Abstract
Cancer accounted for 16% of all death worldwide in 2018. Significant progress has been made in understanding tumor occurrence, progression, diagnosis, treatment, and prognosis at the molecular level. However, genomics changes cannot truly reflect the state of protein activity in the body due to the poor correlation between genes and proteins. Quantitative proteomics, capable of quantifying the relatively different protein abundance in cancer patients, has been increasingly adopted in cancer research. Quantitative proteomics has great application potentials, including cancer diagnosis, personalized therapeutic drug selection, real-time therapeutic effects and toxicity evaluation, prognosis and drug resistance evaluation, and new therapeutic target discovery. In this review, the development, testing samples, and detection methods of quantitative proteomics are introduced. The biomarkers identified by quantitative proteomics for clinical diagnosis, prognosis, and drug resistance are reviewed. The challenges and prospects of quantitative proteomics for personalized medicine are also discussed.
Keywords: quantitative proteomics, cancer, biomarker, diagnostic marker, therapeutic target
Graphical abstract
Yang et al. introduce the development, samples tested, and detection methods of quantitative proteomics. Biomarkers for clinical diagnosis, prognosis, and drug resistance identified by quantitative proteomics are reviewed. The challenges and prospects of quantitative proteomics for personalized medicine are also discussed.
Introduction
Cancer accounted for 16% of global deaths according to the 2018 American Association for Cancer Research (AACR) Cancer Progress Report. Global cancer incidence will continue to increase with the aging of the population. By 2040, the number of new cancer cases is expected to reach 27.5 million worldwide, and the number of deaths is expected to reach 16.3 million. The Cancer Genome Atlas (TCGA) database has published the genomic landscapes of 33 tumor types from 11,000 patients, which could help to promote the understanding of tumor occurrence, progression, diagnosis, treatment, and prognosis at the molecular level.1, 2, 3, 4, 5 As the direct executor of life activities, protein participates in almost all life processes, such as heredity, development, reproduction, material and energy metabolism, and stress.6 However, the correlation between genomics changes and protein abundance is very poor, especially for low-abundance proteins.7 Therefore, proteomics could be the bridge between genome information and functional proteins that helps to further the understanding of cancer.8,9
Proteomics is based on the protein composition and changing of cells, tissues, or organisms. Proteomics studies the characteristics of proteins on a large scale, including protein expression levels, post-translational modifications (PTMs), and protein-protein interactions, to gain a comprehensive understanding of disease occurrence, cell metabolism, and other processes at the protein level.10, 11, 12 Quantitative proteomics provides comprehensive information on the protein interactions, signal pathways, and biomarkers of human disease by detecting the relative changes in protein abundance in diseased tissue samples.13,14 In this review, we discuss the application of quantitative proteomics in cancer research and the discovery of tumor biomarkers, as well as its potential significance in early clinical diagnosis, prognosis, and targeted therapy.
Development of quantitative proteomics
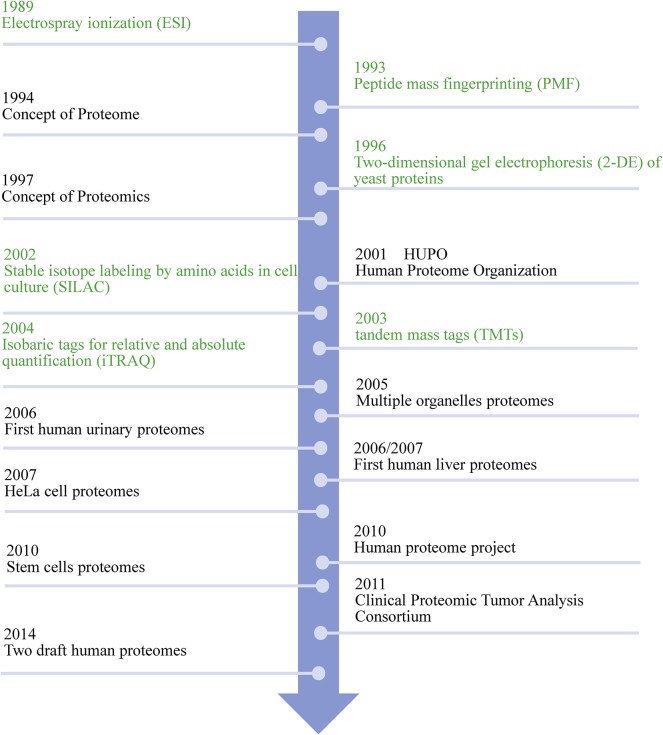
The concept of the proteome was first put forward by Australian scientist Mark Wilkins in 1994, and the concept of proteomics was put forward in 1997 as a science that studies the composition and changes of proteins in cells, tissues, or organs.15 In 2001, the International Human Proteome Organization (HUPO) officially announced the promotion of proteomics research. In the past 20 years, proteomics technology has improved continuously, which has enabled the application of quantitative analysis methods in proteomics.4,16, 17, 18, 19, 20 In 2014, Nature published two papers on human proteome for the first time.21,22 The application of proteomics has extensively promoted the progress of natural science research (Figure 1).
Figure 1.
The development of quantitative proteomics
Green indicates technical MS advances; black indicates MS-identified human proteomes.
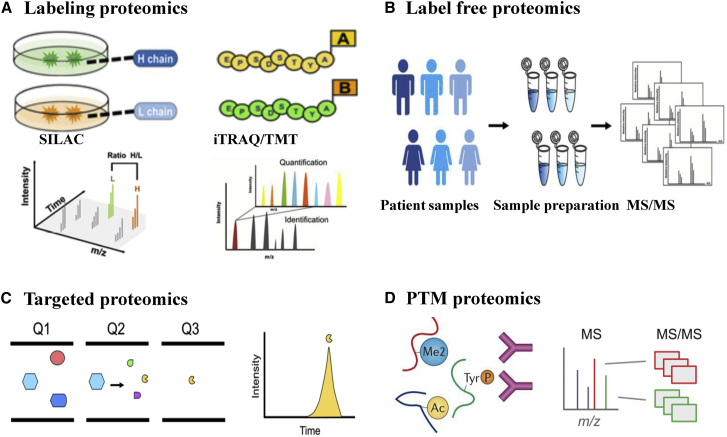
Currently, there are four main quantitative proteomics methods, that is, labeling, label-free, targeted, and PTM, widely used in cancer research (Figure 2). Stable isotope labeling by amino acids in cell culture (SILAC) technology, isobaric tags for relative and absolute quantification (iTRAQ) technology, and tandem mass tags (TMTs) technology are the main methods used for the labeling quantitative proteomics.23, 24, 25, 26 SILAC technology is suitable for analyzing living cells in culture with accurate quantification and good repeatability.27 SILAC removes the false positives in protein-interaction studies, reveals the large-scale kinetics of proteomes, and directly uncovers the important points in the cellular signaling pathways as a quantitative phosphoproteomics technology. The triple-label SILAC proteomic profiles have been used to reveal the deregulation of key cell cycle regulators in long intergenic non-coding RNA-nucleotide metabolism regulator (lincNMR)-depleted cells, such as the key 2′-deoxynucleoside 5′-triphosphate (dNTP) synthesizing enzymes RRM2, TYMS, and TK1, which implicated lincNMR in regulating nucleotide metabolism.28 The iTRAQ/TMT technology has high sensitivity, high throughput, and good reproducibility.29,30 Keller et al.31 reported that secretome analysis using iTRAQ proteomics revealed the caspase-1-mediated secretion of other leaderless proteins with known or unknown extracellular functions. Without labeling processing, label-free quantification is simple to conduct, but it requires high stability and repeatability of experimental operations. It is suitable for large-scale quantitative comparison and experimental design that cannot be realized with labeling quantification.32 Wepr et al.33 presented a label-free mass spectrometry-based strategy for the absolute quantification of protein complex components isolated through affinity purification and quantitatively analyzed the interaction stoichiometries in the human protein phosphatase 2A network.
Figure 2.
A comparison of detection methods used in quantitative proteomics
(A) Labeling proteomics: SILAC is used for cell lines, iTRAQ/TMT is used for labeling in vitro, and MS/MS spectra are assigned to peptides for identification and quantitation. (B) Label-free proteomics is used to quantify the protein expression across different samples. (C) Targeted proteomics, selected from three quadrupoles (Q1, Q2, Q3), is suitable for identifying and quantitating target peptides within complex mixtures. (D) PTM proteomics: using antibody-based immunoprecipitation (IP) to enrich peptides containing modifications (phosphorylation [P], dimethyl [Me2], or acetylation [Ac]), LC-MS/MS is used for peptide identification and quantitation.
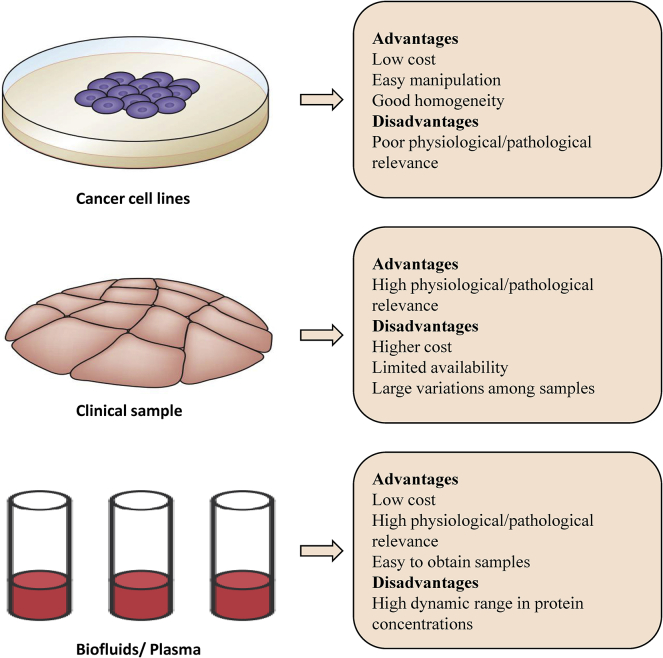
Targeted quantitative proteomics is essentially a mass spectrum scanning mode based on the selection of specific target protein ion and product ion pairs.34, 35, 36 Targeted quantitative proteomics could detect the relative or absolute quantities of various target proteins in complex samples (Table 1). PTM is an important component of protein activity regulation.39, 40, 41, 42 Phosphorylation modification is the most common and most important PTM regulating the protein kinase and other protein activities.43 Quantitative phosphoproteomics is widely used for proteomic stratification and drug target identification. Jiang et al.43 adopted proteomic and phosphoproteomic profiling and characterized 110 paired tumor and non-tumor tissues of clinical early-stage hepatocellular carcinoma (HCC) related to hepatitis B virus (HBV) infection. The quantitative proteomics data highlighted the heterogeneity in early stage HCC. Many analytical methods of proteomics have been developed for different samples, including cell lines, clinical samples, and body fluids.13,37,38 Each type of sample has advantages and disadvantages (Figure 3). The choice of sample type depends on the purpose of the research.
Table 1.
A comparison of detection methods for quantitative proteomics
| Methods of label | Applicable samples | Clinical samples | Advantages | Disadvantages | Application | Ref. | |
|---|---|---|---|---|---|---|---|
| SILAC | in vivo metabolic incorporation of lysine or arginine | tissue culture cells | no | high sensitivity | high cost limited to living samples | biomarker screening in cell lines | 27,28 |
| high accuracy | |||||||
| high repeatability | |||||||
| closely reflect the state of samples | |||||||
| high sensitivity | |||||||
| iTRAQ / TMT | in vitro N terminus and lysine side chains of peptide | non-living samples | yes | compare 2–10 samples in parallel | poor to low-abundance proteins | biomarker screening | 29, 30, 31 |
| high coverage | |||||||
| high throughput | |||||||
| high accuracy | |||||||
| Label-free proteomics | no | non-living samples | yes | low cost | poor stability and repeatability | biomarker screening | 32,33 |
| simple manipulation | |||||||
| not limited by samples | |||||||
| high throughput | |||||||
| closely reflect the state of samples | |||||||
| high sensitivity | |||||||
| Targeted proteomics | no | non-living samples | yes | high accuracy | poor to higher protein complexity and complex analysis | intestinal flora screening | 34, 35, 36 |
| high repeatability | |||||||
| wider dynamic range | |||||||
| PTM proteomics | no | non-living samples | yes | closely reflect the state of samples | high requirements for peptide enrichment | biomarker and drug target screening | 13,37,38 |
| kinase target screening |
Figure 3.
A comparison of the biological samples used in quantitative proteomics
There are three samples for quantitative proteomics analysis, as shown on the left. Each type of sample has its advantages and disadvantages, as shown on the right.
Quantitative proteomics classification of tumor subtype
In clinical practice, there is an urgent need for the early detection of cancer and the differentiation of tumor subtypes to improve the existing treatment. Proteome-informed classification could distinguish the clinical features of early-stage non-smoker lung adenocarcinoma.44,45 Mass spectrometry-based proteomic profiling could classify the pan-cancer molecular subtypes of 532 cancers.46 Quantitative proteomics could identify and quantify the specific signaling pathways from the tumor tissues and corresponding para-tumor tissues of 24 patients at different stages of triple-negative breast cancer (TNBC).47 Quantitative proteomics could be used for the accurate classification of TNBC subtypes.48 Furthermore, the sub-network identified through quantitative phosphoproteomics was highly correlated with clinically identified breast cancer subtypes.49, 50, 51 SWATH/DIA-MS (state-of-the-art sequential windowed acquisition of all theoretical fragment ion/data-independent acquisition mass spectrometry) presented a promising complement for the stable classification of ovarian cancer subtypes.52,53 Quantitative proteomics of reverse-phase protein array (RPPA) could be used to classify diffuse large B cell lymphoma.54, 55, 56
Identifying potential biomarkers with quantitative proteomics
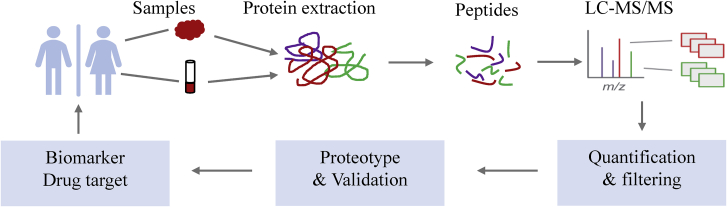
With the development of mass spectrometry, quantitative proteomics has become an important method to discover tumor biomarkers. Increasing amounts of tumor biomarkers have been discovered by quantitative proteomics.57, 58, 59 Samples from tumor tissue and paired adjacent tissue or patients and healthy people were prepared, digested into peptides, and then analyzed with liquid chromatography-tandem mass spectrometry (LC-MS/MS). After quantification and filtration, tumor biomarkers were identified (Figure 4).
Figure 4.
Integrated view of LC-MS/MS proteomics workflow for cancer biomarker discovery
Step 1: cancer tissues and adjacent tissues for protein extraction are prepared. Step 2: the proteins are enzymatically digested into peptides. Step 3: the peptides are analyzed with LC-MS/MS. Step 4: databases are mapped to peptides and proteins through quantification and filtering. Step 5: proteotype-like PPI interactomes are generated by further data validation. Step 6: candidate biomarkers and drug targets are identified. Step 7: after functional verification, biomarkers and drug targets are recommended to clinical medicine.
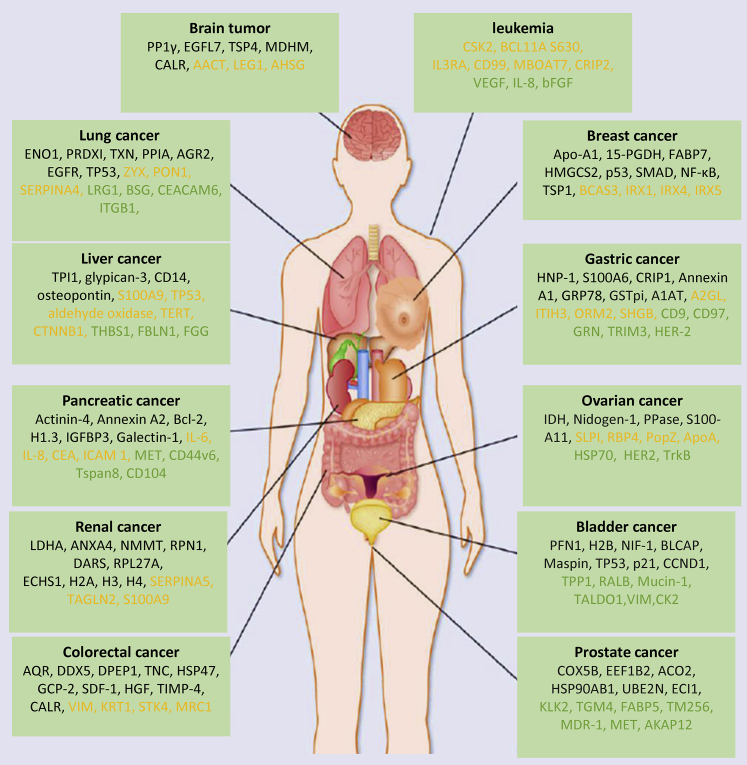
The comprehensive classification of lung adenocarcinoma provided bioinformatics resources for clinical treatment, drug development, and precision medicine.60, 61, 62 Quantitative analysis of control, HBV, cirrhotic, and HCC tissue showed CD14 as a promising biomarker.63 The DIA quantitative proteomics analysis of 10 paired tumor and non-tumor samples verified three oxidative phosphorylation biomarkers (UQCRQ, NDUFB7, and UQCRC2) in gastric cancer.37 By the quantitative analysis of tumor tissues against normal adjacent tissues (NATs), AQR, DDX5, DPEP1, and TNC were identified as biomarkers in colorectal cancer. Through the proteomics approach, triosephosphate isomerase 1 (TPI1) was identified as a biomarker for predicting the recurrence of intrahepatic cholangiocarcinoma.64 Quantitative proteomics is increasingly adopted for identifying biomarkers of early pancreatic cancer, such as actinin-4, annexin A2, Bcl-2, H1.3, IGFBP2, IGFBP3, and galectin-1 (Figure 5).65, 66, 67, 68, 69, 70, 71
Figure 5.
Quantitative proteomics adopted in the discovery of various cancer biomarkers
Many biomarkers for different types of cancer are identified through quantitative proteomics. Biomarkers were found from cancer tissue (black), plasma/serum (orange), and exosome (green).
Discovering drug targets with quantitative proteomics
Quantitative proteomics is a promising tool for revealing the molecular mechanisms of drug action.58,63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75 Proteomics drug maps have greatly promoted the discovery of drug targets. After the quantitative analysis for 10,000 proteins and 55,000 phosphorylation sites (p-sites) from 125 cancer cell lines, the proteome activity landscapes were obtained. Adenylate kinase isoenzyme 1 (AK1) was discovered as a promising drug target for acute myeloid leukemia patients.76 The anaplastic lymphoma kinase (ALK) inhibitor ceritinib was found to be capable of modulating the protein-trafficking and degradation-related process of autophagy after the quantitative analysis of five lung cancer cell lines in response to more than 50 drugs.77 The proto-oncogene serine/threonine-protein kinase PIM3 has been widely used as a drug target. Quantitative phosphoproteomics revealed that PIM3 activated RhoA to promote migration and invasion of hepatoma cells.78 By comprehensive phosphoproteomics characterization of 110 tumors and 101 matched NATs, three candidate drug targets were identified for lung adenocarcinoma (LUAD), including SOS1 inhibition in KRAS mutant, PTPN11/Shp2 inhibition in both ALK fusion and EGFR mutant tumors, and STK11 mutation in neutrophil degranulation.62 Quantitative proteomics was adopted to characterize 200 paired EGFR-positive and EGFR-negative glioma tissues of all pathological types, and EGF-like domain multiple 7 (EGFL7) was identified as a potential diagnostic biomarker and therapeutic target.79
Discovering drug resistance biomarkers with quantitative proteomics
Drug resistance and recurrence are the main obstacles to the long-term survival of cancer patients. It is crucial to understand the mechanisms and identify the biomarkers of drug resistance. Quantitative proteomics could help to identify the proteins related to drug resistance.80, 81, 82
Tamoxifen resistance is one of the unsolved problems in breast cancer treatment. Through proteomics analysis of tumor tissues from tamoxifen therapy-sensitive and tamoxifen therapy-resistant breast cancer patients, high expression of ectonucleotide pyrophosphatase/phosphodiesterase-1 (ENPP1) and extracellular matrix metalloproteinase inducer (EMMPRIN) were found relevant to tamoxifen resistance. However, low expression of eukaryotic translation initiation factor 3 subunit 6/E (EIF3E) and guanine nucleotide-binding protein β subunit 4 (GNB4) were relevant to tamoxifen resistance.83 Moreover, EMMPRIN-negative tumors were more sensitive to neoadjuvant chemotherapy in bladder cancer (BC).84 Using label-free quantitative proteomics analysis of trastuzumab-resistant MKN45/R cells and parental MKN45 human gastric cancer cells, WNT signaling was identified as a potential target in trastuzumab-resistant cancer. Quantitative proteomics analysis of the anti-HCC efficacy of dihydroartemisinin (DHA) combined with sorafenib may help to understand the related molecular mechanism of anti-HCC.85 The drug resistance of ovarian cancer cell lines was evaluated with iTRAQ LC-MS/MS, and 28 biomarkers that might lead to cisplatin resistance were identified.86 Radio resistance biomarkers in several cancers, such as breast cancer, prostate cancer, and lung cancer, were identified with MS-based proteomics approaches.87
Protein kinases are primary molecular drug targets, and phosphorylation regulation is a key mechanism in cancer drug resistance. Through integrated proteomics and phosphoproteomics analysis of cisplatin-sensitive (T24S) and cisplatin-resistant (T24R) T24 human BC cell lines, CDK2 was identified as a potential chemoresistance biomarker in BC.88 Through phosphoproteomics analysis of lapatinib-sensitive (SKBR3) and lapatinib-resistant (SKBR3-LR) breast cell lines, p21-activated kinase 2 (PAK2) was identified as an effective therapeutic target to overcome acquired lapatinib resistance in HER2-positive breast cancer.89 The success in identifying cancer drug resistance biomarkers could help to develop biomarker-guided targeted therapy.
The application of quantitative proteomics in clinical diagnosis and treatment
Liquid biopsy is increasingly recognized as a promising non-invasive identification method of clinical biomarkers. Many studies have shown that exosomes, i.e., 40- to 100-nm vesicles containing nucleic acids, proteins, and lipids, could be used as tumor biomarkers.90, 91, 92 Various biomarkers for different types of cancer have been identified with exosome proteomics (Figure 5). Thrombospondin-1 (THBS1), fibulin-1 (FBLN1), and fibrinogen gamma chain (FGG) were identified as clinical biomarkers for liver cancer.93, 94, 95 Leucine-rich alpha-2-glycoprotein 1 (LRG1), basigin (BSG), carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), and integrin beta-1 (ITGB1) were identified as clinical biomarkers for lung cancer.96, 97, 98, 99 Plasma or serum is an important component for liquid biopsy. The plasma protein level of HSP90β was validated as a potential prognostic biomarker in LUAD after a comprehensive proteomics analysis of 103 cases in China.60 The serum amyloid A protein was identified as a biomarker for renal cancer by comparing 119 patients with clear cell RCC and 69 healthy controls. BCAS3, IRX1, IRX4, and IRX5 were identified in breast cancer plasma samples through label-free quantitative proteomics.100, 101, 102, 103 S100P and aldehyde oxidase were identified as potential liver cancer biomarkers from human serum through quantitative proteomics (iTRAQ).104 SOD2 was identified as a potential salivary biomarker in liver cancer through iTRAQ-based proteomics.105
Conclusions
With the development of mass spectrometry technology, quantitative proteomics has been widely applied for studying cancer mechanisms. Many biomarkers of different cancers identified with quantitative proteomics could help in the early diagnosis, prognosis, and drug resistance analysis.106,107 Three types of samples, including cell lines, clinical samples, and body fluids, are used in quantitative proteomics research. Clinical samples and body fluids are widely used in cancer research. Several biomarkers of 12 types of cancers identified from clinical samples and body fluids are listed in Figure 5. Liquid biopsy is increasingly recognized as a promising non-invasive identification method of clinical biomarkers. Many tumor-related biomarkers have been found in serum, urine, saliva, and exosomes.100, 101, 102, 103 Due to high protein complexity and wide dynamic range, quantitative proteomics for liquid biopsy face significant challenges. Future research may focus on developing mass spectrometry technology with wider coverage and dynamic range.
A series of proteomics technologies have been developed for the comprehensive understanding of cancer occurrence and development mechanisms, including PTM proteomics, spatiotemporal proteomics, single-cell proteomics, and multi-omics. Since the functional diversity of proteins is achieved through PTMs, many protein kinases have been identified as drug targets through quantitative phosphorylation proteomics.108, 109, 110 Spatiotemporal proteomics allows the identification of proteins that change subcellular localization under different experimental conditions using quantitative proteomics.111 As a topic of frequent discussion in the past decade, single-cell proteomics evaluates the heterogeneity and rare types of cells based on cell types and the state of a single cell.112,113 Multi-omics approaches have become promising in the study of human diseases.60, 61, 62 HSP90β was identified as a potential prognostic biomarker for lung cancer through integrative analysis of proteome, phosphoproteome, transcriptome, and whole-exome sequencing data.60 A complicated regulatory map of the SLC2A2 gene with 16 candidate enhancers was identified for HCC by coupling transcriptome and proteome.114 The effective integration of all of these technologies eventually promotes accurate diagnosis and personalized medicine.
Acknowledgments
This study was supported in part by grants from the National Natural Science Foundation of China (81772932, 81472202, 81201535, 81302065, 81671716, 81301993, 81702243, 81372175, and 81472209); the Hunan Provincial Natural Science Foundation of China (2020JJ4278); the Key Program of Hunan Provincial Department of Science and Technology (2020WK2020 and 2019NK2111); the Scientific Research Fund Project of Anhui Medical University (2018xkj058); the Fundamental Research Funds for the Central Universities (22120170212 and 22120170117); the Shanghai Natural Science Foundation (12ZR1436000 and 16ZR1428900); the Shanghai Municipal Commission of Health and Family Planning (201540228 and 201440398); the Program of Shanghai Academic/Technology Research Leader (18XD1403000); the Construction of Clinical Medical Center for Tumor Biological Samples in Nantong (HS2016004); The Peak of Six Personnel Foundation in Jiangsu Province (WSW-009); the Jiangsu 333 Program (BRA2017205); the Nantong Medical Key Talents Training Plan (Key 43); the Nantong Science and Technology Project (yyz15026); and by the Wu Jieping Medical Foundation (320.6750.14326).
Author contributions
X.-L.Y., W.L., Y.-S.M., and D.F. designed the study; X.-L.Y., Y.S., D.-D.Z., R.X., T.-M.W., H.-M.W., P.-Y.W., and D.F. conducted the study; X.-L.Y., J.D., J.-B.L., Y.-S.M., and D.F. collected data; X.-L.Y., J.-B.L., W.L., Y.-S.M., and D.F. performed the statistical analyses and interpreted the data. X.-L.Y., Y.-S.M., and D.F. wrote the manuscript. All authors contributed to the final version of the manuscript and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Wen Li, Email: liwendream@163.com.
Yu-Shui Ma, Email: mayushui2006@126.com.
Da Fu, Email: fu800da900@126.com.
References
- 1.Kristensen V.N., Lingjærde O.C., Russnes H.G., Vollan H.K., Frigessi A., Børresen-Dale A.L. Principles and methods of integrative genomic analyses in cancer. Nat. Rev. Cancer. 2014;14:299–313. doi: 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- 2.Borrebaeck C.A. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer. 2017;17:199–204. doi: 10.1038/nrc.2016.153. [DOI] [PubMed] [Google Scholar]
- 3.Du R., Shen W., Liu Y., Gao W., Zhou W., Li J., Zhao S., Chen C., Chen Y., Liu Y. TGIF2 promotes the progression of lung adenocarcinoma by bridging EGFR/RAS/ERK signaling to cancer cell stemness. Signal Transduct. Target. Ther. 2019;4:60. doi: 10.1038/s41392-019-0098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Min L., Zhu S., Wei R., Zhao Y., Liu S., Li P., Zhang S. Integrating SWATH-MS proteomics and transcriptome analysis identifies CHI3L1 as a plasma biomarker for early gastric cancer. Mol. Ther. Oncolytics. 2020;17:257–266. doi: 10.1016/j.omto.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu W., Xu M., Wang L., Zhou W., Xiang R., Shi Y., Zhang Y., Piao Y. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct. Target. Ther. 2019;4:55. doi: 10.1038/s41392-019-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karczewski K.J., Snyder M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang K., Zeng L., Ge A., Pan X., Bao T., Long Z., Tong Q., Yuan M., Zhu X., Ge J., Huang Z. Integrating systematic biological and proteomics strategies to explore the pharmacological mechanism of danshen yin modified on atherosclerosis. J. Cell. Mol. Med. 2020;24:13876–13898. doi: 10.1111/jcmm.15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu M., Dang Y., Yang Z., Liu Y., Zhang L., Xu Y., Zhou W., Ji G. Comprehensive RNA sequencing in adenoma-cancer transition identified predictive biomarkers and therapeutic targets of human CRC. Mol. Ther. Nucleic Acids. 2020;20:25–33. doi: 10.1016/j.omtn.2020.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang B., Zhu Y., Ni J., Thompson J., Malouf D., Bucci J., Graham P., Li Y. Extracellular vesicles: the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics. 2020;10:2309–2326. doi: 10.7150/thno.39486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan M., Sun L., Li J., Yu H., Lin H., Yu T., Zhao F., Zhu M., Liu L., Geng Q. RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2019;38:478. doi: 10.1186/s13046-019-1479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chee N.T., Lohse I., Brothers S.P. mRNA-to-protein translation in hypoxia. Mol. Cancer. 2019;18:49. doi: 10.1186/s12943-019-0968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao M., Zhang Y., Jiang Y., Wang K., Wang X., Zhou D., Wang Y., Yu R., Zhou X. YAP promotes autophagy and progression of gliomas via upregulating HMGB1. J. Exp. Clin. Cancer Res. 2021;40:99. doi: 10.1186/s13046-021-01897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes E., Sores J., Cotton S., Peixoto A., Ferreira D., Freitas R., Reis C.A., Santos L.L., Ferreira J.A. Esophageal, gastric and colorectal cancers: Looking beyond classical serological biomarkers towards glycoproteomics-assisted precision oncology. Theranostics. 2020;10:4903–4928. doi: 10.7150/thno.42480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan S., Brentnall T.A., Chen R. Proteome alterations in pancreatic ductal adenocarcinoma. Cancer Lett. 2020;469:429–436. doi: 10.1016/j.canlet.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D., Adams A.M., Johnsen R.D., Fletcher S., Wilton S.D. Morpholino oligomer-induced dystrophin isoforms to map the functional domains in the dystrophin protein. Mol. Ther. Nucleic Acids. 2020;22:263–272. doi: 10.1016/j.omtn.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Li H., Fan X., Zhao C., Ye K., Zhao Z., Hu L., Ma H., Wang H., Fang Z. Protein palmitoylation regulates cell survival by modulating XBP1 activity in glioblastoma multiforme. Mol. Ther. Oncolytics. 2020;17:518–530. doi: 10.1016/j.omto.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quesnel A., Karagiannis G.S., Filippou P.S. Extracellular proteolysis in glioblastoma progression and therapeutics. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188428. doi: 10.1016/j.bbcan.2020.188428. [DOI] [PubMed] [Google Scholar]
- 18.Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Heller L., Thinard R., Chevalier M., Arpag S., Jing Y., Greferath R., Heller R., Nicolau C. Secretion of proteins and antibody fragments from transiently transfected endothelial progenitor cells. J. Cell. Mol. Med. 2020;24:8772–8778. doi: 10.1111/jcmm.15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSouza L., Diehl G., Rodrigues M.J., Guo J., Romaschin A.D., Colgan T.J., Siu K.W. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 21.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 23.Choi S., Goswami N., Schmidt F. Comparative proteomic profiling of 3T3-L1 adipocyte differentiation using SILAC quantification. J. Proteome Res. 2020;19:4884–4900. doi: 10.1021/acs.jproteome.0c00475. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.R. ncRNA-encoded peptides or proteins and cancer. Mol. Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griss J., Vinterhalter G., Schwämmle V. IsoProt: A complete and reproducible workflow to analyze iTRAQ/TMT experiments. J. Proteome Res. 2019;18:1751–1759. doi: 10.1021/acs.jproteome.8b00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warrier S., Patil M., Bhansali S., Varier L., Sethi G. Designing precision medicine panels for drug refractory cancers targeting cancer stemness traits. Biochim. Biophys. Acta Rev. Cancer. 2021;1875:188475. doi: 10.1016/j.bbcan.2020.188475. [DOI] [PubMed] [Google Scholar]
- 27.Li N., Li J., Desiderio D.M., Zhan X. SILAC quantitative proteomics analysis of ivermectin-related proteomic profiling and molecular network alterations in human ovarian cancer cells. J. Mass Spectrom. 2021;56:e4659. doi: 10.1002/jms.4659. [DOI] [PubMed] [Google Scholar]
- 28.Gandhi M., Groß M., Holler J.M., Coggins S.A., Patil N., Leupold J.H., Munschauer M., Schenone M., Hartigan C.R., Allgayer H. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1-RRM2 axis in cancer. Nat. Commun. 2020;11:3214. doi: 10.1038/s41467-020-17007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Q., Xu Y., Deng C., Cheng C., Dai Z., Yang Z., Chen X., Liu C., Su J. Differential proteomic analysis to identify proteins associated with apomeiosis in Boehmeria tricuspis (Hance) Makino using an iTRAQ-based strategy. J. Proteome Res. 2021;20:661–669. doi: 10.1021/acs.jproteome.0c00586. [DOI] [PubMed] [Google Scholar]
- 30.Yun H., Wu X., Ding Y., Xiong W., Duan X., Gao H., Yang Y., Chen Z. Explore the mechanism of Swertia mussotii Franch. for hepatoprotective effects with iTRAQ-LC-MS/MS. Comb. Chem. High Throughput Screen. 2020;23:1. doi: 10.2174/1386207323666201020111301. [DOI] [PubMed] [Google Scholar]
- 31.Keller M., Rüegg A., Werner S., Beer H.D. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 32.Di Meo A., Sohaei D., Batruch I., Alexandrou P., Prassas I., Diamandis E.P. Proteomic profiling of the human tissue and biological fluid proteome. J. Proteome Res. 2021;20:444–452. doi: 10.1021/acs.jproteome.0c00502. [DOI] [PubMed] [Google Scholar]
- 33.Wepf A., Glatter T., Schmidt A., Aebersold R., Gstaiger M. Quantitative interaction proteomics using mass spectrometry. Nat. Methods. 2009;6:203–205. doi: 10.1038/nmeth.1302. [DOI] [PubMed] [Google Scholar]
- 34.Valenzuela V., Jackson K.L., Sardi S.P., Hetz C. Gene therapy strategies to restore ER proteostasis in disease. Mol. Ther. 2018;26:1404–1413. doi: 10.1016/j.ymthe.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y., Fillmore T.L., Munoz N., Bentley G.J., Johnson C.W., Kim J., Meadows J.A., Zucker J.D., Burnet M.C., Lipton A.K. High-throughput large-scale targeted proteomics assays for quantifying pathway proteins in Pseudomonas putida KT2440. Front. Bioeng. Biotechnol. 2020;8:603488. doi: 10.3389/fbioe.2020.603488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J., CampbellPalmer L., Vinqvist-Tymchuk M., Fillmore S., Forney C., Luo H., Zhang Z. Proteomic changes in antioxidant system in strawberry during ripening. Front. Plant Sci. 2020;11:594156. doi: 10.3389/fpls.2020.594156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su F., Zhou F.F., Zhang T., Wang D.W., Zhao D., Hou X.M., Feng M.H. Quantitative proteomics identified 3 oxidative phosphorylation genes with clinical prognostic significance in gastric cancer. J. Cell. Mol. Med. 2020;24:10842–10854. doi: 10.1111/jcmm.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ocaña A., Pandiella A. Proteolysis targeting chimeras (PROTACs) in cancer therapy. J. Exp. Clin. Cancer Res. 2020;39:189. doi: 10.1186/s13046-020-01672-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z. Featuring advanced translational strategies: Principles, techniques, devices and applications. Cancer Lett. 2020;489:133–134. doi: 10.1016/j.canlet.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taddei M.L., Pardella E., Pranzini E., Raugei G., Paoli P. Role of tyrosine phosphorylation in modulating cancer cell metabolism. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188442. doi: 10.1016/j.bbcan.2020.188442. [DOI] [PubMed] [Google Scholar]
- 41.Gao F., Li M., Yu X., Liu W., Zhou L., Li W. Licochalcone A inhibits EGFR signalling and translationally suppresses survivin expression in human cancer cells. J. Cell. Mol. Med. 2021;25:813–826. doi: 10.1111/jcmm.16135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaffer L.V., Millikin R.J., Shortreed M.R., Scalf M., Smith L.M. Improving proteoform identifications in complex systems through integration of bottom-up and top-down data. J. Proteome Res. 2020;19:3510–3517. doi: 10.1021/acs.jproteome.0c00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y., Sun A., Zhao Y., Ying W., Sun H., Yang X., Xing B., Sun W., Ren L., Hu B., Chinese Human Proteome Project (CNHPP) Consortium Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257–261. doi: 10.1038/s41586-019-0987-8. [DOI] [PubMed] [Google Scholar]
- 44.Stewart P.A., Welsh E.A., Slebos R.J.C., Fang B., Izumi V., Chambers M., Zhang G., Cen L., Pettersson F., Zhang Y. Proteogenomic landscape of squamous cell lung cancer. Nat. Commun. 2019;10:3578. doi: 10.1038/s41467-019-11452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhuo H., Zhao Y., Cheng X., Xu M., Wang L., Lin L., Lyu Z., Hong X., Cai J. Tumor endothelial cell-derived cadherin-2 promotes angiogenesis and has prognostic significance for lung adenocarcinoma. Mol. Cancer. 2019;18:34. doi: 10.1186/s12943-019-0987-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velloso F.J., Campos A.R., Sogayar M.C., Correa R.G. Proteome profiling of triple negative breast cancer cells overexpressing NOD1 and NOD2 receptors unveils molecular signatures of malignant cell proliferation. BMC Genomics. 2019;20:152. doi: 10.1186/s12864-019-5523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lastiri-Pancardo G., Mercado-Hernández J.S., Kim J., Jiménez J.I., Utrilla J. A quantitative method for proteome reallocation using minimal regulatory interventions. Nat. Chem. Biol. 2020;16:1026–1033. doi: 10.1038/s41589-020-0593-y. [DOI] [PubMed] [Google Scholar]
- 49.Noblejas-López M.D.M., Nieto-Jiménez C., Galán-Moya E.M., Tebar-García D., Montero J.C., Pandiella A., Burgos M., Ocaña A. MZ1 co-operates with trastuzumab in HER2 positive breast cancer. J. Exp. Clin. Cancer Res. 2021;40:106. doi: 10.1186/s13046-021-01907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yim H.E., Kim J.H., Ahn M.S., Jung Y., Roh J., Park S.H., Kim T.G., Choi J.H., Kang S.Y. Clinicopathological and molecular analysis of 45 cases of pure mucinous breast cancer. Front. Oncol. 2021;10:558760. doi: 10.3389/fonc.2020.558760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas R., Al-Khadairi G., Decock J. Immune checkpoint inhibitors in triple negative breast cancer treatment: Promising future prospects. Front. Oncol. 2021;10:600573. doi: 10.3389/fonc.2020.600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas S.N., Friedrich B., Schnaubelt M., Chan D.W., Zhang H., Aebersold R. Orthogonal proteomic platforms and their implications for the stable classification of high-grade serous ovarian cancer subtypes. iScience. 2020;23:101079. doi: 10.1016/j.isci.2020.101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poulos R.C., Hains P.G., Shah R., Lucas N., Xavier D., Manda S.S., Anees A., Koh J.M.S., Mahboob S., Wittman M. Strategies to enable large-scale proteomics for reproducible research. Nat. Commun. 2020;11:3793. doi: 10.1038/s41467-020-17641-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki M., Muroi A., Nojima M., Numata A., Takasaki H., Sakai R., Yokose T., Miyagi Y., Koshikawa N. Utility of a reverse phase protein array to evaluate multiple biomarkers in diffuse large B-cell lymphoma. Proteomics Clin. Appl. 2020;14:e1900091. doi: 10.1002/prca.201900091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saberi Hosnijeh F., van der Straten L., Kater A.P., van Oers M.H.J., Posthuma W.F.M., Chamuleau M.E.D., Bellido M., Doorduijn J.K., van Gelder M., Hoogendoorn M. Proteomic markers with prognostic impact on outcome of chronic lymphocytic leukemia patients under chemo-immunotherapy: Results from the HOVON 109 study. Exp. Hematol. 2020;89:55–60.e6. doi: 10.1016/j.exphem.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Panner Selvam M.K., Finelli R., Baskaran S., Agarwal A. Dysregulation of key proteins associated with sperm motility and fertility potential in cancer patients. Int. J. Mol. Sci. 2020;21:6754. doi: 10.3390/ijms21186754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chanukuppa V., Taware R., Taunk K., Chatterjee T., Sharma S., Somasundaram V., Rashid F., Malakar D., Santra M.K., Rapole S. Proteomic alterations in multiple myeloma: A comprehensive study using bone marrow interstitial fluid and serum samples. Front. Oncol. 2021;10:566804. doi: 10.3389/fonc.2020.566804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu H.F., Xu W.W., Li Y.J., He Y., Zhang W.X., Liao L., Zhang Q.H., Han L., Yin X.F., Zhao X.X. Anti-allergic drug azelastine suppresses colon tumorigenesis by directly targeting ARF1 to inhibit IQGAP1-ERK-Drp1-mediated mitochondrial fission. Theranostics. 2021;11:1828–1844. doi: 10.7150/thno.48698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H., Zhang X.Y., Niu M., Li F.F., Gao S., Wei W., Li S.W., Zhang X.D., Liu S.L., Pang D. Isobaric tags for relative and absolute quantitation in proteomic analysis of potential biomarkers in invasive cancer, ductal carcinoma in situ, and mammary fibroadenoma. Front. Oncol. 2020;10:574552. doi: 10.3389/fonc.2020.574552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J.Y., Zhang C., Wang X., Zhai L., Ma Y., Mao Y., Qian K., Sun C., Liu Z., Jiang S. Integrative proteomic characterization of human lung adenocarcinoma. Cell. 2020;182:245–261.e17. doi: 10.1016/j.cell.2020.05.043. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y.J., Roumeliotis T.I., Chang Y.H., Chen C.T., Han C.L., Lin M.H., Chen H.W., Chang G.C., Chang Y.L., Wu C.T. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–244.e17. doi: 10.1016/j.cell.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 62.Gillette M.A., Satpathy S., Cao S., Dhanasekaran S.M., Vasaikar S.V., Krug K., Petralia F., Li Y., Liang W.W., Reva B., Clinical Proteomic Tumor Analysis Consortium Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182:200–225.e35. doi: 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo J., Jing R., Zhong J.H., Dong X., Li Y.X., Liu Y.K., Huang T.R., Zhang C.Y. Identification of CD14 as a potential biomarker of hepatocellular carcinoma using iTRAQ quantitative proteomics. Oncotarget. 2017;8:62011–62028. doi: 10.18632/oncotarget.18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu W.L., Yu G., Dong H., Chen K., Xie J., Yu H., Ji Y., Yang G.S., Li A.J., Cong W.M., Jin G.Z. Proteomics analysis identified TPI1 as a novel biomarker for predicting recurrence of intrahepatic cholangiocarcinoma. J. Gastroenterol. 2020;55:1171–1182. doi: 10.1007/s00535-020-01729-0. [DOI] [PubMed] [Google Scholar]
- 65.Nweke E.E., Naicker P., Aron S., Stoychev S., Devar J., Tabb D.L., Omoshoro-Jones J., Smith M., Candy G. SWATH-MS based proteomic profiling of pancreatic ductal adenocarcinoma tumours reveals the interplay between the extracellular matrix and related intracellular pathways. PLoS ONE. 2020;15:e0240453. doi: 10.1371/journal.pone.0240453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guerrero P.E., Duran A., Ortiz M.R., Castro E., Garcia-Velasco A., Llop E., Peracaula R. Microfibril associated protein 4 (MFAP4) is a carrier of the tumor associated carbohydrate sialyl-Lewis x (sLex) in pancreatic adenocarcinoma. J. Proteomics. 2021;231:104004. doi: 10.1016/j.jprot.2020.104004. [DOI] [PubMed] [Google Scholar]
- 67.O’Rourke M.B., Sahni S., Samra J., Mittal A., Molloy M.P. Data independent acquisition of plasma biomarkers of response to neoadjuvant chemotherapy in pancreatic ductal adenocarcinoma. J. Proteomics. 2021;231:103998. doi: 10.1016/j.jprot.2020.103998. [DOI] [PubMed] [Google Scholar]
- 68.Melchionna R., Spada S., Di Modugno F., D’Andrea D., Di Carlo A., Panetta M., Mileo A.M., Sperduti I., Antoniani B., Gallo E. The actin modulator hMENA regulates GAS6-AXL axis and pro-tumor cancer/stromal cell cooperation. EMBO Rep. 2020;21:e50078. doi: 10.15252/embr.202050078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coleman O., Henry M., O’Neill F., Roche S., Swan N., Geoghegan J., Conlon K., McVey G., Moriarty M., Meleady P., Clynes M. Proteomic analysis of cell lines and primary tumors in pancreatic cancer identifies proteins expressed only in vitro and only in vivo. Pancreas. 2020;49:1109–1116. doi: 10.1097/MPA.0000000000001633. [DOI] [PubMed] [Google Scholar]
- 70.Flick K.F., Yip-Schneider M.T., Sublette C.M., Simpson R.E., Colgate C.L., Wu H., Soufi M., Dewitt J.M., Mosley A.L., Ceppa E.P. A quantitative global proteomics approach identifies candidate urinary biomarkers that correlate with intraductal papillary mucinous neoplasm dysplasia. Pancreas. 2020;49:1044–1051. doi: 10.1097/MPA.0000000000001628. [DOI] [PubMed] [Google Scholar]
- 71.Guo Z., Wang X., Yang Y., Chen W., Zhang K., Teng B., Huang C., Zhao Q., Qiu Z. Hypoxic tumor-derived exosomal long noncoding RNA UCA1 promotes angiogenesis via miR-96-5p/AMOTL2 in pancreatic cancer. Mol. Ther. Nucleic Acids. 2020;22:179–195. doi: 10.1016/j.omtn.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinaiphat A., Low J.K., Yeoh K.W., Chng W.J., Sze S.K. Application of advanced mass spectrometry-based proteomics to study hypoxia driven cancer progression. Front. Oncol. 2021;11:559822. doi: 10.3389/fonc.2021.559822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang J.W., Ding Y., Tahir Ul Qamar M., Shen Y., Gao J., Chen L.L. A deep learning model based on sparse auto-encoder for prioritizing cancer-related genes and drug target combinations. Carcinogenesis. 2019;40:624–632. doi: 10.1093/carcin/bgz044. [DOI] [PubMed] [Google Scholar]
- 74.Guan N.N., Zhao Y., Wang C.C., Li J.Q., Chen X., Piao X. Anticancer drug response prediction in cell lines using weighted graph regularized matrix factorization. Mol. Ther. Nucleic Acids. 2019;17:164–174. doi: 10.1016/j.omtn.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y., Jeon J., Park J.H. Hypoxia-responsive nanoparticles for tumor-targeted drug delivery. Cancer Lett. 2020;490:31–43. doi: 10.1016/j.canlet.2020.05.032. [DOI] [PubMed] [Google Scholar]
- 76.Frejno M., Meng C., Ruprecht B., Oellerich T., Scheich S., Kleigrewe K., Drecoll E., Samaras P., Hogrebe A., Helm D. Proteome activity landscapes of tumor cell lines determine drug responses. Nat. Commun. 2020;11:3639. doi: 10.1038/s41467-020-17336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruprecht B., Di Bernardo J., Wang Z., Mo X., Ursu O., Christopher M., Fernandez R.B., Zheng L., Dill B.D., Wang H. A mass spectrometry-based proteome map of drug action in lung cancer cell lines. Nat. Chem. Biol. 2020;16:1111–1119. doi: 10.1038/s41589-020-0572-3. [DOI] [PubMed] [Google Scholar]
- 78.Dang Y., Jiang N., Wang H., Chen X., Gao Y., Zhang X., Qin G., Li Y., Chen R. Proto-oncogene serine/threonine kinase PIM3 promotes cell migration via modulating Rho GTPase signaling. J. Proteome Res. 2020;19:1298–1309. doi: 10.1021/acs.jproteome.9b00821. [DOI] [PubMed] [Google Scholar]
- 79.Wang F.Y., Wang-Gou S.Y., Cao H., Jiang N., Yang Q., Huang Q., Huang C.H., Li X.J. Proteomics identifies EGF-like domain multiple 7 as a potential therapeutic target for epidermal growth factor receptor-positive glioma. Cancer Commun. (Lond.) 2020;40:518–530. doi: 10.1002/cac2.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mu Q., Annapragada A., Srivastava M., Li X., Wu J., Thiviyanathan V., Wang H., Williams A., Gorenstein D., Annapragada A., Vigneswaran N. Conjugate-SELEX: A high-throughput screening of thioaptamer-liposomal nanoparticle conjugates for targeted intracellular delivery of anticancer drugs. Mol. Ther. Nucleic Acids. 2016;5:e382. doi: 10.1038/mtna.2016.81. [DOI] [PubMed] [Google Scholar]
- 81.Zhou X.T., Ding J., Li H.Y., Zuo J.L., Ge S.Y., Jia H.L., Wu J. Hedgehog signalling mediates drug resistance through targeting TAP1 in hepatocellular carcinoma. J. Cell. Mol. Med. 2020;24:4298–4311. doi: 10.1111/jcmm.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng Y., Ren M., Li Y., Liu Y., Chen C., Su J., Su B., Xia H., Liu F., Jiang H. Knockdown of RhoGDI2 represses human gastric cancer cell proliferation, invasion and drug resistance via the Rac1/Pak1/LIMK1 pathway. Cancer Lett. 2020;492:136–146. doi: 10.1016/j.canlet.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 83.Umar A., Kang H., Timmermans A.M., Look M.P., Meijer-van Gelder M.E., den Bakker M.A., Jaitly N., Martens J.W., Luider T.M., Foekens J.A., Pasa-Tolić L. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol. Cell. Proteomics. 2009;8:1278–1294. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hemdan T., Malmström P.U., Jahnson S., Segersten U. Emmprin expression predicts response and survival following cisplatin containing chemotherapy for bladder cancer: A validation study. J. Urol. 2015;194:1575–1581. doi: 10.1016/j.juro.2015.06.085. [DOI] [PubMed] [Google Scholar]
- 85.Hou C., Guo D., Yu X., Wang S., Liu T. TMT-based proteomics analysis of the anti-hepatocellular carcinoma effect of combined dihydroartemisinin and sorafenib. Biomed. Pharmacother. 2020;126:109862. doi: 10.1016/j.biopha.2020.109862. [DOI] [PubMed] [Google Scholar]
- 86.Li S.L., Ye F., Cai W.J., Hu H.D., Hu P., Ren H., Zhu F.F., Zhang D.Z. Quantitative proteome analysis of multidrug resistance in human ovarian cancer cell line. J. Cell. Biochem. 2010;109:625–633. doi: 10.1002/jcb.22413. [DOI] [PubMed] [Google Scholar]
- 87.Chang L., Graham P., Hao J., Bucci J., Malouf D., Gillatt D., Li Y. Proteomics discovery of radioresistant cancer biomarkers for radiotherapy. Cancer Lett. 2015;369:289–297. doi: 10.1016/j.canlet.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 88.Jung J.H., You S., Oh J.W., Yoon J., Yeon A., Shahid M., Cho E., Sairam V., Park T.D., Kim K.P., Kim J. Integrated proteomic and phosphoproteomic analyses of cisplatin-sensitive and resistant bladder cancer cells reveal CDK2 network as a key therapeutic target. Cancer Lett. 2018;437:1–12. doi: 10.1016/j.canlet.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang Y., Park K.H., Lee J.E., Han K.C. Phosphoproteomic analysis reveals PAK2 as a therapeutic target for lapatinib resistance in HER2-positive breast cancer cells. Biochem. Biophys. Res. Commun. 2018;505:187–193. doi: 10.1016/j.bbrc.2018.09.086. [DOI] [PubMed] [Google Scholar]
- 90.Wang Z., Sun H., Provaznik J., Hackert T., Zöller M. Pancreatic cancer-initiating cell exosome message transfer into noncancer-initiating cells: the importance of CD44v6 in reprogramming. J. Exp. Clin. Cancer Res. 2019;38:132. doi: 10.1186/s13046-019-1129-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Fu M., Gu J., Jiang P., Qian H., Xu W., Zhang X. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol. Cancer. 2019;18:41. doi: 10.1186/s12943-019-1001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo W., Li Y., Pang W., Shen H. Exosomes: A potential therapeutic tool targeting communications between tumor cells and macrophages. Mol. Ther. 2020;28:1953–1964. doi: 10.1016/j.ymthe.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin Q., Zhou C.R., Bai M.J., Zhu D., Chen J.W., Wang H.F., Li M.A., Wu C., Li Z.R., Huang M.S. Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am. J. Transl. Res. 2020;12:1080–1095. [PMC free article] [PubMed] [Google Scholar]
- 94.Chen W., Quan Y., Fan S., Wang H., Liang J., Huang L., Chen L., Liu Q., He P., Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 95.Lu Y., Duan Y., Xu Q., Zhang L., Chen W., Qu Z., Wu B., Liu W., Shi L., Wu D. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J. Cell. Mol. Med. 2020;24:1311–1318. doi: 10.1111/jcmm.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cao H., Zhu X., Chen X., Yang Y., Zhou Q., Xu W., Wang D. Quantitative proteomic analysis to identify differentially expressed proteins in the persistent atrial fibrillation using TMT coupled with nano-LC-MS/MS. Am. J. Transl. Res. 2020;12:5032–5047. [PMC free article] [PubMed] [Google Scholar]
- 97.Munagala R., Aqil F., Jeyabalan J., Kandimalla R., Wallen M., Tyagi N., Wilcher S., Yan J., Schultz D.J., Spencer W., Gupta R.C. Exosome-mediated delivery of RNA and DNA for gene therapy. Cancer Lett. 2021;505:58–72. doi: 10.1016/j.canlet.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li M.Y., Liu L.Z., Dong M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol. Cancer. 2021;20:22. doi: 10.1186/s12943-021-01312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fan T., Sun N., He J. Exosome-derived lncRNAs in lung cancer. Front. Oncol. 2020;10:1728. doi: 10.3389/fonc.2020.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y.Y., Hung A.C., Lo S., Yuan S.F. Adipocytokines visfatin and resistin in breast cancer: Clinical relevance, biological mechanisms, and therapeutic potential. Cancer Lett. 2021;498:229–239. doi: 10.1016/j.canlet.2020.10.045. [DOI] [PubMed] [Google Scholar]
- 101.Thomas R., Al-Rashed F., Akhter N., Al-Mulla F., Ahmad R. ACSL1 regulates TNFα-induced GM-CSF production by breast cancer MDA-MB-231 cells. Biomolecules. 2019;9:555. doi: 10.3390/biom9100555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma H.Y., Li Y., Yin H.Z., Yin H., Qu Y.Y., Xu Q.Y. TNFAIP8 promotes cisplatin chemoresistance in triple-negative breast cancer by repressing p53-mediated miR-205-5p expression. Mol. Ther. Nucleic Acids. 2020;22:640–656. doi: 10.1016/j.omtn.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Fujiyuki T., Amagai Y., Shoji K., Kuraishi T., Sugai A., Awano M., Sato H., Hattori S., Yoneda M., Kai C. Recombinant SLAMblind measles virus is a promising candidate for nectin-4-positive triple negative breast cancer therapy. Mol. Ther. Oncolytics. 2020;19:127–135. doi: 10.1016/j.omto.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ko C.H., Cheng C.F., Lai C.P., Tzu T.H., Chiu C.W., Lin M.W., Wu S.Y., Sun C.Y., Tseng H.W., Wang C.C. Differential proteomic analysis of cancer stem cell properties in hepatocellular carcinomas by isobaric tag labeling and mass spectrometry. J. Proteome Res. 2013;12:3573–3585. doi: 10.1021/pr4004294. [DOI] [PubMed] [Google Scholar]
- 105.Ding F., Sun K., Sun N., Jiang Q., Cao M., Wu Z. iTRAQ-based proteomics reveals SOD2 as a potential salivary biomarker in liver cancer. Int. J. Biol. Markers. 2019;34:221–231. doi: 10.1177/1724600819841619. [DOI] [PubMed] [Google Scholar]
- 106.Wu C., Zheng L. Proteomics promises a new era of precision cancer medicine. Signal Transduct. Target. Ther. 2019;4:13. doi: 10.1038/s41392-019-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma Y.S., Yang X.L., Xin R., Liu J.B., Fu D. Power and promise of exosomes as clinical biomarkers and therapeutic vectors for liquid biopsy and cancer control. Biochim. Biophys. Acta Rev. Cancer. 2021;1875:188497. doi: 10.1016/j.bbcan.2020.188497. [DOI] [PubMed] [Google Scholar]
- 108.Zhou J., Le K., Xu M., Ming J., Yang W., Zhang Q., Lu L., Xi Z., Ruan S., Huang T. CXCR4 antagonist AMD3100 reverses the resistance to tamoxifen in breast cancer via inhibiting AKT phosphorylation. Mol. Ther. Oncolytics. 2020;18:161–170. doi: 10.1016/j.omto.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen S., Wang J., Zheng B., Tao Y., Li M., Wang Y., Ni X., Suo T., Liu H., Liu H., Zhang J. LINC01714 enhances gemcitabine sensitivity by modulating FOXO3 phosphorylation in cholangiocarcinoma. Mol. Ther. Nucleic Acids. 2020;19:446–457. doi: 10.1016/j.omtn.2019.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bai D., Wu Y., Deol P., Nobumori Y., Zhou Q., Sladek F.M., Liu X. Palmitic acid negatively regulates tumor suppressor PTEN through T366 phosphorylation and protein degradation. Cancer Lett. 2021;496:127–133. doi: 10.1016/j.canlet.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stenström L., Mahdessian D., Gnann C., Cesnik A.J., Ouyang W., Leonetti M.D., Uhlén M., Cuylen-Haering S., Thul P.J., Lundberg E. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol. Syst. Biol. 2020;16:e9469. doi: 10.15252/msb.20209469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Casado J., Lehtonen O., Rantanen V., Kaipio K., Pasquini L., Häkkinen A., Petrucci E., Hynninen J., Hietanen S., Carpén O. Agile workflow for interactive analysis of mass cytometry data. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa946. Published online November 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen K., Rong N., Wang S., Luo C. A novel two-layer-integrated microfluidic device for high-throughput yeast proteomic dynamics analysis at the single-cell level. Integr. Biol. 2020;12:241–249. doi: 10.1093/intbio/zyaa018. [DOI] [PubMed] [Google Scholar]
- 114.Cavalli M., Diamanti K., Pan G., Spalinskas R., Kumar C., Deshmukh A.S., Mann M., Sahlén P., Komorowski J., Wadelius C. A multi-omics approach to liver diseases: integration of single nuclei transcriptomics with proteomics and HiCap bulk data in human liver. OMICS. 2020;24:180–194. doi: 10.1089/omi.2019.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]