Abstract
The recognition of homologous recombination deficiency (HRD) as a frequent feature of high-grade serous ovarian cancer (HGSOC) has transformed treatment paradigms. Poly(ADP-ribose) polymerase inhibitors (PARPis), developed based on the rationale of synthetic lethality that predicates antitumor efficacy in tumors harboring underlying HRD, now represents an important class of therapy for HGSOC. Recent data have drawn attention to the assessment of homologous recombination DNA repair (HRR) as a prognostic and predictive biomarker in HGSOC, leading to increasing debate on the optimal means of defining and evaluating HRD, both genotypically and phenotypically. At present, clinical-grade assays such as myChoice CDx and FoundationOne CDx are approved companion diagnostics which can identify patients with HRD-positive HGSOC by diagnosing a ‘genomic scar’ reflecting underlying genomic instability. Yet despite the rapid maturation of this field, tumoral HRD status has been recognized to be dynamic over time and with treatment pressure. In practice, this means that restoration of HRR through mechanisms of platinum and PARPi resistance are not adequately represented by genomic scar assays, and contribute toward discordance with clinical PARPi response, or lack-thereof. It is thus critical that HRD testing is optimized to address the controversies of diverse HRD testing methodology, appropriate thresholds for HRD identification, and relevant timepoints for HRD testing, in order to realize the potential for PARPis to maximally benefit patients with HGSOC. Here, we discuss the premise of HRD testing in HGSOC, current methodologies for HRD identification and their performance in the clinic, highlight upcoming strategies, and discuss the challenges faced in moving this field forward.
Key words: homologous recombination deficiency, ovarian cancer, poly(ADP-ribose) polymerase inhibitors, biomarkers
Highlights
-
•
The assessment of HRD is as an important prognostic and predictive biomarker in HGSOC.
-
•
HRD assays diagnosing a ‘genomic instability scar’ can identify patients with HGSOC likely to benefit from frontline PARPi.
-
•
Future HRD assays must address diverse testing methodology, optimal thresholds, and functional HRR status.
Introduction
The relevance of homologous recombination deficiency in ovarian cancer
DNA damage is repaired by multiple interconnected pathways. Of these, homologous recombination repair (HRR) represents a central high-fidelity DNA damage repair system responsible for reparation of DNA double-stranded breaks (DSBs) and interstrand crosslinks in a slow, specific, complex, and accurate fashion.1 Functional defects in HRR, termed homologous recombination deficiency (HRD), lead to over-reliance of DSB repair on nonhomologous end joining, single-strand annealing, or microhomology-mediated end joining pathways,2 which represent low-fidelity and error-prone alternate DNA repair systems. The Cancer Genome Atlas (TCGA) project has described that ∼50% of high-grade serous ovarian cancer (HGSOC) exhibit HRD, through a myriad of underlying mechanisms,3 some of which remain poorly defined. Most commonly, loss-of-function mutations and epigenetic modification in BRCA1/2 or genes encoding other key players in the HRR pathway, including RAD51C/D, PALB2, ATM, H2AX, MRE11, RPA, BRIP1, BARD1, RAD51, and Fanconi anemia genes, have been recognized as key causes of HRD in HGSOC. Over time, unrepaired or inaccurately repaired DSBs ultimately lead to the accumulation of genomic aberrations such as insertions and deletions, copy number alterations, or structural chromosomal rearrangements, manifesting as genomic instability which drives carcinogenesis and progression, meanwhile leaving a footprint that may be detected as a ‘genomic scar’.4
As a biomarker, HRD holds both predictive and prognostic value in HGSOC. Poly(ADP-ribose) polymerase inhibitors (PARPis) were developed based on their predicated synthetic lethality in the context of HRD-positive cells. Poly(ADP-ribose) polymerase 1 (PARP1) is an enzyme with pleiotropic cellular functions5 but is best known for its role in the base excision pathway repair of single-strand DNA breaks.2 PARPis trap PARP1 at the sites of single-strand DNA breaks, preventing efficient repair and causing protein–DNA adducts to be processed into DSBs, leading to further genomic instability and cellular death in BRCA1/2-mutant or other HRD-affected cells that are already impaired in their DSB repair capacity. Based on this rationale, HRD has been identified as potential predictive biomarker for PARPi therapy in HGSOC, breast, and prostate cancers.6, 7, 8 In addition, in newly diagnosed advanced ovarian cancer, higher HRD scores have been associated with improved progression-free survival (PFS)7,9 (Table 1), indicating a prognostic significance to this marker.
Table 1.
Key results from reported prospective trials investigating HRD status as a biomarker for PARPi response
| Trial name/study number | PARPi used | Treatment setting | Study population | HRD testing method | Results |
|---|---|---|---|---|---|
| SOLO1 (NCT01844986) | Olaparib | Maintenance therapy after CR/PR to frontline platinum-based chemotherapy | Advanced newly diagnosed OC patients with a deleterious or suspected deleterious BRCA1/2 mutation | Germline or somatic BRCA1/2 sequencing | Median PFS for olaparib versus placebo: 56.0 versus 13.8 months (HR 0.33, 95% CI 0.25-0.43) |
| PRIMA (NCT02655016) | Niraparib | Advanced newly-diagnosed OC patients at high risk for recurrence | Myriad myChoice CDx (HRD+ GIS ≥42) |
Intention-to-treat: Median PFS for niraparib versus placebo: 13.8 versus 8.2 months (HR 0.62, 95% CI 0.50-0.76) BRCAm: Median PFS for niraparib versus placebo: 22.9 versus 10.9 months (HR 0.40, 95% CI 0.27-0.62) HRD+/BRCAwt: Median PFS for niraparib versus placebo: 19.6 versus 8.2 months (HR 0.50, 95% CI 0.31-0.83) HRp: Median PFS for niraparib versus placebo: 8.1 versus 5.4 months (HR 068, 95% CI 0.49-0.94) HRnd: Median PFS for niraparib versus placebo: not reported (HR 085, 95% CI 0.51-1.43) |
|
| PAOLA1 (NCT02477644) | Olaparib + bevacizumab | Advanced newly diagnosed patients with OC | Myriad myChoice CDx (HRD+ GIS ≥42) |
Intention-to-treat: Median PFS for olaparib versus placebo: 22.1 versus 16.6 months (HR 0.59, 95% CI 0.49-0.72) BRCAm: Median PFS for olaparib versus placebo: 37.2 versus 21.7 months (HR 0.31, 95% CI 0.28-0.66) HRD+/BRCAwt: Median PFS for olaparib versus placebo: 28.1 versus 16.6 months (HR 0.43, 95% CI 0.28-0.66) HRp + HRnd: Median PFS for olaparib versus placebo: 16.9 versus 16.0 months (HR 0.92, 95% CI 0.72-1.17) |
|
| VELIA (NCT02470585) | Veliparib | Advanced newly diagnosed patients with OC | Myriad myChoice CDx (HRD+ GIS ≥33) |
Intention-to-treat: Median PFS for veliparib versus placebo: 23.5 versus 17.3 months (HR 0.68, 95% CI 0.56-0.83) BRCAm Median PFS for veliparib versus placebo: 34.7 versus 22 months (HR 0.44, 95% CI 0.28-0.68) HRD+/BRCAwt: Median PFS for veliparib versus placebo: 22.9 versus 19.8 months (HR 0.74, 95% CI 0.52-1.06) HRp: Median PFS for veliparib versus placebo: 15.0 versus 11.5 months (HR 0.81, 95% CI 0.60-1.09) |
|
| ARIEL2 Part 1 (NCT01891344) | Rucaparib | Maintenance therapy after CR/PR to platinum-based chemotherapy for relapsed disease | Recurrent platinum-sensitive OC | Foundation Medicine T5 NGS assay (genomic LOH high ≥14%) |
BRCAm: Median PFS 12.8 months (95% CI 9.0-14.7) LOH high: Median PFS: 5.7 months (95% CI 5.3-7.6) LOH low: Median PFS: 5.2 months (95% CI 3.6-5.5) |
| ARIEL3 (NCT01968213) | Recurrent platinum-sensitive OC | Foundation Medicine T5 NGS assay (genomic LOH high ≥16%) |
Intention-to-treat: Median PFS for rucaparib versus placebo: 10.8 versus 5.4 months (HR 0.36, 95% CI 0.30-0.45) BRCAm: Median PFS for rucaparib versus placebo: 16.6 versus 5.4 months (HR 0.23, 95% CI 0.16-0.34) LOH high: Median PFS for rucaparib versus placebo: 13.6 versus 5.4 months (HR 0.32, 95% CI 0.24-0.42) |
||
| NOVA (NCT01847274) | Niraparib | Recurrent platinum-sensitive OC | Myriad myChoice CDx (HRD+ GIS ≥42) |
BRCAm: Median PFS for niraparib versus placebo: 21.0 versus 5.5 months (HR 0.27, 95% CI 0.17-0.41) HRD+/BRCAwt: Median PFS for niraparib versus placebo: 12.9 versus 3.8 months (HR 0.38, 95% CI 0.24-0.59) BRCAwt: Median PFS for niraparib versus placebo: 9.3 versus 3.9 months (HR 0.45, 95% CI 0.34-0.61) |
|
| Study19 (NCT00753545) | Olaparib | Recurrent platinum-sensitive OC | N/A |
Intention-to-treat: Median PFS for olaparib versus placebo: 8.4 versus 4.8 months (HR 0.35, 95% CI 0.25-0.49) |
|
| SOLO2 (NCT01874353) | Olaparib | Recurrent platinum-sensitive OC | Germline or somatic BRCA1/2 sequencing |
Intention-to-treat: Median PFS for olaparib versus placebo: 19.1 versus 5.5 months (HR 0.30, 95% CI 0.22-0.41) |
|
| QUADRA (NCT02354586) | Niraparib | Treatment of relapsed disease after three or more prior chemotherapy regimens | Relapsed OC that has received three or more prior chemotherapy regimens, irrespective of platinum-sensitivity status | Myriad myChoice CDx (≥42) and germline BRCA status testing |
Platinum-sensitive, BRCAm: ORR 39% Platinum-sensitive, HRD+: ORR 26% Platinum-sensitive, HRDp or HRDnd: ORR 4% Platinum-resistant/refractory, BRCAm: ORR 27% Platinum-resistant/refractory, HRD+: ORR 10% Platinum-resistant/refractory, HRDp or HRDnd: ORR 3% |
| Study42 (NCT01078662) | Olaparib | Relapsed germline BRCA1/2-mutated OC that has received three or more prior chemotherapy regimens | Germline BRCA testing | ORR 34% Median duration of response: 7.9 months (95% CI 5.6-9.6) |
|
| Study10 (NCT01482715) | Rucaparib | Treatment of relapsed disease after two to four prior chemotherapy regimens | Relapsed germline BRCA1/2-mutated OC that has received two to four prior chemotherapy regimens | Germline BRCA testing | ORR 59.5% |
BRCAm, BRCA-mutant; BRCAwt, BRCA wild type; CI, confidence interval; CR, complete response; HR, hazard ratio; HRnd, HR not detected; HRp, HR proficient; NGS, next-generation sequencing; OC, ovarian cancer; ORR, objective response rate; PFS, progression-free survival; PR, partial response.
Shifting treatment paradigms for PARPi therapy in ovarian cancer
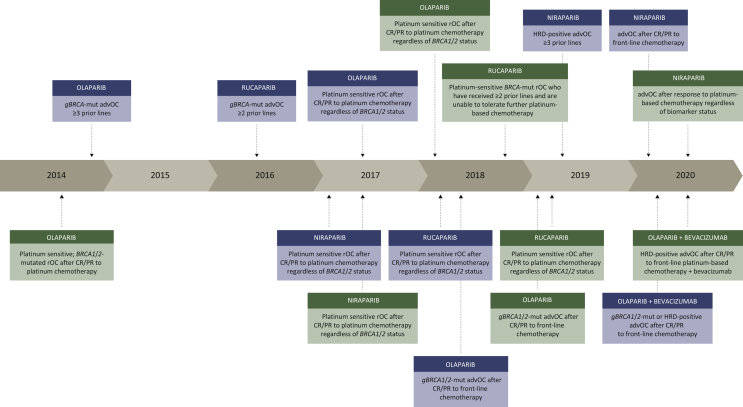
Olaparib was the first-in-class PARPi to receive United States Food and Drug Administration (US FDA) approval in 2014 for the treatment of patients with advanced ovarian cancer with deleterious or suspected deleterious germline BRCA1/2 mutation, who had been treated with three or more prior lines of chemotherapy (Figure 1). In this population, the objective response rate (ORR) was 34% and median duration of response was 7.9 months [95% confidence interval (CI) 5.6-9.6].10 As monotherapy in heavily pretreated patients, rucaparib was similarly approved for the treatment of patients with BRCA-mutated recurrent ovarian cancer after two or more prior treatment lines, based on phase II data showing an ORR of 59.5% and median duration of response of 7.8 months (95% CI 5.6-10.5)11 (Table 1 and Figure 1).
Figure 1.
Timeline of US Food and Drug Administration (US FDA) PARPi approvals.
PARPi approvals by the US FDA (described in blue boxes) and European Medicines Agency (EMA) (described in green boxes) between 2014 and 2020.
AdvOC, advanced ovarian cancer; CR, complete response; gBRCA-mut, germline BRCA1/2 mutation; HRD, homologous recombination deficiency; PR, partial response; rOC, recurrent ovarian cancer.
Since then, significant strides have been made in advancing the use of PARPis to an earlier timepoint in treatment paradigms for ovarian cancer. Platinum sensitivity has been recognized as an indicator of PARPi efficacy, expanding the group of patients benefiting from PARPi use outside those with BRCA1/2 mutations. The first key trial, Study19, evaluated olaparib monotherapy versus placebo as maintenance treatment in patients with relapsed ovarian cancer in complete response (CR) or partial response (PR) after platinum-based chemotherapy, confirming PFS improvement by 65%12 and led to early European Medicines Agency (EMA) approval for this indication in 2014 (Figure 1). Similar findings were reported for the use of niraparib13 and rucaparib14 compared with placebo, and led to their respective EMA and US FDA approvals between 2017 and 2019 (Figure 1). Although all patients in the intention-to-treat populations of Study19, ARIEL3, and NOVA benefited from the use of PARPi maintenance therapy versus placebo in this setting, an evident trend emerged where the magnitude of PFS benefit differed incrementally between HRD-negative, HRD-positive, and BRCA1/2-mutant subgroups (Table 1).
In the frontline setting for BRCA1/2-mutated advanced ovarian cancer, at a median of 5 years follow-up, the SOLO1 trial demonstrated significant improvements in median PFS [56 versus 13.8 months, hazard ratio (HR) 0.33, 95% CI 0.25-0.43]15 and time to second objective disease progression (PFS2) (not reached versus 41.9 months, HR 0.50, 95% CI 0.35-0.72)16 (Table 1), when patients received maintenance olaparib compared with placebo after frontline chemotherapy. Overall, >50% of patients who were in CR after frontline chemotherapy remained free from relapse 5 years later, making it reasonable to believe that some BRCA1/2-mutated patients may indeed achieve cure by the early introduction of olaparib. Furthermore, the PAOLA-1 trial randomized patients to olaparib plus bevacizumab maintenance therapy versus placebo plus bevacizumab while the PRIMA trial randomized patients to niraparib versus placebo, as maintenance therapy after first-line chemotherapy.7,17 Both trials utilized myChoice CDx genomic instability score (GIS) with a cut point ≥42 to determine HRD status.7,17 Collectively, results from these trials have led to EMA and US FDA approvals of PARPi maintenance therapy after frontline chemotherapy in advanced ovarian cancer (Figure 1).
Selecting patients for PARPis: how can we best identify HRD?
To date, no uniformly accepted gold standard for HRD assessment exists.18 Present clinical methods for detecting HRD are limited to assessing for genomic perturbations within tumors resulting from mutations within the HRR pathway, or by detecting a genomic scar reflecting underlying genomic instability. Yet, patients who are found to be HRR proficient/HRD negative have also been found to respond to PARPi therapy,12, 13, 14 with the implication that current HRD assays are imperfect selective biomarkers at best. In this section, we discuss the current and emerging methods for HRD detection, and their relevance to current practice.
Genomic perturbations
BRCA1/2 mutations
In trials investigating maintenance PARPi in frontline and platinum-sensitive relapsed ovarian cancer, the subgroup of patients harboring BRCA1/2 mutation has consistently emerged as those who derive the greatest magnitude of benefit from PARPi addition (Table 1). Germline mutations in BRCA1/2 are highly penetrant mutations which are found in 13%-15% of ovarian cancers,19 leading to germline genetic testing and counselling being universally recommended for all women with nonmucinous epithelial ovarian cancer.20 Germline genetic testing by direct sequencing or panel testing is relatively inexpensive and has acceptable turnaround time but has ostensible limitations in its narrow scope of HRD identification, as it will overlook epigenetic modifications and inactivation of other HRR pathway genes, as well as HRD attributable to somatic events or other poorly defined non-BRCA mechanisms. Somatic BRCA1/2 mutations are found in an additional 5%-7% of ovarian cancer3 and have been described to be early events in carcinogenesis, based on retrospective analysis from Study19 showing that majority of cases had clonal, biallelic inactivation.21 Across several phase III trials and meta-analyses,22 somatic BRCA1/2 mutations have been associated with similar clinical outcomes compared with germline BRCA1/2 mutations. For example, on the ARIEL2 trial, response rates (74% and 85%, respectively) and PFS for patients with somatic and germline BRCA1/2 mutation were similar.23
Non-BRCA HR pathway mutations
Beyond BRCA1/2, germline or homozygous somatic aberrations in genes encoding HRR pathway proteins such as RAD51B/C/D, BRIP1, PALB2, NBN, ATM, CHK1/2, CDK12, and Fanconi Anemia genes, among others, are thought to potentially confer an HRD or ‘BRCAness’ phenotype in view of their known cooperative role in HRR.1 Up to 30% of ovarian cancers may harbor mutations related to the HRR pathway.3 Preclinical data have been reported suggesting that RAD51C/D deficiency and mutations in genes including ATM and CHK1/2 may confer synthetic lethality to strategies targeting effective DNA repair, presumably through HRD.24,25 Yet, recent conflicting data have been reported by Takaya and colleagues based on their analyses of ovarian cancer TCGA data which found that the presence of ATM, ATR, FANCA, FANCD2, FANCM, or PALB2 mutation was not associated with high tumor loss of heterozygosity (LOH) scores, HRD, or platinum sensitivity.26 Interestingly, only homozygous deletions in CHK1 and PTEN led to high HRD-associated LOH scores, outside of BRCA1/2 germline or somatic mutations.26
Despite this, clinical data consisting of anecdotal reports as well as retrospective analyses of the Study10 and ARIEL2 trials have described that mutations or methylation of RAD51C were associated with long-term responses to PARPi therapy.27, 28, 29 Similar findings were reported from Study19, whereby the retrospectively defined cohort of patients harboring mutations in HRR-related non-BRCA1/2 genes such as CDK12, RAD51B, and BRIP1 benefitted similar (HR for PFS 0.21, 95% CI 0.04-0.86) to those harboring a BRCA1/2 mutation (HR for PFS 0.18, 95% CI 0.10-0.31).30 However, owing to the rarity of individual mutations, studies evaluating the predictive relevance for non-BRCA/HRR pathway mutations of PARPi response have been inadequately powered for firm conclusions to be drawn, and these studies while intriguing, should be interpreted with caution. Overall, it is likely that individual HRR gene mutations have distinct sensitivities to platinum and PARPi therapy, and further data on the appropriateness of PARPi addition in specific genomic contexts are required.
Epigenetic modifications
Epigenetic silencing of BRCA1 and other HRR-related genes such as RAD51C accounts for a further 11%-15% of HRD-positive HGSOC, through aberrant methylation of cytosine residues of cytosine–phosphate–guanine (CpG) dinucleotides residing within promoter regions, leading to reduced gene expression.31 Corroborative immunohistochemistry (IHC) studies have validated that epigenetic BRCA1 silencing led to a lack of expression of BRCA1 protein.31 Both BRCA1-methylated HGSOC and RAD51C-methylated HGSOC harbor high HRD scores.32,33 BRCA1 epigenetic modifications have also been associated with BRCA-deficient genomic signatures and observed to have similar effects on HRR as BRCA1/2 mutations.34 Such epigenetic modifications would not be picked up on present-day next-generation sequencing methods. Furthermore, the clinical implications of such epigenetic modifications of HRR-related genes appear to be more variable. Conflicting data exist for the prognostic and predictive significance of epigenetic HRR-related gene modification. Although some studies have reported that BRCA1/RAD51C methylation is associated with a good prognosis,35, 36, 37 other studies have described contradictory findings and poor reliability of this as a biomarker for PARPi response.35,38,39 In a recent study of TCGA samples, epigenetically modified HRD cases were noted to have a similarly poor prognosis as HRD-negative cases.26
Studies have, however, called to attention that zygosity of BRCA1 promoter methylation is a critical factor in determining its relationship with PARPi response.40 In HGSOC patient-derived xenograft models, it was noted that demethylation of a single methylated BRCA1 copy was able to restore HRR proficiency and reduce sensitivity to PARPi.40 Exposure to chemotherapy has also been shown to result in demethylation of previously methylated BRCA1 copies,41,42 and this may occur more easily than the mechanisms of resistance described in the context of pathogenic BRCA1/2 mutations, such as the acquisition of a reversion mutation.43 This sheds light on the importance of using precise techniques to assign BRCA1 methylation status, including its zygosity, as well as the need to consider the timepoint (e.g. before or after chemotherapy) when the sample being analyzed was taken, in future studies evaluating HRR promoter methylation as a potential biomarker.
Genomic signatures
Clinical-grade HRD assays detecting ‘genomic scars’
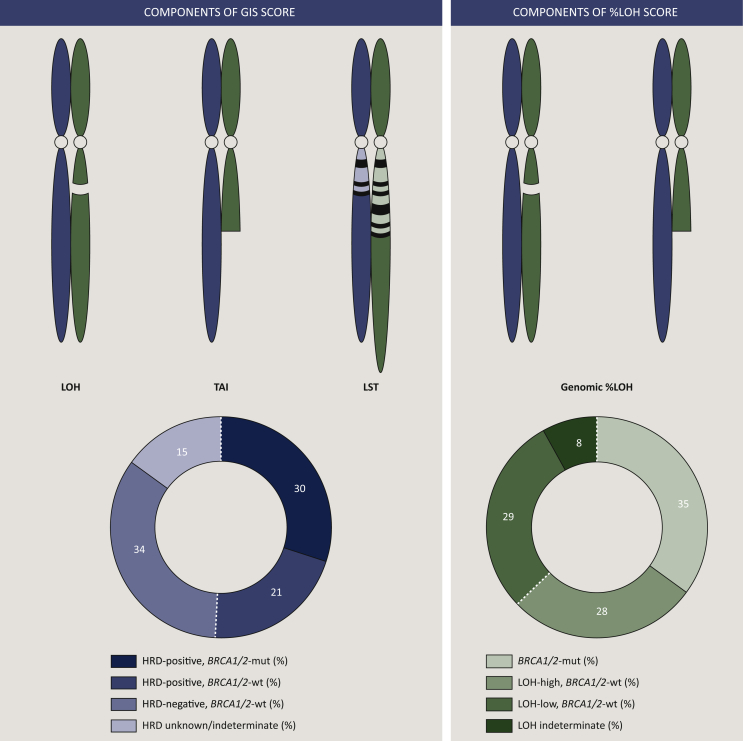
‘Genomic scars’ serve as an indirect measure of HRD as they represent a permanent historical footprint of genomic changes induced by DNA repair deficiency,18 irrespective of the underlying etiology. Prospectively validated HRD genomic scar assays evaluate for the percentage of genomic regions with LOH determined through tumor single-nucleotide polymorphism (SNP) sequencing (FoundationOne CDx, Foundation Medicine) or through a GIS calculated by combining three factors obtained from allele-specific copy number profiles for SNP–LOH, telomeric allelic imbalance (TAI), and large-scale transitions (LSTs) (myChoice CDx, Myriad Genetics) (Figure 2).
Figure 2.
Current clinical-grade genomic scar assays to determine HRD positivity.
Left panel: myChoice CDx (Myriad Genetics) uses a proprietary formula to calculate a genomic instability score (GIS) based on three genomic elements: LOH, TAI, and LST. GIS ≥42 and/or BRCA1/2 mutation status would be denoted as homologous recombination deficiency (HRD) positivity. On the PRIMA trial (NCT02655016), investigating frontline maintenance niraparib in patients with advanced ovarian cancer who have complete or partial response to frontline chemotherapy, the proportion of enrolled patients who were denoted HRD positive, negative, and indeterminate/unknown is presented. Right panel: FoundationOne CDx (Foundation Medicine) includes the %LOH score measuring the percentage of genomic LOH as a marker of HRD positivity, %LOH ≥16 is denoted as %LOH-high. On the ARIEL3 trial (NCT01968213), investigating switch maintenance rucaparib in patients with platinum-sensitive relapsed ovarian cancer who have complete or partial response to the most recent line of platinum-based chemotherapy, the proportion of enrolled patients who were denoted BRCA1/2 mutant, %LOH-high, -low, and -indeterminate are presented.
LOH, loss of heterozygosity; LST, large-scale transition; mut, mutant; TAI, telomeric allelic imbalance; wt, wild type.
Prior studies evaluated specific genomic scarring patterns in patients with known BRCA1/2 mutation, and identified LOH, TAI, and LST to be independently correlated with BRCA1/2 deficiency and platinum sensitivity. LOH, LST, and TAI are highly correlated with each other and reflect increasing genomic instability. LOH refers to permanent loss of one parent's contributed allele copy at a specific locus, leading to homozygosity at that genomic site.44 In HGSOC, LOH regions >15 megabases (Mb) but less than the entire chromosomal length was correlated with BRCA1/2 defects.33 TAI refers to allelic imbalance extending to the subtelomeric region >11 Mb in size and has been correlated with sensitivity to platinums in patients with HGSOC who are BRCA1/2 wild type.45 LSTs refer to allelic imbalance >10 Mb in size between adjacent genomic regions due to translocations or copy gains/losses, and these have similarly been found to be increased in BRCA1-inactivated basal-like breast cancers.46 The combination of these three measures of genomic instability is associated with greater prognostic value than each of the individual components47 (Figure 2).
The GIS from myChoice CDx represents the sum of LOH, LST, and TAI using Myriad's proprietary score. A GIS score of ≥42 has been chosen as the threshold to define HRD-positivity as this represents the 5th percentile of a set of biallelic inactivated BRCA1/2 tumors.48 In addition, myChoice CDx provides BRCA1/2 mutation and rearrangement analysis within its report. Tumors that have GIS ≥42 and/or pathogenic BRCA1/2 alterations are considered to be HRD positive. By contrast, HRD positivity may be inferred using FoundationOne CDx as %LOH greater than the initially predefined threshold of ≥14% based on genomic data from the TCGA.49 This cut point was prospectively evaluated on the ARIEL2 trial23 but later on adjusted to %LOH ≥16% when applied to the ARIEL3 trial.14 As FoundationOne CDx includes tumor next-generation sequencing of 315 cancer-related genes, pathogenic mutations in BRCA1/2 and many HRR-related genes are reported simultaneously.
To date, a number of randomized clinical trials of advanced ovarian cancer have incorporated the myChoice CDx or Foundation Medicine %LOH to define tumor HRD status (Table 1). Of note, trial subgroup analyses based on HRD status was often performed as predefined exploratory endpoints which were not adequately powered or adjusted, precluding definitive conclusions to be reached. Yet, across various treatment settings, patients with BRCA1/2-mutation consistently retrieved the greatest degree of benefit from PARPi therapy, compared with subgroups of patients found to be BRCA1/2 wild type/GIS-high or BRCA1/2 wild type/%LOH-high, which in turn appeared to benefit more than subgroups of patients who were BRCA1/2 wild type/HRD negative7,9,12, 13, 14,17,23,28,50 (Table 1). In the platinum-sensitive relapsed setting, despite the ability for %LOH and myChoice CDx to molecularly define the magnitude of patient benefit from maintenance PARPi in this setting, these assays were unable to discriminate an HRD-negative subgroup of patients that failed to benefit from PARPi.13,14
Mutational signatures
Mutational signatures are another means of evaluating the impact of HRD on the genome by quantifying the type of mutations found and the patterns of nucleotide transitions created as a result.51 Signature3 is a mutational signature based on single base substitutions and is characterized by a high number of larger deletions with overlapping microhomology at breakpoint junctions. Signature3 has been associated with BRCA1/2 mutation and BRCA1 promoter methylation in a number of cancer types51,52 and has been shown to correlate with prognosis and response to platinum therapy53 in HGSOC, leading to it being proposed as a potential HRD biomarker.54 Other mutational signature-based assays such as HRDetect may provide improved sensitivity and specificity. Here, whole-genome sequencing is used, and an algorithm incorporates a weighted aggregate of six HRD-associated signatures predictive of BRCA1/2 deficiency into a single score (microhomology-mediated deletions, base-substitution signature3, rearrangement signature3, rearrangement signature5, HRD index, base-substitution signature8).55 This was initially established in ∼600 breast cancer samples and later validated in breast, ovarian, and pancreatic cancer samples.55 Within the cohort of ovarian cancer samples, HRDetect had a sensitivity approaching 100% to identify BRCA1/2 null cancers, whereas GIS had only 60% sensitivity.48 However, similar to other genomic scar HRD assays, HRDetect and signature3 remain historical representations of HRD effects and will not reflect acquired platinum or PARPi-resistance mechanisms. Clinical validation is still wanted to see if this approach truly holds more promise for improved precision in HRD assessment compared with current clinical assays.
Functional measures of HRD
Platinum-sensitivity status
Real-time HRD assessments may overcome the limitation of genomic scar-based assays by providing a dynamic and current readout of tumor HRR. Sensitivity to platinum salts has been observed to be a surrogate marker of HR proficiency and in itself a predictive biomarker for PARPi. As described earlier, platinum-sensitivity status was a superior biomarker to predict PARPi benefit in the NOVA, ARIEL3, and Study19 trials, compared with myChoice CDx or %LOH.10,12,23 On the phase II QUADRA study which evaluated niraparib monotherapy in patients with recurrent HGSOC after three or more lines of treatment, platinum-sensitive, -resistant and -refractory patients were enrolled. Among BRCA-wild-type patients, who were found to be HRD positive based on myChoice CDx (GIS ≥42), ORR in the platinum-sensitive subgroup of patients was 20% but only 2.4% in patients who were platinum resistant/refractory, which approximated that of the BRCA-wild-type/HRD-negative subgroup (ORR 3%).50
RAD51 foci
Preclinically, the inability to form nuclear RAD51 foci is commonly used to estimate HRD. RAD51 is a downstream protein in the HRR pathway that is loaded onto sites of DSB to facilitate DNA strand invasion into the sister chromatid in cooperation with mediator protein complexes, especially BRCA1 and 2. In the presence of DNA damage, RAD51 colocalization at sites of DSBs with breast cancer 2 (BRCA2) can be visualized in vitro as distinct subnuclear foci.56 BRCA1/2 gene defects and HRD cells are characterized by the reduction of, or inability to form RAD51 foci, which is a functional phenotype irrespective of the underlying cause of HRD.57 Among 39 HGSOC samples on one study, a functional HR assay based on RAD51 foci detection found 26% of samples to be HRD, while ovarian cancers of nonserous histotypes were all HR proficient.58 Several preclinical and small clinical studies have also described that reduced RAD51 foci is associated with PARPi response in ovarian cancer.59,60 Furthermore, in the presence of acquired reversion mutations that restore HRR in the context of germline BRCA1/2-mutated breast cancer samples, RAD51 foci were noted to be present, indicating functional HRR restoration61 and the potential for this assay to overcome a major limitation of genomic scar assays. However, translating a real-time RAD51 foci assay into clinical practice remains a challenge. Urgent retrospective and prospective validation of RAD51 foci visualization protocols is required to better understand the clinical validity of this approach.
HRD testing in the clinic—practical considerations
Choice of HRD genomic scar test
At present, the myChoice CDx and FoundationOne CDx assays are the only prospectively validated commercially available tests for assessment of HRD status. It is important to recognize that these are distinct assays that differ in their methodology for detecting a genomic scar, and they are unlikely to capture an identical patient set. Prospective, head-to-head comparative data between the performance of the two assays are still lacking. One study has examined the interchangeability of the myChoice CDx assay with %LOH alone in identifying HRD-positive tumors.62 From 3336 commercial ovarian cancer samples in the MyChoice laboratory and 176 ovarian cancer samples from the SCOTROC4 trial, GIS profiles were reconstructed using the Myriad proprietary algorithm and paired %LOH was calculated using published methods. Among 3209 samples which were BRCA1/2 wild type, the 53% of MyChoice GIS-positive tumors were called HRD negative by %LOH (presumed false negatives), while only 4% of %LOH-positive tumors were called HRD negative by MyChoice GIS. It is important to note that this retrospective study had chosen to use the GIS cut point of ≥33 rather than ≥42. When the GIS cut point of ≥42 was used and concordance between the tests was compared across all tumors (both BRCA1/2 mutated and BRCA1/2 wild type), the percentage positive agreement between MyChoice GIS and %LOH was 64.9% and 82.5% (for the commercial sample cohort and SCOTROC4 cohorts, respectively), while the percentage negative agreement was 96.6% and 95.8%, respectively.62 Another published retrospective exploratory analysis of BRCA1/2-mutant tumor samples from the SOLO1 trial showed that 23% of evaluable samples would have been classified as %LOH <16% by FoundationOne CDx, despite the presence of a germline or tumor BRCA1/2 mutation in all enrolled patients.57 These data underscore the fact that a degree of overlapping sensitivity exists between these two genomic scar detection methods, but that they cannot be considered equivalent in routine clinical practice.
Although FoundationOne CDx %LOH has yet to be clinically validated as a selective biomarker for PARPi maintenance in the frontline setting, FoundationOne CDx may be useful in providing additional tumor genomic information to the ordering physician. Aside from next-generation sequencing results of important HRR-related genes, this assay would also provide the tumor mutational burden and microsatellite instability status, as well as optional programmed death-ligand 1 (PD-L1) IHC testing. In recent years, the US FDA has approved the use of pembrolizumab for microsatellite instability-high and tumor mutational burden-high (defined as ≥10 mutations/Mb) unresectable or metastatic solid tumors. Furthermore, pembrolizumab monotherapy has been evaluated in patients with advanced ovarian cancer in the KEYNOTE-100 trial, whereby a PD-L1 combined positive score ≥10 was associated with improved ORR (18.2%, 95% CI 5.2-40.3) and OS.63 Therefore, information on these three immunotherapy biomarkers, particularly in the later-line setting, may allow additional treatment options to become available to patients.
Determining the choice of maintenance therapy in a resource-limited context
Both myChoice CDx and FoundationOne CDx remain costly assays, with reimbursement policies varying from country to country. Likewise, there is a significant financial challenge for patients who have to partially or fully cover the cost of maintenance PARPis with or without bevacizumab following frontline chemotherapy, which will be further exacerbated when both are used in combination. Test availability and turnaround times may also differ between countries depending on country-specific test roll-out and logistical arrangements. There is a lack of data regarding the cost-effectiveness of HRD genomic scar testing, particularly in countries with limited resources.
From a practical perspective, the crucial distinction is between functionally HRD-positive versus HRD-negative tumors, because this could help distinguish the patients that are more likely to benefit from a PARPi or bevacizumab-based maintenance strategy in the frontline setting. In situations where access to HRD genomic scar testing is either unavailable or unaffordable to patients, evaluation of platinum sensitivity in patients receiving neoadjuvant chemotherapy for advanced HGSOC could be a viable strategy to select patients that are phenotypically more likely to benefit from PARPi maintenance. Platinum sensitivity as a functional biomarker has already been validated and shown to be superior to genomic scar assays in predicting PARPi benefit in the recurrent platinum-sensitive setting,12, 13, 14 and it would seem reasonable to extrapolate this to the frontline setting. For example, patients who have exquisite response to neoadjuvant platinum-based chemotherapy may be more likely to harbor HRD and potentially benefit from maintenance PARPi. Conversely, patients with upfront inoperable disease who did not achieve RECIST confirmed CR or PR to initial neoadjuvant chemotherapy, or have a poor chemotherapy response score64 following histopathological evaluation at the time of interval debulking surgery, may be less likely to harbor HRD. In the context of stage 4 disease and suboptimally debulked disease with poor neoadjuvant platinum response, these patients would also fall into the high-risk disease category as defined by the ICON7 trial,65 and may be more likely to benefit from bevacizumab addition. However, in patients who have achieved upfront optimal or suboptimal debulking with minimal residual disease that is not measurable on postoperative computed tomography scans, this approach will not be feasible and HRD testing will still be required to determine the appropriate maintenance therapy.
Germline and/or somatic testing?
A small proportion of germline BRCA1/2-mutant patients may have negative tumor BRCA1/2 status66 and hence exclusively performing tumor BRCA1/2 testing could potentially exclude patients who would benefit from frontline PARPi maintenance. Ideally, patients should be offered both tumor BRCA1/2 and HRD testing as well as germline BRCA testing. From a therapeutic perspective, however, if only one test were to be available, then a tumor BRCA/HRD test would be the most practical one in ensuring the most extensive means of identifying all patients for whom PARPi treatment is likely to be beneficial. Germline testing clearly has important implications on the patient's family members who would benefit from familial cascade testing, prophylactic interventions, and enhanced cancer screening with this genetic information. Yet, barriers remain in many countries that limit the uptake of germline genetic testing, such as the lack of protective legislation against genetic discrimination.67, 68, 69 Somatic testing would also potentially help to reduce testing hesitancy in patients who perceive a burden from the potential socioeconomic implications to themselves and their relatives from a positive germline BRCA1/2 test result.67,69
Considering these factors, a reasonable approach would be to conduct targeted germline and/or tumor BRCA1/2 mutation testing, and use HRD assays to guide treatment in patients who are subsequently found to be BRCA1/2 wild type. As early knowledge of BRCA1/2 mutation and HRD status is crucial to discuss and select an appropriate management plan for patients with advanced ovarian cancer, such testing should be performed as soon as possible after the diagnosis is made. On the PAOLA-1 trial, patients had received at least three cycles of bevacizumab concurrent with frontline platinum-based chemotherapy prior to commencing maintenance olaparib plus bevacizumab.17 Therefore, for patients who are found to be HRD positive, early receipt of this information will allow bevacizumab to be promptly added to the treatment regimen. Furthermore, to facilitate early tumor HRD testing in patients receiving neoadjuvant chemotherapy, adequate pretreatment tumor biopsies should be collected. This would also mitigate situations of dramatic tumor response which may lead to minimal residual tumor at interval debulking surgery and preclude adequate tissue for further testing.
Challenges and future directions
One important limitation of existing HRD testing options would be the optimal threshold for scores to distinguish HRD status. As cut points for these scores were developed based on retrospective analyses of BRCA1/2-mutant patients and their response to chemotherapy, further optimization of these thresholds may be required in the future.70 Furthermore, PARPis have differential PARP1/2 trapping potency, which may reflect the need for different cut point thresholds when a higher- versus a lower-potency PARPi is being utilized. Another concerning observation is the proportion of patients in whom the test is not able to determine a result (HRD-not determined),7 particularly given the cost of purchasing the test, and the time-sensitive nature of these results in guiding maintenance therapy in the frontline setting. The discordant results between the PAOLA-1 and PRIMA with respect to benefit of PARPi maintenance in HRD-negative and HRD-not determined subgroups also means the optimal maintenance strategy in these subgroups remains unknown. For patients with HRD-negative/HRD-not determined status receiving neoadjuvant chemotherapy, the degree of platinum response preoperatively could be an additional biomarker to help physicians decide between PARPi or bevacizumab maintenance, although this has yet to be formally tested in the context of a clinical trial.
Most importantly, discordant clinical responses to PARPi with HRD genomic scar assay results have been observed, which is likely due to composite reasons which are incompletely understood. HR function is dynamic over time and may vary with treatment pressure, leading to critical limitations in genomic scar assays and their ability to reflect the current functional HRD status of cancer cells. Acquired resistance mechanisms to PARPi are not adequately captured by these assays, such as BRCA1/2, RAD51C/D, or PALB2 reversion mutations that restore HRR competency,40,43,71 nor secondary somatic mutations that confer resistance to PARPi or platinum therapy,72,73 leading to apparent inconsistencies between clinical response and assay findings. Other mechanisms of resistance to PARPi have been described,71 such as heat shock protein 90 (HSP90) stabilization of BRCA1 C-terminal domains,74 upregulation of P-glycoprotein cellular efflux pumps, restoration of polyADP-ribosylation (PARylation) and replication fork protection, among others,71 all of which will not be discernable by genomic scar tests. Spatial tumor heterogeneity may be a contributing factor and future studies may need to evaluate whether multiple biopsies in a single patient show differential HRD scores. Repeat sequencing of tumors in later-line therapy may help to better address the presence or absence of reversion mutations; however, they would still not indicate the presence or absence of HRR-independent mechanisms of PARPi resistance.75 Given that real-time HRD testing will likely need repeated sampling timepoints to achieve its purpose, liquid biopsy is another approach that is deservedly being explored on ongoing trials (ATHENA NCT03522246).
On the horizon, the role of PARPis in ovarian cancer therapy is likely to continue expanding, as ongoing trials combine PARPis with immune checkpoint inhibitors, phosphoinositide 3-kinase inhibitors as well as novel inhibitors targeting the DNA damage response pathway [e.g. ataxia telangiectasia and Rad3 related (ATR), Wee1-like protein kinase (WEE1), checkpoint kinase 1 (CHK1)].76 The TOPACIO trial investigating the niraparib plus pembrolizumab combination in a mostly platinum-resistant population of ovarian cancer found that MyChoice GIS score, BRCA1/2 status, and even RAD51 foci by IHC failed to correlate with treatment response, yet mutation signature3 showed correlation with clinical benefit, though the reasons for this were unclear.77 Application of further tumor immunogenomic profiling added to the predictive potential of signature3.77 At present, it is unclear how the current HRD assays or other HRD biomarkers will fare in predicting treatment response given the potential for additive or synergistic effects between these and other classes of drugs, and it is possible that combinatorial indices may improve predictive potential.
Conclusions
Undoubtedly, HRD genomic scar tests have been useful in teasing apart the genomic heterogeneity of HGSOC as a disease. Recent trials have also brought to attention the utility of HRD testing to select patients with advanced ovarian cancer most likely to benefit from PARPi treatment in various settings. Based on current data, universal HRD testing using genomic scar tests would ideally be useful to understand individualized maintenance treatment options for patients with advanced ovarian cancer after response to frontline platinum-based chemotherapy. Yet, limited access to HRD testing in many countries, and the socioeconomic challenges related to widespread genomic testing are real-life obstacles preventing this ideal from being attained. The use of targeted tumor and germline sequencing to identify pathogenic BRCA1/2 mutation carriers and to refine, stepwise, the population of patients with advanced ovarian cancer most in need of HRD testing is one viable strategy. Use of platinum sensitivity to neoadjuvant chemotherapy as an alternative biomarker in situations where testing is not available could be another practical compromise to the ideal testing algorithm. Aside from HRD genomic scar tests, newer methods of HRD testing which consider the functional HRR status are forthcoming and eagerly awaited with the hope of expanding PARPi benefit to a wider group of patients with ovarian cancer.
Acknowledgments
Funding
DSPT is supported by the National Medical Research Council, Singapore [grant number CSAINV16may008] and Pangestu Family Foundation Gynaecological Cancer Research Fund. NYLN is supported by the National Medical Research Council, Singapore [grant number MOH-FLWSHP19may-0006].
Disclosures
DSPT reports research support from AstraZeneca, Karyopharm Therapeutics, Bayer, Roche, National Medical Research Council Singapore, Pangestu Family Foundation Gynaecological Cancer Research Fund, and Cancer Science Institute Singapore; serves on the advisory board of AstraZeneca, MSD, Roche, Bayer, Experimental Drug Development Centre (EDDC) – A∗Star Singapore, Genmab, Tessa Therapeutics, and Eisai; receives honoraria/travel support from AstraZeneca, Novartis, Roche, Merck Serono, MSD, Bayer, Genmab, Takeda, Eisai, and Clovis. NYLN reports honoraria/travel support from AstraZeneca and Janssen.
References
- 1.Lord C.J., Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 2.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H., Liu T., Zhang Z. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins J.A., Irshad S., Grigoriadis A., Tutt A. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver A.N., Yang E.S. Beyond DNA repair: additional functions of PARP-1 in cancer. Front Oncol. 2013;3:290. doi: 10.3389/fonc.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robson M.E., Tung N., Conte P. OlympiAD final overall survival and tolerability results: olaparib versus chemotherapy treatment of physician's choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann Oncol. 2019;30:558–566. doi: 10.1093/annonc/mdz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Martin A., Pothuri B., Vergote I. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 8.de Bono J., Mateo J., Fizazi K. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 9.Coleman R.L., Fleming G.F., Brady M.F. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. 2019;381:2403–2415. doi: 10.1056/NEJMoa1909707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domchek S.M., Aghajanian C., Shapira-Frommer R. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol. 2016;140:199–203. doi: 10.1016/j.ygyno.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristeleit R., Shapiro G.I., Burris H.A. A phase I-II study of the oral PARP inhibitor rucaparib in patients with germline BRCA1/2-mutated ovarian carcinoma or other solid tumors. Clin Cancer Res. 2017;23:4095–4106. doi: 10.1158/1078-0432.CCR-16-2796. [DOI] [PubMed] [Google Scholar]
- 12.Ledermann J.A., Pujade-Lauraine E. Olaparib as maintenance treatment for patients with platinum-sensitive relapsed ovarian cancer. Ther Adv Med Oncol. 2019;11 doi: 10.1177/1758835919849753. 1758835919849753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Campo J.M., Matulonis U.A., Malander S. Niraparib maintenance therapy in patients with recurrent ovarian cancer after a partial response to the last platinum-based chemotherapy in the ENGOT-OV16/NOVA trial. J Clin Oncol. 2019;37:2968–2973. doi: 10.1200/JCO.18.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman R.L., Oza A.M., Lorusso D. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee S., Moore K.N., Colombo N. 811MO – maintenance olaparib for patients (pts) with newly diagnosed, advanced ovarian cancer (OC) and a BRCA mutation (BRCAm): 5-year (y) follow-up (f/u) from SOLO1. Ann Oncol. 2020;31(suppl 4):S551–S589. [Google Scholar]
- 16.Oaknin A., Moore K., Colombo N. 4350 – Time to second progression (PFS2) and second subsequent therapy (TSST) for patients (pts) with newly diagnosed, advanced ovarian cancer (OC) and a BRCA mutation (BRCAm) treated with maintenance (mt) olaparib (ola) – Phase III SOLO1 trial. Ann Oncol. 2019;30(suppl 5):v403–v434. [Google Scholar]
- 17.Ray-Coquard I., Pautier P., Pignata S. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 18.Ledermann J.A., Drew Y., Kristeleit R.S. Homologous recombination deficiency and ovarian cancer. Eur J Cancer. 2016;60:49–58. doi: 10.1016/j.ejca.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Lin K.K., Harrell M.I., Oza A.M. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9:210–219. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 20.Konstantinopoulos P.A., Norquist B., Lacchetti C. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol. 2020;38:1222–1245. doi: 10.1200/JCO.19.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dougherty B.A., Lai Z., Hodgson D.R. Biological and clinical evidence for somatic mutations in BRCA1 and BRCA2 as predictive markers for olaparib response in high-grade serous ovarian cancers in the maintenance setting. Oncotarget. 2017;8:43653–43661. doi: 10.18632/oncotarget.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohyuddin G.R., Aziz M., Britt A. Similar response rates and survival with PARP inhibitors for patients with solid tumors harboring somatic versus germline BRCA mutations: a meta-analysis and systematic review. BMC Cancer. 2020;20:507. doi: 10.1186/s12885-020-06948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swisher E.M., Lin K.K., Oza A.M. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 24.Loveday C., Turnbull C., Ramsay E. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennington K.P., Walsh T., Harrell M.I. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaya H., Nakai H., Takamatsu S., Mandai M., Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep. 2020;10:2757. doi: 10.1038/s41598-020-59671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngoi N.Y.L., Tay D., Heong V. Reversal of bowel obstruction with platinum-based chemotherapy and olaparib in recurrent, platinum-free interval, RAD51C germline mutation–associated ovarian cancer. JCO Precision Oncology. 2018;2:1–8. doi: 10.1200/PO.18.00008. [DOI] [PubMed] [Google Scholar]
- 28.McNeish I.A., Oza A.M., Coleman R.L. Results of ARIEL2: a phase 2 trial to prospectively identify ovarian cancer patients likely to respond to rucaparib using tumor genetic analysis. J Clin Oncol. 2015;33:5508. [Google Scholar]
- 29.Swisher E.M., Kristeleit R., Oza A.M. Characterization of patients (pts) with long-term responses to rucaparib in recurrent ovarian cancer (OC) J Clin Oncol. 2020;38:6015. doi: 10.1016/j.ygyno.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 30.Hodgson D.R., Dougherty B.A., Lai Z. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br J Cancer. 2018;119:1401–1409. doi: 10.1038/s41416-018-0274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moschetta M., George A., Kaye S.B., Banerjee S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol. 2016;27:1449–1455. doi: 10.1093/annonc/mdw142. [DOI] [PubMed] [Google Scholar]
- 32.Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., D'Andrea A.D. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abkevich V., Timms K.M., Hennessy B.T. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George J., Alsop K., Etemadmoghadam D. Nonequivalent gene expression and copy number alterations in high-grade serous ovarian cancers with BRCA1 and BRCA2 mutations. Clin Cancer Res. 2013;19:3474–3484. doi: 10.1158/1078-0432.CCR-13-0066. [DOI] [PubMed] [Google Scholar]
- 35.Bernards S.S., Pennington K.P., Harrell M.I. Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated ovarian carcinoma. Gynecol Oncol. 2018;148:281–285. doi: 10.1016/j.ygyno.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Ruscito I., Dimitrova D., Vasconcelos I. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients – a study of the tumour bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD) Eur J Cancer. 2014;50:2090–2098. doi: 10.1016/j.ejca.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M., Silva J.M., Dominguez G. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 38.Sun T., Ruscito I., Dimitrova D. Genetic versus epigenetic BRCA1 silencing pathways: clinical effects in primary ovarian cancer patients: a study of the tumor bank ovarian cancer consortium. Int J Gynecol Cancer. 2017;27:1658–1665. doi: 10.1097/IGC.0000000000001071. [DOI] [PubMed] [Google Scholar]
- 39.Zhu X., Zhao L., Lang J. The BRCA1 methylation and PD-L1 expression in sporadic ovarian cancer. Int J Gynecol Cancer. 2018;28:1514–1519. doi: 10.1097/IGC.0000000000001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondrashova O., Nguyen M., Shield-Artin K. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017;7:984–998. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patch A.M., Christie E.L., Etemadmoghadam D. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 42.Prieske K., Prieske S., Joosse S.A. Loss of BRCA1 promotor hypermethylation in recurrent high-grade ovarian cancer. Oncotarget. 2017;8:83063–83074. doi: 10.18632/oncotarget.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai W., Swisher E.M., Karlan B.Y. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timms K.M., Abkevich V., Hughes E. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014;16:475. doi: 10.1186/s13058-014-0475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkbak N.J., Wang Z.C., Kim J.Y. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popova T., Manié E., Rieunier G. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 47.Mills G.B., Timms K.M., Reid J.E. Homologous recombination deficiency score shows superior association with outcome compared with its individual score components in platinum-treated serous ovarian cancer. Gynecol Oncol. 2016;141(2016):2–208. [Google Scholar]
- 48.Telli M.L., Timms K.M., Reid J. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore K.N., Secord A.A., Geller M.A. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019;20:636–648. doi: 10.1016/S1470-2045(19)30029-4. [DOI] [PubMed] [Google Scholar]
- 51.Alexandrov L.B., Nik-Zainal S., Wedge D.C. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polak P., Kim J., Braunstein L.Z. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hillman R.T., Chisholm G.B., Lu K.H., Futreal P.A. Genomic rearrangement signatures and clinical outcomes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2018;110:265–272. doi: 10.1093/jnci/djx176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gulhan D.C., Lee J.J.K., Melloni G.E.M., Cortés-Ciriano I., Park P.J. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat Genet. 2019;51:912–919. doi: 10.1038/s41588-019-0390-2. [DOI] [PubMed] [Google Scholar]
- 55.Davies H., Glodzik D., Morganella S. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukhopadhyay A., Elattar A., Cerbinskaite A. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res. 2010;16:2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
- 57.Fuh K., Mullen M., Blachut B. Homologous recombination deficiency real-time clinical assays, ready or not? Gynecol Oncol. 2020;159:877–886. doi: 10.1016/j.ygyno.2020.08.035. [DOI] [PubMed] [Google Scholar]
- 58.van Wijk L.M., Vermeulen S., Meijers M. The RECAP test rapidly and reliably identifies homologous recombination-deficient ovarian carcinomas. Cancers (Basel) 2020;12:2805. doi: 10.3390/cancers12102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tumiati M., Hietanen S., Hynninen J. A functional homologous recombination assay predicts primary chemotherapy response and long-term survival in ovarian cancer patients. Clin Cancer Res. 2018;24:4482–4493. doi: 10.1158/1078-0432.CCR-17-3770. [DOI] [PubMed] [Google Scholar]
- 60.Naipal K.A.T., Verkaik N.S., Ameziane N. Functional ex vivo assay to select homologous recombination-deficient breast tumors for PARP inhibitor treatment. Clin Cancer Res. 2014;20:4816–4826. doi: 10.1158/1078-0432.CCR-14-0571. [DOI] [PubMed] [Google Scholar]
- 61.Waks A.G., Cohen O., Kochupurakkal B. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann Oncol. 2020;31:590–598. doi: 10.1016/j.annonc.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Timms K.M., Mills G.B., Perry M. Comparison of genomic instability test scores used for predicting PARP activity in ovarian cancer. J Clin Oncol. 2020;38:1586. [Google Scholar]
- 63.Matulonis U.A., Shapira-Frommer R., Santin A.D. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol. 2019;30:1080–1087. doi: 10.1093/annonc/mdz135. [DOI] [PubMed] [Google Scholar]
- 64.Cohen P.A., Powell A., Bohm S. Pathological chemotherapy response score is prognostic in tubo-ovarian high-grade serous carcinoma: a systematic review and meta-analysis of individual patient data. Gynecol Oncol. 2019;154:441–448. doi: 10.1016/j.ygyno.2019.04.679. [DOI] [PubMed] [Google Scholar]
- 65.Oza A.M., Cook A.D., Pfisterer J. Standard chemotherapy with or without bevacizumab for women with newly diagnosed ovarian cancer (ICON7): overall survival results of a phase 3 randomised trial. Lancet Oncol. 2015;16:928–936. doi: 10.1016/S1470-2045(15)00086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calléns C., Vaur D., Soubeyran I. Concordance between tumor and germline BRCA status in high-grade ovarian carcinoma patients in the phase III PAOLA-1/ENGOT-ov25 trial. J Natl Cancer Inst. 2020 doi: 10.1093/jnci/djaa193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Courtney E., Chok A.K.-L., Ting Ang Z.L. Impact of free cancer predisposition cascade genetic testing on uptake in Singapore. NPJ Genom Med. 2019;4:22. doi: 10.1038/s41525-019-0096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon S.Y., Thong M.K., Taib N.A.M., Yip C.H., Teo S.H. Genetic counseling for patients and families with hereditary breast and ovarian cancer in a developing Asian country: an observational descriptive study. Fam Cancer. 2011;10:199–205. doi: 10.1007/s10689-011-9420-7. [DOI] [PubMed] [Google Scholar]
- 69.Cheung E.L., Olson A.D., Yu T.M., Han P.Z., Beattie M.S. Communication of BRCA results and family testing in 1103 high-risk women. Cancer Epidemiol Biomarkers Prev. 2010;19:2211–2219. doi: 10.1158/1055-9965.EPI-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stover E.H., Fuh K., Konstantinopoulos P.A., Matulonis U.A., Liu J.F. Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol Oncol. 2020;159:887–898. doi: 10.1016/j.ygyno.2020.09.029. [DOI] [PubMed] [Google Scholar]
- 71.Pilié P.G., Tang C., Mills G.B., Yap T.A. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81–104. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Norquist B., Wurz K.A., Pennil C.C. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–3015. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swisher E.M., Sakai W., Karlan B.Y., Wurz K., Urban N., Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drost R., Dhillon K.K., van der Gulden H. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J Clin Invest. 2016;126:2903–2918. doi: 10.1172/JCI70196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mirza M.R., Coleman R.L., Gonzalez-Martin A. The forefront of ovarian cancer therapy: update on PARP inhibitors. Ann Oncol. 2020;31:1148–1159. doi: 10.1016/j.annonc.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 76.Ngoi N.Y., Sundararajan V., Tan D.S. Exploiting replicative stress in gynecological cancers as a therapeutic strategy. Int J Gynecol Cancer. 2020;30:1224–1238. doi: 10.1136/ijgc-2020-001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farkkila A., Gulhan D.C., Casado J. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat Commun. 2020;11:1459. doi: 10.1038/s41467-020-15315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]