SUMMARY
Gut microbiota educate the local and distal immune system in early life to imprint long-term immunological outcomes while maintaining the capacity to dynamically modulate the local mucosal immune system throughout life. It is unknown whether gut microbiota provide signals that dynamically regulate distal immune responses following an extra-gastrointestinal infection. We show here that gut bacteria composition correlated with the severity of malaria in children. Using the murine model of malaria, we demonstrate that parasite burden and spleen germinal center reactions are malleable to dynamic cues provided by gut bacteria. Whereas antibiotic-induced changes in gut bacteria have been associated with immunopathology or impairment of immunity, the data demonstrate that antibiotic-induced changes in gut bacteria can enhance immunity to Plasmodium. This effect is not universal but depends on baseline gut bacteria composition. These data demonstrate the dynamic communications that exist among gut bacteria, the gut-distal immune system, and control of Plasmodium infection.
Graphical abstract

In brief
Mandal et al. demonstrate that gut bacteria composition correlates with the severity of malaria in children and, through the use of the murine model of malaria, examine the dynamic communications that exist among gut bacteria, the systemic immune system, and the plasticity of spleen germinal center reactions during Plasmodium infection.
INTRODUCTION
Gut microbiota, the community of microorganisms living in the gastrointestinal tract, have diverse effects on host biology. One important role of gut microbiota in early life is to educate the mucosal and systemic immune system to reduce the risk of immunopathology that may occur later in life. In mice, neonatal exposure to gut bacteria protects adult mice against experimental inflammatory bowel disease (IBD) via exposure to sphingolipids that inhibit the accumulation of mucosal invariant natural killer T (iNKT) cells (An et al., 2014; Olszak et al., 2012). Early-life exposure to high-diversity gut microbiota also prevents elevated serum immunoglobulin (Ig) E in mice and reduces the risk of oral-induced anaphylaxis (Cahenzli et al., 2013). The reported increase in serum IgE after weaning is consistent with gut microbiota composition in mice programming the immune system during weaning through induction of regulatory T cells to resist inflammatory pathologies into adulthood (Al Nabhani et al., 2019). Perturbations of the microbiota during this critical weaning window alter immune education, which results in increased susceptibility to immunopathologies (colitis, allergic inflammation, and cancer) into adulthood (Al Nabhani et al., 2019) and reduced responsiveness in adolescent mice to numerous adjuvanted and non-adjuvanted vaccines (Lynn et al., 2018). Gut-microbiota-dependent education of the immune system also occurs in humans. The type of lipopolysaccharide (LPS) present in early life, which is influenced by gut bacteria composition, can educate the immune system to be resistant or susceptible to autoimmune diseases, including type 1 diabetes, later in life (Vatanen et al., 2016). In addition, events in early life that can alter gut microbiota composition (e.g., caesarean section, formula feeding, infection/inflammation, and antibiotic exposure) can pathologically imprint the immune system and increase the risk of childhood asthma and atopy (Arrieta et al., 2015; Fujimura et al., 2016; Shao et al., 2019). Consistent with gut microbiota educating or priming the host immune system to respond in a prescribed nature, mice treated with antibiotics 2–4 weeks before viral infection (influenza or lymphocytic choriomeningitis virus [LCMV]) or administration of the seasonal influenza vaccine had reduced adaptive immune responses (Abt et al., 2012; Ichinohe et al., 2011; Oh et al., 2014), resulting in impaired control of the virus (Abt et al., 2012; Ichinohe et al., 2011). It has also been shown that humans treated with antibiotics before administration of the seasonal influenza vaccine exhibited reduced antibody titers to one of three viral strains present in the vaccine (Hagan et al., 2019). Although antibiotics are extremely beneficial at eliminating pathogenic bacteria, these data support the idea that antibiotic-induced changes to gut bacteria populations can be detrimental to host immunity.
The interaction between the gut microbiome and the intestinal mucosal immune system is also dynamic and extends beyond early life (Wang et al., 2019). For example, imbalanced gut microbiota composition leads to altered interactions between the gut microbiome and the gut mucosal immune system; as a consequence, the host becomes susceptible to bacterial translocation, gut-derived infection, and detrimental clinical outcomes like IBD, autoimmunity, and allergy (Khan et al., 2019; Round and Mazmanian, 2009; Schuijt et al., 2013). However, it is not known whether gut microbiota also provide dynamic modulation of a gut-distal immune response to an extra-intestinal infection.
Plasmodium species are the causative agents of malaria and were responsible for 228 million episodes of malaria and 405,000 deaths in 2018 (WHO, 2019). Plasmodium infections are initiated when an infected mosquito delivers sporozoites during a blood meal. Sporozoites infect hepatocytes, initiating the liver-stage infection at which they differentiate into merozoites that are released into the circulation and establish cyclical infection of red blood cells (Cowman et al., 2016). The blood-stage infection is controlled by the formation of germinal center (GC) reactions that result in the production of high-affinity antibodies that mediate clearance of the parasite (Figueiredo et al., 2017; Guthmiller et al., 2017; Obeng-Adjei et al., 2015; Pérez-Mazliah et al., 2015, 2017; Ryg-Cornejo et al., 2016).
Recent research has identified a connection between gut microbiota and malaria. For example, gut microbiota, in particular Escherichia coli O86:B7, can induce cross-reactive antibodies that confer protection against sporozoites, although these antibodies did not provide protection against Plasmodium blood-stage infection (Yilmaz et al., 2014). Another study, in which Malian children were longitudinally tracked from the end of the dry season (i.e., no malaria transmission) through the ensuing malaria transmission season, demonstrated a correlation between stool bacteria composition at the end of the dry season and rate at which children were infected with Plasmodium falciparum (Yooseph et al., 2015); for ethical reasons, this report did not assess severe malaria. Finally, the composition of gut microbiota has been shown to shape the severity of malaria in mice (Denny et al., 2019; Stough et al., 2016; Villarino et al., 2016) by fine-tuning the magnitude and quality of the humoral immune response to Plasmodium (Waide et al., 2020). Yet numerous knowledge gaps remain in understanding malaria-gut microbiome interactions. It is not known whether the gut microbiome affects the severity of malaria in humans or, if it does, which of the gut microbiota constituents (bacteria, viruses, fungi, etc.) affect the severity of malaria. It is also unknown how gut microbiota interact with the host immune system to ultimately affect the parasite burden and the severity of malaria.
Here, we identify differences in stool bacteria composition between Ugandan children with asymptomatic Plasmodium falciparum infection and those with severe malarial anemia. We also report that among gut microbiota constituents, gut bacteria modulate the severity of malaria in mice. We show in mice that gut bacteria provide cues that dynamically modulate parasite burden and spleen GC reactions, providing a potential explanation for the protection from severe malaria associated with stool bacteria compositions in the Ugandan children. Collectively, these data provide new insight into the dynamic communications that exist between the gut bacteria and the systemic immune system and show that the effect of antibiotic-induced changes in gut bacteria populations on host immunity depends on the baseline bacteria composition. These data also support the possibility that human gut bacteria may affect the severity of malaria in children.
RESULTS
Stool bacteria populations correlate with severity of malaria in Ugandan children
To determine whether gut bacteria composition is associated with severity of malaria in humans, bacteria populations were analyzed by Multiple 16S Variable Region Species-Level Identification (MVRSION) (Schriefer et al., 2018) in stool collected from Ugandan children who had severe malaria anemia (SMA; n = 40) or community control children who were otherwise healthy but had asymptomatic Plasmodium falciparum (Pf) infections (Pf Pos; n = 7) or were Pf negative (Pf Neg; n = 28) (Figure 1A). Although there was a relatively low number of samples of children with asymptomatic Pf parasitemia (n = 7), the bacterial species accumulation curve showed a plateau in the curve within this group (Figure S1A), suggesting that additional samples may not have dramatically increased the total number of bacterial species identified in this group.
Figure 1. Gut microbiota composition is associated with severity of malaria in humans.
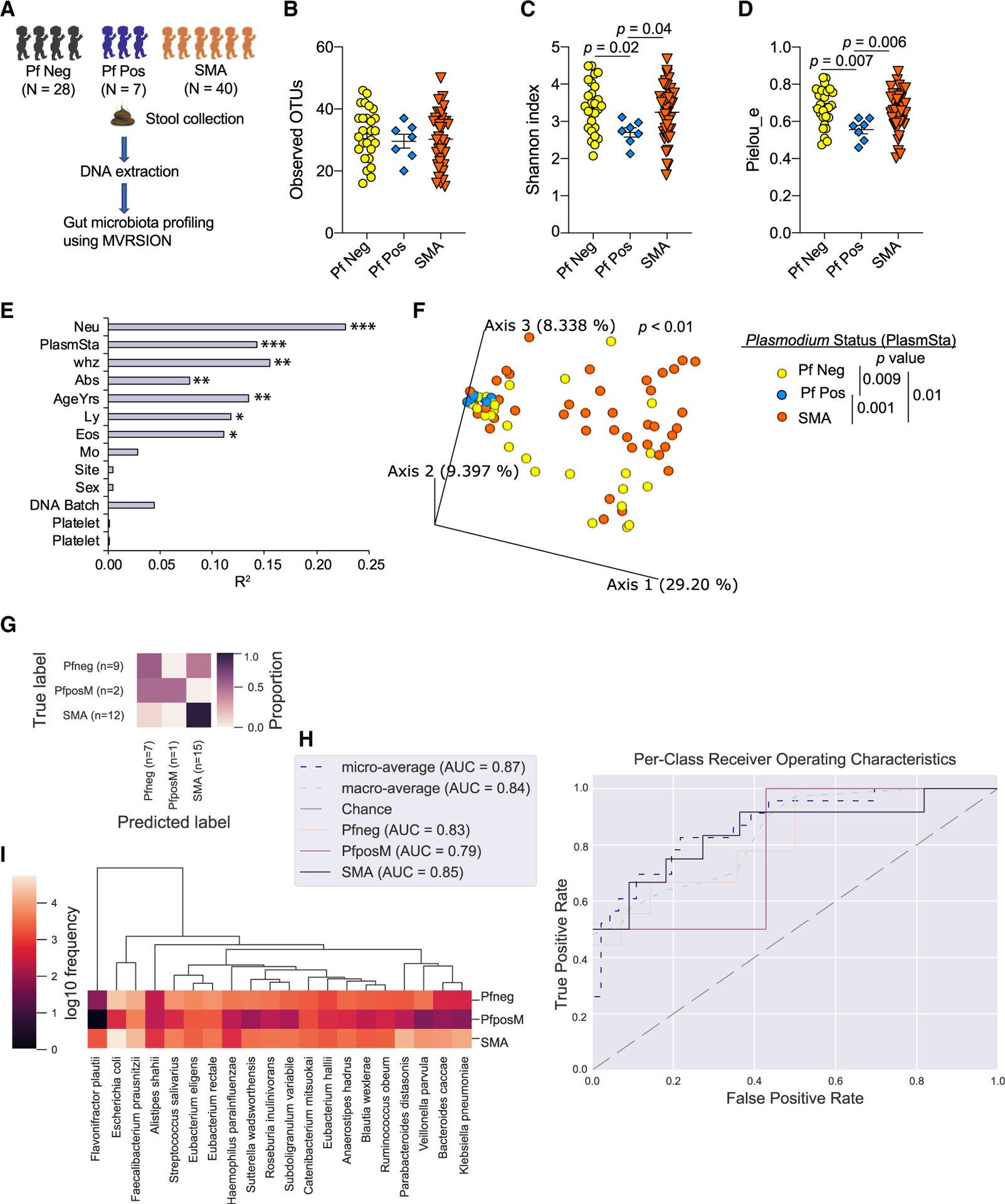
(A) Stool samples were collected from children diagnosed with severe malaria anemia (SMA, n = 40) at the time of hospitalization but before antimalarial treatment and from community control children, Pf negative (Pf Neg) children (n = 28), and asymptomatic Pf positive (Pf Pos) children (n = 7) at the time of enrollment; they stored at −80°C. Stools were subjected to gut microbiome analysis using 16S rRNA gene sequencing using MVRSION. One Pf Neg sample was removed from alpha and beta diversity analysis because of low sequencing depth.
(B–D) Alpha diversity analysis of Plasmodium status measured using observed OTUs (B), the Shannon index (C), and pielou_e (D).
(E) Beta diversity variation explained by different covariates using the Bray-Curtis distance matrix.
(F) Principal-coordinate analysis (PCoA) plot shows clustering of based on Plasmodium status using the Bray-Curtis distance.
(G–I) Predicting categorical sample (PlasmSta) with supervised machine-learning classifiers using random forest. (G) Result of prediction with test set data. (H) Per-class receiver operating characteristics curve. (I) Top 20 species that are the most predictive of the three groups.
Data are means ± SEM. The following statistical tests were used to analyze the data: pairwise Kruskal-Wallis test (B–D), the envfit function of the vegan package implemented in R (E), and the adonis function of the vegan package implemented in R, accounting for the covariates that significantly explained the variation of gut microbiota composition in Figure 2E (F). PlasmSta, Plasmodium status (SMA, Pf Pos, or Pf Neg); whz, weight for height Z score; Neu, Eos, Ly, Mo, and Plt, absolute count (K/mcl) of blood parameter neutrophils, eosinophils, lymphocytes, monocytes, and platelets, respectively; Abx, antibiotic use; AgeYrs, age in years; Site, study area; Sex, sex of participants; DNAbatch, batch of DNA extraction; MCV, mean corpuscular volume (in femtoliters [fl]). *p < 0.05, **p < 0.01, ***p < 0.001.
There were no differences in the stool bacterial alpha diversity (richness and abundance of bacterial species) as measured by observed operational taxonomic units (OTUs; defined using multiple 16S rRNA amplicons), which measure species richness; the Shannon index, which measures both species richness and their abundance; and pielou_e, which measures relative evenness of species richness between children based on categorical variables of interest, such as antibiotic usage, enrollment site, and sex—except for Plasmodium status, but only for pielou_e (Figure S1B). There were no significant linear correlations (−0.9 > r > 0.9 and p < 0.05) among the various continuous covariates that were investigated (Figure S1C). There were also no significant associations in observed OTUs, the Shannon index, or pielou_e and continuous covariates except for age (Figure S1D). Pielou_e was negatively correlated with age (Figure S1E). In contrast to a prior publication (Bokulich et al., 2016) but similar to our previous publication in Kenyan infants (Mandal et al., 2019), there was no significant increase in alpha diversity, as measured by observed OTUs or the Shannon index, as age increased (Figure S1D). There was no difference in observed OTUs based on the Plasmodium status of children (Figure 1B); however, the Shannon index and pielou_e were significantly different between Pf Neg versus Pf Pos and Pf Pos versus SMA (Figure 1C). These data identify differences in stool bacterial alpha diversity based on Plasmodium status of children.
Stool bacterial beta diversity (bacteria composition compared between samples) was also evaluated for the Pf Neg, Pf Pos, and SMA groups. The envfit function (vegan package) identified seven covariates that explained the significant variation in beta diversity measured by the Bray-Curtis dissimilarity distance. These included absolute neutrophil count, Plasmodium status, weight for height Z score (whz), antibiotic usage, age, absolute eosinophils, and lymphocyte count (Figure 1E; Figures S1F–S1K). Stool bacteria composition was significantly different among Plasmodium status groups (p < 0.01), accounting for all six covariates measured by the Bray-Curtis dissimilarity distance (Figure 1F). Pairwise permutational multivariate analysis of variance (PERMANOVA) analysis showed significant differences for Pf Neg versus Pf Pos (p = 0.009), Pf Neg versus SMA (p = 0.01), and Pf Pos versus SMA (p = 0.001) (Figure 1F). Age and antibiotics are known to affect the gut microbiota composition (Hasan and Yang, 2019). However, there was no significant interaction among Plasmodium status, age of child, and antibiotic usage (p > 0.05, ANOVA; Figure S2). Of note, Pf Pos versus SMA were more significantly different compared with Pf Neg versus SMA and Pf Neg versus Pf Pos (Figure 1F). Although stool samples from SMA children were collected at varying days (0–22) after hospitalization and enrollment, gut bacteria populations measured by beta and alpha diversity were not different across the varying days (Figures S2B and S2C; Table S1). This supports that the differential stool bacteria populations observed in SMA children compared with Pf Pos children were not simply products of the severe infection causing changes in gut bacteria. Further supporting an association between gut bacteria compositions and severity of Pf infections, a supervised machine-learning model using random forest was able to predict the Plasmodium status with 74% overall accuracy (Figures 1G and 1H; Figure S3A). Both higher and lower relative abundant bacterial species were predictive of Plasmodium status (Figure 1I; Figure S3B). These data identify different stool bacteria populations between Ugandan children who have an asymptomatic P. falciparum infection and children who have SMA.
Gut bacteria are mediators of gut-microbiota-dependent modulation of malaria
To gain insight into how gut microbiota modulate the severity of malaria and host immunity and to develop approaches to manipulate gut microbiota to address these knowledge gaps, reciprocal ceca content transplants (Figure S4A) were performed between C57BL/6 mice that are either resistant (Taconic Biosciences [Tac]) or susceptible (Charles River Laboratories [CR]) to developing hyperparasitemia and severe anemia following Plasmodium yoelii 17XNL infection (Villarino et al., 2016). Control Tac and CR mice that received ceca contents from Tac and CR mice, respectively, exhibited the expected low (Tac→Tac) and high (CR→CR) parasite burden following P. yoelii 17XNL infection (Figures S4B and S4C). CR ceca content transplants into Tac mice (CR→Tac) resulted in the Tac mice developing a high parasite burden similar to CR control mice, whereas Tac ceca content transplants into CR mice (Tac→CR) did not reduce their elevated parasite burden (Figures S4B and S4C). 16S rRNA gene sequence analysis was done using MVRSION to analyze gut bacteria population beta diversity, as measured by the Bray-Curtis dissimilarity distance. Consistent with the parasite burden data, the Tac→Tac mice displayed different bacterial communities from the CR→Tac, Tac→CR, and CR→CR mice (Figures S4D and S4E). The Tac and CR donor bacterial communities differentiate from the Tac→Tac and CR→CR samples because they were from the ceca contents and fecal pellets, respectively. These data demonstrated that the bacteria in CR mice are ecologically dominant over the bacteria in Tac mice. They also revealed that although ceca content transplants from CR→Tac mice provide an approach to probe how gut microbiota affect host immunity in Tac mice, they do not allow for manipulation of gut microbiota-host immunity interactions in CR mice.
Currently, it is not known which constituent of gut microbiota (fungi, virus, bacteria, etc.) affects the severity of malaria. Because stool bacteria correlated with the severity of malaria in Ugandan children (Figure 1), the contribution of gut bacteria toward severity of malaria was assessed. Mice were treated with one of five antibiotics in drinking water for 14 days before and continuously after infection with P. yoelii (Figures 2A and 2B). Each of the antibiotics resulted in a decrease in parasite burden in CR mice compared with the control CR mice, with ampicillin, gentamicin, metronidazole, and vancomycin treatment exerting the greatest effect (Figures 2C and 2D). In the Tac mice, the effect of antibiotic treatment on P. yoelii parasite burden was less pronounced, but those antibiotic treatments that had minimal effects (ampicillin, gentamicin, and vancomycin) trended toward increased parasite burden compared with the control Tac mice (Figures 2E and 2F). This effect was in contrast to the decreased parasite burden that was observed in antibiotic-treated CR mice. Given that gentamicin and vancomycin have little to no intestinal absorption (Figure 2B) (Koga et al., 2006; Rao et al., 2011) and that treatment with ampicillin, gentamicin, and vancomycin resulted in the parasite burden shifting in opposite directions compared with the control CR and Tac mice, these data suggest that these antibiotics did not have a direct effect on P. yoelii; rather, they exerted an indirect effect on parasitemia through changes in gut bacteria composition.
Figure 2. Bacteria are critical gut microbiota constituents that modulate the severity of malaria.

(A) C57BL/6 mice from Charles River (CR) and Taconic (Tac) were treated with one of five antibiotics in drinking water from two weeks before infection until the clearance of infection. Mice were infected with Plasmodium yoelii 17XNL on day 0, and parasitemia was tracked every other day from day 5 post-infection (p.i.) as indicated.
(B) Spectrum of antibiotics against bacteria and intestinal absorption (Int. abs). Gm+, Gram-positive bacteria; Gm−, Gram-negative bacteria; An, anaerobic.
(C and D) Parasitemia and area under curve (AUC) of CR mice.
(E and F) Parasitemia and AUC of Tac mice.
(G and H) Gut microbiota analysis from fecal pellets at day 0 using MVRSION. (G) Alpha diversity analysis of CR and Tac. (H) Beta diversity analysis of CR and Tac using the Bray-Curtis distance.
(I and J) Bray-Curtis distance comparison between groups of CR mice (I) and groups of Tac mice (J). The box ends show the lower and upper quartiles, and the horizontal line inside the box is the median. The y axis shows the Bray-Curtis dissimilarity distance of groups on the x axis to groups on the top of the vertical columns. Statistical significance is compared between groups on the top and groups on the x axis.
Data are means ± SEM. Statistical significance was analyzed by (D and F) one-way ANOVA with Tukey’s multiple comparison test, (G) pairwise Kruskal-Wallis test, and (I and J) pairwise PERMANOVA with 999 permutations. Significance level is compared between the group at the top of the column and the group on the x axis. (G) Only comparisons with control are shown. Data are cumulative from 2 independent experiments, with n = 4 in each trial. *p < 0.05, **p < 0.01, ****p < 0.0001.
To address the possibility of inbred strain differences as underlying contributors to the differential effect of antibiotic treatment in CR and Tac mice, Tac mice received CR fecal microbiota transplantation (FMT), or control saline treatment, for three consecutive days. Mice within each group were then left untreated or treated with vancomycin in drinking water and infected with P. yoelii the day after the third FMT. Mice remained on vancomycin treatment through clearance of P. yoelii. Following infection, Tac mice treated with CR FMT and treated with vancomycin (CR→Tac+Van) exhibited significantly lower parasite burden than untreated CR/Tac mice, with all control mice exhibiting the anticipated high or low parasite burden (Figures S4F–S4H). These data demonstrate that the differential effects of antibiotic treatment in CR and Tac mice (Figures 2C–2F) were not attributed to differences among the inbred strains of C57BL/6 mice.
MVRSION analysis of gut bacteria populations identified that each antibiotic treatment changed the richness and abundance of bacterial species (alpha diversity), as measured by OTUs and the Shannon index (Figure 2G), and composition of the bacteria populations (beta diversity), as measured by the Bray-Curtis dissimilarity distance (Figure 2H), compared with the control CR and Tac mice. Bacteria populations were also significantly different across individual antibiotic-treated groups within CR mice (Figure 2I) and Tac mice (Figure 2J). A supervised machine-learning model was able to classify the Plasmodium susceptibility phenotype (high versus low parasitemia) in mice with 97% overall accuracy (Figures S5A–S5C). Metronidazole-treated mice were excluded from the model because of the inability to attribute decreased parasitemia to antibiotic-induced changes in gut microbiota versus the known anti-Plasmodium effects of metronidazole (James, 1985). Intriguingly, differential abundance of just 8 bacterial species was able to classify malaria susceptibility in these mice (Figures S5D and S5E).
Collectively, these data suggest that gut bacteria can profoundly affect the severity of malaria in mice. Moreover, they identify antibiotic treatment as an effective tool to probe how gut bacteria affect the severity of malaria in CR mice. In particular, vancomycin treatment is useful in this model system, given the pronounced effect of treatment on bacteria populations and the inability of vancomycin to be absorbed through the intestinal tract, thereby limiting the observed effect of vancomycin treatment on P. yoelii parasite burden to the effect of this antibiotic on gut bacteria. These data also suggest that the effect of antibiotic-induced changes in gut bacteria compositions on host immunity is not always detrimental; this effect can be beneficial for host protection. Therefore, the immunological effect of antibiotic treatment on an infection or vaccine likely depends on the baseline gut bacteria composition and nature of the particular pathogen or vaccine.
Vancomycin treatment causes long-term changes in adult mouse gut bacteria populations that affect the severity of malaria
Gut microbiota in infancy are capable of conferring long-term imprinting of the host immune system (Yang et al., 2016). It has also been shown that gut microbiota dynamically engage the mucosal immune system (Wang et al., 2019). In the previous experiment (Figure 2), mice were continuously treated with antibiotics, which precluded the ability to assess whether changes in gut bacteria before or after P. yoelii infection resulted in changes in the severity of malaria. To determine the extent of gut microbiota imprinting on host immune response to Plasmodium infection, groups of adult CR mice were provided vancomycin drinking water 4 weeks before P. yoelii infection and treated for at least 2 weeks. Vancomycin treatment was stopped 14 days before, 7 days before, 3 days before, or the day of P. yoelii infection (Figure S6A). Regardless of when vancomycin treatment ceased, the vancomycin-treated CR mice displayed significantly lower parasitemia than did the control CR mice (Figures S6B and S6C). These data suggest that the early-life gut microbiota do not imprint programmed immunological responses to Plasmodium that are unchangeable; rather, the gut microbiota may imprint a particular immune response to Plasmodium infection when the gut microbiota are modulated up to the day of P. yoelii infection. Moreover, the 14-day cessation in vancomycin before P. yoelii infection supported that the effect of vancomycin treatment on parasitemia is attributed not to a direct effect of vancomycin in P. yoelii but rather to vancomycin-induced changes in gut bacteria compositions.
The observation that 14 days of vancomycin treatment followed by 14 days of no treatment resulted in a significant decrease in parasitemia led to a question: How long does the protection afforded by antibiotic treatment last after cessation of treatment? To probe this question, CR mice were treated with vancomycin for 14 days and then placed on regular drinking water for 90, 60, 30, or 15 days before P. yoelii infection (Figure 3A). Strikingly, mice that were treated with vancomycin for 14 days and then rested for 90 days showed a profound decrease in parasitemia to levels that were barely detectable, which was in contrast to the age-matched control mice that developed the expected high levels of parasitemia (Figures 3B and 3C).
Figure 3. Vancomycin treatment provides protection against P. yoelii for months and imparts cross-species immunity to experimental cerebral malaria.

(A) C57BL/6 mice from CR (susceptible) were treated with vancomycin for two weeks and put back on regular drinking water for 90 (m90), 60 (m60), 30 (m30), and 15 (m15) days before P. yoelii infection. Mice were challenged with P. berghei ANKA on day 52 post-infection. All mice were procured at the same time.
(B and C) Parasitemia and AUC of P. yoelii infection.
(D) Survival curve after P. berghei ANKA challenge.
(E and F) Parasitemia and AUC after P. berghei ANKA challenge.
Data are means ± SEM. (C) One-way ANOVA with Tukey’s multiple comparison test. (D) Pairwise log-rank (Mantel-Cox) test. Representative of two experiments with 5 mice per group in each trial. ****p < 0.0001. Similar results were obtained from two independent experiments.
P. yoelii-immune mice confer cross-species protection against an otherwise-lethal Plasmodium berghei ANKA infection that causes experimental cerebral malaria (ECM) or, in the rare ECM survivor, hyperparasitemia and severe anemia (Kurup et al., 2017). The extremely low P. yoelii parasite burden in the vancomycin-treated mice led to the question of whether these mice elicited an adaptive immune response that would provide protection against P. berghei ANKA infection. To test this possibility, each of these groups of CR mice, along with age-matched P. yoelii-naive CR mice, were challenged with P. berghei ANKA 52 days after the original P. yoelii infection (Figure 3A). All naive control mice died of ECM within 10 days of infection (Figure 3D), and P. yoelii-naive vancomycin-treated CR mice were susceptible to ECM (Figure S6D). Whereas vancomycin control P. yoelii-immune CR mice remain susceptible to ECM, all groups of CR mice that had been treated with vancomycin and infected with P. yoelii exhibited high rates of survival from ECM (Figure 3D). Moreover, among all P. yoelii-immune, ECM survivor, vancomycin-treated mice (minus 90, 60, 30, and 15 days), 56% (10 of 18) were also protected from P. berghei hyperparasitemia (Figures 3E and 3F).
Antibiotic treatment in early life can have long-term effects on gut bacteria (Nobel et al., 2015). In adult populations, the long-term effect of antibiotic treatments on gut bacteria populations is less clear. Some reports have noted a recovery of baseline gut bacteria within 4 weeks of treatment cessation (Palleja et al., 2018; Suez et al., 2018), whereas others have observed differences that last many months after antibiotic cessation (Haak et al., 2019). To determine whether the long-term effect of 2-week vancomycin treatments on P. yoelii-parasite burden correlated with sustained changes in gut bacteria populations, bacterial community analysis was performed. MVRSION analysis of fecal pellets from age-matched control and vancomycin-treated CR mice (Figure 3A) collected before P. yoelii infection on day 0 post-infection (p.i.) revealed a loss in alpha diversity in the vancomycin-treated mice that lasted up to 90 days after vancomycin treatment had stopped (Figures 4A and 4B). Bacterial community composition, as measured by the Bray-Curtis dissimilarity distance, demonstrated clear differences in bacteria populations between any of the vancomycin-treated mice and the control CR mice (Figures 4C and 4D), with all but two of the −90-day mice, which also exhibited low parasitemia, tightly clustered with minimal differences among any of the vancomycin-treated mice (Figure 4D). Collectively, the data indicate within the context of a Plasmodium infection that the composition of gut bacteria at the time of infection, rather than during early life, will ultimately affect the subsequent severity of infection.
Figure 4. Gut microbiota are not able to recover up to 3 months following vancomycin exposure.

Fecal pellets were collected on day 0 before P. yoelii infection (from Figure 3A) to analyze gut microbiota composition.
(A) Alpha diversity estimated using observed OTUs.
(B) Alpha diversity estimated using the Shannon index.
(C) PCoA plot shows beta diversity measured with the Bray-Curtis dissimilarity distance.
(D) Bray-Curtis dissimilarity distance comparisons between groups. The box ends show lower and upper quartiles, and the horizontal line inside the box is the median. The y axis shows the Bray-Curtis dissimilarity distance of groups on the x axis to groups on the top of the vertical columns.
Data are means ± SEM. (A and B) Pairwise Kruskal-Wallis test. (D) Pairwise PERMANOVA with 999 permutations. Significance level is compared between the group at the top of the column and the group on the x axis. Gut microbiota were analyzed from only one experiment of two independent experiments. *p < 0.05, **p < 0.01.
Gut bacteria provide continuous cues that dynamically modulate the severity of malaria in mice
The data indicate that gut bacteria present at the time of a Plasmodium infection shape the severity of infection. Yet it remains unclear whether the preinfection bacteria composition imprints a scripted immune response that is fixed or a dynamic interaction in which gut bacteria provide continuous signals to the host immune system following Plasmodium infection that allows changes in the adaptive immune response and outcome of infection. Two separate approaches, using either CR or Tac mice, were used to test these two possibilities. First, CR mice were treated with vancomycin drinking water for various lengths of time beginning before P. yoelii infection, beginning on the day of P. yoelii infection, or beginning 7 days after P. yoelii infection (Figure 5A). Consistent with the prior data, CR mice treated with vancomycin at any time before P. yoelii infection resulted in reduced P. yoelii parasite burden (Figures 5B and 5C). Vancomycin treatment started on the day of P. yoelii infection also dramatically reduced parasite burden (Figures 5B and 5C). Strikingly, starting vancomycin treatment 7 days post-infection, when parasitemia had already reached ~10%, prevented further increase in parasitemia, resulting in accelerated parasite clearance and a reduction in parasite burden (Figures 5B and 5C).
Figure 5. Gut microbiota dynamically modulate the Plasmodium parasite burden in susceptible and resistant mice.

(A–C) C57BL/6 mice from CR mice were treated with vancomycin before, at, and after P. yoelii infection (A). n = 4 mice/group. (B and C) Parasitemia and AUC of CR mice treated with vancomycin from (A). Data are means ± SEM and representative of two or more experiments. Control, regular water; D−14 cont, continuous vancomycin water from day −14 until the end of infection; D−7 cont, continuous vancomycin water from day −7 until the end of infection; D−14 to 0, vancomycin water from day −14 to day 0; D−7 to 0, vancomycin water from day −7 to day 0; D0 cont, continuous vancomycin water from day 0 until the end of infection; D7 cont, continuous vancomycin water from day 7 post-infection until the end of infection.
(D) C57BL/6 mice from Tac were gavaged with 3 consecutive cecal contents (FMTs) from CR before, at, and after P. yoelii infection. n = 4 mice/group.
(E–K) Gut microbiota analysis using the MVRSION approach of Tac and CR mice receiving FMTs collected at the respective days. PCoA plots show beta diversity analysis using the Bray-Curtis dissimilarity distance. The vertical dashed line clearly marks the clustering of the Tac type to the left and the CR type to the right.
(L and M) Parasitemia and AUC of Tac mice receiving FMTs from CR mice from (D). Data are means ± SEM and representative of two experiments.
(D–M) FMT day −7 (FMT −D7), FMT on day −7, 6, and −5; FMT D0, FMT on days 0, 1, and 2; FMT D3, FMT on days 3, 4, and 5; FMT D7, FMT on days 7, 6, and 8. (C and M) One-way ANOVA with Tukey’s multiple comparison test. Only comparisons with control are shown. **p < 0.01, ***p < 0.001, and ****p < 0.0001. (L) Two-way ANOVA with Dunnett’s multiple comparison test. a, Tac Con (control) versus FMT D3, p < 0.05; b, Tac Con versus FMT D7, p = 0.002.
In the second approach, Tac mice received CR ceca content transplants for three consecutive days beginning 7 days before P. yoelii infection, beginning on the day of P. yoelii infection, beginning 3 days after P. yoelii infection, or beginning 7 days after P. yoelii infection (Figure 5D). Bacterial community analysis by MVRSION revealed that control Tac mice maintained a differential bacterial community compared with control CR mice through day 14 p.i. (Figures 5E and 5F) and that when stool bacterial communities were compared in all samples, there were distinct Tac and CR microbiota types (Figure 5G). Analysis of each group of Tac mice that received CR FMTs revealed that after the FMT, the bacterial communities transitioned to a CR microbiota type (i.e., transition from the left to the right side of the dashed line; Figures 5H–5K), with the FMT D3 group demonstrating this transition occurred within 4 days (comparison of day 3 to day 7 samples; Figure 5J). As with CR mice, manipulating gut microbiota in Tac mice before or on the day of P. yoelii infection resulted in a profound change in the parasite burden (i.e., increase in Tac mice) compared with control Tac mice (Figures 5L and 5M). Tac mice that received CR cecal content transplants after the P. yoelii infection also showed a shift in the parasitemia kinetics (Figures 5L and 5M). Collectively, these data demonstrate that gut bacteria provide continuous cues that dynamically modulate the severity of malaria in susceptible and resistant mice.
Signals from gut bacteria dynamically regulate germinal center reactions and parasite burden
An adaptive immune response, particularly GC reactions and Plasmodium-specific antibodies, is required for control of Plasmodium blood-stage infections (Del Portillo et al., 2012). The data in Figure 5 led to the hypothesis that GCs are malleable to the continuous cues provided by gut bacteria, which ultimately determines parasite burden. To test this hypothesis, CR mice were infected with P. yoelii (Figure 6A) and left untreated (noVan), treated with vancomycin beginning on day 0 (d0Van), or treated with vancomycin beginning on day 7 p.i. (d7Van). Seven days after P. yoelii infection, by which time mice have induced GC responses (Figures 6D and 6E) (Waide et al., 2020), mice were treated with either an anti-CD40L monoclonal antibody (clone MR1) that disrupts GC reactions (Foy et al., 1993; Noelle et al., 1992) or an isotype control antibody (IgG) (Figure 6A). If vancomycin-induced changes in gut bacteria affect P. yoelii parasite burden independent of GC reactions, then disrupting GCs would have minimal effect on parasite burden in the vancomycin-treated mice. Fecal pellets were collected on day 10 p.i., and MVRSION analysis demonstrated that vancomycin treatment changed the bacterial community (Figure 6B), with only the d0Van group showing a difference in the Bray-Curtis dissimilarity distance between IgG and MR1 mice within any of the three vancomycin treatment groups (Figure 6C).
Figure 6. Blocking CD40-CD40L interactions impairs augmented germinal center reactions observed in vancomycin-treated mice.

(A) C57BL/6 mice from CR mice were continuously treated with vancomycin from day 0 and day 7 post-infection. Mice received either 6 doses of isotype control (IgG) or MR1 intraperitoneally (i.p.) on alternate days from day 7 to day 17 post-infection.
(B) PCoA plot shows beta diversity using the Bray-Curtis dissimilarity of fecal pellets collected at day 10 p.i.
(C) Bray-Curtis distance comparison between the groups. The box ends show the lower and upper quartiles, and the horizontal line inside the box is the median. The y axis shows the Bray-Curtis dissimilarity distance of groups on the x axis to groups on the top of the vertical columns.
(D–F) Gating strategy for the indicated cell subsets. Events in the left gate of each cell population are total splenocytes (i.e., singlets gated from a side scatterplot and forward scatterplot).
(G–O) Spleens were harvested on the indicated days for cellular analysis. In (G), (J), and (M), Tfh cells (CXCR5+PD-1+CD44+CD4+) are shown on days 7, 11, and 19 p.i., respectively. In (H), (K), and (N), GC B cells (CD95+GL7+CD19+) are shown on days 7, 11, and 19 p.i., respectively. In (I), (L), and (O), plasmablasts (CD138+IgD−B220+CD19+) are shown on days 7, 11, and 19 p.i. respectively.
Data are means ± SEM. (C) Pairwise PERMANOVA with 999 permutations. Representative of two independent experiments with similar results. n = 3 mice/group. (G–O) Cumulative results from three experiments. (G–I) t test. (J–O) One-way ANOVA with Sidak correction for selected pairs. noVan IgG, no vancomycin with IgG; noVan MR1, no vancomycin with MR1; d0Van IgG, vancomycin from day 0 and IgG; d0Van MR1, vancomycin from day 0 and MR1; d7Van IgG, vancomycin from day 7 and IgG; d7Van MR1, vancomycin from day 7 and MR1. *p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Within GC reactions, CD4+ follicular helper T cells (Tfh cells) provide necessary signals to GC B cells to support somatic hypermutation and affinity maturation of antibodies (Victora and Nussenzweig, 2012). GC B cells exit the GC as plasma cells, which produce large quantities of antibodies, or memory B cells. In contrast to prior studies that demonstrated antibiotic treatment impaired adaptive immunity (Abt et al., 2012; Hagan et al., 2019; Ichinohe et al., 2011; Lynn et al., 2018; Oh et al., 2014), vancomycin treatment beginning on the day of P. yoelii infection had no effect (noVan versus d0Van) on the number of splenic Tfh cells (Figures 6D and 6G), GC B cells (Figures 6E and 6H), or plasmablasts (Figures 6F and 6I) on day 7 p.i. In addition, vancomycin treatment beginning on the day of P. yoelii infection or day 7 p.i. (noVan IgG versus d0Van IgG or d7Van IgG, respectively) did not result in fewer numbers of splenic Tfh cells (Figure 6J), GC B cells (Figure 6K), or plasmablasts (Figure 6L) on day 11 p.i. Intriguingly, over the course of infection, vancomycin treatment (d0Van IgG or d7Van IgG compared with noVan IgG) resulted in increased numbers of Tfh cells and GC B cells, with a more modest trend observed within the plasmablasts (Figures 6M–6O; Figure S7). Collectively, the data demonstrate that the effect of antibiotics is not necessarily detrimental to host immunity but in certain contexts may support a more robust adaptive immune response.
To assess the efficiency of MR1 treatment that began day 7 p.i. at blocking GC reactions, the number of Tfh cells, GC B cells, and plasmablasts were quantified in mice that received MR1. At day 11 p.i., MR1 treatment had little effect on the number of Tfh cells except between d7Van IgG- and d7Van MR1-treated mice (Figure 6J). In contrast, MR1 treatment resulted in significant decreases in both GC B cells (Figure 6K) and plasmablasts (Figure 6L) in all three groups of mice (noVan, d0Van, and d7Van) when measured at day 11 p.i. On day 19 p.i., MR1 treatment, compared with IgG, resulted in fewer Tfh cells (Figure 6M) and a sustained loss in GC B cells (Figure 6N) among the noVan, d0Van, and d7Van groups of mice, with no significant effect on numbers of plasmablasts (Figure 6O). Collectively, these data demonstrate that vancomycin treatment, even up to 1 week after P. yoelii infection, rapidly changes gut bacteria populations and increases numbers of GC B cells and that MR1 treatment 1 week after infection was sufficient at ablating the elevated GC responses observed in vancomycin-treated mice.
Finally, parasite burden was analyzed to determine whether signals from gut bacteria affect the severity of malaria through dynamic modulation of GC reactions in CR mice (Figure 7A). Consistent with GCs required for control of Plasmodium-infected red blood cells, MR1 treatment in the noVan group resulted in a continuous increase in parasite burden through the end of MR1 treatment followed by a pronounced delay in clearance (Figures 7B and 7C). Mice treated with vancomycin on day 0 showed the expected low parasite burden; however, disruption of GC reactions in these mice beginning on day 7 p.i., even though parasite burden was already quite low compared with that of noVan mice, resulted in a significant increase in parasite burden (Figures 7D and 7E). Moreover, MR1 treatment ablated the low parasite burden observed in d7Van IgG-treated mice (Figures 7F and 7G). These data suggest that gut bacteria dynamically interact with GC reactions to affect Plasmodium infection and that certain microbiota compositions can lead to enhanced host protection.
Figure 7. Gut bacteria dynamically interact with germinal center reactions to modulate Plasmodium parasite burden.

(A) Experiment design.
(B–G) Parasitemia on the indicated days in (B), (D), and (F). AUC analysis in (C), (E), and (G).
Data are means ± SEM. (C, E, and G) t test. Data are cumulative from two independent experiments. n = 8 mice/group.
DISCUSSION
These results provide an intriguing observation that gut microbiota modulated before or during active Plasmodium infection can alter host protection and disease prognosis. This stems from the malleable nature of GC reactions following Plasmodium infection and the ability of GC reactions to respond to cues from gut bacteria, which ultimately affect parasite burden. Because a Plasmodium infection is blood-borne, most immune responses involved in the clearance of the parasite occur in the spleen (Del Portillo et al., 2012). GCs form in the spleen of mice by day 6 after Plasmodium infection (Ryg-Cornejo et al., 2016). The development of GC reactions during the first week of infection, through the continuous cues provided by gut bacteria, is quite dynamic and is critical to disease outcome, as evidenced by these data. Important questions remain, including the following: What cells of the immune system do gut microbiota continuously signal through to modulate GC reactions? What components of gut bacteria (proteins, polysaccharides, biochemical metabolites, small RNAs, etc.) regulate GC reactions? Prior work has shown that short-chain fatty acids from gut bacteria can affect GC B cells (Kim et al., 2016). Yet Tac and CR mice do not show differences in serum short-chain fatty acid levels (Chakravarty et al., 2019), suggesting that they are not responsible for dynamic modulation of GC reactions during Plasmodium infection.
Vancomycin treatment beginning the day of P. yoelii infection resulted in low parasitemia (Figures 5B, 7B, and 7D), despite no increase in GC-associated cell numbers through day 11 p.i. (Figures 6G–6L). This suggests that vancomycin-induced decreases in parasitemia early during infection may be the result of changes in the quantity or quality of the P. yoelii-induced antibodies. In support of this possibility, we have shown that low parasite burden in mice mediated by gut microbiota composition is correlated with increased GC B cell numbers, parasite-specific antibody, better GC structure, and quality antibody response that also provided robust protection against lethal rodent Plasmodium challenge (Waide et al., 2020). Alternatively (Zhuang et al., 2019), the reduction in parasitemia following vancomycin treatment may have occurred through augmentation of other aspects of the host immune response. Given the complexity of the innate and adaptive immune response to Plasmodium, assessing whether and how vancomycin-induced changes in gut microbiota modulate additional components of the immune response to Plasmodium will require future investigation. Our data do not exclude the possibility that vancomycin treatment dynamically alters parasitemia independent of host immunity.
The present data indicate that gut bacteria are the critical gut microbes that effect parasitemia. However, other gut microbiota components, like fungi, viruses, and archaea, might contribute to the Plasmodium parasite burden. Curiously, the effect of gut bacteria on the blood-stage parasite burden was not attributed specifically to Gram-positive, Gram-negative, or anaerobic bacteria, because antibiotics that target these different classes of bacteria were capable of decreasing parasite burden in CR mice. Therefore, it appears that the consortium of gut bacteria affects parasitemia, rather than a single class of bacteria. Alternatively, a single bacterial species could be responsible for the changes in parasitemia whose presence, and ability to modulate the outcome of Plasmodium infection, could be influenced by the presence or absence of other bacteria.
Random forest was able to predict with high accuracy the malaria phenotype based on the abundance of 8 bacteria populations. Among those bacteria, Bacteroides uniformis and Lactobacillus johnsonii, which were abundant in malaria-resistant mice, are well known for probiotic potential (Morita et al., 2020). Conversely, the Alistipes genus and Lactobacillus reuteri, which are abundant in malaria-susceptible mice, are associated with detrimental health effects like depression, anhedonia-like phenotypes, inflammation of spinal cords, cancer, and mental health (Miyauchi et al., 2020; Parker et al., 2020; Wang et al., 2020). In addition, L. reuteri is associated with higher plasma interleukin-6 (IL-6) levels in mice, and circulating IL-6 levels correlate with the severity of blood-stage malaria in humans and mice (Wang et al., 2020; Wunderlich et al., 2012).
We report that antibiotic-induced changes in gut bacteria were beneficial to CR mice, as shown by the decreased blood-stage Plasmodium parasite burden. In contrast, numerous studies have shown that oral antibiotics lead to gut bacteria dysbiosis, which has detrimental effects on host health. For example, antibiotic-induced gut microbiota perturbation leads to increased inflammation in blood and alters immunity to vaccines in humans (Hagan et al., 2019). A single dose of intraperitoneal clindamycin leads to sustained susceptibility to Clostridioides difficile-induced diarrhea and colitis in mice because of significant alterations of intestinal microbiota (Buffie et al., 2012). Similarly, antibiotic use in neonates and adults predisposes them to a range diseases, such as diabetes, obesity, inflammatory bowel diseases, asthma, depression, and autism (Zhang and Chen, 2019). In contrast to these scenarios, and in agreement with this report, a cocktail of antibiotics (ampicillin, metronidazole, neomycin, and vancomycin) was advantageous to the host, because it prevented motor dysfunction and limited axon damage in a murine model of progressive multiple sclerosis (Mestre et al., 2019). Yet this cocktail of antibiotics, which eliminates most gut bacteria, creates a scenario different from the one in the present study, in which single antibiotics were used to induce altered bacteria compositions.
It was recently shown that Kenyan children who have had two episodes of malaria have different gut microbiota compositions than children who have had only one episode (Mandal et al., 2019). This observation supports the possibility that gut bacteria in humans may affect the severity or outcome of Plasmodium infection. Excitingly, for the first time, to our knowledge, we have reported that gut bacteria composition in Uganda children differs between asymptomatic P. falciparum infection and severe malarial anemia. The observation that stool bacteria populations in Ugandan children with SMA collected at varying times after enrollment and treatment were not different is consistent with a prior study in Kenya that showed malaria episodes did not alter stool bacteria populations (Mandal et al., 2019). Collectively, these observations provide tantalizing support that the gut microbiome may shape the severity of malaria in children. Nevertheless, future studies incorporating longitudinal analysis (i.e., collection human stool samples before and through malaria transmission seasons) are necessary to demonstrate the potential for baseline gut microbiota to modulate the severity of malaria in children. Finally, shotgun metagenomics, culturomics, clinical trials, etc. need to be performed to fully decipher links between specific gut bacteria and severity of malaria in humans (Mukherjee et al., 2020). Because the decline in the number of malaria deaths has largely plateaued in recent years, and because the gut microbiome significantly affects host health and is amendable to change via diet, prebiotics, and/or probiotics, gut-microbiota-based therapeutics may represent novel approaches to prevent severe malaria and deaths.
This report demonstrates that gut bacteria in mice continuously interact with gut-distal germinal center reactions during an ongoing extra-gastrointestinal tract infection to shape the severity of disease. Importantly, gut bacteria composition was also shown to associate with the severity of malaria in humans, suggesting that human gut microbiota may represent a novel target to ameliorate severe malaria. Finally, these data demonstrate that antibiotic-induced changes in gut bacteria do not uniformly impair systemic adaptive immunity but can in some cases result in elevated immune responses. These results highlight the need for ongoing research to investigate the diverse interactions between gut microbiota and host immunity.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nathan W. Schmidt (nwschmid@iu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The 16S rRNA gene sequencing datasets generated and analyzed from mice and humans in this study are publicly available and deposited in the NCBI Sequence Read Archive under the BioProject IDs PRJNA643559 and PRJNA642859 respectively.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and housing
Genetically similar 6–8 weeks old female C57BL/6N mice were obtained from Taconic Biosciences (Isolated Barrier Unit IBU050401C; Hudson, NY) and Charles River Laboratories (barrier room R01; Wilmington, MD) and housed conventionally in a specific pathogen-free (SPF) facility. Mice were kept on NIH-31 Modified Open Formula Mouse/Rat Irradiated diet (Harlan 7913) (Harlan, Indianapolis, IN) and non-acidified autoclaved reverse osmosis water ad libitum upon arrival. Mice were acclimatized for a minimum of one week prior to performing experiments. Mice were kept on a 12-hour light (6 AM – 6 PM) and 12-hour dark (6 PM – 6 AM) cycle. Animal handling and experiment protocols were approved by the University of Louisville and Indiana University Institutional Animal Care and Use Committees (IACUC).
Plasmodium infection and parasitemia analysis
Donor mice were injected intravenously (IV) with thawed 105 infected red blood cells (iRBCs) with either Plasmodium yoelii 17XNL or Plasmodium berghei ANKA. Blood was collected on day 5 post infection (p.i.) via retro-orbital bleed. The blood was counted for iRBCs with Giemsa stain and total RBCs (using Hemocytometer) and diluted in 0.9% saline (Teknova, Hollister, CA) at a concentration of 105 iRBCs / 200 μl. Experimental mice were infected with 105 iRBCs IV unless indicated.
For parasitemia, approximately 5 μl whole blood was collected from tail snip in cold 100 μl 1X PBS in a 96 well plate on ice. The cells were fixed in 0.00625% glutaraldehyde, stained with conjugated antibodies, and subjected flow-cytometry analysis for evaluation of percent parasitemia. The conjugated antibodies for staining panel were CD45.2-APC clone 104; Biolegend, San Diego, CA), Ter 119-APC/Cy7 clone TER-119; Biolegend, San Diego, CA), dihydroethidium MilliporeSigma, St. Louis, MO), and Hoechst 33342 MilliporeSigma, St. Louis, MO). Forward and side scatter singlets were gated on Ter119+CD45.2− for RBC. RBCs were gated on Hoechst+Dihydroethidium+ to calculate the percent of iRBC (% Parasitemia). Parasitemia was tacked every other day beginning day 5 to clearance of parasite unless indicated.
METHOD DETAILS
Stool and blood collection from Ugandan children
The study was reviewed and approved by the Makerere University School of Medicine Research and Ethics Committee, Indiana University School of Medicine Institutional Review Board, and the Ugandan National Council for Science and Technology. Written and informed consent was obtained from parents or guardians prior to enrolment.
Children between the ages of 0.5 to 4 years old, with 5 most common clinical manifestations of severe malaria (cerebral malaria, CM; respiratory distress, RD; severe malarial anemia, SMA; malaria with complicated seizures, M/S; and prostration) were enrolled in a prospective longitudinal cohort study at two sites: 1) the Pediatric Acute Care Unit at Mulago National Referral Hospital in Kampala, Uganda, and 2) the Pediatric Emergency Ward at the Jinja Regional Referral Hospital in Jinja, Uganda. Children from the same neighborhoods without active illness or fever at time of enrollment were enrolled in the healthy community control (CC) group. SMA was defined as P. falciparum smear or RDT positive and serum hemoglobin level ≤ 5 g/dL. All study participants (SMA or CC) were tested for parasitemia by microscopy or RDT and those determined to be parasite positive were treated immediately, according to the Ugandan Ministry of Health treatment guideline at the time of diagnosis. CC children that were P. falciparum microscopy negative were classified as Pf Neg, while CC children that were P. falciparum microscopy positive were classified as Pf pos. One Pf Neg subject (Study ID: 1551; Table S1) was blood smear positive for P. malariae. Of note, P. malariae is not associated with severe malaria. Stool samples from Pf Neg (n = 28), Pf Pos (n = 7), and SMA (n = 40) children from the parent study were included in this gut microbiome study. Stool samples were collected various days after enrollment from SMA children (Table S1) and during enrollment from CC (Pf Pos + Pf Neg) children and frozen immediately at −80°C. Stool samples were kept frozen until DNA extraction. For blood parameters and parasite detection, peripheral blood was collected by venipuncture on hospital admission from SMA and as an outpatient from CC. A complete blood count (CBC) with differential and reticulocyte count and a blood smear for malaria parasite count were conducted. HIV testing was done on any participants whose parents/guardians did not opt out of testing. There were only two HIV positive participants, which were in the SMA group. No power outage or other situations were noted during samples handling and processing to affect the stability of samples.
Gut Microbiota Analysis
DNA from feces was extracted using QIAamp PowerFecal DNA kit (QIAGEN, Germantown, MD) according to the manufacturer’s instructions. Extracted DNA samples were shipped overnight on ice packs to the Genome Technology Access Center (GTAC; Washington University, St. Louis, MO) for 16S rRNA sequencing unless indicated. rRNA were sequenced using an approach, Multiple 16S Variable Region Species-level Identification (MVRSION), that can sequence all the 9 hypervariable regions of 16S rRNA gene with 12 primer pairs (Schriefer et al., 2018). Observed operational taxonomic unit (OTU) table were constructed by GTAC. OTU table was imported inside QIIME2 for core-diversity and statistical analysis.
For analysis of bacteria communities in the reciprocal ceca transplants (Figure 1), DNA samples were shipped overnight on ice packs to the Integrated Microbiome Resource within the Centre for Comparative Genomics and Evolutionary Bioinformatics at Dalhousie University (IMR-CGEB, Halifax, NS, Canada). V6-V8 regions of 16S rRNA were sequenced and analyzed as described previously (Denny et al., 2019). Briefly, paired end sequences were stitched together, low quality reads and chimera were removed, and OTU table was constructed using open reference method using SortMeRNA v2.0.
Sample metadata values were predicted with supervised machine learning algorithm using random forest with q2-sample-classifier plugin using QIIME2. One-third of the samples were used as test set data. 15 and 20 K-fold cross-validation were performed in mice and human study, respectively.
Cecal microbiota transplantation
Cecal donor mice were anesthetized with isoflurane and cervical dislocation was performed inside a laminar flow hood. Mice were laid on the back and ventral side were doused with 70% ethanol. Ceca were aseptically removed, and contents were squeezed on Petri plate with 2 mL saline. Cecal content was mixed with 1ml syringe plunger. The diluted ceca contents were drawn into a 1ml syringe and 200 μl content were gavaged per mouse. A sterile gavage needle and syringe was changed between each group.
Antimicrobials
Mice were treated with antimicrobials in drinking water. Antibiotics used were ampicillin (0.5 mg/ml) (Sigma Aldrich; St. Louis, MO), gentamicin sulfate (0.5 mg/ml) (Corning; Manassas, VA), metronidazole (0.5 mg/ml) (Spectrum Chemical, Gardena, CA), neomycin sulfate (0.5 mg/ml) (EMD Millipore; Billerica, MA), and vancomycin (0.25 mg/ml) (VWR Life Science). All of the antimicrobials were dissolved in non-acidified autoclaved reverse osmosis drinking water. Antimicrobial water was changed every week.
Spleen immune cell analysis
RP10 media: RPMI 1640 media (Thermo Fisher Scientific Inc., Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Inc., Lawrenceville, GA), 1.19 mg/ml HEPES (Thermo Fisher Scientific Inc., Waltham, MA), 0.2 mg/ml L-glutamine (Research Products International Corp., Mt. Prospect, IL), 0.05 units/ml & 0.05 mg/ml penicillin/streptomycin (Invitrogen, Grand Island, NY), 0.05 mg/ml gentamicin sulfate (Invitrogen, Grand Island, NY), and 0.05 μM 2-Mercaptoethanol (Thermo Fisher Scientific Inc., Waltham, MA). Spleens on indicated days were harvested in RP10 media and smashed on a wire sheath with flat side of 10 mL syringe plunger to make single cell suspension. Single cell suspensions were washed once with RP10 and treated with ACK lysis buffer (NH4Cl – 150 mM, KHCO3 – 1 mM, and Na2EDTA – 0.1 mM in distilled water) to lyse red blood cells. Single cell suspensions were counted and resuspended in RP10 at 2×107 cells/ml.
Cells (2×106) were stained in FACS buffer (1X PBS, 1%FBS, and 0.02% sodium azide) for 30 min at 4C with the following fluorescence-conjugated antibodies (CD45.2, clone 104; CD4, clone RM4-5; CD19, clone 6D5; Ter119, clone Ter-119; PD-1, clone 29F.1A12, CD95, clone Jo2; GL7, clone GL7; biotion-CXCR5, clone 2G8; CD44, clone IM7; B220, clone RA3-6B2; CD138, clone 281-2; IgD, clone 11-26c.2a) purchased from Biolegend (San Diego, AC) and BD Biosciences (San Diego, CA). For CXCR5 staining, cells were first stained with biotinylated CXCR5 at room temperature for 30 min and then stained with fluorescence-conjugated streptavidin. Cells were fixed and permeabilized with fixation buffer (Biolegend, San Diego, CA) after staining. Cells were acquired with a BD LSRFortessa (BD Biosciences, San Jose, CA). Data were analyzed by FlowJo software (Tree Star, Ashland, OR)
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad), QIIME2, and R packages. Specific statistical tests are described in the figure legends. For area under the parasitemia curve (AUC) analyses, the trapezoidal rule was used for equation:
where “p” is percent parasitemia at the designated time point “t” (Méndez et al., 2006).
Significance level of alpha diversity between Pf Pos and SMA (malaria severity) were tested with ANOVA using following model accounting for age (AgeYrs) and sex.
Significance level of beta diversity between Pf Pos and SMA (malaria severity) were tested with “Adonis” function with “bray” method of vegan package implemented in R with following model accounting for age, antibiotics use, percent neutrophil, and weight for height z-score (whz).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45.2-APC, Clone 104 | Biolegend | Cat# 109814; RRID:AB_389211 |
| Ter119-APC/Cy7, Clone TER-119 | Biolegend | Cat# 116223; RRID:AB_2137788 |
| Dihydroethidium | Sigma Aldrich | Cat# 37291 |
| Hoechst 33342 | Sigma Aldrich | Cat# 875756-97-1; RRID:AB_10626776 |
| CD4-PE-Cy7, clone RM4-5 | Biolegend | Cat# 100527; RRID:AB_312728 |
| CD4-PE, clone RM4-4 | Biolegend | Cat# 116005; RRID:AB_313690 |
| CD11a-FITC, clone M17/4 | Biolegend | Cat# 101106; RRID:AB_312779 |
| CXCR5-biotin, clone L138D7 | Biolegend | Cat# 145509; RRID:AB_2562125 |
| Streptavidin-PE-Cy7 | Biolegend | Cat# 405206; RRID:AB_10116480 |
| CD44-FITC and APC-Cy7, clone IM7 | Biolegend | Cat# 103021; RRID:AB_493684 |
| PD-1 APC, clone 29F.1A12 | Biolegend | Cat# 135209; RRID:AB_2251944 |
| GL-7 Pac Blue, clone GL-7 | Biolegend | Cat# 144613;RRID:AB_2563291 |
| CD19 PerCP-Cy5.5, clone 6D5 | Biolegend | Cat# 115534; RRID:AB_313654 |
| CD138-BV421, Clone 281-2 | Biolegend | Cat# 142507; RRID:AB_11204257 |
| IgD APC-Cy7, clone 11-26c2a | Biolegend | Cat# 405716; RRID:AB_10662544 |
| CD95-PE, clone SA367H8 | Biolegend | Cat# 152608; RRID:AB_2632902 |
| Anti-CD40L mAB (MR1) | BioXcell | Cat# BE0017-1; RRID:AB_1107601 |
| Armenian hamster IgG | BioXcell | Cat# BE0091; RRID:AB_1107773 |
| Chemicals, peptides, and recombinant proteins | ||
| QIAamp PowerFecal DNA kit | QIAGEN | Cat# 12830-50 |
| Qbit dsDNA BR Assay Kit | InvitrogenTM | Cat# Q32850 |
| Ampicillin | Sigma Life Science | Cat# A1593-25G |
| Gentamicin sulfate | Corning | Cat# 30-005-CR |
| Metronidazole | Spectrum Chemical | Cat# M1511 |
| Neomycin sulfate | EMD Millipore | Cat# 4801-25GM |
| Vancomycin | VWR Life Science | Cat# 97062-554 |
| Deposited data | ||
| Raw sequence data | This paper | BioProject: PRJNA643559 |
| Raw sequence data | This paper | BioProject: PRJNA642859 |
| Analyzed data | This paper | N/A |
| Experimental models: cell lines | ||
| Plasmodium yoelii 17XNL | BEI Resources Repository/MR4/ATCC, NIAID, DC Kaslow | N/A |
| Plasmodium berghei ANKA | BEI Resources Repository/MR4/ATCC, NIAID, DC Kaslow | N/A |
| Experimental models: organisms/strains | ||
| C57BL/6N Conventional mice | Taconic Biosciences | Barrier room; IBU504 |
| C57BL/6N Conventional mice | Charles River Laboratories | Barrier room; R01 |
| Software and algorithms | ||
| QIIME2 | Bolyen et al., 2019 | https://qiime2.org/ |
| Graphpad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| MVRSION | Schriefer et al., 2018 | N/A |
| Vegan: R package | http://sortie-admin.readyhosting.com/lme/R%20Packages/vegan.pdf | |
INCLUSION AND DIVERSITY.
We worked to ensure gender balance in the recruitment of human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Highlights.
Human gut bacteria composition correlates with severity of malaria
Gut bacteria dynamically modulate spleen GC reactions to systemic infection
Modulating gut bacteria can halt escalating parasitemia
Antibiotic-induced changes in gut bacteria can boost anti-Plasmodium host immunity
ACKNOWLEDGMENTS
The authors thank Dr. Martin Richer for critical review of this manuscript. This work was supported by grants from the National Institute of Allergy and Infectious Disease of the National Institutes of Health (NIH) (R01AI123486 and R01AI148525 to N.W.S. and R01NS055349 to C.C.J.) and funds from the University of Louisville (to N.W.S.). Support provided by the Herman B. Wells Center (to N.W.S. and C.C.J.) was in part from the Riley Children’s Foundation. The project described was supported by the Indiana University Health-Indiana University School of Medicine Strategic Research Initiative (to N.W.S.). The Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility is funded in part by NIH National Cancer Institute (grant P30 CA082709), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (grant U54 DK106846), and NIH (instrumentation grant 1S10D012270). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109094.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, Sparwasser T, Bérard M, Cerf-Bensussan N, and Eberl G (2019). A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, and Kasper DL (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med 7, 307ra152. [DOI] [PubMed] [Google Scholar]
- Bokulich, Chung NA, Battaglia J, Henderson T, Jay N,M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Y, et al. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med 8, 343ra382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, and Asnicar F (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, and Pamer EG (2012). Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun 80, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahenzli J, Köller Y, Wyss M, Geuking MB, and McCoy KD (2013). Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 14, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S, Mandal RK, Duff ML, and Schmidt NW (2019). Intestinal short-chain fatty acid composition does not explain gut microbiota-mediated effects on malaria severity. PLoS ONE 14, e0214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Healer J, Marapana D, and Marsh K (2016). Malaria: Biology and Disease. Cell 167, 610–624. [DOI] [PubMed] [Google Scholar]
- Del Portillo HA, Ferrer M, Brugat T, Martin-Jaular L, Langhorne J, and Lacerda MV (2012). The role of the spleen in malaria. Cell. Microbiol 14, 343–355. [DOI] [PubMed] [Google Scholar]
- Denny JE, Powers JB, Castro HF, Zhang J, Joshi-Barve S, Campagna SR, and Schmidt NW (2019). Differential sensitivity to Plasmodium yoelii infection in C57BL/6 mice impacts gut-liver axis homeostasis. Sci. Rep 9, 3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo MM, Costa PAC, Diniz SQ, Henriques PM, Kano FS, Tada MS, Pereira DB, Soares IS, Martins-Filho OA, Jankovic D, et al. (2017). T follicular helper cells regulate the activation of B lymphocytes and antibody production during Plasmodium vivax infection. PLoS Pathog. 13, e1006484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy TM, Shepherd DM, Durie FH, Aruffo A, Ledbetter JA, and Noelle RJ (1993). In vivo CD40-gp39 interactions are essential for thymus-dependent humoral immunity. II. Prolonged suppression of the humoral immune response by an antibody to the ligand for CD40, gp39. J. Exp. Med 178, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, et al. (2016). Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med 22, 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller JJ, Graham AC, Zander RA, Pope RL, and Butler NS (2017). Cutting edge: IL-10 is essential for the generation of germinal center B cell responses and anti-plasmodium humoral immunity. J. Immunol 198, 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak BW, Lankelma JM, Hugenholtz F, Belzer C, de Vos WM, and Wiersinga WJ (2019). Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on the gut microbiota in healthy humans. J. Antimicrob. Chemother 74, 782–786. [DOI] [PubMed] [Google Scholar]
- Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng N-Y, et al. (2019). Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178, 1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan N, and Yang H (2019). Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 7, e7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, and Iwasaki A (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 108, 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RF (1985). Malaria treated with emetine or metronidazole. Lancet 2, 498. [DOI] [PubMed] [Google Scholar]
- Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, Che T, and Zhang C (2019). Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 8, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Qie Y, Park J, and Kim CH (2016). Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga K, Kusawake Y, Ito Y, Sugioka N, Shibata N, and Takada K (2006). Enhancing mechanism of Labrasol on intestinal membrane permeability of the hydrophilic drug gentamicin sulfate. Eur. J. Pharm. Biopharm 64, 82–91. [DOI] [PubMed] [Google Scholar]
- Kurup SP, Obeng-Adjei N, Anthony SM, Traore B, Doumbo OK, Butler NS, Crompton PD, and Harty JT (2017). Regulatory T cells impede acute and long-term immunity to blood-stage malaria through CTLA-4. Nat. Med 23, 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, Young GP, Marshall HS, Wesselingh SL, Rogers GB, and Lynn DJ (2018). Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe 23, 653–660. [DOI] [PubMed] [Google Scholar]
- Mandal RK, Crane RJ, Berkley JA, Gumbi W, Wambua J, Ngoi JM, Ndungu FM, and Schmidt NW (2019). Longitudinal analysis of infant stool bacteria communities before and after acute febrile malaria and artemether-lumefantrine treatment. J. Infect. Dis 220, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez F, Munoz A, and Plowe CV (2006). Use of area under the curve to characterize transmission potential after antimalarial treatment. Am. J. Trop. Med. Hyg 75, 640–644. [PubMed] [Google Scholar]
- Mestre L, Carrillo-Salinas FJ, Mecha M, Feliú A, Espejo C,Álvarez-Cermeño JC, Villar LM, and Guaza C (2019). Manipulation of gut microbiota influences immune responses, axon preservation, and motor disability in a model of progressive multiple sclerosis. Front. Immunol 10, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi E, Kim S-W, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, and Ohno H (2020). Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 585, 102–106. [DOI] [PubMed] [Google Scholar]
- Morita H, Kano C, Ishii C, Kagata N, Ishikawa T, Uchiyama Y, Hara S, Nakamura T, and Fukuda S (2020). Bacteroides uniformis enhances endurance exercise performance through gluconeogenesis. bioRxiv. 10.1101/2020.03.04.975730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Chora ÂF, and Mota MM (2020). Microbiota, a Third Player in the Host-Plasmodium Affair. Trends Parasitol. 36, 11–18. [DOI] [PubMed] [Google Scholar]
- Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, et al. (2015). Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun 6, 7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noelle RJ, Roy M, Shepherd DM, Stamenkovic I, Ledbetter JA, and Aruffo A (1992). A 39-kDa protein on activated helper T cells binds CD40 and transduces the signal for cognate activation of B cells. Proc. Natl. Acad. Sci. USA 89, 6550–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng-Adjei N, Portugal S, Tran TM, Yazew TB, Skinner J, Li S, Jain A, Felgner PL, Doumbo OK, Kayentao K, et al. (2015). Circulating Th1-cell-type Tfh cells that exhibit impaired B cell help are preferentially activated during acute malaria in children. Cell Rep. 13, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, et al. (2014). TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, and Blumberg RS (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, Hansen TH, Liang S, Feng Q, Zhang C, et al. (2018). Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol 3, 1255–1265. [DOI] [PubMed] [Google Scholar]
- Parker BJ, Wearsch PA, Veloo ACM, and Rodriguez-Palacios A (2020). The genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol 11, 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mazliah D, Ng DHL, Freitas do Rosário AP, McLaughlin S, Mastelic-Gavillet B, Sodenkamp J, Kushinga G, and Langhorne J (2015). Disruption of IL-21 signaling affects T cell-B cell interactions and abrogates protective humoral immunity to malaria. PLoS Pathog. 11, e1004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mazliah D, Nguyen MP, Hosking C, McLaughlin S, Lewis MD, Tumwine I, Levy P, and Langhorne J (2017). Follicular helper T cells are essential for the elimination of Plasmodium infection. EBioMedicine 24, 216–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Kupfer Y, Pagala M, Chapnick E, and Tessler S (2011). Systemic absorption of oral vancomycin in patients with Clostridium difficile infection. Scand. J. Infect. Dis 43, 386–388. [DOI] [PubMed] [Google Scholar]
- Round JL, and Mazmanian SK (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol 9, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryg-Cornejo V, Ioannidis LJ, Ly A, Chiu CY, Tellier J, Hill DL, Preston SP, Pellegrini M, Yu D, Nutt SL, et al. (2016). Severe malaria infections impair germinal center responses by inhibiting T follicular helper cell differentiation. Cell Rep. 14, 68–81. [DOI] [PubMed] [Google Scholar]
- Schriefer AE, Cliften PF, Hibberd MC, Sawyer C, Brown-Kennerly V, Burcea L, Klotz E, Crosby SD, Gordon JI, and Head RD (2018). A multi-amplicon 16S rRNA sequencing and analysis method for improved taxonomic profiling of bacterial communities. J. Microbiol. Methods 154, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijt TJ, van der Poll T, de Vos WM, and Wiersinga WJ (2013). The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol. 21, 221–229. [DOI] [PubMed] [Google Scholar]
- Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, et al. (2019). Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stough JM, Dearth SP, Denny JE, LeCleir GR, Schmidt NW, Campagna SR, and Wilhelm SW (2016). Functional characteristics of the gut microbiome in C57BL/6 mice differentially susceptible to Plasmodium yoelii. Front. Microbiol 7, 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Brik RB-Z, Federici S, et al. (2018). Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423. [DOI] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen A-M, et al. ; DIABIMMUNE Study Group (2016). Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, and Nussenzweig MC (2012). Germinal centers. Annu. Rev. Immunol 30, 429–457. [DOI] [PubMed] [Google Scholar]
- Villarino NF, LeCleir GR, Denny JE, Dearth SP, Harding CL, Sloan SS, Gribble JL, Campagna SR, Wilhelm SW, and Schmidt NW (2016). Composition of the gut microbiota modulates the severity of malaria. Proc. Natl. Acad. Sci. USA 113, 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waide ML, Polidoro R, Powell WL, Denny JE, Kos J, Tieri DA, Watson CT, and Schmidt NW (2020). Gut Microbiota Composition Modulates the Magnitude and Quality of Germinal Centers during Plasmodium Infections. Cell Rep. 33, 108503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Q, and Ren J (2019). Microbiota-Immune Interaction in the Pathogenesis of Gut-Derived Infection. Front. Immunol 10, 1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ishima T, Zhang J, Qu Y, Chang L, Pu Y, Fujita Y, Tan Y, Wang X, and Hashimoto K (2020). Ingestion of Lactobacillus intestinalis and Lactobacillus reuteri causes depression- and anhedonia-like phenotypes in antibiotic-treated mice via the vagus nerve. J. Neuroinflammation 17, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2019). World Malaria Report 2018 (World Health Organization; ). [Google Scholar]
- Wunderlich CM, Delić D, Behnke K, Meryk A, Ströhle P, Chaurasia B, Al-Quraishy S, Wunderlich F, Brüning JC, and Wunderlich FT (2012). Cutting edge: Inhibition of IL-6 trans-signaling protects from malaria-induced lethality in mice. J. Immunol 188, 4141–4144. [DOI] [PubMed] [Google Scholar]
- Yang I, Corwin EJ, Brennan PA, Jordan S, Murphy JR, and Dunlop A (2016). The infant microbiome: implications for infant health and neurocognitive development. Nurs. Res 65, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B, Portugal S, Tran TM, Gozzelino R, Ramos S, Gomes J, Regalado A, Cowan PJ, d’Apice AJ, Chong AS, et al. (2014). Gut microbiota elicits a protective immune response against malaria transmission. Cell 159, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, Kirkness EF, Tran TM, Harkins DM, Jones MB, Torralba MG, O’Connell E, Nutman TB, Doumbo S, Doumbo OK, et al. (2015). Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics 16, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, and Chen D-C (2019). Facing a new challenge: the adverse effects of antibiotics on gut microbiota and host immunity. Chin. Med. J. (Engl.) 132, 1135–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, Zhang MC, Li YG, Zhao JB, Gao YN, et al. (2019). Dysbiosis of the Gut Microbiome in Lung Cancer. Front. Cell. Infect. Microbiol 9, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequencing datasets generated and analyzed from mice and humans in this study are publicly available and deposited in the NCBI Sequence Read Archive under the BioProject IDs PRJNA643559 and PRJNA642859 respectively.


