Using mutations associated with cardiomyopathy, Sitbon et al. study the transition between the super relaxed state of myosin and the disordered-relaxed state. Their findings suggest that the N terminus of myosin essential light chain controls the transition between these two states.
Abstract
In this study, we assessed the super relaxed (SRX) state of myosin and sarcomeric protein phosphorylation in two pathological models of cardiomyopathy and in a near-physiological model of cardiac hypertrophy. The cardiomyopathy models differ in disease progression and severity and express the hypertrophic (HCM-A57G) or restrictive (RCM-E143K) mutations in the human ventricular myosin essential light chain (ELC), which is encoded by the MYL3 gene. Their effects were compared with near-physiological heart remodeling, represented by the N-terminally truncated ELC (Δ43 ELC mice), and with nonmutated human ventricular WT-ELC mice. The HCM-A57G and RCM-E143K mutations had antagonistic effects on the ATP-dependent myosin energetic states, with HCM-A57G cross-bridges fostering the disordered relaxed (DRX) state and the RCM-E143K model favoring the energy-conserving SRX state. The HCM-A57G model promoted the switch from the SRX to DRX state and showed an ∼40% increase in myosin regulatory light chain (RLC) phosphorylation compared with the RLC of normal WT-ELC myocardium. On the contrary, the RCM-E143K–associated stabilization of the SRX state was accompanied by an approximately twofold lower level of myosin RLC phosphorylation compared with the RLC of WT-ELC. Upregulation of RLC phosphorylation was also observed in Δ43 versus WT-ELC hearts, and the Δ43 myosin favored the energy-saving SRX conformation. The two disease variants also differently affected the duration of force transients, with shorter (HCM-A57G) or longer (RCM-E143K) transients measured in electrically stimulated papillary muscles from these pathological models, while no changes were displayed by Δ43 fibers. We propose that the N terminus of ELC (N-ELC), which is missing in the hearts of Δ43 mice, works as an energetic switch promoting the SRX-to-DRX transition and contributing to the regulation of myosin RLC phosphorylation in full-length ELC mice by facilitating or sterically blocking RLC phosphorylation in HCM-A57G and RCM-E143K hearts, respectively.
Introduction
Cardiac muscle contraction results from the ATP-coupled interactions between myosin cross-bridges and actin/tropomyosin-troponin (Tm-Tn) filaments. It is regulated by Ca2+ ions and controlled by myosin filament–based mechanisms, including phosphorylation of myosin regulatory light chain (RLC) and myosin binding protein C (MyBP-C; Brunello et al., 2020). Myosin binds ATP and actin in the motor domain, and its lever arm region amplifies small conformational changes generated in the catalytic domain into the large movements needed to produce force and muscle contraction (Rayment et al., 1993; Geeves and Holmes, 2005). The myosin lever arm is structurally supported by myosin RLC and essential light chain (ELC), two important myosin subunits that bind to their respective IQ motifs within the myosin heavy chain lever arm region. During muscle relaxation, myosin head domains of paired myosin heavy chains appear to form an interacting-head motif that structurally characterizes two different myosin states, a disordered relaxed (DRX) state, in which myosin heads protrude into the interfilament space, and a super relaxed (SRX) state, in which they interact with the myosin backbone and each other (Alamo et al., 2017). Compared with DRX, the SRX state is characterized by low ATP turnover rates, typical of an energy-conserving state that serves as a modulator of energy utilization in cardiac muscle (Hooijman et al., 2011; McNamara et al., 2015). The normal cardiac relaxation phase is highly significant for normal heart function, and the disruption of the SRX state of myosin has been thought to increase the possibility for inefficient myocardial performance and poor clinical outcomes (Toepfer et al., 2020).
The myosin ELC constitutes an essential part of the lever arm structure, but it also plays a role in the contractile force development and the ATP-dependent kinetics of actin-myosin interactions (VanBuren et al., 1994; Timson, 2003; Miller et al., 2005; Kazmierczak et al., 2009). A functionally important domain of the cardiac ELC is its 43-amino-acid-long N-terminal extension (N-ELC) shown by many to directly interact with the C-terminal region of actin (Trayer et al., 1987; Milligan et al., 1990; Aydt et al., 2007; Kazmierczak et al., 2009; Wang et al., 2018). We previously reported that transgenic mice expressing the truncated N-ELC (Δ43 mice) display near-physiological cardiac remodeling with an increase in left ventricular (LV) wall thickness but no changes in cardiac morphology or function or in myofilament contraction/relaxation parameters are observed (Sitbon et al., 2020). On the other hand, pathological hypertrophy due to hypertrophic cardiomyopathy (HCM) or restrictive cardiomyopathy (RCM) is characterized by interstitial fibrosis, myofilament disarray, reduced LV cavity, and impaired heart’s relaxation phase and can lead to diastolic dysfunction and sudden cardiac death (Seidman and Seidman, 1998; Spirito et al., 2000; Arad et al., 2002; Ip et al., 2013). RCM is additionally manifested by biatrial enlargement, increased stiffness of the LV wall with no increase in wall thickness (Kushwaha et al., 1997; Yuan et al., 2017). Both HCM and RCM can result from genetic mutations in major sarcomeric proteins such as β-myosin heavy chain, cardiac MyBP-C, titin, actin, Tm, TnT, TnI, TnC, and both myosin RLC and ELC.
In this report, we focus on the A57G and E143K mutations in the ventricular ELC, the products of the mutated MYL3 gene that were shown by population studies to cause human HCM or RCM, respectively (Lee et al., 2001; Olson et al., 2002; Caleshu et al., 2011). We previously reported that transgenic mice expressing the A57G ELC mutation (HCM-A57G model) exhibited characteristic HCM remodeling that included increased LV wall thickness, fibrosis, and diastolic abnormalities manifested by prolongation of isovolumetric relaxation time (Sitbon et al., 2020). Skinned and intact papillary muscle (PM) experiments demonstrated increased Ca2+ sensitivity of force, decreased maximal tension, and delayed Ca2+ transients in HCM-A57G mice (Kazmierczak et al., 2013; Sitbon et al., 2020). Likewise, transgenic mice expressing the E143K ELC mutation (RCM-E143K model) displayed a typical RCM phenotype with a decrease in LV inner diameter and increased LV stiffness, fibrosis, and diastolic dysfunction, as judged by prolongation of isovolumetric relaxation time and an increase in Ca2+ sensitivity of force (Yuan et al., 2017). Interestingly, in contrast to HCM-A57G mice, RCM-E143K animals exhibited decreased cardiac output and stroke work in vivo (Yuan et al., 2017) and downregulation of myosin power and a reduced force–velocity relationship in the in vitro motility assay (Wang et al., 2018).
These in vivo and in vitro functional characteristics of ELC transgenic animal models built the scientific premise of the current investigation aimed at the comprehensive assessment of the bioenergetic signatures present in HCM-A57G, RCM-E143K, and Δ43 mice. We first focused on the ATP-dependent interaction of myosin and actin assessed in skinned and intact PMs to provide insight into the energetic states of myosin and cross-bridge kinetics at the level of myofilaments. Specifically, we looked at the mechanism by which HCM and RCM ELC mutations modulate the SRX-to-DRX transition and how myosin energetic states in the hearts of HCM-A57G, RCM-E143K, and Δ43 mice correlate with myofilament protein phosphorylation. Because RLC-mediated regulation of myosin motor activity via myosin light chain kinase–dependent RLC phosphorylation has long been acknowledged to be critical for heart function in health and disease (reviewed in Yadav and Szczesna-Cordary, 2017), the emphasis was put on testing the myosin RLC phosphorylation in the hearts of mice. We thoroughly analyzed the interplay between myosin energetic states, SRX↔DRX equilibrium, and the degree of myosin RLC phosphorylation in all investigated ELC models.
Finally, because of our recently published proteomic data on HCM-A57G and Δ43 relative to WT-ELC hearts (Gomes et al., 2015) and on RCM-E143K versus WT-ELC mice (Yuan et al., 2017) that revealed significant differences between the models in biogenesis, cellular organization, and biological regulation, we also investigated ELC mutation-dependent mitochondrial and metabolic remodeling, oxidative phosphorylation (OXPHOS), and ATP respiration.
Materials and methods
Transgenic mice
All animal procedures and experiments were performed following the “Guide for the Care and Use of Laboratory Animals” (National Institutes of Health publication 85-23, revised 2011). All the protocols were approved by the Institutional Animal Care and Use Committee at the University of Miami Miller School of Medicine (protocol #18-110), which has an Animal Welfare Assurance on file with the Office of Laboratory Animal Welfare, National Institutes of Health (assurance number #A-3224-01, approved through November 30, 2019). We are registered with US Department of Agriculture Animal and Plant Health Inspection Service (registration #58-R-007, approved through December 3, 2023). We have full accreditation with the Association for Assessment and Accreditation of Laboratory Animal Care (site 001069, latest effective date, November 8, 2016). Mice were euthanized through CO2 inhalation followed by cervical dislocation.
We have previously generated transgenic mice models expressing the mutated human cardiac ELC in HCM-A57G, RCM-E143K, and physiological-like model Δ43 mice as well as transgenic WT mice expressing the human ventricular ELC (Kazmierczak et al., 2009; Muthu et al., 2011; Yuan et al., 2017). Multiple crosses of transgenic ELC mice with B6SJL/F1 mice were performed before the animals were used for experiments. For this study, two lines of WT (L1, 76%; and L4, 71%), two of A57G (L1, 80%; and L5, 74%), and two of Δ43 (L8, 34–39%; and L9, 35–40%) were used. We also used L2 of E143K mice expressing 55% of mutant ELC protein (Yuan et al., 2017). All experiments were performed on the hearts of 8 ± 1.5-mo-old male and female mice.
Skinned PM experiments and the SRX state of myosin
N-methylanthraniloyl (mant)-ATP assays were performed on skinned PM fibers from HCM-A57G, RCM-E143K, Δ43, and WT mice using the IonOptix instrumentation as described in Yadav et al. (2019a). Fibers were incubated in a rigor solution containing 250 µM mant-ATP, and decay in fluorescence versus time curves were collected when mant-ATP was rapidly exchanged with 4 mM nonlabeled ATP. Decay isotherms were fitted to a two-state exponential equation, and the P1 and P2 and T1 and T2 parameters were calculated to describe the proportion of myosin heads in the SRX (P2) or DRX (P1) states and assess their respective T2 and T1 lifetimes (in seconds; Yadav et al., 2019a; Sitbon et al., 2020).
Intact PM fiber mechanics and force/calcium transients
Intact PM fiber studies were performed according to protocols described previously (Yuan et al., 2018; Sitbon et al., 2020). Briefly, intact PMs were carefully dissected from the LVs of mice in preoxygenated (95% O2, 5% CO2) 2,3-butanedione monoxime (BDM)-Krebs-Ringer solution and incubated in BDM–Krebs-Ringer buffer containing 50 µM Fura-2 AM (Life Technologies) at room temperature for 1.5 h in the dark, followed by several washes in BDM–Krebs-Ringer solution to remove excess Fura-2 AM. Fibers were then mounted in a flow chamber between a hook connected to a length-controlled micromanipulator and a force transducer. Preoxygenated Krebs-Ringer solution (95% O2, 5% CO2, pH 7.4) was perfused into the chamber at a flow rate of ∼2 ml/min. The temperature in the muscle chamber was kept constant at 25°C by a heat exchanger at the inflow line and a circulating water bath. The muscle was stretched to an optimum length at which the developed tension was maximal during an isometric twitch. Sarcomere length, monitored by a camera, was maintained at ∼2.1 µm for another 10–15 min to allow the force development to stabilize. The stimulus was applied through platinum hooks, and strength was adjusted to 20% above threshold (∼7–8 V) at a frequency of 1 Hz for PMs. Normalized developed force, which allowed for the comparison between muscles of different diameters, was calculated using the physiological cross-sectional area, PM mass (mg) / [PM density (mg/mm3) × PM length (mm)], and expressed in millinewtons per square millimeter. The HyperSwitch dual excitation light source with sub-millisecond switching times, an inverted fluorescence microscope equipped with a calcium photometry objective, and MyoCam-S digital variable field rate charge-coupled device video system were used to detect and record Ca2+ transients of the PMs loaded with Fura-2 AM, a dual excitation indicator dye. Fluorescence intensity of Fura-2 AM was collected by a charge-coupled device camera from 340- and 380-nm channels, followed by background subtraction. The dynamic measurement allowed up to 1,000 length recordings per second. Data were averaged from ∼20 force/calcium transient peaks and analyzed using system IonWizard software (Yuan et al., 2018; Sitbon et al., 2020).
Analysis of sarcomeric protein phosphorylation
Phosphorylation of cardiac myosin RLC, TnI, and cMyBP-C in all investigated animal models was measured in cardiac myofibrils (CMFs) isolated from six to eight hearts per group as described previously (Kazmierczak et al., 2013). Specifically, 4 M and 3 F (7.5–10-mo-old) WT, 3 M and 5 F (8–10-mo-old) HCM-A57G, 3 M and 3 F (6–9-mo-old) RCM-E143K, and 4 M and 2 F (7.5–10-mo-old) Δ43 mice were used. After euthanasia, the hearts were excised and placed in liquid nitrogen. Before the experiment, the tissue was thawed in CMF buffer consisting of 5 mM NaH2PO4, 5 mM Na2HPO4, pH 7.0, 0.1 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, 5 mM ATP, 5 nM microcystin, 0.1% Triton X-100, 10 µl/ml of phosphatase inhibitor cocktails 2 and 3 (P5726 and P0044; Sigma-Aldrich), 5 mM DTT, and 1 µl/ml protease inhibitor cocktail (P8340; Sigma-Aldrich) and homogenized in a Mixer-Mill MM301. The homogenate was centrifuged for 4 min at 8,000 ×g, and the supernatant was discarded. After centrifugation, the pellets were left on ice for 2 min. This step was repeated three times with CMF buffer without Triton X-100. The pellets were then resuspended in the CMF buffer, and the samples were subsequently mixed at a 1:1 ratio with Laemmli buffer and 5% β-mercaptoethanol and heated at 100°C for 5 min. Samples were loaded onto 12% or 4–20% gradient gels (Mini-Protean TGX; Bio-Rad), and phosphoproteins were detected using Pro-Q Diamond/Sypro system (Invitrogen, Thermo Fisher Scientific). Total protein was also detected using Coomassie staining as described previously (Kazmierczak et al., 2014). RLC and cMyBP-C phosphorylation was tested by Western blots and protein-specific antibodies. To detect phosphorylated RLC, ∼30 µg of protein per lane was loaded into 15% SDS-PAGE, and the membranes were probed with phospho-specific RLCmse antibody (gift from Dr. Neal Epstein, National Institutes of Health, Bethesda, MD), which recognizes the phosphorylated form of the RLC and does not react with nonphosphorylated RLC. Total RLC protein was detected with rabbit polyclonal CT-1 antibody, produced in this laboratory (Szczesna-Cordary et al., 2005), which served as a loading control. Phosphorylation of cMyBP-C was tested at Ser282. Approximately 20 µg of protein per lane was loaded into 12% SDS-PAGE, and the membranes were probed with phospho-specific cMyBP-CSer282 antibody (gift from Dr. Sakthivel Sadayappan, University of Cincinnati, Cincinnati, OH). Goat polyclonal antibody was used to detect total cMyBP-C. Myofilament protein phosphorylation was calculated based on the density of bands of phosphorylated protein/total protein. The bands were analyzed using ImageJ software (National Institutes of Health).
Statistical analyses
All values are shown as mean ± SD. Differences between multiple groups were assessed using one-way ANOVA followed by Tukey’s multiple comparison test. To detect statistical differences between the genotypes that were sex-dependent, two-way ANOVA followed by Tukey’s or Sidak’s multiple comparison test was used with significance defined as P < 0.05 (GraphPad Prism v7.05).
Online supplemental material
In the supplemental text (at the end of the PDF), the Materials and methods section addresses experimental procedures regarding OXPHOS complexes, enzymatic activities, and mitochondrial respiration. The Results section provides assessment of calcium transients presented in Fig. S1 and Table S1; mitochondrial remodeling in HCM-A57G, RCM-E143K, and Δ43 versus WT-ELC mice (mitochondrial protein expression [Fig. S2], enzymatic activities [Fig. S3], and cellular respiration and oxygen consumption rates [OCRs; Fig. S4 and Table S2]).
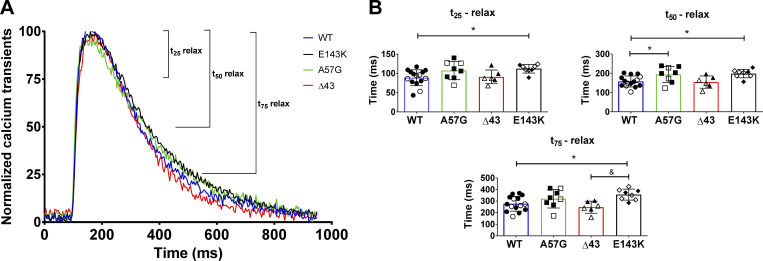
Figure S1.
Calcium transients in intact PMs from HCM-A57G, RCM-E143K, and Δ43 versus WT-ELC hearts. Data for HCM-A57G and Δ43 were taken, with permission, from Sitbon et al. (2020). (A) Normalized calcium transients for HCM-A57G (green), RCM-E143K (black), Δ43 (red), and WT (blue) mice. (B) Time points t25, t50, and t75, depicting the time (in milliseconds) from peak [Ca2+] to 25%, 50%, and 75% baseline for all four genotypes. Open symbols depict female mice and closed symbols depict male mice. The hearts of n = 8 (5 M, 3 F) HCM-A57G, n = 6 (3 M, 3 F) Δ43, n = 8 (3 M, 5 F) RCM-E143K, and n = 15 (10 M, 5 F) WT-ELC mice were tested. Values are mean ± SD and were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. Significance was depicted as *, P < 0.05 for mutant versus WT-ELC; and &, P < 0.05 for RCM-E143K versus Δ43. Note delayed relaxation times for HCM-A57G (t50) and RCM-E143K (t25, t50, and t75) compared with WT-ELC mice.
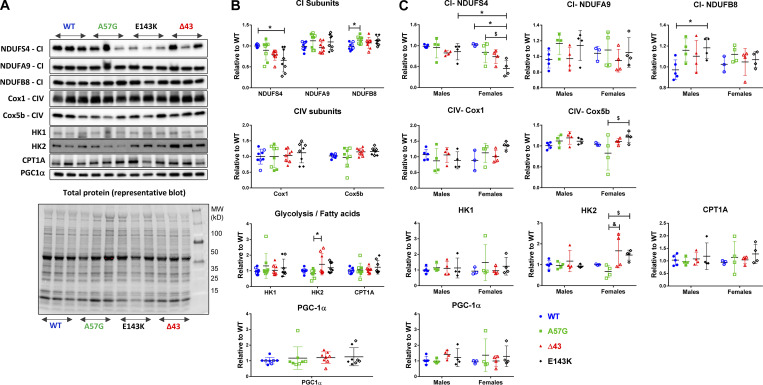
Figure S2.
Assessment of mitochondrial protein expression in heart homogenates from 7–11-mo-old HCM-A57G, Δ43, and RCM-E143K relative to WT-ELC mice. (A) Representative Western blots of mitochondrial and metabolism proteins in heart homogenates of mice. (B and C) Expression of CI, CIV, glycolysis, fatty acid subunits (HK1, HK2, and CPT1A), and peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α; PPAR-γ coactivator and master regulator of mitochondrial biogenesis) for all four genotypes (B), and separately for female versus male mice (C). Values are mean ± SD of n = 8 (5 M, 3 F) WT-ELC; n = 8 (4 M, 4 F) HCM-A57G; n = 8 (4 M, 4 F) Δ43; and n = 8 (4 M, 4 F) RCM-E143K animals with significance depicted as *, P < 0.05 for mutant versus WT or between genders. $, P < 0.05 for HCM-A57G versus RCM-E143K; and &, P < 0.05 between HCM-A57G versus Δ43 analyzed by one-way ANOVA with Tukey’s multiple comparison test (B) and two-way ANOVA with two nominal variables (genotype, sex) followed by Tukey’s or Sidak’s multiple comparison test (C). Optical density was assessed with ImageJ. Each protein band was normalized to total protein content assessed for each blot before the primary antibody was applied. HK, hexokinase (glucose metabolism); NDUFS4, NDUFA9, and NDUFB8, CI subunits (NADH dehydrogenase); Cox5b and Cox1, CIV subunits; CPT1A, carnitine palmitoyltransferase 1A (fatty acid metabolism).
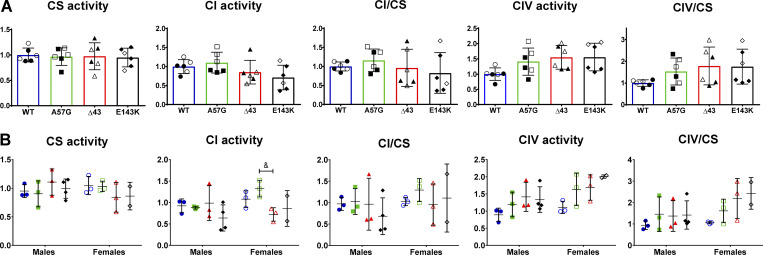
Figure S3.
Enzymatic activities in the hearts of ELC mice. (A and B) Spectrophotometric assessment of enzymatic activities in heart homogenates from HCM-A57G (3 M, 3 F), Δ43 (3 M, 3 F), and RCM-E143K (4 M, 2 F) relative to WT (3 M, 3 F) mice (A), and separated by sex (B). Values are mean ± SD and analyzed using two-way ANOVA with Tukey’s or Sidak’s multiple comparisons test with significance depicted as &, P < 0.05 between HCM-A57G versus Δ43. Open symbols depict female mice and closed symbols depict male mice.
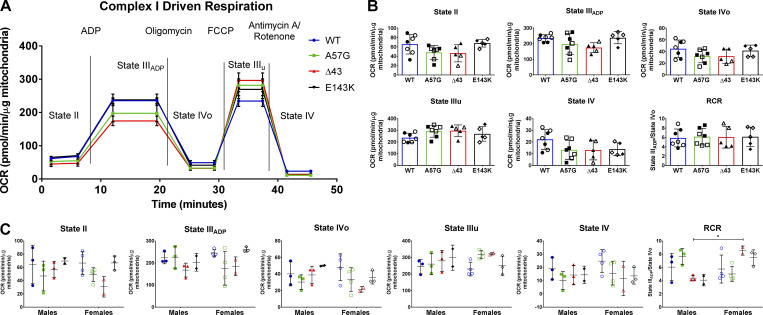
Figure S4.
CI-driven respiration profiles in the hearts of 8–10-mo-old WT, HCM-A57G, Δ43, and RCM-E143K mice. (A) Averaged traces of CI OCR in isolated mitochondria from WT-ELC (n = 7 mice), HCM-A57G (n = 7 mice), Δ43 (n = 5 mice), and RCM-E143K (n = 5 mice) using Seahorse XFp analyzer. (B) Characterization of mitochondrial OCR for state II, state IIIADP, state IVo, state IIIu, state IV, and the RCR, calculated as OCR-state IIIADP/OCR-state IVo. (C) Data from B separated by sex: WT (3 M, 4 F), HCM-A57G (3 M, 4 F), Δ43 (3 M, 2 F), and RCM-E143K (2 M, 3 F). Values are mean ± SD analyzed by two-way ANOVA with Tukey’s or Sidak’s multiple comparisons test with significance *, P < 0.05 for female versus male Δ43 mice. Open symbols depict female mice and closed symbols depict male mice.
Results
Energetic remodeling in HCM-A57G, RCM-E143K, and Δ43 ELC mice at the level of myofilaments
SRX state of myosin
The cardiac relaxation phase is critically important for normal heart function, and disruption of SRX↔DRX equilibrium has been postulated to increase the risk for inefficient heart performance that can lead to adverse clinical outcomes (Toepfer et al., 2020). Both the HCM-A57G and RCM-E143K ELC mutations have been shown to cause disease-specific abnormalities in patients (Lee et al., 2001; Olson et al., 2002; Caleshu et al., 2011) and in transgenic mice (Kazmierczak et al., 2013; Yuan et al., 2017), but little is understood about the mechanistic basis by which these mutations confer distinct in vivo cardiac function. To learn about the effect of ELC mutations on SRX↔DRX equilibrium and the proportion of myosin heads existing in the SRX (P2) versus DRX (P1) conformations, we performed ATP binding/exchange assays in skinned PM fibers from HCM-A57G and RCM-E143K mice. The effects due to these two pathological mutations were compared with WT-ELC mice and to Δ43 animals that manifest a time-dependent hypertrophic growth with no obvious abnormalities in heart function (Muthu et al., 2011). Fibers were incubated in a solution containing 250 µM mant-ATP, and decay fluorescence curves as a function of time were collected during a rapid mant-ATP exchange with 4 mM nonlabeled ATP. Averaged fluorescence decay curves were fitted to a double-exponential equation (Yadav et al., 2019a; Sitbon et al., 2020):
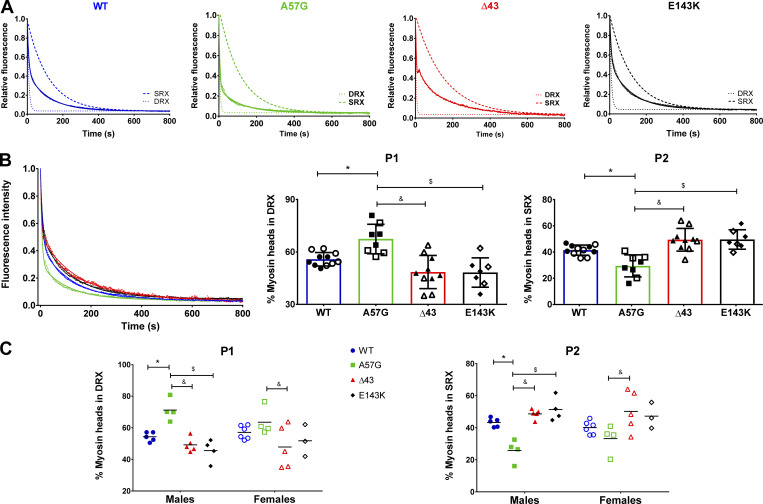
where P1 and P2 (in %) are the amplitudes of fast and slow phases of fluorescence intensity decay, indicating percentage of heads in the DRX versus SRX state, respectively. Averaged fluorescence decay curves and their simulated single-exponential traces for the fast and slow phases of fluorescence decay for fibers from all genotypes are shown in Fig. 1 A. Fig. 1 B and Table 1 compare the fluorescence intensity decay isotherms and depict the fraction of heads existing in either the DRX or SRX state, and Fig. 1 C analyzes the effect of ELC mutations on SRX↔DRX equilibrium separately for male and female mice. To complete the comparison between all genotypes studied, previously reported SRX data for HCM-A57G and Δ43 mice (Sitbon et al., 2020) were included in the analysis of the newly acquired results on HCM-A57G and Δ43 mice as well as RCM-E143K and WT mice (Fig. 1 and Table 1).
Figure 1.
The SRX state of myosin assessed in skinned PMs from 7–10-mo-old mouse models of hypertrophic (HCM-A57G) and restrictive (RCM-E143K) cardiomyopathy and in a near-physiological Δ43 ELC model compared with WT-ELC mice. (A) Fluorescence decay curves and simulated single-exponential curves of mant-ATP release. (B) Comparison of fluorescence decay curves for WT-ELC (11 mice, 5 M and 6 F), HCM-A57G (8 mice, 4 M and 4 F), Δ43 (10 mice, 5 M and 5 F), and RCM-E143K (7 mice, 4 M and 3 F) animals (left), redistribution of cross-bridges (in %) between the DRX (P1) state (middle) and the SRX (P2) state (right). Open symbols depict female mice, and closed symbols depict male mice. Data are mean ± SD; *, P < 0.05 for mutant versus WT-ELC; $, P < 0.05 for E143K versus A57G; and &, P < 0.05 for Δ43 versus A57G calculated with one-way ANOVA with Tukey’s multiple comparison test. Note that the percentage of DRX is increased in HCM-A57G compared with WT-ELC, Δ43, and RCM-E143K mice, and this increase is coupled with a decrease in percentage of SRX for HCM-A57G mice. (C) Myosin head redistribution between the DRX (P1) versus SRX (P2) states plotted as a function of genotype and sex. Data are mean ± SD of male or female mice analyzed by two-way ANOVA with two nominal variables (genotype, sex) followed by Tukey’s or Sidak’s multiple comparison test.
Table 1. The SRX state of myosin measured by mant-ATP/ATP chase assays in skinned PMs from the hearts of HCM-A57G, RCM-E143K, Δ43, and WT-ELC mice.
| Parameter | WT | HCM-A57G | Δ43 | RCM-E143K |
|---|---|---|---|---|
| P1 (%) | 55.8 ± 3.8 | 67.4 ± 8.4a | 48.5 ± 9.5b | 48.3 ± 8.4c |
| P2 (%) | 41.5 ± 3.8 | 29.5 ± 8.4a | 49.4 ± 8.7b | 49.6 ± 7.4c |
| T1 (s) | 8.2 ± 4.3 | 6.2 ± 3.4 | 4.9 ± 3.5 | 9.0 ± 5.2 |
| T2 (s) | 182.8 ± 126.3 | 109.0 ± 71.5 | 197.3 ± 108.7 | 186.3 ± 123.9 |
| Number of animals | 11 (5 M, 6 F) | 8 (4 M, 4 F) | 10 (5 M, 5 F) | 7 (4 M, 3 F) |
Data are mean ± SD. P1 and P2 represent the percentage of myosin cross-bridges in the DRX and SRX states, respectively; T1 and T2 depict their respective lifetimes. P values were calculated with one-way ANOVA with Tukey’s multiple comparison test.
P < 0.05 for mutant versus WT-ELC.
P < 0.05 for Δ43 versus A57G.
P < 0.05 for E143K versus A57G.
The HCM-A57G mutation significantly reduced the SRX state of myosin and promoted the SRX-to-DRX transition (Fig. 1, A and B), usually associated with greater ATP consumption (Alamo et al., 2016). The ratio of myosin heads existing in the DRX to SRX (P1:P2) states was 56:42 for WT-ELC and changed to 67:30 in HCM-A57G mice (Table 1). The RCM-E143K and Δ43 models promoted the SRX conformation and exhibited a significantly larger proportion of myosin heads occupying the SRX (P2) state compared with P2 observed in HCM-A57G mice (Fig. 1 B and Table 1). Interestingly, male mice of all genotypes demonstrated a more pronounced phenotype compared with female counterparts (Fig. 1 C), indicating sexual dimorphism in the effect of the ELC mutation on SRX↔DRX equilibrium. No changes between the genotypes were noted in T1 or T2 lifetimes (Table 1).
Overall, these results explain the opposing effects of two pathological ELC variants as the results of a disturbance in SRX↔DRX equilibrium and underscore the importance of studying the energetic states of myosin at the level of myofilaments. The HCM-A57G mutation promoted the SRX-to-DRX transition that mechanistically signifies the HCM clinical phenotype of hypercontractility, while the RCM-E143K mutation favored the energy-saving SRX state. Similar to RCM-E143K, the near-physiological Δ43 model also stabilized the sequestered energy-saving conformation. Larger SRX populations may be cardioprotective in normal Δ43 myocardium and during times of stress in cardiomyopathic RCM-E143K mice. The data on Δ43 mice favoring the SRX conformation suggests a likely involvement of the N-ELC (missing in Δ43 mice) in regulation of the SRX-to-DRX transition in full-length ELC mutant mice. The results also revealed sex dimorphism among the ELC models and the dominant phenotype of male mice.
Kinetics of force development
Our previous in vivo investigations demonstrated major differences in function between the pathological HCM-A57G and RCM-E143K models and suggested distinct mechanisms by which they influence cardiac function. In contrast to HCM-A57G (Kazmierczak et al., 2013), RCM-E143K animals exhibited decreased cardiac output and stroke work in vivo (Yuan et al., 2017). These differences most likely originated from different effects of mutations on myosin motor power, with A57G upregulating and E143K downregulating myosin power as observed in the in vitro motility assay (Wang et al., 2018). Interestingly, despite such different in vivo and in vitro behaviors due to these two ELC variants, the mutations resulted in a common phenotype of diastolic dysfunction in mice (Yuan et al., 2017; Sitbon et al., 2020) and humans (Lee et al., 2001; Olson et al., 2002; Caleshu et al., 2011).
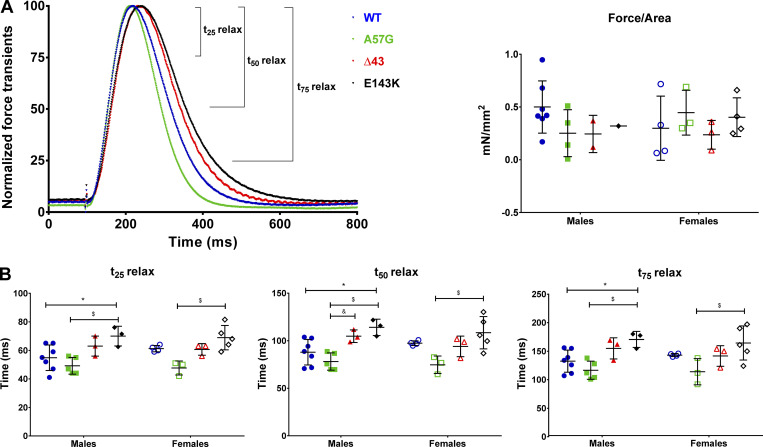
To understand the origin of physiological differences between the HCM-A57G and RCM-E143K models, we compared their ability to produce force in electrically stimulated intact PMs from these mice. Unlike the RCM-E143K model that has not been characterized before, HCM-A57G and Δ43 were tested for their effects on the kinetics of the force and calcium transients during twitch contractions (Sitbon et al., 2020). To compare the effect of all ELC mutations on the kinetics of force/calcium transients and relate the results to the distribution of myosin energetic states in these models, the data previously reported for HCM-A57G and Δ43 mice (Sitbon et al., 2020) were merged with the new results on RCM-E143K animals and compared with sex- and age-matched WT controls (Fig. 2). We observed no differences in the effects of the two genotypes on maximal force (in millinewtons) or the peak force/cross-section area (in millinewtons per square millimeter; Fig. 2 A and Table 2). However, differences between the two pathogenic models were noted in force relaxation kinetics, with t25, t50, and t75 times (in milliseconds) at 25, 50, and 75% peak force to baseline being significantly longer in RCM-E143K compared with HCM-A57G and WT fibers (Fig. 2 B and Table 2). Interestingly, as in SRX state experiments, male mice presented with a more pronounced functional phenotype than female counterparts (Fig. 2 B). Despite differences in force transient kinetics, both pathological models, HCM-A57G and RCM-E143K, displayed delayed calcium transients compared with WT mice, indicating diastolic abnormalities in these mice (Fig. S1 and Table S1). The findings of longer versus shorter force transients observed for RCM-E143K and HCM-A57G models, respectively, are consistent with the SRX results showing slower cross-bridge kinetics and stabilization of the sequestered SRX conformation in RCM-E143K mice versus faster cross-bridge relaxation kinetics and larger DRX populations in the HCM-A57G mice.
Figure 2.
Force transients in intact PMs from 8–10-mo-old HCM-A57G (5 M and 3 F), Δ43 (3 M and 3 F), and RCM-E143K (3 M and 5 F) compared with WT-ELC (7 M and 4 F). The data for A57G and Δ43 animals were taken, with permission, from Sitbon et al. (2020). (A) Left: Normalized force transients for WT-ELC (blue), HCM-A57G (green), Δ43 (red), and RCM-E143K (black) mice. Right: Force (in millinewtons per square millimeter) was calculated as a ratio of peak force to physiological cross-sectional area (see Materials and methods) and plotted as a function of genotype and sex. (B) Time points t25, t50, and t75 depicting the time (in milliseconds) from peak force to 25%, 50%, and 75% baseline for male and female mice of all genotypes. Open symbols depict female mice, and closed symbols depict male mice. Data are mean ± SD of male or female mice analyzed by two-way ANOVA followed by Tukey’s or Sidak’s multiple comparison test. *, P < 0.05 for mutant versus WT; $, P < 0.05 for A57G versus E143K; and &, P < 0.05 for A57G versus Δ43.
Table 2. Force transients in intact PMs from the hearts of RCM-E143K versus WT-ELC mice.
| Parameter | WT | HCM-A57G | Δ43 | RCM-E143K |
|---|---|---|---|---|
| Number of animals | 11 (7 M, 4 F) | 8 (5 M, 3 F) | 6 (3 M, 3 F) | 8 (3 M, 5 F) |
| t25 relax (ms) | 57.2 ± 7.7 | 48.6 ± 5.2a | 61.8 ± 5.3b | 69.3 ± 7.4a,c |
| t50 relax (ms) | 92.6 ± 11.5 | 78.1 ± 8.6a | 98.8 ± 11.0b | 111.8 ± 13.9a,c |
| t75 relax (ms) | 136.5 ± 16.0 | 115.8 ± 17.3 | 148.3 ± 17.90b | 166.8 ± 21.0a,c |
| Force/area (mN/mm2) | 0.42 ± 0.27 | 0.34 ± 0.23 | 0.24 ± 0.13 | 0.39 ± 0.16 |
Data are mean ± SD. P values were calculated with one-way ANOVA with Tukey’s multiple comparison test. For comparisons between the RCM, HCM, and Δ43 phenotypes, the data for HCM-A57G and Δ43 animals (columns 3 and 4) were taken, with permission, from Sitbon et al. (2020).
P < 0.05 for mutant versus WT-ELC.
P < 0.05 for Δ43 versus A57G.
P < 0.05 for E143K versus A57G.
Protein phosphorylation in HCM-A57G, RCM-E143K, and Δ43 ELC mice
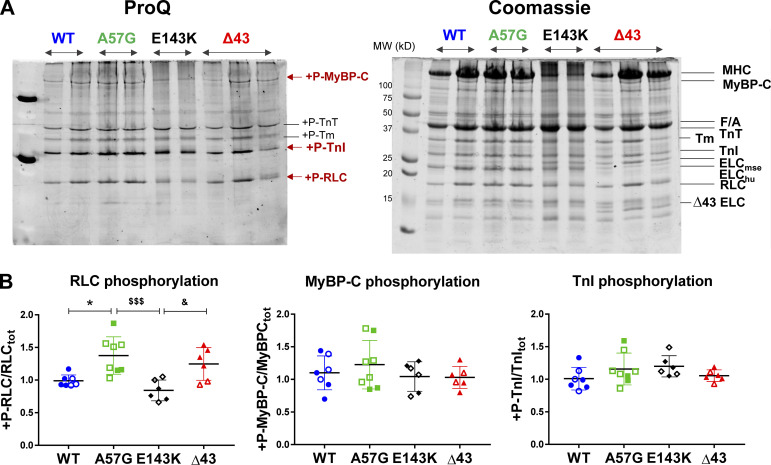
The structure of cardiac thick and thin filaments is known to undergo dynamic changes produced by posttranslational modifications such as altered phosphorylations of myosin RLC, cMyBP-C, or TnI that may lead to alterations in energy utilization and cardiac output (Moss and Fitzsimons, 2010). To test this idea, we assessed the phosphorylation status of myofilament proteins in the hearts of female and male WT, HCM-A57G, RCM-E143K, and Δ43 mice. The phosphorylation of myosin RLC, cMyBP-C, and TnI was determined in CMFs prepared from the hearts of mice that were run on SDS-PAGE and stained with ProQ/Coomassie (Fig. 3) or blotted with protein-specific antibodies (Fig. 4).
Figure 3.
Myosin RLC, cMyBP-C, and TnI phosphorylation in ventricular CMFs purified from the hearts of WT, HCM-A57G, RCM-E143K, and Δ43 mice. The hearts of 4 M and 3 F (7.5–10-mo-old) WT-ELC, 3 M and 5 F (8–10-mo-old) A57G, 3 M and 3 F (6–9-mo-old) E143K, and 4 M and 2 F (7.5–10-mo-old) Δ43 mice were used. (A) SDS-PAGE and sarcomeric protein phosphorylation monitored by ProQ/Coomassie. (B) Quantification of myosin RLC, cMyBP-C, and TnI phosphorylation in CMFs from all genotypes. Open symbols depict female mice and closed symbols depict male mice. Data are mean ± SD and were analyzed using one-way ANOVA with Tukey’s comparison test with significance depicted as *, P < 0.05 between mutant versus WT; $$$, P < 0.001 for A57G versus E143K; and &, P < 0.05 for E143K versus Δ43 mice. Abbreviations: +P-RLC, phosphorylated form of mouse ventricular RLC; +P-MyBP-C, phosphorylated form of mouse cMyBP-C; +P-TnI, phosphorylated form of TnI; F/A, F-actin; ELCmse, mouse ventricular essential light chain; ELChu, human transgenic ELC; Δ43 ELC, N-terminally truncated ELC protein standard.
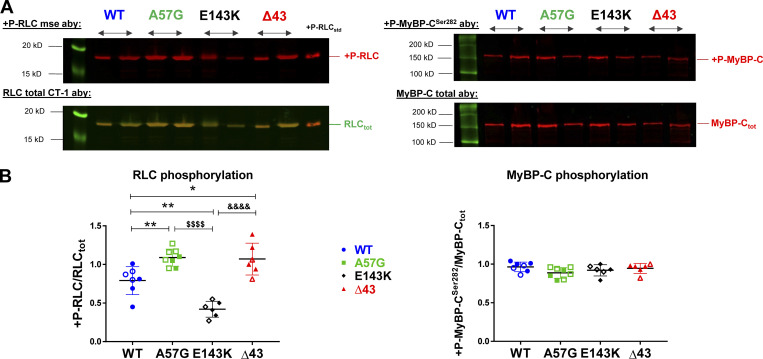
Figure 4.
Myosin RLC and cMyBP-CSer282 phosphorylation in myofibrils from heart ventricles of WT-ELC, HCM-A57G, RCM-E143K, and Δ43 mice detected by Western blotting. (A) SDS-PAGE and sarcomeric protein phosphorylation monitored by Western blots and RLC-specific (left) or cMyBP-C–specific (right) antibodies (Aby). Phosphorylated forms of the mouse RLC and cMyBP-C (upper panels) were detected with +P-RLCmse or +P-MyBP-CSer282 antibodies, while total proteins were assessed with RLCtot (CT-1) or MyBP-Ctot antibodies (lower panels). (B) Quantification of myosin RLC and cMyBP-CSer282 phosphorylation in CMFs purified from WT-ELC (4 M, 3 F), HCM-A57G (3 M, 5 F), RCM-E143K (3 M, 3 F), and Δ43 (4 M, 2 F) mice. Open symbols depict female mice and closed symbols depict male mice. Data are mean ± SD and were analyzed using one-way ANOVA with Tukey’s comparison test. Significance was depicted as *, P < 0.05 and **, P < 0.01 between mutant versus WT-ELC; $$$$, P < 0.0001 for HCM-A57G versus RCM-E143K; and &&&&, P < 0.0001 for RCM-E143K versus Δ43 mice.
ProQ/Coomassie data revealed a significant upregulation of myosin RLC phosphorylation in HCM-A57G compared with the RLC of WT-ELC (P < 0.05) and RCM-E143K (P < 0.001) mice (Fig. 3). A significantly higher RLC phosphorylation level was also observed in Δ43 CMFs compared with RCM-E143K hearts (P < 0.05). No changes in cMyBP-C or TnI phosphorylation were noted in HCM-A57G, RCM-E143K, or Δ43 CMFs compared with WT-ELC (Fig. 3). A more quantitative analysis of RLC and MyBP-CSer282 phosphorylation assessed with Western blotting and protein-specific antibodies (Fig. 4) showed ∼40% upregulation of myosin RLC phosphorylation in HCM-A57G versus WT-ELC mice (P < 0.01) and ∼2.7-fold greater RLC phosphorylation in HCM-A57G versus RCM-E143K (P < 0.0001) CMFs. No changes in cMyBP-C phosphorylation seen by ProQ/Coomassie (Fig. 3) were further confirmed by Western blotting using cMyBP-CSer282 specific antibodies (Fig. 4). On the other hand, an approximately twofold lower level of myosin RLC phosphorylation was observed in RCM-E143K versus WT-ELC mice (P < 0.01) and an ∼2.6-fold lower level when compared with the RLC of Δ43 (P < 0.0001) CMFs (Fig. 4). Remarkably, upregulation and downregulation of myosin RLC phosphorylation seen in HCM-A57G and RCM-E143K hearts, respectively, were correlated with an increase in DRX populations in HCM-A57G mice and a larger SRX population in RCM-E143K animals (Fig. 1), further highlighting the consistency in phenotypic differences between the two pathological models.
Upregulation of RLC phosphorylation was also observed in Δ43 versus WT-ELC hearts (Fig. 4), which corresponded to larger SRX populations and presumably greater energy conservation in this model (Fig. 1). These data imply that the N-ELC that is missing in the hearts of Δ43 mice not only binds to actin during muscle contraction (Timson et al., 1998) but may also interact with myosin RLC and modulate its phosphorylation in HCM-A57G (hypercontractile) or RCM-E143K (obstructed output) hearts, which express the full-length ELC. Combined SRX and RLC phosphorylation results on Δ43 mice suggest that the SRX conformation of myosin is stabilized by the lack of N-ELC, which is required for the SRX-to-DRX redistribution, and its role in the regulation of myosin energetic states can be further facilitated by myosin RLC phosphorylation.
Mitochondrial respiratory function in HCM-A57G, RCM-E143K, and Δ43 mice
Cardiomyopathy is commonly associated with an increased energy cost of tension generation through inefficient or excessive ATP usage (Ashrafian et al., 2003). This may come about as a result of shifting myosin heads out of the energy-saving SRX state into the DRX or active states, in which ATP usage is greatly increased (McNamara et al., 2015). Our published proteomic studies performed on hearts of transgenic HCM-A57G and Δ43 ELC mice revealed that 30 proteins were differentially expressed in the pathological versus physiological-like heart models and that significant differences occurred in biogenesis, cellular organization, and biological regulation (Gomes et al., 2015). Similarly, proteomic analyses of RCM-E143K versus WT-ELC samples also indicated that major affected proteins were those involved in metabolic heart remodeling (Yuan et al., 2017). Combining data on proteomic profiling of HCM-A57G, RCM-E143K, and Δ43 versus WT hearts, and because of the differences in myosin energetic states observed between the models (Fig. 1), we evaluated the mutant-specific metabolic protein/enzyme expression in heart homogenates from all ELC mice (Fig. S2 A). The analysis of OXPHOS subunits revealed that the two pathological models differently regulated the expression of complex I (CI) subunits of the mitochondrial electron transport chain. Compared with WT-ELC mice, the RCM-E143K model downregulated the expression of the NDUFS4 subunit while the HCM-A57G model upregulated the β subunit of NADH dehydrogenase, NDUFB8 (Fig. S2 B). Decreased expression of NDUFS4 may be indicative of a decreased ability of RCM-E143K to properly use NADH to fuel the mitochondrial electron transport chain, leading to less ATP production in RCM-E143K hearts, while increased expression of NDUFB8 in HCM-A57G myocardium may suggest some abnormalities in CI function due to HCM.
We also studied the expression of complex IV (CIV; cytochrome c oxidase [COX]), which contributes to the proton gradient across the inner mitochondrial membrane to drive ATP synthesis (Bourens and Barrientos, 2017). Except for small but significant alterations in Cox5b expression observed in female HCM-A57G mice (lower) versus female RCM-E143K animals (higher; Fig. S2 C), no differences in any of the CIV subunits were observed. Compared with HCM-A57G mice, the Δ43 ELC model of near-physiological hypertrophic remodeling showed a significant upregulation of hexokinase 2 (HK2), an enzyme involved in glucose utilization pathways (Fig. S2 B). Similar to alterations in CI/CIV expression, the differences in HK2 expression between HCM-A57G and Δ43 mice were also observed in females (Fig. S2 C).
We then investigated the enzymatic activities of the OXPHOS complexes, as any alterations in CI or CIV activities could be indicative of defective mitochondrial respiration, uncoupled proton gradient, and altered ATP production (Diaz, 2010). Spectrophotometric assessment of heart homogenates from HCM-A57G, RCM-E143K, and Δ43 relative to WT mice showed no changes of CI/CIV enzymatic activities in any of the investigated models (Fig. S3 A). Interestingly, analysis of sex differences in CI activity between the genotypes revealed that females of the pathological HCM-A57G model had significantly higher activity compared with Δ43 females (Fig. S3 B).
Finally, oxygen consumption profiles were determined in mitochondria isolated from HCM-A57G, RCM-E143K, and Δ43 hearts relative to WT hearts in the presence of specific substrates and inhibitors (Fig. S4 A). Determination of basal OCR or state II respiration using pyruvate and malate as substrates showed that OCR rates were similar among all genotypes (Fig. S4 B and Table S2), and no differences were noted between females and males of all genotypes (Fig. S4 C). No changes were observed in ADP-dependent state III respiration, state IVo and state IIIu respiration, or respiratory control ratio (RCR), calculated as state IIIADP to state IVo ratio (Rogers et al., 2011), and delineating the mitochondrial quality control (Khalifa et al., 2017; Fig. S4). Overall, these data indicated that no mitochondrial respiratory dysfunction occurred in the hearts of investigated ELC models and that the hearts of mice were supplied with enough ATP to sustain their contractile function.
Discussion
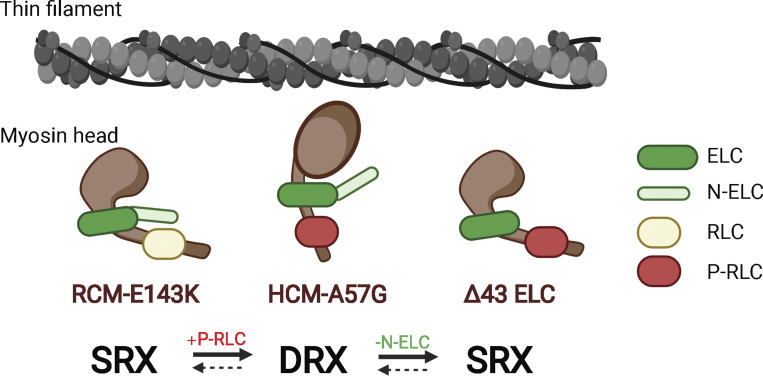
Cardiac muscle contraction depends on the ATP-driven cyclical interaction of myosin with thin filaments comprising actin and Tm-Tn (Geeves and Holmes, 2005). The myosin head or cross-bridge is the molecular motor of the heart and contains a motor domain with actin-activated MgATPase activity and the lever-arm region where both myosin ELC and RLC are attached to their respective myosin heavy chain IQ motifs (Rayment et al., 1993). The outstanding question in myosin ELC biology regards the function of the cardiac-specific 43-amino-acid-long N-ELC, comprising a lysine-rich actin-binding region that can bridge the ELC core of the myosin head with the actin filament (reviewed in Yadav et al., 2019b). Based on extensive research of myosin ELC function in the heart, we hypothesized that the cardiac N-ELC works as a molecular linker and/or energetic switch of the actin-myosin interaction that can regulate the SRX-to-DRX transition and the proportion of myosin heads that exist in either of these two energetically different states (Fig. 5). In DRX, the myosin heads protrude into the interfilament space, but they are restricted from binding to actin, while in the SRX state, myosin heads are neatly ordered along the thick filament axis but cycle very slowly at a correspondingly low ATP turnover rate (Hooijman et al., 2011; McNamara et al., 2015). Because variations in the size of the myosin SRX population are considered central to modulating sarcomeric force production and energy utilization in cardiac muscle, we aimed to explore the mechanism by which the two pathological models of cardiomyopathy, HCM-A57G and RCM-E143K, as well as the structural Δ43 ELC mutation, affect the stability of the sequestered state and SRX↔DRX equilibrium and whether these processes can be regulated by N-ELC and myosin RLC phosphorylation (Fig. 5).
Figure 5.
Schematic representation of the antagonistic mechanism by which the HCM-linked A57G and RCM-associated E143K mutations in cardiac myosin ELC regulate the SRX state of myosin and myosin RLC phosphorylation. The HCM-A57G mutation promotes the SRX-to-DRX transition, and this process is coupled with a ∼40% increase in myosin RLC phosphorylation compared with the level of RLC phosphorylation in WT-ELC myocardium. The RCM-E143K model favors the SRX conformation, and its RLC phosphorylation is approximately twofold less than in WT-ELC hearts, as judged by Western blotting and RLC-specific antibodies. Mechanistically, it is proposed that N-ELC, which is missing in the Δ43 model, regulates the SRX-to-DRX transition and contributes to sterically blocking (RCM-E143K) or facilitating (HCM-A57G) myosin RLC phosphorylation. Fig. 5 was created with https://BioRender.com.
Myosin energetic states and cross-bridge kinetics in HCM-A57G, RCM-E143K, and Δ43 ELC mouse models
Mant-ATP chase assays revealed that the two models, HCM-A57G and RCM-E143K, exhibited antagonistic effects on SRX↔DRX equilibrium, with HCM-A57G favoring the DRX state and RCM-E143K favoring the energy-saving SRX state (Fig. 1). Interestingly, fluorescence decay isotherms of RCM-E143K were similar to those produced by Δ43 animals, with a larger number of heads occupying the SRX state (P2) compared with the SRX (P2) of the HCM-A57G model (Table 1). The fact that the structural Δ43 ELC mutation that brings about the physiological-like hypertrophy phenotype in mice also promotes the formation of SRX suggests that the N-ELC, which is lacking in Δ43 mice, may be involved in regulating the myosin sequestered state and the SRX-to-DRX transition (Fig. 5). All models demonstrated sex dimorphism, showing greater effects and significant differences in myosin energetic states in male mice (Fig. 1 C).
Force production and energy use within the sarcomere are intricately regulated by the SRX mechanism (Schmid and Toepfer, 2021). We observed a strong correlation between myosin energetic states and the kinetics of force transients measured in electrically stimulated LV PMs from mice (Fig. 2). Faster relaxation rates calculated in HCM-A57G fibers were related to the SRX-to-DRX transition, while the stabilization of the SRX state correlated with slower force transients measured in RCM-E143K samples compared with WT controls (Table 2). While both pathological variants demonstrated significant differences in comparison to WT, no changes in force transient kinetics were observed in Δ43 versus WT-ELC mice. Notably, in agreement with myosin SRX/DRX experiments, the kinetics of force transients were significantly different between male mice, indicating sexual dimorphism (Fig. 2 B). Considering their distinct clinical phenotypes, and despite showing differences in force transient kinetics, both models of ELC-related cardiomyopathy displayed delayed calcium transients compared with WT-ELC mice (Fig. S1 and Table S1), suggesting the potential for diastolic dysfunction. Taken together, the A57G and E143K ELC mutations that were shown to cause human HCM (Lee et al., 2001) or RCM (Olson et al., 2002; Caleshu et al., 2011), respectively, presented clear differences in myosin energetic states and force transients kinetics (Figs. 1 and 2). They showed an upregulation (HCM-A57G) or downregulation (RCM-E143K) of myosin power in vitro (Wang et al., 2018) and exhibited opposite effects on cardiac output in vivo (Kazmierczak et al., 2013; Yuan et al., 2017).
Myosin RLC phosphorylation is upregulated in HCM-A57G and Δ43 hearts and downregulated in RCM-E143K hearts
Myosin-based regulation of cardiac muscle is increasingly recognized as supplementing actin-based mechanisms in controlling the strength and time course of contraction (Brunello et al., 2020). The RLC-mediated modulation of myosin motor activity via myosin light chain kinase–dependent RLC phosphorylation has long been known to be crucial for normal heart function and was also shown to be perturbed in heart disease (Scruggs et al., 2009; Chang et al., 2015). The myocardium containing dephosphorylated myosin was shown to have limited ability to produce force and maintain cardiac function at physiological levels (Muthu et al., 2012; Yuan et al., 2015), suggesting myosin RLC phosphorylation as a promising target for the development of novel therapies (Yadav et al., 2019c). Structurally, RLC phosphorylation is proposed to control the myosin head conformation by disrupting a compact OFF conformation of myosin heads, which is equivalent to an energy-saving SRX state (Fig. 5). Disruption of the OFF state and promoting the DRX cross-bridge conformation is linked to potentiated contractility by increasing the population of myosin heads that are available for interaction with actin and force production in muscle (Jung et al., 2008; Kampourakis et al., 2016; Alamo et al., 2017).
Comparison of the results from the two cardiomyopathy models strongly suggests that RLC phosphorylation can regulate SRX↔DRX equilibrium in cardiac muscle and the proportion of force-producing myosin heads. Myosin RLC phosphorylation promoted the SRX-to-DRX redistribution in HCM-A57G myocardium, while the absence of RLC phosphorylation in RCM-E143K mice contributed to stabilization of the sequestered SRX state (Fig. 5). The interplay between the level of myosin RLC phosphorylation and the energetic states of myosin further explains the mechanisms of phenotypic differences between the HCM-A57G and RCM-E143K models. Upregulation of RLC phosphorylation was also observed in Δ43 versus WT hearts and, as we showed before (Sitbon et al., 2020) and in this study, the Δ43 myosin favored the energy-conserving SRX conformation. One can speculate that the N-ELC that is truncated in the hearts of Δ43 mice can assume different conformations in the full-length ELC models to either promote or sterically hinder myosin RLC phosphorylation in HCM-A57G or RCM-E143K mice, respectively (Fig. 5).
The interrelation between myosin N-ELC and RLC phosphorylation has been addressed before: the Burghardt group, using single-molecule approaches and quantum dot motility assays elegantly demonstrated that N-ELC is involved in stimulus frequency modulation to affect power output (Wang et al., 2016) and that this process is highly regulated by RLC phosphorylation (Wang et al., 2014). After RLC phosphorylation, which increases at higher stimulus frequencies, an 8-nm step was the dominant myosin step size resulting in a significant gain in the average step size and leading to increased work produced per ATPase cycle (Wang et al., 2014). Of note, in our recent work, we determined that the deletion of N-ELC in Δ43 mice increased the prevalence of 5-nm step sizes with coincidental loss of the 8-nm step as compared with native full-length ELC (Wang et al., 2016; Wang et al., 2018). Ensemble motility and single myosin mechanical characteristics were also consistent with the notion that HCM-A57G mutation impairs N-ELC–actin binding, while RCM-E143K mutation leads to weakening of myosin lever arm stability (Wang et al., 2018). High-resolution structural approaches (e.g., x-ray crystallography and in silico analysis; Robert-Paganin et al., 2018) coupled with time-resolved biophysical assessments (e.g., Guhathakurta et al., 2017) of the ELC and RLC conformational transitions associated with HCM-A57G and RCM-E143K mutations in myosin ELC ± RLC phosphorylation would provide a still greater understanding of the effects of these mutations on the prevalence of the sequestered state of myosin and how the mutations modify myosin power and the number of heads available for force production.
Mitochondrial energy supply is not affected by myosin ELC mutations
Cardiac function is highly dependent on ample energy supply, and the regulation of heart muscle contraction and relaxation relies heavily on mitochondria, which produce energy in the form of ∼6 kg of ATP daily (Hall et al., 2014; Ritterhoff and Tian, 2017). Most of the cell’s ATP is produced via the OXPHOS system, comprising five multi-subunit complexes embedded in the mitochondrial inner membrane. Oxidative metabolism in mitochondria provides most of the energy consumed by the heart, and the inability to generate and/or transfer energy may trigger abnormalities in contractile function (Tian et al., 2019). Considering the clear differences between the ELC mutations in the myosin energetic states and force transient kinetics, it was somewhat surprising not to observe any significant alterations in the OXPHOS complexes or mitochondrial respiration.
In summary, our study revealed that under normal conditions, the overall respiratory function assessed in all investigated ELC models was normal, and the hearts of HCM-A57G, RCM-E143K, and Δ43 mice were supplied with enough ATP to sustain their contractile function. However, the minute differences in CI subunit expression noted between the two pathological models (Fig. S2) may become more evident under stress conditions (Garnier et al., 2003; Dai et al., 2012) that may exacerbate mutation-dependent deficits in energy transfer and mitochondrial function.
Significance and innovation
Our study contributes new information on myosin energetic states associated with hypertrophic (A57G) and restrictive (E143K) cardiomyopathy phenotypes and provides insight into the interplay between myosin SRX and DRX conformations and the phosphorylation of myosin RLC (Fig. 5). The hypercontractility that results from the SRX-to-DRX transition in the HCM-A57G myocardium is also associated with increased phosphorylation of the myosin RLC, while stabilization of the SRX energy-conserving state of cross-bridges in RCM-E143K hearts is related to the downregulation of myosin RLC phosphorylation. We observed a strong correlation between the increased or decreased degree of RLC phosphorylation in HCM-A57G and RCM-E143K mice, respectively, with their enhanced or depressed in vivo function monitored by echocardiography (Kazmierczak et al., 2013; Yuan et al., 2017) and upregulation or downregulation of myosin power in the in vitro motility assay (Wang et al., 2018).
We propose a new regulatory function for N-ELC, which is missing in Δ43 mice, as being involved in the modulation of myosin energetic states and myosin RLC phosphorylation in full-length ELC mutant mice (Fig. 5). The N-ELC is presumed to adopt different conformations relative to myosin heavy chain and myosin RLC to either facilitate (HCM-A57G) or sterically block (RCM-E143K) myosin RLC phosphorylation and is considered a key regulator of SRX↔DRX equilibrium in these two pathogenic mouse models of cardiomyopathy (Fig. 5). Taken together, our findings show that both mechanisms, RLC phosphorylation and N-ELC, could be crucial targets in therapeutic interventions for ELC-associated cardiomyopathy.
Supplementary Material
shows calcium transients in intact PMs from RCM-E143K versus WT mice.
shows CI-driven mitochondrial respiratory states in isolated mitochondria from the hearts of WT-ELC, HCM-A57G, RCM-E143K, and Δ43 mice using Seahorse XFp analyzer.
Acknowledgments
Henk L. Granzier served as editor.
The authors thank Dr. Manish Bharadwaj for his expert advice regarding mitochondria isolation and seahorse experiments.
This work was supported by the National Institutes of Health (grants R01-HL143830 and R56-HL146133 to D. Szczesna-Cordary).
The authors declare no competing financial interests.
Author contributions: Y.H. Sitbon designed and performed experiments, analyzed and interpreted data, and wrote and edited the manuscript. F. Diaz designed and performed experiments, analyzed and interpreted data, and wrote and edited the manuscript. K. Kazmierczak designed and performed experiments and analyzed and plotted the data. J. Liang designed and performed experiments and analyzed data. M. Wangpaichitr designed experiments and analyzed the data. D. Szczesna-Cordary conceived the study, analyzed and interpreted data, and wrote and revised the manuscript. All authors read, edited, and approved the revised version of the manuscript.
Footnotes
This work is part of a special collection on myofilament function and disease.
References
- Alamo, L., Qi D., Wriggers W., Pinto A., Zhu J., Bilbao A., Gillilan R.E., Hu S., and Padrón R.. 2016. Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. J. Mol. Biol. 428:1142–1164. 10.1016/j.jmb.2016.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Ware J.S., Pinto A., Gillilan R.E., Seidman J.G., Seidman C.E., and Padrón R.. 2017. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 6:e24634. 10.7554/eLife.24634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arad, M., Seidman J.G., and Seidman C.E.. 2002. Phenotypic diversity in hypertrophic cardiomyopathy. Hum. Mol. Genet. 11:2499–2506. 10.1093/hmg/11.20.2499 [DOI] [PubMed] [Google Scholar]
- Ashrafian, H., Redwood C., Blair E., and Watkins H.. 2003. Hypertrophic cardiomyopathy:a paradigm for myocardial energy depletion. Trends Genet. 19:263–268. 10.1016/S0168-9525(03)00081-7 [DOI] [PubMed] [Google Scholar]
- Aydt, E.M., Wolff G., and Morano I.. 2007. Molecular modeling of the myosin-S1(A1) isoform. J. Struct. Biol. 159:158–163. 10.1016/j.jsb.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Barrientos, A., Fontanesi F., and Díaz F.. 2009. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet. Chapter 19:Unit19.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, M.S., Tyrrell D.J., Lyles M.F., Demons J.L., Rogers G.W., and Molina A.J.A.. 2015. Preparation and respirometric assessment of mitochondria isolated from skeletal muscle tissue obtained by percutaneous needle biopsy. J. Vis. Exp. (96):e52350. 10.3791/52350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens, M., and Barrientos A.. 2017. Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J. Biol. Chem. 292:7774–7783. 10.1074/jbc.M117.778514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, M.D., and Nicholls D.G.. 2011. Assessing mitochondrial dysfunction in cells. Biochem. J. 435:297–312. 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunello, E., Fusi L., Ghisleni A., Park-Holohan S.J., Ovejero J.G., Narayanan T., and Irving M.. 2020. Myosin filament-based regulation of the dynamics of contraction in heart muscle. Proc. Natl. Acad. Sci. USA. 117:8177–8186. 10.1073/pnas.1920632117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caleshu, C., Sakhuja R., Nussbaum R.L., Schiller N.B., Ursell P.C., Eng C., De Marco T., McGlothlin D., Burchard E.G., and Rame J.E.. 2011. Furthering the link between the sarcomere and primary cardiomyopathies: restrictive cardiomyopathy associated with multiple mutations in genes previously associated with hypertrophic or dilated cardiomyopathy. Am. J. Med. Genet. A. 155A:2229–2235. 10.1002/ajmg.a.34097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A.N., Battiprolu P.K., Cowley P.M., Chen G., Gerard R.D., Pinto J.R., Hill J.A., Baker A.J., Kamm K.E., and Stull J.T.. 2015. Constitutive phosphorylation of cardiac myosin regulatory light chain in vivo. J. Biol. Chem. 290:10703–10716. 10.1074/jbc.M115.642165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, D.F., Hsieh E.J., Liu Y., Chen T., Beyer R.P., Chin M.T., MacCoss M.J., and Rabinovitch P.S.. 2012. Mitochondrial proteome remodelling in pressure overload-induced heart failure: the role of mitochondrial oxidative stress. Cardiovasc. Res. 93:79–88. 10.1093/cvr/cvr274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, F. 2010. Cytochrome c oxidase deficiency: patients and animal models. Biochim. Biophys. Acta. 1802:100–110. 10.1016/j.bbadis.2009.07.013 [DOI] [PubMed] [Google Scholar]
- Galati, D., Srinivasan S., Raza H., Prabu S.K., Hardy M., Chandran K., Lopez M., Kalyanaraman B., and Avadhani N.G.. 2009. Role of nuclear-encoded subunit Vb in the assembly and stability of cytochrome c oxidase complex: implications in mitochondrial dysfunction and ROS production. Biochem. J. 420:439–449. 10.1042/BJ20090214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier, A., Fortin D., Deloménie C., Momken I., Veksler V., and Ventura-Clapier R.. 2003. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J. Physiol. 551:491–501. 10.1113/jphysiol.2003.045104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves, M.A., and Holmes K.C.. 2005. The molecular mechanism of muscle contraction. Adv. Protein Chem. 71:161–193. 10.1016/S0065-3233(04)71005-0 [DOI] [PubMed] [Google Scholar]
- Gomes, A.V., Kazmierczak K., Cheah J.X., Gilda J.E., Yuan C.C., Zhou Z., and Szczesna-Cordary D.. 2015. Proteomic analysis of physiological versus pathological cardiac remodeling in animal models expressing mutations in myosin essential light chains. J. Muscle Res. Cell Motil. 36:447–461. 10.1007/s10974-015-9434-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhathakurta, P., Prochniewicz E., Roopnarine O., Rohde J.A., and Thomas D.D.. 2017. A Cardiomyopathy Mutation in the Myosin Essential Light Chain Alters Actomyosin Structure. Biophys. J. 113:91–100. 10.1016/j.bpj.2017.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, C.J., Sanderson L.E., Crosier K.E., and Crosier P.S.. 2014. Mitochondrial metabolism, reactive oxygen species, and macrophage function-fishing for insights. J. Mol. Med. (Berl.). 92:1119–1128. 10.1007/s00109-014-1186-6 [DOI] [PubMed] [Google Scholar]
- Hooijman, P., Stewart M.A., and Cooke R.. 2011. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys. J. 100:1969–1976. 10.1016/j.bpj.2011.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iommarini, L., Ghelli A., and Diaz F.. 2020. Methods and models for functional studies on mtDNA mutations. In The Human Mitochondrial Genome. Gasparre G., and Porcelli A.M., editors. Academic Press, Cambridge, MA; 305–349. 10.1016/B978-0-12-819656-4.00013-9 [DOI] [Google Scholar]
- Ip, J.E., Seidman C.E., Liu C.F., Cheung J.W., Thomas G., Markowitz S.M., and Lerman B.B.. 2013. Conundrum of sudden cardiac death: making sense of missense. Circ. Arrhythm. Electrophysiol. 6:e58–e63. 10.1161/CIRCEP.113.000553 [DOI] [PubMed] [Google Scholar]
- Jung, H.S., Komatsu S., Ikebe M., and Craig R.. 2008. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell. 19:3234–3242. 10.1091/mbc.e08-02-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampourakis, T., Sun Y.B., and Irving M.. 2016. Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proc. Natl. Acad. Sci. USA. 113:E3039–E3047. 10.1073/pnas.1602776113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P.T., Chen C.L., Lin P., Zhang L., Zweier J.L., and Chen Y.R.. 2018. Mitochondrial complex I in the post-ischemic heart: reperfusion-mediated oxidative injury and protein cysteine sulfonation. J. Mol. Cell. Cardiol. 121:190–204. 10.1016/j.yjmcc.2018.07.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, K., Paulino E.C., Huang W., Muthu P., Liang J., Yuan C.C., Rojas A.I., Hare J.M., and Szczesna-Cordary D.. 2013. Discrete effects of A57G-myosin essential light chain mutation associated with familial hypertrophic cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 305:H575–H589. 10.1152/ajpheart.00107.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, K., Xu Y., Jones M., Guzman G., Hernandez O.M., Kerrick W.G.L., and Szczesna-Cordary D.. 2009. The role of the N-terminus of the myosin essential light chain in cardiac muscle contraction. J. Mol. Biol. 387:706–725. 10.1016/j.jmb.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, K., Yuan C.-C., Liang J., Huang W., Rojas A.I., and Szczesna-Cordary D.. 2014. Remodeling of the heart in hypertrophy in animal models with myosin essential light chain mutations. Front. Physiol. 5:353. 10.3389/fphys.2014.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa, A.R.M., Abdel-Rahman E.A., Mahmoud A.M., Ali M.H., Noureldin M., Saber S.H., Mohsen M., and Ali S.S.. 2017. Sex-specific differences in mitochondria biogenesis, morphology, respiratory function, and ROS homeostasis in young mouse heart and brain. Physiol. Rep. 5:e13125. 10.14814/phy2.13125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha, S.S., Fallon J.T., and Fuster V.. 1997. Restrictive cardiomyopathy. N. Engl. J. Med. 336:267–276. 10.1056/NEJM199701233360407 [DOI] [PubMed] [Google Scholar]
- Lee, W., Hwang T.H., Kimura A., Park S.W., Satoh M., Nishi H., Harada H., Toyama J., and Park J.E.. 2001. Different expressivity of a ventricular essential myosin light chain gene Ala57Gly mutation in familial hypertrophic cardiomyopathy. Am. Heart J. 141:184–189. 10.1067/mhj.2001.112487 [DOI] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Dos Remedios C.G., and Cooke R.. 2015. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 7:5–14. 10.1007/s12551-014-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.S., Palmer B.M., Ruch S., Martin L.A., Farman G.P., Wang Y., Robbins J., Irving T.C., and Maughan D.W.. 2005. The essential light chain N-terminal extension alters force and fiber kinetics in mouse cardiac muscle. J. Biol. Chem. 280:34427–34434. 10.1074/jbc.M508430200 [DOI] [PubMed] [Google Scholar]
- Milligan, R.A., Whittaker M., and Safer D.. 1990. Molecular structure of F-actin and location of surface binding sites. Nature. 348:217–221. 10.1038/348217a0 [DOI] [PubMed] [Google Scholar]
- Moss, R.L., and Fitzsimons D.P.. 2010. Regulation of contraction in mammalian striated muscles--the plot thick-ens. J. Gen. Physiol. 136:21–27. 10.1085/jgp.201010471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthu, P., Kazmierczak K., Jones M., and Szczesna-Cordary D.. 2012. The effect of myosin RLC phosphorylation in normal and cardiomyopathic mouse hearts. J. Cell. Mol. Med. 16:911–919. 10.1111/j.1582-4934.2011.01371.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthu, P., Wang L., Yuan C.C., Kazmierczak K., Huang W., Hernandez O.M., Kawai M., Irving T.C., and Szczesna-Cordary D.. 2011. Structural and functional aspects of the myosin essential light chain in cardiac muscle contraction. FASEB J. 25:4394–4405. 10.1096/fj.11-191973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien, L.C., Wade R.C., Segal L., Chen Q., Savas J., Lesnefsky E.J., and Gorgey A.S.. 2017. Mitochondrial mass and activity as a function of body composition in individuals with spinal cord injury. Physiol. Rep. 5:e13080. 10.14814/phy2.13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, T.M., Karst M.L., Whitby F.G., and Driscoll D.J.. 2002. Myosin light chain mutation causes autosomal recessive cardiomyopathy with mid-cavitary hypertrophy and restrictive physiology. Circulation. 105:2337–2340. 10.1161/01.CIR.0000018444.47798.94 [DOI] [PubMed] [Google Scholar]
- Rayment, I., Rypniewski W.R., Schmidt-Bäse K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., and Holden H.M.. 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 261:50–58. 10.1126/science.8316857 [DOI] [PubMed] [Google Scholar]
- Ritterhoff, J., and Tian R.. 2017. Metabolism in cardiomyopathy: every substrate matters. Cardiovasc. Res. 113:411–421. 10.1093/cvr/cvx017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Paganin, J., Auguin D., and Houdusse A.. 2018. Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 9:4019. 10.1038/s41467-018-06191-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, D.J., and Miyamoto S.. 2015. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 22:248–257. 10.1038/cdd.2014.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.W., Brand M.D., Petrosyan S., Ashok D., Elorza A.A., Ferrick D.A., and Murphy A.N.. 2011. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 6:e21746. 10.1371/journal.pone.0021746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, G.C., Jiang A., and Arany Z.. 2010. PGC-1 coactivators in cardiac development and disease. Circ. Res. 107:825–838. 10.1161/CIRCRESAHA.110.223818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla, R.C. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88:611–638. 10.1152/physrev.00025.2007 [DOI] [PubMed] [Google Scholar]
- Schmid, M., and Toepfer C.N.. 2021. Cardiac myosin super relaxation (SRX): a perspective on fundamental biology, human disease and therapeutics. Biol. Open. 10:bio057646. 10.1242/bio.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs, S.B., Hinken A.C., Thawornkaiwong A., Robbins J., Walker L.A., de Tombe P.P., Geenen D.L., Buttrick P.M., and Solaro R.J.. 2009. Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J. Biol. Chem. 284:5097–5106. 10.1074/jbc.M807414200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman, C.E., and Seidman J.G.. 1998. Molecular genetic studies of familial hypertrophic cardiomyopathy. Basic Res. Cardiol. 93:s13–s16. 10.1007/s003950050196 [DOI] [PubMed] [Google Scholar]
- Sitbon, Y.H., Kazmierczak K., Liang J., Yadav S., Veerasammy M., Kanashiro-Takeuchi R.M., and Szczesna-Cordary D.. 2020. Ablation of the N terminus of cardiac essential light chain promotes the super-relaxed state of myosin and counteracts hypercontractility in hypertrophic cardiomyopathy mutant mice. FEBS J. 287:3989–4004. 10.1111/febs.15243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzi, M., Casarin A., Pertegato V., Salviati L., and Angelini C.. 2012. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 7:1235–1246. 10.1038/nprot.2012.058 [DOI] [PubMed] [Google Scholar]
- Spirito, P., Bellone P., Harris K.M., Bernabo P., Bruzzi P., and Maron B.J.. 2000. Magnitude of left ventricular hypertrophy and risk of sudden death in hypertrophic cardiomyopathy. N. Engl. J. Med. 342:1778–1785. 10.1056/NEJM200006153422403 [DOI] [PubMed] [Google Scholar]
- Szczesna-Cordary, D., Guzman G., Zhao J., Hernandez O., Wei J., and Diaz-Perez Z.. 2005. The E22K mutation of myosin RLC that causes familial hypertrophic cardiomyopathy increases calcium sensitivity of force and ATPase in transgenic mice. J. Cell Sci. 118:3675–3683. 10.1242/jcs.02492 [DOI] [PubMed] [Google Scholar]
- Tian, R., Colucci W.S., Arany Z., Bachschmid M.M., Ballinger S.W., Boudina S., Bruce J.E., Busija D.W., Dikalov S., Dorn G.W. II, et al. 2019. Unlocking the Secrets of Mitochondria in the Cardiovascular System: Path to a Cure in Heart Failure—A Report from the 2018 National Heart, Lung, and Blood Institute Workshop. Circulation. 140:1205–1216. 10.1161/CIRCULATIONAHA.119.040551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timson, D.J. 2003. Fine tuning the myosin motor: the role of the essential light chain in striated muscle myosin. Biochimie. 85:639–645. 10.1016/S0300-9084(03)00131-7 [DOI] [PubMed] [Google Scholar]
- Timson, D.J., Trayer H.R., and Trayer I.P.. 1998. The N-terminus of A1-type myosin essential light chains binds actin and modulates myosin motor function. Eur. J. Biochem. 255:654–662. 10.1046/j.1432-1327.1998.2550654.x [DOI] [PubMed] [Google Scholar]
- Toepfer, C.N., Garfinkel A.C., Venturini G., Wakimoto H., Repetti G., Alamo L., Sharma A., Agarwal R., Ewoldt J.F., Cloonan P., et al. 2020. Myosin Sequestration Regulates Sarcomere Function, Cardiomyocyte Energetics, and Metabolism, Informing the Pathogenesis of Hypertrophic Cardiomyopathy. Circulation. 141:828–842. 10.1161/CIRCULATIONAHA.119.042339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trayer, I.P., Trayer H.R., and Levine B.A.. 1987. Evidence that the N-terminal region of A1-light chain of myosin interacts directly with the C-terminal region of actin. A proton magnetic resonance study. Eur. J. Biochem. 164:259–266. 10.1111/j.1432-1033.1987.tb11019.x [DOI] [PubMed] [Google Scholar]
- VanBuren, P., Waller G.S., Harris D.E., Trybus K.M., Warshaw D.M., and Lowey S.. 1994. The essential light chain is required for full force production by skeletal muscle myosin. Proc. Natl. Acad. Sci. USA. 91:12403–12407. 10.1073/pnas.91.26.12403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Ajtai K., and Burghardt T.P.. 2014. Ventricular myosin modifies in vitro step-size when phosphorylated. J. Mol. Cell. Cardiol. 72:231–237. 10.1016/j.yjmcc.2014.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Ajtai K., Kazmierczak K., Szczesna-Cordary D., and Burghardt T.P.. 2016. N-Terminus of Cardiac Myosin Essential Light Chain Modulates Myosin Step-Size. Biochemistry. 55:186–198. 10.1021/acs.biochem.5b00817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Yuan C.C., Kazmierczak K., Szczesna-Cordary D., and Burghardt T.P.. 2018. Single cardiac ventricular myosins are autonomous motors. Open Biol. 8:170240. 10.1098/rsob.170240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., and Szczesna-Cordary D.. 2017. Pseudophosphorylation of cardiac myosin regulatory light chain: a promising new tool for treatment of cardiomyopathy. Biophys. Rev. 9:57–64. 10.1007/s12551-017-0248-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., Kazmierczak K., Liang J., Sitbon Y.H., and Szczesna-Cordary D.. 2019a. Phosphomimetic-mediated in vitro rescue of hypertrophic cardiomyopathy linked to R58Q mutation in myosin regulatory light chain. FEBS J. 286:151–168. 10.1111/febs.14702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., Sitbon Y.H., Kazmierczak K., and Szczesna-Cordary D.. 2019b. Hereditary heart disease: pathophysiology, clinical presentation, and animal models of HCM, RCM, and DCM associated with mutations in cardiac myosin light chains. Pflugers Arch. 471:683–699. 10.1007/s00424-019-02257-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., Yuan C.C., Kazmierczak K., Liang J., Huang W., Takeuchi L.M., Kanashiro-Takeuchi R.M., and Szczesna-Cordary D.. 2019c. Therapeutic potential of AAV9-S15D-RLC gene delivery in humanized MYL2 mouse model of HCM. J. Mol. Med. (Berl.). 97:1033–1047. 10.1007/s00109-019-01791-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C.C., Kazmierczak K., Liang J., Kanashiro-Takeuchi R., Irving T.C., Gomes A.V., Wang Y., Burghardt T.P., and Szczesna-Cordary D.. 2017. Hypercontractile mutant of ventricular myosin essential light chain leads to disruption of sarcomeric structure and function and results in restrictive cardiomyopathy in mice. Cardiovasc. Res. 113:1124–1136. 10.1093/cvr/cvx060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C.C., Kazmierczak K., Liang J., Zhou Z., Yadav S., Gomes A.V., Irving T.C., and Szczesna-Cordary D.. 2018. Sarcomeric perturbations of myosin motors lead to dilated cardiomyopathy in genetically modified MYL2 mice. Proc. Natl. Acad. Sci. USA. 115:E2338–E2347. 10.1073/pnas.1716925115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C.C., Muthu P., Kazmierczak K., Liang J., Huang W., Irving T.C., Kanashiro-Takeuchi R.M., Hare J.M., and Szczesna-Cordary D.. 2015. Constitutive phosphorylation of cardiac myosin regulatory light chain prevents development of hypertrophic cardiomyopathy in mice. Proc. Natl. Acad. Sci. USA. 112:E4138–E4146. 10.1073/pnas.1505819112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows calcium transients in intact PMs from RCM-E143K versus WT mice.
shows CI-driven mitochondrial respiratory states in isolated mitochondria from the hearts of WT-ELC, HCM-A57G, RCM-E143K, and Δ43 mice using Seahorse XFp analyzer.