Abstract
Epithelial-mesenchymal transition (EMT) promotes tumorigenesis and metastasis and increases tumor tolerance to treatment intervention. Abnormal activation of transforming growth factor β (TGF-β) and Wnt pathway induces EMT. Long non-coding RNAs (lncRNAs) significantly influence EMT regulation. Herein, we show that MIR210HG is overexpressed in endometrial cancer tissues, which is associated with poor prognosis. MIR210HG silencing significantly inhibited proliferation, migration, invasion, and EMT phenotype formation in vitro as well as tumorigenesis in vivo. Mechanistically, bioinformatics analyses, RNA binding protein immunoprecipitation (RIP) assays, and luciferase assays showed that MIR210HG acts as a molecular sponge of miR-337-3p and miR-137 to regulate the expression of HMGA2. Additionally, MIR210HG overexpression significantly enriched the Wnt/β-catenin and TGF-β/Smad3 signaling pathway genes, while MIR210HG or HMGA2 knockdown suppressed the Wnt/β-catenin and TGF-β/Smad3 signaling pathway. Our findings on the MIR210HG-miR-337-3p/137-HMGA2 axis illustrate its potential as a target for endometrial cancer therapeutic development.
Keywords: MIR210HG, miR-137, EMT, endometrial cancer, prognosis
Graphical abstract
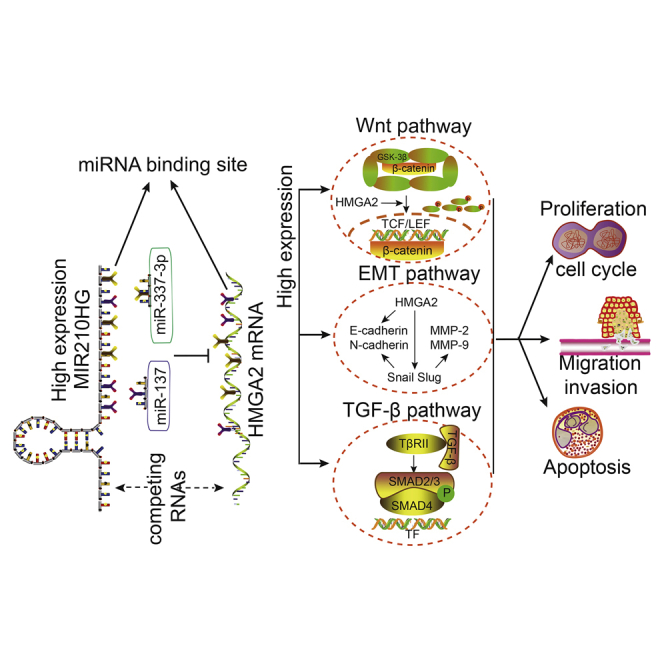
In this study, we identified that lncRNA MIR210HG promotes endometrial cancer via the miR-337-3p/137-HMGA2 axis. In summary, our research provides new molecular prognostic markers for endometrial cancer as well as potential therapeutic targets.
Introduction
Endometrial cancer is the fifth-most-common fatal malignancy among women globally and first among gynecological malignancies in Europe and the United States.1 With improvements in social and economic conditions, the incidence of endometrial cancer has also increased, while the age of onset has decreased.2 Metastatic or recurrent endometrial cancer is incurable in most cases, and management prolongs the patient’s life and relieves symptoms.3 With the elucidation of the molecular mechanism of endometrial cancer, drug-targeted therapies have proven efficacy.4,5 Nonetheless, patients with advanced endometrial cancer may develop drug resistance with adverse consequences.6 Therefore, further research on the molecular mechanism of endometrial cancer is warranted to identify potential drug targets for better clinical treatment and outcomes.
Long non-coding RNAs (lncRNAs) are involved in almost all tumorigenesis processes.7,8 The lncRNA can be used as competing endogenous RNAs (ceRNAs) that compete with microRNA (miRNA) sponges to regulate multiple target genes.9 Abnormal expression levels of various lncRNAs have been reported in endometrial cancer and affect patient prognosis.10,11
The lncRNA MIR210 (MIR210HG), an miR-210 host gene located in 21q13.3, is 567 nucleotides in length,12 highly expressed in hepatocellular carcinoma (HCC) and colon cancer, and associated with poor prognosis.13,14 In invasive breast and osteosarcoma cells, MIR210HG regulates cell invasion and migration through the epithelial-mesenchymal transition (EMT) pathway.15,16 The expression and the underlying mechanism of MIR210HG in endometrial cancer remains unknown.
Metastasis is the leading cause of cancer-related deaths. Cancer cell metastasis is a multi-step process involving local infiltration and colonization.17 EMT is the process by which epithelial cells are transformed into mesenchymal cells, which are involved in the malignant progression of tumors.18 Aberrant transforming growth factor beta (TGF-β) and Wnt signaling is the most important factor in EMT induction in cancer and other pathological conditions.19,20 Additionally, they co-regulate EMT progression in multiple tumors,21 such as colorectal cancer,22 HCC,23 and non-small cell lung cancer.24
The high mobility group protein A2 (HMGA2) is an architectural transcription factor that belongs to the HMGA protein family. HMGA2 has a characteristic adenine-thymidine (AT) hook structure, which can preferentially bind the A- and T-rich regions of DNA molecular hooks, thereby changing the spatial structure of DNA molecules; AT hooks have been shown to interact with other nuclear proteins. HMGA2 influences various biological processes, while its overexpression is a feature of malignancy.25 We have previously shown that high expression of HMGA2 is associated with poor endometrial cancer prognosis and that overexpression of HMGA2 promotes the proliferation, migration, and invasion of endometrial cancer cells.26 In addition, HMGA2 can promote the metastatic phenotype of cancer cells27 and cancer cell EMT through the TGF-β/SMAD pathway and Wnt/β-catenin signaling in bladder cancer.28
In this study, we identified a novel aberrantly expressed lncRNA, MIR210HG, and demonstrated that it is more highly expressed in endometrial cancer than in normal endometrial tissues. High MIR210HG expression was associated with poor prognosis in patients with endometrial cancer. As a ceRNA, MIR210HG sponges miR-337-3p/137 to increase HMGA2 expression to modulate the malignant behavior of endometrial carcinoma cells. This study provides potential biomarkers and therapeutic targets for endometrial cancer.
Results
MIR210HG is associated with poor survival in patients with endometrial cancer
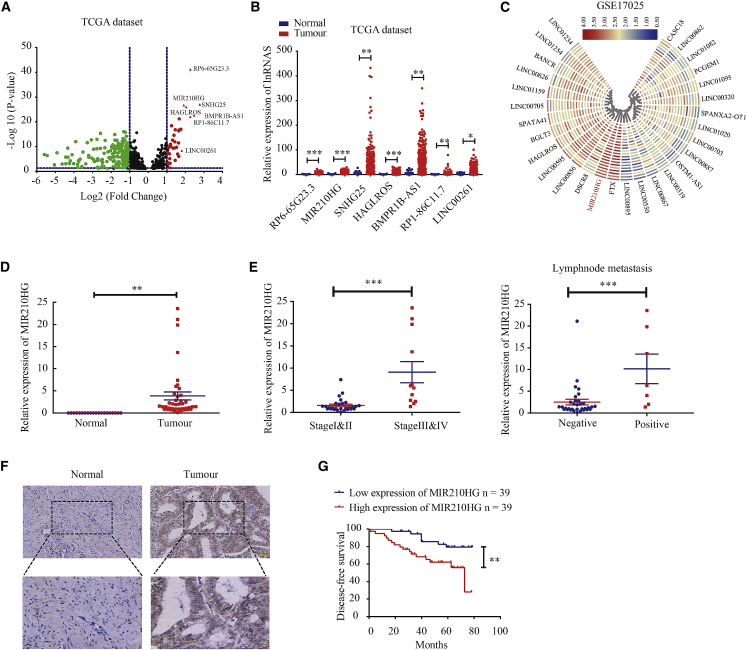
To identify lncRNAs abnormally expressed in the endometrial cancer group, we analyzed data from The Cancer Genome Atlas (TCGA). Expression array analysis revealed 332 lncRNAs with a statistically significant difference in expression. The expression of seven lncRNAs was upregulated with a fold change of >1.9 (Figures 1A and 1B). We then analyzed Gene Expression Omnibus (GEO) datasets and found that MIR210HG expression was also upregulated in GSE17025 endometrial cancer cohorts (Figure 1C). Moreover, quantitative real-time PCR showed that the expression of MIR210HG was significantly higher in endometrial cancer than in normal endometrial tissues (Figure 1D). In addition, we analyzed the association of MIR210HG expression with the clinicopathologic parameters of patients with endometrial cancer and showed that MIR210HG was increased during the progression from stages I and II to III and IV. MIR210HG expression was significantly associated with lymph node metastasis (Figure 1E; Table S1). The results of the in situ hybridization assay showed that the expression level of MIR210HG was higher in endometrial cancer than in normal endometrial tissues (Figure 1F). In situ hybridization analysis also showed that MIR210HG was expressed in 63/78 (80.8%) of endometrial cancer tissues, far higher than the positive MIR210HG expression rate in normal endometrial tissues (5/19, 26.3%, p < 0.0001). In patients with advanced (stages III and IV) endometrial cancer, MIR210HG expression was significantly elevated and possibly associated with lesion depth (Table S2). Endometrial cancer patients with high MIR210HG expression appear to have lower survival rates (Figure 1G).
Figure 1.
MIR210HG may be the core molecule that induces endometrial cancer
(A) The volcano plot obtained using data from TCGA database. The red points represent upregulated genes screened based on fold change >1.9 and a corrected p value of <0.05. (B) mRNA expression of RP6-65G23.3, MIR210HG, SNHG25, HAGLROS, BMPR1B-AS1, RP1-86C11.7, and LINC00261 in TCGA cohort of endometrial tissues, tumor n = 548, compared with normal endometrial tissue (n = 35), (the controls were collected from paracancerous tissues in patients with endometrial cancer). (C) The heatmap shows that lncRNA overexpression was the most in GSE17025 cohorts. (D) Expression of MIR210HG in 40 endometrial cancer tissues and 20 normal tissues determined using quantitative real-time PCR. (E) Relative expression of MIR210HG in endometrial cancer tissues analyzed according to the tumor stage and lymph node metastasis. (F) Expression of MIR210HG in 78 endometrial cancer tissues and 19 normal tissues analyzed using in situ hybridization assay. (G) Disease-free survival curves for endometrial carcinoma patients (n = 78) indicate that high expression of MIR210HG correlates with poor clinical outcomes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
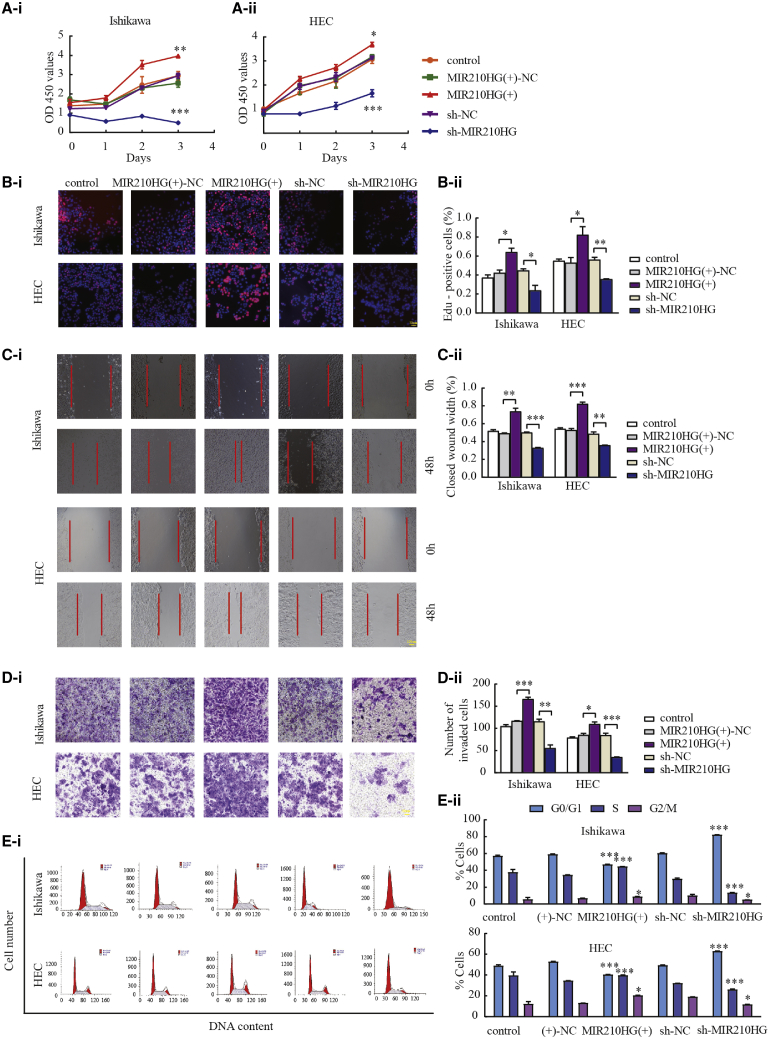
Knockdown of MIR210HG inhibited the proliferation, migration, and invasion of endometrial cancer cells
Using quantitative real-time PCR, we investigated the effects of the overexpression and knockdown of MIR210HG. We found that one construct, LV-MIR210HG-RNAi-1, decreased the MIR210HG expression (Figure S1A), while MIR210HG knockdown effectively reduced the proliferation of Ishikawa and HEC-1A cells (Figures 2A and 2B). Wound-healing and Transwell experiments were used to investigate the effects of MIR210HG on tumor migration and invasion. Compared with the sh-negative control (NC) group, cells transfected with sh-MIR210HG exhibited reduced migration and invasion (Figures 2C and 2D). Flow cytometry analysis was used to determine the cell cycle distribution (Figure 2E). The above results suggest that MIR210HG functions as an oncogene in endometrial cancer.
Figure 2.
MIR210HG plays an oncogenic role in Ishikawa and HEC-1A cell lines
(A and B) The effect of MIR210HG expression on proliferation determined using CCK-8 and EdU assays in Ishikawa and HEC-1A cells. (C) Wound-healing assays to evaluate the cell migration capacity. (D) Transwell assays to determine the number of invading cells. (E) Flow cytometry of Ishikawa and HEC-1A cells to determine the effect of MIR210HG on the cell cycle. Data are presented as the mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the NC group.
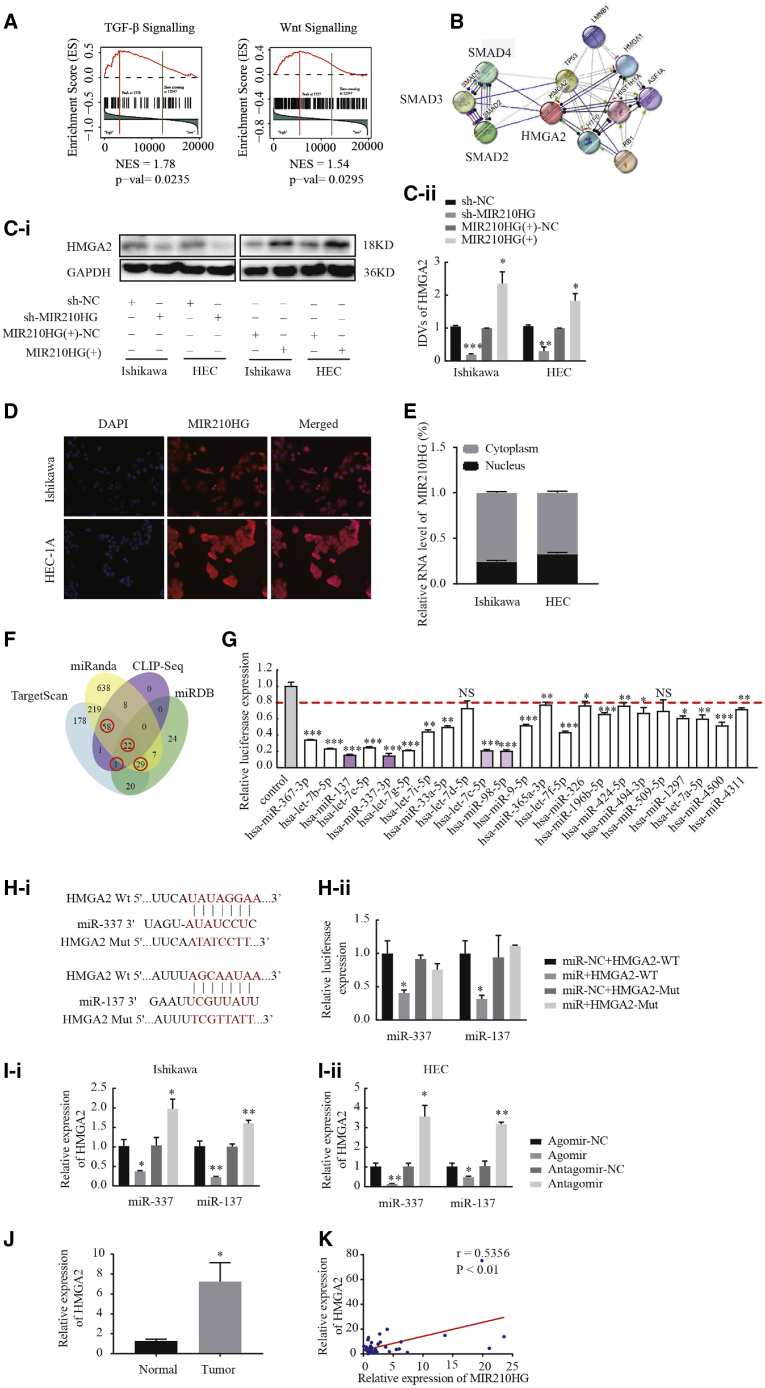
MIR210HG may be involved in endometrial malignant biological behavior mediated by an lncRNA-miRNA-mRNA regulatory network
Gene set enrichment analysis (GSEA) of endometrial cancer samples was performed using the TCGA dataset to explore the biological pathways by which MIR210HG is involved in regulating endometrial cancer. MIR210HG expression was significantly associated with the TGF-β and Wnt pathways, and the Smad3 gene strongly contributed to these associations (Figure 3A). Search Tool for Recurring Instances of Neighboring Genes (STRING) analysis also showed that SMAD3 and HMGA2 are strongly associated (Figure 3B). We explored whether HMGA2 is a downstream gene of MIR210HG and showed that the protein expression levels of HMGA2 were reduced after MIR210HG knockdown. Conversely, HMGA2 expression was significantly increased when MIR210HG was highly expressed (Figure 3C). This finding indicates that MIR210HG may upregulate HMGA2 expression to promote malignant biological behavior in endometrial cancer cells. Fluorescence in situ hybridization (FISH) and nuclear cytoplasmic RNA fraction experiment results showed that MIR210HG is located in both the nucleus and cytoplasm of Ishikawa and HEC-1A cells (Figures 3D and 3E). We hypothesized that MIR210HG acts as a sponge for miRNAs. To explore whether MIR210HG is involved in a novel MIR210HG-miRNA-HMGA2 regulatory network, TargetScan, miRanda, CLIP-Seq, and the miRDB bioinformatics database were used to predict miRNAs that bind HMGA2 (Figure 3F). miRNAs at the intersection of analyses with 3 software programs were selected as candidate miRNAs. In total, 110 miRNAs were identified and screened via the co-transfection of HEK293T cells with wild-type dual-luciferase vector-mediated HMGA2 constructs (Figures S2A–S2E). After transfection, the luciferase activities in cells transfected with 23 miRNAs were significantly decreased (Figure 3G). Among the above-mentioned 23 miRNAs, a significant decrease in luciferase activity was observed in response to transfection with miR-337-3p, miR-137, let-7c-5p, and miR-98-5p, and the luciferase activity was most decreased in response to transfection with miR-337-3p and miR-137. The bioinformatics database TargetScan was used to predict the sites in miR-337-3p and miR-137 that bind HMGA2. The results of the dual-luciferase gene reporter assay indicated that miR-337-3p and miR-137 bind HMGA2 at their predicted binding sites (Figure 3H). Quantitative real-time PCR showed that miR-337-3p and miR-137 negatively regulated the expression of HMGA2 (Figure 3I); the transfection efficiency is shown in Figure S1B. The correlation between the expression of MIR210HG and HMGA2 were analyzed using Pearson’s rank correlation method, and we found that HMGA2 expression correlated positively with MIR210HG (Figures 3J and 3K). These results indicate that MIR210HG may be involved in the new MIR210HG-miR-337-3p/137-HMGA2 regulatory network.
Figure 3.
lncRNA MIR210HG regulates HMGA2 expression via miR-337-3p and miR-137
(A and B) GSEA analyses showed that MIR210HG is primarily enriched in the TGF-β and Wnt pathways. STRING database-based analysis shows the functional association between the genes. (C) Western blotting assay was performed to analyze the expression of HMGA2. (D) FISH to determine the MIR210HG localization in Ishikawa and HEC-1A. (E) The mRNA level of MIR210HG in the nuclear and cytoplasmic fractions of Ishikawa and HEC-1A cells using quantitative real-time PCR. (F) Venn diagrams showing the bioinformatics prediction of miRNA interaction with seeded sequences from the 3′ UTR of HMGA2 using four different algorithms. (G) Among the 110 miRNAs co-transfected with the reporter vector HMGA2-WT in HEK293T cells, the relative luciferase activities of 23 miRNAs were most inhibited. (H) Predicted miR-337-3p/137 binding sites in the 3′ UTR of wild-type (WT) (MIR210HG-3′ UTR-WT) and mutant (MIR210HG-3′ UTR-Mut) MIR210HG sequences and the luciferase reporter assay using HEK293T cells co-transfected with MIR210HG-WT (or MIR210HG-Mut) and the indicated miRNA. (I) Changes in miR-337-3p/137 and HMGA2 expression were analyzed using quantitative real-time PCR in Ishikawa and HEC-1A cell lines. (J) Expression of HMGA2 in 40 endometrial cancer tissues and 20 normal tissues determined using quantitative real-time PCR. (K) Positive correlation between MIR210HG and HMGA2 expression in endometrial carcinoma patients (Pearson’s correlation coefficient analysis). Data are presented as the mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
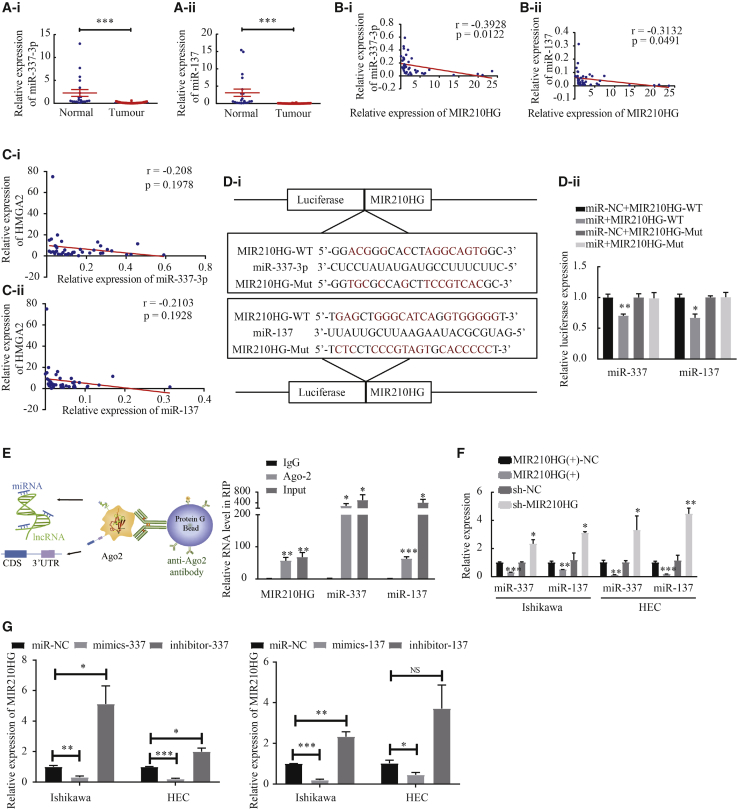
The expression of miR-337-3p/miR-137 is low in endometrial cancer tissues, and miR-337-3p/miR-137 are targets of MIR210HG
The results of quantitative real-time PCR showed that the expressions of miR-337-3p and miR-137 were significantly downregulated in tumor tissue (Figure 4A). In addition, the correlation between the expression of miR-337-3p and miR-137 and that of MIR210HG were analyzed using the Pearson’s rank correlation method (miR-337-3p: r = −0.3928, p = 0.0122; miR-137: r = −0.3132, p = 0.0491) (Figure 4B). We then analyzed the relationship between HMGA2 and miR-337-3p/137 expression (miR-337-3p: r = −0.208, p = 0.1978; miR-137: r = −0.2103, p = 0.1928) (Figure 4C). We also evaluated the association between the expression level of miR-337-3p/137 and clinicopathological parameters (Tables S3 and S4). miR-337-3p and miR-137 expression were associated with clinical stage and lymphatic metastasis in patients with endometrial cancer. These results suggest that miR-337-3p and miR-137 contribute to the development of endometrial cancer and that they are novel prognostic markers in endometrial cancer. A luciferase assay was performed to determine whether MIR210HG could directly target miR-337-3p and miR-137 at the predicted binding sites (Figure 4D). To determine whether MIR210HG binds with miRNA ribonucleoprotein complex, an RNA immunoprecipitation (RIP) experiment was conducted using anti-AGO2 antibody. lncRNA MIR210HG and miR-337-3p/miR-137 were significantly enriched in AGO2-containing immunoprecipitates compared with control immunoglobulin G (IgG) immunoprecipitates (Figure 4E). When MIR210HG was overexpressed in Ishikawa and HEC-1A cells, miR-337-3p and miR-137 expression levels were decreased. In contrast, when cells were transfected with sh-MIR210HG (the efficiency of which was determined using quantitative real-time PCR), the expression levels of miR-337-3p and miR-137 were significantly increased, indicating that MIR210HG can regulate miR-337-3p and miR-137 expressions (Figure 4F). Subsequently, we investigated whether miR-337-3p and miR-137 negatively regulated MIR210HG expression and found that overexpressed miR-337-3p/137 downregulated MIR210HG expression, while knocked-down miR-337-3p/137 upregulated MIR210HG expression (Figure 4G).
Figure 4.
miR-337-3p and miR-137 were targets of MIR210HG
(A) miR-337-3p and miR-137 were downregulated in endometrial carcinoma tissues (n = 40) compared with normal tissues (n = 20). (B) Negative correlation was found between MIR210HG and miR-337-3p and miR-137 expression in endometrial carcinoma patients (Pearson’s correlation coefficient analysis). (C) Pearson’s correlation coefficient analysis was used to analyze the relationship between HMGA2 and miR-337-3p/137 expression. (D) Sequence alignment of lncRNA MIR210HG 3-UTR with WT versus mutant potential miR-337-3p and miR-137 targeting sites. (E) Schematic diagram of the Ago2-RIP process. The association between MIR210HG, miR-337-3p/137, and Ago2 was ascertained by analyzing Ishikawa cell lysates. The level of lncRNA MIR210HG and miR-337-3p/137 analyzed using real-time PCR from Ago2 precipitate. ∗p < 0.05 versus IgG control. (F and G) MIR210HG or miR-337-3p/137 expression analyzed using quantitative real-time PCR in Ishikawa and HEC-1A cell lines. Data are presented as the mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
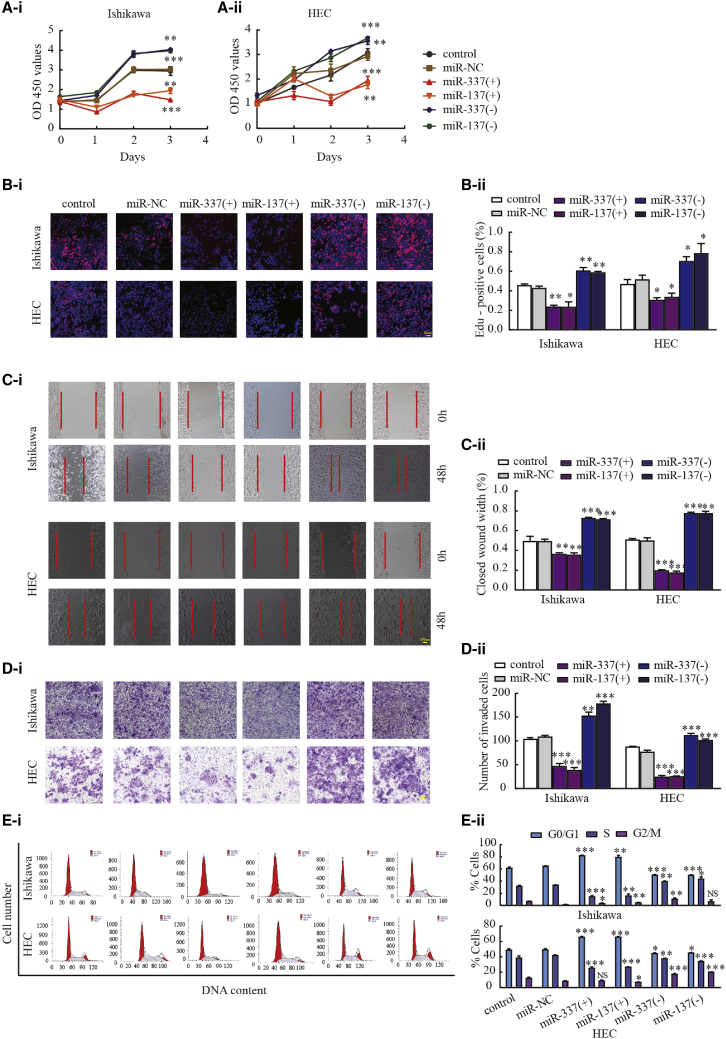
Overexpression of miR-337-3p and miR-137 inhibited the malignant biological behavior of endometrial cancer cells
To investigate the possible roles of miR-337-3p and miR-137 in the biological behavior of endometrial cancer, Ishikawa and HEC-1A cells were transfected with agomir-337-3p/137 and antagomir-337-3p/137, and their transfection was confirmed via quantitative real-time PCR (Figure S1B). The Cell Counting Kit-8 (CCK-8) and EdU assays were used to detect cell proliferation (Figures 5A and 5B). Wound-healing and Transwell assays were used to evaluate cell migration and invasion, respectively. Overexpression of miR-337-3p and miR-137 decreased the migration and invasion of Ishikawa and HEC-1A cells (Figures 5C and 5D). Flow cytometry was used to detect and analyze cell cycle distribution (Figure 5E). The above experimental results confirmed that miR-337-3p and miR-137 overexpression significantly inhibited proliferation, invasion, and migration compared with those in the miR-NC group.
Figure 5.
miR-337-3p and miR-137 inhibit malignant biological behavior of Ishikawa and HEC-1A cell lines
(A and B) CCK-8 and EdU assays were used to evaluate the proliferation effect of miR-337-3p/137 on Ishikawa and HEC-1A cell lines. (C and D) Representative images of the wound-healing assay and Transwell assays in Ishikawa and HEC-1A cell lines in response to changes in the expression of miR-337-3p/137. (E) Effects of miR-337-3p/137 on cell cycle distribution in Ishikawa and HEC-1A cell lines. Data are presented as the mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the miR-NC group.
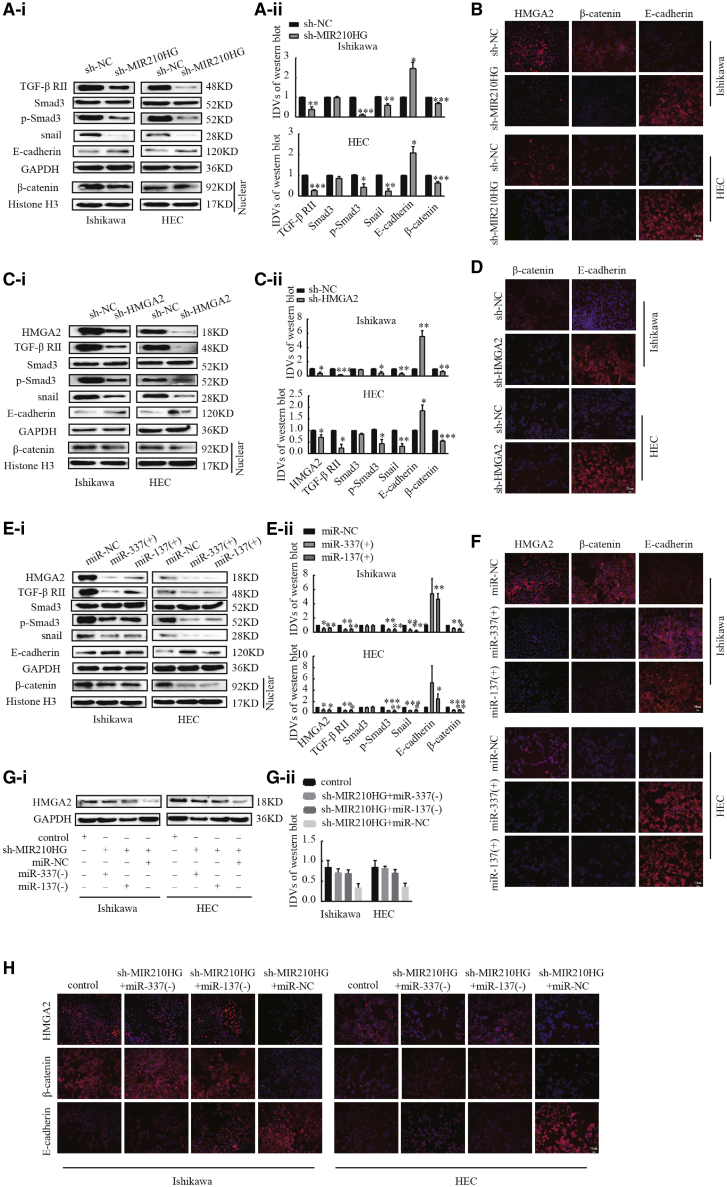
Silencing MIR210HG blocked TGF-β and Wnt signaling pathways through the miR-337-3p/137-HMGA2 axis
Previous GSEA indicated that MIR210HG is associated with the TGF-β/Smad3 and Wnt/β-catenin signaling pathways. To further investigate the mechanism underlaying MIR210HG regulation of endometrial cancer cell progression, we examined the regulatory effects of MIR210HG, HMGA2, miR-337-3p, and miR-137 on the protein levels associated with the TGF-β, Wnt, and EMT pathways. Knockdown of MIR210HG decreased the expression of TGF-βII, p-Smad3, and Snail and the distribution of β-catenin in the nucleus (Figures 6A and 6B). Based on these results, we suspected that MIR210HG may inhibit the EMT, TGF-β, and Wnt signaling pathways by targeting the miR-337-3p/137-HMGA2 regulatory axis. Interference with HMGA2 significantly reduced the expression of TGF-βII, p-Smad3, and Snail; the distribution of β-catenin in the nucleus (Figures 6C and 6D); and the expression of p-GSK-3β and c-myc reduced (Figure S3A). We have previously reported that HMGA2 regulates the expression of Snail, Slug, N-cadherin, MMP-2, and MMP-9 in Ishikawa and HEC-1A cell lines.26 We next investigated whether the induction of miR-337-3p/137 expression affects the levels of TGF-β, Wnt, and EMT pathway-associated proteins. The results showed that following miR-337-3p and miR-137 overexpression, the expression of HMGA2, TGF-βII, p-Smad3, and Snail was inhibited (Figures 6E and 6F). The distribution of β-catenin in the nucleus was reduced in agomir-337-3p/137-treated and sh-MIR210HG- or sh-HMGA2-treated Ishikawa and HEC-1A cells, and the expression of E-cadherin was increased. Then, we examined whether MIR210HG affects EMT progression by altering the miR-337-3p/137-HMGA2 axis. Western blotting and immunofluorescence showed that the knockdown of MIR210HG reduced expression of the HMGA2 protein in Ishikawa and HEC-1A cells, and upon the addition of miR-337-3p/137 inhibitor, the inhibitory effect on HMGA2, β-catenin, and E-cadherin expression was reversed (Figures 6G and 6H).
Figure 6.
HMGA2 is a functional target of the MIR210HG-miR-337-3p/137 axis in regulating EMT
(A) Western blot analysis of the expression of EMT, TGF-β, and Wnt-pathway-related proteins in response to MIR210HG in Ishikawa and HEC-1A cell lines. (B) HMGA2, β-catenin, and E-cadherin analyzed using immunofluorescence staining in Ishikawa and HEC-1A. (C) Western blot analysis of the expression of EMT, TGF-β, and Wnt-related pathway proteins in response to HMGA2 in Ishikawa and HEC-1A cell lines. (D) immunofluorescence experiment. (E) Western blot analysis of the expression of EMT, TGF-β, and Wnt- pathway-related proteins in response to miR-337-3p/137 in Ishikawa and HEC-1A cell lines. (F) immunofluorescence experimental results. (G) Protein level of HMGA2 determined using western blot analysis after co-transfection with sh-MIR210HG and miR-337-3p/137 inhibitor. (H) Immunofluorescence experiment. Data are presented as the mean ± SEM, (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
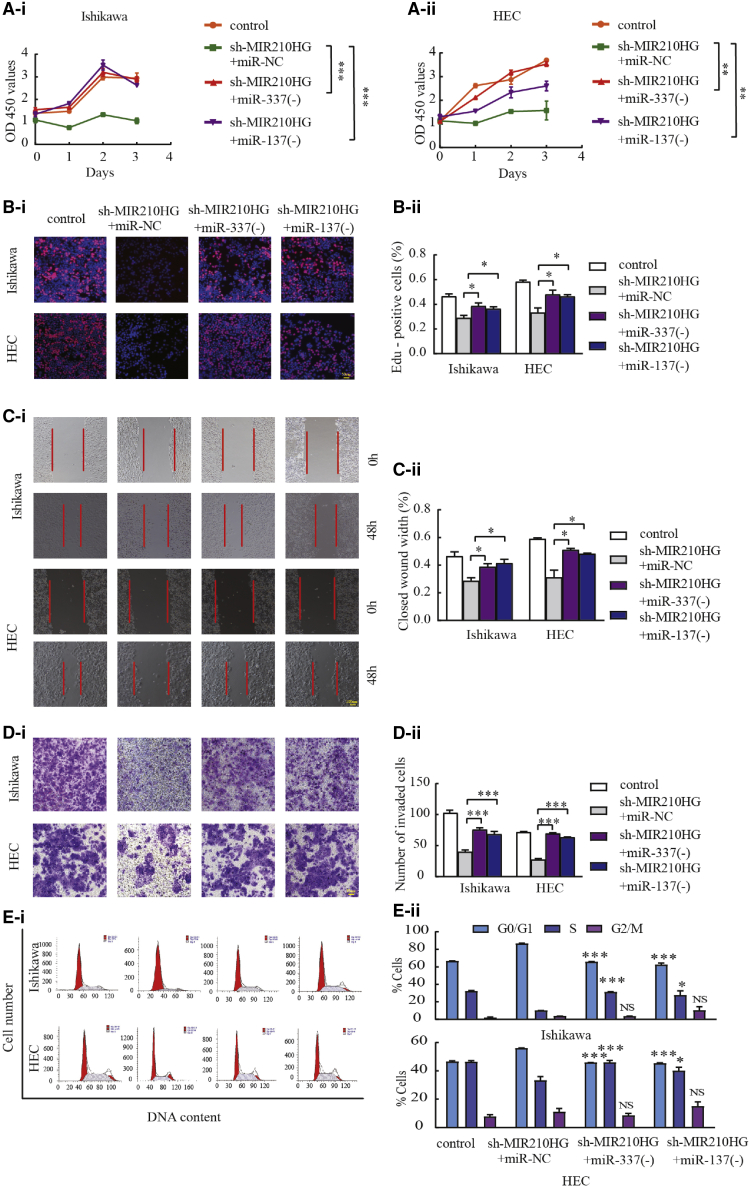
miR-337-3p/137-HMGA2 axis is responsible for the tumor-promoting effects of MIR210HG in endometrial cancer cells
To determine whether the inhibitory effects of MIR210HG knockdown on malignant behaviors in the Ishikawa and HEC-1A cell lines are mediated through the MIR210HG-miR-337-3p/137-HMGA2 axis, functional rescue experiments were conducted. The results confirmed that the malignant inhibitory effects of MIR210HG silencing on the migration and invasion of Ishikawa and HEC-1A cells were significantly reversed by miR-337-3p/137 knockdown (Figures 7A–7E). Restoration of HMGA2 expression partially rescued the suppressive effects of MIR210HG knockdown on Ishikawa and HEC-1A cell proliferation, migration, and invasion (Figures S4A–S4C). Moreover, rescue experiments showed that HMGA2 overexpression increased the expression of E-cadherin, and the distribution of β-catenin in the nucleus was inhibited by MIR210HG downregulation (Figure S4D). This demonstrates that MIR210HG regulates the malignant biological behavior of Ishikawa and HEC-1A cells via the MIR210HG-miR-337-3p/137-HMGA2 axis.
Figure 7.
MIR210HG promotes tumor progression through miR-337-3p and miR-137
(A and B) CCK-8 and EdU assays were used to evaluate proliferation in Ishikawa and HEC-1A cell lines. (C) Wound-healing assays to investigate the migratory ability of the cells. (D) Transwell assays to determine quantities of invading cells. Representative images and statistical plots are presented. (E) Representative flow diagrams and graphs showing cell cycle distribution. Data are presented as the mean ± SEM, (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus sh-MIR210HG + miR-NC.
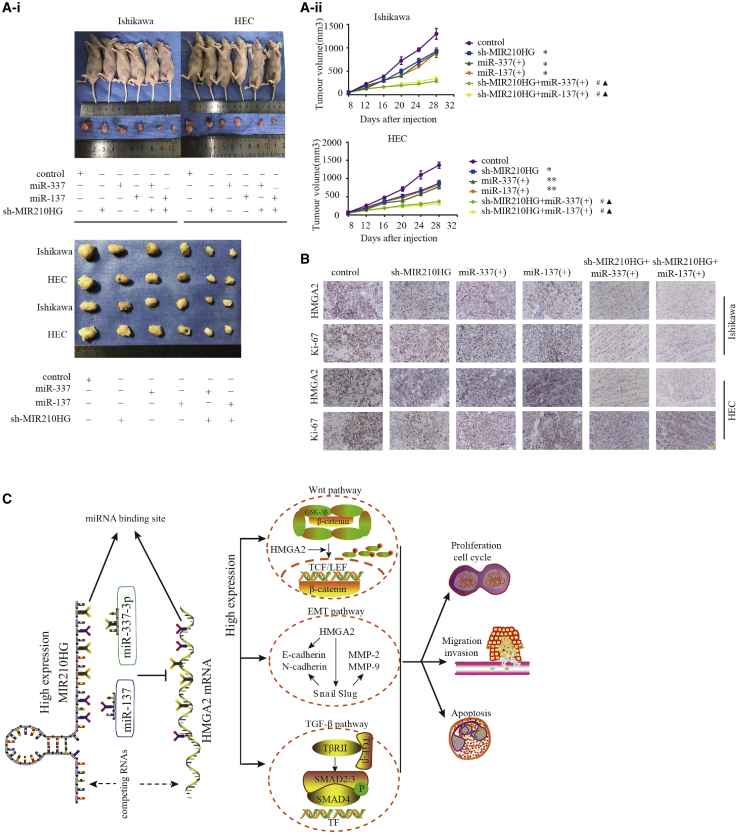
Knockdown of MIR210HG combined with miR-337-3p/137 overexpression suppressed tumor growth
We next determined the function of MIR210HG and miR-337-3p/137 in an in vivo tumor model. In the experiment of nude mice, there were 3 experimental mice in each group. The results showed that sh-MIR210HG transfection and miR-337-3p and miR-137 overexpression decreased tumor volume compared with the control group (Figure 8A). Additionally, in the group transfected with sh-MIR210HG and overexpressing both miR-337-3p and miR-137, the tumor volume was smallest. Immunohistochemical analysis showed that the expression of HMGA2 and Ki-67 was lower in the co-transfection group compared with that in the groups transfected with agomir-337-3p/137 or sh-MIR210HG alone (Figure 8B); a working model is shown in (Figure 8C).
Figure 8.
In vivo tumor xenografts
(A) Nude mice bearing tumors, and a specimen from each respective group, are shown. Tumor volume was measured every 4 days after injection. Twenty-eight days later, the mice were sacrificed, and the tumors were excised and weighed (n = 3 per group). (B) Expression levels of HMGA2 and Ki-67. Positive cells are stained brown. Data are presented as the mean ± SEM (n = 3 per group). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the control group, #p < 0.001 versus the agomir group, ▴p < 0.001 versus the sh-MIR210HG group. (C) A working model showing the interaction between MIR210HG and the miR-337-3p/137-HMGA2 axis in Ishikawa and HEC-1A cells.
Discussion
In recent years, the role of lncRNAs in various cancers has been demonstrated through analytical and functional assays.29 Cumulative key evidence has illustrated the role of lncRNAs in the development and progression of endometrial cancer30 and that lncRNAs are molecular tumor markers for the early diagnosis and prognosis of endometrial cancer31 and serve as potential therapeutic targets for the molecular treatment of endometrial cancer.
In this study, we focused on the mechanisms by which the lncRNA MIR210HG regulates endometrial cancer. Analysis of data from TCGA and GEO datasets showed a significant increase in MIR210HG expression in tumor compared to normal samples, and the expression level of MIR210HG in clinical endometrial cancer samples was also confirmed. Analysis of clinicopathological parameters suggested that MIR210HG expression was increased with clinical stage and that the high expression of MIR210HG in endometrial cancer was associated with tumor lymph node metastasis and tumor invasion. Patients expressing high levels of MIR210HG had lower survival rates. These results indicate that MIR210HG may be a prognostic biomarker for patients with endometrial cancer. Subsequent in vitro experiments showed that MIR210HG silencing inhibited the proliferation, invasion, and migration of endometrial cancer cells, while MIR210HG overexpression promoted the malignant phenotype of endometrial cancer cells. Studies have shown that the abnormal expression of MIR210HG in invasive breast cancer is associated with poor prognosis and decreased survival. MIR210HG was shown to regulate mucin-1c expression and the EMT pathway in invasive breast cancer by acting as a ceRNA for miR-1226-3p, thereby promoting tumor metastasis,15 and high MIR210HG expression was reported as an independent and unfavorable prognostic factor for the overall survival of patients with HCC. In vitro studies showed that silencing MIR210HG inhibited the proliferation, migration, and invasion of HCC cells.13 However, the role of MIR210HG in endometrial cancer cells remains largely unknown, and the mechanism of action of MIR210HG in endometrial cancer warrants further investigation. We used GSEA to analyze an endometrial cancer sample dataset from TCGA, which indicated that MIR210HG is enriched primarily in the TGF-β and Wnt pathways and that the SMAD gene family, especially the Smad3 gene, strongly contributes to this enrichment. STRING protein interaction analysis showed that Smad3 and HMGA2 interact. Our previous studies have shown that overexpression of HMGA2 promoted the malignant behaviors of endometrial cancer cells.26 In this study, we found that MIR210HG could positively regulate the expression of HMGA2. We hypothesized that MIR210HG regulates the malignant phenotype of endometrial cancer by affecting the HMGA2 expression. lncRNAs play a variety of roles in the regulation of tumors; herein we showed that the lncRNA MIR210HG is located mostly in the cytoplasm. Through bioinformatics analysis, we also found that miR-337-3p and miR-137 prominently mediate MIR210HG and HMGA2. Further investigations showed that MIR210HG targets miR-337-3p- and miR-137-specific sequences. miR-337-3p/137 was shown to target HMGA2 through specific binding sites, while miR-337-3p/137 overexpression reduced HMGA2 expression. The RIP assays showed that endogenous MIR210HG was preferentially enriched in Ago2-RIPs compared to control IgG-RIPs. These findings suggested that MIR210HG functions as a ceRNA to enhance HMGA2 expression by competitively sponging miR-337-3p/137.
miR-337-3p, which plays an essential role in the development of various tumors, could be used as an intermediate between circ_0132266 and probable transcription factor, affecting the progression of chronic lymphocytic leukemia.32 In cervical cancer, miR-337-3p expression was shown to be decreased, while the upregulation of miR-337-3p inhibited the proliferation, migration, and invasion of cervical cancer cells by downregulating Rap1A.33 Notably, low miR-337-3p expression was associated with poor liver cancer34 and gastric cancer35 prognoses. miR-337-3p has been suggested to predict tumor prognosis and that miR-337-3p is a candidate for treatment. miR-337-3p is expressed at a low level in endometrioid endometrial cancer,36 and the expression of miR-337-3p was more frequent in white endometrial cancer patients than in African American endometrial cancer patients.37 However, the phenotypic effects of miR-337-3p in endometrial cancer and its involved mechanisms remain unknown. The loss of miR-137 expression was found in both endometrioid and serosal endometrial cancer, and miR-137 methylation was abnormally high. Overexpression of miR-137 inhibited the malignant biological behavior of tumor cells.38 However, in endometrial cancer, the relationships between miR-137 and clinicopathological parameters as well as the mechanism by which miR-137 affects downstream target genes, especially the role of lncRNA in these effects, are poorly understood.
Our results showed that miR-337-3p and miR-137 were downregulated in endometrial cancer tissues, while the expression of miR-337-3p and miR-137 was correlated with clinical stage and lymph node metastasis. Pearson analysis showed a negative correlation between miR-337-3p/137 and MIR210HG expression. In vitro studies showed that the overexpression of miR-337-3p and miR-137 inhibited the proliferation, migration, and invasion of endometrial cancer cells. In vitro, restoration of HMGA2 or miR-337-3p/137 expression partially rescued the suppressive effects of MIR210HG knockdown on Ishikawa and HEC-1 cell proliferation, migration, and invasion. Taken together, the above results indicated that MIR210HG upregulates HMGA2 expression by competitively sponging miR-337-3p and miR-137. The MIR210HG-miR-337-3p/137-HMGA2 pathway forms a surveillance axis that plays a role in regulating the malignant biological behaviors of endometrial cancer.
TGF-β regulates cell proliferation and differentiation through a series of signaling cascades.39 In the classic SMAD pathway, TGF-β signaling activates Smad2 and Smad3 via TbRI and TbRII to form a Smad complex, which is transferred to the nucleus where it regulates the transcriptional induction or inhibition of the target gene.40 In liver cancer,41 lung adenocarcinoma,42 and cervical cancer,43 TGF-β is induced and regulated by Smad3 transcription to promote EMT progression. In the Wnt signaling pathway, when the Ser-9 site of GSK3 β is phosphorylated, GSK-3β activity was inhibited, and resulting in an increase in the amount of β-catenin entering the nucleus, where it interacts with the T cell factor (TCF/LEF) to activate the transcription of downstream target genes (such as c-Myc, Cyclin D1, and Wisp) to induce EMT.44 Wnt/β-catenin regulates cellular EMT processes in HCC,45 colon cancer,46 and endometrial cancer.47 We examined the relevant pathway proteins to study the mechanism by which MIR210HG affects endometrial cancer cell invasion and migration and showed that interfering with MIR210HG expression inhibited the TGF-β and Wnt pathways and EMT pathway, while overexpressing miR-337-3p/137 or interfering with HMGA2 expression inhibited the EMT pathway. The HMGA2 protein plays a central role in regulating EMT, since it can directly regulate the expression of EMT-related transcription factors, including Snail and Slug.48 In addition, HMGA2 can promote EMT by activating TGF-β/Smad3 signaling, thereby increasing the invasion and metastasis of cancer cells.49 Furthermore, HMGA2 can directly induce β-catenin expression, affecting the EMT process and thus the ability of cells to invade and migrate. The above evidence indicates that HMGA2 acts as a hub gene in the process by which EMT is regulated. The results of the recovery experiments indicate that MIR210HG behaves as an oncogene in endometrial cancer cells to promote proliferation and metastasis, partly by sponging miR-337-3p and miR-137, thereby regulating HMGA2 levels. The mechanism of ceRNAs at the gene level is very complicated. An lncRNA can simultaneously regulate a group of miRNAs, while a single miRNA can regulate multiple genes.50 In endometrial cancer, the lncRNA MIR210HG alone cannot regulate cell proliferation and invasion through the miR-337-3p/137-HMGA2 pathway. The oncogene HMGA family plays a central role in various carcinogenic pathways. Epigenetic mechanisms of non-coding RNA that participate in the regulation of HMGA family members should be further investigated. Research showed that HMGA1 pseudogenes may play a role in the regulation of expression of HMGA family members in endometrioid endometrial carcinoma.51 In this study, we found that HMGA1P1 positively regulates HMGA2 expression through western blot experiments (Figure S5). Since the regulatory network of endometrial cancer involves numerous molecules, to better understand endometrial cancer pathogenesis and tumor progression it may be necessary to identify more key regulatory molecules.
In summary, MIR210HG is an independent prognostic factor in patients with endometrial cancer, and interference with MIR210HG expression both in vivo and in vitro inhibits the malignant phenotype of endometrial cancer cells. MIR210HG-miR-337-3p/137 was found to mediate the ability of HMGA2 to regulate the malignant behavior of endometrial carcinoma cells. Thus, we suggest new molecular prognostic markers for endometrial cancer as well as potential therapeutic targets.
Materials and methods
Bioinformatics prediction
To predict miRNAs with potential binding sites for HMGA2, four types of bioinformatics software were used, namely TargetScan 6.0 (http://www.targetscan.org/), miRanda (http://www.microrna.org/microrna/home.do), miRDB (http://mirdb.org/miRDB/index.html), and clip-seq (http://lulab.life.tsinghua.edu.cn/clipdb/). To analyze the expression of lncRNA in endometrial cancer, TCGA data were searched and analyzed. To predict potential binding sites for lncRNA, miR-337-3p, and miR-137, we used the bioinformatics software RNA hybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) and starBase (http://starbase.sysu.edu.cn/). To analyze the interaction between HMGA2 and Smad3, the data were retrieved and analyzed using STRING database (https://string-db.org/).
Patients and samples
Tissue samples used in all experiments were obtained from patients undergoing surgery in the Department of Obstetrics and Gynecology at Shengjing Hospital affiliated to China Medical University. Fresh tissues included 40 endometrial cancer tissues and 20 normal endometrial tissues. There were 97 paraffin-embedded specimens, including 78 endometrial cancer tissues and 19 normal endometrial tissues. Patient specimens were collected between 2011 and 2017; the patients had not received radiotherapy or chemotherapy prior to surgery. According to the International Federation of Obstetrics and Gynecology (FIGO 2009) staging criteria, the diagnosis of endometrial cancer was confirmed by histopathological observation of postoperative paraffin sections. The control group comprised samples of normal endometrial proliferation and secretion. All experiments received ethical approval from the institution and obtained the patients’ informed consent.
Cell culture
Endometrial cancer is divided into two types from the perspective of pathogenesis; type I is estrogen-dependent, and type II is non-estrogen-dependent, with the cell lines Ishikawa from type I and HEC-1A from type II tumors.52 The endometrial cancer cell line Ishikawa was cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA), and HEC-1A was cultured in McCoy’s 5A medium (Gibco). High-glucose DMEM (Gibco) was used to culture human embryonic kidney cell line HEK293T. All cells were cultured in a humidified incubator at 37°C in a 5% CO2 atmosphere. A 10% fetal bovine serum (FBS) (Gibco) was added to the respective medium of the cells for cell culture. The cells were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China).
Transfection of cells
MIR210HG interference lentivirus vector and its corresponding NC were purchased from GenePharma (Shanghai, China). DNA fragments of lncRNA MIR210HG short hairpin RNA (shRNA) were subcloned into a lentiviral vector with endonucleases Agel and EcoRI to form a sh-MIR210HG construct; the sh-MIR210HG or NC lentivirus was packaged and harvested from 293 T cells and then infected Ishikawa and HEC-1A cell lines. The cells were transfected at an MOI of 50; puromycin was used to select the stably transfected cells. MIR210HG overexpression plasmid (pCDNA3.1-MIR210HG), its corresponding NC (pCDNA3.1-NC), agomir, antagomir, and NC of the control group were purchased from GenePharma (Shanghai, China). Cells were transfected with Lipofectamine 3000 (Invitrogen) at the transfection dose according to the manufacturer’s instructions. Sequences of primers used for cloning, shRNA, agomir, and antagomir are listed in Table S5.
RNA extraction and quantitative real-time PCR
RNA was extracted from tissues and cells using Trizol (Takara, Dalian, China) and reverse transcribed into cDNA dose according to the manufacturer’s instructions. The PCR-specific primer for MIR210HG was designed by Sangon Biotech (Shanghai, China), and the RNA was converted to cDNA using the PrimeScript RT-Polymerase (Takara) reverse transcription kit to determine MIR210HG mRNA expression. The RNA was converted into cDNA using the Mir-X miRNA First-Strand Synthesis Kit (Clontech, Dalian, China) to determine miR-337-3p and miR-137 expression. PCR-specific primers for miR-337-3p and miR-137 were designed by Sangon Biotech (Shanghai, China) and used according to the manufacturer’s instructions. The fold change was calculated using the 2−ΔΔCt method using GAPDH and U6 as internal controls. The primer sequences are listed in Table S6.
Western blotting assay
Total protein was extracted using a total protein extraction kit (Beyotime Biotechnology, Shanghai, China), and nuclear protein was extracted using a nuclear protein extraction kit (Beyotime Biotechnology, Shanghai, China), processed according to the manufacturer’s instructions. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) using a 10% gel was used for electrolytic separation of proteins, which were then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Burlington, MA, USA). After the PVDF membrane was blocked with skimmed milk, the membrane was next incubated with the primary antibody at the concentration recommended by the manufacturer. It was incubated with the primary antibody for 16 h at 4°C (the primary antibodies used are listed in Table S7), following which secondary antibody was added and incubated for 1.5 h at 25°C. After repeated washing, the film was developed using the Quantum One imaging software (Bio-Rad, Hercules, CA, USA). The intensity of the protein bands was quantified using ImageJ software v.1.48 (https://imagej.nih.gov/ij/download.html).
In situ hybridization
The MIR210HG in situ hybridization probe was customized by China Nanjing Bosite Biotechnology and used to determine the expression of MIR210HG in paraffin-embedded tissues of endometrial cancer. The probe sequences are shown in Table S6. The experiment was carried out according to the instructions of the in situ hybridization kit manufacturer. After the sections were subjected to conventional dehydration, the exposed mRNA fragments were inactivated using the endogenous enzyme. After pre-hybridization-hybridization, following the manufacturer’s instructions, they were observed under a microscope (original magnification: ×20).
FISH
To identify the mRNA localization of MIR210HG in endometrial cancer cells Ishikawa and HEC-1A, FISH probe (China Nanjing Bosite Biotechnology) was constructed. The probe sequences are shown in Table S6. Following these steps, the cells were treated with 1% paraformaldehyde. After pre-hybridizing at 37°C for 3 h, hybridization probes were added to the cells and then placed in the incubator for hybridization at 37°C for 16 h. The next day, after the anti-digoxin rhodamine conjugate SABC, images were taken using a fluorescence inversion microscope (20× magnification) and analyzed.
RNA isolation of nuclear and cytoplasmic fractions
The nuclear/cytoplasmic RNA from cells extracted using the Isolation Kit (Beyotime Biotechnology, Shanghai, China) was applied to isolate and collect cytosolic and nuclear fractions. Then, all lysates were treated with Trizol to extract RNA.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 15 min; 0.5% Triton X-100 was permeated for 20 min, washed with PBS, and then added with serum for 1.5 h. At 4°C, the primary antibody was blocked for 16 h (the primary antibody is listed in Table S7). Fluorescent dye conjugated secondary antibody was added the next day, and cells were stained with DAPI for 5 min. Images were taken using a fluorescence inversion microscope (20× magnification) and analyzed.
Luciferase assay
HEK293T cells with a good fold-growth index were evenly spread in 96-well plates, and the 110 selected target miRNAs (RiboBio, Guangzhou, China) and 100 ng of wild-type cells containing HMGA2 gene fragment were co-transfected into cells. miR-NC was used for the control group. The mimic-337-3p and HMGA2 wild-type and mutant dual-luciferase reporter vector were co-transfected. The above-mentioned products were all customized by RiboBio, Guangzhou, China. The mimics-337-3p and MIR210HG wild-type and mutant dual-luciferase reporter vectors were co-transfected; miR-NC was used as a control group. The wild-type and mutant dual-luciferase reporter vector and miRNAs were purchased from GenePharma (Shanghai, China). The verification of miR-137 binding with MIR210HG and HMGA2 is as above. After 48 h of co-transfection, the luciferase assay kit (Promega, Madison, WI, USA) was used to determine activity.
RIP assay
RIP assay was performed according to the instructions of the EZ-Magna RIP kit (Millipore). Briefly, cells were lysed in complete RIP lysis buffer. Then, A/G magnetic beads were used and incubated with the anti-Ago2 antibody (Abcam) in RIP buffer. The magnetic bead binding complex was fixed with a magnetic stand, following which unbound reagents were washed off. Proteinase K buffer was prepared, and finally RNA was extracted. Subsequently, the purified RNA was subjected to quantitative real-time PCR analysis.
Cell proliferation assay
Ishikawa and HEC-1A cells were seeded in a 96-well plate and incubated for 24 h, 48 h, and 72 h in an incubator cultured in a humidified incubator at 37°C in a 5% CO2 atmosphere. To measure cell proliferation, 100 μL of a mixed reagent containing 10 μL of CCK-8 (Dojindo, Japan) was added to the wells. The cells were then incubated for 3 h before measuring the optical density (OD) of each well at 450 nm using a microplate reader.
Cell proliferation was also checked using the (RiboBio) EdU Cell Proliferation Assay Kit. According to the manufacturer’s instructions, a 50 μM EdU mixed reagent was prepared, and the cells were incubated with the reagent for 2 h. After the cells were immobilized, DNA staining was performed, following which the cell stain was placed and the cells were washed with PBS. Image acquisition and analysis were then performed using a (Nikon, Japan) fluorescence microscope at an original magnification of 20×.
Wound-healing assay
The cells in the 6-well plate were grown to a density of 90%. Using a smooth and flat 100-μL-pipette tip, a straight wound was evenly made in the cell plate. The culture solution was then aspirated and replaced with fresh culture medium. Images were obtained daily at a fixed time using a microscope (10× magnification) (Nikon, Japan). The pictures taken at 0 h and 48 h were analyzed and have been displayed.
Cell invasion assay
Cell invasion ability was determined using the Transwell assay. After pre-coating Matrigel in the upper chamber (pore size 8 μM; Corning, Corning, NY, USA), the cells were washed with serum-free medium, suspended in 200 μL of serum-free medium, and placed in the upper chamber. Then, 700 μL of 10% FBS medium was added in the lower chamber for induction. After several hours, some suspended cells were observed in the lower chamber. Further, the chamber pores were washed, and non-invasive cells on the upper surface of the membrane were wiped with a cotton swab. After fixation and staining, images were obtained using a fluorescence inversion microscope (20× magnification) and analyzed.
Cell cycle analyses
After 48 h of cell transfection, 1 × 107 cells were harvested from each group. After washing, the cells were fixed in 70% ethanol and placed at 4°C for 16 h, according to the manufacturer’s instructions (KeyGen Biotech, Nanjing, China). Further, cells were treated with RNase and DNA staining was performed using propidium. The cell cycle analysis was performed using flow cytometry (BD FACSCalibur, Franklin Lakes, NJ, USA). All experiments were performed in triplicate.
Tumor xenografts in nude mice
The experiment was approved by the Ethics Committee of Shengjing Hospital affiliated to China Medical University, and the experiments on nude mice were conducted strictly in accordance with ethical standards. Athymic BALB/c nude mice (4–6 weeks old) were purchased from HFK Bioscience (Beijing, China). Then, the transfected cells were suspended in 106/100 μL PBS and injected into the nude mice. According to the protocol, cells were injected into the underarm tumor of each mouse every 4 days. Tumor volume was measured every 4 days until 28 days, and the mice were then sacrificed. After the tumor was identified, photos were obtained, and the tumor cells were quickly placed in 4% paraformaldehyde and embedded in paraffin. Paraffin specimens were then sectioned for immunohistochemical analysis.
Immunohistochemistry
Immunohistochemistry was used to determine the expression of proteins in paraffin specimens. Routine dewaxing, sodium citrate buffer for antigen retrieval (operation was according to the kit’s instructions) DAB (ZSGB-BIO, China) color development, hematoxylin counterstaining, tap water washing blue, gradient. The alcohol was dehydrated and dried, the xylene was transparent, and the neutral gum was fixed. The results of immunohistochemistry were determined using a double-blind scoring method and analyzed by two senior pathologists to ensure the credibility of the experiment. Staining was observed using an optical microscope and scored as the percentage of positive cells as follows: no staining, 0; light yellow, 1; yellow, 2; and brown and sepia, 3. According to the percentage of positive cells, the mean value after scoring with negative count as 0, the scores were assigned as follows: percentage of positive cells less than 10%, 1; ≥10% to 50%, 2; >50% to 75%, 3; and ≥75%, 4. The product of two scores was considered as the total score, and the results were interpreted as follows: ≤2, negative; 3 to 4, weak positive (+); 5 to 8, medium positive (++); and 9 to 12, strong positive (+++). Low and high expression were indicated by −/+ and ++/+++, respectively. Original magnification: ×40.
Statistical analysis
The data were presented as the mean ± SEM. Student’s t test and the chi-square test were used for comparisons between two independent groups, and one-way ANOVA was used to compare differences among more than two groups. Data analysis was performed using GraphPad Prism 7.0 (La Jolla, CA, USA) and SPSS 21.0 (Abbott Laboratories, Chicago, IL, USA). Pearson’s correlation coefficient analysis was used to determine correlations. Survival curves were plotted using the results of Kaplan-Meier analysis, and disease-free survival was defined as the time from the date of diagnosis to the time of progression/death or the last follow-up. p <0.05 was defined as statistically significant.26
Ethics approval and consent to participate
The study protocol was reviewed and approved by the Scientific Research and New Technology Ethical Committee of the Shengjing Hospital of China Medical University (ethical number: human: 2018PS251K; animal: 2018PS136K).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We sincerely thank Yixue Xue for critical comments regarding experiments described in this article. This work was supported by the National Natural Science Foundation of China (grant numbers 81872123 and 81472438 to X.X.M.).
Author contributions
J.M. performed most of the experiments and contributed to the writing of the manuscript. J.M. and X.X.M. conceived of the study, participated in its design and coordination, and helped draft the manuscript. J.M., F.F.K., D.Y., H.Y., C.W., and R.C. performed the cell culture experiments. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.04.011.
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Monsivais D., Peng J., Kang Y., Matzuk M.M. Activin-like kinase 5 (ALK5) inactivation in the mouse uterus results in metastatic endometrial carcinoma. Proc. Natl. Acad. Sci. USA. 2019;116:3883–3892. doi: 10.1073/pnas.1806838116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neri M., Peiretti M., Melis G.B., Piras B., Vallerino V., Paoletti A.M., Madeddu C., Scartozzi M., Mais V. Systemic therapy for the treatment of endometrial cancer. Expert Opin. Pharmacother. 2019;20:2019–2032. doi: 10.1080/14656566.2019.1654996. [DOI] [PubMed] [Google Scholar]
- 4.Fader A.N., Roque D.M., Siegel E., Buza N., Hui P., Abdelghany O., Chambers S.K., Secord A.A., Havrilesky L., O’Malley D.M. Randomized Phase II Trial of Carboplatin-Paclitaxel Versus Carboplatin-Paclitaxel-Trastuzumab in Uterine Serous Carcinomas That Overexpress Human Epidermal Growth Factor Receptor 2/neu. J. Clin. Oncol. 2018;36:2044–2051. doi: 10.1200/JCO.2017.76.5966. [DOI] [PubMed] [Google Scholar]
- 5.Mileshkin L., Edmondson R., O’Connell R.L., Sjoquist K.M., Andrews J., Jyothirmayi R., Beale P., Bonaventura T., Goh J., Hall M., PARAGON study group Phase 2 study of anastrozole in recurrent estrogen (ER)/progesterone (PR) positive endometrial cancer: The PARAGON trial - ANZGOG 0903. Gynecol. Oncol. 2019;154:29–37. doi: 10.1016/j.ygyno.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Ding N., Zhang H., Su S., Ding Y., Yu X., Tang Y., Wang Q., Liu P. Emodin Enhances the Chemosensitivity of Endometrial Cancer by Inhibiting ROS-Mediated Cisplatin-resistance. Anticancer. Agents Med. Chem. 2018;18:1054–1063. doi: 10.2174/1871520618666171219113036. [DOI] [PubMed] [Google Scholar]
- 7.Botti G., Scognamiglio G., Aquino G., Liguori G., Cantile M. LncRNA HOTAIR in Tumor Microenvironment: What Role? Int. J. Mol. Sci. 2019;20:9. doi: 10.3390/ijms20092279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begolli R., Sideris N., Giakountis A. LncRNAs as Chromatin Regulators in Cancer: From Molecular Function to Clinical Potential. Cancers (Basel) 2019;11:10. doi: 10.3390/cancers11101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Wu Z., Fu X., Han W. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. Mutat. Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Tang H., Wu Z., Zhang Y., Xia T., Liu D., Cai J., Ye Q. Identification and Function Analysis of a Five-Long Noncoding RNA Prognostic Signature for Endometrial Cancer Patients. DNA Cell Biol. 2019;38:1480–1498. doi: 10.1089/dna.2019.4944. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang D., Li R., Li Y., Zhu X. A 7-lncRNA signature predict prognosis of Uterine corpus endometrial carcinoma. J. Cell. Biochem. 2019;120:18465–18477. doi: 10.1002/jcb.29164. [DOI] [PubMed] [Google Scholar]
- 12.Voellenkle C., Garcia-Manteiga J.M., Pedrotti S., Perfetti A., De Toma I., Da Silva D., Maimone B., Greco S., Fasanaro P., Creo P. Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci. Rep. 2016;6:24141. doi: 10.1038/srep24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Li W., Chen X., Li Y., Wen P., Xu F. MIR210HG predicts poor prognosis and functions as an oncogenic lncRNA in hepatocellular carcinoma. Biomed. Pharmacother. 2019;111:1297–1301. doi: 10.1016/j.biopha.2018.12.134. [DOI] [PubMed] [Google Scholar]
- 14.Ruan Z., Xu Z., Li Z., Lv Y. Integral analyses of survival-related long non-coding RNA MIR210HG and its prognostic role in colon cancer. Oncol. Lett. 2019;18:1107–1116. doi: 10.3892/ol.2019.10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X.Y., Zhou L.Y., Luo H., Zhu Q., Zuo L., Liu G.Y., Feng C., Zhao J.Y., Zhang Y.Y., Li X. The long noncoding RNA MIR210HG promotes tumor metastasis by acting as a ceRNA of miR-1226-3p to regulate mucin-1c expression in invasive breast cancer. Aging (Albany NY) 2019;11:5646–5665. doi: 10.18632/aging.102149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., Wu Q.M., Wang X.Q., Zhang C.Q. Long Noncoding RNA miR210HG Sponges miR-503 to Facilitate Osteosarcoma Cell Invasion and Metastasis. DNA Cell Biol. 2017;36:1117–1125. doi: 10.1089/dna.2017.3888. [DOI] [PubMed] [Google Scholar]
- 17.Xia W., Hu C. Progress in Research on Tumor Metastasis Inhibitors. Curr. Med. Chem. 2020;27:5758–5772. doi: 10.2174/0929867326666190927120847. [DOI] [PubMed] [Google Scholar]
- 18.Scheel C., Eaton E.N., Li S.H., Chaffer C.L., Reinhardt F., Kah K.J., Bell G., Guo W., Rubin J., Richardson A.L., Weinberg R.A. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Guo L., Li Y., Feng G.H., Teng F., Li W., Zhou Q. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol. Cancer. 2018;17:1. doi: 10.1186/s12943-017-0753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez D.M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Lu W., Yin T., Lu L. Calycosin suppresses TGF-β-induced epithelial-to-mesenchymal transition and migration by upregulating BATF2 to target PAI-1 via the Wnt and PI3K/Akt signaling pathways in colorectal cancer cells. J. Exp. Clin. Cancer Res. 2019;38:240. doi: 10.1186/s13046-019-1243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinway S.N., Zañudo J.G., Ding W., Rountree C.B., Feith D.J., Loughran T.P., Jr., Albert R. Network modeling of TGFβ signaling in hepatocellular carcinoma epithelial-to-mesenchymal transition reveals joint sonic hedgehog and Wnt pathway activation. Cancer Res. 2014;74:5963–5977. doi: 10.1158/0008-5472.CAN-14-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Sun Y., Wu Y., Tang D., Ding X., Xu W., Su B., Gao W. Downregulation of miR-3127-5p promotes epithelial-mesenchymal transition via FZD4 regulation of Wnt/β-catenin signaling in non-small-cell lung cancer. Mol. Carcinog. 2018;57:842–853. doi: 10.1002/mc.22805. [DOI] [PubMed] [Google Scholar]
- 25.Su L., Deng Z., Leng F. The Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2): Biochemical and Biophysical Properties, and Its Association with Adipogenesis. Int. J. Mol. Sci. 2020;21:10. doi: 10.3390/ijms21103710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Li D., Kong F.F., Yang D., Yang H., Ma X.X. miR-302a-5p/367-3p-HMGA2 axis regulates malignant processes during endometrial cancer development. J. Exp. Clin. Cancer Res. 2018;37:19. doi: 10.1186/s13046-018-0686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan E.J., Kahata K., Idås O., Thuault S., Heldin C.H., Moustakas A. The high mobility group A2 protein epigenetically silences the Cdh1 gene during epithelial-to-mesenchymal transition. Nucleic Acids Res. 2015;43:162–178. doi: 10.1093/nar/gku1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Z., Li X., Wu D., Tang R., Chen R., Xue S., Sun X. Silencing of HMGA2 suppresses cellular proliferation, migration, invasion, and epithelial-mesenchymal transition in bladder cancer. Tumour Biol. 2016;37:7515–7523. doi: 10.1007/s13277-015-4625-2. [DOI] [PubMed] [Google Scholar]
- 29.Xie Y., Dang W., Zhang S., Yue W., Yang L., Zhai X., Yan Q., Lu J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer. 2019;18:37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B.J., Byrne F.L., Takenaka K., Modesitt S.C., Olzomer E.M., Mills J.D., Farrell R., Hoehn K.L., Janitz M. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecol. Oncol. 2017;147:654–662. doi: 10.1016/j.ygyno.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Liu J., Nie S., Liang J., Jiang Y., Wan Y., Zhou S., Cheng W. Competing endogenous RNA network of endometrial carcinoma: A comprehensive analysis. J. Cell. Biochem. 2019;120:15648–15660. doi: 10.1002/jcb.28831. [DOI] [PubMed] [Google Scholar]
- 32.Wu W., Wu Z., Xia Y., Qin S., Li Y., Wu J., Liang J., Wang L., Zhu H., Fan L. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging (Albany NY) 2019;11:3561–3573. doi: 10.18632/aging.101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X.M. Role of miR-337-3p and its target Rap1A in modulating proliferation, invasion, migration and apoptosis of cervical cancer cells. Cancer Biomark. 2019;24:257–267. doi: 10.3233/CBM-181225. [DOI] [PubMed] [Google Scholar]
- 34.Zuo X.L., Chen Z.Q., Wang J.F., Wang J.G., Liang L.H., Cai J. miR-337-3p suppresses the proliferation and invasion of hepatocellular carcinoma cells through targeting JAK2. Am. J. Cancer Res. 2018;8:662–674. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z., Wang J., Yang Y., Hao B., Wang R., Li Y., Wu Q. Loss of has-miR-337-3p expression is associated with lymph node metastasis of human gastric cancer. J. Exp. Clin. Cancer Res. 2013;32:76. doi: 10.1186/1756-9966-32-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong H., Li Q., Liu S., Wang F., Xiong Z., Chen J., Chen H., Yang Y., Tan X., Luo Q. Integrated microRNA and mRNA transcriptome sequencing reveals the potential roles of miRNAs in stage I endometrioid endometrial carcinoma. PLoS ONE. 2014;9:e110163. doi: 10.1371/journal.pone.0110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maxwell G.L., Shoji Y., Darcy K., Litzi T., Berchuck A., Hamilton C.A., Conrads T.P., Risinger J.I. MicroRNAs in endometrial cancers from black and white patients. Am. J. Obstet. Gynecol. 2015;212:191.e1–191.e10. doi: 10.1016/j.ajog.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W., Chen J.H., Shan T., Aguilera-Barrantes I., Wang L.S., Huang T.H., Rader J.S., Sheng X., Huang Y.W. miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation. Lab. Invest. 2018;98:1397–1407. doi: 10.1038/s41374-018-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu S., Huang W., Huang C., Luo Z., Yan X. Contextual Regulation of TGF-β Signaling in Liver Cancer. Cells. 2019;8:1235. doi: 10.3390/cells8101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung B., Staudacher J.J., Beauchamp D. Transforming Growth Factor β Superfamily Signaling in Development of Colorectal Cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Yang B., Zhou Q., Wu Y., Shang D., Guo Y., Song Z., Zheng Q., Xiong J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- 42.Jiang L., Wang R., Fang L., Ge X., Chen L., Zhou M., Zhou Y., Xiong W., Hu Y., Tang X. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9:2460–2474. doi: 10.7150/thno.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Q., Qiu M.T., Zhu Z., Zhou J.H., Chen L., Zhou Y., Gu W., Wang L.H., Li Z.N., Xu Y. Twist induces epithelial-mesenchymal transition in cervical carcinogenesis by regulating the TGF-β/Smad3 signaling pathway. Oncol. Rep. 2015;34:1787–1794. doi: 10.3892/or.2015.4143. [DOI] [PubMed] [Google Scholar]
- 44.Ghahhari N.M., Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur. J. Cancer. 2015;51:1638–1649. doi: 10.1016/j.ejca.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Qu C., He D., Lu X., Dong L., Zhu Y., Zhao Q., Jiang X., Chang P., Jiang X., Wang L. Salt-inducible Kinase (SIK1) regulates HCC progression and WNT/β-catenin activation. J. Hepatol. 2016;64:1076–1089. doi: 10.1016/j.jhep.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Qi J., Yu Y., Akilli Öztürk Ö., Holland J.D., Besser D., Fritzmann J., Wulf-Goldenberg A., Eckert K., Fichtner I., Birchmeier W. New Wnt/β-catenin target genes promote experimental metastasis and migration of colorectal cancer cells through different signals. Gut. 2016;65:1690–1701. doi: 10.1136/gutjnl-2014-307900. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Sun D., Gao J., Shi Z., Chi P., Meng Y., Zou C., Wang Y. MicroRNA-373 promotes the development of endometrial cancer by targeting LATS2 and activating the Wnt/β-Catenin pathway. J. Cell. Biochem. 2018;39 doi: 10.1002/jcb.28149. BSR20190680. [DOI] [PubMed] [Google Scholar]
- 48.Han W., Zhang Y., Niu C., Guo J., Li J., Wei X., Jia M., Zhi X., Yao L., Meng D. BTB and CNC homology 1 (Bach1) promotes human ovarian cancer cell metastasis by HMGA2-mediated epithelial-mesenchymal transition. Cancer Lett. 2019;445:45–56. doi: 10.1016/j.canlet.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Morishita A., Zaidi M.R., Mitoro A., Sankarasharma D., Szabolcs M., Okada Y., D’Armiento J., Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K., Jin W., Song Y., Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol. Cancer. 2017;16:166. doi: 10.1186/s12943-017-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palumbo J.A., de Sousa V.P.L., Esposito F., De Martino M., Forzati F., Moreira F.C.B., Simão T.A., Nasciutti L.E., Fusco A., Ribeiro P.L.F. Overexpression of HMGA1 Figures as a Potential Prognostic Factor in Endometrioid Endometrial Carcinoma (EEC) Genes (Basel) 2019;10:372. doi: 10.3390/genes10050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albitar L., Pickett G., Morgan M., Davies S., Leslie K.K. Models representing type I and type II human endometrial cancers: Ishikawa H and Hec50co cells. Gynecol. Oncol. 2007;106:52–64. doi: 10.1016/j.ygyno.2007.02.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.