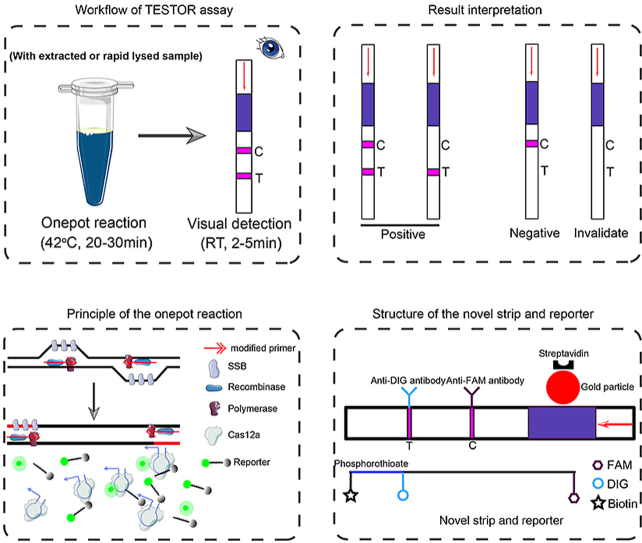
Abstract
CRISPR-Cas12a system has been shown promising for nucleic acid diagnostics due to its rapid, portable and accurate features. However, cleavage of the amplicons and primers by the cis- and trans-activity of Cas12a hinders the attempts to integrate the amplification and detection into a single reaction.
Through phosphorothioate modification of primers, we realized onepot detection with high sensitivity using plasmids of SARS-CoV-2, HPV16 and HPV18. We also identified the activated Cas12a has a much higher affinity to C nucleotide-rich reporter than others. By applying such reporters, the reaction time required for a lateral-flow readout was significantly reduced. Furthermore, to improve the specificity of the strip-based assay, we created a novel reporter and, when combined with a customized gold-nanopaticle strip, the readout was greatly enhanced owing to the elimination of the nonspecific signal. This established system, termed Targeting DNA by Cas12a-based Eye Sight Testing in an Onepot Reaction (TESTOR), was validated using clinical cervical scrape samples for human papillomaviruses (HPVs) detection.
Our system represents a general approach to integrating the nucleic acid amplification and detection into a single reaction in CRISPR-Cas systems, highlighting its potential as a rapid, portable and accurate detection platform of nucleic acids.
Keywords: CRISPR-Cas12a, Onepot, Visual detection
Graphical abstract
Highlights
-
•
The nucleic acid amplification and Cas12a detection were integrated into a single reaction.
-
•
By using the C nucleotide-rich reporter, the reaction time required for a sensitive detection was significantly reduced.
-
•
A novel lateral flow system was created, which significantly enhanced the readout.
1. Introduction
Recent outbreak of SARS-CoV-2 has highlighted the challenges of detecting viral infections, especially in areas where specialized equipment is not available [[1], [2], [3]]. Polymerase chain reaction (PCR) is the most commonly used method and has been considered as a “gold standard” for nucleic acid diagnosis due to high sensitivity and specificity [[4], [5], [6]]. However, the requirement of expensive equipment and well-trained personnel as well as long reaction times (normally more than 2 h) makes it unsuitable for point-of-care test (POCT) diagnostics. These limitations hinder its applications in many cases and whereby delay the prescription and administration of antiviral agents to patients. In contrast, serology tests are rapid and require minimal equipment but have lower sensitivity and specificity [7,8]. It may need several days to weeks following symptom onset for a patient to mount a detectable antibody response [9].
CRISPR-Cas systems are adaptive immune systems in archaea and bacteria [10,11]. Some Cas nucleases display strong collateral activities after binding to their specific cis targets, which has been fully evaluated for diagnostic use [[12], [13], [14]]. By combination with recombinase polymerase amplification (RPA), Cas12 and Cas13 have been shown to permit a single molecule detection in a given reaction [13,15,16]. Although both of them are RNA-guided nucleases [16], CRISPR/Cas13a may be less promising because it uses single stranded RNA (ssRNA) as reporters, which could be false-positive-prone, as RNases are common and highly stable in the environment. In addition to the widely used RPA method, LAMP (loop-mediated isothermal amplification) and PCR approaches have also been reported to facilitate the Cas12a-based nucleic acid detection [17,18]. However, all of these methods have to separate the amplification step from the detection step owing to the fact that cleavage of the amplicons and primers by cis- and trans-activities of Cas12a could prevent the chain reactions [14]. The requirement of opening the reaction tube after pre-amplification makes this system more time-consuming and manpower demanding as well as aerosol generation that often causes false positive [16,19].
Recently, Zhang et al. has reported a onepot method using extracted nucleic acids for the detection of SARS-CoV-2 by combination of LAMP and Cas12b endonuclease [20]. This system seems not very sensitive with the LoD of 100 copies for a given reaction, and also it needs to be performed at higher temperature, which may dampen its applications.
By combining RPA amplification with Cas12a detection, we herein report a onepot assay, termed Targeting DNA by Cas12a-based Eye Sight Testing in a Onepot Reaction (TESTOR), with a better sensitivity and can be performed at 37 °C for rapid, ultrasensitive and specific detection of nucleic acids. The primers were modified with phosphorothioate at certain sites to prevent the degradation by activated Cas12a. The crRNAs were designed to allow the cleavage on amplicons occur at the modified sites, whereby the amplicon is just nicked, but not cleaved, leading to continuation of the chain reaction. We also optimized the reporters and identified that activated Cas12a has the highest affinity to C nucleotide-rich reporters, with which the reaction time was significantly reduced. Furthermore, we designed a novel reporter labeled with FAM, DIG and Biotin, and modified with phosphorothioate at the sites between DIG and Biotin. When combined with a strip that can capture DIG and Biotin at the control and test line, respectively, the unspecific signal at the test line was completely eliminated.
This Cas12a-based nucleic acid detection platform (TESTOR) holds the potential to address the key challenges for viral diagnostics and will undoubtedly have a great clinical potential.
2. Materials and methods
2.1. Nucleic acid preparation
The synthetic DNA fragment of ORF1ab or N gene was ligated into a pUC19 vector (BIOLIGO, Shanghai, China) and amplified in E.coli. system. Plasmid DNA was extracted with a commercially available kit (Tiangen, Wuxi, China) and the concentration was quantified using a spectrophotometer (Thermo Fisher, NJ, USA). Copy number of the plasmid was calculated based on the concentration using the following equation: DNA copy number = (M × 6.022 × 1023)/(n × 1 × 109 x 650), in which M represents the amount of DNA in nanograms, n is the length of the plasmid in base pair, and the average weight of a base pair is assumed to be 650 Da.
2.2. Primer selection
RPA primers for SARS-CoV-2 and HPVs detections were designed according to a protocol described by Twist-Dx (Maidenhead, UK). Primer pairs were screened using a single forward primer against all reverse primers, where the best reverse primer was selected and then used to screen all the forward primers. The best performance primer pairs were used in subsequent experiments.
2.3. Cas12a detection reaction
The RPA or PCR product (5 μL) was mixed with 20 μL of the 1x Cas12a reaction mixture containing 50 nM Cas12a (NEB, Ipswich, UK), 100 nM crRNA (BIOLIGO, Shanghai, China), and 250 nM ssDNA reporter (Sangon, Shanghai, China). Then, signal was collected using a fluorescence plate reader (Molecular Devices, California, USA) or a Real-Time PCR Detection System (Bio-Rad, Watford, UK) for up to 120 min at 37 °C. The fluorescence intensity data was expressed as Relative fluorescence units (RFU) or baseline subtracted RFU.
2.4. Fluorescence TESTOR assay
Each lyophilized pellet from the Basic DNA RPA kit was resuspended in a solution comprising 29.4 μL of rehydration buffer, 3.2 μL of primer mix with each at 5 μM, 10.1 μL of H2O. This mix was then divided into 2 aliquots, with each comprising a volume of 21.75 μL, followed by addition of 2 μL of template DNA, 1.25 μL magnesium acetate (720 mM) and 5 μL of 5x Cas12a mixture containing NEB 2.1 reaction buffer (5x), 250 nM Cas12a enzyme, 500 nM crRNA, and 1.25 μM ssDNA reporters. Regarding to the ratio study, 5x Cas12a and RPA mixtures was prepared with each ingredient described above. The two components were then mixed to form a 30 μL reaction with the ratios of volume varying from 1:1 to 1:6. Reactions were incubated in a Real-Time PCR Detection System for up to 120 min at 37 °C with fluorescent signals collected every 30 s (ssDNA FQ reporter = λex: 485 nm; λem: 535 nm). For all endpoint analysis, fluorescence was taken at 30 min after reaction. The fluorescence value was exported as Relative fluorescence units (RFU) or baseline subtracted RFU.
2.5. Lateral flow assay using DNase digested reporter
A 50 μL reaction containing 5 μL of 10x NEB 2.1 reaction buffer, 2U DNase I (NEB, Ipswich, UK) and the novel reporter at the concentration of 0.5 μM or 1 μM was incubated at 37 °C for 30 min. The digested product was subjected to 1: 5 dilution using Tris-HCl buffer (0.05 M, pH = 7.6) and then the strip (Bioustar, Hangzhou, China) was inserted and incubated for 2 min at room temperature for color development.
2.6. Lateral flow assay using two-step RPA-Cas12a assay
The N gene of SARS-CoV-2 was amplified with RPA reaction according to the manufacturer's protocol (Amp-Future, Weifang, China). Briefly, the reaction was performed in a total volume of 12.5 μL comprising the RPA enzymes, 1 μL of sample, 7.35 μL of rehydration buffer, 28 mM magnesium acetate and 0.4 μM of each primer. Next, the Cas12a detection assay as described above was performed with 0.5 μM novel reporter at 37 °C for 20 min. The cleavage product was diluted at the ratio of 1: 5 or 1: 10, and the strip was inserted and incubated for 2 min at room temperature for color reaction.
2.7. Lateral flow TESTOR assay
The components of strip-based TESTOR reaction was similar to that for fluorescence-based TESTOR assay, except replacing the FQ reporter with a ssDNA labeled with Biotin, DIG and FAM at the concentration of 0.5 μM. The reaction was incubated at 37 °C for 30 min, followed by dilution at the indicated ratio in Tris-HCl buffer, and then a strip (TwistDx, Cambridge, UK; Bioustar, Hangzhou, China) was inserted and incubated at room temperature. After 2–5 min of incubation, the strip were removed and photographed with a smartphone camera.
2.8. Specimen collection and DNA extraction for HPV detection
A conventional cytological scrape was taken with a cytobrush from women visiting the gynecological outpatient clinic of the Shenzhen Luohu People's Hospital in China. The specimens were placed into tubes containing 3 mL of cell collection medium (Yaneng Bio, Shenzhen, China) and stored at −20 °C until use. Total DNA was released by using a lysis buffer containing 50 mM guanidine hydrochloride, 50 mM Tris-HCl, 1% Triton X-100, 0.5% Tween-20 and 100 mM NaOH. The pellets from 1 mL of the liquid-based cervical cytology samples after centrifugation were mixed with 200 μL of the lysis buffer and heated for 10 min at 95 °C. The supernatant was used for HPV detection assays following a centrifugation step.
2.9. qPCR assay for HPV testing
HPV DNA was detected using a HPV test kit (Yaneng Bio, Shenzhen, China) according to the instruction of manufacturer. The assay was performed using the Biorad CFX96 instrument, with the following program: pre-denaturation at 95 °C for 10 min, denaturation at 95 °C for 10 s, and annealing and extension as well as signal detection at 55 °C for 45 s.
All the primers, crRNAs and reporters used in this study are listed in Table 1.
Table 1.
Primers, crRNAs and reporters used in this study.
| Oligo name | Oligo Sequence(5′-3′) | Usage |
|---|---|---|
| STD-N-F1 | CCAGGCAGCAGTAGGGGAACTTCTCCTGCTAGAAT | Fig. 1a |
| STD-N-F2 | CAGGCAGCAGTAGGGGAACTTCTCCTGCTAGAAT | Fig. 1a |
| STD-N-F3 | TCAACTCCAGGCAGCAGTAGGGGAACTTCTCC | Fig. 1a |
| STD-N-R1 | GTTGGCCTTTACCAGACATTTTGCTCTCAAGCTGG | Fig. 1a |
| STD-N-R2 | TGGCCTTTACCAGACATTTTGCTCTCAAGCTG | Fig. 1a |
| STD-N-R3 | TTACCAGACATTTTGCTCTCAAGCTGGTTCAA | Fig. 1a |
| N–F0 | G*G*AACTTCTCCTGCTAGAATGGCTGGCAAT*G*G | Fig. 1e and g; Fig. S1a; throughout the paper when N gene was examined unless otherwise stated |
| N-R0 | G*G*CCTTTACCAGACATTTTGCTCTCAAGCT*G*G | Fig. 1e and g; Fig. S1a; throughout the paper when N gene was examined unless otherwise stated |
| N–F01 | G*GAACTTCTCCTGCTAGAATGGCTGGCAATG*G | Fig. 2c |
| N-R01 | G*GCCTTTACCAGACATTTTGCTCTCAAGCTG*G | Fig. 2c |
| N–F02 | G*G*A*ACTTCTCCTGCTAGAATGGCTGGCAA*T*G*G | Fig. 2c |
| N-R02 | G*G*C*CTTTACCAGACATTTTGCTCTCAAGC*T*G*G | Fig. 2c |
| N–F03 | G*G*A*A*CTTCTCCTGCTAGAATGGCTGGCA*A*T*G*G | Fig. 2c |
| N-R03 | G*G*C*C*TTTACCAGACATTTTGCTCTCAAG*C*T*G*G | Fig. 2c |
| N–F1 | C*C*AGGCAGCAGTAGGGGAACTTCTCCTGCTAGA*A*T | Fig. S1b-S1d |
| N–F2 | C*A*GGCAGCAGTAGGGGAACTTCTCCTGCTAGA*A*T | Fig. S1b-S1d |
| N–F3 | T*C*AACTCCAGGCAGCAGTAGGGGAACTTCT*C*C | Fig. S1b-S1d |
| N–F4 | A*G*AAATTCAACTCCAGGCAGCAGTAGGGGAA*C*T | Fig. S1b-S1d |
| N–F5 | G*C*AGTAGGGGAACTTCTCCTGCTAGAATGG*C*T | Fig. 1f; Fig. S1c-S1e |
| N–F6 | G*T*TCAAGAAATTCAACTCCAGGCAGCAGTA*G*G | Fig. 1f; Fig. S1e |
| N-R1 | G*T*TGGCCTTTACCAGACATTTTGCTCTCAAGCT*G*G | Fig. 1f; Fig. S1b-S1e |
| N1–F | A*G*AGGCGGCAGTCAAGCCTCTTCTCGTTCCTC*A*T | Fig. 1h and i |
| N1-R | C*A*GGAGAAGTTCCCCTACTGCTGCCTGGAG*T*T | Fig. 1h and i |
| ORF1ab-F11 | T*T*GCCTGGCACGATATTACGCACAACTAATG*G*T | Fig. S2a, S2b |
| ORF1ab-F18 | A*T*GGTGACTTTTTGCATTTCTTACCTAGAGT*T*T | Fig. 2d–f, 2h; Fig. S2a, S2c |
| ORF1ab-R2 | A*A*GTCAGTGTACTCTATAAGTTTTGATGGTGT*G*T | Fig. 2d–f, 2h; Fig. S2a-S2c |
| HPV16–F1 | G*T*TACAACGAGCACAGGGCCACAATAAT*G*G | Fig. S4a, S4b |
| HPV16–F2 | C*C*CAAATATTCAATAAACCTTATTGGTTACA*A*C | Fig. S4b |
| HPV16–F3 | A*A*TATTCAATAAACCTTATTGGTTACAACGAGC*A*C | Fig. S4b |
| HPV16–F4 | G*G*TTACAACGAGCACAGGGCCACAATAATGG*C*A | Fig. S4b, S4e; throughout the paper when HPV16 was examined unless otherwise stated |
| HPV16-R1 | C*C*CATGTCGTAGGTACTCCTTAAAGTTAGTATT*T*T | Fig. S4a |
| HPV16-R2 | A*C*TCCTTAAAGTTAGTATTTTTATATGTAG*T*T | Fig. S4a |
| HPV16-R3 | T*A*CTGCGTGTAGTATCAACAACAGTAACAA*A*T | Fig. S4a, S4b, S4e; throughout the paper when HPV16 was examined unless otherwise stated |
| HPV16-R4 | T*A*TTTGTACTGCGTGTAGTATCAACAACAG*T*A | Fig. S4a |
| HPV16-R5 | T*A*GTATCAACAACAGTAACAAATAGTTGGT*T*A | Fig. S4a |
| HPV18–F1 | G*C*ATAATCAATTATTTGTTACTGTGGTAGATACC*AC*T | Fig. S4c, S4d |
| HPV18–F2 | A*T*AACAATGGTGTTTGCTGGCATAATCAATTAT*T*T | Fig. S4d |
| HPV18–F3 | T*G*GTTACATAAGGCACAGGGTCATAACAAT*G*G | Fig. S4d |
| HPV18–F4 | A*A*TTATTTGTTACTGTGGTAGATACCACTCGC*A*G | Fig. S4d, S4f; throughout the paper when HPV18 was examined unless otherwise stated |
| HPV18–F5 | C*A*CAGGGTCATAACAATGGTGTTTGCTGGCAT*A*A | Fig. S4d |
| HPV18-R1 | A*A*ATTTGGTAGCATCATATTGCCCAGGTAC*A*G | Fig. S4c, S4d |
| HPV18-R2 | T*G*GTAGCATCATATTGCCCAGGTACAGGAGAC*T*G | Fig. S4c, S4d, S4f; throughout the paper when HPV18 was examined unless otherwise stated |
| HPV18-R3 | T*A*GCATCATATTGCCCAGGTACAGGAGACTG*T*G | Fig. S4c |
| HPV18-R4 | T*T*GGTAGCATCATATTGCCCAGGTACAGGAGAC*T*G | Fig. S4c |
| HPV18-R5 | T*T*GGTAGCATCATATTGCCCAGGTACAGGAG*A*C | Fig. S4c |
| qPCR primers and probes | Proprietary | Fig. 4b and d; Fig. S5-S8 |
| crRNA-N0 | UAAUUUCUACUAAGUGUAGAUcugcugcuugacagauugaacc | throughout the paper when N gene was examined unless otherwise stated |
| crRNA-N1 | UAAUUUCUACUAAGUGUAGAUuugaacuguugcgacuacgugau | Fig. 1h and i |
| crRNA-ORF1ab | UAAUUUCUACUAAGUGUAGAUgugcaguugguaacaucuguuac | Fig. 2d–f, 2h; Fig. S2 |
| crRNA-HPV16 | UAAUUUCUACUAAGUGUAGAUuugggguaaccaacuau | throughout the paper when HPV16 was examined |
| crRNA-HPV18 | UAAUUUCUACUAAGUGUAGAUacaauaugugcuucuacaca | throughout the paper when HPV18 was examined |
| n-reporter-0 | /56-FAM/TTATT/3Bio/ | Fig. 3a–c |
| FQ1 | /56-FAM/TTATT/3BHQ1/ | Fig. 1a, 1e-1i; Fig. 2a and c; Fig. S1; Fig. S2; Fig. S3; Fig. S4 |
| FQ3 | /56-FAM/TTCCTT/3BHQ1/ | Fig. S3 |
| FQ4 | /56-FAM/TTCCCTT/3BHQ1/ | Fig. S3 |
| FQ5 | /56-FAM/TTCCCCCTT/3BHQ1/ | Fig. S3 |
| FQ7 | /56-FAM/TTCCCCTT/3BHQ1/ | Fig. S3 |
| FQ8 | /56-FAM/AAAAAA/3BHQ1/ | Fig. 2b |
| FQ9 | /56-FAM/TTTTTT/3BHQ1/ | Fig. 2b |
| FQ10 | /56-FAM/GGGGGG/3BHQ1/ | Fig. 2b |
| FQ11 | /56-FAM/CCCCCC/3BHQ1/ | Fig. 2b; Fig. 4; Fig. S3; Fig. S5; Fig. S6; Fig. S7; Fig. S8 |
| reporter-1 | /56-FAM/T(dT-BHQ1)ATT | Fig. 3g |
| reporter-2 | /56-FAM/T*(dT-BHQ1)ATT | Fig. 3g |
| reporter-3 | /56-FAM/T(dT-BHQ1)CCCCTT | Fig. 3h |
| reporter-4 | /56-FAM/T*(dT-BHQ1)CCCCTT | Fig. 3h |
| n-reporter-1 | /56-Bio/T(dT-DIG)ATT/3FAM/ | Fig. 3e |
| n-reporter-2 | /56-Bio/T*(dT-DIG)ATT/3FAM/ | Fig. 3e and i, Fig. S3d, S3e |
| n-reporter-3 | /56-Bio/T*(dT-DIG)CCCCTT/3FAM/ | Fig. 3i; Fig. 5 |
2.10. Statistical analyses
P values were calculated using one-way ANOVA (multiple groups). Data were expressed as mean ± SD. Differences with p values < 0.05 were considered significant. All statistical analysis was performed using GraphPad Prism software.
2.11. Ethical statement
The research on rapid diagnostic technique for HPVs using clinical samples was approved by the ethical committee of Shenzhen Luohu People's Hospital. Oral consent was obtained from all enrolled patients.
3. Result
3.1. Development of Cas12a-based TESTOR detection system
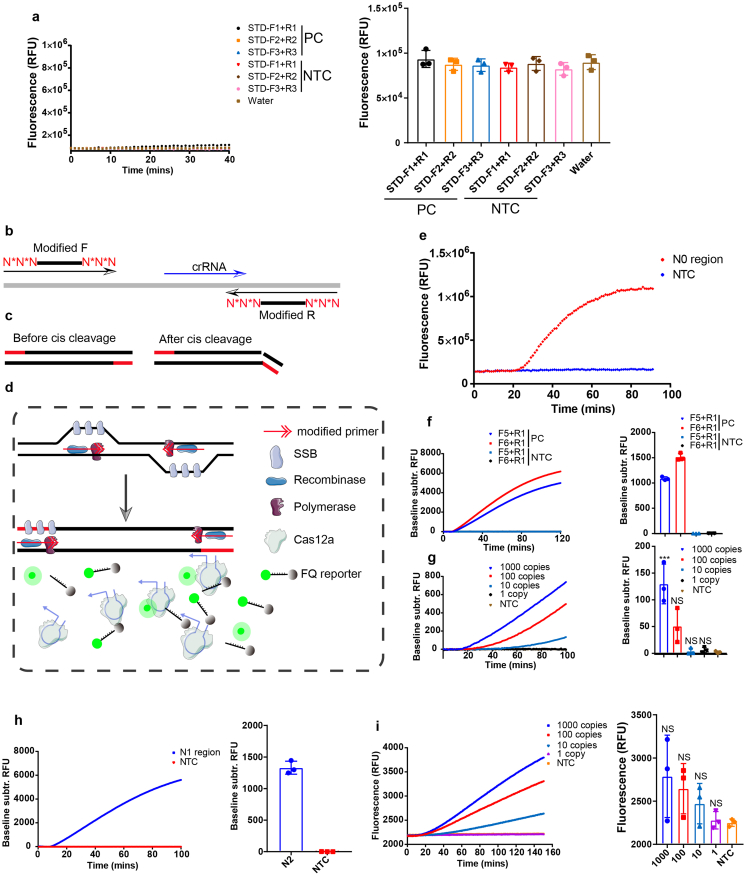
Studies of Cas12a-based diagnostics previously applied a separate pre-amplification step prior to Cas12a-mediated detection [14,17,18,21], which inevitably complicates the procedures and brings about contaminations. To simplify the operations, we attempted to develop an assay in which the components of RPA and Cas12a enzyme were added all together in a single reaction. As an application example, we first designed primers and crRNA targeting N gene (referred to as N0 region) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus responsible for the COVID-19 global pandemic. After incubation of the RPA with Cas12a at 37 °C for 40 min, however, there was no signal detected (Fig. 1a).
Fig. 1.
Detecting N gene of SARS-CoV-2 with a onepot method using phosphorothioate modified primers. a, Representative plot of fluorescence intensity versus time for onepot detection of N gene of SARS-CoV-2 plasmid using three unmodified-primer pairs (left panel). Fluorescent signal was obtained at 30 min after reaction (right panel). b, Primers were modified with phosphorothioate on the first two phosphate backbones proximity to 5′ and 3′ end. crRNA was designed to have two nucleotides overlapping with the reverse primer (upper panel). Modified F: forward primer modified with phosphorothioate; modified R: reverse primer modified with phosphorothioate. c, Intact amplicons derived from the modified primers (left panel) and nicked dsDNA products after Cas12a cis cleavage (right panel). d, Schematic of TESTOR workflow. SSB, single-stranded DNA binding protein; F, fluorophore; Q, quencher. e, Real-time fluorescence detection of the TESTOR assay for N gene of SARS-CoV-2 (N0 region) and 105 copies of plasmid DNA was used. f, Fluorescence kinetics of two primer pairs for N gene of SARS-CoV-2 (N0 region) detection (left panel) in a closed-tube. Fluorescent signal was measured at 30 min after reaction (right panel) using 105 copies of plasmid DNA. g, Analytical sensitivity of TESTOR for N gene of SARS-CoV-2 (N0 region) detection (left panel). Fluorescent signal was measured at 30 min after reaction (right panel) using 105 copies of plasmid DNA. h, Another region of N gene of SARS-CoV-2 (N1 region) was detected using 105 copies of plasmid DNA template. i, Analytical sensitivity of TESTOR for N1 gene of SARS-CoV-2 detection. Signals were obtained using a plate reader in an uncapped 96-well plate (a, e) or using an real-time PCR detection system in a capped PCR tube (f, g, h, i). Error bars represent the mean ± s.d., where n = 3 replicates (a, f, g, h, i). ***, P < 0.001; NS, not significant. One-way ANOVA followed by a Tukey's multiple comparison post-test (f, g, i).
Previous study showed that Cas12 family enzymes generate double-strand DNA breaks in a sequential dependent manner [18,22]. The non-target DNA strand (NTS) cleavage is known to precede target DNA strand cleavage. The cleavage of the NTS in dsDNA substrates activates the collateral activity of Cas12a and initiates cleavage of non-target substrates [23]. As the single-stranded primers could also be a substrate of the activated Cas12a, we hypothesized that both cleavage of the amplicons and degradation of the primers contribute to the failure of detection for the target gene. To verify this, we performed phosphorothioate modifications at multiple sites of primers (Fig. 1b). Such modifications are typically resistant to the cleavage by divalent cation-dependent nucleases [22,24,25]. A crRNA overlapping multiple nucleotides with the primer was designed, which enables the cleavage by Cas12a-crRNA complex occur at the modified sites [22]. As shown in Fig. 1c, the non-target strand without modifications would be cleaved while the target strand would keep intact after reaction. Although the detailed mechanisms are unclear, the nicked structure might serve as a template for the next round of DNA amplification, enabling continuous production of amplicons. When incubating the components of RPA and Cas12a in a single reaction at 37 °C, RPA amplification is first initiated and the amplified target is subsequently recognized by Cas12a-crRNA complex. The Cas12a endonuclease is then activated and cleaves the nearby FQ reporters (a single stranded DNA labeled with a FAM fluorophore on its 5′ end and a BHQ1 quencher on its 3’ end) to generate fluorescence (Fig. 1d).
In initial experiments with the modified primers, we were surprised to find a strong signal accumulation over time in presence of the target gene (Fig. 1e, S1a). Nevertheless, when we tried to repeat this finding with other primer pairs, the signals became extremely weak (Fig. S1b, S1c), suggesting instability of this system. To investigate either failed amplification of the nucleic acid or unsuccessful cleavage of the amplicons by Cas12a led to the faint signal, we resolved the reactions by agarose electrophoresis. Consistent with the fluorescent signals, three out of five reactions showed obvious bands whereas the rest two had little products (Fig. S1d). To identify whether the observed bands were the intended products, 5 μL of the reaction was added into 20 μL of 1x Cas12a mixture. Similar trends were observed but the signals were much stronger in comparison with that of the onepot assay (Fig. S1d). We wondered if the RPA reaction system impeded the collateral activity of activated Cas12a, because RPA solution is rather viscous [26]. By adding additional 5 μL of water into the system, we found a remarkable increase of signals generated by cleaved reporters (Fig. S1e). These observations drove us to think that whether the inhibited cleavage of reporter was resulted from the liquid evaporation in the reaction as an uncapped 96-well plate was used during the monitoring process. We therefore switched to a capped PCR detection system for signal monitoring. As expected, a strong fluorescence increasing was observed after incubation at 37 °C for around 10 min (Fig. 1f). Additionally, we found the ratio of the components of RPA to Cas12a was extremely important for the onepot detection. We made a 5x Cas12a solution and mixed it with the RPA solution in different proportions in volume. As shown in Fig. S1f, S1g, the reaction gave the strongest signal when the ratio was 1: 5. Thus, the following assays were conducted with 1: 5 ratio in volume of Cas12a mixture and RPA solution.
We next evaluated the limit of detection (LoD) of the TESTOR system. Whereas this assay lacked the sensitivity to detect 1, 10 and 100 copies of molecule in the reaction, it unambiguously identified 1000 copies within 30 min (Fig. 1g). To assess whether this primer-modified method can serve as a general approach to integrating the amplification and detection into a single reaction, we designed other primer sets targeting a different region of N (referred to as N1 region) and ORF1ab gene of SARS-CoV-2. In the presence of high number of targets, all reactions for both N1 and ORF1ab could readily distinguish their corresponding targets from NTC (non-target control) within a short incubation time (Fig. 1h, S2a). However, when copy numbers in the system were low, the fluorescence-differential time was a bit long, and most of the cases failed to detect the targets at 30 min of reaction (Fig. 1i, S2b, S2c). These results together reveal the feasibility of Cas12a-based onepot detection using purified nucleic acids by phosphorothioate modifications on the backbone of primers, even with limited sensitivity.
3.2. Optimization of the TESTOR assay
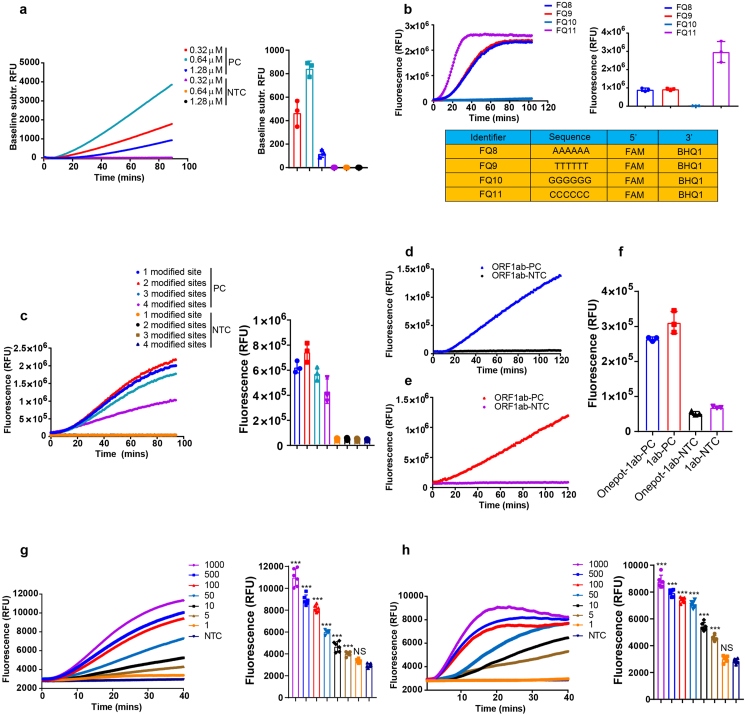
To achieve a better sensitivity in a shorter time, we first evaluated the effect of primer concentration on the efficiency of our assay. As shown in Fig. 2a, we found the optimal concentration was 0.64 μM. Cleavage efficiency of CRISPR/Cas9 systems seemed to be influenced by GC content and purine residues in the gRNA end [27]. We were therefore interested in knowing whether the trans activity of Cas12a was dependent on the sequence of reporters. The C nucleotide-rich reporters exhibited the strongest fluorescence signal when the same amount of RPA product for N0 gene of SARS-CoV-2 was used for the Cas12a cleavage assay, indicating the highest affinity of this reporter to crRNA-Cas12a complex (Fig. 2b, S3a, S3c). Interestingly, the G nucleotide reporter showed negligible increase of fluorescence, implying there was almost no cleavage induced by crRNA-Cas12a complex (Fig. 2b). We further tested the specificity of these reporters, and found none of them showed obvious signal increasing over time in NTC groups (Fig. S3b, S3c). Besides, we observed two sites of modification on the ends of forward and reverse primers is sufficient for a strong detection. Too many modifications seemed to inhibit the reaction (Fig. 2c), which is in agreement with a previous study [25]. We thus used a condition of two-site modified primers at a concentration of 0.64 μM and combined with the C nucleotide-rich reporter in a reaction for the following experiments.
Fig. 2.
Optimization of TESTOR system. a, Real-time (left panel) and end point (right panel) fluorescence detection using primers specific to the N0 gene at the indicated concentration. b, Reporters with A, T, G, or C nucleotide sequence was screened to identify the one with the best affinity to Cas12a. The same amount of RPA product of N0 gene was added to a Cas12a mixture with different reporter, and fluorescence was monitored by real-time or taken at 30 min after incubation at 37 °C. c, Primers modified with phosphorothioate on different phosphate backbones were compared for reaction efficiency by real-time (left panel) or endpoint (right panel) method. d, TESTOR approach for detection of ORF1ab gene of SARS-CoV-2. e, Fluorescence kinetics of Cas12a cleavage using product of RPA for ORF1ab gene as input. f, Quantification of the fluorescence intensity of TESTOR method or routine two-step method (from Fig. 2d and 2e) after 30 min of incubation at 37 °C. g, h, Determination of LoDs for N0 (g) and ORF1ab (h) genes using the optimized conditions for TESTOR system. Representative plot of fluorescence intensity over time for N0 and ORF1ab genes of SARS-CoV-2 (left panel) or fluorescent signal was taken at 30 min after reaction (right panel). Error bars represent the mean ± s.d., where n = 3–6 replicates (a, b, c, f, g, h). ***, P < 0.001; NS, not significant. One-way ANOVA followed by a Tukey's multiple comparison post-test (g, h).
With these optimizations, we compared the canonical two-step method with TESTOR assay in the presence of 100 copies of target in a given reaction. The former of which began to produce signal upon incubation at 37 °C, while the latter showed a lag around 10 min before signal climbing (Fig. 2d–f). This phenomenon could be explained by the required time for amplicon accumulation before recognition and cleavage by crRNA-Cas12a complex. Moreover, we examined the analytic performance of this optimized system for detection of N0 and ORF1ab gene of SARS-CoV-2 and demonstrated it could detect as low as five copies per reaction in 30 min (Fig. 2g and h, Table S1, S2).
3.3. Development of lateral flow TESTOR assay
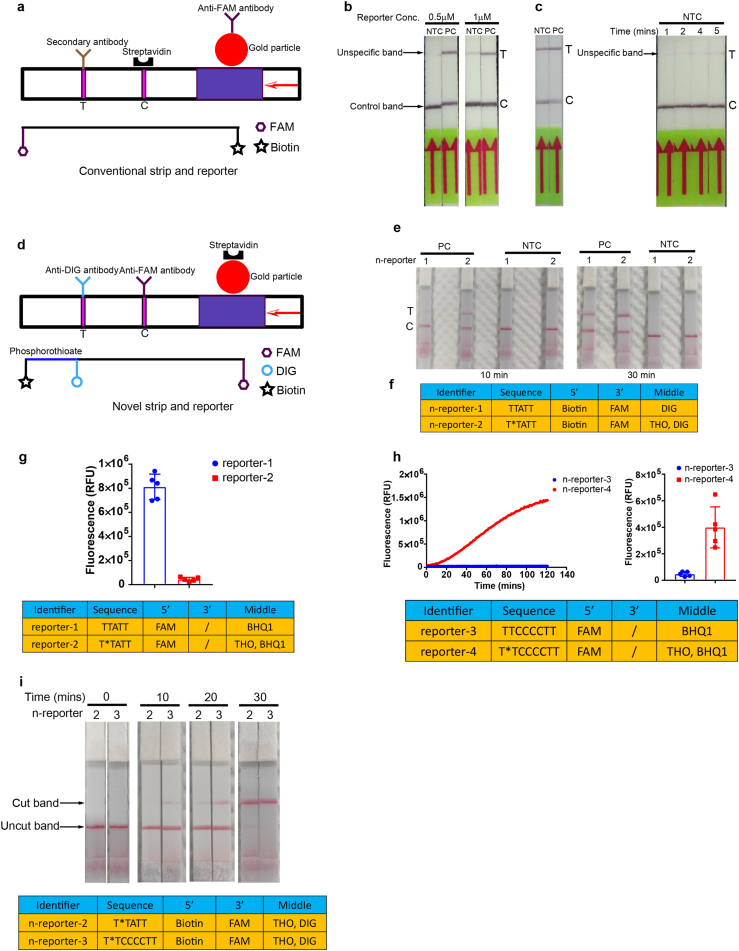
For any on site detection assays, pairing with a visual readout is crucial especially in situations when instruments are not available. We herein developed a method enabling colorimetric readout with a lateral flow strip. We designed a reporter with a dye label (FAM) on the 5′ end and a biotin on the 3’ end. When coupled with a ready-to-use strip (Fig. 3a), destruction of the reporter is visible by naked eye. The control line captures abundant reporter that binds to anti-FAM antibody-gold conjugates with streptavidin molecules, which prevents accumulation of the antibody-gold conjugates to secondary antibodies on the test line; the cleaved reporter-antibody-gold conjugates flow over the control line and will be fixed there by species-specific antibodies on the test line (Fig. 3a) [28]. While the positive samples displayed an easy-to-see band after 5 min of lateral flow in the diluted reaction solution (1:5), a faint signal, in the absence of target, was also present at the test line (Fig. 3b). Increased concentration of reporter displayed more aggravation of non-specificity (Fig. 3b) and this unspecific signal was even more obvious when the dilution ratio increased from 1:5 to 1:10 (Fig. 3c). Time point monitoring demonstrated the unspecific band was produced as early as 1 min of incubation at room temperature (Fig. 3c).
Fig. 3.
Development of the novel lateral flow assay. a, Schematic of conventional strip and reporter used for Cas12a-based nucleic acid detection. b, Lateral flow strip readout of 1:5 diluted TESTOR reactions with 0.5 μM or 1 μM reporter in the presence or absence of N0 gene target using conventional strip and reporter. Strip was incubated at room temperature for 5 min following 30 min of TESTOR reaction at 37 °C. c, Lateral flow strip readouts of 1:10 diluted TESTOR reactions with 1 μM reporter in the presence or absence of N0 gene target using conventional strip and reporter (left panel). Time course of lateral flow strip readouts using 1:5 diluted TESTOR reactions with 1 μM reporter in the absence of N0 gene target (right panel). d, Schematic of the novel strip and reporter. The reporter is labeled with a biotin on 5′ end, a FAM molecule on its 3′ end and a DIG in the middle. Anti-FAM and Anti-DIG antibodies are immobilized at the control and test line, respectively. e, Lateral flow strip readouts using novel strip and reporter at indicated conditions. The onepot reactions were performed at 37 °C for 10 min or 30 min. Novel reporters with or without the phosphorothioate modification between biotin and DIG were used to perform the lateral flow assay. f, Sequences of novel reporters; * and THO represent phosphorothioate modification. g, Fluorescence obtained at 30 min after reaction using two different reporters with or without phosphorothioate modification (upper panel). Error bars represent the mean ± s.d., where n = 3 replicates. Sequences and modification of reporters; * and THO represent phosphorothioate modification (bottom panel). h, Representative plot of fluorescence intensity versus time (upper left) and its quantification (upper right) after 30 min of reaction using C nucleotide-rich reporters. Error bars represent the mean ± s.d., where n = 3 replicates. Sequences and modification of reporters; * and THO represent phosphorothioate modification (bottom panel). i, Comparison of cleavage efficiency for C nucleotide-rich and –lacking reporters at specified conditions (upper panel). Sequences and modification of reporters; * and THO represents phosphorothioate modification (bottom panel).
In clinical applications, presence of an nonspecific faint band at the test line would make judgement of positivity rather difficult, since weak positive samples can also give a faint signal. The nonspecific band might be generated by the antibody-gold conjugates that did not bind to intact or destroyed reporter, making it a problem rather tough to be addressed [17]. Here, we designed a novel reporter labeled with biotin, FAM and DIG (Fig. 3d), and modified with phosphorothioate at the sites between DIG and Biotin. Besides, we created a dipstick on which the gold particles were coupled with the biotin-ligand molecules in the sample application area, and anti-FAM antibodies and anti-DIG antibodies were immobilized on the control and test line, respectively. When reacted solution with the novel reporter was applied onto the strip, intact reporter will be trapped by anti-FAM antibodies at the first line while the degraded reporter without FAM fluorophore on its 3’ end will be captured by anti-DIG antibodies at the second line.
As a proof-of-concept assay, we digested the novel reporter with DNase I to mimic the Cas12a cleavage and applied the digested solution to the strip. The non-digested reporter at concentrations of 0.5 μM and 1 μM did not show any signal at the test line, while the digested ones produced strong bands there (Fig. S3d). We next evaluated this system using RPA-Cas12a assay for the detection of N0 gene of SARS-CoV-2. To simplify the procedure, we used the two-step method. Target was first amplified by RPA reaction with unmodified primers and then the product was added into the Cas12a mixture for cleavage assay. The reactions diluted at the ratio of 1:5 or 1:10 were then examined with the novel strip and neither of them displayed signal at the test line in NTC group (Fig. S3e). In addition, it seemed that 1:5 dilution could give a stronger signal than that of 1:10 dilution (Fig. S3e).
We then tested this new lateral flow assay with the TESTOR assay, and in agreement with the DNase-digested assay and the two-step RPA-Cas12a assay, no signal was observed at the test line in NTC group (Fig. 3e and f). Notably, the signal was much stronger when the reporter was modified with phosphorothioate compared to that without such modification (Fig. 3e). The inhibited cleavage at the site between biotin and DIG, which led to increased accumulation of ssDNA-antibody-gold conjugates at the test line, may contribute to this result. To confirm it, we applied a phsphorothioate modified reporter that was labeled with a FAM molecule on its 5’ end and a BHQ1 quencher in the middle, and found the fluorescence produced by destruction of the reporter was completely abolished (Fig. 3g and h). Furthermore, we reconfigured this new lateral flow assay with the C-nucleotide-rich reporter for the detection of amplified-product of N0 gene. In accordance with the fluorescent assay, such reporter was able to significantly shorten the reaction time (Fig. 3i). These results together indicate that the TESTOR system for lateral flow assay is of high rapidity and specificity.
3.4. TESTOR validation with clinical samples for detection of HPV16 and HPV18
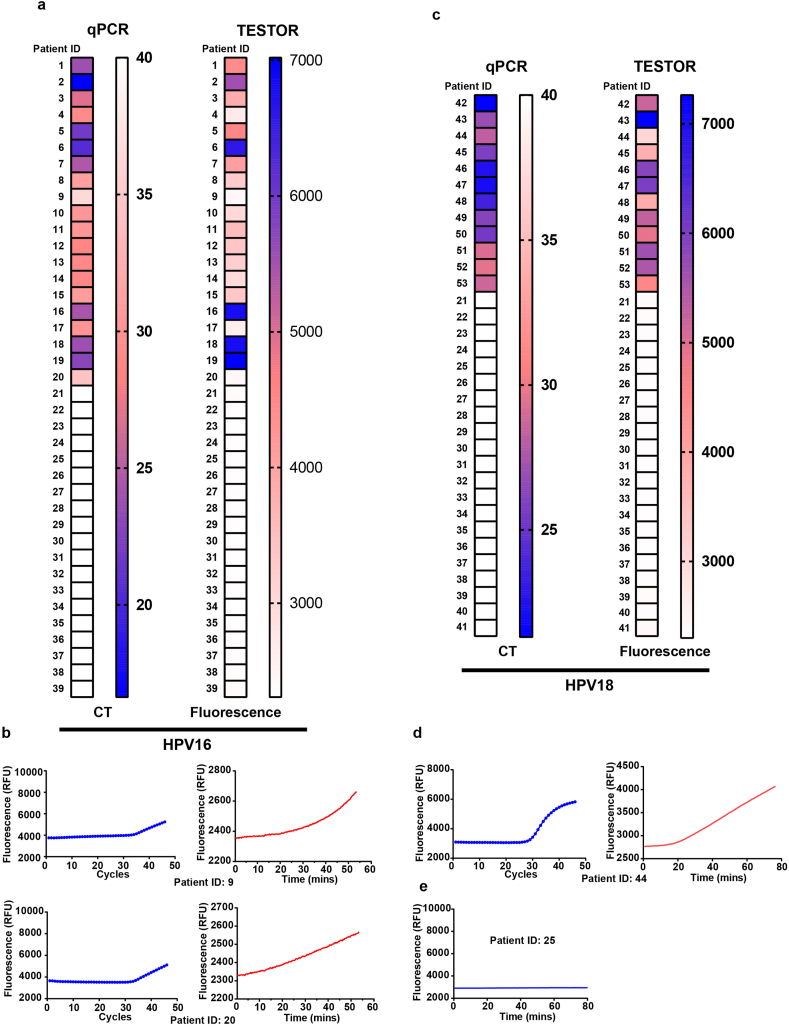
Human papillomaviruses (HPVs) are the major causative agents of cervical carcinomas with types 16 (HPV16) and 18 (HPV18) accounting for most precancerous lesion [29]. We designed sets of primers targeting L1 region of HPV16 or HPV18, and selected the best performance primer pairs for a rapid and sensitive detection (Fig. S4a-S4d). High sensitivities (5 copies per reaction) for detection of HPV16 and HPV18 plasmids were achieved with the selected primer pairs (Fig. S4e, S4f). We then tested unextracted DNA released with a rapid lysis method from 53 cervical scrape samples Of these, 20 and 12 were positive for HPV16 and HPV18 infection, respectively, by qPCR testing, and 21 were negative for HPV16 or HPV18 infection but either positive for other type of HPVs or negative by all testing. All positive samples for HPV16 or HPV18 by qPCR were positive in our assay, confirming that the false negative rate for TESTOR is very low (Fig. 4a–d, S5, S7). Two samples for HPV16 and one sample for HPV18 were found to display weak signals at 30 min. Nonetheless, the fluorescence kept increasing over time and the fluorescent values at 30 min exceeded the threshold of 2400 for a typical positive call in the context of HPV16 testing (pre-determined by testing several negative samples using the primer set and crRNA for HPV16) (Fig. 4b and d, Table S3, S4). While all the HPV18 negative samples by qPCR were negative by TESTOR (Fig. S8), 1 out of 20 PCR-negative samples for HPV16 displayed slight increase in fluorescence after more than 40 min of reaction (Fig. S6). To explore whether the signal was generated by specific activation of Cas12a, we performed PCR amplification using the product. The amplified solution was then subjected to Cas12a detection but, as shown in Fig. 4e, no signal was detected, indicating the signal was generated by unspecific reporter cleavage, which may ascribe to the long incubation time. To make the data more convincing, we blindly tested 20 more HPV16 positive and negative samples with qPCR and TESTOR. We set up a cutoff value of 2400 for a positive call after 30 min of reaction, and with this criteria, all positive and negative samples by qPCR were identified as the same by TESTOR (Table S5). Taken together, our TESTOR assay had 100% positive and negative agreements relative to the qPCR assay, for detection of the HPV16 and HPV18 in a total of 93 clinical samples.
Fig. 4.
Detection of HPV in clinical samples using fluorescence TESTOR assay. a, Heatmaps showing the CT values by qPCR (left panel) and fluorescence at 30 min by TESTOR assay (right panel) for HPV16 detection. Two out of twenty clinical samples were qPCR positive but showed weak signal at 30 min by TESTER assay (Patient ID: 9, 20). b, Fluorescence kinetics of the two samples showing late CT values by qPCR or weak signals by TESTER. c, Results of the qPCR (left panel) and fluorescence TESTOR assay at 30 min (right panel) for HPV18 detection. One out of thirteen clinical samples was positive by qPCR but showed weak signal at 30 min by fluorescence TESTER assay (Patient ID: 44). d, Fluorescence kinetics of the clinical sample (Patient ID: 44) showing late CT value by qPCR or weak signal by TESTOR assay. e, Fluorescence curve of re-examination by Cas12a for one patient negative for HPV18 by qPCR but showing slight signal increase by TESTOR. The yield of TESTOR from the patient (ID: 25) was amplified by PCR and then the PCR product was detected by Cas12a reaction.
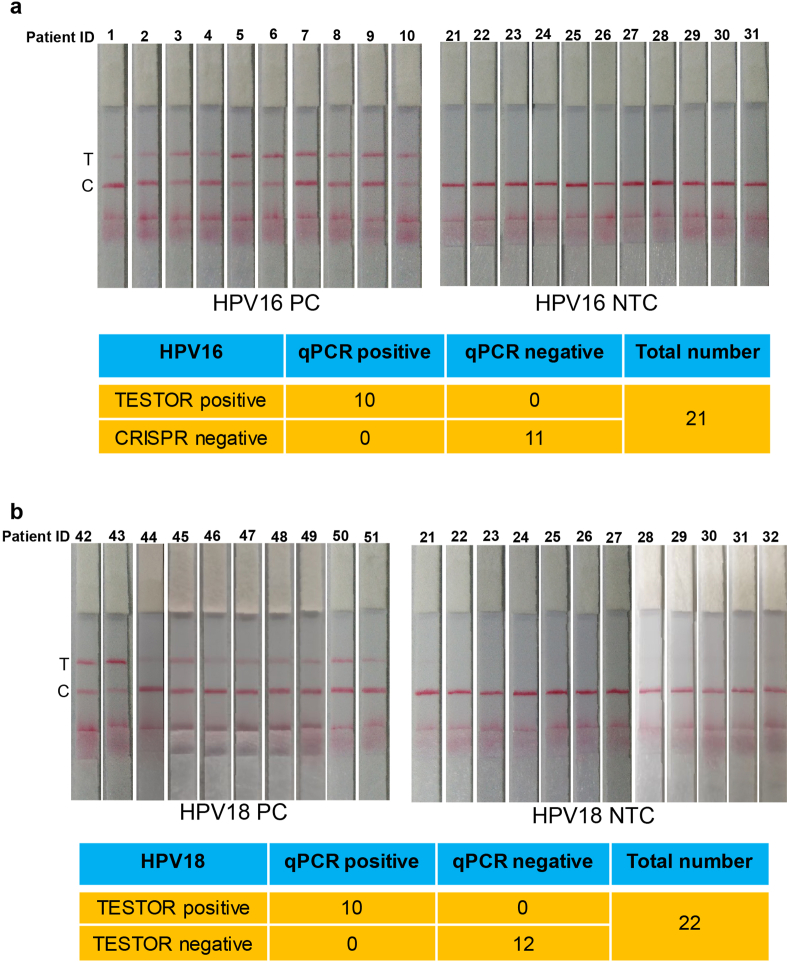
Next, we evaluated our established lateral flow system using 20 positive and 20 negative cervical scrape samples (Fig. 5). The positive and negative agreements of the TESTOR assay relative to the qPCR assay were 100%, for detection of both HPV16 and HPV18. One sample with HPV18 infection gave a very faint band at the test line, which is consistent with its qPCR result, revealing the advantages of our established lateral flow system over the traditional one in detecting samples with low viral load, as there was no nonspecific signal at all at the test line in negative samples. These data illustrate the high sensitivity and specificity of the lateral flow TESTOR assay.
Fig. 5.
Detection of HPV in clinical samples using lateral flow TESTOR assay. a, Lateral flow strips showing HPV16 TESTOR assay results (upper panel). Ten qPCR-positive and eleven qPCR-negative samples were used for HPV16 detection. The Cas12a detection assays were run on lateral flow strips and imaged after 5 min. Performance characteristics of lateral flow TESTOR assay (bottom panel). A total of 21 clinical samples were evaluated using the lateral flow version of the TESTOR assay. Both the positive agreement and negative agreements are 100%. NTC, no-template control; T, test line; C, control line. b, Lateral flow strip readouts for HPV18 detection using clinical samples. A total of 21 clinical samples were evaluated (10 HPV18 positives and 12 negatives). The reactions were 1:5 diluted after incubation at 37 °C for 30 min and then run on lateral flow strips and imaged after 5 min.
4. Discussion
In present study, we provide a general onepot method for Cas12a-based nucleic acid detection using extracted/rapidly released nucleic acids by modifying the primers with phosphorothioate and designing crRNA allowing the cleavage occur at the modified sites. With these innovations, we integrate the amplification and detection into a single reaction system, which is highly valuable because avoidance of uncapping is important for prevention of aerosol generation that easily causes false positivity. Previous studies adopted a physical separation method to combine nucleic acid amplification and Cas12a detection into a single tube [30,31]. Wang et al. added Cas12a on the inner wall of the reaction tube, and a centrifugation step was followed to initiate the cis- and trans-cleavage of Cas12a after RPA reaction [31]. However, this method is troublesome and also unreliable. The TESTOR system applies all components into a single common-used tube, which completely circumvents the pre-amplification of target nucleic acids and the requirement of special tubes or devices [32]. We also report here, for the first time, that the trans activity of the activated Cas12a prefers a C-nucleotide sequence. By leveraging this feature, the reaction time for a sensitive detection was significantly reduced when C-nucleotide rich reporters were used. To make it more field deployable, we created a novel lateral flow assay, which surmounts the shortcoming of the routine lateral flow method, and provided a more specific strip-based system.
Our TESTOR system needs minimal instrumentation and can be performed by lay users, making it suitable for diagnostic tests in resource-restricted areas. As proof-of-concept assays, TESTOR has been developed for SARS-CoV-2 detection. The infection cases of SARS-CoV-2 are increasing rapidly around the world and it seems now to be driven by community transmission [33,34]. As numerous infections are asymptomatic, nucleic acid testing is vital to differentiate infected from healthy individuals [35]. Therefore, a rapid diagnostic method for SARS-CoV-2 is urgently needed. Our Cas12a-based TESTOR technology can be reconfigured within days to detect SARS-CoV-2 and has the promise to address the key challenges for this global pandemic. This system has also been validated using clinical specimens for HPV detection. By combining with a rapid sample processing method, the sample-to-result can be achieved in 30 min with high sensitivity (5 copies per reaction). Our system reported here could be an alternative to qPCR test as it is faster, simpler and highly specific. However, although the TESTOR system was developed using the example sequences of SARS-CoV-2, HPV16 and HPV18, we only validated it to detect clinical samples of HPV16 and HPV18. Therefore, the results for SARS-CoV-2 should be carefully interpreted. Testing genuine SARS-CoV-2 samples would be crucial to better understand the sensitivity and specificity and thus provide confidence that this assay is adequate to address the pandemic's needs.
To facilitate routine surveillance of pathogens and other comprehensive applications, integration of the TESTOR with microfluidic system would be necessary because in many cases there is a need for diagnostic technologies to be able to test many samples while simultaneously testing for many targets. A recent study developed a massive multiplexing system for nucleic acid detection by combining a micro-well array that harnesses solution-based fluorescent color codes with CRISPR-Cas13 detection. This novel system is capable of testing >4500 crRNA-target pairs on a single array [36]. Another appealing development would be accommodation of a portable cartridge to streamline the workflow and to enable point-of-care testing in diverse environments, such as airports, clinics, local communities and other locations. The cartridge could also reduce the risk of aerosol contamination as all processes including sample manipulation, onepot reaction and lateral flow strip visualization could be completed in a closed-environment [20].
Funding
This work was supported by the Discipline Construction Ability Promotion Project of Shenzhen Health and Population Family Planning Commission (NO.SZXJ2018031 (Kan L.J.)), Sanming Project of Medicine in Shenzhen (NO. SZSM201601062 (Zhang X.M.)) and Shenzhen Key Medical Discipline Construction Fund (NO.SZXK054 (Zhang X.M.)). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Jiaojiao Gong: Data curation, Formal analysis, Investigation, Validation. Lijuan Kan: Data curation, Investigation, Resources. Xiuming Zhang: Resources. Ying He: Investigation, Resources. Jiaqiang Pan: Investigation. Liping Zhao: Investigation, Resources. Qianyun Li: Methodology, Resources. Menghao Liu: Investigation. Jie Tian: Methodology. Sili Lin: Methodology. Zhouyu Lu: Methodology. Liang Xue: Conceptualization, Funding acquisition. Chaojun Wang: Funding acquisition, Project administration. Guanghui Tang: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Declaration of competing interest
G.T. and L.X. are the technical directors of the department of R&D in Yaneng Biotech, Co., Ltd, and receive research support funding from this company. J.P., J.T., S.L., Z.L., J.G. are employees of Yaneng Biotech, Co., Ltd.
Acknowledgements
We thank Wei He and Jing Qu for comments and discussion and Yuanyuan Chang for preparation of the HPV plasmids.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.05.005.
Contributor Information
Liang Xue, Email: xuel0620@sina.com.
Chaojun Wang, Email: wangchaojundf@hotmail.com.
Guanghui Tang, Email: shtangguanghui@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radmard S., Reid S., Ciryam P., Boubour A., Ho N., Zucker J., Sayre D., Greendyke W.G., Miko B.A., Pereira M.R. Clinical utilization of the FilmArray meningitis/encephalitis (ME) multiplex polymerase chain reaction (PCR) assay. Front. Neurol. 2019;10:281. doi: 10.3389/fneur.2019.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang A.M., Doyle M.V., Mark D.F. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding X., Yin K., Li Z., Liu C.J.b. All-in-One dual CRISPR-cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. 2020 doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch I., de Puig H., Hiley M., Carre-Camps M., Perdomo-Celis F., Narvaez C.F., Salgado D.M., Senthoor D., O'Grady M., Phillips E. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balmaseda A., Stettler K., Medialdea-Carrera R., Collado D., Jin X., Zambrana J.V., Jaconi S., Cameroni E., Saborio S., Rovida F. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:8384–8389. doi: 10.1073/pnas.1704984114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L.J.E.m. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. J Emerging microbes infections. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.J.S. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini L.A., Sontheimer E.J.J.s. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F.J.S. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F., Abudayyeh O.O., Gootenberg JS. Broadinstitute; 2020. A Protocol for Detection of COVID-19 Using CRISPR Diagnostics. [Google Scholar]
- 16.Wang B., Wang R., Wang D., Wu J., Li J., Wang J., Liu H., Wang Y. Cas12aVDet: a CRISPR/cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 2019;91:12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- 17.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S.-Y., Cheng Q.-X., Liu J.-K., Nie X.-Q., Zhao G.-P., Wang J.J.C.r. CRISPR-Cas12a has both cis-and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong D., Dai W., Gong J., Li G., Liu N., Wu W., Pan J., Chen C., Jiao Y., Deng H. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.-l.W., Kim N.-G., Yu X., Li J., Walker B.D.J.m. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 [Google Scholar]
- 21.Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., Cao R.-B., Zhao G.-P., Wang J.J.C.d. CRISPR-Cas12a-assisted nucleic acid detection. Cell discovery. 2018;4:1–4. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swarts D.C., Jinek M.J.M.c. Mechanistic insights into the cis-and trans-acting DNase activities of Cas12a. 2019;73:589–600. doi: 10.1016/j.molcel.2018.11.021. e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swarts D.C., Jinek M.J.M.c. Mechanistic insights into the cis-and trans-acting DNase activities of Cas12a. Mol. Cell. 2019;73:589–600. doi: 10.1016/j.molcel.2018.11.021. e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckstein F., Gish G. Phosphorothioates in molecular biology. Trends Biochem. Sci. 1989;14:97–100. doi: 10.1016/0968-0004(89)90130-8. [DOI] [PubMed] [Google Scholar]
- 25.Li B., Zeng C., Li W., Zhang X., Luo X., Zhao W., Zhang C., Dong Y. Synthetic oligonucleotides inhibit CRISPR-cpf1-mediated genome editing. Cell Rep. 2018;25:3262–3272. doi: 10.1016/j.celrep.2018.11.079. e3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A.J.P.B. DNA detection using recombination proteins. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruegmann T., Deecke K., Fladung M. Evaluating the efficiency of gRNAs in CRISPR/Cas9 mediated genome editing in poplars. Int. J. Mol. Sci. 2019;20:3623. doi: 10.3390/ijms20153623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zur Hausen H., De Villiers E.M. Human papillomaviruses. Annu. Rev. Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Ke Y., Liu W., Sun Y., Ding X.J.A.s. A one-pot toolbox based on Cas12a/crRNA enables rapid foodborne pathogen detection at attomolar level. ACS Sens. 2020;5:1427–1435. doi: 10.1021/acssensors.0c00320. [DOI] [PubMed] [Google Scholar]
- 31.Wang B., Wang R., Wang D., Wu J., Li J., Wang J., Liu H., Wang Y. Cas12aVDet: a CRISPR/cas12a-based platform for rapid and visual nucleic acid detection. ACS Chem. 2019;91:12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- 32.Wu H., He J.-s., Zhang F., Ping J., Wu J.J.A.C.A. Contamination-free visual detection of CaMV35S promoter amplicon using CRISPR/Cas12a coupled with a designed reaction vessel: rapid, specific and sensitive. Anal. Chim. Acta. 2020;1096:130–137. doi: 10.1016/j.aca.2019.10.042. [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Liao X., Qian S., Yuan J., Wang F., Liu Y., Wang Z., Wang F.-S., Liu L., Zhang Z. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2606.200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghinai I., Woods S., Ritger K.A., McPherson T.D., Black S.R., Sparrow L., Fricchione M.J., Kerins J.L., Pacilli M., Ruestow P.S. Community transmission of SARS-CoV-2 at two family gatherings—chicago, Illinois, February–March 2020. MMWR (Morb. Mortal. Wkly. Rep.) 2020;69:458–464. doi: 10.15585/mmwr.mm6915e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaman J., Galanti M.J.m. Direct measurement of rates of asymptomatic infection and clinical care-seeking for seasonal coronavirus. MedRxiv. 2020 [Google Scholar]
- 36.Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., Simon H.Y., Boehm C.K., Kosoko-Thoroddsen T.-S.F., Kehe J.J.N. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.