells, functioning as a key suppressor for DSS-triggered excessive inflammatory response in the gut. Interestingly, Xiao et al31 identified a subset of B cells (CD5hi CD24-/+ CD27 hi/+ CD38dim) in advanced-stage hepatocellular carcinoma, which promoted tumor progression. The roles and mechanisms of these B-cell subsets in circadian clock disorder–elicited colitis and colorectal cancer progression still are unclear. More studies are needed to identify more subsets of circadian clock–regulated B cells in inflammatory response and tumor progression.
In addition, we found that IL33 is a target of Bmal1 transcriptionally and IL33 in the intestinal microenvironment is key for Bmal1-regulated PDL1+ Breg cells. It also has been reported that experimental animals have shown that IL33 can regulate a type of Breg cells that produce IL10, thereby effectively reducing intestinal inflammation.32 In a mouse sepsis model, a study reported that deletion of Bmal1 up-regulated Pyruvate Kinase M2 and then influenced PDL1 expression through the signal transducer and activator of transcription 1 pathway in macrophages.33 In our study, IL33, an innate cytokine, played an important role in mucosal organs,34, 35, 36, 37 and was expressed mostly and down-regulated significantly in the intestinal microenvironment of either WT or Bmal1-/- mice treated with or without DSS. It has been suggested that IL33 is secreted by intestinal epithelial cells and is highly unregulated in UC,38 and IL33 can regulate the number of Breg cells in mouse colitis models.32 Our results show that there were E-box classic binding sites in the mouse IL33-promoter regions and Bmal1 can bind directly to and regulate IL33 expression in lymphocytes. In addition, we found that IL33 increased the expression of PDL1 in B cells from both WT mice and Bmal1-/- mice, and IL33-neutralizing antibody decreased the expression of PDL1 in B cells from both WT mice and Bmal1-/- mice.
Moreover, our results showed that dysregulated PDL1+ B cells induce cell death of activated CD4+ T cells in DSS-treated Bmal1-/- mice. A previous study reported that UC patients and DSS-induced experimental IBD in mice were characterized as decreased the BCL2/Bcl-2 Associated X Protein ratio of CD4+ T cells.39 Interestingly, among tumor microenvironments, macrophages and tumor cells expressing PDL1 effectively inhibit functions of infiltrating T cells and promote tumor progression,40,41 suggesting the key role of macrophages and tumor cells in the regulation of T-cell function. Notably, in the early stage of colitis-associated CRC, we found that Bmal1 depletion in B cells also inhibited functions of T cells, indicated as decreased numbers of GranzB secreted by CD4+ and CD8+ T cells. More studies are needed to establish the interactions between these cell types and mechanisms involved. Notably, our findings also uncovered that circadian clock disorders are characterized as decreased numbers of Breg+ PDL1+ cells and dysfunction of CD4+ T cells promotes IBD-associated CRC in mice. Interestingly, there are some studies that have shown that defects of Clock Circadian Regulator1, BMAL1, and PER and Cryptochrome Circadian Regulator proteins in IELs play the critical roles in the development and progression of CRC by increasing or activating MYC Proto-Oncogene/p21 and Wingless-Type MMTV Integration Site Family/β-catenin pathways.42 Our results show that abnormal circadian clocks in B cells in either jet lag mouse models or mouse models with defective clock core genes can promote pathogenesis of CRC. In an experimental IBD-related CRC mouse model, we found that CD4+ T cells in abnormal biological clock mice were dramatically fewer than those in WT mice. Among these CD4+ cells, PD1+CD4+ cells were much higher in abnormal biological clock mice than in WT mice.
In summary, we identified the role of the biological clock, or biological clock genes regulated a class of PDL1+Breg+ cells in experimental IBD and IBD-associated CRC in mice. We found that circadian clock disorders led to high risks for IBD and IBD-associated CRC by decreasing the number of Breg+PDL1+ cells and dysfunction of CD4+ T cells.
Materials and Methods
Animal Models
Bmal1-/- mice were kindly gifted by Dr Ying Xu from Soochow University. All mice were housed under specific pathogen-free conditions of Soochow University. Approval for this research was obtained from the ethic committees of Animal Ethics of Soochow University.
CJ mice were established as described before43,44 (Figure 1A). C57BL/6 mice kept in a steady 12:12-hour light–dark cycle were used as the control group. Zeitgeber time is a unit of time based on the period of a zeitgeber, such as the 12:12-hour light–dark cycle. We labeled the period when the lights were turned on or off on in Figure 12.
To establish DSS-induced colitis models, mice were fed 1.0% or 1.5% DSS dissolved in distilled drinking water for 4, 7, or 11 days (Figure 1).
To establish AOM/DSS-induced CRC, Bmal1-/- and WT mice were injected intraperitoneally with AOM (A5486; Sigma, St. Louis, MO) once a week for the first 3 weeks, fed with 1% DSS for 1 week, and drinking water for the following 2 weeks. The cycle was repeated 3 times. From the eighth week the mice were fed with drinking water (Figure 1B). At the 19th week, blood in the stool and prolapse of the anus were found in most of the Bmal1-/- mice. Mice were killed, and the colorectums were isolated and fixed in formaldehyde before further processing and immunohistochemistry procedures. In some mice, tissues were used for isolation of infiltrating leukocytes, and the function of them was examined by flow cytometry.
Patients and Specimens
Four UC patients, 29 CD patients (Table 1), and paracancerous specimens from 10 CRC patients were collected from Guangdong General Hospital (Table 1).
Cohorts of 91 CRC patients without lymph node metastasis who underwent curative resection at the Department of Pathology of Sun Yat-Sen Memorial Hospital (Guangzhou, China) (Table 2) between November 2016 and November 2017 were used for analyses of expressions of Clock genes and immune cells. None of the patients had received anticancer therapy before the sampling. Ninety-one patients’ cohorts including areas of CT, the IM, and paracancer were obtained in an unbiased manner (according to Mlecnik et al45). Approval for this research was obtained from the Ethic Committees of the Sun Yat-Sen Memorial Hospital.
Flow Cytometry
Single-cell suspensions prepared from livers, spleens, and intestinal IELs were generated by macerating the tissues through a 70-mm nylon mesh. Blood was collected with an anticoagulant tube, transferred into a centrifuge tube, and diluted with phosphate-buffered saline (PBS). For lymphocyte isolation from liver and spleen, tissues were cut into small pieces and incubated in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS). For intestines, Peyer’s patches and fat tissue were carefully removed. The small intestine and colon was opened longitudinally, washed in RPMI 1640 medium, and cut into 5-mm pieces, which were transferred to a 50-mL conical tube (Falcon 2070, Becton, Dickinson and Company) containing 25 mL EDTA-RPMI medium including 10 mmol/L/L HEPES, 25 mmol/L/L NaHCO3, 1 mmol/L/L EDTA, 1 mmol/L/L dithiothreitol, 2% FBS, and 40 μg/mL penicillin and streptomycin. The tube was shaken at 37°C for 30 minutes (horizontal position; orbital shaker at 280 rpm). Cell suspensions were collected in a new 50-mL conical tube and passed through a 70-μm strainer to deplete cell debris and sticky cells (crude cell preparation), and centrifuged at 1200 rpm at room temperature for 6 minutes. Cell pellets were resuspended with EDTA-RPMI medium, filtered through 70-μm nylon mesh, and then centrifuged at 1200 rpm at room temperature for 6 minutes. Subsequently, the pelleted cells were suspended in 40% Percoll (17089101; GE, Uppsala, Sweden). Percoll was diluted with PBS to make 1× Percoll. Cells were suspended with 16 mL of 40% Percoll in 15-mL tubes. Two milliliters of 75% Percoll was added into the lower layer of 40% Percoll in each 15-mL tube. IELs were collected by harvesting the center layer between 40% and 70% Percoll after being centrifuged at 2000 rpm for 15 minutes at 20ºC with free-brake and low-accelerator conditions.
Aliquots of 1 × 106 cells of these tissues were stained with fluorochrome-conjugated antibodies (Table 3). Flow cytometric acquisition was performed using FACScan (BD Biosciences, Franklin Lakes, NJ), and data were analyzed using FlowJo software (version X.0.7; Tree Star, Inc). For intracellular cytokine GranzB, IFNγ, IL10, and IL33 staining, cells were restimulated. Fixation and Permeabilization Solution (554722, BD Cytofix/Cytoperm, Becton, Dickinson and Company), Perm/Wash Buffer (554723, BD Perm/Wash), protein transport inhibitor (containing monensin) (554724, BD GolgiStop), PMA (16561-29-8; Sigma), and ionomycin calcium salt (5608212, Peprotech, BioGems, Cranbury, NJ) were used. For measurement of Ki67, the eBioscience Transcription Factor staining buffer set (00-5523; Invitrogen) was used.
Table 3.
Antibodies Used for Flow Cytometry
| Name | Company | Catalog number | Clone |
|---|---|---|---|
| APC-Cy7 anti-mouse B220 antibody | BD Pharmingen San Diego, CA | 552094 | RA3-6B2 |
| FITC anti-mouse CD45 antibody | BD Pharmingen | 553080 | 30-F1 |
| PE rat anti-mouse IgD antibody | BD Pharmingen | 558597 | 11-26c.2a |
| BV421 anti-mouse CD138 antibody | BD Horizon, Piscataway, NJ. | 562610 | 281-2 |
| PE-CF594 anti-mouse CD3e antibody | BD Horizon | 562332 | 145-2C11 |
| Alexa Fluor 647 anti-mouse CD1d antibody | BD Pharmingen | 564706 | 1B1 |
| BB700 rat anti-mouse CD5 antibody | BD OptiBuild, Becton, Dickinson and Company | 742083 | 53-7.3 |
| APC anti-mouse CD11b antibody | BD Pharmingen | 553312 | M1/70 |
| PerCP-Cy5.5 anti-mouse CD8a antibody | BD Pharmingen | 551162 | 53-6.7 |
| APC anti-mouse CD4 antibody | BD Pharmingen | 553051 | RM4-5 |
| BV421 mouse anti-Ki67 antibody | BD Horizon | 562899 | B56 |
| PE anti-mouse granzyme B antibody | eBioscience, San Diego, CA | 12-8898-80 | NGZB |
| APC-Cy7 anti-mouse IFNγ antibody | BD Pharmingen | 561479 | XMG1.2 |
| BV421 anti-mouse CD279 antibody | BD Horizon | 562584 | J43 |
| PerCP-Cy5.5 anti-mouse CD11c antibody | BD Pharmingen | 560584 | HL3 |
| PE anti-mouse CD49b antibody | BD Pharmingen | 553858 | DX5 |
| PE anti-mouse CD274 antibody | BD Pharmingen | 745135 | MIH5 |
| Alexa Fluor 647 anti-mouse IL33 antibody | RD, Mckinley Place NE | IC3626R | 396118 |
| APC anti-mouse IL10 antibody | BD Pharmingen | 561059 | JES5-16E3 |
| Annexin V apoptosis detection kit | BD | 559763 |
APC, Allophycocyanin; FITC, Fluoresceine Isothiocyanate; PE, Phycoerythrin.
Isolating B Cells and Adoptive Lymphocyte Transfer Treatment
Spleen of donor Bmal1-/- and WT mice were treated with 1.5% DSS for 4 days, and some donors that were untreated were isolated. We used the BD IMag Mouse B Lymphocyte Enrichment Set (557792, BD Pharmingen, San Diego, CA, USA) and followed the kit instructions to isolate B cells (cells of >90% purity and >90% viability). A total of 1 × 106 sorted B cells in 200 μL PBS were injected into the recipient mice in an intravenous injection every other day. Throughout the process, recipient mice received 1.5% DSS. Bmal1-/- and WT mice with 1.5% DSS treatment only were used as the control group (Figure 1C).
For adoptive B cells activated in vitro experiment, B cells from spleen of naive Bmal1-/- and WT mice were isolated and activated by 10 μg/mL lipopolysaccharide (Sigma) for 72 hours. A total of 1 × 106 sorted B cells in 200 μL PBS were injected into the recipient mice in an intravenous injection every other day. Throughout the process, recipient mice received 1% DSS (Figure 1D).
For some experiments, donor WT mice were injected with anti-PDL1 (BE0101, 100 μg/mice, intraperitoneally; Bioxcell, NH) on the fourth day after they received DSS treatment (Figure 1E). For some experiments, Bmal1-/- and WT mice were injected with IL33-neutralizing antibody (AF3626, 1 μg/mice, intraperitoneally; RD) every 3 days, 4 times in total. After that, both groups were treated with 0.5% DSS (Figure 1F). To analyze whether cytokines regulated B cells by Bmal1, spleen of donor mice that were treated with 1.5% DSS or from untreated mice were isolated. B cells were isolated, counted for 4 × 106 per well, and added into the DMEM medium containing 10% FBS. Lipopolysaccharide (10 μg/mL; Sigma) was added to the medium to activate B cells for 72 hours. For others, 5 ng/mL TGFβ (7666-MB; RD) was added in DMEM medium for 72 hours, 50 ng/mL IL21 (210-21; Peprotech) for 72 hours, 30 ng/mL anti-IL-33 neutralizing antibody (AF3626; RD) for 48 hours, and 10 ng/mL IL33 (3626-ML/CF; RD) for an hour, respectively. B cells subsequently were collected for Western blot.
Isolation of CD4+ and CD8+ T cells
The mouse CD4+ T-Cell Isolation Kit (130-104-454; Miltenyi Biotec, Bergisch Gladbach, Germanywas used for isolating CD4+ T cells from spleen of mice. The mouse CD8+ T Lymphocyte Enrichment Set - DM (558471; BD) was used for isolating CD8+ T cells from spleen of mice. CD3 (B257866; Biolegend), CD28 (B256182; Biolegend), and IL2 (402-ML; R&D Systems) were used to activate CD8+ T cells in vitro.
Apoptosis Analysis
Apoptosis was determined using the Annexin V Apoptosis Detection Kit (559763; BD). Briefly, cells were incubated with buffer containing phycoerythrin-conjugated Annexin V and 7-amino-actinomycin D antibody for 15 minutes at room temperature. After incubation, cells were washed and resuspended in binding buffer. After 1 hour, cells were analyzed by flow cytometry.
Histologic Analyses and Scorings of Colitis and Colitis-Associated CRC
Mice colorectums were fixed in 10% neutral-buffered formalin or 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with H&E. The colitis severity of the mouse models was estimated by measuring body weight loss, colon length, and colon weight. Histology was scored by 2 pathologists in a blinded fashion as a combination of the presence and severity of inflammatory cell infiltration, hyperplasia, or epithelial damage according to Seregi et al.46 For each lesion, a weighted average percentage was calculated as follows: (1 × the number of fields with score = 1) + (2 × the number of fields with score = 2) + (3 × the number of fields with score = 3)]/3 × the total number of fields.
Assessing the severity of enteritis-associated bowel cancer in the mouse is usually quantitative by counting the number of intestinal polyps.
Immunohistochemical Staining
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded samples. Sections were dewaxed and rehydrated through a graded series of ethanol. Endogenous peroxidase was quenched by incubation in hydrogen peroxide (3% w/v in methanol for 15 minutes). Antigen retrieval was performed by boiling in a pressure cooker for 10 minutes in citrate buffer. Sections were blocked for 30 minutes with 10% normal goat serum (Life Technologies, Carlsbad, CA) before incubation with primary antibody overnight at 4°C. Sections and slides were stained for Bmal1 and CD19 (Table 4). Histology was scored by 2 pathologists in a blinded fashion. At low power, the tissue sections were screened, and the 5 most representative fields were selected. Thereafter, to evaluate the density of these genes, the respective areas were measured at a magnification of 400×, and the Bmal1+ lymphocytes and CD19+ cells in each area were counted.
Table 4.
Antibodies Used for Western Blot, ChIP, and Immunohistochemistry
| Name | Company | Catalog number |
|---|---|---|
| Anti-SBNO1 | Atlas, Mount Prospect, IL | HPA042388 |
| BMAL1 (D2L7G) rabbit mAb | CST, Danfoss, MA | 14020 |
| Mouse PD-L1/B7-H1 antibody | RD, Mckinley Place NE | AF1019 |
| Cleaved caspase-3 (Asp175) (5A1E) rabbit mAb | CST | 9664 |
| Bax antibody | CST | 2772 |
| Mouse anti-Bcl2 | Bioss Beijing, China | Bsm33411 |
| Anti-CD19 antibody | Abcam, Cambridge, MA | Ab31947 |
Bax, Bcl-2 Associated X Protein; mAb, monoclonal antibody.
Western Blot
B cells, CD4+ T cells, and CD8+ T cells were collected in RIPA lysis buffer (1% Triton X-100, Sigma-Aldrich, St. Louis, MO), 20 mmol/L Tris, pH 7.5, 137 mmol/L NaCl, 1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1.5 mmol/L MgCl2, and protease inhibitor mixture and phosphatase inhibitors (Roche Products Limited, Welwyn Garden City, UK). Lysates were sonicated and centrifuged at 4°C. Per lane, whole-cell lysate was separated on 12% sodium dodecyl sulfate–acrylamide gels and transferred on Immobilon polyvinylidene difluoride membranes (Millipore). The membranes were probed with primary antibodies (Table 4) overnight at 4°C and incubated for 1 hour with secondary peroxidase-conjugated antibodies. For detecting the apoptotic pathway, anti–caspase 3, anti-Bcl-2 Associated X Protein, and anti-Bcl2 (Table 4) were used. Chemiluminescent signals then were developed with Lumiglo reagent (Cell Signaling Technology) and detected by the ChemiDoc XRS gel documentation system (Bio-Rad).
ChIP Assay
The ChIP assay was performed using the EZ-Magna ChIP A/G Chromatin Immunoprecipitation Kit (17-10086; Millipore, Merck, Bedford, MD). The experiment was performed according to the manufacturer’s instructions.
The immunoprecipitated DNA was used in PCR. Results were shown as a percentage of input. The Bmal1 antibody used for ChIP (Table 4) and the primers (Table 5) used for DNA quantification are listed.
Table 5.
Primers Used for qRT-PCR, ChIP, and Plasmid Construction
| Gene | Forward primer | Reverse primer |
|---|---|---|
| PDL1 | GCTCCAAAGGACTTGTACGTG | TGATCTGAAGGGCAGCATTTC |
| Bmal1 | ACAGTCAGATTGAAAAGAGGCG | GCCATCCTTAGCACGGTGAG |
| Per1 | TTCGTGGACTTGACACCTCTT | GGGAACGCTTTGCTTTAGAT |
| Per2 | ACAGAGGCAGAGCACAACCC | TAGCCTTCACCTGCTTCACG |
| IL4 | GGTCTCAACCCCCAGCTAGT | GCCGATGATCTCTCTCAAGTGAT |
| IL6 | TCTATACCACTTCACAAGTCGGA | GAATTGCCATTGCACAACTCTTT |
| IL10 | GGCCCAGAAATCAAGGAG | CCT TGT AGA CAC CTT GGT |
| IL21 | GGACCCTTGTCTGTCTGGTAG | TGTGGAGCTGATAGAAGTTCAGG |
| IL33 (for RT-PCR) | TCCAACTCCAAGATTTCCCCG | CATGCAGTAGACATGGCAGAA |
| IL33 (for ChIP) | GCCAGAAAGGATGACTGACC | AGCTGCAAGCATTCAAAACC |
| Nonsense IL33 (for ChIP) | TGCCTTGGTCAGCTATTGTG | TTGGAGCGAGAGCTACAGGT |
| TGFβ | CTCCCGTGGCTTCTAGTGC | GCCTTAGTTTGGACAGGATCTG |
| MCP-1 | ATCCCAATGAGTAGGCTGGAGAGC | CAGAAGTGCTTGAGGTGGTTGTG |
| IL33 (for expression vector) | CCGCTCGAGGGCAAAGAATAAAGGCCACCATAC | CCCAAGCTTCCAACTATATGGACCTGAAGTCATC |
| IL33 without E-box sites (for expression vector) | CCGCTCGAGGCTTGCAGCTTCTTAGCTTTGTG | CCCAAGCTTGAAGCAACCTTGGCTATCTCACC |
| PDL1 (for expression vector) | CTAGCTAGCCAGGCGTGGTGGCGCACACCTT | CCCAAGCTTGGAGACTGGCCCGCGACGTGC |
RNA Isolation and qRT-PCR Analysis
To test whether the clock genes, cytokines, and immune checkpoints fluctuate in mice models with circadian rhythm, we isolated hepatic lymphocytes and IELs from each group (5 mice) for qRT-PCR every 4 hours within 24 hours.
RNA was isolated from lymphocytes using TRIzol (Invitrogen) according to the manufacturer’s instructions. For mRNA, complementary DNA was generated from 1 μg total RNA per sample using the Transcriptor First Strand Complementary DNA Synthesis Kit (Roche). qRT-PCR was performed using the ABI StepOne Plus and the SYBR Select Master Mix (Applied Biosystem, CA). mRNA expression was normalized using detection of 18S ribosomal RNA. Results are represented as fold induction using the ΔΔCt method with the control set to 1. The sequences of qRT-PCR primers are listed in Table 5.
Plasmid Construction
To construct the expression vector of mouse IL33, a 2.8-kb DNA fragment containing E-Box of the 5’ region IL33 was amplified by PCR from mouse genomic DNA (primers are listed in Table 5). The PCR fragment was digested with Xanthomonas holcicola I and HindIII, and then ligated to the multiple cloning sites of a pGL4.17[luc2/Neo] Vector (Promega, Madison, WI) to generate the IL33:luc reporter plasmid. In addition, a fragment without the E-box of the 5’ region IL33 also was cloned into the reporter vector. For expression vector of mouse PDL1, a 1.8-kb DNA fragment containing E-Box was digested with Neisseria mucosa heidelbergensis I and Hemophilus influenzae D III, and then ligated to the multiple cloning sites of a pGL4.17[lutetium carbide 2/Neo] Vector.
Luciferase Assay
Two days after 293T cells had been transfected with the plasmids they were treated under normoxic or hypoxic conditions for 24 hours and then harvested in passive lysis buffer (Promega) for the luciferase assay. The luciferase assay was performed using the Dual Luciferase Assay System (Promega) according to the manufacturer’s instructions. Each experiment was conducted in triplicate.
Statistical Analysis
To detect the transcription levels of Bmal1 and other clock genes in enteritis samples, we used the expression profile data from GSE22307 and obtained the mRNA expression values of Bmal1, Clock, Cry1, Cry2, Per1, and Per2 in normal mouse colon and colons of mice treated with DSS through GEO2R of NCBI.15 A box plot with P value (calculated by the Wilcox test) was used.
The results are presented as means ± SD, and analyses were performed with SPSS version 17.0 (SPPS Inc, Chicago, IL). Differences between the groups were evaluated by 1-way analysis of variance and correlation analysis was performed using Spearman rank correlation. A P value less than .05 was considered significant. JTK_CYCLE (https://openwetware.org/wiki/HughesLab: JTK_Cycle, HughesLab Ohio State University, University of Pennsylvania School of Medicine, Philadelphia, PA) was used to distinguish the rhythmic transcripts.13,14 All data come from the results of experiments that were repeated at least twice. Relevance and prognostic analysis were performed by GraphPad Prism 8.0 (Graphpad Software, Inc, San Diego, CA).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants from the National Key R&D Program of China (2019YFA0802400), the National Natural Science Foundation of China (81525020 and U1801282), and the Guangzhou Science and Technology Plan Projects (Health Medical Collaborative Innovation Program of Guangzhou) (201803040019). This work also was supported by a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Contributor Information
Tong Shen, Email: tshen@suda.edu.cn.
Jian-Ming Li, Email: jianmingli@suda.edu.cn.
References
- 1.Bamias G., Nyce M.R., De La Rue S.A., Cominelli F. New concepts in the pathophysiology of inflammatory bowel disease. Ann Intern Med. 2005;143:895–904. doi: 10.7326/0003-4819-143-12-200512200-00007. [DOI] [PubMed] [Google Scholar]
- 2.Magnusson M.K., Brynjolfsson S.F., Dige A., Uronen-Hansson H., Borjesson L.G., Bengtsson J.L., Gudjonsson S., Ohman L., Agnholt J., Sjovall H., Agace W.W., Wick M.J. Macrophage and dendritic cell subsets in IBD: ALDH+ cells are reduced in colon tissue of patients with ulcerative colitis regardless of inflammation. Mucosal Immunol. 2016;9:171–182. doi: 10.1038/mi.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mauri C., Gray D., Mushtaq N., Londei M. Prevention of arthritis by interleukin 10-producing B cells. J Exp Med. 2003;197:489–501. doi: 10.1084/jem.20021293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fillatreau S., Sweenie C.H., McGeachy M.J., Gray D., Anderton S.M. B cells regulate autoimmunity by provision of IL-10. Nat Immunol. 2002;3:944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- 5.Mizoguchi A., Mizoguchi E., Takedatsu H., Blumberg R.S., Bhan A.K. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 2002;16:219–230. doi: 10.1016/s1074-7613(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 6.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 7.Mantis N.J., Forbes S.J. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol Invest. 2010;39:383–406. doi: 10.3109/08820131003622635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 9.Koike S., Morimoto H., Kozakai C., Arimoto I., Soga M., Yamazaki K., Koganezawa M. The role of dung beetles as a secondary seed disperser after dispersal by frugivore mammals in a temperate deciduous forest. Acta Oecologica. 2012;41:8. [Google Scholar]
- 10.Wang S., Lin Y., Yuan X., Li F., Guo L., Wu B. REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat Commun. 2018;9:4246. doi: 10.1038/s41467-018-06568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson G.R., Burgess H.J., Keshavarzian A. Sleep disturbances and inflammatory bowel disease: a potential trigger for disease flare? Expert Rev Clin Immunol. 2011;7:29–36. doi: 10.1586/eci.10.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzoccoli G., Palmieri O., Corritore G., Latiano T., Bossa F., Scimeca D., Biscaglia G., Valvano M.R., D'Inca R., Cucchiara S., Stronati L., Annese V., Andriulli A., Latiano A. Association study of a polymorphism in clock gene PERIOD3 and risk of inflammatory bowel disease. Chronobiol Int. 2012;29:994–1003. doi: 10.3109/07420528.2012.705935. [DOI] [PubMed] [Google Scholar]
- 13.Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masri S., Patel V.R., Eckel-Mahan K.L., Peleg S., Forne I., Ladurner A.G., Baldi P., Imhof A., Sassone-Corsi P. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang K., Bruce M., Pattillo C.B., Zhang S., Stone R., 2nd, Clifford J., Kevil C.G. Temporal genomewide expression profiling of DSS colitis reveals novel inflammatory and angiogenesis genes similar to ulcerative colitis. Physiol Genomics. 2011;43:43–56. doi: 10.1152/physiolgenomics.00138.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali T., Madhoun M.F., Orr W.C., Rubin D.T. Assessment of the relationship between quality of sleep and disease activity in inflammatory bowel disease patients. Inflamm Bowel Dis. 2013;19:2440–2443. doi: 10.1097/MIB.0b013e3182a0ea54. [DOI] [PubMed] [Google Scholar]
- 17.Ananthakrishnan A.N., Long M.D., Martin C.F., Sandler R.S., Kappelman M.D. Sleep disturbance and risk of active disease in patients with Crohn's disease and ulcerative colitis. Clin Gastroenterol Hepatol. 2013;11:965–971. doi: 10.1016/j.cgh.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graff L.A., Vincent N., Walker J.R., Clara I., Carr R., Ediger J., Miller N., Rogala L., Rawsthorne P., Lix L., Bernstein C.N. A population-based study of fatigue and sleep difficulties in inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:1882–1889. doi: 10.1002/ibd.21580. [DOI] [PubMed] [Google Scholar]
- 19.Ranjbaran Z., Keefer L., Farhadi A., Stepanski E., Sedghi S., Keshavarzian A. Impact of sleep disturbances in inflammatory bowel disease. J Gastroenterol Hepatol. 2007;22:1748–1753. doi: 10.1111/j.1440-1746.2006.04820.x. [DOI] [PubMed] [Google Scholar]
- 20.Swanson G.R., Burgess H.J. Sleep and Circadian hygiene and inflammatory bowel disease. Gastroenterol Clin North Am. 2017;46:881–893. doi: 10.1016/j.gtc.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Preuss F., Tang Y., Laposky A.D., Arble D., Keshavarzian A., Turek F.W. Adverse effects of chronic circadian desynchronization in animals in a "challenging" environment. Am J Physiol Regul Integr Comp Physiol. 2008;295:R2034–R2040. doi: 10.1152/ajpregu.00118.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyoko O.O., Kono H., Ishimaru K., Miyake K., Kubota T., Ogawa H., Okumura K., Shibata S., Nakao A. Expressions of tight junction proteins Occludin and Claudin-1 are under the circadian control in the mouse large intestine: implications in intestinal permeability and susceptibility to colitis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokes K., Cooke A., Chang H., Weaver D.R., Breault D.T., Karpowicz P. The Circadian clock gene BMAL1 coordinates intestinal regeneration. Cell Mol Gastroenterol Hepatol. 2017;4:95–114. doi: 10.1016/j.jcmgh.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauri C., Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- 25.Michaud D., Steward C.R., Mirlekar B., Pylayeva-Gupta Y. Regulatory B cells in cancer. Immunol Rev. 2020 doi: 10.1111/imr.12939. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger T.L., Abt M.C., Gasteiger G., Firth M.A., O'Connor M.H., Geary C.D., O'Sullivan T.E., van den Brink M.R., Pamer E.G., Hanash A.M., Sun J.C. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seillet C., Rankin L.C., Groom J.R., Mielke L.A., Tellier J., Chopin M., Huntington N.D., Belz G.T., Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W., Domingues R.G., Fonseca-Pereira D., Ferreira M., Ribeiro H., Lopez-Lastra S., Motomura Y., Moreira-Santos L., Bihl F., Braud V., Kee B., Brady H., Coles M.C., Vosshenrich C., Kubo M., Di Santo J.P., Veiga-Fernandes H. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep. 2015;10:2043–2054. doi: 10.1016/j.celrep.2015.02.057. [DOI] [PubMed] [Google Scholar]
- 29.Yu X., Wang Y., Deng M., Li Y., Ruhn K.A., Zhang C.C., Hooper L.V. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014;3 doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song M.Y., Hong C.P., Park S.J., Kim J.H., Yang B.G., Park Y., Kim S.W., Kim K.S., Lee J.Y., Lee S.W., Jang M.H., Sung Y.C. Protective effects of Fc-fused PD-L1 on two different animal models of colitis. Gut. 2015;64:260–271. doi: 10.1136/gutjnl-2014-307311. [DOI] [PubMed] [Google Scholar]
- 31.Xiao X., Lao X.M., Chen M.M., Liu R.X., Wei Y., Ouyang F.Z., Chen D.P., Zhao X.Y., Zhao Q.Y., Li X.F., Liu C.L., Zheng L.M., Kuang D.M. PD-1(hi) Identifies a novel regulatory B-cell population in human hepatoma that promotes disease progression. Cancer Discov. 2016;6:546–559. doi: 10.1158/2159-8290.CD-15-1408. [DOI] [PubMed] [Google Scholar]
- 32.Sattler S., Ling G.S., Xu D., Hussaarts L., Romaine A., Zhao H., Fossati-Jimack L., Malik T., Cook H.T., Botto M., Lau Y.L., Smits H.H., Liew F.Y., Huang F.P. IL-10-producing regulatory B cells induced by IL-33 (Breg(IL-33)) effectively attenuate mucosal inflammatory responses in the gut. J Autoimmun. 2014;50:107–122. doi: 10.1016/j.jaut.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng W., Zhu S., Zeng L., Liu J., Kang R., Yang M., Cao L., Wang H., Billiar T.R., Jiang J., Xie M., Tang D. The Circadian clock controls immune checkpoint pathway in sepsis. Cell Rep. 2018;24:366–378. doi: 10.1016/j.celrep.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bamias G., Corridoni D., Pizarro T.T., Cominelli F. New insights into the dichotomous role of innate cytokines in gut homeostasis and inflammation. Cytokine. 2012;59:451–459. doi: 10.1016/j.cyto.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besnard A.G., Togbe D., Guillou N., Erard F., Quesniaux V., Ryffel B. IL-33-activated dendritic cells are critical for allergic airway inflammation. Eur J Immunol. 2011;41:1675–1686. doi: 10.1002/eji.201041033. [DOI] [PubMed] [Google Scholar]
- 36.Carriere V., Roussel L., Ortega N., Lacorre D.A., Americh L., Aguilar L., Bouche G., Girard J.P. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobori A., Yagi Y., Imaeda H., Ban H., Bamba S., Tsujikawa T., Saito Y., Fujiyama Y., Andoh A. Interleukin-33 expression is specifically enhanced in inflamed mucosa of ulcerative colitis. J Gastroenterol. 2010;45:999–1007. doi: 10.1007/s00535-010-0245-1. [DOI] [PubMed] [Google Scholar]
- 38.Seidelin J.B., Bjerrum J.T., Coskun M., Widjaya B., Vainer B., Nielsen O.H. IL-33 is upregulated in colonocytes of ulcerative colitis. Immunol Lett. 2010;128:80–85. doi: 10.1016/j.imlet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Ina K., Itoh J., Fukushima K., Kusugami K., Yamaguchi T., Kyokane K., Imada A., Binion D.G., Musso A., West G.A., Dobrea G.M., McCormick T.S., Lapetina E.G., Levine A.D., Ottaway C.A., Fiocchi C. Resistance of Crohn's disease T cells to multiple apoptotic signals is associated with a Bcl-2/Bax mucosal imbalance. J Immunol. 1999;163:1081–1090. [PubMed] [Google Scholar]
- 40.Kuang D.M., Zhao Q.Y., Peng C., Xu J., Zhang J.P., Wu C.Y., Zheng L.M. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206:1327–1337. doi: 10.1084/jem.20082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escors D., Gato-Canas M., Zuazo M., Arasanz H., Garcia-Granda M.J., Vera R., Kochan G. The intracellular signalosome of PD-L1 in cancer cells. Signal Transduct Target Ther. 2018;3:26. doi: 10.1038/s41392-018-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karantanos T., Theodoropoulos G., Pektasides D., Gazouli M. Clock genes: their role in colorectal cancer. World J Gastroenterol. 2014;20:1986–1992. doi: 10.3748/wjg.v20.i8.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hadden H., Soldin S.J., Massaro D. Circadian disruption alters mouse lung clock gene expression and lung mechanics. J Appl Physiol (1985) 2012;113:385–392. doi: 10.1152/japplphysiol.00244.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papagiannakopoulos T., Bauer M.R., Davidson S.M., Heimann M., Subbaraj L., Bhutkar A., Bartlebaugh J., Vander Heiden M.G., Jacks T. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 2016;24:324–331. doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mlecnik B., Van den Eynde M., Bindea G., Bindea G., Church S.E., Vasaturo A., Fredriksen T., Lafontaine L., Haicheur N., Marliot F., Debetancourt D., Pairet G., Jouret-Mourin A., Gigot J.F., Hubert C., Danse E., Dragean C., Carrasco J., Humblet Y., Valge-Archer V., Berger A., Pages F., Machiels J.P., Galon J. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J Natl Cancer Inst. 2018;110 doi: 10.1093/jnci/djx123. [DOI] [PubMed] [Google Scholar]
- 46.Seregin S.S., Golovchenko N., Schaf B., Chen J.C., Pudlo N.A., Mitchell J., Baxter N.T., Zhao L.L., Schloss P.D., Martens E.C., Eaton K.A., Chen G.Y. NLRP6 protects Il10(-/-) mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep. 2017;19:733–745. doi: 10.1016/j.celrep.2017.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]