Graphical abstract
Keywords: COVID-19, CT images, Deep learning, Multiple instance learning, Unsupervised complementary loss
Abstract
Chest computed tomography (CT) based analysis and diagnosis of the Coronavirus Disease 2019 (COVID-19) plays a key role in combating the outbreak of the pandemic that has rapidly spread worldwide. To date, the disease has infected more than 18 million people with over 690k deaths reported. Reverse transcription polymerase chain reaction (RT-PCR) is the current gold standard for clinical diagnosis but may produce false positives; thus, chest CT based diagnosis is considered more viable. However, accurate screening is challenging due to the difficulty in annotation of infected areas, curation of large datasets, and the slight discrepancies between COVID-19 and other viral pneumonia. In this study, we propose an attention-based end-to-end weakly supervised framework for the rapid diagnosis of COVID-19 and bacterial pneumonia based on multiple instance learning (MIL). We further incorporate unsupervised contrastive learning for improved accuracy with attention applied both in spatial and latent contexts, herein we propose Dual Attention Contrastive based MIL (DA-CMIL). DA-CMIL takes as input several patient CT slices (considered as bag of instances) and outputs a single label. Attention based pooling is applied to implicitly select key slices in the latent space, whereas spatial attention learns slice spatial context for interpretable diagnosis. A contrastive loss is applied at the instance level to encode similarity of features from the same patient against representative pooled patient features. Empirical results show that our algorithm achieves an overall accuracy of 98.6% and an AUC of 98.4%. Moreover, ablation studies show the benefit of contrastive learning with MIL.
1. Introduction
The coronavirus disease 2019 (COVID-19), first recognized in Wuhan, China has spread to a global scale infecting millions and causing death to hundreds of thousands. As of August, 2020 infections surpassed 18 million, with reported deaths reaching over 690,000 globally. Caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), COVID-19 is highly contagious with increasing infections each day. Despite having a relatively lower fatality rate Mahase (2020) than SARS and Middle East Respiratory Syndrome (MERS), COVID-19 has already caused more deaths. Consequently, there is an urgent need for rapid diagnosis to improve prevention while an effective vaccine is being developed.
Reverse transcription polymerase chain reaction (RT-PCR) is the current gold standard for COVID-19 diagnosis based on viral nucleic acid (VNA) (Zu et al., 2020). However, low sensitivity, high number of false positives and lengthy test to diagnosis times pose a challenge for early identification and treatment of patients (Ai et al., 2020). Moreover, potential patients left unattended increase the risk of spreading the infection. As an easy non invasive imaging alternative, chest computed tomography (CT) is viable for fast diagnosis (Ai et al., 2020). It can detect key imaging characteristics manifested in infected areas such as ground glass opacity (GGO), multifocal patchy consolidation and/or bilateral patchy shadows (Wang et al., 2020a). However, image characteristics between COVID-19 and other pneumonia types may possess similarities, making accurate diagnosis challenging. Also, automated screening sensitivity is limited and not on par with radiologist level performance (Wang et al., 2021b). Therefore, there is an urgent need to improve and/or develop robust screening methods based on chest CT.
On the other hand, deep learning (LeCun et al., 2015) based solutions have shown success in medical image analysis (Litjens et al., 2017) due to the ability to extract rich features from clinical datasets, and include a wide range of application areas such as organ segmentation (Ronneberger et al., 2015) and disease diagnosis, etc. Deep learning has been employed for the diagnosis of COVID-19 in chest CT (Song, Zheng, Li, Zhang, Zhang, Huang, Chen, Wang, Zhao, Zha, Shen, Chong, Yang, 2021, Gozes, Frid-Adar, Greenspan, Browning, Zhang, Ji, Bernheim, Siegel, Gozes, Frid-Adar, Sagie, Zhang, Ji, Greenspan) and community acquired pneumonia (CAP) (Kermany et al., 2018). For example, Ouyang et al. (2020) proposed a 3D convolutional neural network (CNN) with online attention refinement to diagnose COVID-19 from CAP and introduced a sampling strategy to mitigate the imbalanced distribution of infected regions between COVID-19 and CAP. Song et al. (2021) proposed DeepPneumonia for localization and detection of COVID-19 pneumonia; attention was also applied to detect key regions with impressive results on a large cohort. Despite showing promising performance, most methods are supervised and require considerable labeling efforts. Notably, even without annotated examples of infection areas, some works use existing deep learning models (Shan et al., 2021) to extract infection regions and/or manually select slices in CT that show key characteristics. However, taking into consideration that during the pandemic experts have had limited time to perform labeling of CT volumes for supervised methods, unsupervised or weakly supervised learning methods that do not heavily rely on extensive data pre-processing and/or strong prior knowledge are a preferred option for accurate diagnosis.
Recently, several works focused on accurate diagnosis under weak supervision have been proposed. Notably, we consider approaches that use (a) patch-based (Wang, Jin, Yan, Xu, Luo, Wei, Zhao, Hou, Ma, et al., 2021, Shi, Xia, Shan, Song, Wu, Wei, Yuan, Jiang, He, Gao, et al., 2021) (b) slice-based (Gozes, Frid-Adar, Sagie, Zhang, Ji, Greenspan, Gozes, Frid-Adar, Greenspan, Browning, Zhang, Ji, Bernheim, Siegel, Hu, Gao, Niu, Jiang, Li, Xiao, Wang, Fang, Menpes-Smith, Xia, et al., 2020), and (c) 3D CT-based (Han, Wei, Hong, Li, Cong, Zhu, Wei, Zhang, 2020, Wang, Deng, Fu, Zhou, Feng, Ma, Liu, Zheng, 2020) methods for diagnostic decisions. The first often uses prior segmented infection regions as input to train classifiers in a two-stage setup. The second performs slice-wise inference directly, whereas 3D based methods use the entire 3D CT scans as input with 3D convolutional neural networks (CNN). For the patch and slice-based approaches to be effective, infected regions must be well selected for training. Also, 3D CNN models are inherently slow during inference due to bigger model size and may lack interpretability.
In this work, we propose a novel end-to-end attention-based weakly supervised framework using multiple instance learning (MIL) (Carbonneau et al., 2018) and self-supervised contrastive learning (Chen et al., 2020a) of features towards accurate diagnosis of COVID-19 from bacterial pneumonia. We refer to this framework as DA-CMIL. The goal of DA-CMIL is to assign patients a single category label i.e. (COVID-19 or bacterial pneunomia) given as input a CT volume of multiple 2D slices. In general, each patient CT scan is considered as a bag of instances that may be positive or negative. Moreover, it would be beneficial to identify which slices/instances contribute to the final patient diagnosis with the potential to localize infected regions. Herein, we propose an attention based permutation-invariant MIL method for the pooling of slices to obtain a single representative feature of patients. In addition, spatial attention is jointly applied to learn spatial features key for infection area discovery. We incorporate contrastive learning at the instance level to encourage instance features from the same patient to be semantically similar to the patient level aggregated feature in an unsupervised manner. To achieve this, an unsupervised contrastive loss is employed alongside patient category labels for the supervised loss during training.
Existing works using MIL applied in different domains often decouple instance and bag level learning into a two-step procedure i.e. first learn instance level encoders, then learn aggregation models for inference using the trained encoders with MIL pooling (Hashimoto, Fukushima, Koga, Takagi, Ko, Kohno, Nakaguro, Nakamura, Hontani, Takeuchi, 2020, Hou, Samaras, Kurc, Gao, Davis, Saltz, 2016). However, due to the ambiguity of the instance labels and noise, learning a robust encoder can be challenging. Thus, the proposed framework aims to address the aforementioned challenges via end-to-end learning; instance selection is implicity achieved via attention based pooling of CT slices with model optimization focused only on accurate patient labels. Moreover, by jointly using a supervised and constrastive loss, our model can avoid overfitting when trained on smaller datasets and improve feature robustness at the instance level without sacrificing accuracy. We empirically show the benefit of DA-CMIL on a recently collected dataset, with interpretable results and competitive performance against state-of-the-art methods.
The main contributions of this study include:
-
•
We propose a novel end-to-end model for weakly supervised classification of COVID-19 from bacterial pneumonia.
-
•
We show that joint contrastive learning of instance features and patient level features in the MIL setting is viable. A novel setting of learning instance level features without inferring labels.
-
•
Towards interpretability, we show that dual attention, in particular spatial attention can be used to assess and visualize model decisions.
-
•
We empirically show that DA-CMIL is robust to different CT sizes when instance (i.e. slice/patch) count varies via ablation studies.
The rest of the article is arranged as follows. In Section 2, we review recent works related to computer aided diagnosis with artificial intelligence for COVID-19 and relevant methodologies under weak supervision. We introduce the relavant background and details regarding DA-CMIL in Section 3. In Section 4, we provide descriptions on experimental settings and datasets employed. Experimental results are discussed in Sections 5 and 6. We conclude this study in Section 7.
2. Related works
This section presents related works in terms of COVID-19 screening, methods for weak supervision and self-supervised learning.
2.1. Deep learning for COVID-19 diagnosis
The success of deep learning based techniques applied to medical image analysis have shown promising results for several application areas such as segmentation and disease detection. Several pioneering methods (Shi, Wang, Shi, Wu, Wang, Tang, He, Shi, Shen, 2021, Gozes, Frid-Adar, Greenspan, Browning, Zhang, Ji, Bernheim, Siegel, Xie, Jacobs, Charbonnier, van Ginneken, 2020, Wang, Jin, Yan, Xu, Luo, Wei, Zhao, Hou, Ma, et al., 2021, Kang, Hong, Chikontwe, Luna, Jang, Park, Shin, Park, Ahn, 2021, Kang, Chikontwe, Luna, Hong, Ahn, Park) have been proposed for the analysis of COVID-19 in both X-ray and CT images. COVID-19 lesion segmentation (Gozes, Frid-Adar, Greenspan, Browning, Zhang, Ji, Bernheim, Siegel, Xie, Jacobs, Charbonnier, van Ginneken, 2020), automated screening (Wang, Jin, Yan, Xu, Luo, Wei, Zhao, Hou, Ma, et al., 2021, Song, Zheng, Li, Zhang, Zhang, Huang, Chen, Wang, Zhao, Zha, Shen, Chong, Yang, 2021, Han, Wei, Hong, Li, Cong, Zhu, Wei, Zhang, 2020) and severity assessment (Huang et al., 2020) have been key areas of research. Notably, a recent review (Shi et al., 2021a) shows that automated screening is predominant and continues to receive much interest. Moreover, given that chest CT best shows key image characteristics for COVID-19 diagnosis, CT is preferred over X-ray despite being a low cost solution. Ng et al. (2020) recently claimed that consolidative and/or ground glass opacities (GGO) on CT are often undetectable in chest radiography and highlighted the pros and cons of each imaging modality.
Accordingly, Oh et al. (2020) recently proposed a patch-based CNN for COVID-19 diagnosis applied to chest radiography with limited datasets. They show that statistically significant differences in patch-wise intensity distributions can serve as biomarkers for diagnosis of COVID-19; with existing correlations to current radiological findings of chest X-ray. Alom et al. (2020) introduced a multi-task deep model that jointly considers chest CT and X-ray for diagnosis. They showed impressive results in both modalities for both detection and localization of infected regions. Song et al. (2021) developed DeepPneumonia, a deep learning system with rapid diagnosis to aide clinicians. Mei et al. (2020) used deep learning to integrate chest CT findings with clinical information such as laboratory tests and exposure history to rapidly diagnose COVID-19 patients. From a technical standpoint, most methods require pre-segmented lesions prior to training, and/or include multi-stages in the frameworks. Moreover, patch-based methods may suffer from noisy samples in scans, often requiring careful manual selection of slices for efficiency.
2.2. Weak supervision and multiple instance learning
MIL is a form of weakly supervised learning where labels/categories are provided only for a bag of instances i.e. training instances arranged in sets and the labels of instances contained in the bags are unknown (Carbonneau et al., 2018). In this study, we consider a patient CT scan as a bag with unlabeled slices (instances), having only the diagnostic label for training. In general, existing algorithms can be categorized as instance-level (Hou et al., 2016), bag-level (Hashimoto et al., 2020), embedding-based, and joint methods that combine several approaches such as attention mechanisms (Hashimoto, Fukushima, Koga, Takagi, Ko, Kohno, Nakaguro, Nakamura, Hontani, Takeuchi, 2020, Ilse, Tomczak, Welling, 2018, Han, Wei, Hong, Li, Cong, Zhu, Wei, Zhang, 2020). In literature, MIL has been applied to several domains including object detection (Zhang et al., 2016a), image classification (Yao, Zhu, Huang, 2019, Hou, Samaras, Kurc, Gao, Davis, Saltz, 2016, Zhang, Meng, Han, 2016), and object tracking (Hu et al., 2017).
Also, several works have been applied in the medical imaging domain (Wang, Deng, Fu, Zhou, Feng, Ma, Liu, Zheng, 2020, Hu, Gao, Niu, Jiang, Li, Xiao, Wang, Fang, Menpes-Smith, Xia, et al., 2020, Han, Wei, Hong, Li, Cong, Zhu, Wei, Zhang, 2020, Wang, Deng, Fu, Zhou, Feng, Ma, Liu, Zheng, 2020, Campanella, Hanna, Geneslaw, Miraflor, Silva, Busam, Brogi, Reuter, Klimstra, Fuchs, 2019). Hashimoto et al. (2020) recently introduced a novel CNN for the classification of malignant lymphoma in histopathology slides. Notably, they combined domain adaptation and multi-scale approaches with MIL for improved performance. Ilse et al. (2018) proposed attention-based pooling for MIL in an end-to-end framework; impressive results are shown across different domain problems including cancer region detection in histopathology. Weakly supervised detection of COVID-19 infection regions in chest CT is presented by Hu et al. (2020) with multi-scale learning applied for localization. More recently, Wang et al. (2020c) proposed (DeCoVNet), a method applied to 3D CT volumes using 3D CNNs with weak labels. DeCoVNet takes as input a CT volume and its lung mask for COVID-19 classification. Han et al. (2020) proposed AD3DMIL, a 3D MIL method with attention for COVID-19 screening with a deep instance generation module based on 3D latent features inspired by the pioneering work of Feng and Zhou (2017).
2.3. Self supervised learning
Self Supervised Learning (SSL) is a form of unsupervised learning where the data provides the supervision, and the network is trained to solve auxiliary tasks with a proxy loss. This is highly beneficial, especially in medical imaging where supervision is limited and the existing difficulty of curating annotations. Auxiliary tasks include context prediction (van den Oord et al., 2019), automatic colorization (Zhang et al., 2016b), and image inpainting (Pathak et al., 2016). Most recently, Chen et al. (2020a) introduced a simple framework for contrastive learning (SimCLR) that uses extensive data-augmentation for defining predictive tasks, which achieves comparable performance to state-of-the-art supervised methods. For medical imaging tasks, He et al. (2020) recently proposed (Self-Trans) a method that combines contrastive self-training with transfer learning for COVID-19 diagnosis. Notably, the authors focus on establishing robust strategies for transfer learning with limited data, and/or when using external datasets for COVID-19 Chest CT analysis.
Inspired by recent works both for MIL and SSL, we propose to synergistically integrate contrastive self-supervision with MIL in an end-to-end framework. Though previous works such as AD3DMIL Han et al. (2020) have shown impressive results; the model is based on 3D CNN and considerably has a larger model size. Also, Self-Trans (He et al., 2020) follows a two-step approach by first pre-training the network via self-supervision using (Chen et al., 2020b); then performs fine-tuning or transfer learning. We believe that joint self supervised training with transfer learning is still underexplored. Thus, we aim to extend the current scope of the literature regarding COVID-19 via a novel formulation of MIL and self-supervised contrastive learning.
3. Methods
This sections presents the necessary notations and overall objectives of the task of COVID-19 diagnosis, including details of the relative modules of the proposed method.
3.1. Preliminaries
We consider a chest CT dataset where the model receives a set of labeled example scans drawn from the joint distribution defined by . is a patient CT scan with instances (i.e. 2D CT slices or patches) and is the label set of patient-level labels, wherein is for binary classification of COVID-19 and other. Also, is considered as a bag of instances with where denotes the total number of instances in the bag. It can be assumed that each instance has a label , however not all instances may be negative or positive. Moreover, not all slices in a scan may show infection regions vital for diagnosis, as others may be noisy artifacts not useful for learning.
Accordingly, MIL must satisfy the following constraints: if a bag is negative, then it can be assumed that all corresponding instances should be negative. In the case of positive bags, at least one instance is assumed to be positive. Formally, it follows that
| (1) |
In this work, this assumption may not hold given that both sets of bags (COVID-19 and other pneumonia) considered contain both negative and positive instances (lesions). Thus, we consider a relaxed version of this constraint wherein an attention mechanism is applied to implicitly weight instances and learn their labels.
3.2. Proposed approach
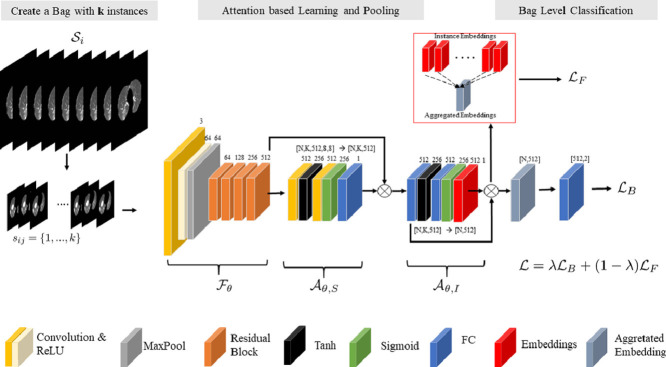
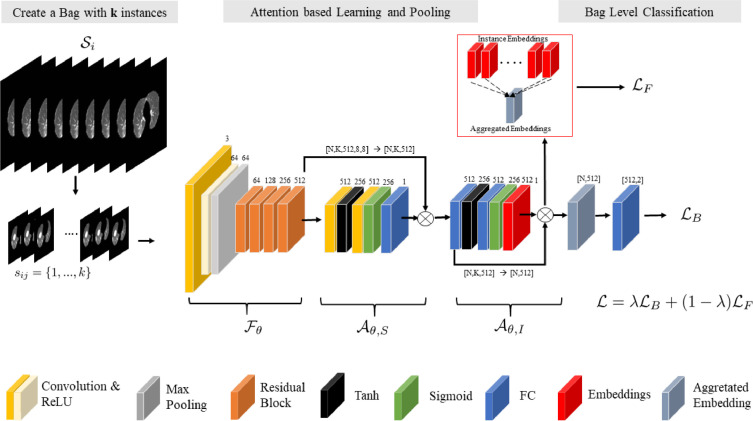
We developed a CNN model for patient CT scan level diagnosis between COVID-19 and other pneumonia in a single end-to-end framework. Herein, a dual-attention multi-instance learning deep model with unsupervised contrastive learning (DA-CMIL) is proposed. As presented in Figure 1 , our method takes a CT scan with unlabeled instances as input and learns key semantic representations. It further uses an attention-based pooling method to transform patient instances into a single bag representation for final prediction (see Section 3.3). Unsupervised contrastive learning is employed to encourage instances in a bag to be semantically similar to the bag representation during training (see Section 3.4).
Fig. 1.
Overview of the proposed framework. For a given patient CT scan, we sample instances during training to create a bag as input and feed them through the backbone to obtain feature maps. Modules and learn spatial and instance attention, then perform permutation invariant pooling via on feature maps (from ) to obtain a single patient representative feature. The backbone features are spatially pooled via the 1st operator after , whereas the instance feature aggregation is done with 2nd in , respectively. Prior to pooling (aggregation of all features via ), attention-weighted instance features from are employed for unsupervised contrastive learning as well as patient level learning to obtain the final predictions and update the model.
In the proposed framework, a backbone CNN model is implemented as a feature extractor to transform the -th instance from a CT bag into a low dimension embedding with spatial dimensions of shape , where , and are the channel size, height and width, respectively. Following, is feed to a spatial attention module in order to learn spatial representative features and output spatial attention maps of size per instance with . The obtained maps highlight key regions and are further used to weight all the initial instances features to obtain a single spatial pooled feature1 , with , where is the feature dimension size (see Section 3.3). To aggregate the instance features for each CT scan, we implement a second module that performs attention-based permutation invariant pooling to obtain a single bag representation , with having the same dimension for consistency. Following, is passed to the patient level classifier to obtain predictions for the entire bag , where is the probability of a CT scan being labeled as COVID-19 or other pneumonia. Formally, we employ the bag loss using cross-entropy. It follows that
| (2) |
3.3. Dual attention based learning
In recent works (Hashimoto, Fukushima, Koga, Takagi, Ko, Kohno, Nakaguro, Nakamura, Hontani, Takeuchi, 2020, Ilse, Tomczak, Welling, 2018, Han, Wei, Hong, Li, Cong, Zhu, Wei, Zhang, 2020) attention has shown to be vital for learning robust features, especially under the MIL setting. In particular, attention-based pooling (Ilse et al., 2018) is preferred over existing pooling methods such as max or mean, since they are not differentiable/applicable for end-to-end model updates. In this work, we implemented both spatial () and latent embedding () based attention pooling via the respective modules. In the spatial module, given the input , we employ two convolutional layers each followed by hyperbolic tangent (tanh) and sigmoid (sigm) non-linearities, respectively. Feature maps are passed to each module successively, then to the final convolutional layer having a single channel output representing the presence of infection. In particular, we performed element-wise multiplication between the output of each branch of the convolutional layers before passing it the final layer to obtain spatial scores . Following, the spatial scores are normalized by a softmax operation, with the final spatially pooled features obtained by a summed matrix multiplication across both height and weight dimensions i.e. , where , though for consistency we refer to as . It is worth noting that we simply implemented gated spatial attention (Dauphin et al., 2017) instead of the commonly applied global average pooling (GAP) on the initial backbone features . Moreover, the initial normalized spatial maps can be used to visually show the regions the model focuses on to make decisions.
In order to aggregate the features , we employ attention based pooling proposed by Ilse et al. (2018) in the instance attention module . Formally, we consider the same architectural design previously applied for gated spatial attention on the initial backbone features, except all convolutional layers are replaced with fully connected layers since attention is applied to instance embeddings. We denote , with as a bag with instance features. Then, attention based pooling MIL with gating mechanism is defined as
| (3) |
with,
| (4) |
where , , and are trainable parameters. and are element wise non-linearities, with representing element-wise multiplication. In addition, is considered as the attention score per instance indicating the relevance of a given instance to the overall bag prediction. From a technical standpoint, attention based pooling allows different weights to be assigned to instances alleviating the need for explicit instance selection. Moreover, the final bag representation will be more informative. The synergistic combination of spatial and attention based pooling allows for improved training towards learning robust and interpretable features.
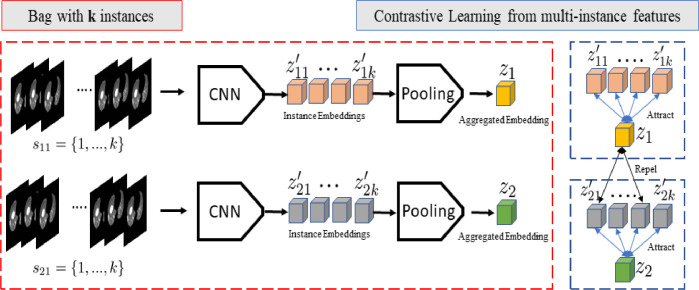
3.4. Contrastive MIL
Inspired by recently proposed self-supervised learning methods (Chen, Kornblith, Norouzi, Hinton, 2020, Chen, Fan, Girshick, He), we integrate an unsupervised contrastive loss with the proposed MIL method for improved learning of instance level features. Formally, our model learns representations that maximize the agreement between instance features and an aggregated bag feature of the same patient via a contrastive loss (Chen et al., 2020a) in the latent space. Fig. 2 shows the overall concept of the applied technique.
Fig. 2.
Illustration of contrastive learning applied to the MIL setting during model training (Blue section).(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
According to the previously proposed self-supervised framework that uses contrastive loss, stochastic data augmentation is applied on 2D data samples to create two correlated views of the same example (Chen, Fan, Girshick, He, Chen, Kornblith, Norouzi, Hinton, 2020). Augmentations include random cropping, color distortions and random Gaussian bluring. Moreover, the contrastive loss is proposed to define contrastive predictive tasks on unlabeled samples, wherein positive and negative pairs are identified for given samples. To incorporate this idea, stochastic data augmentation is omitted in our study since contrastive loss is applied in the latent space. In addition, for any given patient CT scan; we infer that each slice can be considered as a pseudo augmentation of the overall patient characteristics. Thus, we consider that stochastic augmentation is implicitly applied (i.e. different views of the same patient).
Let be the latent instance level feature of patient, and the patient bag-level feature obtained via the proposed modules. Then, following normalization of and features, a contrastive loss can be defined as
| (5) |
where is an indicator function that evaluates to 1 iff and denotes a temperature parameter. is a similarity function i.e. cosine similarity. The loss is computed across all patient slice features and respective bag-level features, herein considered as augmentations per mini-batch. The final loss function of the entire framework is defined as:
| (6) |
where is a parameter to weight the contribution of the bag and constrastive losses, respectively. The detailed algorithm is presented in Algorithm 1 .
Algorithm 1.
DA-CMIL Algorithm
4. Experiments
We evaluate the proposed method on a recently collected dataset and compare diagnostic performance against existing methods similar to ours. We present details on evaluation settings and any pre-processing applied.
4.1. Datasets
In this study, we collected a chest CT dataset comprised of 173 samples at Yeungnam University Medical Center (YUMC), in Daegu, South Korea. The dataset includes 75 CT examples for patients with COVID-19, and 98 examples from patients with bacterial pneumonia collected between February and April, 2020. The study was approved by the Institutional Review Board (IRB) of Yeungnam University Hospital. COVID-19 patients were confirmed by RT-PCR assay of nasal and pharyngeal swab samples.
Further, we designed variants of YUMC CT dataset to fairly assess the performance of our method and others such as 3D based approaches. Namely, based on the original YUMC CT data using CT slices per patient, we processed a patch-based version of the dataset. In the MIL framework, 2D CT slice or patches can be used as instances, thus we evaluate our method on both cases. In addition, a 3D CT volume dataset is also processed for training/testing 3D based methods under fully-supervised settings.
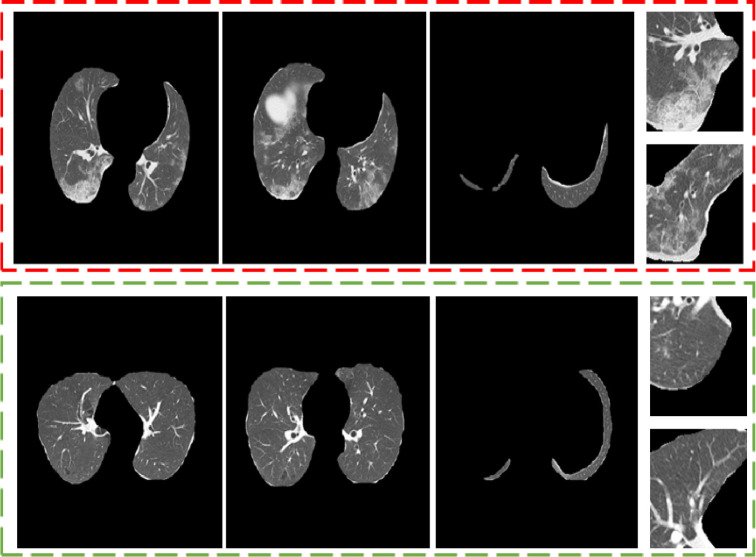
For pre-processing, lung regions were segmented for all CT examples. To achieve this, we employed a ResNeSt (Zhang et al., 2020a) model for segmentation training and inference. The model was trained on two public datasets i.e. non-small cell lung cancer (NSCLC) (Aerts et al., 2014) and COVID-19 lung infection dataset (Jun et al., 2020). Herein, a total of 50,756 lung slices were used for training and evaluated on 1,222 independent slices. Fig. 3 shows examples of CT slices and patches employed.
Fig. 3.
Pre-processed CT examples. (Red-section) COVID-19 CT slice and patch samples. (Green-section) bacterial pneumonia samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.2. Experimental settings
Accordingly, all the datasets were split into training, validation and testing by patient IDs with ratios 0.5, 0.1, and 0.4, respectively. The same split was used across all the dataset variants with all versions using only cropped lung regions. CT examples were , and in size for the slices, patches and 3D CT volume sets, respectively. Each CT slice was resized from to and patch slices were resized to 256 from 128. In particular, the slices set consisted of approximately 14,000 slices, whereas the patch version yielded 64,000 patches that mainly showed of lung tissue. In the case of 3D CT volumes, all slices belonging to a patient were used to construct a volume with nearest neighbor sampling applied to obtain the desired input sizes.
The proposed model was implemented in Pytorch. A ResNet-34 (He et al., 2016) finetuned from imageNet pretrained weights was used as the feature extraction module , with a single fully connected (FC) layer employed as the bag classifier . The dimension of the features was fixed to 512; this includes the feature maps obtained from which had , with . Following spatial pooling, features were reshaped back to 512.
During training, data augmentation consisting of random transformations such as flipping were applied for both 2D and 3D based methods. All models were trained for 30 epochs except 3D based methods with an initial learning rate of for , and for the other modules with Adam optimization and a batch size of 8. On the other hand, 3D CNN based methods used a batch size of 4, learning rate of and were trained for 60 epochs.
For the proposed method, a bag was constructed with instances during training following step 4 of our algorithm, though for inference all available instances per patient were used to obtain the final prediction. We also evaluated the efficacy of our method based on varying during training via ablation studies. For stable training, the learning rate was annealed by a factor of 0.1 at epochs 10, 15 and 25, respectively. We empirically set the loss hyper-parameters and to 0.5 and 1.0, respectively.
4.3. Comparison methods
To evaluate the efficacy of the proposed method, we compared against recent MIL based methods i.e. DeepAttentionMIL (Ilse et al., 2018), ClassicMIL (Campanella et al., 2019) and JointMIL (Chikontwe et al., 2020). Also, recent 3D based methods DeCovNet (Wang et al., 2020c) and Zhang3DCNN (Zhang et al., 2020b) were included for comparison. For a fair evaluation, the same backbone feature extractor is used in all methods except for the 3D methods as we used the publicly available implementations.
In particular, ClassicMIL follows the traditional assumption of the MIL setting and focuses on instance level learning wherein only the top instance per bag is considered for the final patient level prediction. DeepAttentionMIL uses attention-based pooling for bag level learning. In constrast, JointMIL combines both instance and bag level learning with bag feature clustering during training. Lastly, DeCoVNet and Zhang3DCNN both use all available CT slices in a constructed volume under the fully supervised setting. The later methods serve as an upper-bound over the weakly supervised methods evaluated in this study.
5. Results
We present both quantitative and qualitative results of the proposed methods. Also, ablation studies on the effect of bag size, attention modules with/without contrastive learning and the weighting parameter are presented.
5.1. Quantitative results
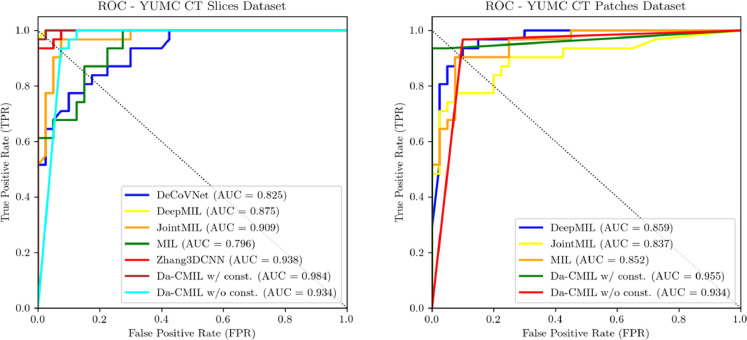
Diagnostic performance was evaluated on YUMC CT slice, patch and CT volume based datasets using accuracy, area under the curve (AUC), f1-score, specificity and sensitivity, respectively. Tables 1 and 2 show the performance of the evaluated methods on the datasets.
Table 1.
Evaluation of the proposed methods on YUMC dataset including results of using DA-CMIL with/without contrastive loss .
| Method | Accuracy | AUC | F1 | Specificity | Sensitivity |
|---|---|---|---|---|---|
| DeCoVNet Wang et al. (2020c) | 0.831 | 0.825 | 0.8 | 0.875 | 0.774 |
| MIL Campanella et al. (2019) | 0.803 | 0.796 | 0.767 | 0.85 | 0.742 |
| DeepAttentionMIL Ilse et al. (2018) | 0.859 | 0.875 | 0.861 | 0.75 | 1 |
| JointMIL Chikontwe et al. (2020) | 0.901 | 0.909 | 0.896 | 0.85 | 0.968 |
| Zhang3DCNN Zhang et al. (2020b) | 0.93 | 0.938 | 0.925 | 0.875 | 1 |
| DA-CMIL (w/o ) | 0.76 | 0.72 | 0.62 | 1.0 | 0.45 |
| DA-CMIL (w/o ) | 0.93 | 0.934 | 0.923 | 0.9 | 0.968 |
| DA-CMIL (w/ ) | 0.986 | 0.984 | 0.984 | 0.975 | 1 |
Table 2.
Evaluation of the proposed methods on YUMC patch dataset.
| Method | Accuracy | AUC | F1 | Specificity | Sensitivity |
|---|---|---|---|---|---|
| MIL Campanella et al. (2019) | 0.845 | 0.852 | 0.836 | 0.8 | 0.903 |
| DeepAttentionMIL Ilse et al. (2018) | 0.845 | 0.859 | 0.845 | 0.75 | 0.968 |
| JointMIL Chikontwe et al. (2020) | 0.845 | 0.837 | 0.814 | 0.9 | 0.774 |
| DA-CMIL (w/o ) | 0.718 | 0.728 | 0.714 | 0.65 | 0.806 |
| DA-CMIL (w/o ) | 0.873 | 0.88 | 0.866 | 0.825 | 0.935 |
| DA-CMIL (w/ ) | 0.958 | 0.955 | 0.951 | 0.975 | 0.935 |
In Table 1, DA-CMIL with contrastive loss achieves the best overall performance of 98.6% accuracy and an AUC of 98.4%. Notably, even when was not applied during training, our method still reports 93%(+2.9) and 93.4%(+2.5) in terms of accuracy and AUC over the best weakly supervised method JointMIL. MIL reports the lowest performance among all methods, which is expected since it only considers the top instance among multiple 2D slices in bag for inference. Interestingly, our method outperformed both Zhang3DCNN and DeCoVNet which are fully supervised methods even without used in the training stage. Though both DA-CMIL and DeepAttentionMIL employ attention based pooling, the proposed method shows improved performance via dual-attention pooling, validating the architectural design.
To further validate the proposed method, we applied DA-CMIL to randomly cropped patches of the CT samples. As shown in Table 2, performance was consistently better than the compared methods. All weakly supervised method showed similar accuracy with considerable margins observed for sensitivity. DeepAttentionMIL reported the best sensitivity at 96.8% with accuracy consistent with other methods. However, DA-CMIL showed an improvement of +11.3% in accuracy over the best compared method with an equally larger margin without employed. The effect of using attention and contrastive loss was more pronounced in the case of patches as not applying showed reduced performance (-2.8%).
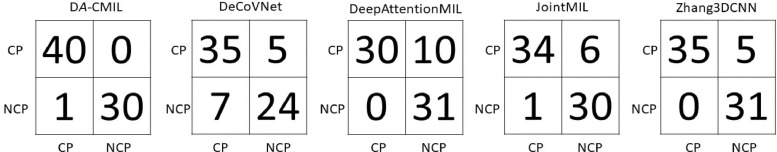
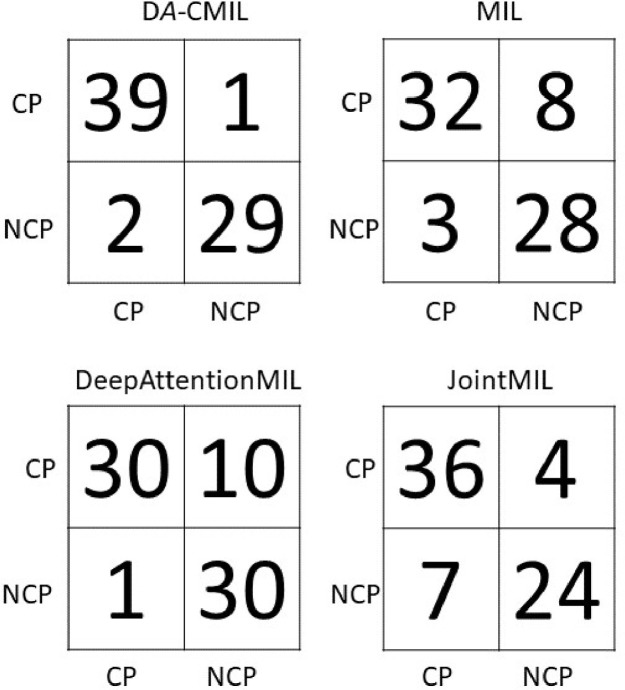
Fig. 4 shows the receiver operating characteristic(ROC) curves of the compared methods on different datasets. Overall, the proposed method shows a high TPR and lower FPR across all settings. This is further evidenced in the summaries of the confusion matrices of the comparison methods as presented in Figs. 5 and 6 . This indicates DA-CMIL can be viable option for accurate and robust screening of COVID-19.
Fig. 4.
The Receiver operating characteristic (ROC) curves of compared methods on the YUMC CT slices and patch datasets.
Fig. 5.
Confusion matrices of compared methods on YUMC CT Slices dataset. CP represent Pneumonia and NCP implies COVID-19, respectively.
Fig. 6.
Confusion matrices of compared methods on YUMC CT patch dataset.
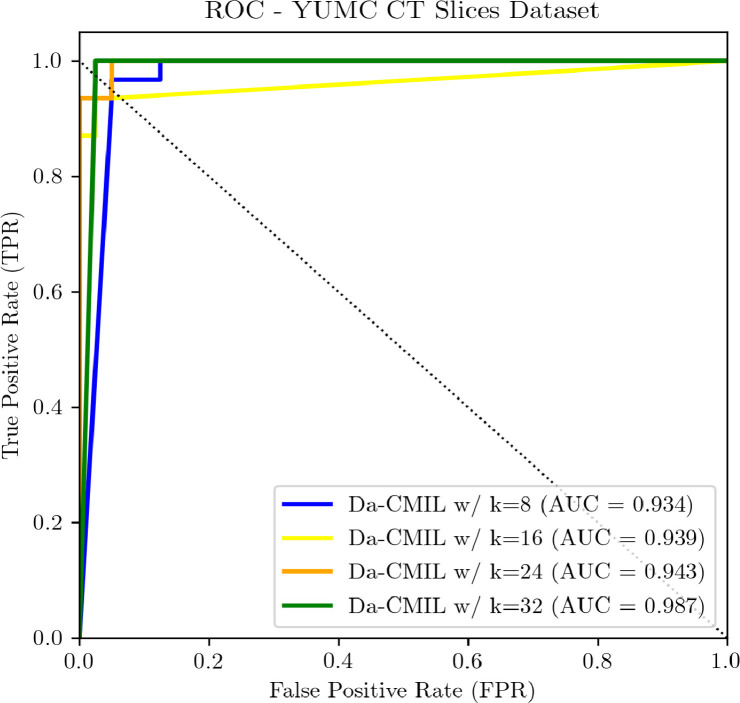
5.2. Effect of the bag size
To assess the effect of bag size during training on the proposed method, we performed an ablation study where the bag was constructed by varying i.e. each bag consisted of max instances (slices/patches). As shown in Table 3 and Fig. 7 , as the bag size increases DA-CMIL performance improves. The best result was achieved when with a considerable margin across all metrics. We limited evaluation to due to computational limitations. Moreover, it worth noting that contrastive methods benefit from large batch sizes; as evidenced from the reported results, our findings are consistent with existing observations based on self-supervised methods applied to general vision datasets. However, as is increased the relative batch size based on bags should be reduced to compensate for training time and memory requirements. In general, results show that our method is not limited/affected by the number of instances available per CT scan and can benefit from using more instances for training, though during the inference stage all instances are used.
Table 3.
Evaluation of varying bag sizes with the proposed method on YUMC CT slices dataset.
| Method | Accuracy | AUC | F1 | Specificity | Sensitivity |
|---|---|---|---|---|---|
| DA-CMIL (w/ ) | 0.93 | 0.934 | 0.923 | 0.9 | 0.968 |
| DA-CMIL (w/ ) | 0.944 | 0.939 | 0.933 | 0.975 | 0.903 |
| DA-CMIL (w/ ) | 0.944 | 0.943 | 0.935 | 0.95 | 0.935 |
| DA-CMIL (w/ ) | 0.986 | 0.988 | 0.984 | 0.975 | 1 |
Fig. 7.
ROC curves of DA-CMIL on YUMC CT Slide dataset when is varied during training.
5.3. Effect of the weight parameter
DA-CMIL uses contrastive feature learning of multiple instances with a weighting parameter to balance the effect of the losses. When , has no effect on learning and showed a lower performance of 93% compared to using i.e. when . Though similar performance was noted across different values of , the most significant is when contrastive loss was not applied entirely as presented in Table 4 .
Table 4.
Evaluation on varying in the cost function on the YUMC Dataset.
| =0.1 | =0.5 | =0.9 | =1.0 | |
|---|---|---|---|---|
| Accuracy | 0.972 | 0.986 | 0.986 | 0.932 |
5.4. Effect of dual-attention modules on learning
In order to assess the effect of attention in the proposed framework, we consider several settings where both contrastive and attention modules are either employed or not (Tables 1 and 2). Formally, when attention is excluded, the framework would require modification in two aspects; (1) without spatial attention-based pooling of features (), we default to using global average pooling (GAP) of instance features for simplicity, and (2) without attention based bag-level feature aggregation via (), one may opt for using the mean of instance features to obtain the overall bag-level feature alongside . Following these modifications, evaluation can be easily performed.
Evidently, the best performance was achieved when both and were part of learning. On the other hand, when the attention modules were excluded, significant reductions in the overall performance were noted i.e. -20% compared to the best performing method (Table 1). Similar performance drops were noted on the CT patch dataset (Table 2). Using the contrastive feature loss alone without any attention modules highlighted worsened results without any performance gains over the compared methods. This serves to show the benefit of the combination of the proposed techniques (i.e. both attention and the feature loss), reporting improved results via complementary learning.
5.5. Qualitative Results
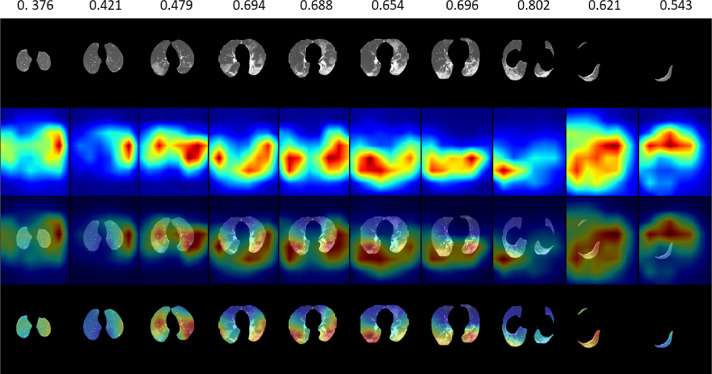
In Figs. 8 and 9 , qualitative results are presented based on spatial attention maps and attention scores, respectively. This demonstrates that DA-CMIL is able to find key slices related to infected areas with coarse maps (Fig. 8). Interestingly, low attention scores were observed for slices such as noisy slices/artifacts with no infected areas further indicating the utility of our method. Moreover, attention maps focus on key areas such as ground-glass opacities and consolidations, both consistent with clinical findings.
Fig. 8.
Qualitative examples of DA-CMIL spatial attention maps with attention scores on CT samples from a single patient with COVID-19. The top row shows the attention value of each slice with the spatial maps normalized to focus on the lung regions only.
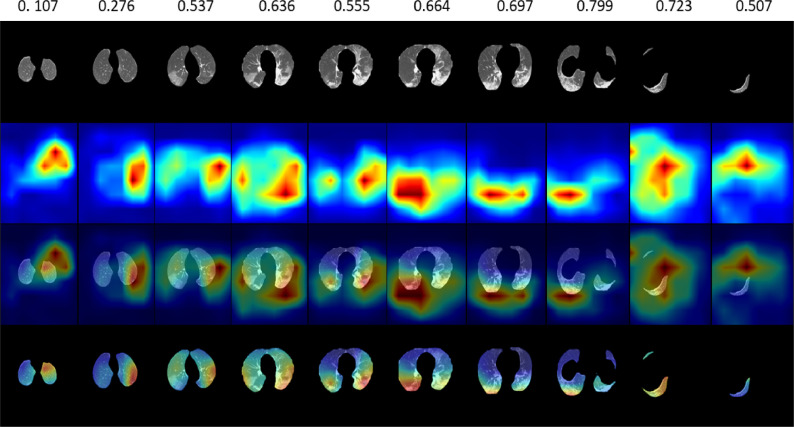
Fig. 9.
Examples of DA-CMIL spatial attention maps with attention scores without contrastive learing on CT samples from a single patient with COVID-19.
In Fig. 9, we also highlight attention maps when contrastive learning is not applied during. In general, results show similar maps as with the case when the loss is applied. However, localization of key regions is slightly degraded, especially with huge differences in the attention scores, whereas for some CT slices, both spatial maps and scores had marginal changes. This is largely expected since the contrastive loss is aimed at encouraging similarity between representative features of a subject. The benefit of using both losses is better verified via quantitative assessment of classification performance. We infer that the proposed mechanism of attention is still relevant in both cases and can be highly beneficial in clinical evaluation.
In addition, according to clinical literature on similar studies (Xu, Yu, Qu, Zhang, Jiang, Huang, Chen, Zhang, Guan, Ling, et al., 2020, Gozes, Frid-Adar, Sagie, Zhang, Ji, Greenspan, Gozes, Frid-Adar, Greenspan, Browning, Zhang, Ji, Bernheim, Siegel, Shi, Xia, Shan, Song, Wu, Wei, Yuan, Jiang, He, Gao, et al., 2021): Bilateral multi-focal ground-glass opacities (GGO) in the lower lobes are the most common initial findings on CT, with other characteristics such as pleural thickening less commonly observed in imaging manifestations depending on the severity stage. This is consistent with the most of the spatial attention maps being largely focused in the lower regions. In general, Class Activation Maps (CAM) may not indicate exact lesion locations due to the resolution issue. Moreover, we wish to note that enforcing the model to produce tissue-constrained maps is challenging especially in the weakly supervised setting; without access to the actual lesion locations, it is non-trivial for the model. Herein, we are confident in the current results even when the maps are normalized to the tissue region only, it is evident that the high-density regions are clinically relevant regions corresponding to lesions and/or GGOs.
6. Discussion
Though RT-PCR is the gold standard for COVID-19 diagnosis, it is still hindered by lengthy test times, as it can take days to obtain the results. Accordingly, CT has been considered as a reasonable alternative for current testing methods as it can produce results within minutes. We showed a novel approach to the application of deep CNNs for COVID-19 diagnosis under weak supervision with clinical implications (Xu et al., 2020). It is important to have a fully automated and interpretable method in actual settings for rapid evaluation. Moreover, given the subtleties that exist between COVID-19 and other pneumonia in terms of imaging characteristics that field experts find hard to differentiate, accurate diagnosis is highly relevant.
Our method was evaluated on recently curated dataset wherein only patient diagnostic labels are available without lesion infected regions of interest as is common in existing methods. To further validate our approach, we qualitatively showed the regions that are focused on by our model via coarse attention maps alongside attention scores. Our method achieved an AUC of 98.4%, accuracy of 98.6% and a true positive rate (TPR) of 96.8%. In addition, attention maps obtained highlight key infection areas in the majority of samples with attention scores corresponding to key slices.
We also empirically showed the benefit of using an unsupervised contrastive loss to complement the supervised learning of patient labels and may serve as a base for more complex methods. Moreover, the proposed method surpassed 3D based methods by large margins. We infer this may be due to the limited size of the dataset employed as most recent methods applied to 3D CT volumes report using large cohorts in literature. In addition, since DeCoVNet was trained from scratch and has an custom deep architecture, performance was subpar. Though ZhangCNN’s performance was considerably better than the later, it still did not achieve comparable performance even when the model was trained for more epochs. It is also worth noting that models trained with extensive augmentation did not achieve any considerable improvements across the evaluation metrics, since COVID-19 and bacterial pneumonia present similar characteristics.
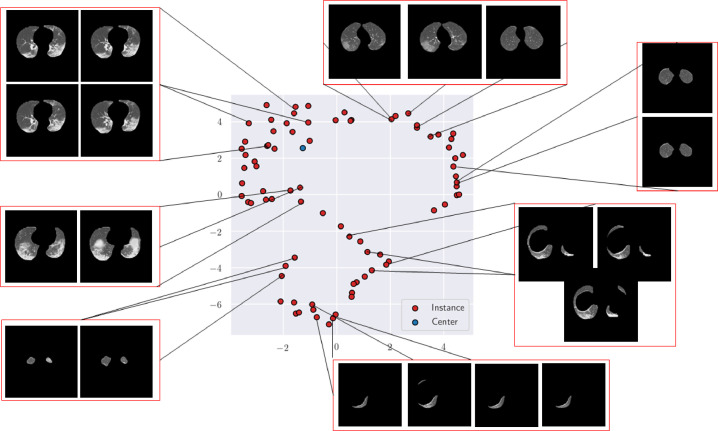
To further show the benefit of the proposed technique in the capturing overall statistics of a single subject, Fig. 10 presents the instance and representative features plotted in 2D space. Notably, the aggregated feature (top-left of the figure: blue dot) captures features of key slices (red) that are well clustered together and ignores noisy artifacts in other slices. This shows that though no explicit labels are employed for instance discovery, our model is able to effectively learn which slices are useful for patient classification.
Fig. 10.
Illustration of instance (red) and representative (center-blue) features of CT slices of a COVID subject. The aggregated center feature captures the key statistics of the most informative slices with similar characteristics and ignores noisy slices.
There exist a few limitations with regard to the proposed method. Though attention maps could show interpretability and explainability for COVID-19 diagnosis, there exist some failure cases where the attention map do not correctly indicate an infected region as shown in Fig. 8. Second, we found that extensive data augmentation such as color jittering lead to reduced performance and was largely negligible compared to the benefit of using a contrastive loss which showed consistent improvements across all evaluation settings. This motivates us to consider using more complex attention modes for better diagnostic interpretability. Lastly, since no healthy scans were employed in this study; it is reasonable to assume this approach would produce unfavorable results when supplied with healthy inputs as it is limited to the sub-typing scenario. Nevertheless, we believe our approach can be feasibly integrated into existing systems that address the later first (i.e. lesion/healthy slice detection) with accurate sub-typing for diagnosis as the final goal. Along this line of thought, we infer that even without such samples, there is a possibility that healthy scans could be considered noisy artifacts and later ignored similar to observations made via Fig. 10. Though we leave this for future research as well as the viability of unsupervised pre-training using the proposed method both in 2D or 3D settings.
7. Conclusion
In this study, we developed a 2D CNN framework with dual attention modules and contrastive feature learning under the multiple instance learning (MIL) framework to distinguish COVID-19 and a bacterial sub-type of pneumonia in chest CTs. We verified performance on both CT patch and slice based versions of the datasets and report results comparable to state-of-the-art methods. In addition, ablation experiments show the benefit of using large bag sizes during training and the effect of weighting losses correctly for stable learning. Through this study, we hope to add valuable contribution to the current literature on weakly supervised methods for COVID-19 screening.
CRediT authorship contribution statement
Philip Chikontwe: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Visualization, Software, Writing - original draft, Writing - review & editing. Miguel Luna: Investigation, Methodology, Writing - review & editing. Myeongkyun Kang: Investigation, Methodology. Kyung Soo Hong: Resources, Data curation. June Hong Ahn: Resources, Data curation, Supervision, Writing - review & editing. Sang Hyun Park: Funding acquisition, Methodology, Project administration, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT)(No. 2019R1C1C1008727).
Footnotes
Please note, for efficiency a summation operation between the attention maps and backbone features is achieved via Einstein summation. https://pytorch.org/docs/stable/generated/torch.einsum.html
References
- Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S., Bussink J., Monshouwer R., Haibe-Kains B., Rietveld D., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5(1):1–9. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in Coronavirus Disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32101510
- Alom, M. Z., Rahman, M. M. S., Nasrin, M. S., Taha, T. M., Asari, V. K., 2020. COVID_MTNet: COVID-19 detection with multi-task deep learning approaches. 2004.03747.
- Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Silva V.W.K., Busam K.J., Brogi E., Reuter V.E., Klimstra D.S., Fuchs T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25(8):1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonneau M.-A., Cheplygina V., Granger E., Gagnon G. Multiple instance learning: a survey of problem characteristics and applications. Pattern Recognit. 2018;77:329–353. [Google Scholar]
- Chen T., Kornblith S., Norouzi M., Hinton G.E. Proceedings of the 37th International Conference on Machine Learning, ICML 2020, 13-18 July 2020, Virtual Event. PMLR; 2020. A simple framework for contrastive learning of visual representations; pp. 1597–1607. [Google Scholar]
- Chen, X., Fan, H., Girshick, R., He, K., 2020b. Improved baselines with momentum contrastive learning. 2003.04297.
- Chikontwe P., Kim M., Nam S.J., Go H., Park S.H. Medical Image Computing and Computer Assisted Intervention - MICCAI 2020 - 23rd International Conference, Lima, Peru, October 4-8, 2020, Proceedings, Part V. Springer; 2020. Multiple instance learning with center embeddings for histopathology classification; pp. 519–528. [DOI] [Google Scholar]
- Dauphin Y.N., Fan A., Auli M., Grangier D. Proceedings of the 34th International Conference on Machine Learning, ICML 2017, Sydney, NSW, Australia, 6-11 August 2017. PMLR; 2017. Language modeling with gated convolutional networks; pp. 933–941. [Google Scholar]
- Feng J., Zhou Z. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, February 4-9, 2017, San Francisco, California, USA. AAAI Press; 2017. Deep MIML network; pp. 1884–1890. [Google Scholar]
- Gozes, O., Frid-Adar, M., Greenspan, H., Browning, P. D., Zhang, H., Ji, W., Bernheim, A., Siegel, E., 2020a. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis. 2003.05037.
- Gozes, O., Frid-Adar, M., Sagie, N., Zhang, H., Ji, W., Greenspan, H., 2020b. Coronavirus detection and analysis on chest CT with deep learning. 2004.02640.
- Han Z., Wei B., Hong Y., Li T., Cong J., Zhu X., Wei H., Zhang W. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- Hashimoto N., Fukushima D., Koga R., Takagi Y., Ko K., Kohno K., Nakaguro M., Nakamura S., Hontani H., Takeuchi I. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. Multi-scale domain-adversarial multiple-instance CNN for cancer subtype classification with unannotated histopathological images; pp. 3852–3861. [Google Scholar]
- He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2016, Las Vegas, NV, USA, June 27-30, 2016. IEEE Computer Society; 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- He, X., Yang, X., Zhang, S., Zhao, J., Zhang, Y., Xing, E., Xie, P., 2020. Sample-efficient deep learning for covid-19 diagnosis based on ct scans. 10.1101/2020.04.13.20063941
- Hou L., Samaras D., Kurc T.M., Gao Y., Davis J.E., Saltz J.H. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2016. Patch-based convolutional neural network for whole slide tissue image classification; pp. 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Liu Z., Zhang J., Zhang G. Robust object tracking via multi-cue fusion. Signal Process. 2017;139:86–95. [Google Scholar]
- Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J., et al. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. [Google Scholar]
- Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest CT assessment of COVID-19: Deep-learning approach. Radiol. Cardiothorac. Imaging. 2020;2(2):e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilse M., Tomczak J.M., Welling M. Proceedings of the 35th International Conference on Machine Learning, ICML 2018, Stockholmsmässan, Stockholm, Sweden, July 10-15, 2018. PMLR; 2018. Attention-based deep multiple instance learning; pp. 2132–2141. [Google Scholar]
- Jun, M., Cheng, G., Yixin, W., Xingle, A., Jiantao, G., Ziqi, Y., Minqing, Z., Xin, L., Xueyuan, D., Shucheng, C., Hao, W., Sen, M., Xiaoyu, Y., Ziwei, N., Chen, L., Lu, T., Yuntao, Z., Qiongjie, Z., Guoqiang, D., Jian, H., 2020. COVID-19 CT lung and infection segmentation dataset. 10.5281/zenodo.3757476
- Kang, M., Chikontwe, P., Luna, M., Hong, K. S., Ahn, J. H., Park, S. H., 2021a. Mixing-AdaSIN: Constructing a de-biased dataset using adaptive structural instance normalization and texture mixing. 2103.14255.
- Kang M., Hong K.S., Chikontwe P., Luna M., Jang J.G., Park J., Shin K.C., Park S.H., Ahn J.H. Quantitative assessment of chest CT patterns in COVID-19 and bacterial pneumonia patients: a deep learning perspective. J. Korean Med. Sci. 2021;36(5):e46. doi: 10.3346/jkms.2021.36.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., Van Der Laak J.A., Van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Mahase, E., 2020. Coronavirus COVID-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. 10.1136/bmj.m641 [DOI] [PubMed]
- Mei X., Lee H.-C., Diao K.-y., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.-Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., Lui M.M.-s., Lo C.S.-Y., Leung B., Khong P.-L., et al. Imaging profile of the COVID-19 infection: Radiologic findings and literature review. Radiol. Cardiothorac. Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- van den Oord, A., Li, Y., Vinyals, O., 2019. Representation learning with contrastive predictive coding. 1807.03748.
- Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- Pathak D., Krähenbühl P., Donahue J., Darrell T., Efros A.A. 2016 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2016, Las Vegas, NV, USA, June 27-30, 2016. IEEE Computer Society; 2016. Context encoders: feature learning by inpainting; pp. 2536–2544. [DOI] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. International Conference on Medical iImage Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shi Y. Lung infection quantification of COVID-19 in CT images with deep learning. Med. Phys. 2021;48(4):16331645. doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2021;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- Shi F., Xia L., Shan F., Song B., Wu D., Wei Y., Yuan H., Jiang H., He Y., Gao Y., et al. Large-scale screening to distinguish between COVID-19 and community-acquired pneumonia using infection size-aware classification. Phys. Med. Biol. 2021;66(6):065031. doi: 10.1088/1361-6560/abe838. [DOI] [PubMed] [Google Scholar]
- Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Wang R., Zhao H., Zha Y., Shen J., Chong Y., Yang Y. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol.Bioinform. 2021:1. doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Jin S., Yan Q., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. Appl. Soft Comput. 2021;98:106897. doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, china. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., Xu B. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) Eur. Radiol. 2021 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- Xie W., Jacobs C., Charbonnier J.-P., van Ginneken B. Relational modeling for robust and efficient pulmonary lobe segmentation in CT scans. IEEE Trans. Med. Imaging. 2020;39(8):2664–2675. doi: 10.1109/TMI.2020.2995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Chen B., Zhang Z., Guan W., Ling Z., et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol.Imaging. 2020;47:1275–1280. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Zhu X., Huang J. Medical Image Computing and Computer Assisted Intervention - MICCAI 2019 - 22nd International Conference, Shenzhen, China, October 13-17, 2019, Proceedings, Part I. Springer; 2019. Deep multi-instance learning for survival prediction from whole slide images; pp. 496–504. [DOI] [Google Scholar]
- Zhang D., Meng D., Han J. Co-saliency detection via a self-paced multiple-instance learning framework. IEEE Trans. Pattern Anal. Mach.Intell. 2016;39(5):865–878. doi: 10.1109/TPAMI.2016.2567393. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Wu, C., Zhang, Z., Zhu, Y., Lin, H., Zhang, Z., Sun, Y., He, T., Mueller, J., Manmatha, R., Li, M., Smola, A., 2020a. Resnest: split-attention networks. 2004.08955.
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433.e11. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Isola P., Efros A.A. Computer Vision - ECCV 2016 - 14th European Conference, Amsterdam, The Netherlands, October 11-14, 2016, Proceedings, Part III. Springer; 2016. Colorful image colorization; pp. 649–666. [DOI] [Google Scholar]
- Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from china. Radiology. 2020;296(2):E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32083985