Summary
Increased control of biological growth and form is an essential gateway to transformative medical advances. Repairing of birth defects, restoring lost or damaged organs, normalizing tumors, all depend on understanding how cells cooperate to make specific, functional large-scale structures. Despite advances in molecular genetics, significant gaps remain in our understanding of the meso-scale rules of morphogenesis. An engineering approach to this problem is the creation of novel synthetic living forms, greatly extending available model systems beyond evolved plant and animal lineages. Here, we review recent advances in the emerging field of synthetic morphogenesis, the bioengineering of novel multicellular living bodies. Emphasizing emergent self-organization, tissue-level guided self-assembly, and active functionality, this work is the essential next generation of synthetic biology. Aside from useful living machines for specific functions, the rational design and analysis of new, coherent anatomies will greatly increase our understanding of foundational questions in evolutionary developmental and cell biology.
Subject areas: developmental biology, bioengineering, synthetic biology
Graphical abstract
Developmental biology; Bioengineering; Synthetic biology
Introduction
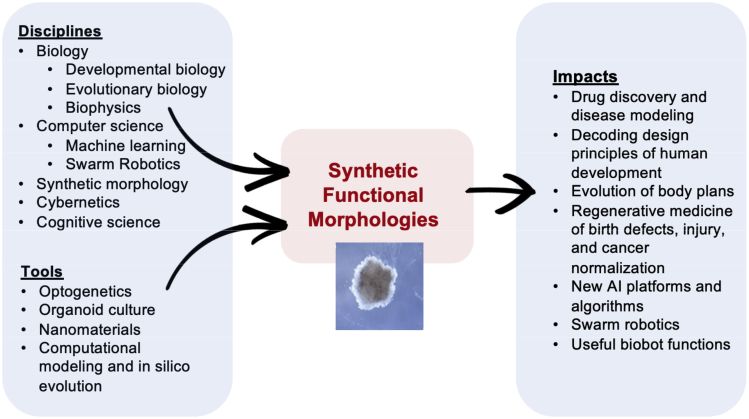
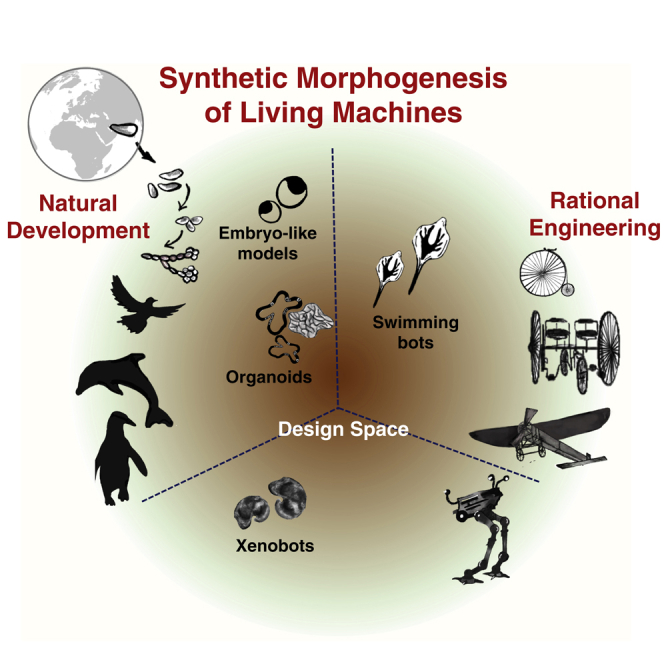
As observational knowledge grows, and models of the underlying patterns are inferred, it becomes possible to begin constructing systems that exploit natural laws in new ways. The trajectories of computer science and physics in the last two centuries clearly demonstrate the value of the engineering approach—efforts to build specific novel devices led to fundamental advances in thermodynamics and theory of computation. Biology is embarking on its own exciting journey via the emerging field of synthetic morphology (expansion of cell-level synthetic biology tools toward reprogramming the anatomy and behavior of multicellular collectives from tissue to organism levels). Moving beyond the reverse engineering of natural systems such as embryogenesis (Beloussov and Grabovsky, 2006; Levin and Martinez Arias, 2019) to the construction of novel ones provides useful technologies with broad applications for society and improves the fundamental understanding of basic mechanisms (Figure 1). A key concept in this framework is that of guided self-assembly: understanding the system-level properties of cellular collectives and using this knowledge to design material and informational inputs to control the emergent form and function.
Figure 1.
Disciplines and tools that are enabling for development of synthetic functional morphologies and their related impacts on life
Continuous feedback between analysis and modeling of surprising biological phenomena, and the rational engineering of living systems with novel structure and function, is a powerful driver of exponential progress (Kamm et al., 2018; Sample et al., 2019). Below, we highlight recent advances in the bioengineering of synthetic morphology and discuss ways in which the resulting new examples of form and function are advancing biological science. These novel living systems constitute a pivotal emerging technology at the intersection of several fields that provides new “model species” in which to develop and test meso-scale morphogenetic theories. This is a quintessentially multidisciplinary undertaking, integrating input of techniques and deep ideas from cell and developmental biology, physics, computer science, and neuroscience.
Design, known as a specification or process for the construction of a system in engineering disciplines, has been less often practiced in generating biological models. Recent approaches in biological engineering and synthetic biology have tried to implement such processes at different scales from synthesis of gene circuits (Brophy and Voigt, 2014) to artificial cell-cell communication (Toda et al., 2020). Standard model system organisms developed as a result of evolutionary processes produced through tinkering, rather than rational design. The promise of building living constructs with pre-defined topology and functionality enables an implementation of rational design approaches to control developmental processes or manufacture systems with close proximity to in vivo counterparts. It also can result in novel systems (not designed before) with new set of specifications and objectives with medical and environmental applications.
Like in other areas of engineering, biology makes significant use of modular decomposition and analysis of parts. The recombination of sophisticated components into novel systems can occur at multiple levels. For example, organ transplants and parabiosis experiments have a long history as tools to understand and control body-level physiology by constructing novel hybrid bodies at the large scale. At the other end of the size range, genomic editing, transgenics, and nanomaterials are extensively used to produce novel biology by modulating the lowest level components. Here, we focus on a critical new direction at the meso-scale: taking synthetic biology to the next level beyond reprogramming single-cell phenotypes to the rational control of growth and form—employing genetic circuits, growth factors, and materials, together with top-down external stimuli to implement multicellular morphogenesis to desired structural and functional specifications. An important aspect of this developing field is gaining predictive control over system-level outcomes; inference of the required starting configurations and real-time stimuli is facilitated by computational modeling approaches and in silico experiments in virtual embryogeny (Delile et al., 2017; Faure et al., 2016; Pascalie et al., 2016; Varenne et al., 2015; Villoutreix et al., 2016). Such computational efforts are an essential part of a quantitative, rigorous understanding of the rules of morphogenesis and how they relate to the rules governing cell behavior, deriving, for the first time, the equivalent of, for example, Boyle's Law, to enable the guidance of the activity of organized cellular collectives toward specific anatomical outcomes.
State of the art
One feature of bioengineering at the meso-scale that is unique (with notable exceptions [Brambilla et al., 2013]) is the fact that bioengineers build out of parts that are themselves highly competent (Levin, 2019), for example, cells that have their own internal homeostatic and signaling systems. Thus, the experiments that are done with biological parts have the potential to help understand how swarm intelligence (Couzin, 2007; Valentini et al., 2018) plays out at the tissue level to solve morphogenetic problems. Such advances not only contribute to improved control of anatomy (the holy grail of regenerative medicine) (Pezzulo and Levin, 2015) and its disorders such as injury and cancer (Deisboeck and Couzin, 2009) but also act as an inspiration for novel architectures in machine learning, artificial intelligence, and resilient autonomous swarm robotics (Slavkov et al., 2018). The integrated focus on controlling the emergent self-assembly of artificial proto-bodies (Doursat, 2013; Doursat and Sanchez, 2014; Doursat et al., 2012, 2013; Fernandez et al., 2012; Olle-Vila et al., 2016), as well as of their computational and behavioral capacities (Macia et al., 2012; Sole et al., 2016; Sole and Macia, 2013, 2014; Urrios et al., 2016), parallels the biological study of natural model systems and forms an important, embodied branch of the field of Artificial Life (Bedau, 2005).
Significant advances so far
Here, we focus on efforts aiming toward developing new living systems using cells as starting building blocks. This “build-to-understand” approach enables decoding of novel design principles as well as generation of useful technological platforms (Elowitz and Lim, 2010; Raspopovic et al., 2014; Schaerli et al., 2014; Slavkov et al., 2018). We loosely catalog two sets of studies. In one set, the main objective is to reconstitute or emulate developmental mechanism in vitro either in cell lines with no developmental potentials or in stem cells with unique self-organization properties. For instance, genetic circuits and artificial cell-cell communication were engineered to induce cell sorting and pattern formation in cell lines (Davies and Glykofrydis, 2020). Exploiting the innate self-organization properties of stem cells could also generate mimetic of early developmental tissues or later human organogenesis in vitro. As a result, these efforts have produced a myriad of living models to study and understand the principle of human development.
The other approach has been less driven by naturally developed human tissues and developmental events, but rather follows designs that are mainly based on a set of specifications for applications of interest. Such synthetic living systems can represent a new form of artificial life that is heavily built on morphology and function by design, examples of which are the artificial stingray and Xenobots. All these efforts are paving a path toward development of complex biological living machines with adaptive and autonomous operation, sensory inputs, signal processing, and actuation in the form of organismal behaviors. Additionally, both sets of studies teach us key lessons for a better practice in engineering form and function.
Synthetic multicellularity
When it comes to engineering synthetic multicellularity, lessons learned from past evolutionary and ecological processes of multicellular development on earth will provide important principles for a successful outcome (Sole et al., 2018). The transition from single cells to multicellular entities has been a key invention in life that has fascinated scientists from diverse fields. Likewise, how a single zygote can robustly produce a whole organism with diverse cell types, function, and controlled organization has been a central question. Seminal works across distinct scientific disciplines, from evolutionary biology to bioengineering, have focused to reconstruct these processes in silico or at bench (Sole et al., 2018).
After million years of evolution in existing animals, the organismal forms and anatomies show certain degree of prevalence in stereotypical morphological motifs such as folds, segments, and cavities. To explain the conserved nature of such structures, the presence of physiogenetic processes to control pattern, shape, or structures was suggested (Newman, 2012). The generic physical properties (e.g., viscoelasticity), active in both living and non-living systems, promote generation of such morphological motifs (Newman, 2012, 2014, 2019). The GRNs governing cellular interaction, initiation, and development of multicellular entities operate within this framework, by fine-tuning, stabilizing, and mobilizing the core generic properties (Newman, 2012).
Cell-cell communication and pattern formation are one of the early events during development of multicellular systems. Hence, understanding and engineering different modes of communication and patterning in prokaryotic and eukaryotic cells have been a quest for developmental and synthetic biologists (Basu et al., 2005; Davies and Glykofrydis, 2020; Green and Sharpe, 2015; Levin and Martinez Arias, 2019; Matsuda et al., 2012; Weber et al., 2007; You et al., 2004). Inspired by pioneering works by Steinberg (1963), seminal studies used cadherins to probe differential adhesion, cell sorting, and pattern formations (Cachat et al., 2016; Davies and Glykofrydis, 2020). Combining cadherin-mediated cell sorting with artificial cell-cell communication also produced multi-layer self-organization and multicellular patterning (Toda et al., 2018, 2020). Instead Skine et al., showed synthetic mammalian reaction-diffusion patterns via reconstituting Nodal-Lefty network in mammalian cells (Sekine et al., 2018). Future efforts will focus on engineering other morphological motifs such as controlled tube formation and folding (Cachat et al., 2014). It is not clear how far one can go in reconstituting fully new forms and whether our ability to invent is limited by inherent constraints that are imposed by natural physical properties within biological systems (Sole et al., 2018).
Synthetic embryo-like entities for early developmental engineering
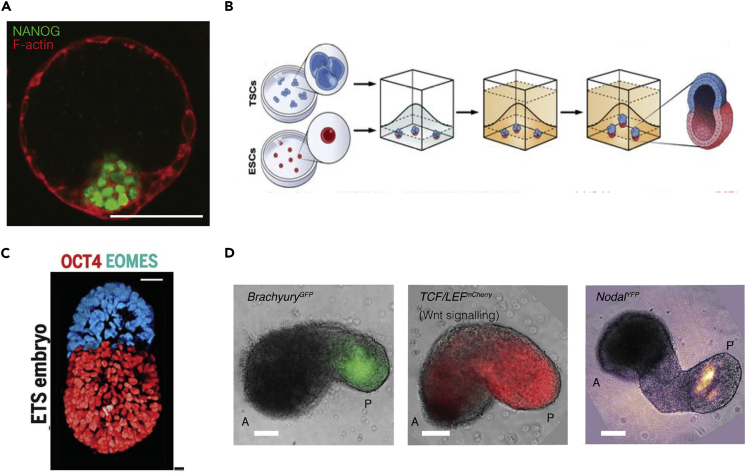
The study of human embryos in vitro is constrained by broad ethical consensus restricting their study to maximum period of 14 days after their creation. This consensus has resulted in a broad gap in knowledge regarding the genetic and morphological changes that are involved in gastrulation, the specification of human germ layers, and the subsequent early stages of body plan segregation and organ specification. Models of early development encompassing living systems, entitled “embryoids,” “blastoids,” and “gastruloids,” produced from both human and murine stem cells, have become an important means to gain insights on the processes underlying this “black box” of human development (Figures 2A–2D) (Fu et al., 2020).
Figure 2.
Synthetic embryo-like entities
(A) Blastoids co-stained with F-actin (red) and Nonog (green). Nanog shows ESC identity (taken with permission from Rivron et al., 2018).
(B) Schematic to show strategy for generation of mouse embryonic-like structures via combining mouse ESC (red) and TSCs (blue). Cells are suspended in 3D extracellular matrices and allowed to self-organize (taken with permission from Harrison et al., 2017).
(C) ETS-embryo structure. Red shows Oct4 expression from ESCs and cyan is Eomes in TSCs (taken with permission from Harrison et al., 2017).
(D) Gastruloids at 120 h after aggregation showing features of mouse embryo at embryonic day 1 and spatially confined presence of signaling cues and markers associated with axis formation such as Brachyury-GFP, WNT signaling activity (TCF/LEF-mCherry), and Nodal-YFP (taken with permission from [Beccari et al., 2018]).
Pioneering studies used human embryonic stem cells subjected to geometric confinement in circular micropatterns. In response to key developmental cues, the cells self-organize and pattern into fates similar to the three germ layers (Warmflash et al., 2014). These three cell types arrange themselves in circular layers that are ordered in an equivalent arrangement to the embryo, with a core-to-edge arrangement of concentric rings of ectoderm, mesoderm, endoderm, and extraembryonic ectoderm. This phenomenon has been used to study the patterns of signal diffusion in a quantitatively reproducible way, and even showed how chirality of single cells leads to a large-scale asymmetry of tissue-level structure (Chen et al., 2012; Wan et al., 2011, 2016; Wan and Vunjak-Novakovic, 2011).
Subsequent to these efforts, major 3D embryonic stem cell models emerged that focused on the integration of early embryonic and extraembryonic mouse stem cells to drive the formation of self-organizing embryo-like structures. The combination of these cells in tightly defined ratios resulted in the formation of structures that surprisingly resembled the early pre-implantation embryo, including a compacted epiblast surrounded by a blastocyst cavity created by trophoblast cells (Figure 2A) (Rivron et al., 2018). These blastocyst-like constructs (blastoids) are capable of supporting the development and decidualization of trophoblast stem cells when implanted into the mouse uterus; however, they fail to achieve later developmental milestones (Rivron et al., 2018). These blastoids offer a high-throughput way to investigate the morphological and genetic features of early embryo-like structures.
Other methodologies also resulted in novel synthetic embryo-like structures using combination of embryonic and extraembryonic stem cells in 3D culture (Figure 2B) (Harrison et al., 2017; Sozen et al., 2018, 2019; Zhang et al., 2019). These studies have demonstrated the capability to spatially organize into a structure that sorts into polarized embryonic compartments and form lumens in a manner that replicates the organization of a post-implantation embryo (Sozen et al., 2018). These “ESC + TSC + XEN” embryos (ETX-embryo) (Figure 2C) develop anterior and posterior structures, recapitulating the spatial opposition of anterior visceral endoderm and primitive streak (Sozen et al., 2018; Zhang et al., 2019). Similar to the murine blastoids described previously, these embryoids can implant and decidualize in utero, although they also exhibit limited ability to develop into more advanced embryonic structures (Sozen et al., 2019). Additionally, gastruloids mimicking the posterior portion of embryo were developed using aggregates of defined size for mouse embryonic stem cell that are pulsed with a Wnt agonist (Figure 2D) (Beccari et al., 2018). The cell aggregates elongate and polarize, forming structures similar to the post-implantation embryonic somites (Beccari et al., 2018). In addition to the formation of T-brachyury+ and Gata6+ poles, these structures show spatial segregation of Hox genes. These 3D models of embryogenesis provide a set of models to build and understand tissue development in vitro.
In a parallel effort, microfluidic devices were used to recapitulate more controlled signaling environments that could effectively mimic the signaling roles of extraembryonic tissues. For instance, by exposing pluripotent cells to a polar signaling environment, these cells organize and differentiate in a manner that mimics Epiblast-Amnion morphogenetic events (Zheng et al., 2019). Human pluripotent stem cells seeded into a pocketed two-channel microfluidic device creating a directional BMP4 signal show the development of a lumen and segregation of amnion from epiblast (Zheng et al., 2019). The epiblast-like cells subsequently differentiate into primitive streak-like and primordial germ cell-like cells. Development of these channeled microfluidic devices could allow the recapitulation of more complex signaling environments, up to and including the post-implantation uterine environment.
Organoids and synthetic tissues to mimic human organ-like structures
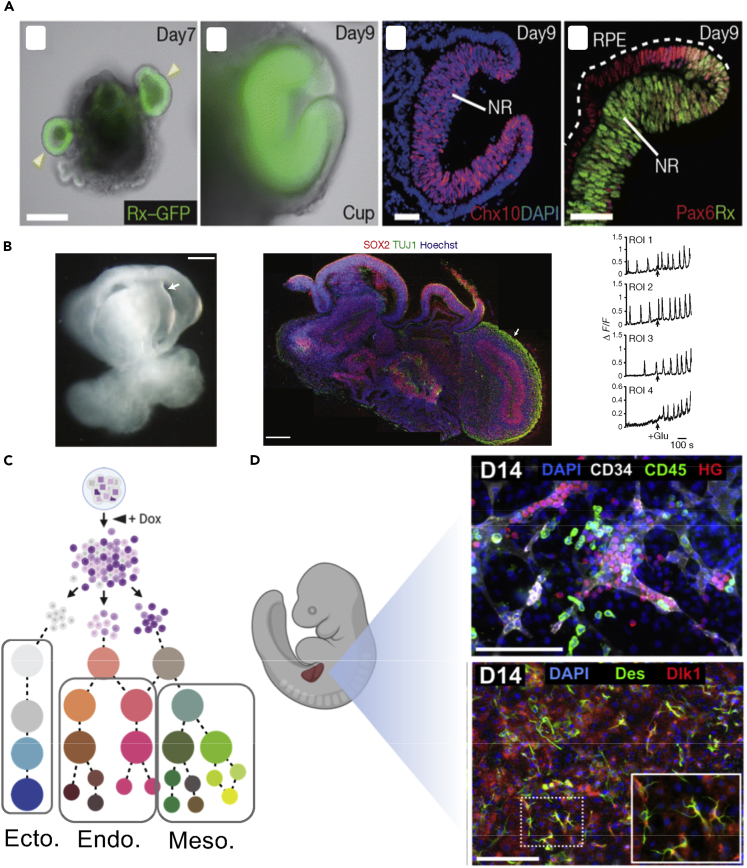
A class of synthetic living systems dubbed organoids represent 3D partial mini replicas of human organs, which were made by combining stem cell and developmental biology with bioengineering (Sasai, 2013). Most of the studies focused mainly on recapitulating the interaction of the stem cells and the niche in laminin-rich 3D extracellular matrices using defined developmental cues. Such interaction controls the processes of self-renewal, differentiation, and generation of diverse cell types with a variety of states that self-organize to functional forms mimicking organogenesis (Zinner et al., 2020). Seminal studies by Sasai and colleagues showed self-organized formation of apico-basally polarized cortical tissues from human pluripotent stem cells (Eiraku et al., 2008) or self-formation of optic cup using designed 3D conditions with tightly controlled medium and cell number (Figure 3A) (Nakano et al., 2012). Parallel studies by Sato and by Clevers and colleagues showed formation of mini-gut organoids using adult stem cells in Matrigel with ever-expanding potential (Clevers, 2013; Sato et al., 2009). A landmark study in 2013 presented the generation of cerebral organoids from human pluripotent stem cells that showed features of cortical development, brain region identities, and organization with a subset of cells maturing to form functional cortical neurons with calcium oscillation (Figure 3B) (Lancaster et al., 2013). Within a short period, these tissue replicas rapidly emerged as tools to decode principles of organogenesis and model human disease with a long-term perspective for regenerative therapies. For a comprehensive history of organoids, we refer readers to several excellent reviews (Del Dosso et al., 2020; Dutta et al., 2017; Kim et al., 2020; Lancaster and Huch, 2019; Lancaster and Knoblich, 2014; Simian and Bissell, 2017; Takebe and Wells, 2019). Endoderm- (e.g., intestine, liver, lung), mesoderm- (e.g., kidney, heart), and ectoderm- (e.g., brain, optic cup, mammary gland) derived organoids have been generated (Kim et al., 2020; Lancaster and Huch, 2019). More complex behavior was also successfully modeled; for example, traveling wave-like expression patterns representative of the segmentation clock, which is central to rhythmic emergence of somites and the axial skeleton, was shown (Diaz-Cuadros et al., 2020; Matsuda et al., 2020b). Each one of these models provides a window through which to study the black box of human development, enabling us to understand broad questions in the contexts of evolution, aging, and organismal differences. For instance, human laboratory-grown brain organoids taught us about the temporal cell atlas of great ape forebrain and shed light on molecular pathways that are unique to humans (Kanton et al., 2019). Ebisuya and colleagues also used a human segmentation clock model to understand species-specific differences in developmental timing via kinetics of biochemical reactions (Matsuda et al., 2020a).
Figure 3.
Examples of organoids and multicellular tissues
(A) Self-organization of an optic-cup-like structure with differentiating epithelial cells from ES cell aggregates in 3D. Rx-GFP shows retinal analge (taken with permission from Nakano et al., 2012).
(B) Left: Bright-field view of cerebral organoid with cavities (arrow) representing ventricle-like structures of brain. Middle: Immunostained cerebral organoid shows tissue morphology with neural progenitors (SOX2, red) and neurons (TUJ1, green). Right: Recorded neural activity using calcium dye (taken with permission from Lancaster et al., 2013).
(C) Schematic for genetically guided engineering by using stem cells engineered with inducible genetic circuit to pre-program cell fates associated with different germ layers. Doxycycline (Dox) turns on the expression.
(D) Generation of liver bud-like tissue using genetic programming. Immunostained tissue exhibits the presence of subset of cells similar to fetal liver. CD34 marks vasculature, CD45 and HG mark blood cell lineages, DLK1 and Des mark hepatocyte progenitor and pericyte populations, respectively (taken with permission from [Guye et al., 2016]). Part of this figure is created using BioRender (BioRender.com).
Although quite promising, one key challenge with multicellular morphogenesis in vitro centers on the ability to control the process of morphogenesis and self-organization (Brassard and Lutolf, 2019; Ebrahimkhani and Ebisuya, 2019; Garreta et al., 2020; Johnson et al., 2017). Additionally, tools for quantitative and universal assessment of tissue function, identity, and cellular composition across studies is missing. The ability to both control and quantitate cell fate and tissue morphogenesis allows improved robustness and reproducibility in manufacturing (Velazquez et al., 2018). Many studies have added a cocktail of growth factors to cell culture medium to reconstitute niche-mediated signaling cues. However, the application of growth factors in media can encounter certain challenges such as supraphysiological concentration. It is also challenging to identify and reconstitute spatiotemporal dynamic and combinatorial cues displayed in native developing tissues. Microfluidic approaches as well as engineered matrices offer a cell-extrinsic based control for signaling cues (Brassard and Lutolf, 2019). For instance, shape-morphing materials dubbed kinomorphs can rationally control the shape and size of multicellular networks (Viola et al., 2020). Additionally, self-organization of intestinal organoids-on-a-chip using scaffold was shown to enable regulation of morphological specifications. Subsequently, externally guided forms can improve maturation and composition of cell types, providing mini-intestines with physiological hallmarks of the in vivo organ (Nikolaev et al., 2020).
Engineering synthetic genetic circuits offers a cell intrinsic-based approach that can be used to enable control over cell fate, communication, and self-organization properties (Santorelli et al., 2019; Velazquez et al., 2018). Employing cells as the niche-associated signaling centers is a powerful method to reconstitute developmental cues and re-create the niche-stem cell cross talk. For instance, genetically engineered Sonic Hedgehog (SHH)-expressing sender cells were used to establish a gradient of the morphogen and improved cell fate and self-organization along dorsoventral and anteroposterior axes in forebrain organoids (Cederquist et al., 2019). Genetically embedded gene circuits can be also used to drive multilineage co-differentiation to produce genetically pre-programmed tissues and organoids (Figure 3C). For instance, inducible and transient expression of GATA6 transcription factor was used to trigger symmetry breaking, co-development of endoderm and mesoderm germ layers, and formation of human fetal liver bud (Figure 3D) (Guye et al., 2016). In a follow-up study, multilineage maturation was also achieved through the synthetic control of GRNs (Velazquez et al., 2020). Recent studies have also used orthogonal differentiation of genetically engineered stem cells to produce multicellular tissues such as vascularized brain organoids (Skylar-Scott et al., 2020). The use of transcription factor-based cell fate programming offers less dependence on signaling cues present in the medium, alleviating the challenge of finding a supporting medium for generation of multiple cell types. Additionally, synthetic genetic programs can interface with cell state (McDonald et al., 2020; Xie et al., 2011) as well as mechanical cues (Nims et al., 2021), light (Johnson et al., 2020), and heat (Corbett et al., 2020). How the engineering of genetic circuits might alter mechanical and electrical characteristics of cells to modulate morphogenetic events is an attractive area that warrants future studies.
Medusoids and optogenetic stingrays: the swimming biobots
There have been several efforts to understand and apply the design principles governing biomechanics of marine organisms, to generate novel technologies such as bio-inspired autonomous underwater vehicles (Raj and Thakur, 2016; Russo et al., 2015). A group of studies combined living and non-living systems to achieve this objective. For instance, medusa jelly fish were reverse-engineered to identify key specifications and design in the context of structure-function, stroke kinematics, and animal-fluid interactions (Nawroth et al., 2012). Jellyfish stroke is composed of a fast contraction and slow elastic recoil. To build in this feature, a bilayer of muscle tissue and soft synthetic elastomer material from silicone polymer was engineered to be controlled by an electrical field and named a medusoid. The muscle provided the force to contract, whereas the elastomer could restore the original shape. Further examination of jellyfish muscle contraction showed importance of highly ordered myofibril organization paired with spatiotemporal coordination to function. The authors found that rat cardiac tissue, evolved to deliver ventricular heart function, can generate a self-organized sheet of tissue with well-defined axes of force generation and cell-to-cell electromechanical coupling with spatiotemporal coordinated contraction. When medusoids with the cardiac tissue are stimulated by an external electric field, their muscles contract in a way that follows the pattern of jellyfish's power stroke and is followed by a pullback of the elastic silicone to the initial flat shape. This contraction and relaxation give the medusoid biobot the ability to move very similarly to a jellyfish, while still missing several features of jellyfish movement such as full control over navigating turns, directions, or sense-response features. Design and computational simulations with experiments were used to find optimum parameters at each step. Medusoids show a successful generation of macroscale functional morphologies via careful division of complex behavior into mechanistic components, exploiting self-organization ability of cells, with the engineering design algorithm and quantitative benchmarking of performance. The similarity of mechanical behaviors between these biobots with other synthetic muscular pumps such as human heart enable the application of this framework to model and build therapeutically valuable muscular pumps at macroscale size.
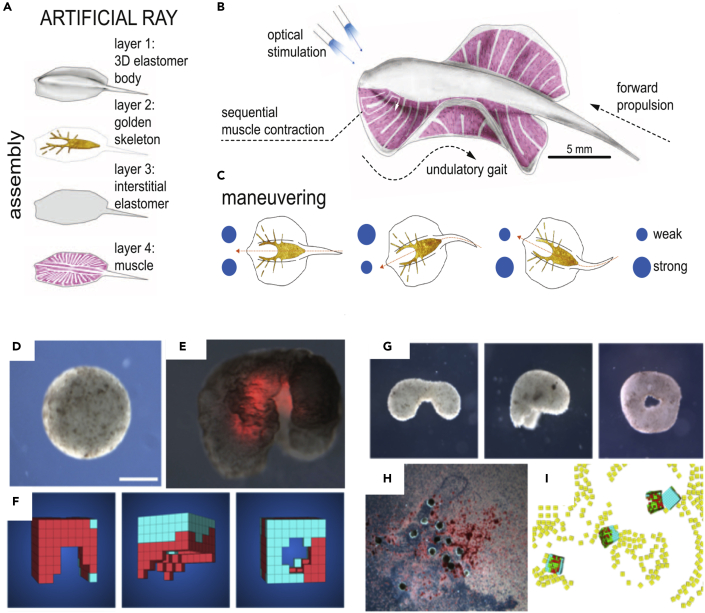
The swimming behavior in medusoids is limited in control with no ability to execute turning or maneuvering. A follow-up study aimed to address this limitation. Bathoid fish such as skate and ray show a unique anatomy such as flattened body, extended pectoral fin, swimming pattern characterized by spanwise bending deformations, and chordwise front-to-rear undulatory motion. These design parameters collectively enable forward movement, swim stability, and high-energy-efficiency maneuvers. Additionally, asymmetrical actuation of pectoral fins provides turning capacity during the swim. To this end, a system-level design and engineering was applied for bottom-up composition of a synthetic ray. The developed artificial ray is within the size of a nickel, a 10-fold reduction in size when compared with live animal (Figures 4A–4C) (Park et al., 2016). It integrates optogenetic technology, rat muscle cells, and three-dimensional elastomer (polydimethylsiloxane) body with a rudimentary skeleton made of gold (Figure 4A). This biobot shows front-to-rear undulating motion that is controlled via light stimulation and displays the same rhythmic motion as a real stingray (Figure 4B). The muscle cells reacted to optical stimuli using a genetically engineered light-sensitive ion channel. These cells were electrically coupled via gap junctions, which enabled propagation of action potential and contractility. The speed and direction of movement is controlled by altering light frequency and the synchrony of stimulation between right and left fins. For example, stimulation of both sides results in forward navigation and asynchronous light-mediated stimulation produces counterclockwise and clockwise turns (Figure 4C). In this way, the synthetic ray can navigate a curved obstacle course with a speed of about 1.5 mm/s over a distance of roughly 15 times longer than its length (250 mm). However, the biobot moves and turns much slower than its naturally evolved counterpart and requires a laminar flow regime, in contrast to the living ray that can navigate turbulent settings. Integration of multiple cell types with novel materials with sense-process-response circuits can equip the synthetic rays with diverse behavior appropriate for each environmental condition. An example of this design is a recent study that combined computational design and optogenetic and neurovascular unit for neuron-enabled biohybrid machines (Aydin et al., 2019). Overall, the synthetic ray couples sensory information to coordinated motor function and behavior; this sets the stage for adaptive living machines with increasingly sophisticated primitive cognition, as additional processing circuits (either bioelectrical or biochemical) are added in the future between the sensory and effector components.
Figure 4.
Biobots and Xenobots
(A) Four layers of body architecture for engineered artificial ray as an example of a biobot.
(B and C) (B) Concept of operation and (C) phototactic control via optical stimulation that triggers muscle activation and produces undulatory locomotion and swimming. Asymmetric stimulation controls the direction (taken with permission from Park et al., 2016).
(D) Ectodermal and muscle cells from Xenopus embryos are extracted, allowed to re-associate, and then micro-sculpted to remove some cells.
(E–H) (E) The remaining cells self-organize a large-scale pattern (with muscle inside, shown by red fluorescent protein signal) to enable forward locomotion of the Xenobot using the available features of the structure. The sculpting is done in accordance with a simulated Xenobot evolved in a virtual computer environment (F), and many different shapes with diverse emergent movement profiles are possible (G). The movement of these synthetic organisms alters their environment by moving materials (H and I) in ways exactly as predicted by the computational model of the swarm behavior.
Walking biological machines
Another type of locomotive biobots was made by exploiting various designs (Chan et al., 2012; Feinberg et al., 2007; Xi et al., 2005) that combined contractility of muscle cells as actuators with materials to bestow the moving functions. A silicon-based microdevice was used to grow and self-assemble cardiac muscle bundles on appropriate substrate (Au films) in poly-N-isopropylacrylamide (PNIPAAm), a thermally responsive polymer. After being cooled to ~32°C, PNIPAAm undergoes a solid-liquid phase transition, and the hybrid structures (about 0.5 mm size) can then be controllably released (Xi et al., 2005). This approach allowed assembly, growth, and maturation within the device to form functional structures, enable control of tight bindings in key location and joints, and then release the structure for operation. The manufactured hybrid (biotic/abiotic) structure operates as a result of collective cooperative motion of muscle bundles. The contraction of muscle bundles governs the device movement, with a force that must overcome opposing forces such as friction. The speed of this biobot was about 38 μm per second, a function of step size and contraction frequency. Living machines with more efficient locomotion were recently developed with velocities in the range of 150–500 μm per second (Chan et al., 2012; Feinberg et al., 2007; Pagan-Diaz et al., 2018). These studies demonstrated the integration of 3D printing, hydrogel scaffold, and living muscle tissue with computational design and engineering (Chan et al., 2012; Pagan-Diaz et al., 2018).
Xenobots
Metazoan cells with a standard genome normally undergo embryogenesis and regeneration that reliably builds or repairs a body with a species-specific morphology and behavior. However, it has been argued that the genetically specified cellular components can implement physiological networks that provide considerable morphological and functional flexibility and plasticity (Levin, 2014; Sullivan et al., 2016). An engineering approach asks: what else are normal cells able to build besides their genomic default outcomes? Can cellular collectives cooperate to form novel target morphologies in the absence of genetic change? The study of artificial embryogenesis (Metzger et al., 2018; Ouyang and Chen, 2010; Rosado-Olivieri and Brivanlou, 2021; Shao and Fu, 2020; Tomoda and Kime, 2021) provides a large option space of novel model systems where the relationship between evolutionary forces, genomic information, and structural/behavioral outcomes can be studied. Examples include two recent types of novel living constructs called “Xenobots” (a portmanteau word joining “biobot” and “Xenopus,” as the cells were sourced from embryos of the frog Xenopus laevis) (Kriegman et al., 2020) (Figure 4D-4I).
The first type consists of skin and muscle cells. A computational process (an evolutionary search over possible architectures) identified specific patterns in which these cells can be layered together and then subtractively sculpted (Kriegman et al., 2020). Such Xenobots exploit the “legs” revealed by the removal of some of the cellular mass to exhibit forward movement (a kind of “walking”). These constructs require no external stimulation, the muscle cells contract in synchrony on their own and the skin cells conduct electrical signals and calcium waves that are synchronized to produce a coherent forward motion (Figure 4D-4I). The in silico evolution was used to identify patterns of cell removal that was predicted to result in forward locomotion, whereas the rest of the cellular signaling, force transmission, and pacing occurred as emergent features.
The second type of Xenobot is even more self-organized. These result from prospective ectoderm cells isolated from embryos (related, but not identical to, the classic animal cap technique of developmental biologists [Sive et al., 2007]). Dissociated cells, liberated from the rest of the embryo, reboot their multicellularity along a novel path: they coalesce and assemble into a spherical proto-body that has a morphology, behavior, and developmental sequence different from typical Xenopus embryos (Blackiston et al., 2021). These exhibit a wide range of diverse swimming behavior driven by ciliary motion (including traversal of several different kinds of mazes and dynamic interactions with features in their environment), ability to repair after mechanical damage (to their new Xenobot forms), and collective behavior that rearranges particles in their milieu into patterns that was effectively predicted by a multi-scale computational model.
One way to view these constructs, which blur traditional definitions attempting to cleanly demarcate categories of living beings, robots, and machines (Bongard and Levin, 2021), is as a platform for bio-robotics. For example, the current implementation demonstrated a simple photo-memory functionality, where a synthetic protein was used to implement a long-term, externally readable engram of fluorescent light exposure experience, as well as movement and collective behavior that was predictably derived from a computational approach. Clearly, there is an opportunity for much future work to program the morphogenesis and behavior of these constructs into biobots with highly diverse outcomes by including existing synthetic biology circuits (Nandagopal et al., 2018; Slusarczyk and Weiss, 2012; Toda et al., 2018) in the tractable Xenopus cells. However, another aspect of this model highlighted by the first two Xenobot studies is their endogenous plasticity, because these functions were obtained from genetically wild-type cells—their shape and function would not be guessable from inspection of the genome alone, which is 100% Xenopus laevis.
The individual cells of Xenopus of course evolved in the Earth's biosphere, but the skin cells' genome was selected for their ability to sit on the outer surface of the embryo and keep out the pathogens. Skin cells, in a novel configuration, can self-organize structure and function toward that of a coherent proto-organism in hours—without the benefit of evolutionary timescales that provided specific selection pressure for these kinds of organism-scale anatomy and behaviors in the normal life cycle of the frog. Instead, their evolution literally took place in a virtual world inside the computers at the University of Vermont. This raises fascinating questions about the real-time plasticity of cellular collectives and their ability to reach areas of morphospace that differ from those reached by normal embryogenesis. Although the study of Xenobot behaviors (and the ability of cells from diverse species to self-organize novel functional anatomies) is just beginning, it is clear that a rich field of discoveries awaits engineering approaches that merge novel inputs with endogenous plasticity of swarms in cooperating toward diverse specific anatomical outcomes despite their wild-type genomes.
Developmental modules to build and control synthetic morphogenesis
Synthetic Morphogenesis field aims to systematically understand and engineer developmental process through application of engineering principles to developmental biology (Basu et al., 2005; Davies, 2008; Isalan et al., 2005; Teague et al., 2016). To study existing design principles in organismal developments, it will be useful to perform top-down decomposition of the process to operational subunits or modules, collections of signaling and morphogenetic events that can be triggered in diverse circumstances by simple trigger signals (Jimenez et al., 2017; Lowell and Pollack, 2016; Verd et al., 2019; Wagner et al., 2007). These modules work both independently and interconnected at different scales to sculpt our tissues and organs through generation of morphological motifs. They can be classified as chemical, mechanical, electrical, as well as gene regulatory networks (GRN) modules. Based on the need of organism, they gradually trigger the production of cell types, trigger cellular self-organization, and advance functional maturities. These modules also participate in physiological and pathological processes during aging, degenerative disease, and various tissue responses to injuries. Each one of the modules can be further divided to sense-process-actuation elements that direct morphogenetic processes. Cells use GRN module to differentiate and create distinct cell types with a set of soluble and insoluble signaling cues (Davidson et al., 2002). These cells then form collectives, employing multilineage communication through chemical signals (Sagner and Briscoe, 2019; Zagorski et al., 2017) as well as bioelectrical (Levin and Martyniuk, 2018; Levin et al., 2017) and biomechanical (Davidson, 2012; Miller and Davidson, 2013) mechanisms to coordinate long-range outcomes. The activation of these modules underlies the phenomena of symmetry breaking, growth, and morphogenesis. Recent evidence shows the presence of feedback regulatory controls, within and between the modules, that ensures robustness and resilience during organismal development (Chan et al., 2017; Hannezo and Heisenberg, 2019; Kunche et al., 2016). However, the details of how the distinct modules are linked, and the details of how the hierarchical directionality of connections is shaped, are the subject of ongoing studies. For instance, from the early steps of embryogenesis, cellular GRNs are controlled by signals originating in the cell itself, its neighbors, or distant tissues (Chan et al., 2017; Smith et al., 2018). This enables continuous top-down feedback controls and the establishment of setpoints to operate within the target morphology specs. Examples include the ability of some species (and Xenobots) to regenerate to the same shape after significant damage (Pezzulo and Levin, 2016), and the ability of salamander kidney tubules to form the same overall shape and size, out of cells with radically different sizes, by mechanisms that depend on cell:cell communication or single-cell cytoskeletal shape control, as needed (Fankhauser, 1945). Large-scale anatomical homeostasis also involves bioelectric signaling across all (not just neural) tissues, as recent work has shown that the setpoints of self-assembly (the overall target morphology toward which cell swarms cooperate) can be permanently changed by transiently modifying the bioelectrical information stored in ion channel-driven pattern memory circuits, rather than by rewiring (or modifying) the cells themselves (Durant et al., 2017).
In the context of synthetic living machines, bioengineers are not limited to using only those modules that we know function during the embryogenesis of naturally evolved living systems. There are other means of sensing and communication present in nature that have not had a role in traditional tissue morphogenesis, but that potentially can be utilized in our rational design of new functionality. For instance, optogenetic control of cellular behavior (a light control module) has been widely used to steer cellular functions and morphogenesis in development, regeneration, and cancer (Adams et al., 2014; Baaske et al., 2018; Bonzanni et al., 2020; Chernet et al., 2016; Izquierdo et al., 2018; Jewhurst et al., 2014), and is expected to play a major role in cybernetic control of living machines. A recent study has demonstrated the use of automated optogenetic feedback control for precise and robust regulation of cell growth (Milias-Argeitis et al., 2016). Magnetoreception is another modality used by organisms such as homing pigeons (Wiltschko and Wiltschko, 2019) and fish (Formicki et al., 2019), and new tools for magneto-sensitivity have already been employed for neural modulation (Long et al., 2015).
Future: A rational integrative framework for engineering living systems in vitro
The design principles of organogenesis and regeneration are complex and are still not well understood. Our understanding of how tissue-level computation is executed in controlling cell fate, tissue composition, coordinated cell functions, tissue patterns, and large-scale anatomical morphologies is limited. Our comprehension of how robustness and resilience are incorporated into this process, and of how they can be engineered within synthetic tissue morphologies, is also limited. Reading and writing of the process of morphogenesis, combined with iterative design-build-test cycles, will not only help us understand biology, but also will provide a path for the generation of more advanced human tissues.
Traditional efforts in the past usually have tried to mimic developmental events by exploiting chemical modules in the form of cocktails of growth factors that are sequentially added to cells in culture. This approach, informed by data from model organisms (i.e., mouse, Xenopus) usually exercises a trial-and-error process to determine the proper dosage, timing, and combination of growth factors for generating self-organizing tissues. However, this method encounters key bottlenecks such as limited control of sense and response events in the culture, or challenges in finding media conditions that can initiate and steer development of cell types in multilineage tissues. Therefore, strategies that enable control over the electrical, chemical, and mechanical properties of tissue, transcriptional signatures and cellular states, cell proliferation, and communication will be fundamental to the next generation of morphogenetic engineering. For a successful operation, design strategies should combine computational modeling, quantitative high-resolution analysis, and iterative engineering. Here we provided Box 1 and 2 as examples of how exploring developmental modules can enable advancement in the tissue fates and morphologies.
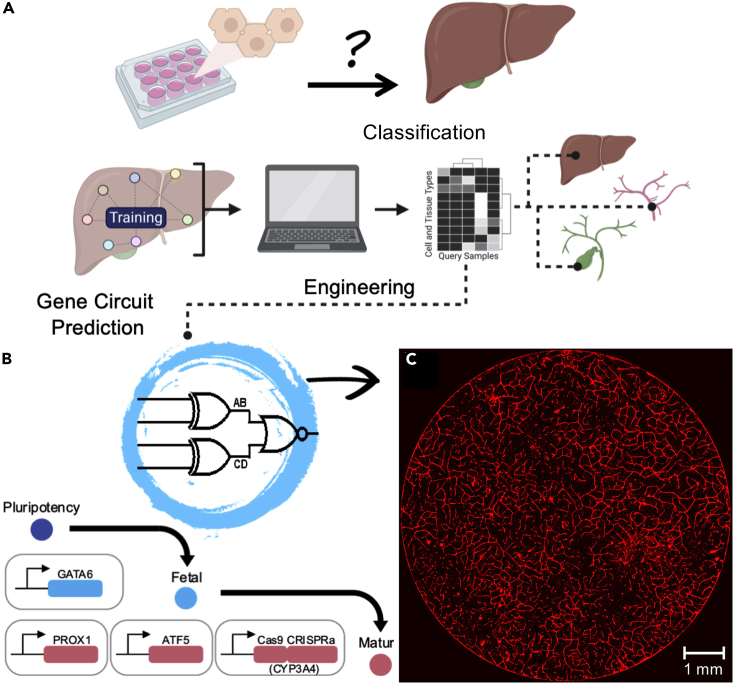
Box 1. Integrated analysis and engineering of GRN modules to program living systems.
A gene regulatory network is a complex dynamic system that evolved to direct cellular processes, fate, and function. From the control standpoint it would be efficient to shape GRNs in ways that can be controlled with master regulators and driver nodes. The reprogramming of somatic cells to a pluripotent state with a small set of transcription factors is an example of such control implemented in naturally designed cellular systems and used by scientists for cell fate conversion.
In the context of multicellular self-organization, it is useful to ask whether GRN-associated driving nodes are present in cells or can be engineered rationally (i.e., as control switches) to initiate not only the cell state but also multicellular interactions and self-organization. In support of this idea, transcription factor-triggered tissue formation has been shown in a number of studies to generate tissues such as the brain, thyroid, or liver (Antonica et al., 2012; Ebrahimkhani and Ebisuya, 2019; Skylar-Scott et al., 2020; Velazquez et al., 2018). A transient and heterogeneous expression of GATA6 transcription factor in human iPSCs was sufficient to generate different germ layers, co-development of progenitors, and self-organization to a fetal liver-like tissue in 14 days (Guye et al., 2016). A systematic approach to exploit GRNs for directing tissue morphogenesis can be instrumental for rational engineering strategies. To implement this approach in multilineage liver tissue development, Velazquez et al. have recently investigated whether a set of drivers can be identified and engineered to direct fetal liver organoids across developmental trajectory and toward adult human livers (Figure 5) (Velazquez et al., 2020). They used a machine learning-based algorithms (Cahan et al., 2014; Roost et al., 2015) to quantitatively compare starting and target GRNs to predict a set of genetic targets (Figure 5A). Subsequently they built and tested genetic circuits to advance tissue development (Figure 5B). This approach enabled synthetic maturation of liver tissue and generation of organoids by a genetic design. Interestingly, the set of identified genetic circuits not only improved epithelial identity but also induced co-maturation of different cell types as well as morphogenesis of the vascular network (Figure 5C). Upon introduction of each circuit, GRN score was improved that resulted in development of tissues that are phenotypically one step closer to native human livers. In summary, by using four genetic circuits, human liver organoids with vascular networks and pericytes were generated. This study suggests the potential presence of key signaling nodes embedded in cellular GRN networks that, if triggered properly, can direct and control multicellular formation, cell-cell communication, and self-organization. It also offers a framework for iterative GRN assessment and engineering to navigate the development of in vitro tissues.
Figure 5.
Integrative analysis and engineering of gene regulatory network
(A) Comparison of synthetic tissues with native tissues via quantitative computational analysis and classification algorithms. Genetic circuits to program cell and tissue fates are predicted, engineered, and delivered to synthetic tissue to program developmental trajectories and cellular interactions.
(B and C) (B) Example of genetic circuits enabled directing development of liver organoids from pluripotent stem cells; (C) self-vascularized human liver organoids generated via genetic design and engineering (taken with permission from Velazquez et al., 2020). Part of this figure is created using BioRender (BioRender.com).
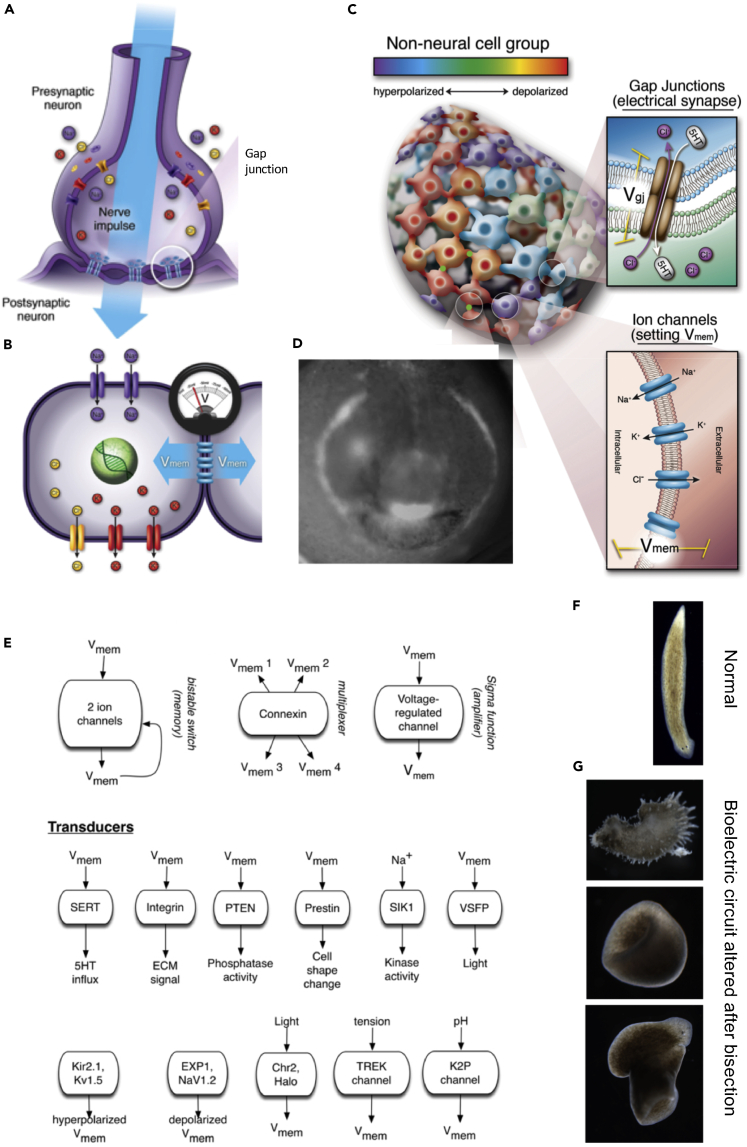
Box 2. Bioelectricity: a new modality to enrich in synthetic morphology.
Many biochemical tools have been created for the synthetic biology field, focused on transcriptional dynamics, protein-protein interactions, gradients, and enzymatic activity. These tools are beginning to be exploited for multicellular coordination in bioengineering (Bashor et al., 2010; Chau et al., 2012; Woodruff et al., 2017). However, there is another important aspect of endogenous morphogenesis that is poised to significantly impact this field: bioelectricity (Levin, 2021; Levin et al., 2019). All cells, not just neurons, communicate electrically by forming networks of cells that set their resting potential by means of ion channels and pumps, and then communicate those states to each other dynamically using gated electrical synapses known as gap junctions (Figures 6A–6C) (Bates, 2015; Levin et al., 2017). Evolution long ago established that bioelectric networks are an ideal way to coordinate across distance and manage complex outcomes long ago. Such dynamics can be seen in bacterial biofilms (Prindle et al., 2015; Yang et al., 2020), can be used for coordination of embryogenesis and regeneration of complex organs (Figure 6D) (McLaughlin and Levin, 2018; Silver and Nelson, 2018), and can be sped up to form specialized computational organs such as brains (Fields et al., 2020). These dynamics can now be tracked by fluorescent voltage reporter dyes (Adams and Levin, 2012a, b) and genetically encoded voltage indicator proteins (Abdelfattah et al., 2016; St-Pierre et al., 2015; Treger et al., 2015), which are especially well suited to the study of synthetic constructs that are easier to image than whole animals. Moreover, the advances in molecular developmental bioelectricity now provide bioengineers with plasmids encoding wild-type, mutant, and dominant-negative ion channels and gap junctions that enable precise control of resting potential and its spatial propagation (Figure 6E) (Levin, 2013; Mustard and Levin, 2014). In addition, techniques like optogenetics can be adapted to non-neural tissues to provide even tighter spatiotemporal control (Adams et al., 2014). Likewise, additional tools targeting transduction machinery help design circuits in which bioelectric computations are linked to transcriptional outcomes. Bioelectricity can contribute to synthetic morphology in three distinct ways. First, it provides increased control over cell-level properties, for example, resting potential is a powerful regulator of differentiation (Hinard et al., 2008; Sundelacruz et al., 2009). Second, it enables computational circuits with specific dynamic properties; non-neural bioelectricity exploits the attractive properties of neural networks (Manicka and Levin, 2019), but it is embedded in deformable media, which allows it to implement morphological computation (Cheney et al., 2014; Rieffel et al., 2010). Finally, control of the bioelectric layer provides a convenient entry point to the design of minimal cognitive agents—synthetic creations that have functional and behavioral repertoires in addition to their morphology (Baluška and Levin, 2016; Macia et al., 2017). By building simple artificial bodies that house tractable neural (and non-neural) information processing dynamics, developmental bioelectricity and synthetic morphology can be combined to better understand the evolution of the mind-body connection (Pezzulo and Levin, 2015).
Figure 6.
Developmental bioelectricity
(A) Neurons maintain a resting potential via ion channels in their membranes and propagate their electric state to neighbors via gap junctions (electrical synapses).
(B and C) (B) Bioelectric signaling using these same molecular components is a property of all cells, which join together into tissues forming networks (C) that enable large-scale electrically mediated computations to regulate distributions of morphogens and control gene expression.
(D) Fluorescent voltage dye image showing an example of an instructive bioelectric prepattern—the frog embryo face, showing the future locations of the eye, mouth, and other organs (taken with permission from(Vandenberg et al., 2011)).
(E–G) (E) A variety of channel, connexin, and neurotransmitter machinery proteins are available as a parts library, complementing canonical transcriptional modules, which enables synthetic biologists to build bioelectric circuits for control of tissue-level morphogenesis. An example of the plasticity of self-assembly beyond genetic default outcomes are shown in planaria, where normal cells can build wild-type forms (F) or highly altered morphologies (G) if the bioelectric circuit states are modified.
Conceptual implications
One of the important contributions of this field to basic science is that it has highlighted problems that have previously been the province of philosophy alone, for the first time allowing empirical approaches to deep questions and pushing us to refine terms that are often used but that have no clear definition. We now have the ability to construct entirely novel functional proto-bodies, by processes that are part rational design and part simulated evolutionary algorithms, using chimerism between natural components at one level (genomes, whole cells) to make synthetic outcomes at another level (novel morphologies beyond the tissue level). This raises profound questions about how to update concepts like organism, animal, body, evolved/designed, and self (Levin, 2019; Rosen, 1972). One example is the notion of “machine.”
Constructs such as Xenobots and mammalian muscle-driven constructs have been called synthetic living machines. Are these constructs machines? Although the use of machine metaphors in biology has been criticized (Boudry and Pigliucci, 2013; Nicholson, 2012, 2013, 2014, 2019; Witzany and Baluska, 2012), it is important to note that many critiques are due to an outdated picture of machines as mechanical, inflexible, and driven entirely by external causes. The existence of designed biobots pushes us to ask what exactly is a machine, what features might life have that are permanently (not just temporarily) beyond the reach of engineers if they have access to the same ingredients and evolutionary methods used by natural life, and what precisely would make something not a machine? A modern understanding of emergence, complexity theory, chaos theory, and computation has spawned a new field studying the behavior of machines (Man and Damasio, 2019), which highlights the surprise, novelty, intelligence, self-control, and flexible behavior that can result from even today's relatively primitive autonomous agents (Braitenberg, 1984; Lehman et al., 2020; Rahwan et al., 2019). This is especially true in biology, where the boundaries between machines and living agents are being erased by progress in bioengineering.
On the one end of the continuum, biobots that are externally actuated and have a precise 3D-engineered morphology are uncontroversially machines, as they were designed in great detail and use muscle cells to generate predictable forces in the way that one might use animal power to drive a mechanical machine. In contrast, biobots with highly emergent morphologies, self-actuation, and surprising behaviors represent our journey toward the other end of the continuum, where all the key capacities of purely natural forms might be encountered. If one strips away prior centuries' assumptions about machines' simplicity and reliance on external control, one is left with a definition of a machine that focuses on the fascinating balance between surprising emergent behaviors and the possibility of rational understanding that enables design for specific shapes and functionality. Thus, we use the machine metaphor for the products of synthetic biology not to reduce or diminish the wonderous capacity of living forms, but instead, to refine and expand our currently meager conception of what a “machine” can be. As biobots and other forms of artificial life have the benefit of both design and evolutionary dynamics (Lehman et al., 2020; Miller, 2004; Stanley and Miikkulainen, 2003), there is no principled reason why future versions could not enjoy the same agency that exclusively evolved lineages do. In fact, one can conjecture that by adding design to the power of evolution, creatures will arise that have improved emergent agency and capacities over what evolution alone has done to date.
As often happens, engineering moves ahead to empirically resolve (solve or redefine) problems that have resisted pure philosophy for centuries. The creation of these constructs cuts the Gordian knot of the life versus machine philosophical debate, because they reveal the smooth continuum of every possible combination of inorganic design and evolved species. They show that there is no sharp line between designed artifacts and the magic of “standard” life forms (those present before the rational age of biology)— every possible mix can be made, and thus we need a mature science of life, and of machines, that does justice to these empirical facts (Levin, 2020).
Several other conceptual areas are highlighted by new work in this field. First is the importance of meso-scale “laws of form” (Beloussov and Grabovsky, 2007; Briscoe and Kicheva, 2017; Graner and Riveline, 2017; Polanyi, 1968): synthetic organisms are an ideal sandbox in which to hone our understanding of system-level morphogenetic rules that are distinct from molecular-level details, akin to what thermodynamics accomplished by giving up the hope of tracking every particle. Such work is a critical complement to today's molecular profiling efforts. Similarly, this work brings up important complementarities in understanding and control of systems from bottom-up (emergentist) and top-down (representationalist) strategies, both of which have important practical techniques to offer (Pezzulo and Levin, 2016). Finally, this field brings up important issues in bioethics (Haase and Freedman, 2020; Lawrence, 2019; Levin et al., 2020): the increasing sophistication of artificial bodies, chimeric constructs with cells from various species tightly integrated with electronic interfaces/processors and engineered neural networks, is inevitably going to give rise to a wide range of creatures with completely novel behaviors and cognitive capacities, which will raise obvious but difficult questions about how we relate to truly diverse intelligences as this field matures in the coming century.
Thus, the next advances in this exciting field will occur on a number of concurrent tracks of technological and conceptual development. Computational modeling of structure and function will increasingly merge biophysically realistic simulation with machine learning of rules and inference of novel stimuli to apply to cells for desired system-level morphogenesis and behavior. At the bench, it will be crucial to develop standardized computer-controlled environments where real-time feedback during morphogenesis and behavior can be provided to synthetic morphologies. The tight integration of synthetic tissues with optogenetic, microfluidic, and electrode interfaces to computational platforms (Hamann et al., 2015; Tsuda et al., 2009; Tsuda et al., 2007; von Mammen et al., 2016; Wahby et al., 2016) will enable real-time closed-loop controls and thus facilitate the refinement of strategies for rational guided self-assembly of novel living bodies and, eventually, primitive cognition. In this way, advances in synthetic morphology and biologically embodied artificial life will lead to a deep unification of several fields of basic science and applied technology, with numerous transformative impacts ranging from biomedicine to artificial intelligence.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Mo R. Ebrahimkhani (mo.ebr@pitt.edu).
Materials availability
No materials were newly generated for this paper.
Data and code availability
No data and codes were newly generated for this paper.
Acknowledgments
M.L. gratefully acknowledges support of the Bailey Family Foundation, the Elisabeth Giauque Trust, and the Templeton World Charity Foundation (TWCF0089/AB55). M.R.E. gratefully acknowledges supports from the National Institute of Biomedical Imaging and Bioengineering (RO1 EB028532) and the National Heart, Lung, and Blood Institute (RO1 HL141805) as well as Pittsburgh Liver Research Center (NIH- NIDDK P30DK120531). We apologize to those colleagues whose works were not cited due to space limitations. We also thank our anonymous reviewers for their constructive comments.
Author contributions
M.L. and M.R.E. discussed together the theme and subsections, implemented the ideas, developed the figures, and wrote the manuscript.
Declaration of interests
M.L. is a co-inventor on U.S. Application 63/136,564 submitted by Tufts University that covers some engineered multicellular organisms. M.R.E. is a co-inventor on US9677085B2 and Application WO2019237124 and PCT/US2020/045926, which are related to engineering multicellular systems.
Contributor Information
Mo R. Ebrahimkhani, Email: mo.ebr@pitt.edu.
Michael Levin, Email: michael.levin@tufts.edu.
References
- Abdelfattah A.S., Farhi S.L., Zhao Y., Brinks D., Zou P., Ruangkittisakul A., Platisa J., Pieribone V.A., Ballanyi K., Cohen A.E. A Bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J. Neurosci. 2016;36:2458–2472. doi: 10.1523/JNEUROSCI.3484-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D.S., Lemire J.M., Kramer R.H., Levin M. Optogenetics in Developmental Biology: using light to control ion flux-dependent signals in Xenopus embryos. Int. J. Dev. Biol. 2014;58:851–861. doi: 10.1387/ijdb.140207ml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D.S., Levin M. General principles for measuring resting membrane potential and ion concentration using fluorescent bioelectricity reporters. Cold Spring Harb. Protoc. 2012;2012:385–397. doi: 10.1101/pdb.top067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D.S., Levin M. Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4(3) and CC2-DMPE. Cold Spring Harb. Protoc. 2012;2012:459–464. doi: 10.1101/pdb.prot067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonica F., Kasprzyk D.F., Opitz R., Iacovino M., Liao X.H., Dumitrescu A.M., Refetoff S., Peremans K., Manto M., Kyba M. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491:66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin O., Zhang X., Nuethong S., Pagan-Diaz G.J., Bashir R., Gazzola M., Saif M.T.A. Neuromuscular actuation of biohybrid motile bots. Proc. Natl. Acad. Sci. U S A. 2019;116:19841–19847. doi: 10.1073/pnas.1907051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baaske J., Gonschorek P., Engesser R., Dominguez-Monedero A., Raute K., Fischbach P., Muller K., Cachat E., Schamel W.W.A., Minguet S. Dual-controlled optogenetic system for the rapid down-regulation of protein levels in mammalian cells. Sci. Rep. 2018;8:15024. doi: 10.1038/s41598-018-32929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F., Levin M. On having No head: cognition throughout biological systems. Front. Psychol. 2016;7:902. doi: 10.3389/fpsyg.2016.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor C.J., Horwitz A.A., Peisajovich S.G., Lim W.A. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu. Rev. Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Gerchman Y., Collins C.H., Arnold F.H., Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Bates E. Ion channels in development and cancer. Annu. Rev. Cell Dev. Biol. 2015;31:231–247. doi: 10.1146/annurev-cellbio-100814-125338. [DOI] [PubMed] [Google Scholar]
- Beccari L., Moris N., Girgin M., Turner D.A., Baillie-Johnson P., Cossy A.C., Lutolf M.P., Duboule D., Arias A.M. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature. 2018;562:272–276. doi: 10.1038/s41586-018-0578-0. [DOI] [PubMed] [Google Scholar]
- Bedau M.A. Artificial life: more than just building and studying computational systems. Artif. Life. 2005;11:1–3. doi: 10.1162/1064546053278928. [DOI] [PubMed] [Google Scholar]
- Beloussov L.V., Grabovsky V.I. Morphomechanics: goals, basic experiments and models. Int. J. Dev. Biol. 2006;50:81–92. doi: 10.1387/ijdb.052056lb. [DOI] [PubMed] [Google Scholar]
- Beloussov L.V., Grabovsky V.I. Information about a form (on the dynamic laws of morphogenesis) Biosystems. 2007;87:204–214. doi: 10.1016/j.biosystems.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Blackiston D., Lederer E., Kriegman S., Garnier S., Bongard J., Levin M. A cellular platform for the development of synthetic living machines. Sci. Robot. 2021;6:eabf1571. doi: 10.1126/scirobotics.abf1571. [DOI] [PubMed] [Google Scholar]
- Bongard J., Levin M. Living things are not (20th Century) machines: updating mechanism metaphors in light of the modern science of machine behavior. Front. Ecol. Evol. 2021;9 doi: 10.3389/fevo.2021.650726. [DOI] [Google Scholar]
- Bonzanni M., Rouleau N., Levin M., Kaplan D.L. Optogenetically induced cellular habituation in non-neuronal cells. PLoS One. 2020;15:e0227230. doi: 10.1371/journal.pone.0227230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry M., Pigliucci M. The mismeasure of machine: synthetic biology and the trouble with engineering metaphors. Stud. Hist. Philos. Biol. Biomed. Sci. 2013;44:660–668. doi: 10.1016/j.shpsc.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Braitenberg V. MIT Press; 1984. Vehicles, Experiments in Synthetic Psychology. [Google Scholar]
- Brambilla M., Ferrante E., Birattari M., Dorigo M. Swarm robotics: a review from the swarm engineering perspective. Swarm Intell. 2013;7:1–41. [Google Scholar]
- Brassard J.A., Lutolf M.P. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Kicheva A. The physics of development 100years after D'arcy thompson's "on growth and form. Mech. Dev. 2017;145:26–31. doi: 10.1016/j.mod.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Brophy J.A., Voigt C.A. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E., Liu W., Hohenstein P., Davies J.A. A library of mammalian effector modules for synthetic morphology. J. Biol. Eng. 2014;8:26. doi: 10.1186/1754-1611-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E., Liu W., Martin K.C., Yuan X., Yin H., Hohenstein P., Davies J.A. 2- and 3-dimensional synthetic large-scale de novo patterning by mammalian cells through phase separation. Sci. Rep. 2016;6:20664. doi: 10.1038/srep20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist G.Y., Asciolla J.J., Tchieu J., Walsh R.M., Cornacchia D., Resh M.D., Studer L. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.J., Heisenberg C.P., Hiiragi T. Coordination of morphogenesis and cell-fate specification in development. Curr. Biol. 2017;27:R1024–R1035. doi: 10.1016/j.cub.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Chan V., Park K., Collens M.B., Kong H., Saif T.A., Bashir R. Development of miniaturized walking biological machines. Sci. Rep. 2012;2:857. doi: 10.1038/srep00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau A.H., Walter J.M., Gerardin J., Tang C., Lim W.A. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell. 2012;151:320–332. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.H., Hsu J.J., Zhao X., Guo C., Wong M.N., Huang Y., Li Z., Garfinkel A., Ho C.M., Tintut Y. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ. Res. 2012;110:551–559. doi: 10.1161/CIRCRESAHA.111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney N., Clune J., Lipson H. Evolved electrophysiological soft robots. ALIFE. 2014;14:222–229. [Google Scholar]
- Chernet B.T., Adams D.S., Lobikin M., Levin M. Use of genetically encoded, light-gated ion translocators to control tumorigenesis. Oncotarget. 2016;7:19575–19588. doi: 10.18632/oncotarget.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Corbett D.C., Fabyan W.B., Grigoryan B., O'Connor C.E., Johansson F., Batalov I., Regier M.C., DeForest C.A., Miller J.S., Stevens K.R. Thermofluidic heat exchangers for actuation of transcription in artificial tissues. Sci. Adv. 2020;6:eabb9062. doi: 10.1126/sciadv.abb9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin I. Collective minds. Nature. 2007;445:715. doi: 10.1038/445715a. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Davidson L.A. Epithelial machines that shape the embryo. Trends Cell Biol. 2012;22:82–87. doi: 10.1016/j.tcb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.A. Synthetic morphology: prospects for engineered, self-constructing anatomies. J. Anat. 2008;212:707–719. doi: 10.1111/j.1469-7580.2008.00896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.A., Glykofrydis F. Engineering pattern formation and morphogenesis. Biochem. Soc. Trans. 2020;48:1177–1185. doi: 10.1042/BST20200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisboeck T.S., Couzin I.D. Collective behavior in cancer cell populations. Bioessays. 2009;31:190–197. doi: 10.1002/bies.200800084. [DOI] [PubMed] [Google Scholar]
- Del Dosso A., Urenda J.P., Nguyen T., Quadrato G. Upgrading the physiological relevance of human brain organoids. Neuron. 2020;107:1014–1028. doi: 10.1016/j.neuron.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delile J., Herrmann M., Peyrieras N., Doursat R. A cell-based computational model of early embryogenesis coupling mechanical behaviour and gene regulation. Nat. Commun. 2017;8:13929. doi: 10.1038/ncomms13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Cuadros M., Wagner D.E., Budjan C., Hubaud A., Tarazona O.A., Donelly S., Michaut A., Al Tanoury Z., Yoshioka-Kobayashi K., Niino Y. In vitro characterization of the human segmentation clock. Nature. 2020;580:113–118. doi: 10.1038/s41586-019-1885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doursat, R. (2013). Bridging the Mind-Brain Gap by Morphogenetic “Neuron Flocking”: The Dynamic Self-Organization of Neural Activity into Mental Shapes. Association for the Advancement of Artificial Intelligence (AAAI) Fall Symposia 2013.
- Doursat R., Sanchez C. Growing fine-grained multicellular robots. Soft Robot. 2014;1:110–121. [Google Scholar]
- Doursat R., Sayama H., Michel O. Morphogenetic engineering: reconciling self-organization and architecture. Underst. Complex Syst. 2012:1–24. [Google Scholar]
- Doursat R., Sayama H., Michel O. A review of morphogenetic engineering. Nat. Comput. 2013;12:517–535. [Google Scholar]
- Durant F., Morokuma J., Fields C., Williams K., Adams D.S., Levin M. Long-term, stochastic editing of regenerative anatomy via targeting endogenous bioelectric gradients. Biophys. J. 2017;112:2231–2243. doi: 10.1016/j.bpj.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Ebrahimkhani M.R., Ebisuya M. Synthetic developmental biology: build and control multicellular systems. Curr. Opin. Chem. Biol. 2019;52:9–15. doi: 10.1016/j.cbpa.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Elowitz M., Lim W.A. Build life to understand it. Nature. 2010;468:889–890. doi: 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser G. Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. J. Exp. Zool. 1945;100:445–455. doi: 10.1002/jez.1401000310. [DOI] [PubMed] [Google Scholar]
- Faure E., Savy T., Rizzi B., Melani C., Stasova O., Fabreges D., Spir R., Hammons M., Cunderlik R., Recher G. A workflow to process 3D+time microscopy images of developing organisms and reconstruct their cell lineage. Nat. Commun. 2016;7:8674. doi: 10.1038/ncomms9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.W., Feigel A., Shevkoplyas S.S., Sheehy S., Whitesides G.M., Parker K.K. Muscular thin films for building actuators and powering devices. Science. 2007;317:1366–1370. doi: 10.1126/science.1146885. [DOI] [PubMed] [Google Scholar]
- Fernandez J.D., Lobo D., Martin G.M., Doursat R., Vico F.J. Emergent diversity in an open-ended evolving virtual community. Artif. Life. 2012;18:199–222. doi: 10.1162/artl_a_00059. [DOI] [PubMed] [Google Scholar]
- Fields C., Bischof J., Levin M. Morphological coordination: a common ancestral function unifying neural and non-neural signaling. Physiology (Bethesda) 2020;35:16–30. doi: 10.1152/physiol.00027.2019. [DOI] [PubMed] [Google Scholar]
- Formicki K., Korzelecka-Orkisz A., Tanski A. Magnetoreception in fish. J. Fish. Biol. 2019;95:73–91. doi: 10.1111/jfb.13998. [DOI] [PubMed] [Google Scholar]
- Fu J., Warmflash A., Lutolf M.P. Stem-cell-based embryo models for fundamental research and translation. Nat. Mater. 2020;20:132–144. doi: 10.1038/s41563-020-00829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreta E., Kamm R.D., Chuva de Sousa Lopes S.M., Lancaster M.A., Weiss R., Trepat X., Hyun I., Montserrat N. Rethinking organoid technology through bioengineering. Nat. Mater. 2020;20:145–155. doi: 10.1038/s41563-020-00804-4. [DOI] [PubMed] [Google Scholar]
- Graner F., Riveline D. The forms of tissues, or cell-aggregates': D'arcy thompson's influence and its limits. Development. 2017;144:4226–4237. doi: 10.1242/dev.151233. [DOI] [PubMed] [Google Scholar]
- Green J.B., Sharpe J. Positional information and reaction-diffusion: two big ideas in developmental biology combine. Development. 2015;142:1203–1211. doi: 10.1242/dev.114991. [DOI] [PubMed] [Google Scholar]
- Guye P., Ebrahimkhani M.R., Kipniss N., Velazquez J.J., Schoenfeld E., Kiani S., Griffith L.G., Weiss R. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase K., Freedman B.S. Once upon a dish: engineering multicellular systems. Development. 2020;147:dev188573. doi: 10.1242/dev.188573. [DOI] [PubMed] [Google Scholar]
- Hamann H., Wahby M., Schmickl T., Zahadat P., Hofstadler D., Stoy K., Risi S., Faina A., Veenstra F., Kernbach S. Ieee Ssci); 2015. Flora Robotica - Mixed Societies of Symbiotic Robot-Plant Bio-Hybrids. 2015 Ieee Symposium Series on Computational Intelligence; pp. 1102–1109. [Google Scholar]
- Hannezo E., Heisenberg C.P. Mechanochemical feedback loops in development and disease. Cell. 2019;178:12–25. doi: 10.1016/j.cell.2019.05.052. [DOI] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., Christodoulou N., Kyprianou C., Zernicka-Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:eaal1810. doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- Hinard V., Belin D., Konig S., Bader C.R., Bernheim L. Initiation of human myoblast differentiation via dephosphorylation of Kir2.1 K+ channels at tyrosine 242. Development. 2008;135:859–867. doi: 10.1242/dev.011387. [DOI] [PubMed] [Google Scholar]
- Isalan M., Lemerle C., Serrano L. Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 2005;3:e64. doi: 10.1371/journal.pbio.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo E., Quinkler T., De Renzis S. Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat. Commun. 2018;9:2366. doi: 10.1038/s41467-018-04754-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewhurst K., Levin M., McLaughlin K.A. Optogenetic control of apoptosis in targeted tissues of Xenopus laevis embryos. J. Cell Death. 2014;7:25–31. doi: 10.4137/JCD.S18368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A., Cotterell J., Munteanu A., Sharpe J. A spectrum of modularity in multi-functional gene circuits. Mol. Syst. Biol. 2017;13:925. doi: 10.15252/msb.20167347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H.E., Djabrayan N.J.V., Shvartsman S.Y., Toettcher J.E. Optogenetic rescue of a patterning mutant. Curr. Biol. 2020;30:3414–3424.e3. doi: 10.1016/j.cub.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.B., March A.R., Morsut L. Engineering multicellular systems: using synthetic biology to control tissue self-organization. Curr. Opin. Biomed. Eng. 2017;4:163–173. doi: 10.1016/j.cobme.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm R.D., Bashir R., Arora N., Dar R.D., Gillette M.U., Griffith L.G., Kemp M.L., Kinlaw K., Levin M., Martin A.C. Perspective: the promise of multi-cellular engineered living systems. APL Bioeng. 2018;2:040901. doi: 10.1063/1.5038337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchis-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegman S., Blackiston D., Levin M., Bongard J. A scalable pipeline for designing reconfigurable organisms. Proc. Natl. Acad. Sci. U S A. 2020;117:1853–1859. doi: 10.1073/pnas.1910837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunche S., Yan H., Calof A.L., Lowengrub J.S., Lander A.D. Feedback, lineages and self-organizing morphogenesis. PLoS Comput. Biol. 2016;12:e1004814. doi: 10.1371/journal.pcbi.1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Huch M. Disease modelling in human organoids. Dis. Model. Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D.R. Advanced bioscience and AI: debugging the future of life. Emerg. Top. Life Sci. 2019;3:747–751. doi: 10.1042/ETLS20180069. [DOI] [PubMed] [Google Scholar]
- Lehman J., Clune J., Misevic D., Adami C., Altenberg L., Beaulieu J., Bentley P.J., Bernard S., Beslon G., Bryson D.M. The surprising creativity of digital evolution: a collection of anecdotes from the evolutionary computation and artificial life research communities. Artif. Life. 2020;26:274–306. doi: 10.1162/artl_a_00319. [DOI] [PubMed] [Google Scholar]
- Levin M. Reprogramming cells and tissue patterning via bioelectrical pathways: molecular mechanisms and biomedical opportunities. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013;5:657–676. doi: 10.1002/wsbm.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration. J. Physiol. 2014;592:2295–2305. doi: 10.1113/jphysiol.2014.271940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. The computational boundary of a “self”: developmental bioelectricity drives multicellularity and scale-free cognition. Front. Psychol. 2019;10:2688. doi: 10.3389/fpsyg.2019.02688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. Life, death, and self: fundamental questions of primitive cognition viewed through the lens of body plasticity and synthetic organisms. Biochem. Biophysical Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.10.077. [DOI] [PubMed] [Google Scholar]
- Levin M. Bioelectric signaling: reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell. 2021;184:1971–1989. doi: 10.1016/j.cell.2021.02.034. [DOI] [PubMed] [Google Scholar]
- Levin M., Bongard J., Lunshof J.E. Applications and ethics of computer-designed organisms. Nat. Rev. Mol. Cell Biol. 2020;21:655–656. doi: 10.1038/s41580-020-00284-z. [DOI] [PubMed] [Google Scholar]
- Levin M., Martinez Arias A. Reverse-engineering Growth and Form in Heidelberg. Development. 2019;146:dev177261. doi: 10.1242/dev.177261. [DOI] [PubMed] [Google Scholar]
- Levin M., Martyniuk C.J. The bioelectric code: an ancient computational medium for dynamic control of growth and form. Biosystems. 2018;164:76–93. doi: 10.1016/j.biosystems.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Pezzulo G., Finkelstein J.M. Endogenous bioelectric signaling networks: exploiting voltage gradients for control of growth and form. Annu. Rev. Biomed. Eng. 2017;19:353–387. doi: 10.1146/annurev-bioeng-071114-040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Selberg J., Rolandi M. Endogenous bioelectrics in development, cancer, and regeneration: drugs and bioelectronic devices as electroceuticals for regenerative medicine. iScience. 2019;22:519–533. doi: 10.1016/j.isci.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Ye J., Zhao D., Zhang S.-J. Magnetogenetics: remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Sci. Bull. 2015;60:2107–2119. doi: 10.1007/s11434-015-0902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell J., Pollack J. Proceedings of the Artificial Life Conference 2016. MIT Press; 2016. Developmental encodings promote the emergence of hierarchical modularity. [Google Scholar]
- Macia J., Posas F., Sole R.V. Distributed computation: the new wave of synthetic biology devices. Trends Biotechnol. 2012;30:342–349. doi: 10.1016/j.tibtech.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Macia J., Vidiella B., Sole R.V. Synthetic associative learning in engineered multicellular consortia. J. R. Soc. Interf. 2017;14:20170158. doi: 10.1098/rsif.2017.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man K., Damasio A. Homeostasis and soft robotics in the design of feeling machines. Nat. Mach. Intell. 2019;1:446–452. [Google Scholar]
- Manicka S., Levin M. Modeling somatic computation with non-neural bioelectric networks. Sci. Rep. 2019;9:18612. doi: 10.1038/s41598-019-54859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Hayashi H., Garcia-Ojalvo J., Yoshioka-Kobayashi K., Kageyama R., Yamanaka Y., Ikeya M., Toguchida J., Alev C., Ebisuya M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020;369:1450–1455. doi: 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Koga M., Nishida E., Ebisuya M. Synthetic signal propagation through direct cell-cell interaction. Sci. Signal. 2012;5:ra31. doi: 10.1126/scisignal.2002764. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Yamanaka Y., Uemura M., Osawa M., Saito M.K., Nagahashi A., Nishio M., Guo L., Ikegawa S., Sakurai S. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 2020;580:124–129. doi: 10.1038/s41586-020-2144-9. [DOI] [PubMed] [Google Scholar]
- McDonald D., Wu Y., Dailamy A., Tat J., Parekh U., Zhao D., Hu M., Tipps A., Zhang K., Mali P. Defining the teratoma as a model for multi-lineage human development. Cell. 2020;183:1402–1419 e1418. doi: 10.1016/j.cell.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin K.A., Levin M. Bioelectric signaling in regeneration: mechanisms of ionic controls of growth and form. Dev. Biol. 2018;433:177–189. doi: 10.1016/j.ydbio.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger J.J., Simunovic M., Brivanlou A.H. Synthetic embryology: controlling geometry to model early mammalian development. Curr. Opin. Genet. Dev. 2018;52:86–91. doi: 10.1016/j.gde.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milias-Argeitis A., Rullan M., Aoki S.K., Buchmann P., Khammash M. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat. Commun. 2016;7:12546. doi: 10.1038/ncomms12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.J., Davidson L.A. The interplay between cell signalling and mechanics in developmental processes. Nat. Rev. Genet. 2013;14:733–744. doi: 10.1038/nrg3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.F. Springer Berlin Heidelberg; 2004. Evolving a Self-Repairing, Self-Regulating, French Flag Organism. [Google Scholar]
- Mustard J., Levin M. Bioelectrical mechanisms for programming growth and form: taming physiological networks for soft body robotics. Soft Robot. 2014;1:169–191. [Google Scholar]
- Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., Saito K., Yonemura S., Eiraku M., Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Nandagopal N., Santat L.A., LeBon L., Sprinzak D., Bronner M.E., Elowitz M.B. Dynamic ligand discrimination in the notch signaling pathway. Cell. 2018;172:869–880 e819. doi: 10.1016/j.cell.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth J.C., Lee H., Feinberg A.W., Ripplinger C.M., McCain M.L., Grosberg A., Dabiri J.O., Parker K.K. A tissue-engineered jellyfish with biomimetic propulsion. Nat. Biotechnol. 2012;30:792–797. doi: 10.1038/nbt.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman S.A. Physico-genetic determinants in the evolution of development. Science. 2012;338:217–219. doi: 10.1126/science.1222003. [DOI] [PubMed] [Google Scholar]
- Newman S.A. Physico-genetics of morphogenesis: the hybrid nature of developmental mechanisms. In: Minelli Alessandro, Pradeu Thomas., editors. Towards a Theory of Development. Oxford Scholarship Online; 2014. pp. 95–113. [Google Scholar]
- Newman S.A. Inherency of form and function in animal development and evolution. Front. Physiol. 2019;10:702. doi: 10.3389/fphys.2019.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson D.J. The concept of mechanism in biology. Stud. Hist. Philos. Biol. Biomed. Sci. 2012;43:152–163. doi: 10.1016/j.shpsc.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Nicholson D.J. Organisms not equal Machines. Stud. Hist. Philos. Biol. Biomed. Sci. 2013;44:669–678. doi: 10.1016/j.shpsc.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Nicholson D.J. The machine conception of the organism in development and evolution: a critical analysis. Stud. Hist. Philos. Biol. Biomed. Sci. 2014;48 Pt B:162–174. doi: 10.1016/j.shpsc.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Nicholson D.J. Is the cell really a machine? J. Theor. Biol. 2019;477:108–126. doi: 10.1016/j.jtbi.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Nikolaev M., Mitrofanova O., Broguiere N., Geraldo S., Dutta D., Tabata Y., Elci B., Brandenberg N., Kolotuev I., Gjorevski N. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- Nims R.J., Pferdehirt L., Ho N.B., Savadipour A., Lorentz J., Sohi S., Kassab J., Ross A.K., O’Conor C.J., Liedtke W.B. A synthetic mechanogenetic gene circuit for autonomous drug delivery in engineered tissues. Sci Adv. 2021;7:eabd9858. doi: 10.1126/sciadv.abd9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olle-Vila A., Duran-Nebreda S., Conde-Pueyo N., Montanez R., Sole R. A morphospace for synthetic organs and organoids: the possible and the actual. Integr. Biol. 2016;8:485–503. doi: 10.1039/c5ib00324e. [DOI] [PubMed] [Google Scholar]
- Ouyang X., Chen J.K. Synthetic strategies for studying embryonic development. Chem. Biol. 2010;17:590–606. doi: 10.1016/j.chembiol.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan-Diaz G.J., Zhang X., Grant L., Kim Y., Aydin O., Cvetkovic C., Ko E., Solomon E., Hollis J., Kong H.J. Simulation and fabrication of stronger, larger, and faster walking biohybrid machines. Adv. Funct. Mater. 2018;28:1801145. [Google Scholar]
- Park S.J., Gazzola M., Park K.S., Park S., Di Santo V., Blevins E.L., Lind J.U., Campbell P.H., Dauth S., Capulli A.K. Phototactic guidance of a tissue-engineered soft-robotic ray. Science. 2016;353:158–162. doi: 10.1126/science.aaf4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascalie J., Potier M., Kowaliw T., Giavitto J.L., Michel O., Spicher A., Doursat R. Developmental design of synthetic bacterial architectures by morphogenetic engineering. ACS Synth. Biol. 2016;5:842–861. doi: 10.1021/acssynbio.5b00246. [DOI] [PubMed] [Google Scholar]
- Pezzulo G., Levin M. Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integr. Biol. 2015;7:1487–1517. doi: 10.1039/c5ib00221d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo G., Levin M. Top-down models in biology: explanation and control of complex living systems above the molecular level. J. R. Soc. Interf. 2016;13:20160555. doi: 10.1098/rsif.2016.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanyi M. Life's irreducible structure. Live mechanisms and information in DNA are boundary conditions with a sequence of boundaries above them. Science. 1968;160:1308–1312. doi: 10.1126/science.160.3834.1308. [DOI] [PubMed] [Google Scholar]
- Prindle A., Liu J., Asally M., Ly S., Garcia-Ojalvo J., Suel G.M. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahwan I., Cebrian M., Obradovich N., Bongard J., Bonnefon J.F., Breazeal C., Crandall J.W., Christakis N.A., Couzin I.D., Jackson M.O. Machine behaviour. Nature. 2019;568:477–486. doi: 10.1038/s41586-019-1138-y. [DOI] [PubMed] [Google Scholar]
- Raj A., Thakur A. Fish-inspired robots: design, sensing, actuation, and autonomy--a review of research. Bioinspir. Biomim. 2016;11:031001. doi: 10.1088/1748-3190/11/3/031001. [DOI] [PubMed] [Google Scholar]
- Raspopovic J., Marcon L., Russo L., Sharpe J. Modeling digits. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science. 2014;345:566–570. doi: 10.1126/science.1252960. [DOI] [PubMed] [Google Scholar]
- Rieffel J.A., Valero-Cuevas F.J., Lipson H. Morphological communication: exploiting coupled dynamics in a complex mechanical structure to achieve locomotion. J. R. Soc. Interf. 2010;7:613–621. doi: 10.1098/rsif.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.C., Korving J., Vivie J., Truckenmuller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Roost M.S., van Iperen L., Ariyurek Y., Buermans H.P., Arindrarto W., Devalla H.D., Passier R., Mummery C.L., Carlotti F., de Koning E.J. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Rep. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado-Olivieri E.A., Brivanlou A.H. Synthetic by design: exploiting tissue self-organization to explore early human embryology. Dev. Biol. 2021;474:16–21. doi: 10.1016/j.ydbio.2021.01.004. [DOI] [PubMed] [Google Scholar]
- Rosen R. Are dynamics of a system operationally determinable. J. Theor. Biol. 1972;36:635. doi: 10.1016/0022-5193(72)90014-8. [DOI] [PubMed] [Google Scholar]
- Russo R.S., Blemker S.S., Fish F.E., Bart-Smith H. Biomechanical model of batoid (skates and rays) pectoral fins predicts the influence of skeletal structure on fin kinematics: implications for bio-inspired design. Bioinspir. Biomim. 2015;10:046002. doi: 10.1088/1748-3190/10/4/046002. [DOI] [PubMed] [Google Scholar]
- Sagner A., Briscoe J. Establishing neuronal diversity in the spinal cord: a time and a place. Development. 2019;146 doi: 10.1242/dev.182154. [DOI] [PubMed] [Google Scholar]
- Sample M., Boulicault M., Allen C., Bashir R., Hyun I., Levis M., Lowenthal C., Mertz D., Montserrat N., Palmer M.J. Multi-cellular engineered living systems: building a community around responsible research on emergence. Biofabrication. 2019;11:043001. doi: 10.1088/1758-5090/ab268c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli M., Lam C., Morsut L. Synthetic development: building mammalian multicellular structures with artificial genetic programs. Curr. Opin. Biotechnol. 2019;59:130–140. doi: 10.1016/j.copbio.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schaerli Y., Munteanu A., Gili M., Cotterell J., Sharpe J., Isalan M. A unified design space of synthetic stripe-forming networks. Nat. Commun. 2014;5:4905. doi: 10.1038/ncomms5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine R., Shibata T., Ebisuya M. Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. Nat. Commun. 2018;9:5456. doi: 10.1038/s41467-018-07847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Fu J. Synthetic human embryology: towards a quantitative future. Curr. Opin. Genet. Dev. 2020;63:30–35. doi: 10.1016/j.gde.2020.02.013. [DOI] [PubMed] [Google Scholar]
- Silver B.B., Nelson C.M. The bioelectric code: reprogramming cancer and aging from the interface of mechanical and chemical microenvironments. Front. Cell Dev. Biol. 2018;6:21. doi: 10.3389/fcell.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simian M., Bissell M.J. Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H.L., Grainger R.M., Harland R.M. Animal cap isolation from Xenopus laevis. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4744. pdb prot4744. [DOI] [PubMed] [Google Scholar]
- Skylar-Scott M.A., Huang J.Y., Lu A., Ng A.H.M., Duenki T., Nam L.L., Damaraju S., Church G.M., Lewis J.A. An orthogonal differentiation platform for genomically programming stem cells, organoids, and bioprinted tissues. bioRxiv. 2020 doi: 10.1101/2020.07.11.198671. [DOI] [Google Scholar]
- Slavkov I., Carrillo-Zapata D., Carranza N., Diego X., Jansson F., Kaandorp J., Hauert S., Sharpe J. Morphogenesis in robot swarms. Sci. Robot. 2018;3:eaau9178. doi: 10.1126/scirobotics.aau9178. [DOI] [PubMed] [Google Scholar]
- Slusarczyk A.L., Weiss R. Understanding signaling dynamics through synthesis. Sci. Signal. 2012;5:pe16. doi: 10.1126/scisignal.2003092. [DOI] [PubMed] [Google Scholar]
- Smith S.J., Rebeiz M., Davidson L. From pattern to process: studies at the interface of gene regulatory networks, morphogenesis, and evolution. Curr. Opin. Genet. Dev. 2018;51:103–110. doi: 10.1016/j.gde.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole R., Amor D.R., Duran-Nebreda S., Conde-Pueyo N., Carbonell-Ballestero M., Montanez R. Synthetic collective intelligence. Biosystems. 2016;148:47–61. doi: 10.1016/j.biosystems.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Sole R., Macia J. Synthetic biology: biocircuits in synchrony. Nature. 2014;508:326–327. doi: 10.1038/nature13224. [DOI] [PubMed] [Google Scholar]
- Sole R., Ollé-Vila A., Vidiella Rocamora B., Duran-Nebreda S., Conde-Pueyo N. The road to synthetic multicellularity. Curr. Opin. Syst. Biol. 2018;7:60–67. [Google Scholar]
- Sole R.V., Macia J. Expanding the landscape of biological computation with synthetic multicellular consortia. Nat. Comput. 2013;12:485–497. [Google Scholar]
- Sozen B., Amadei G., Cox A., Wang R., Na E., Czukiewska S., Chappell L., Voet T., Michel G., Jing N. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 2018;20:979–989. doi: 10.1038/s41556-018-0147-7. [DOI] [PubMed] [Google Scholar]
- Sozen B., Cox A.L., De Jonghe J., Bao M., Hollfelder F., Glover D.M., Zernicka-Goetz M. Self-organization of mouse stem cells into an extended potential blastoid. Dev. Cell. 2019;51:698–712 e698. doi: 10.1016/j.devcel.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F., Chavarha M., Lin M.Z. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 2015;27:31–38. doi: 10.1016/j.cbpa.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley K.O., Miikkulainen R. A taxonomy for artificial embryogeny. Artif. Life. 2003;9:93–130. doi: 10.1162/106454603322221487. [DOI] [PubMed] [Google Scholar]
- Steinberg M.S. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science. 1963;141:401–408. doi: 10.1126/science.141.3579.401. [DOI] [PubMed] [Google Scholar]
- Sullivan K.G., Emmons-Bell M., Levin M. Physiological inputs regulate species-specific anatomy during embryogenesis and regeneration. Commun. Integr. Biol. 2016;9:e1192733. doi: 10.1080/19420889.2016.1192733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundelacruz S., Levin M., Kaplan D.L. Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev. Rep. 2009;5:231–246. doi: 10.1007/s12015-009-9080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T., Wells J.M. Organoids by design. Science. 2019;364:956–959. doi: 10.1126/science.aaw7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague B.P., Guye P., Weiss R. Synthetic morphogenesis. Cold Spring Harb. Perspect. Biol. 2016;8:a023929. doi: 10.1101/cshperspect.a023929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., Blauch L.R., Tang S.K.Y., Morsut L., Lim W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., McKeithan W.L., Hakkinen T.J., Lopez P., Klein O.D., Lim W.A. Engineering synthetic morphogen systems that can program multicellular patterning. Science. 2020;370:327–331. doi: 10.1126/science.abc0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K., Kime C. Synthetic embryology: early mammalian embryo modeling systems from cell cultures. Dev. Growth Differ. 2021;63:116–126. doi: 10.1111/dgd.12713. [DOI] [PubMed] [Google Scholar]
- Treger J.S., Priest M.F., Bezanilla F. Single-molecule fluorimetry and gating currents inspire an improved optical voltage indicator. Elife. 2015;4:e10482. doi: 10.7554/eLife.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda S., Artmann S., Zauner K.-P. The Phi-bot: a robot controlled by a slime mould. In: Adamatzky A., Komosinski M., editors. Artificial Life Models in Hardware. Springer London; 2009. pp. 213–232. [Google Scholar]
- Tsuda S., Zauner K.P., Gunji Y.P. Robot control with biological cells. Bio Syst. 2007;87:215–223. doi: 10.1016/j.biosystems.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Urrios A., Macia J., Manzoni R., Conde N., Bonforti A., de Nadal E., Posas F., Sole R. A synthetic multicellular memory device. ACS Synth. Biol. 2016;5:862–873. doi: 10.1021/acssynbio.5b00252. [DOI] [PubMed] [Google Scholar]
- Valentini G., Moore D.G., Hanson J.R., Pavlic T.P., Pratt S.C., Walker S.I. 2018 Conference on Artificial Life (Alife 2018) 2018. Transfer of information in collective decisions by artificial agents; pp. 641–648. [Google Scholar]
- Vandenberg L.N., Morrie R.D., Adams D.S. V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev Dyn. 2011;240:1889–1904. doi: 10.1002/dvdy.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne F., Chaigneau P., Petitot J., Doursat R. Programming the emergence in morphogenetically architected complex systems. Acta Biotheor. 2015;63:295–308. doi: 10.1007/s10441-015-9262-z. [DOI] [PubMed] [Google Scholar]
- Velazquez J.J., LeGraw R., Moghadam F., Tan Y., Kilbourne J., Maggiore J.C., Hislop J., Liu S., Cats D., Chuva de Sousa Lopes S.M. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 2020;12:41–55.e11. doi: 10.1016/j.cels.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez J.J., Su E., Cahan P., Ebrahimkhani M.R. Programming morphogenesis through systems and synthetic biology. Trends Biotechnol. 2018;36:415–429. doi: 10.1016/j.tibtech.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verd B., Monk N.A., Jaeger J. Modularity, criticality, and evolvability of a developmental gene regulatory network. Elife. 2019;8:e42832. doi: 10.7554/eLife.42832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villoutreix P., Delile J., Rizzi B., Duloquin L., Savy T., Bourgine P., Doursat R., Peyrieras N. An integrated modelling framework from cells to organism based on a cohort of digital embryos. Sci. Rep. 2016;6:37438. doi: 10.1038/srep37438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola J.M., Porter C.M., Gupta A., Alibekova M., Prahl L.S., Hughes A.J. Guiding cell network assembly using shape-morphing hydrogels. Adv. Mater. 2020;32:e2002195. doi: 10.1002/adma.202002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mammen S., Hamann H., Heider M. 22nd Acm Conference on Virtual Reality Software and Technology (Vrst 2016. 2016. Robot gardens: an augmented reality prototype for plant-robot biohybrid systems; pp. 139–142. [Google Scholar]
- Wagner G.P., Pavlicev M., Cheverud J.M. The road to modularity. Nat. Rev. Genet. 2007;8:921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- Wahby M., Hofstadler D.N., Heinrich M.K., Zahadat P., Hamann H. 10th IEEE International Conference on Self-Adaptive and Self-Organizing Systems. 2016. An evolutionary robotics approach to the control of plant growth and motion: modeling plants and crossing the reality gap; pp. 21–30. [Google Scholar]
- Wan L.Q., Chin A.S., Worley K.E., Ray P. Cell chirality: emergence of asymmetry from cell culture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L.Q., Ronaldson K., Park M., Taylor G., Zhang Y., Gimble J.M., Vunjak-Novakovic G. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc. Natl. Acad. Sci. U S A. 2011;108:12295–12300. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L.Q., Vunjak-Novakovic G. Micropatterning chiral morphogenesis. Commun. Integr. Biol. 2011;4:745–748. doi: 10.4161/cib.17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W., Daoud-El Baba M., Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc. Natl. Acad. Sci. U S A. 2007;104:10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltschko R., Wiltschko W. Magnetoreception in birds. J. R. Soc. Interf. 2019;16:20190295. doi: 10.1098/rsif.2019.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzany G., Baluska F. Life's code script does not code itself. The machine metaphor for living organisms is outdated. EMBO Rep. 2012;13:1054–1056. doi: 10.1038/embor.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff L.B.A., Gorochowski T.E., Roehner N., Mikkelsen T.S., Densmore D., Gordon D.B., Nicol R., Voigt C.A. Registry in a tube: multiplexed pools of retrievable parts for genetic design space exploration. Nucleic Acids Res. 2017;45:1553–1565. doi: 10.1093/nar/gkw1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Schmidt J.J., Montemagno C.D. Self-assembled microdevices driven by muscle. Nat. Mater. 2005;4:180–184. doi: 10.1038/nmat1308. [DOI] [PubMed] [Google Scholar]
- Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- Yang C.Y., Bialecka-Fornal M., Weatherwax C., Larkin J.W., Prindle A., Liu J., Garcia-Ojalvo J., Suel G.M. Encoding membrane-potential-based memory within a microbial community. Cell Syst. 2020;10:417–423.e3. doi: 10.1016/j.cels.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L., Cox R.S., 3rd, Weiss R., Arnold F.H. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- Zagorski M., Tabata Y., Brandenberg N., Lutolf M.P., Tkacik G., Bollenbach T., Briscoe J., Kicheva A. Decoding of position in the developing neural tube from antiparallel morphogen gradients. Science. 2017;356:1379–1383. doi: 10.1126/science.aam5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Chen T., Chen N., Gao D., Shi B., Kong S., West R.C., Yuan Y., Zhi M., Wei Q. Implantation initiation of self-assembled embryo-like structures generated using three types of mouse blastocyst-derived stem cells. Nat. Commun. 2019;10:496. doi: 10.1038/s41467-019-08378-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinner M., Lukonin I., Liberali P. Design principles of tissue organisation: how single cells coordinate across scales. Curr. Opin. Cell Biol. 2020;67:37–45. doi: 10.1016/j.ceb.2020.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data and codes were newly generated for this paper.