Abstract
Coronaviruses are deadly and contagious pathogens that affects people in different ways. Researchers have increased their efforts in the development of antiviral agents against coronavirus targeting Mpro protein (main protease) as an effective drug target. The present study explores the inhibitory potential of characteristic and non-characteristic Withania somnifera (Indian ginseng) phytochemicals (n ≈ 100) against SARS-Cov-2 Mpro protein. Molecular docking studies revealed that certain W. somnifera compounds exhibit superior binding potential (−6.16 to −12.27 kcal/mol) compared to the standard inhibitors (−2.55 to −6.16 kcal/mol) including nelfinavir and lopinavir. The non-characteristic compounds (quercetin-3-rutinoside-7-glucoside, rutin and isochlorogenic acid B) exhibited higher inhibitory potential in comparison to characteristic W. somnifera compounds withanolide and withanone. Molecular dynamics (MD) simulation studies of the complex for 100 ns confirm favorable and stable binding of the lead molecule. The MMPBSA calculation of the last 10 ns of the protein-ligand complex trajectory exhibited stable binding of quercetin-3-rutinoside-7-glucoside at the active site of SARS-Cov-2 Mpro. Taken together, the study demonstrates that the non-characteristic compounds present in W. somnifera possess enhanced potential to bind SARS-Cov-2 Mpro active site. We further recommend in vitro and in vivo experimentation to validate the anti-SARS-CoV-2 potential of these lead molecules.
Keywords: Withania somnifera, SARS-CoV-2, In silico studies, Mpro, Phytochemicals, Antiviral
1. Introduction
COVID-19 is an infectious disease caused by a newly discovered SARS-CoV-2 virus. The number of infections has significantly exceeded globally and threatens human population worldwide. On January 30, 2020 World Health Organization (WHO) announced the COVID-19 epidemic as a public health emergency of international concern. Researchers from all over the world attempted to develop treatment against SARS-CoV-2 infection and identify effective agents to combat this global threat [1]. Studies from several laboratories have identified some SARS-CoV-2 proteins which are potential drug target(s) having ability to block viral-host interaction, virulence and pathogenesis [[2], [44]]. SARS-CoV-2 main protease (Mpro), a cysteine protease plays a pivotal role in viral gene expression, transcription, and replication. It mediates its function through a highly complex cascade of proteolytic processing of replicase proteins [3]. [4] developed activity-based probes for the Mpro protein in diagnosing COVID-19 infected patients, providing a framework for drug design against this molecule [4]. Thus, targeting viral Mpro protein could be a potential and attractive strategy against SARS-CoV-2 antiviral drug development. Moreover, synthetic antiviral drugs show common to life threatening side-effects such as nausea, diarrhea, hepatotoxicity, hyperglycemia etc. [5]. In contrast, natural agents show antiviral potential by inhibiting viral attachment, and targeting critical enzymes (helicase, protease and polymerase) involved in the pathogenesis of the disease [6]. Cost effectiveness and low side effects of phytochemicals make them excellent candidates for antiviral drug discovery.
Accumulating research demonstrates that medicinal plants possess therapeutic potential against microbes and pathogen [[7], [8], [9]]. Recent reports suggest that natural-like products and their metabolites present in several plant spices exhibit remarkable potential to block SARS-CoV-2 Mpro receptor [[10], [11], [12]]. Withania somnifera (Solanaceae) also known as Indian Ginseng is a well-known medicinal plant that possess antiviral properties. Phytochemicals present in W. somnifera are known to possess antiviral activity against H1N1 influenza virus, HIV and infectious bursal disease. Studies indicate that the antiviral mode of action of W. somnifera compounds include viral protein inhibition, mitigation of viral infection induced inflammation as well as immunomodulatory effects [[13], [14], [15]]. W. somnifera possess both characteristic and non-characteristic (present in other plant also) phytochemicals. Recent literature data shows SARS-CoV-2 Mpro inhibitory potential in W. somnifera characteristic compounds especially wihanolides [16]. However, the non-characteristic compounds of the plant have not been screened against SARS-CoV-2 Mpro protein. The present study aimed to identify lead W. somnifera phytochemical from a list of approximately hundred compounds having potential to inhibit the SARS-CoV-2 Mpro protein using computer aided drug discovery approach.
2. Materials and methods
2.1. Withania somnifera compound retrieval and preparation
A total of 93 compounds present in W. somnifera plant were obtained from different literature and search engine platforms such as PubMed, Google Scholar, Web of Science, Science Direct, Scopus, Semantic Scholar, Medline, and PubMed Central [17,18]. The structures of these compounds present in W. somnifera were prepared using Marvin Sketch software [19]. The 2D or 3D structure of standard compounds against target proteins were retrieved from the NCBI PubChem in. sdf format [20]. Open Babel molecule format converter was used to perform conversion of 2D to 3D conformation and their conversion from. sdf to. mol file [21]. Ligand energy was minimized by applying mmff94 force field and conjugate gradients optimization algorithm using PyRx-Python prescription 0.8 for 200 steps [22].
2.2. Receptor retrieval
3D structure of SARS-CoV-2 main protease (PDB ID: 5RFS) receptor was obtained from protein data bank (https://www.rcsb.org/) [24]. The resolution of the retrieved structures was 1.70 Å.
2.3. Receptor preparation
3D structure of SARS-CoV-2 main protease was loaded onto UCSF Chimera for molecular docking preparation [25]. Protein models were cleaned and optimized by removing ligands and other heteroatoms including water. After this step, the energy minimization of protein structures was performed by steepest descent method having 100 steps (step size 0.02 Å), and a conjugate gradient method with 10 steps (step size 0.02 Å) using UCSF Chimera.
2.4. Molecular docking studies
Auto Dock Tools v.1.5.6 (ADT) was used to dock the test ligands on targeted protein [26,27]. Gestgeiger partial charges assigned to the ligands and docking calculations were performed. Polar hydrogen atoms, Kollman charges, and solvation parameters were applied using appropriate Auto Dock tools. Lamarckian Genetic Algorithm (LGA) was used to explore the active binding region in this study. The grid box included the entire binding site of the protein providing enough space for the ligands translational and rotational maneuverability. Docking was performed by keeping the grid dimensions 9.05, −1.87 and 22.09 in X, Y and Z dimension with 10 Å grid spacing for the target protein. For each of the 30 independent runs, a maximum number of 27,000 GA operations generated on a single population of 150 individuals. Operator weights for the rate of crossover, gene mutation, and elitism were set as 0.80, 0.02, and 1, respectively. LigPlot+ (v.1.4.5) and UCSF Chimera (v.1.10.2) online tools were used for protein-ligand interaction visualization [28].
2.5. Molecular dynamics simulation
MD simulations for all atoms were performed for Mpro protein of SARS-CoV-2 before and after the binding of the known inhibitors and identified compounds. GROMOS 54A7 force field was used to perform the molecular dynamics simulation using the GROMACS 5.1.1 package. Ligand topologies and other parameters generated using PRODRG server and combined with the protein topology to make simulation complexes. All systems were solvated using the SPC water model in a cubic box. Energy minimization was achieved to get rid of steric clashes by using 1500 steps of the steepest descent approach. Following energy minimization the temperature of all systems was increased from 0 to 300 K in equilibration phase of 100 ps at a constant volume and stable pressure of 1 bar. Pressure of the systems was maintained by using Parrinello-Rahman barostat [19]. Conservation of bond lengths was achieved using LINear Constraint Solver (LINCS) [29] algorithm. Long range interactions were computed using Particle Mesh Ewald (PME) [30]. The final MD run was set to 100,000 ps for all six systems. Trajectory analysis was performed using various GROMACS utilities such as gmx rms, gmx rmsf, gmx gyrate, gmx sasa, gmx hbond and gmx do_dssp for the analysis of rmsd, rmsf, radius of gyration, solvent accessible surface area, hydrogen bonds and secondary structure respectively. Principal component analysis (PCA) was performed to determine the correlated motions of proteins. For PCA analysis, gmx covar module of GROMACS was utilized for the generation and diagonalization of covariance matrix. Further gmx anaeig was used to determine the overlap between the principal components. Free energy surface (FES) was generated to show the energy of conformations adopted by the protein throughout the 100 ns MD simulation. gmx sham module was applied for the generation of FES [31].
2.6. g-mmpbsa analysis
The binding free energies of the protein-ligand complexes of Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complexes were estimated by Molecular Mechanics-Poisson–Boltzmann Solvent-Accessible surface area (MMPBSA) method using g-mmpbsa package [32]. g-mmpbsa is a widely used method to determine the free energy of biomolecular interactions. Binding free energy was determined by using following equations:
| ΔGBind=GComp - (GProt + GLig) |
Where ΔGComp signifies the energy of protein-ligand complex, GProt and GLig signify the individual energy of protein and ligand respectively. The MMPBSA calculation was performed for last 10 ns (90–100 ns) of each simulation trajectories.
3. Result and discussion
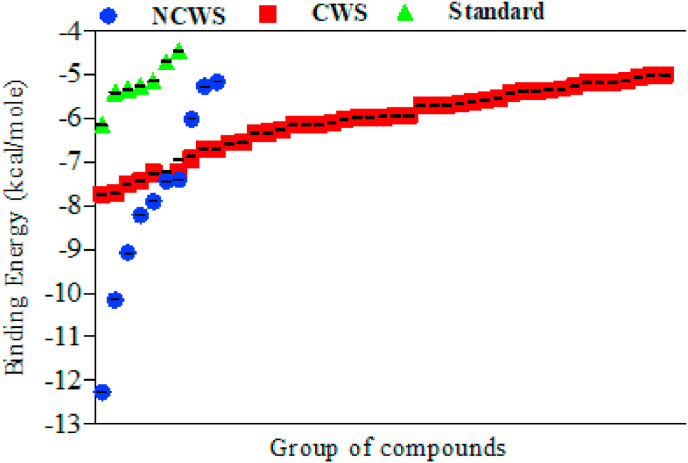
A total of 93 phytochemicals were extracted after comprehensive literature survey on Withania somnifera. These phytochemicals were further categorized on the basis of characteristic and non-characteristic presence in W. somnifera (Supplementary Table). Molecular docking screening of 93 phytochemicals against Mpro protein revealed 23 lead compounds exhibiting higher binding potential than standard/reference compounds. These test phytochemicals showed potential binding energy ranging from −6.16 to −12.27 kcal/mol at Mpro active site, compared to the standard protease inhibitors (nelfinavir, lopinavir, etc.) in the range of −2.55 to −6.16 kcal/mol (Supplementary Table 1). The name and structure of lead compounds (cutoff less than −6.16 kcal/mol binding energy) are shown in Fig. 2. The ADME of the compounds used in the present study are provided in Table 1 . Quercetin-3-rutinoside-7-glucoside and rutin showed low binding score (−12.27 and −10.16 kcal/mol respectively) against Mpro protein, compared to nelfinavir (−6.16 kcal/mol). Type of amino acid residue involved in the protein-ligand interactions are summarized in Table 2 . The binding energy of the characteristic and non-characteristic compounds were plotted in a grouped manner to compare the binding potential. The result showed that the W. somnifera non-characteristic compound have better potential than the characteristic phytochemicals in terms of binding energy at the active site of the SARS-CoV-2 Mpro protein (Fig. 1 ).
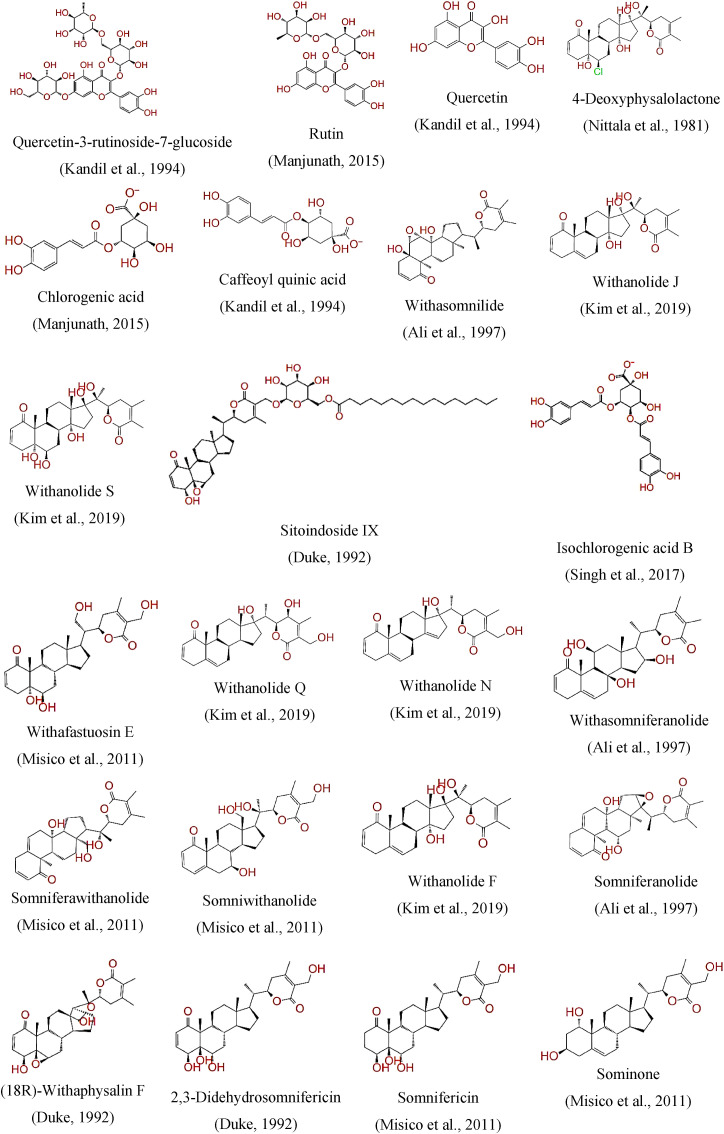
Fig. 2.
List of Withania somnifera phytochemicals lead SARS-CoV-2 main protease inhibitors with less than −6.00 kcal/J docking score cut-off value [[34], [35], [37], [38], [39], [41], [43], [45]].
Table 1.
ADME of the compounds from W. somnifera.
| Compounds | MF | LS(E)S | WSC | BBBP | P-gpS | GIA | BS | NHBA | NHBD | TPSA (Å2) | MR | LKP(SP) (cm/s) | LV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Quercetin-3-rutinoside-7-glucoside | C33H40O21 | −2.91 | S | No | Yes | low | 0.17 | 21 | 13 | 348.58 | 173.50 | −12.53 | 3*** |
| Rutin | C27H30O16 | −3.30 | S | No | Yes | low | 0.17 | 16 | 10 | 269.43 | 141.38 | −10.26 | 3*** |
| Quercetin | C15H10O7 | −3.16 | S | No | No | High | 0.55 | 7 | 7 | 131.36 | 78.04 | −7.05 | 0# |
| 4-Deoxyphysalolactone | C28H39ClO7 | −4.23 | MS | No | Yes | High | 0.55 | 7 | 4 | 124.29 | 135.75 | −8.05 | 1* |
| Chlorogenic acid | C16H17O9 | −1.61 | VS | No | No | Low | 0.11 | 9 | 5 | 167.58 | 81.56 | −8.75 | 0# |
| Caffeoyl quinic acid | C16H17O9 | −1.61 | VS | No | No | Low | 0.11 | 9 | 5 | 167.58 | 81.56 | −8.75 | 0# |
| Withasomnilide | C28H38O6 | −4.67 | MS | No | Yes | High | 0.55 | 6 | 2 | 96.36 | 127.53 | −6.87 | 0# |
| Withanolide J | C28H38O6 | −4.14 | MS | No | Yes | High | 0.55 | 6 | 3 | 104.06 | 129.28 | −7.46 | 0# |
| Withanolide S | C28H40O8 | −3.31 | S | No | Yes | Low | 0.55 | 8 | 5 | 144.52 | 132.12 | −8.85 | 1* |
| Sitoindoside IX | C34H48O11 | −4.43 | MS | No | Yes | Low | 0.17 | 11 | 5 | 175.51 | 159.87 | −8.97 | 2** |
| Isochlorogenic acid B | C25H23O12 | −3.64 | S | No | Yes | Low | 0.11 | 12 | 6 | 214.11 | 124.95 | −8.37 | 3*** |
| Withafastuosin E | C28H40O7 | −4.32 | MS | No | Yes | High | 0.55 | 7 | 4 | 124.29 | 130.84 | −7.35 | 0# |
| Withanolide Q | C28H38O6 | −4.37 | MS | No | Yes | High | 0.55 | 6 | 3 | 104.06 | 129.21 | −7.13 | 0# |
| Withanolide N | C28H36O5 | −4.62 | MS | No | Yes | High | 0.55 | 5 | 2 | 83.83 | 127.57 | −6.62 | 0# |
| Withasomniferanolide | C28H38O6 | −4.24 | MS | No | Yes | High | 0.55 | 6 | 3 | 104.06 | 129.21 | −7.35 | 0# |
| Somniferawithanolide | C28H38O6 | −3.86 | S | No | Yes | High | 0.55 | 6 | 3 | 104.06 | 129.24 | −7.71 | 0# |
| Somniwithanolide | C28H38O7 | −3.78 | S | No | Yes | High | 0.55 | 7 | 4 | 124.29 | 130.37 | −7.93 | 0# |
| Withanolide F | C28H38O6 | −4.14 | MS | No | Yes | High | 0.55 | 6 | 3 | 104.06 | 129.28 | −7.46 | 0# |
| Somniferanolide | C28H36O6 | −4.19 | MS | No | Yes | High | 0.55 | 6 | 2 | 96.36 | 127.05 | −7.38 | 0# |
| (18R)-Withaphysalin F | C28H36O7 | −4.15 | MS | No | Yes | High | 0.55 | 7 | 2 | 105.59 | 126.50 | −7.71 | 0# |
| 2,3-Didehydrosomnifericin | C28H40O7 | −4.45 | MS | No | Yes | High | 0.55 | 7 | 4 | 124.29 | 130.84 | −7.27 | 0# |
| Somnifericin | C28H42O7 | −4.47 | MS | No | Yes | High | 0.55 | 7 | 4 | 129.29 | 131.32 | −7.28 | 0# |
| Sominone | C28H42O5 | −5.46 | MS | No | Yes | High | 0.55 | 5 | 3 | 86.99 | 129.44 | −5.75 | 0# |
***=(No; 3 violations: MW > 500, NorO>10, NHorOH>5); **= (No; 2 violations: MW > 500, NorO>10); *= (Yes; 1 violation: MW > 500); #= (Yes; 0 violation); S=Soluble; MS = Moderately soluble; VS=Very soluble; LS(E)S = Log S (ESOL) solubility; WSC=Water solubility class; BBBP BBB permeant; P-gpS = P-gp substrate; NHBD=Num. H-bond donors; MR = Molar refractivity; NHBA=Num. H-bond acceptors; LKP(SP) = log Kp (skin permeation); MF = Molecular formula; LV = Lipinski violation; GIA = Gastrointestinal absorption; BS=Bioavailability score.
Table 2.
DockScore and list of amino residue involved in the interaction with standard inhibitors and W. somnifera phytochemicals with −6.16 < cutoff value.
| S.No. | Ligand | Dock Score (kcal/mol) | Hydrogen bond interaction amino acid | Hydrophobic interaction amino acid |
|---|---|---|---|---|
| S1 | Nelfinavir | −6.16 | Asn142, His41 | Leu141, His163, Cys145, Met165, Glu166, Gln189, His164, Asp187, Arg188, Ser46, Thr25, Met49, Leu27 |
| S2 | Lopinavir | −5.33 | Cys145, Glu166 | Met165, Gln189, Arg188, His41, Thr26, Gly143, Thr24, Cys44, Thr25, Thr45, Ser46, Leu141, Asn142 |
| 1 | Quercetin-3-rutinoside-7-glucoside | −12.27 | Thr25, Thr24, Ser46, Thr26, Asn142, Glu166, His164 | Gly143, Leu27, Thr45, Met165, His41, Met49, Arg188, Leu167, Pro168 |
| 2 | Rutin | −10.16 | Thr25, Thr24, Gly143, His164, Asn142 | Thr26, Cys145, Arg188, Met165, Asp187, Met49, Gln189, Glu166 |
| 3 | Isochlorogenic acid B | −9.08 | Gly143, His41, Phe140, Glu166, | His163, His164, Met49, Ser46, Gln189, Met165, Leu141, Asn142 |
| 4 | Caffeoyl quinic acid | −8.22 | Asn142, Gly143, His164, Thr190 | Cys145, His41, Glu166, Met165, Gln189, Leu167, Pro168, Gln192 |
| 5 | 4-Deoxyphysalolactone | −7.91 | His41, Gly143, Gln189, Glu166, | Thr26, Thr25, Asn142, Met49, Met165, Thr190, Pro168 |
| 6 | Sitoindoside IX | −7.74 | Thr26, Cys145, Asn142, Leu141, Glu166 | Gly143, Thr25, His164, met49, Gln189, Met165, His163 |
| 7 | Withanolide J | −7.72 | Glu166, Gly143 | Asn142, Thr25, Leu27, Cys145, Thr26, His41, Met165, Met49, Gln189, Thr190, pro168 |
| 8 | Withanolide S | −7.51 | Gln189, Gly143, His41 | Glu166, Thr190, Arg188, Met165, Pro168, Met49, Thr26, Asn142, Cys145, His164 |
| 9 | Withanolide Q | −7.45 | Gly143, Glu166, Thr190 | Met149, Thr25, Leu27, His41, Cys145, Asn142, Gln189, Pro168 |
| 10 | Quercetin | −7.44 | Thr190 | His164, His41, Met49, Met165, Gln192, Pro168, Glu166 |
| 11 | Chlorogenic acid | −7.41 | Gly143, Glu166, Thr190 | Cys145, His41, Asn142, Met165, Gln189, Pro168 |
| 12 | Withafastuosin E | −7.25 | Thr26, Cys44, Glu166, | Gly143, Thr25, Ser46, Thr45, Met49, Gln189 |
| 13 | Withanolide N | −7.21 | Gly143, Glu166 | Asn142, Cys145, Leu27, Thr26, Thr25, His41, Met165, Gln189, Thr190, Pro168 |
| 14 | Somniferawithanolide | −6.96 | Asn142, Glu166 | Met165, pro168, Gln189, Ser46Thr25, Met49, Cys44, Thr45, His41, His164, Cys145 |
| 15 | Somniwithanolide | −6.89 | Gly143, His164, Glu166 | Thr45, His41, Thr25, Pro168, Met165, Cys145, Asn142 |
| 16 | Withasomniferanolide | −6.7 | Glu166, His164, Gly143 | Cys145, His41, Thr26, Thr25, Asn142, Gln189, Pro168, Met165 |
| 17 | Somnifericin | −6.7 | Thr24, Glu166 | Met165, His41, Gly143, Asn142, Thr25, Ser46, Thr45, Gln189 |
| 18 | Withanolide F | −6.57 | Glu166, Asn142 | Ser46, His41,Met49, Thr25, Gln189, Arg188, Thr190, Met165 |
| 19 | Somniferanolide | −6.55 | Glu166 | Pro168, Gln189, leu27, Asn142, His41, Gly143, Thr25, Thr26, His164, Met165 |
| 20 | (18R)-Withaphysalin F | −6.35 | Gly143 | Asn142, Thr26, Thr25, His41, Glu166, Gln189, Thr190, Met165, Pro168, Cys145 |
| 21 | 2,3-Didehydrosomnifericin | −6.33 | Thr190, Gly143, Thr26 | Gln189, Pro168, Asn142, His41, Thr25, Cys145, Met165, Glu166 |
| 22 | Sominone | −6.27 | Thr24, Thr25, Ser46, Glu166 | Cys44, Thr45, Met49, His41, Gln189, Asn142 |
| 23 | Withasomnilide | −6.16 | Cys145 | His41, His164, Met165, Glu166, Pro168, Gln189, Met49, Asn142, Thr25, Gly143 |
S1-Standard 1, S2-Standard 2.
Fig. 1.
Analysis of W. somnifera characteristic, non-characteristic phytochemicals and standard inhibitor versus binding energy of targeted SARS-CoV-2 Mpro protein. NCWS-Non-characteristic W. somnifera phytochemicals, CWS-Characteristic W. somnifera phytochemicals.
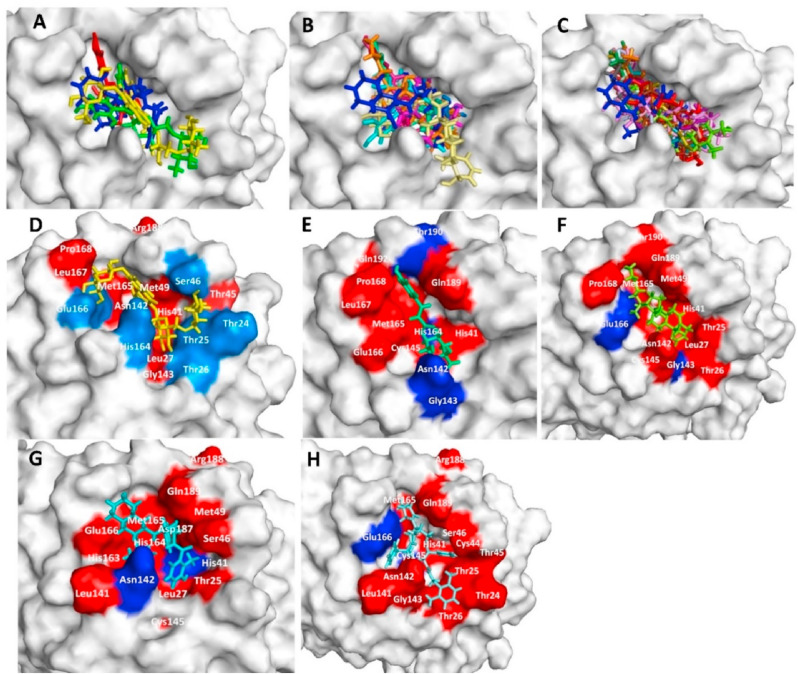
Binding pose of lead phytochemicals at SARS-CoV-2 Mpro active site was predicted and depicted (DockScore less than −6.16 kcal/mol) in Fig. 3 . The lead compounds were categorized into three major groups. First group comprised quercetin and its associated derivative structures such as rutin, and quercetin-3-rutinoside-7-glucoside. Second group consists of compounds reported in W. somnifera but also found in other plants i.e. non-characteristic compounds. The third group of compounds was W. somnifera specific i.e. characteristic compounds. Result showed that all the three groups of W. somnifera compounds as well as standard Mpro inhibitor binds in the same pose at the active site of the enzyme (Fig. 3A–C). Amino acids involved in the binding with quercetin-3-rutinoside-7-glucoside, caffeoyl quinic acid and withanolide J as well as standard inhibitor (nelfinavir and lopinavir) are depicted in Fig. 3D–H. Asn142, His41, Cys145, and Glu166 residue of Mpro were involved in hydrogen bond formation with standard inhibitors (nilfinavir and lopavir). Besides several amino acid residues were involved in hydrophobic interaction with the protease (Table 2). Hydrogen bond length between the quercetin-3-rutinoside-7-glucoside and Thr25, Thr24, Ser46, Thr26, Asn142, Glu166, His164 residues were 2.74; 2.89; 3.00; 3.03 and 2.80; 3.22; 3.12 2.79 and 2.81; and 2.91 Å respectively (Supplementary Figure 1). Gly143, Leu27, Thr45, Met165, His41, Met49, Arg188, Leu167, and Pro168 amino acid residues Mpro protein interacted with Quercetin-3-rutinoside-7-glucoside by hydrophobic interaction (Table 2). Rutin showed hydrogen bonding with Thr25, Thr24, Gly143, His164, and Asn142 residue and interacted with Cys145 and Glu166 amino acids via hydrophobic interaction. Further an important compound caffeoyl quinic acid formed four hydrogen bonds (with Asn142, Gly143, His164, and Thr190) and several hydrophobic interactions at the active site of the protein (Supplementary Figure 1 and Table 2). The top three lead molecules (Quercetin-3-rutinoside-7-glucoside, Rutin, Caffeoyl quinic acid) showed H-bond interaction with Gly143, His164 and Asn142 amino acid residues. Besides the lead showed hydrophobic interaction with some of the common amino acids (Supplementary Figure 1 and Table 2) at the active site of the protein.
Fig. 3.
Superimposed docking pose, and interaction of test compounds at SARS-CoV-2 main protease active site (A) Docking pose of quercetin-3-rutinoside-7-glucoside (yellow), rutin (green), quercetin (red) and nelfinavir (blue). (B) Docking pose of sitoindoside IX (pale yellow), chlorogenic acid (lime green), isochlorogenic acid B (cyan), caffeoyl quinic acid (hotpink), 4-Deoxyphysalolactone (orange) and nelfinavir (blue). (C) Withanolide F (lightblue), somniwithanolide (grey), somniferawithanolide (Purple), somniferanolide (wheat), withasomniferanolide (firebrick), withasomnilide (deapteal), somnifericin (chartreuse), withanolide Q (orange), 2,3-didehydrosomnifericin (chocolate), sominone (violet), withanolide N (forest), withanolide J (palecyan), (18R)-withaphysalin F (pink) and nelfinavir (blue) (D) Surface structure of Mpro protein interacted with lead compound quercetin-3-rutinoside-7-glucoside. (E) Surface structure of Mpro protein interacted with lead compound Caffeoyl quinic acid. (F) Surface structure of Mpro protein interacted with lead compound withanolide J. Green color represents the mentioned amino acid involved in hydrogen bonding (G) Surface structure of Mpro protein interacted with nelfinavir. (H) Surface structure of Mpro protein interacted with lopinavir. Green color represents the amino acid involved in hydrogen bonding.
W. somnifera characteristic compounds such as withanolide J (−7.72 kcal/mol) showed tight binding with Mpro active site in comparison to nilfinavir (−6.16 kcal/mol). It should be noted that Rutin, Caffeoyl quinic acid, Withanolide J, Withanolide Q, Chlorogenic acid, Withanolide N, Somniferawithanolide, Somniwithanolide, Withasomniferanolide, 2,3-Didehydrosomnifericin, and Withasomnilide showed similar type of interaction with Cys145, His41, Glu166 and Asn142 amino acid residues (Table 2). Type of interaction, hydrogen bond length, amino acids involved in hydrogen bonding and hydrophobic interaction for the lead phytochemicals-Mpro complex binding are summarized in Table 2 and supplementary figure 1.
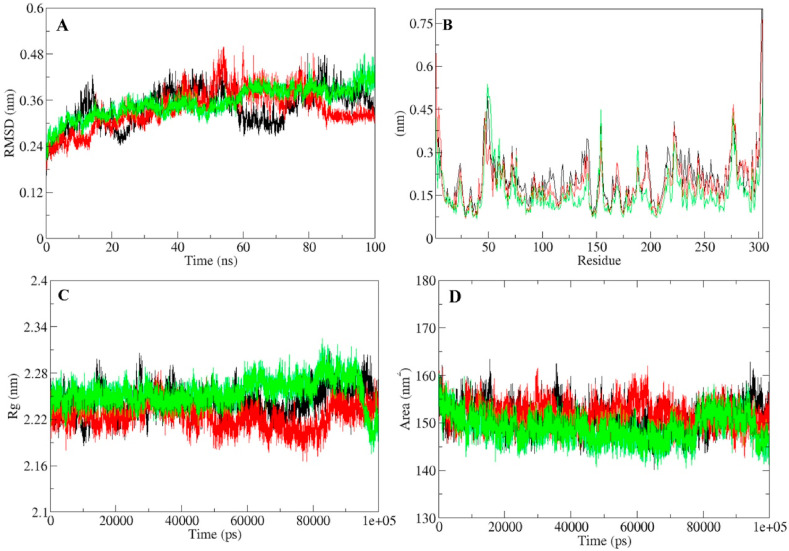
The three systems, Mpro protein (MP), Mpro-lopinavir complex (MLC), and Mpro-quercetin-3-rutinoside-7-glucoside complex (MQC) were subjected to the molecular dynamics (MD) simulations for 100 ns. Dynamic properties of the system such as temperature, pressure, density, and total energy were further calculated to ensure the equilibration and stability of the systems during the simulation period. Result showed that there were no significant alterations in the parameters of free protein and protein bound with standard/lead inhibitor during the simulation (Supplementary Figure 2). Root-mean-square deviation (RMSD) is widely used to get insights of the structural dynamics of proteins. For the assessment of the structural dynamics of Mpro protein in free and complex states, we evaluated the RMSD of MP, MLC and MQC. The average RMSD of MP, MLC and MQC was ~0.29, ~0.28, and ~0.30 nm respectively (Fig. 4 A). Backbone RMSD of all the three systems showed that Mpro gets stabilized after ligand binding (Fig. 4A). RMSD of Mpro-lead complexes were very close to the unbound protein. The stable RMSD showed that the ligand bound complex was equilibrated throughout the simulation trajectory and the binding was stable.
Fig. 4.
Molecular dynamic simulation trajectory analysis of SARS-CoV-2 Mpro protein and protein-ligand complexes during 100 ns simulation (A) RMSD of solvated SARS-CoV-2 Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex during 100 ns MD simulation (B) RMSF values of Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex (C) Rg during 100 ns MD simulation of Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. (D) SASA during 100 ns MD simulation of Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. Unbound protein parameters are depicted in black color. Parameters for Mpro-lopinavir complex and Mpro-quercetin-3-rutinoside-7-glucoside complex are represented in red and green color respectively.
To determine the residual vibrations in the free and ligand bound Mpro protein, the fluctuation of each residue was evaluated as root-mean-square fluctuation (RMSF). Random fluctuations present in Mpro protein in bound and unbound states were evident from N- to C- termini of the protein (Fig. 4B). The fluctuation in the protein was compared for each residue after the binding of lopinavir and quercetin-3-rutinoside-7-glucoside. The fluctuations were found to be minimum at several residues in the case of MLC and MQC complexes. However there is a slight increase in RMSF of MQC which might be induced by the larger size of bound inhibitor. Another important parameter radius of gyration (Rg) is associated with the structural volume of the protein tertiary structure which indicates the structural compactness and folding behavior in the biological system. Elevated Rg value indicate loose packing of amino acid residues in protein. In the present study, the average values of Rg for MP, MLC and MQC were ~2.24, ~2.22, and ~2.25 nm, respectively (Fig. 4C). The Rg of test complex and unbound protein showed a minor deviation which suggests that the binding of compounds did not affected the compactness of protein structure. SASA is the surface area of proteins which is available for the surrounding water to interact with the protein. It is a fundamental parameter which provides insights into the structural folding−unfolding dynamics of a protein in the aqueous environment. SASA values for MP, MLC and MQC were ~151.48, ~151.76, and ~148.93 nm2, respectively (Fig. 4D).
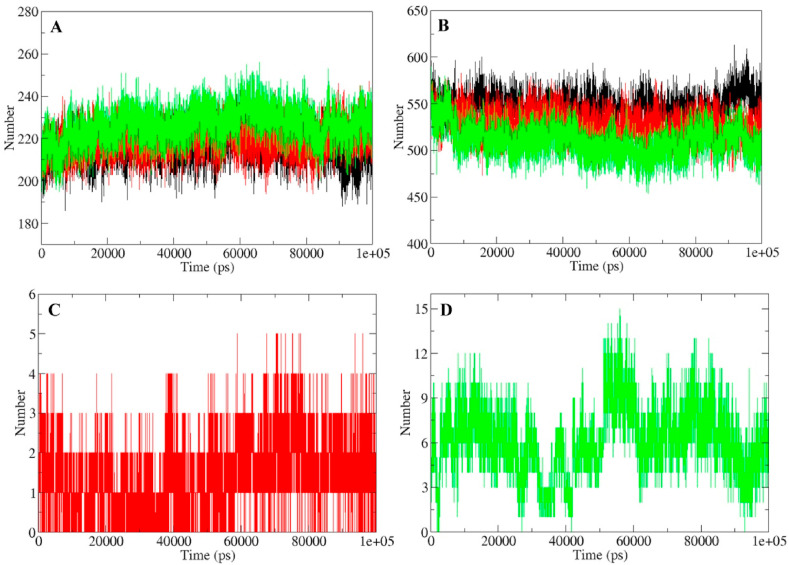
Intramolecular hydrogen bonding network of a protein is an essential parameter in the determination of its three dimensional structure and conformation. For the assessment of stability of Mpro protein in unbound and bound (with standard/lead inhibitor) conformations, number of intramolecular hydrogen bonds were calculated. MQC had an average of 226 hydrogen bonds in comparison to MLC and MP which had 219 and 215 hydrogen bonds (Fig. 5 A). Based on this observation, it is evident that binding of quercetin-3-rutinoside-7-glucoside to Mpro protein stabilizes its internal conformation. Result showed no significant changes in hydrogen bonding network of Mpro protein in unbound and ligand (standard/lead inhibitor) bound state (Fig. 5A). Furthermore number of hydrogen bonds in Mpro protein remained similar throughout the trajectory which also suggests minimal fluctuation in the protein structure. Similarly the hydrogen bonding interaction of the complexes (MP, MLC, and MQC) with surrounding solvent showed stable pattern (5B). MQC had an average of 510 hydrogen bonds with surrounding solvent in comparison to the 527 and 546 hydrogen bonds formed by MLC and MP respectively. Assessment of the hydrogen bonds between the protein and ligand provides direction and specificity of the interaction which are important criterion in protein-ligand binding. To get an insight of stability of binding among MLC and MQC, hydrogen bonds between protein and inhibitor were calculated. Binding of quercetin-3-rutinoside-7-glucoside to Mpro protein encompasses significantly higher number of hydrogen bonds (≈6) in comparison to lopinavir binding (≈1.5) (Fig. 5C and D). These results indicate the stable binding of quercetin-3-rutinoside-7-glucoside to the Mpro protein and was comparable to lopinavir binding.
Fig. 5.
Hydrogen bond analyses of SARS-CoV-2 Mpro protein and protein-ligand complexes during 100 ns simulation. (A) Number of hydrogen bonds in the SARS-CoV2 Mpro protein in unbound, bound with lopinavir and quercetin-3-rutinoside-7-glucoside. (B) Hydrogen bond between Mpro protein and solvent in unbound state, bound with lopinavir and quercetin-3-rutinoside-7-glucoside. (C) Number of hydrogen bonds formed between Mpro and lopinavir (D) Number of hydrogen bonds formed between Mpro protein and quercetin-3-rutinoside-7-glucoside. Unbound protein parameters are depicted in black color. Parameters for Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex are represented in red and green color respectively.
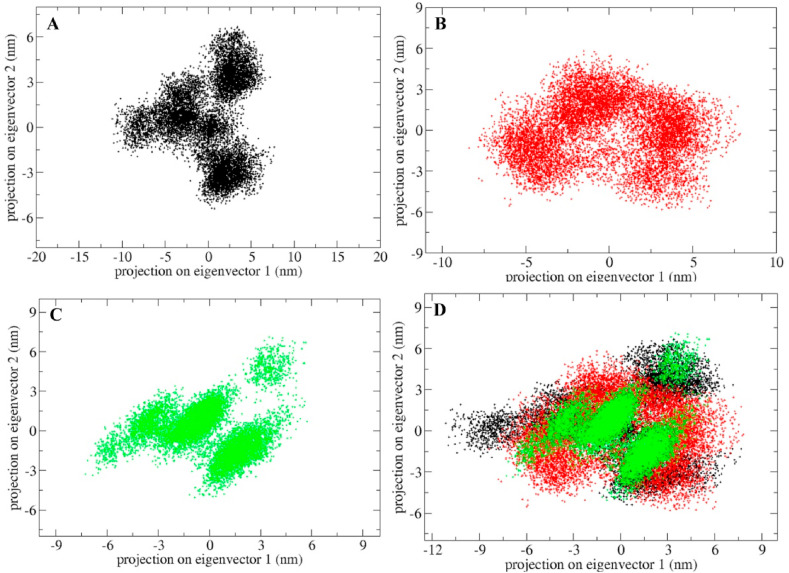
Protein exerts its function by collective motions of atoms. Principle component analysis (PCA) was performed to understand the conformational dynamics of the MP, MLC and MQC by utilizing their collective motions using essential dynamics method. The conformational sampling along the eigenvector 1 and eigenvector 2 was projected by backbone atoms of Mpro protein in unbound and standard/lead inhibitor bound complex (Fig. 6 A–D). The results showed MQC possess less collective motions in comparison to MP and MLC suggesting stable conformation.
Fig. 6.
Principal component analyses of Mpro protein in unbound and bound state (A) Projection of backbone atoms of Mpro protein in phase space along the first two principal eigenvectors in unbound state. (B) Projection of backbone atoms of Mpro protein in complex with lopinavir in phase space along the first two principal eigenvectors (C) Projection of backbone atoms of Mpro protein in complex with quercetin-3-rutinoside-7-glucoside in phase space along the first two principal eigenvectors (D) Superimposition of Plots of PCA analysis for Mpro protein in unbound state and bound with lopinavir and quercetin-3-rutinoside-7-glucoside. Unbound protein parameters are depicted in black color. Parameters for Mpro-lopinavir complex and Mpro-quercetin-3-rutinoside-7-glucoside complex are represented in red and green color respectively.
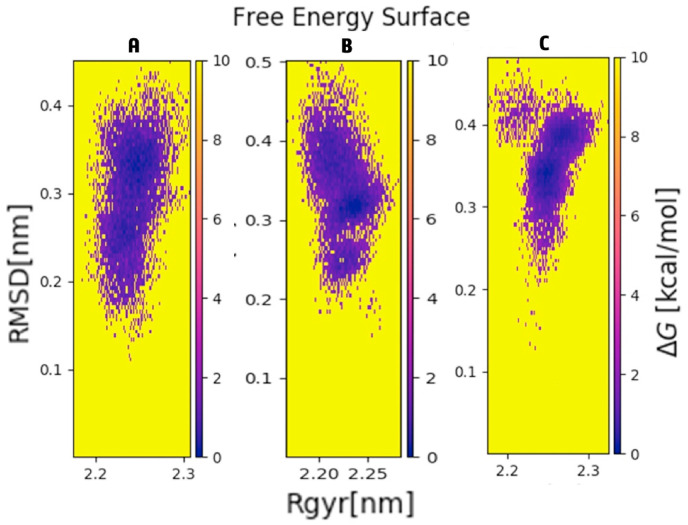
To visualize of minimum energy state of the MP, MLC, and MQC, next we studied the free energy surface (FES) against the PC1 (Radius of gyration, Rg) and PC2 (Root mean square deviation, RMSD) principal components (Fig. 7 A–C). The size and shape of the minimum energy area (shown in blue color) illustrated the stability of the protein in unbound and ligand bound conformation (Fig. 7A–C). FES plot of MQC was more compact and had dense energy minima in comparison to the MP and MLC indicating the stability of the complex.
Fig. 7.
Free energy surface of the first two principal components for SARS CoV-2 (A) Mpro protein (B) Mpro-lopinavir complex and (C) Mpro-quercetin-3-rutinoside-7-glucoside complex.
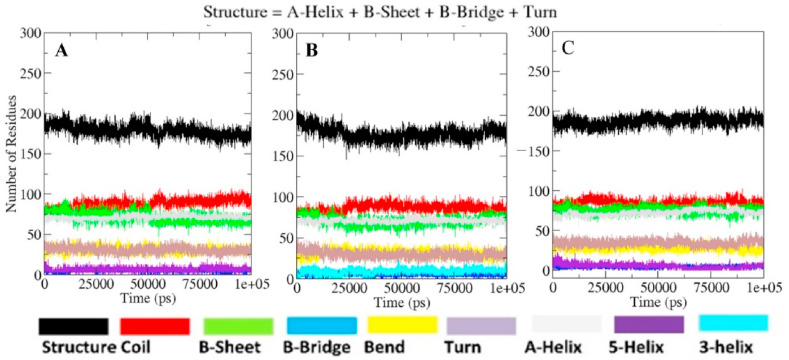
The percentage of secondary structure content in a protein is an important parameter in the study of its structural behavior. There were no significant change in the secondary structure content of MP, MLC and MQC (Fig. 8 A–C). However binding of lead phytochemical to the Mpro protein appears to result in a modest increase in the residue number in structured region. The involvement of more amino residues at the binding site might be responsible for the stability of the ligand bound conformations of targeted protein, compared to unbound conformation (Fig. 8C).
Fig. 8.
DSSP analysis for the secondary structure fluctuations as a function of time from 0 to 100 ns for SARS-CoV-2 Mpro protein in unbound and ligand bound state at 300 K (A) SARS-CoV-2 Mpro unbound (B) SARS-CoV-2 Mpro complexed with lopinavir (C) SARS-CoV-2 Mpro complexed with quercetin-3-rutinoside-7-glucoside.
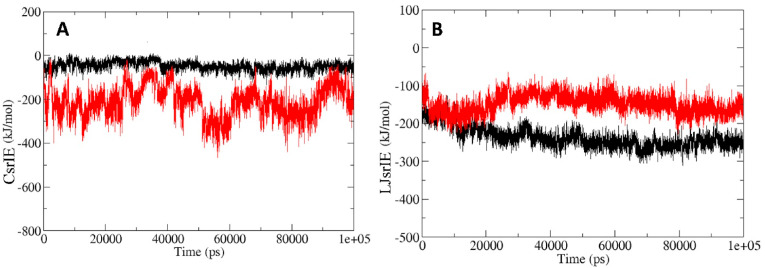
Protein-ligand interaction energy was calculated to find out the ligand interaction and stability of the complexes (MLC and MQC). Interaction energy between protein and inhibitor mainly consist of two short range interactions known as Coulombic short range interaction energy (CsrIE) and Lennard Jones short range interaction energy (LJsrIE) [23]. Interaction energy plot of MLC and MQC clearly indicate that the high interaction energy for the lead phytochemical bound Mpro protein complex in comparison to Mpro-lopinavir complex (Fig. 9 A and B). The CsrIE and LJsrIE for MLC and MQC were ~-47.92 kJ/mol and ~-237.25 K, ~-212.26 and ~-149.32 kJ/mol respectively.
Fig. 9.
Short range interaction analysis of Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. (A) Short range Coulombic interaction energy of Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. (B) Short range Lennard jones interaction energy of Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. Parameters for Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex are depicted in black and red color respectively. CsrIE = coulombic short range interaction energy; LJsrIE = lennard jones short range interaction energy.
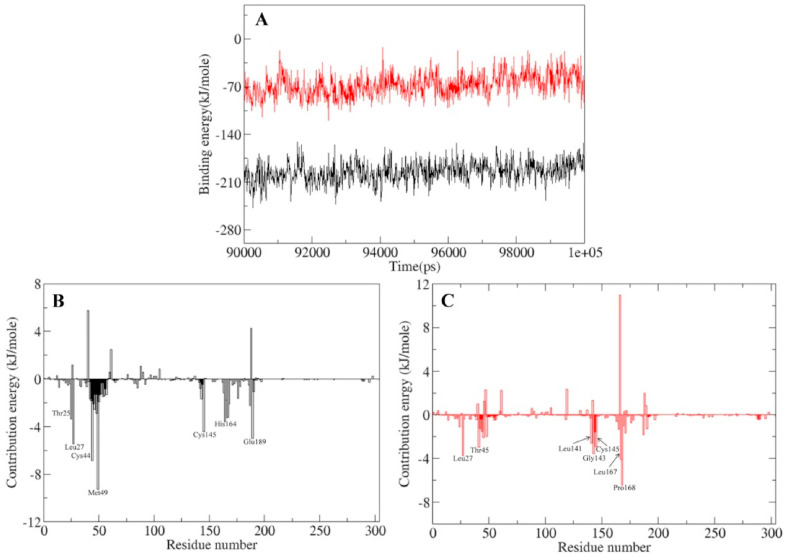
The last 10 ns (90–100 ns) of the trajectory obtained from 100 ns MD simulations of the ligand bound Mpro were utilized to estimate the binding free energy of the MLC and MQC. Binding free energy (ΔEbinding), van der Waals energy (Evdw), electrostatic energy (Eelec), polar solvation energy (ΔEpolar), and SASA were estimated (Supplementary figure 3, Table 3 and Fig. 10 A). Binding energy for both the complexes was stable throughout the analyzed simulation trajectory (Supplementary figure 3). Further contribution of each residue was evaluated by decomposing the total binding free energy into per residue contribution energy (Fig. 10B and C). In binding of lopinavir to Mpro protein amino acid residues Thr25, Leu27, Cys44, Met49, Cys145, His164, and Glu189 contributed significantly in binding. Furthermore in binding of quercetin-3-rutinoside-7-glucoside to the Mpro protein amino acid residues Leu27, Thr45, Leu141, Gly143, Cys145, Leu167, and Pro168 contributed significantly in binding (Supplementary table 2).
Table 3.
MM-PBSA calculations of binding free energy for Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex.
| Type of Binding energy | Binding energy values (Mpro-Lopinavir complex) | Binding energy values (Mpro-quercetin-3-rutinoside-7-glucoside) |
|---|---|---|
| ΔEbinding (kj/mol) | −195.525 ± 0.481 | −67.767 ± 0.536 |
| SASA (kj/mol) | −25.823 ± 0.052 | −22.548 ± 0.044 |
| ΔEpolar solvation (kj/mol) | 141.280 ± 0.552 | 235.481 ± 0.911 |
| ΔEElectrostatic (kj/mol) | −31.008 ± 0.288 | −89.499 ± 0.892 |
| ΔEVan der Waal (kj/mol) | −279.994 ± 0.485 | −191.207 ± 0.538 |
Fig. 10.
Binding energy and contribution energy in g_mmpbsa analysis of residues. (A) Binding energy plot of Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex (B) Residue contribution plot of Mpro-lopinavir complex (C) Residue contribution plot of Mpro-quercetin-3-rutinoside-7-glucoside complex. Parameters for Mpro-lopinavir complex and Mpro-quercetin-3-rutinoside-7-glucoside are shown in black and red color respectively.
The Mpro protein has been considered as one of the best drugable target for the anti- SARS-CoV-2 drug discovery. The protease activity of the protein is required to process viral protein in the host and thus aid in the viral replication and virulence. Our study revealed the potential binding of the W. somnifera phytochemicals at the active site of the target SARS-CoV-2 Mpro protein in comparison to the known standard inhibitor (Supplementary Table 1). The lead phytochemicals showed better interaction with the targeted protein which was revealed by hydrogen bond formation, hydrophobic interaction and docking pose of the compounds (Supplementary Figure 1 and Table 2). Interaction of the W. somnifera phytochemicals with the key amino acids (Asn142, Gly143, Cys145, and His164 amino acid residues) of the protein indicates that they have ability to lock the active site of the targeted protein. Further, the MD simulations results including (RMSD, RMSF, Rg, SASA and hydrogen bonding pattern) showed the dynamically stable and potential binding of lead W. somnifera phytochemical in comparison to the standard Mpro protein inhibitor (Fig. 4, Fig. 5). Binding free energy calculations of the lead-protein complex showed that the amino residue such as Cys145 contributes significantly in the overall binding energy of the complex (Fig. 10). Taken together, the results of the present study indicates the W. somnifera phytochemicals possess SARS-CoV-2 Mpro protein inhibition potential and could serve as potential source of the lead molecules for anti- SARS-CoV-2 drug discovery. Further, the in vitro and in vivo experimentations are required to assess the viral replication and virulence by inhibiting the Mpro protein by W. somnifera phytochemicals.
4. Conclusion
Natural products are composed of several bioactive constituents and are recognized as multiple targeting agents. This property of the natural compounds makes them better candidates for novel drug discovery. In the present study, we identified the non-characteristic and characteristic phytochemicals from W. somnifera which possess differential potential against SARS-COV-2 Mpro protein in molecular docking studies. The 100ns molecular dynamic and MMPBSA analysis establishes the energetically and structurally stable complex formation ability of the lead phytochemicals with the SARS-COV-2 Mpro protein. Overall, we conclude that non-characteristic compound of W. somnifera either alone or in combination with the characteristic phytochemicals has ability to target Mpro and might be useful in the future anti-SARS-CoV-2 drug discovery.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
PPK and MS acknowledge financial support from Indian Council of Medical Research (ICMR), India in the form of ICMR-Senior Research fellowships (File No. 5/3/8/82/ITR-F/2020 and File No. 5/3/8/80/ITR-F/2020-ITR respectively). SK acknowledges University Grants Commission, India and Department of Science and Technology, India for providing financial support in the form of UGC-BSR Research Start-Up-Grant [No. F.30–372/2017 (BSR)] and DST-SERB Grant [EEQ/2016/000350] respectively. SK acknowledges Central University of Punjab, Bathinda, India for providing Research Seed Money Grant [GP-25]. AKS and KSP acknowledge CSIR-India and DBT-India funding agencies respectively for providing financial assistance in the form of Junior Research Fellowship. SK also acknowledges DST-FIST India, for providing financial support to the Department of Biochemistry, Central University of Punjab, Bathinda, India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2021.104954.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52(1):52–56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr. Pharmaceut. Des. 2006;12(35):4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 4.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Wang X., Jochmans D., Neyts J., Młynarski W., Hilgenfeld R. SARS-CoV-2 M pro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 5.Shibuyama S., Gevorkyan A., Yoo U., Tim S., Dzhangiryan K., Scott J.D. Understanding and avoiding antiretroviral adverse events. Curr. Pharmaceut. Des. 2006;12(9):1075–1090. doi: 10.2174/138161206776055796. [DOI] [PubMed] [Google Scholar]
- 6.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020;(1–12) doi: 10.1080/07391102.2020.1776157. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kushwaha P.P., Vardhan P.S., Kapewangolo P., Shuaib M., Prajapati S.K., Singh A.K., Kumar S. Bulbine frutescens phytochemical inhibits notch signaling pathway and induces apoptosis in triple negative and luminal breast cancer cells. Life Sci. 2019;234(1–15) doi: 10.1016/j.lfs.2019.116783. [DOI] [PubMed] [Google Scholar]
- 8.Kushwaha P.P., Vardhan P.S., Kumari P., Mtewa A.G., Kumar S. Phytochemicals as Lead Compounds for New Drug Discovery. Elsevier; 2020. Bioactive lead compounds and targets for the development of antimalarial drugs; pp. 305–316. [Google Scholar]
- 9.Pandey A.K., Mishra A.K., Mishra A., Kumar S., Chandra A. C. zeylanicum. Int. J. Biol. Med. Res. 2010;1(4):228–233. [Google Scholar]
- 10.Ibrahim M.A., Abdeljawaad K.A., Abdelrahman A.H., Hegazy M.E.F. Natural-like products as potential SARS-CoV-2 Mpro inhibitors: in-silico drug discovery. J. Biomol. Struct. Dyn. 2020;(1–13) doi: 10.1080/07391102.2020.1790037. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim M.A., Abdelrahman A.H., Hegazy M.E.F. In-silico drug repurposing and molecular dynamics puzzled out potential SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020;(1–12) doi: 10.1080/07391102.2020.1791958. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim M.A., Abdelrahman A.H., Hussien T.A., Badr E.A., Mohamed T.A., El-Seedi H.R., Pare P.W., Efferth T., Hegazy M.E.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020;126(1–11) doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Z., Zhang G., Tang B., Liu Y., Fu X., Zhang X. Promising anti-influenza properties of active constituent of Withania somnifera ayurvedic herb in targeting neuraminidase of H1N1 influenza: computational study. Cell Biochem. Biophys. 2015;72(3):727–739. doi: 10.1007/s12013-015-0524-9. [DOI] [PubMed] [Google Scholar]
- 14.Ganguly B., Umapathi V., Rastogi S.K. Nitric oxide induced by Indian ginseng root extract inhibits Infectious Bursal Disease virus in chicken embryo fibroblasts in vitro. J. Anim. Sci. Technol. 2018;60(1):1–5. doi: 10.1186/s40781-017-0156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain J., Narayanan V., Chaturvedi S., Pai S., Sunil S. In vivo evaluation of withania somnifera–based Indian traditional formulation (Amukkara choornam), against chikungunya virus–induced morbidity and arthralgia. J. Evid-Based Integr Med. 2018;23(1–7) doi: 10.1177/2156587218757661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar V., Dhanjal J.K., Kaul S.C., Wadhwa R., Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J. Biomol. Struct. Dyn. 2020;(1–13) doi: 10.1080/07391102.2020.1772108. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poolsup N., Suksomboon N., Kurnianta P.D.M., Deawjaroen K. Effects of curcumin on glycemic control and lipid profile in prediabetes and type 2 diabetes mellitus: a systematic review and meta-analysis. PloS One. 2019;14(4):1–18. doi: 10.1371/journal.pone.0215840. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Tiwari A., Kushwaha P.P., Kumar S. vol. 180. Manipur University; Imphal: 2018. Future scope for Withania somnifera in anticancer drug discovery. (Proceedings of the 105th Indian Science Congress, Section of New Biology (Including Biochemistry, Biophysics & Molecular Biology and Biotechnology) at). 3-7 January, 2018. [Google Scholar]
- 19.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. [Google Scholar]
- 20.Xie X.Q.S. Exploiting PubChem for virtual screening. Expet Opin. Drug Discov. 2010;5(12):1205–1220. doi: 10.1517/17460441.2010.524924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3(1):1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dallakyan S. The Scripps Research Institute; 2010. PyRx-python Prescription V. 0.8. [Google Scholar]
- 23.Londhe A.M., Gadhe C.G., Lim S.M., Pae A.N. Investigation of molecular details of Keap1-Nrf2 inhibitors using molecular dynamics and umbrella sampling techniques. Molecules. 2019;24(22):1–22. doi: 10.3390/molecules24224085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., Fagan P. The protein data bank. Acta. Crystallogr. D Biol. Crystallogr. 2002;58(6):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dash S., Kushwaha P.P., Kumar S. Phytochemistry. Apple Academic Press .2018; 2018. Targeting cancer cell carbohydrate metabolism by phytochemicals; pp. 537–560. [Google Scholar]
- 28.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 29.Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. [Google Scholar]
- 30.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- 31.Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Gupta S., Kumar S. Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1pp domain inhibitor: an in silico study. Comp. boil. Med. 2021;130(1–10) doi: 10.1016/j.compbiomed.2020.104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumari R., Kumar R., Open Source Drug Discovery Consortium and Lynn, A. g_mmpbsa: a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 34.Ali M., Shuaib M., Ansari S.H. Withanolides from the stem bark of Withania somnifera. Phytochemistry. 1997;44(6):1163–1168. [Google Scholar]
- 35.Duke J.A. CRC Press; 1992. Database of Phytochemical Constituents of GRAS Herbs and Other Economic Plants. [Google Scholar]
- 37.Kandil F.E., El Sayed N.H., Abou-Douh A.M., Ishak M.S., Mabry T.J. Flavonol glycosides and phenolics from Withania somnifera. Phytochemistry. 1994;37(4):1215–1216. [Google Scholar]
- 38.Kim S., Yu J.S., Lee J.Y., Choi S.U., Lee J., Kim K.H. Cytotoxic withanolides from the roots of Indian Ginseng (Withania somnifera) J. Nat. Prod. 2019;82(4):765–773. doi: 10.1021/acs.jnatprod.8b00665. [DOI] [PubMed] [Google Scholar]
- 39.Manjunath M.J. Standardized extract of Withania somnifera (Ashwagandha) markedly offsets rotenone-induced locomotor deficits, oxidative impairments and neurotoxicity in Drosophila melanogaster. J. Food Sci. Technol. 2015;52(4):1971–1981. doi: 10.1007/s13197-013-1219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misico R.I., Nicotra V.E., Oberti J.C., Barboza G., Gil R.R., Burton G. vol. 94. Springer; 2011. Withanolides and related steroids; pp. 127–229. (Progress in the Chemistry of Organic Natural Products). [DOI] [PubMed] [Google Scholar]
- 43.Nittala S.S., Velde V.V., Frolow F., Lavie D. Chlorinated withanolides from Withania somnifera and Acnistus breviflorus. Phytochemistry. 1981;20(11):2547–2552. [Google Scholar]
- 44.Senapati S., Kumar S., Singh A.K., Banerjee P., Bhagavatula S. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J. Genet. 2020;99(1–5) doi: 10.1007/s12041-020-01217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh V., Singh B., Sharma A., Kaur K., Gupta A.P., Salar R.K., Hallan V., Pati P.K. Leaf spot disease adversely affects human health‐promoting constituents and withanolide biosynthesis in Withania somnifera (L.) Dunal. J. Appl. Microbiol. 2017;122(1):153–165. doi: 10.1111/jam.13314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.