Summary
Transcriptional changes happen within minutes; however, RNAi or genetic deletion requires days to weeks before transcription networks can be analyzed. This limitation has made it challenging to distinguish direct from indirect targets of sequence-specific transcription factors. This inability to define direct transcriptional targets hinders detailed studies of transcriptional mechanisms. This protocol combines rapid degradation of endogenous transcription factors with nascent transcript analysis to define the earliest, and likely direct, regulatory targets of transcription factors.
For complete details on the use and execution of this protocol, please refer to Stengel et al., 2021).
Subject areas: Cell Biology, Flow Cytometry/Mass Cytometry, Genomics, Molecular Biology, CRISPR, Protein Biochemistry
Graphical abstract

Highlights
-
•
A chemical genetic approach for small-molecule-induced degradation of endogenous proteins
-
•
Genomic analysis identifies direct targets of transcriptional regulators
-
•
Powerful system for interrogating mechanisms of transcriptional control
-
•
Step-by-step protocol from sgRNA design to integration of genomic data sets
Transcriptional changes happen within minutes; however, RNAi or genetic deletion requires days to weeks before transcription networks can be analyzed. This limitation has made it challenging to distinguish direct from indirect targets of sequence-specific transcription factors. This inability to define direct transcriptional targets hinders detailed studies of transcriptional mechanisms. This protocol combines rapid degradation of endogenous transcription factors with nascent transcript analysis to define the earliest, and likely direct, regulatory targets of transcription factors.
Before you begin
We have developed a chemical-genetic workflow that combines the insertion of a degron tag into the endogenous alleles of a transcription factor with established techniques for transcriptional analysis. Using this workflow, precision nuclear run-on sequencing (PRO-seq) is combined with a short degradation time course (0–6 h) of the factor to identify changes in nascent transcription to define the direct gene targets of the factor. Changes in nascent transcription are then compared to genomic localization data to confirm direct regulation. This workflow yields a rigorously defined set of direct transcriptional target genes that can then be used to identify mechanisms by which the transcription factor regulates gene expression.
A limitation of this approach is that it may underestimate the total number of transcriptional targets. For example, for a transcriptional repressor (such as the AML1-ETO fusion protein found in acute myeloid leukemia), targets that were silenced by secondary effects such as H3K27 methylation were not reactivated upon degradation of the repressor in the first 6 h of dTAG-47 treatment. However, because the expression of direct gene targets, which are themselves transcriptional regulators, can be affected within 2 h of transcription factor degradation, changes as early as 6 h post-degradation could represent secondary transcriptional changes resulting from the altered expression of these factors. Therefore, the short time frame is key to identify only direct targets, even though some targets may be missed.
This protocol describes the incorporation of a C-terminal FKBP12F36V degron tag (dTAG) (Nabet et al., 2018), which may not be suitable for all targets. For example, if there are alternative last exons, an N-terminal tag may be required. It is also possible to exogenously express the tagged cDNA while deleting the endogenous gene. Note that most exogenous expression systems express the cDNA of interest at a higher level than the endogenous locus, which could lead to incomplete degradation of the “dTAGged” protein.
Cell line considerations
Cell lines should be carefully chosen before attempting this workflow. Important factors to consider when choosing a cell line include:
-
1.
Expression levels of your protein of interest: This protocol relies on fluorescence activated cell sorting (FACS) and western blotting to establish the edited cell line. The expression level of your protein of interest must be high enough to separate the unedited cell population from the successfully edited, fluorophore positive, cell population by FACS. Additionally, protein levels must be high enough to detect by western blot using antibodies available for the untagged protein. Highly expressed proteins may require further optimization to ensure complete degradation.
-
2.
Expression status of the E3 ubiquitin ligase cereblon (CRBN): This protocol was developed using the dTAG-47 proteolysis targeting chimera (PROTAC), which binds to the FKBP12F36V degron tag and the CRBN E3 ubiquitin ligase (Nabet et al., 2018; Weintraub et al., 2017). If your cell line does not express CRBN, additional PROTACs are available that bind to the Von Hippel-Lindau (VHL) E3 ubiquitin ligase. Moreover, even if both CRBN and VHL ligases are expressed, some targets seem to show a preference for degradation by one E3 ligase over the other (Lai et al., 2016). However, this would need to be determined empirically. Importantly, a PROTAC capable of linking FKBP12F36V to the VHL E3 ubiquitin ligase has recently been developed and is commercially available (Nabet et al., 2020). Thus, by varying the choice of PROTAC (dTAG-47 vs dTAGV-1), FKBP12F36V-tagged proteins can be targeted for either CRBN- or VHL-mediated degradation.
-
3.
Ploidy of your cell line and/or copy number of your target of interest should be considered (see Homology Directed Repair Donor Plasmid Design and Preparation).
gRNA selection
Timing: [1–2 h]
The Type II CRISPR system is used in this protocol. This system uses the Cas9 nuclease, a crRNA encoding the 20-nucleotide guide sequence that targets the ribonucleoprotein (RNP) complex to specific locations in the genome, and a trans-activating crRNA (tracrRNA) (Ran et al., 2013). The crRNA and tracrRNA will be annealed to form a single-guide RNA (sgRNA).
-
4.
The targeting sequence encoded in the crRNA must be immediately 5’ of a NGG PAM sequence.
-
5.
Wild-type Cas9 typically induces cut sites 3–4 bp upstream of the PAM sequence (Ran et al., 2013).
-
6.
sgRNA cutting efficiency and cutting location will determine the success of endogenous integration of the FKBP12F36V degron tag. Optimal cut sites are within 30 bp of the stop codon; the closer the cut site is to the insertion site, the higher the efficiency (Singh et al., 2015).
-
7.
There are a number of publicly available web tools to help design crRNAs for CRISPR/Cas9-directed genome editing, we suggest CHOPCHOP (https://chopchop.cbu.uib.no/) (Labun et al., 2019) as a starting point (Figure 1/Methods video S1). tracrRNAs are typically universal and will work with any crRNA if ordered from the same company.
Figure 1.
gRNA selection
Representative Screenshot from the UCSC genome browser of the RUNX1T1 locus. A custom track of crRNA generated by ChopChop and the NCBI RefSeq tracks have been loaded into the browser. Blue shaded area indicates 30 bp +/- the stop codon. crRNAs are colored according to a ranking metric calculated by CHOPCHOP. Arrows within gRNA indicated strandedness. (∗) marks three candidate crRNA that are most likely to be successful based on proximity of the predicted cut sites to the desire insertion site.
Note: While it may be useful to test two or three crRNAs for cutting efficiency, we have found the location of the crRNA relative to the insertion site is more predictive of successful editing.
Note: The PAM sequence should not be included in the crRNA sequence when ordering oligos.
Homology directed repair donor plasmid design and preparation
Timing: 1–2 weeks
This protocol utilizes dsDNA donor plasmids to facilitate homology directed repair (HDR) (Addgene #104370), in which YY1-specific homology arms were removed to create an “empty vector”. A key component of this cassette is the P2A “self-cleaving peptide” sequence, which causes ribosome skipping. The P2A sequence is located between the 2xHA tag and the mCherry fluorophore creating a single mRNA transcript that is translated into two independent proteins (the dTAGged protein of interest and mCherry). This is important, because mCherry expression requires the integration to be in frame without requiring the fluorophore to be fused to your protein of interest. It is important to note that the following protocol was used to tag a fusion protein in a cell line in which the non-translocated allele is not expressed, thus only one fluorophore was needed. When the targeting of two alleles is required, the addition of a second fluorophore is helpful for the selection of cells which have undergone homozygous editing. This eliminates the need for single cell cloning and more laborious methods for distinguishing homozygous from heterozygous clones. We have had success combining plasmids containing either BFP (Addgene #104371) or eGFP with mCherry in a 1:1 ratio for diploid targets.
-
8.
Generation of the targeting vector involves the cloning of target-specific 5’ and 3’ homology arms to flank the FKBP12F36V-2xHA-P2A-mCherry cassette (Weintraub et al., 2017) (Figure 2).
-
9.Gibson assembly can be used to clone the homology arms. Two genomic PCRs of the sequence flanking the stop codon should be sufficient to generate the homology arms; however, gene blocks can be used as an alternative if necessary.
-
a.The 5’ homology arm should be a minimum of 200 bp and the 3’ homology arm should be ∼ 2–3 times the size of the 5’ homology arm.
-
b.The homology arms can be extended if the gRNA is cutting the locus, but the HDR template is not integrating.
-
a.
-
10.
The PAM sequence should be mutated using site directed mutagenesis to prevent additional cutting after the template is inserted. If the gRNA sequence spans both of the homology arms, it is not necessary to mutate the PAM sequence, because the insertion of the FKBP12F36V-2xHA-P2A-mCherry cassette will disrupt sgRNA binding.
Figure 2.
Homology directed repair donor plasmid design
Schematic depicting insertion of the FKBP12F36V-2xHA-P2A-flourescent protein tag in to the endogenous locus of the target of interest. (UTR = untranslated region, H. Arm = homology arm, FP = fluorescent protein)
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Zombie NIR Fixable Viability Kit | BioLegend | CAT # 423105 |
| α-HA tag [HA.C5] (WB) | Abcam | CAT# ab18181; RRID: AB_444303 |
| αGAPDH (G-9) | Santa Cruz | CAT# sc-365062; RRID: AB_10847862 |
| α-HA-Tag (C29F4) Rabbit mAB (C&R) | Cell Signaling Technology | CAT# 3724S; RRID: AB_1549585 |
| Donkey α-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody | Invitrogen | CAT# 31238; RRID: AB_429690 |
| α-Histone H3 (acetyl K27) antibody-ChIP Grade | Abcam | CAT# ab4729; RRID: AB_2118291 |
| α-H3K4me3 | Abcam | CAT# ab12209; RRID: AB_442957 |
| α-RUNX1 | Santa Cruz | CAT# sc-365644; RRID: AB_10843207 |
| α-PU.1 | Santa Cruz | CAT# sc-390405 |
| Rabbit α-Mouse IgG | Abcam | CAT# ab46540 |
| a-Histone H3 (acetyl K27) antibody-ChIP Grade | Abcam | CAT# ab4729; RRID: AB_2118291 |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI | Fisher Scientific | CAT#MT15040CV |
| FetalPlex | Gemini Bio-Products | CAT#100-602 |
| Corning Penicillin/Streptomycin | Fisher Scientific | CAT#MT30002CI |
| Corning L-glutamine Solution | Fisher Scientific | CAT#MT25005CI |
| dTAG-47 | Vanderbilt Chemical Synthesis Core | Custom dTAG-47 Synthesis |
| DMSO | MilliporeSigma | CAT#D8418; CAS: 67-68-5 |
| Deposited data | ||
| Raw and analyzed data | (Stengel et al., 2021) | GEO: GSE153281 |
| Experimental models: cell lines | ||
| Kasumi-1 | ATCC | CAT#CRL-2724; RRID: CVCL_0589 |
| Kasumi-1-AML1-ETO-FKBP12F36V | (Stengel et al., 2021) | |
| Oligonucleotides | ||
| Alt-R® CRISPR-Cas9 tracrRNA | Integrated DNA Technologies | CAT#1072534 |
| crRNA Runx1t1 (ETO): TCTGAGTTCACGTCTAGCGA | Integrated DNA Technologies | CAT#Alt-R® CRISPR-Cas9 crRNA |
| Recombinant DNA | ||
| pAW62.YY1.FKBP.knockin.mCherry | Addgene | Plasmid #104370 |
| pAW63.YY1.FKBP.knockin.BFP | Addgene | Plasmid #104371 |
| Software and algorithms | ||
| Bedtools (v. 2.22.1) | Quinlan and Hall, 2010 | N/A |
| Bowtie2 (v. 2.2.2) | Langmead and Salzberg, 2012 | N/A |
| Cuffdiff (v. 2.1.1) | Trapnell et al., 2013 | N/A |
| deepTools (v. 3.4.3) | Ramirez et al., 2016 | N/A |
| DESeq2 | Love et al., 2014 | N/A |
| DiffBind | Ross-Innes et al., 2012 | N/A |
| edgeR | McCarthy, Chen and Smyth, 2012 | N/A |
| FASTX toolkit (v. 0.0.13) | hannonlab.cshl.edu | N/A |
| HOMER | Heinz et al., 2010 | N/A |
| Integrative Genomics Viewer | Thorvaldsdottir, Robinson and Mesirov, 2018 | N/A |
| MACS2 peak caller (v. 2.0.10.20131216) | Feng et al., 2012 | N/A |
| Nascent RNA Sequencing Anaylsis (NRSA) | Wang et al., 2018 | N/A |
| Samtools (v. 0.1.19-44428cd) | Li et al., 2009 | N/A |
| TopHat (v. 2.0.11) | Trapnell et al., 2009 | N/A |
| Trimmomatic-0.32 | Bolger et al., 2014 | N/A |
| Venny 2.1 | https://bioinfogp.cnb.csic.es/tools/venny/index.html | N/A |
Step-by-step method details
Electroporation optimization (neon transfection system)
Timing: 1–2 days
Determining the optimal electroporation conditions for each cell line is important to ensure efficient transfection and reduce damage to the cells. This protocol utilizes a fluorophore for selection purposes. Any reference to antibiotics refers to the antibiotics typically included in cell culture media (i.e., Penicillin/Streptomycin) and not antibiotics used for selection. This protocol was optimized for the Kasumi-1 cell line; spin times and speeds should be adjusted for your cell line. We used the Neon electroporation system, and the Thermo Fisher website contains useful hints for optimization (https://tools.thermofisher.com/content/sfs/manuals/neon_device_man.pdf)
Optimization 1
-
1.
Warm E2 buffer, R buffer, and culture medium without antibiotics to 37°C.
-
2.
Prepare a 24-well plate with 1 mL of culture medium without antibiotics per well to transfer electroporated cells to. Place plate in incubator to allow media to equilibrate.
-
3.
Count cells and aliquot 30 million cells to a 50 mL conical tube.
-
4.
Spin down for 5 min at 100 × g ∼20°C.
-
5.
Wash cells twice with 50 mL ∼20°C PBS.
-
6.
Load a Neon tube with 3 mL of 37°C E2 buffer into the Neon Pipette Station.
-
7.
Resuspend cells in 3 mL of 37°C R buffer.
CRITICAL: Do not store the cell suspension in R buffer for more than 30 mins, because this can reduce cell viability and transfection efficiency.
-
8.
Add 60 μg of plasmid expressing a fluorescent protein marker (e.g., GFP) and mix well.
-
9.
Select the optimization option on the Neon and load the optimization protocols. See the Neon website for a chart of the optimization parameters.
-
10.
Slowly load the 100 μL Neon tip with the resuspended cell/plasmid mixture. Make sure there are no air bubbles in the tip before proceeding.
-
11.
Insert the Neon Pipette into the Neon Pipette Station.
-
12.
Select the first protocol in the optimization window and press start to electroporate cells.
-
13.
After electroporation, immediately transfer cells to the 24-well plate with pre-warmed (37°C) culture medium.
-
14.
Repeat steps 9–12 for the remaining optimization protocols.
-
15.
Gently shake plate to evenly distribute cells.
-
16.
Incubate plate in a humidified CO2 incubator at 37°C for 24–48 h.
-
17.Assess cell viability:
-
a.Mix 10 μL Trypan Blue with 10 μL well mixed cell culture.
-
b.Load 10 μL of the mixture onto a hemocytometer.
-
c.Count the number of cells.
-
d.Calculate the percentage of viability (Live Cells/ Total Cells)
-
a.
-
18.Assess transfection efficiency:
-
a.Spin down remaining cells in a micro centrifuge tube.
-
b.Wash once in 500 μL TC-PBS.
-
c.Resuspend in 400 μL 0.5% BSA in PBS.
-
d.Flow for fluorescent marker.
-
a.
Optimization 2 (optional)
Based on the best electroporation conditions from Optimization 1, perform optimization with narrower parameters.
CRISPR/Cas9 electroporation
Timing: [1.5 h]
This step will deliver the CRISPR/Cas9 ribonucleoprotein (RNP) complexes and template DNA to the cells via electroporation. This protocol utilizes fluorophores for selection purposes. Any reference to antibiotics refers to the antibiotics typically included in cell culture media (i.e., Penicillin/Streptomycin) and not antibiotics used for selection.
-
19.In a micro centrifuge tube mix:
-
a.3 μL crRNA (100 μM)
-
b.3 μL tracrRNA (100 μM)
-
c.4 μL of R buffer
-
a.
-
20.
Heat at 95°C for 5 min and let cool to ∼25°C (10–15 min at bench).
-
21.
Prepare RNP Complex by adding 1.5 μL of Cas9 nuclease (10 mg/mL) to cooled gRNA complex.
-
22.Incubate at ∼20°C for 25–30 min to form the RNP complex.
-
a.Use this incubation time to warm Buffer E2 and Buffer R to 37°C, set up the Neon Transfection System, and begin preparing cells for electroporation.
-
a.
-
23.
Aliquot 1.25 ×106 cells per reaction to 15 mL conical tube.
-
24.
Spin down at 100 × g at ∼20°C.
-
25.
Wash twice with ∼20°C PBS.
-
26.
Add 3 mL of 37°C E2 buffer to the Neon tube.
-
27.Set the electroporation parameters:
-
a.Kasumi-1: 1300 V, 30 ms, 1 pulse
-
a.
-
28.
Resuspend cells in 105 μL 37°C R buffer per 1.25 ×106 cells (avoid making air bubbles when pipetting up and down.)
CRITICAL: Do not store the cell suspension in R buffer for more than 30 min because this can reduce cell viability and transfection efficiency.
-
29.
Add 3 mL of 37°C media without antibiotics per reaction to a 6 well plate.
-
30.
Add 12 μg of the HDR plasmid to tube.
Note: If multiple plasmids are being used to introduce different fluorophores an equal amount of each plasmid should be added to total 12 μg.
-
31.
Aliquot 105 μL of cells to each of the tubes and gently pipette up and down to mix.
-
32.
Using 100 μL Neon tip, draw up the cell/plasmid/RNP mixture
-
33.
Insert the pipette tip directly into the E2 buffer in the plastic cuvette. Do not touch the walls.
-
34.
Press start.
-
35.
After the protocol is complete, remove the pipette and dispense contents into a well with pre-warmed (37°C), equilibrated media without antibiotics.
-
36.
Incubate in a humidified CO2 incubator at 37°C for 24 h, then check viability and replace the media with culture media + penicillin and streptomycin.
-
37.
Expand cells for FACS.
Fluorescence-activated cell sorting
Timing: [1–2 weeks]
The incorporation of fluorescent proteins into the cassette for homology directed repair allows for selection of positive cells by fluorescence-activated cell sorting (Figure 3). Fluorophore positive cells are readily detectable 4 days post-electroporation, with maximal positivity observed by 7 days post-electroporation. If possible, it is recommended to assess editing efficiency by flow cytometry in advance to determine how many cells will need to be sorted. Electroporation can damage cells which may result in increased auto fluorescence while the cells are recovering. Using a viability dye, such as Zombie-NIR, may be beneficial to reduce auto-fluorescence associated with dying cells. Depending on the efficiency of the editing, it may take a few weeks for cells to recover and expand to the point where they can be used for subsequent experiments.
-
38.
Count cells.
-
39.
Spin down cells at 100 × g for 5 min at 4°C.
-
40.
Optional: Zombie Viability Stain
-
i.
Resuspend cells in 100 μL 1:2000 Zombie NIR in PBS per million cells
-
ii.
Incubate for 15 min at 4°C in the dark.
-
iii.
Spin at 100 × g for 5 min at 4°C
-
41.
Resuspend in 1 mL of 0.5% BSA in PBS per 10 million cells
-
42.
Transfer to strainer capped flow tube.
-
43.
Swap strainer cap for standard cap.
-
44.
Sort.
Note: Cells positive for only one fluorophore can also be sorted and saved for use as compensation controls for later flow cytometry experiments.
-
45.
Spin down cells at 100 × g for 5 min @ 4°C.
-
46.
Dilute sorted cells to 0.5×106 cells/mL in culture media.
-
47.
Incubate in a humidified CO2 incubator at 37°C. Expand sorted cells until you have enough to freeze down.
CRITICAL: Following expansion of the cells post-sort, cells should be reanalyzed for fluorophore expression. If the sorted cell population is less than 95% positive for fluorophore expression (95% of the cells should be positive for both fluorophores if targeting both alleles), a second sort is recommended. In these cases, it is important to be very conservative with gating. For example, sorting only the brightest 30% of fluorophore positive cells.
Figure 3.
Fluorescence-activated cell sorting of edited cells
Representative flow cytometry plots of mCherry and BFP expression from endogenous AML1-ETO locus after integration of donor DNA. Note: AML1-ETO is monoallelic and the remaining ETO allele is not expressed, the double positive population is a result of auto fluorescence after electroporation.
Western blot to establish the degradation time course
Timing: [2–3 days]
The PROTAC, dTAG-47, is a heterobifunctional molecule consisting of a thalidomide derivative and a FKBP12F36V selective ligand that is connected by a linker. Thus, treatment with dTAG-47 results in the selective recruitment of the FKBP12F36V degron tagged protein to CRBN to facilitate proteasome-mediated degradation (Figure 4A) (Nabet et al., 2018; Weintraub et al., 2017). dTAG-47 is not yet commercially available; however, the structure and synthesis protocol have been published elsewhere (Weintraub et al., 2017). The related, but less potent, dTAG-13 small molecule is chemically similar to dTAG-47 (Nabet et al., 2018) and is commercially available.
Figure 4.
Establishing the degradation time course
(A) Schematic of the dTAG system showing how the FKBP12F36V tag directs the chimeric protein to the proteosome.
(B) Western blot analysis of whole-cell lysates from Kasumi-1 AML1-ETO-FKBP12F36V-2xHA cells treated with dTAG-47 for 0, 0.5, 1, 2, 6, and 24 h. AML1-ETO- FKBP12F36V-2xHA was rapidly degraded after addition of dTAG-47. GAPDH served as a loading control. A titration of the untreated (0 h) lysate was included for comparison.
The kinetics of protein degradation are highly dependent on the protein target and cell line. Some proteins are completely degraded within 30 min of dTAG-47 addition, while other proteins take up to 6 h for full degradation. This protocol uses dTAG-47 at a final concentration of 500 nM. However, the optimal concentration of dTAG-47 should be determined for each cell line and protein target, as too little dTAG molecule may result in incomplete degradation, while too much can be associated with the “hook effect” and also limit optimal degradation (Pettersson and Crews, 2019).
Once sorted, western blot analysis is used to confirm that the correct alleles were tagged, the protein produced is tagged, and to determine if tag incorporation compromised protein stability. Incorporation of the FKBP12F36V-2xHA-P2A-mCherry cassette adds ∼13 kD to your protein resulting in slower migration on a gel than the untagged protein. A time course of 0, 30 min, 1, 2, 4, 8 and 24 h is recommended to identify 2–3 time points for nascent RNA transcription analysis. A serial dilution (e.g., 10%, 20%, 50%) of the 0 h lysate should be included to determine the lower detection limit of the antibody and approximate protein levels after dTAG-47 treatment (Figure 4B).
-
48.
Harvest 14 ×106 tagged cells (2 ×106 per time point).
-
49.
Dilute to 4×105 cells per mL in culture media.
-
50.
Aliquot 5 mL of diluted cells to 7 T25 flasks.
-
51.
Add 5 μL of 500 μM dTAG-47 (final concentration 500 nm) to the flasks 24 h, 8 h, 4 h, 2 h, 1 h and 30 min before harvesting the protein for western blot analysis (do not treat the 0 h control).
Note: We recommend blotting for both HA (Abcam, 1:1000) and the endogenous protein to ensure that the majority of the protein is tagged as well as to compare expression levels of the endogenous and tagged proteins.
Temporal analysis of transcriptional changes
Timing: [1–7 days]
Mechanistic studies of transcriptional regulators have been complicated by a lack of experimental approaches and reagents that can assay changes in transcription before compensatory or secondary changes in gene expression take effect. Incorporation of the FKBP12F36V tag removes several barriers to defining the direct effects resulting from the loss of a transcriptional regulator. Rapid degradation of endogenous proteins allows for transcriptional analysis immediately after loss of the protein, which circumvents issues with traditional methodologies such as compensation or changes in cell phenotype (e.g., differentiation, cell cycle arrest, apoptosis). Furthermore, the inclusion of the HA epitope tag eliminates the need for a high-quality antibody to the endogenous protein for genome-wide localization analysis such as ChIP-seq or CUT&RUN. Below we describe the ideal set of experiments to define transcriptional targets and interrogate mechanisms of transcriptional control. Detailed protocols for the techniques used have been published elsewhere, and any adjustments made to publicly available protocols are described here.
Note: All sequencing experiments should be performed in biological duplicate.
CRITICAL: For all time courses discussed here, the endpoint is considered to be when the cells are first harvested. Therefore, it is very important to process samples as quickly as possible.
CRITICAL: Method of normalization should be considered while planning an experiment. Spike-in controls are essential for experiments in which you expect global changes in the factor you are measuring. We use nuclei (prepared in house) from the Drosophila melanogaster cell line, Schneider 2 (S2), as spike-in controls for PRO-seq and ChIP-seq experiments and fragmented DNA from Escherichia coli (available from EpiCypher) for CUT&RUN experiments. If you are expecting a small number of changes, normalization may be calculated by using algorithms such as DESeq2 or possibly based on read counts for each experimental condition (e.g., reads per million).
PRO-seq
Precision nuclear run-on transcription coupled with deep sequencing (PRO-seq) (Mahat et al., 2016) was selected to measure changes in nascent transcription over the first 6 h of the degradation time course. PRO-seq was selected because it generates strand-specific maps of transcriptionally engaged RNA polymerases, which can be analyzed to identify changes in gene body transcription as well as changes in promoter-proximal pausing, elongation rates, and eRNA production. Therefore, PRO-seq provides a deep understanding of the mechanisms by which a given protein contributes to transcriptional control. In our initial experiment, PRO-seq was performed at 0, 0.5, 1, 2, and 6 h after addition of dTAG-47. These time points were selected based on western blot analysis of AML1-ETO degradation (Figure 4B). We recommend choosing at least two to three of the earliest time points at which the protein is maximally degraded. We do not recommend performing longer PRO-seq time courses for the purpose of identifying direct transcriptional targets, as indirect transcriptional effects will occur if your factor regulates other transcription factors or epigenomic modifiers.
RNA-seq
While PRO-seq measures changes in nascent transcription rates, RNA-seq measures changes in the cytoplasmic pools of mRNA. There are a number of mechanisms that regulate the processing of nascent transcripts into translatable mRNA. We recommend performing an RNA-seq time course after degradation of your protein target to ensure that transcriptional targets identified by changes in nascent transcription contribute to biologically meaningful changes in mRNA levels. It takes longer for changes in transcription to affect the cytoplasmic pools of mRNA, so a longer time course is needed (e.g., 0, 4, 8, and 24 h after dTAG-47 addition). A degradation time course paired with RNA-seq will still identify waves of gene expression changes but will lack the resolution to make meaningful mechanistic conclusions or separate direct and indirect targets.
CUT&RUN/ChIP-seq
Identification of DNA binding sites by ChIP-seq or CUT&RUN (Skene et al., 2018) can further increase the confidence that identified targets are indeed directly regulated by the protein of interest. Most importantly, these datasets can be overlaid with genomic localization of histone marks, histone modifying proteins, or other transcriptional regulators to begin to dissect the mechanism by which the transcription factor of interest controls transcription. We suggest using CUT&RUN for most genomic localization experiments, because it requires fewer cells and less sequencing depth than ChIP-seq and can be performed without formaldehyde crosslinking. However, currently ChIP-seq remains the best technique for assessing H3K27ac levels throughout the genome. The time points selected for these experiments will depend on the purpose of the experiment. Experiments focused on identifying binding sites for your protein of interest do not require a thorough time course. For this type of experiment, we compare untreated samples to those treated at a time point where your protein is maximally degraded (i.e., 8 or 24 h after addition of dTAG-47). This system is ideal, because it allows the uses the same antibody in the experimental and control samples instead of IgG to identify non-specific binding of the antibody. Experiments focused on identifying mechanisms of transcriptional regulation (e.g., histone marks, histone modifying enzymes, or other transcriptional regulators) should be performed as an extensive time course with early time points matching your PRO-seq time points and extended to 24 h. Experiments using antibodies to targets other than the dTAGged protein should include an IgG control. The purpose of both the dTAG treated and IgG controls is to identify peaks that are not specific to the antibody used. These peaks can correspond to non-specific binding sites or areas of open chromatin that are hyperaccessible to the MNase.
We recommend using the EpiCypher protocol (https://www.epicypher.com/content/documents/protocols/cutana-cut&run-protocol.pdf), which is publicly available and routinely updated. This protocol is based off of the protocol published by Skene, Henikoff, and Henikoff in 2018, but it has been further optimized for reproducibility and is performed in PCR tubes which enables a more rapid workflow. We recommend including a secondary antibody incubation in all CUT&RUN experiments. After the primary antibody incubation (step 15 in EpiCypher CUT&RUN Protocol v1.6):
-
52.
Place the tube on a magnet until slurry clears and pipet to remove supernatant.
-
53.
While beads are on the magnet, add 250 μL cold Digitonin Buffer directly onto beads of each sample, and then pipet to remove supernatant.
-
54.
Repeat previous step for total of two washes.
-
55.
Add 50 μL cold Antibody Buffer to each tube
-
56.
Add 1:200 secondary antibody and incubate for 1 h at 4°C.
-
57.
Proceed with step 16 of EpiCypher CUT&RUN Protocol v1.6.
Antibodies and incubation times that we have successfully used for CUT&RUN and ChIP-seq are listed in Table 1(supplier and catalog information can be found in the Key resources table).
Table 1.
Validated CUT&RUN/ChIP-seq antibodies
| Antibody | Concentration | Incubation Conditions | Technique |
|---|---|---|---|
| α-HA-Tag Rabbit mAB | 1:800 | Overnight, 4°C | CUT&RUN |
| α-PU.1 Mouse mAB | 1:100 | Overnight, 4°C | CUT&RUN |
| α-RUNX1 Mouse mAB | 1:100 | Overnight, 4°C | CUT&RUN |
| Donkey α-Rabbit IgG | 1:200 | 1 h, 4°C | CUT&RUN |
| Rabbit α-Mouse IgG | 1:200 | 1 h, 4°C | CUT&RUN |
| α-Histone H3K4me3 | 1:100 | Overnight, 4°C | CUT&RUN |
| α-Histone H3K27ac | 1 μg/1 million cells | Overnight, 4°C | ChIP-seq |
Note: For CUT&RUN time courses, we recommend including a light crosslinking (0.1% formaldehyde for 5 min.) to prevent additional changes during the overnight primary antibody incubation.
Data analysis
A general workflow for analyzing genomic datasets is depicted in Figure 5. There are a number of publicly available software packages available for each step. We have summarized the software packages we commonly use in Table 2.
Figure 5.
Analyzing genomic data
A general workflow for processing the genomic data sets generated by this protocol. The software programs required to complete each step are listed in Table 2.
Table 2.
Summary of software used to analyze genomic data sets
| Step | Software | Language | PS | RS | C&R/ChIP | Citation |
|---|---|---|---|---|---|---|
| Trim adaptors | Trimmomatic | Command line | X | X | X | (Bolger et al., 2014) |
| Reverse Complement | FASTX | Command line | X | http://hannonlab.cshl.edu | ||
| Alignment | BowTie2 | Command line | X | X | (Langmead and Salzberg, 2012) | |
| Alignment | TopHat | Command line | X | (Trapnell et al., 2009) | ||
| Processing SAM/BAM files | Samtools | Command line | X | X | X | (Li et al., 2009) |
| Peak calling | Macs2 | Command line | X | (Feng et al., 2012) | ||
| Differential analysis | Nascent RNA sequencing analysis (NRSA) | Command line | X | (Wang et al., 2018) http://bioinfo.vanderbilt.edu/NRSA/#pause-PROseq | ||
| Differential analysis | Cuffdiff | Command line | X | (Trapnell et al., 2013) | ||
| Differential analysis | edgeR | R | X | (McCarthy et al., 2012) | ||
| Differential analysis | DEseq2 | R | X | (Love et al., 2014) | ||
| Differential analysis | DiffBind | R | X | (Ross-Innes et al., 2012) | ||
| Peak annotation/motif analysis | HOMER | Command line | X | X | (Heinz et al., 2010) | |
| Processing BED files | Bedtools | Command line | X | (Quinlan and Hall, 2010) | ||
| Data visualization | Venny 2.1 | Web Application | X | X | X | https://bioinfogp.cnb.csic.es/tools/venny/index.html |
| Data visualization | HOMER | Command line | X | X | (Heinz et al., 2010) | |
| Data visualization | deepTools | Command line | X | (Ramirez et al., 2016) | ||
| Data visualization | Integrative Genomics Viewer (IGV) | Desktop Application | X | X | (Thorvaldsdottir, Robinson, and Mesirov, 2018) |
The table describes the software packages we use for each step of analyzing the genomic data sets generated by this protocol, the coding language required to run the software, the techniques each software package can be applied to, and citations for the manuals that describe these packages. (PRO-seq = PS, RNA-seq = RS, CUT&RUN = C&R, and ChIP-seq = ChIP)
Data interpretation: identifying direct transcriptional targets
To identify direct transcriptional targets, we intersect our PRO-seq and CUT&RUN data to identify genes that have changes in nascent transcription, that translate into similar changes in mRNA levels, and that are bound by the transcription factor of interest. This section describes how we identify direct transcriptional targets and can be applied to both genes and enhancers.
PRO-seq: The 0 h control files should be compared to the files from each experimental time point individually when using NRSA. The pause_PROseq.pl command from NRSA performs a number of functions including calculating the differential expression of all expressed genes. The eRNA.pl command performs a similar function to identify intergenic eRNA. These commands generate tables containing differential expression data that are saved to files called gb_change.txt and Enhancer_change.txt, respectively, in the specified output directory (Wang et al., 2018). These files can be loaded into R, Python, and Microsoft Excel for further analysis. We use the gb_change.txt and Enhancer_change.txt files for the following analysis:
Note: The eRNA.pl command also generates an Enhancer_info.txt file which contains annotations to the closest gene as well as intersection with the FANTOM5 and 4DGenome datasets.
-
58.
Identify significantly changed genes or eRNA (fold change > 1.5, padj <0.05, calculated by NRSA) for each time point.
-
59.Overlap the significantly changed genes or eRNA from each time point (Figure 6A).
-
a.Gene body changes: Overlaps in gene body changes can be identified using R or Python to intersect the transcript IDs or gene names of the significantly changed genes identified in step 49. The Venny 2.1 web application provides a nice tool to perform the same analysis for those without knowledge of R or Python.
-
b.eRNA changes: Overlaps in eRNA changes can be identified using the bedtools intersect command or HOMER’s mergePeaks command with option –d given to identify literal overlaps in the genomic regions encoding eRNAs.
-
a.
-
60.
Select genes or eRNA that are significantly changed in at least 2 time points (Figure 6B).
Note: We use two time points here instead of all time points because the dynamics of transcription will vary by loci and it is common for some gene targets to miss the significance thresholds in the earliest time point.
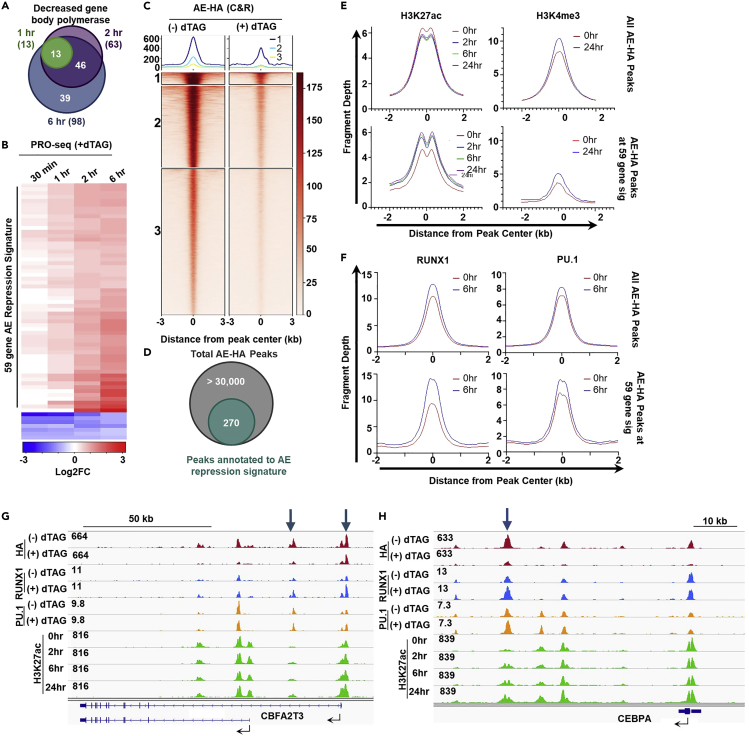
Figure 6.
Temporal analysis of transcriptional changes
Genes identified as differentially expressed (fold change > 1.5, padj < 0.05, calculated by NRSA) in each PRO-seq time point were overlapped (A). Genes that are differentially expressed in at least 2 PRO-seq time points were compiled into a candidate gene set. The heat map (B) shows the log2(Fold Change) of the candidate gene set in each time point. Log2(Fold Change) was calculated by NRSA and the heat map was produced in R. The fragment depth around AML1-ETO CUT&RUN binding sites that are identified as differentially bound (fold change > 1.5, padj < 0.05, calculated by DiffBind) are shown in heat maps made with deepTools (C). Differentially bound sites were annotated with HOMER and overlapped with the 59 gene AML1-ETO repression signature to identify binding sites that correspond to transcriptional changes (D). This identified 270 peaks that correlated with transcriptional changes. The binding sites that were annotated to the AML1-ETO repression signature were overlapped with ChIP-seq/CUT&RUN data (+/- dTAG-47) for histone modifications associated with transcriptional activation, H3K27ac and H3K4me3 (E), and transcription factors RUNX1 and PU.1 (F) to identify mechanisms of transcriptional activation after degradation of AML1-ETO. Fragment depth was calculated using HOMER and graphs were produced in PRISM. We have determined that although multiple peaks may annotate to the same gene, many of these peaks are non-regulatory. For example four peaks are annotated to CBFA2T3 (G) but only the peaks at the promoter and at the intronic enhancer (blue arrows) show recruitment of RUNX1 and corresponding accumulation of H3K27ac following AML1-ETO degradation. Similarly multiple peaks were annotated to CEBPA (H) but only the distal enhancer (blue arrow) shows increased occupancy of RUNX1 and PU. 1 and subsequent accumulation of H3K27ac. Images show bedGraph files produced with HOMER loaded into IGV for visualization. Figure parts reprinted with permission from (Stengel et al., 2021).
CUT&RUN/ChIP-seq: Using the Macs2 callpeak command will generate a peak file that can be loaded into DiffBind with the corresponding BAM files. The 0 h control files should be compared to the files from each experimental time point individually when using DiffBind. After processing the files with DiffBind, you can generate a report containing the differential binding data for each peak using the dba.report function. We use the output of this report for the following analysis:
-
61.
Identify peaks that are significantly (fold change > 1.5, padj <0.05, calculated by DiffBind) reduced after dTAG-47 treatment (Figure 6C).
-
62.Identify significantly changed peaks that correspond to significantly changed genes in at least two time points (identified in step 51) (Figure 6D):
-
a.Determine which gene is most likely to be associated with a given peak by nearest neighbor annotation. (This can be done using HOMER’s annotatepeaks.pl command).
-
b.Overlap nearest neighbor annotation with the significantly changed gene list using R, Python, or Venny.
-
a.
Note: Nearest neighbor annotation is a prediction tool and does not represent empirically validated data.
-
63.Identify significantly changed peaks that correspond to significantly changed eRNAs in at least two time points (identified in step 51):
-
a.Overlaps in significantly changed eRNAs and significantly changed peaks can be identified using the bedtools intersect command or HOMER’s mergePeaks command with option –d given to identify literal overlaps in the genomic regions identified as peaks and those identified as eRNA.
-
a.
RNA-seq
-
64.
Genes that have been determined to be both significantly changed in at least two PRO-seq time points and have identified binding sites can be intersected with genes significantly changed (fold change > 1.5, padj <0.05, calculated by Cuffdiff, DESeq2, edgeR, or similar package) by RNA-seq.
The binding sites identified in steps 53 and 54 can be intersected with CUT&RUN or ChIP-seq time courses for histone modifications (Figure 6E) or other transcriptional regulators (Figure 6F) to gain a mechanistic understanding of how transcription dynamics change after your protein of interest is degraded. Although multiple binding sites may annotate to the same gene, not all of these peaks are regulatory. Studying changes in histone modifications and the occupancy of other transcriptional regulators can help separate regulatory (Figures 6G and 6H, marked by blue arrows) from non-regulatory peaks.
Expected outcomes
Development of edited cell lines
CRISPR/Cas9 mediated homology directed repair with a dsDNA donor plasmid is much less efficient than CRISPR/Cas9 mediated non-homologous end joining. While efficiency will be different for every crRNA, locus, and cell line, the expected pool of positive cells will be 0.5%–15% for single positive populations (Figure 3). Double positive populations (i.e., homozygous integration) will be lower. Please note that if the cassette is not inserted, crRNA/Cas9-induced DNA cutting will continue until non-homologous end joining causes mutations that impair the binding of the crRNA. As such, re-electroporating cell lines with only one allele edited with the same sgRNA is inefficient. Successful integration of the FKBP12F36V-2xHA-P2A-mCherry cassette is required for production of the fluorescent protein and will add approximately 13 kDa to your protein of interest.
Establishing the degradation time course
The kinetics of protein degradation will be dependent on the target protein and the E3 ligase. Certain target proteins may be more efficiently degraded with one E3 ligase (CRBN or VHL) than the other. Once the optimal PROTAC and dose have been chosen, degradation of the target protein should occur quickly with maximal protein degradation within 6 h of treatment.
Transcriptional analysis
The size of the transcriptional network identified will vary depending on the transcription factor. It is critical to use short time points following protein degradation coupled with nascent transcription to identify direct targets and functional DNA binding sites. One should expect many fewer direct targets than one observes using slower approaches (e.g., RNAi, CRISPR-mediated deletion) that will lead to the detection of both direct and indirect transcriptional changes.
Limitations
Development of edited cell lines
While this protocol has worked well for a number of different gene targets, there are some caveats that could limit success. The homology directed repair strategy requires a high-quality gRNA with a cut site near the stop codon. This may limit the feasibility of targeting certain loci. Additionally, some cell lines are extremely sensitive to dsDNA which may impact the practicality of this protocol in those cell lines.
Establishing the degradation time course
Addition of the FKBP12 tag may alter the stability of the tagged protein. The decreased protein levels or disruption of protein function by the addition of a C-terminal tag may cause negative selection with certain proteins of interest. We have targeted 10 different genes with this approach and negative selection has only been a possible issue with one target. We have not tested an N-terminal tagging strategy.
Transcriptional analysis
This approach may underestimate the total number of transcriptional targets. The short time course used in this protocol is critical to separate the direct and indirect transcriptional targets. However, there are many processes that regulate transcription that may occur on different time scales. In our experience, targets that were silenced by secondary effects such as H3K27 methylation were not reactivated upon degradation of the transcriptional repressor AML1-ETO in the first 6 h of dTAG-47 treatment.
Troubleshooting
Problem 1
There are no high quality gRNA that cut close to the stop codon of my target (Before you begin: gRNA selection).
Potential solution
Using the SpCas9D10A nickase can increase the efficiency of homology directed repair when the cut site is farther away from the desired insertion site.
Note: The SpCas9D10A nickase requires two gRNA to induce double stranded breaks (this design strategy is supported by CHOPCHOP).
Problem 2
My cells are dying after electroporation with the RNP complexes and donor plasmids (after electroporation optimization, step 37).
Potential solution
Some cell lines are very sensitive to cytosolic DNA and the donor plasmids can trigger an interferon response. Titrating the total concentration of DNA may decrease the amount of death. Ensuring that buffers are kept warm, spins are performed at ∼20°C, and antibiotics are not present in the media will also promote cell viability following electroporation. If a large number of your cells are dying, you should replace the media routinely with conditioned media from healthy cells. It is also important to ensure that your cells remain at the appropriate density for log phase growth and may need to be moved to a smaller plate in the days following electroporation. Finally, if suitable electroporation conditions cannot be identified, standard transfection of plasmids expressing Cas9 and sgRNAs can be used (Weintraub et al., 2017).
Problem 3
My parental (untagged) cell line has background auto-fluorescence, making it difficult to identify the double positive population (step 44).
Potential solution
Using a viability dye such as Zombie NIR can minimize the amount of auto-fluorescence caused by dead cells. Another option is to include an additional fluorophore when gating cells because most auto-fluorescence is detectable in multiple channels. This will allow you to separate background fluorescence from positive cells. In addition, cells should be reassessed for fluorophore positivity several days following sorting. If the cultures are not at least 95% positive for fluorophore expression, a second sort is recommended. In these cases, it is important to be very conservative with gating. For example, sort only the brightest 30% of fluorophore positive cells.
Problem 4
I don’t have a high quality antibody to my protein of interest or there are off target bands/degradation products at the same molecular weight of the untagged protein. How can I confirm that all alleles are tagged (step 51)?
Potential solution
A genomic PCR of the region around the stop codon can help determine if there are untagged alleles in your sorted double positive population. The primers should be within the 5’ and 3’ homology arms (a minimum of 100 bp away from each other). The FKBP12F36V-2xHA-P2A-mCherry cassette is approximately 1,200 bp. This design will amplify both the tagged and untagged locus, which will allow you to determine if there is a large portion of cells with a wild type allele.
Problem 5
My protein is not fully degraded after dTAG-47 treatment (step 51).
Potential solution
The amount of dTAG-47 needs to be titrated to ensure optimal degradation. In our experience, the degradation is dramatic with maximal degradation occurring within the first 6 h of treatment. Other groups have reported differential substrate selectivity based on the E3 ligase recruited by the PROTAC (Lai et al., 2016), therefore, it is possible that a VHL E3 ligase PROTAC (e.g., dTAGV-1) may work better.
Problem 6
A good, but not perfect, overlap is observed between genes annotated in the CUT&RUN datasets and the PRO-seq degradation time course (step 62/63).
Potential solution
Many annotation algorithms use nearest-neighbor assignments, which may mis-assign enhancer-promoter pairs when genes are clustered. Because this chemical-genetic system yields small, rigorously defined, transcriptional circuits, we manually curate all of our datasets to ensure that the nearest neighbor assignments are correct. If a larger network is identified, chromosome conformation capture analysis may be needed to identify enhancer-promoter pairs.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the technical contact hillary.layden@vanderbilt.edu and the lead contact (kristy.r.stengel@vanderbilt.edu).
Material availability
This study did not generate new reagents.
Data and code availability
This study did not generate/analyze [datasets/code].
Acknowledgments
We thank the members of the Hiebert lab for helpful discussions, reagents, and advice. We thank the VICC Flow Cytometry, Chemical Synthesis, and Genome Sciences Shared Resources for services and support. This work was supported by the T.J. Martell Foundation, the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, the National Institutes of Health (grants RO1- CA178030, RO1-CA164605, and R01-CA64140), core services performed through Vanderbilt Digestive Disease Research grant NIDDK P30DK58404 and Vanderbilt-Ingram Cancer Center support grant NCI P30CA68485, and the National Center for Advancing Translational Sciences (grant 2 UL1 TR000445-06). H.M.L. and K.R.S. were supported by NCI grant 5 T32 CA009582. H.M.L. was supported by NHBLI NRSA fellowship F31HL156565. K.R.S. was supported by American Cancer Society postdoctoral fellowship PF-13-303-01-DMC.
Author contributions
Conceptualization, K.R.S. and S.W.H.; methodology, K.R.S.; investigation, H.M.L. and N.A.E.; writing – original draft, H.M.L.; writing – review & editing, H.M.L, K.R.S., and S.W.H.; visualization, H.M.L; supervision, K.R.S. and S.W.H.; funding acquisition, S.W.H.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100530.
Contributor Information
Hillary M. Layden, Email: hillary.layden@vanderbilt.edu.
Scott W. Hiebert, Email: scott.hiebert@vanderbilt.edu.
Kristy R. Stengel, Email: kristy.r.stengel@vanderbilt.edu.
References
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.C., Toure M., Hellerschmied D., Salami J., Jaime-Figueroa S., Ko E., Hines J., Crews C.M. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat D.B., Kwak H., Booth G.T., Jonkers I.H., Danko C.G., Patel R.K., Waters C.T., Munson K., Core L.J., Lis J.T. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat. Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B., Ferguson F.M., Seong B.K.A., Kuljanin M., Leggett A.L., Mohardt M.L., Robichaud A., Conway A.S., Buckley D.L., Mancias J.D. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-18377-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B., Roberts J.M., Buckley D.L., Paulk J., Dastjerdi S., Yang A., Leggett A.L., Erb M.A., Lawlor M.A., Souza A. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018;14:431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M., Crews C.M. PROteolysis TArgeting Chimeras (PROTACs) — past, present and future. Drug Discov. Today Technol. 2019;31:15–27. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Schimenti J.C., Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene P.J., Henikoff J.G., Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 2018;13:1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- Stengel K., Ellis J., Spielman C., Bomber M., Hiebert S. Definition of a small core transcriptional circuit regulated by AML1-ETO. Mol. Cell. 2021;81:530–545. doi: 10.1016/j.molcel.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2018;19:506–523. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao Y., Zhou X., Hiebert S.W., Liu Q., Shyr Y. Nascent RNA sequencing analysis provides insights into enhancer-mediated gene regulation. BMC Genomics. 2018;19:633. doi: 10.1186/s12864-018-5016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–1588. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze [datasets/code].