Neuronal autophagosomes form in the distal axon and mature via lysosomal fusion during transport to the soma. Dynein regulators JIP1, HAP1, and JIP3 function on autophagosomes depending on location and autophagosomal maturity. In this pathway, transport and maturity are tightly linked to maintain neuronal health.
Abstract
Autophagy is a degradative pathway required to maintain homeostasis. Neuronal autophagosomes form constitutively at the axon terminal and mature via lysosomal fusion during dynein-mediated transport to the soma. How the dynein–autophagosome interaction is regulated is unknown. Here, we identify multiple dynein effectors on autophagosomes as they transit along the axons of primary neurons. In the distal axon, JIP1 initiates autophagosomal transport. Autophagosomes in the mid-axon require HAP1 and Huntingtin. We find that HAP1 is a dynein activator, binding the dynein–dynactin complex via canonical and noncanonical interactions. JIP3 is on most axonal autophagosomes, but specifically regulates the transport of mature autolysosomes. Inhibiting autophagosomal transport disrupts maturation, and inhibiting autophagosomal maturation perturbs the association and function of dynein effectors; thus, maturation and transport are tightly linked. These results reveal a novel maturation-based dynein effector handoff on neuronal autophagosomes that is key to motility, cargo degradation, and the maintenance of axonal health.
Introduction
As some of the longest-living cells in the body, neurons require continuous clearance and recycling of proteins and organelles to maintain homeostasis and function. Macroautophagy is a major conserved degradation pathway essential for neuronal health (Kulkarni et al., 2018). Genetic disruption of autophagy in neurons is sufficient to induce neurodegeneration in mice (Hara et al., 2006; Komatsu et al., 2006). Deficits in neuronal autophagy are observed in most neurodegenerative diseases, and disease-causing mutations occur in proteins involved in autophagy, including PINK1 in Parkinson’s disease, TBK1 in amyotrophic lateral sclerosis, and Huntingtin (Htt) in Huntington’s disease (Wong and Holzbaur, 2015).
The extended and highly polarized morphology of neurons poses challenges for intracellular trafficking pathways, like autophagy. Neuronal autophagosomes form at presynaptic sites and axon terminals then traverse the long distance to the soma via processive retrograde motility along axonal microtubules (MTs; Maday et al., 2012; Stavoe et al., 2016; Neisch et al., 2017). MTs are organized in a polarized array in axons with minus ends directed toward the soma (Heidemann et al., 1981); thus, the primary minus-end–directed MT motor dynein drives their retrograde motility. A new class of dynein regulatory proteins, known as activating adaptors, (1) stabilize the interaction between dynein and its obligate partner dynactin, (2) align dynein’s two motor domains into an active parallel conformation required for efficient stepping along the MT, and (3) link the motor to specific cargos (Gill et al., 1991; McKenney et al., 2014). Activating adaptors for some cargos (e.g., Rab6 vesicles and signaling endosomes; Matanis et al., 2002; Olenick et al., 2019), have been identified, but the activating adaptor required for autophagosomal motility has not. During transport to the soma, autophagosomes mature by fusing with lysosomes (Maday et al., 2012), altering their membrane composition and further complicating motor regulation.
c-Jun N-terminal kinase–interacting protein 1 (JIP1) interacts with the autophagosomal membrane protein light chain 3 (LC3), dynactin, and the plus-end–directed MT motor kinesin-1 and is required for the initiation of autophagosomal transport from the distal tip of dorsal root ganglion (DRG) neurites (Fu and Holzbaur, 2013; Fu et al., 2014). Htt and its interacting partner, Htt-associated protein 1 (HAP1), are also involved in autophagosomal transport in DRGs, though the underlying mechanism is unknown (Wong and Holzbaur, 2014). Htt can bind dynein, dynactin, and LC3, and HAP1 can bind dynactin and kinesin-1 (Engelender et al., 1997; Li et al., 1998; McGuire et al., 2006; Caviston et al., 2007; Ochaba et al., 2014). c-Jun N-terminal kinase–interacting protein 3 (JIP3; Sunday driver/SYD; UNC-16) also interacts with kinesin-1, dynein, and dynactin and was recently shown to facilitate transport of autophagosomes in Caenorhabditis elegans axons (Cavalli et al., 2005; Arimoto et al., 2011; Cockburn et al., 2018; Hill et al., 2019).
Here, we identify multiple dynein effectors bound to autophagosomes during transit along the axon. Surprisingly, different effectors are required depending on location and maturation state. JIP1 colocalizes primarily with nascent autophagosomes in the distal axon, consistent with its role in initiating retrograde transport. HAP1 and Htt are required for autophagosomal motility specifically in the mid-axon. Complementary in vitro and cellular assays show that HAP1 activates dynein and contains canonical and noncanonical determinants for binding dynein–dynactin. JIP3 colocalizes with autophagosomes throughout the axon but regulates the motility of mature autolysosomes. While inhibition of motility is known to disrupt autophagosomal maturation (Wong and Holzbaur, 2014), we find that genetic or pharmacological inhibition of autophagosomal maturation likewise perturbs transport. Together, these results illustrate a novel coordination mechanism between autophagosomal maturation state and motor regulation by effector proteins.
Results
Multiple dynein effectors associate with axonal autophagosomes
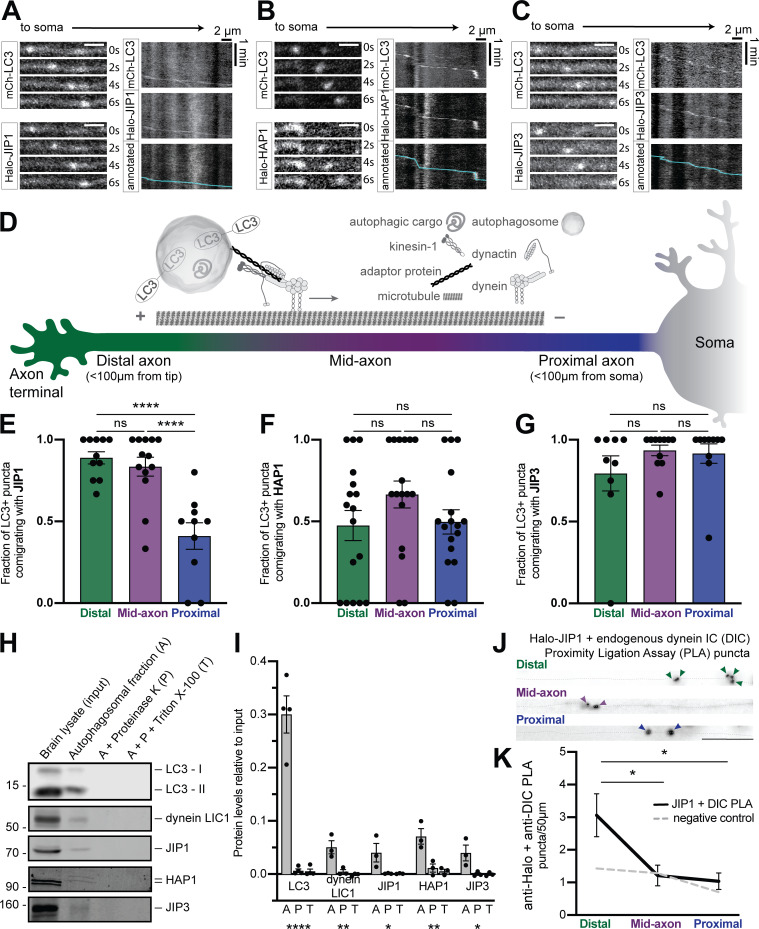
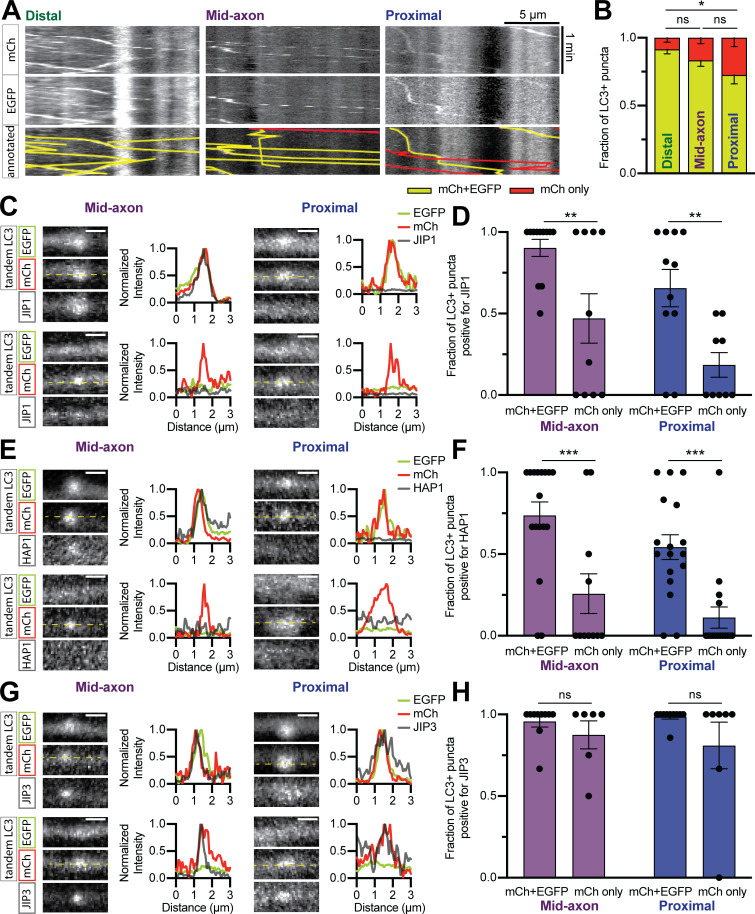
To investigate the association of dynein regulators with axonal autophagosomes, we assayed the colocalization of JIP1, HAP1, and JIP3 with the autophagosome marker LC3 in the axons of primary rat hippocampal neurons. We coexpressed Halo-tagged versions of each effector with mCherry (mCh)-EGFP-LC3B and imaged neurons at 6–8 d in vitro (DIV) using live-cell confocal microscopy. All three effectors were punctate in the axon and comigrated with LC3-positive (+) puncta (Fig. 1, A–C). When we coexpressed EGFP-LC3 concurrently with all three candidates, we frequently observed simultaneous colocalization of all four (Fig. S1, A and B).
Figure 1.
Multiple dynein effectors associate with axonal autophagosomes. (A–C) Time series and kymographs from separate LC3+ puncta demonstrating comigration with JIP1, HAP1, and JIP3. Scale bar, 2 µm. (D) Schematic illustrating neuronal subregions and axonal transport of autophagosomes. (E–G) Quantification of LC3+ puncta comigrating with JIP1, HAP1, and JIP3 in different axonal regions. n = 9–17 neurons; one-way ANOVA with Tukey’s multiple comparisons test (JIP1: distal or mid vs. proximal, P < 0.0001). (H and I) Immunoblotting and quantification (relative to brain lysate, input) of autophagosome isolation illustrating enrichment on the outer membrane. n = 3–4 preps; one-way ANOVA (LC3, P < 0.0001; LIC1, P = 0.0056; JIP1, P = 0.0497; HAP1, P = 0.0058; JIP3, P = 0.0277). (J and K) Micrographs and quantification showing PLA puncta for endogenous DIC with Halo-JIP1 along the axon (dotted gray line). Arrowheads indicate PLA puncta. Scale bar, 10 µm. Dashed gray line indicates negative control (missing primary antibody). n = 7 neurons; one-way ANOVA with Tukey’s multiple comparisons test (distal vs. mid, P = 0.0236; distal vs. proximal, P = 0.0131). Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
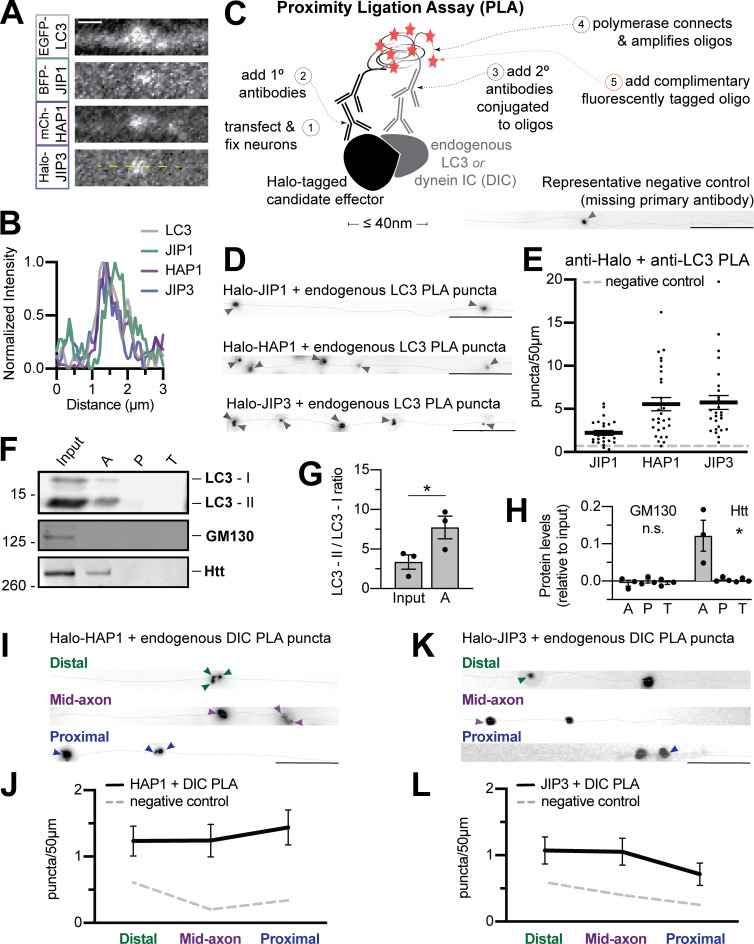
Figure S1.
Dynein effectors associate with autophagosomes in the axon. (A and B) Micrograph and line scan demonstrating LC3 can colocalize with all three candidates simultaneously. Yellow dashed line indicates line scan. Scale bar, 1 µm. (C) Schematic illustrating PLA method. Bottom right; Representative negative control (no primary antibody). (D and E) Representative micrographs and quantifications of PLA puncta for endogenous LC3 with Halo-tagged JIP1, HAP1, and JIP3 along the axon (dotted gray line). Arrowheads indicate PLA puncta. Scale bar, 10 µm. Dashed gray line indicates negative control (missing primary antibody). n = 27–28 neurons. (F) Immunoblotting of autophagosomal isolation. (G) Quantification of lipidated LC3-II isoform compared with cytosolic LC3-I. n = 3 preps; two-tailed paired t test (P = 0.0254). (H) Quantification of enrichment in the autophagosomal fraction, displayed as relative to brain lysate (input). n = 3 preps; one-way ANOVA for each; Golgi protein GM130 (P = 0.2128), Htt (P = 0.0198). (I–L) Representative micrographs and quantifications of PLA puncta between endogenous DIC and Halo-tagged HAP1 (I and J) or JIP3 (K and L) along the axon (dotted gray line). Arrowheads indicate PLA puncta. Scale bar, 10 µm; n = 9 neurons/region; dashed gray line indicates negative control (missing primary antibody); one-way ANOVA. Bars throughout show mean ± SEM. *, P < 0.05.
Next, we performed proximity ligation assays (PLAs) to detect proteins colocalizing within 40 nm (Alam, 2018). Neurons expressing low levels of Halo-JIP1, Halo-HAP1, or Halo-JIP3 were fixed and stained with primary antibodies to HaloTag and endogenous LC3, and then secondary antibodies conjugated to complementary oligonucleotides were added to assess their physical proximity (Fig. S1 C). All three effectors were closely apposed to LC3 in the axon, visible as fluorescent oligonucleotide puncta (Fig. S1, D and E).
Previous work on JIP1 found that it specifically promotes autophagosomal motility in the distal axon (Fu et al., 2014), suggesting that motors may require distinct effectors in different subaxonal regions. We therefore assayed colocalization in the distal (<100 µm from the axon tip), mid-, and proximal axon (<100 µm from the soma; Fig. 1 D). JIP1 comigrated with LC3+ puncta more in the distal and mid-axon than in the proximal axon (Fig. 1 E). HAP1 and JIP3 colocalized to a similar extent with LC3+ puncta in all regions (Fig. 1, F and G).
We isolated autophagosomes from mouse brain via sequential ultracentrifugation (Strømhaug et al., 1998; Maday et al., 2012). To differentiate between cargo engulfed within the autophagosome and membrane-associated proteins on the outer surface, we treated the autophagosomal fraction with Proteinase K to degrade exposed proteins or with Proteinase K and Triton X-100 as a control (Boecker et al., 2021). The autophagosomal fraction was enriched for the lipidated form of LC3 and LC3-II and depleted of markers for other organelles (Fig. S1, F–H). JIP1, JIP3, HAP1, and Htt were all significantly enriched on the cytosolic face of isolated autophagosomes, along with dynein itself (light intermediate chain 1 [LIC1]; Fig. 1, H and I; and Fig. S1, F–H).
Finally, we asked whether the candidate proteins were closely apposed to dynein in the axon. PLA between the Halo-tagged effectors and endogenous dynein intermediate chain (DIC) revealed that JIP1–dynein complexes were significantly enriched in the distal axon compared with other regions (Fig. 1, J and K). HAP1 and JIP3 exhibited proximity to dynein throughout the axon, but showed no regional specificity (Fig. S1, I–L). These results suggest that multiple adaptors interact with dynein on axonal autophagosomes.
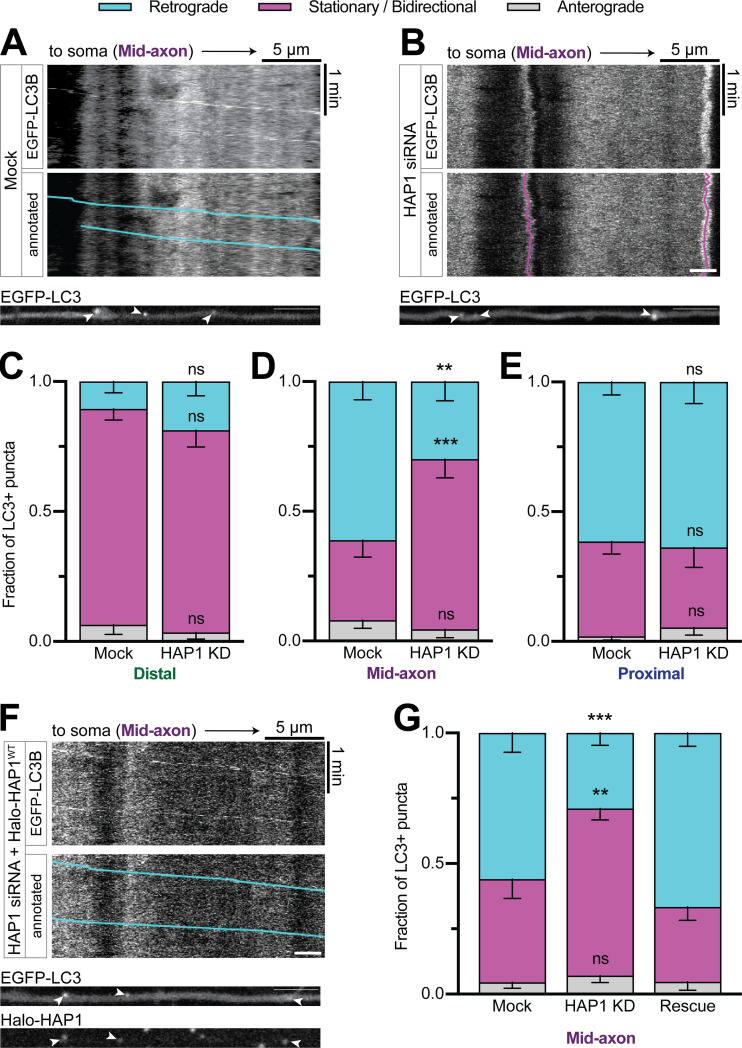
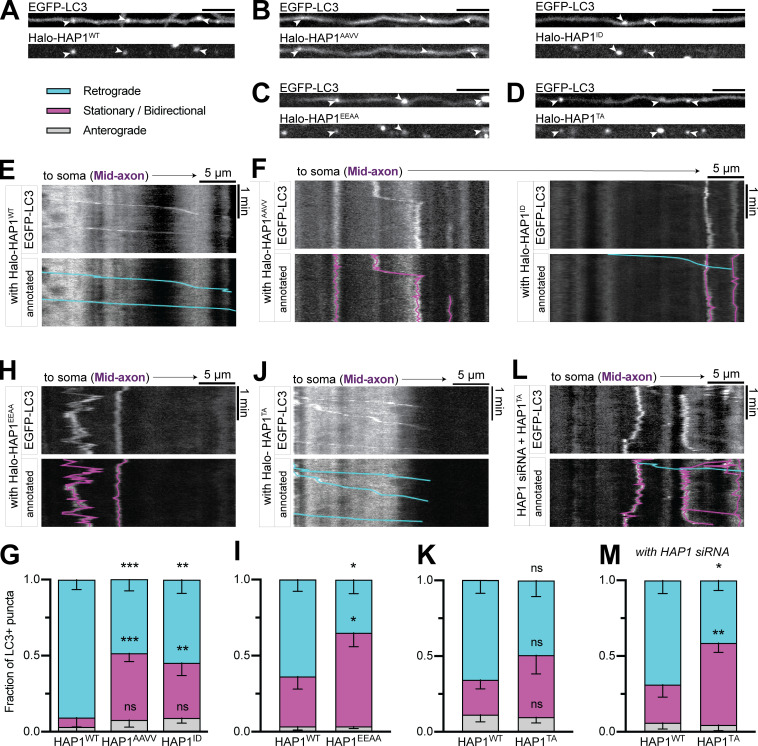
HAP1 regulates autophagosomal motility specifically in the mid-axon
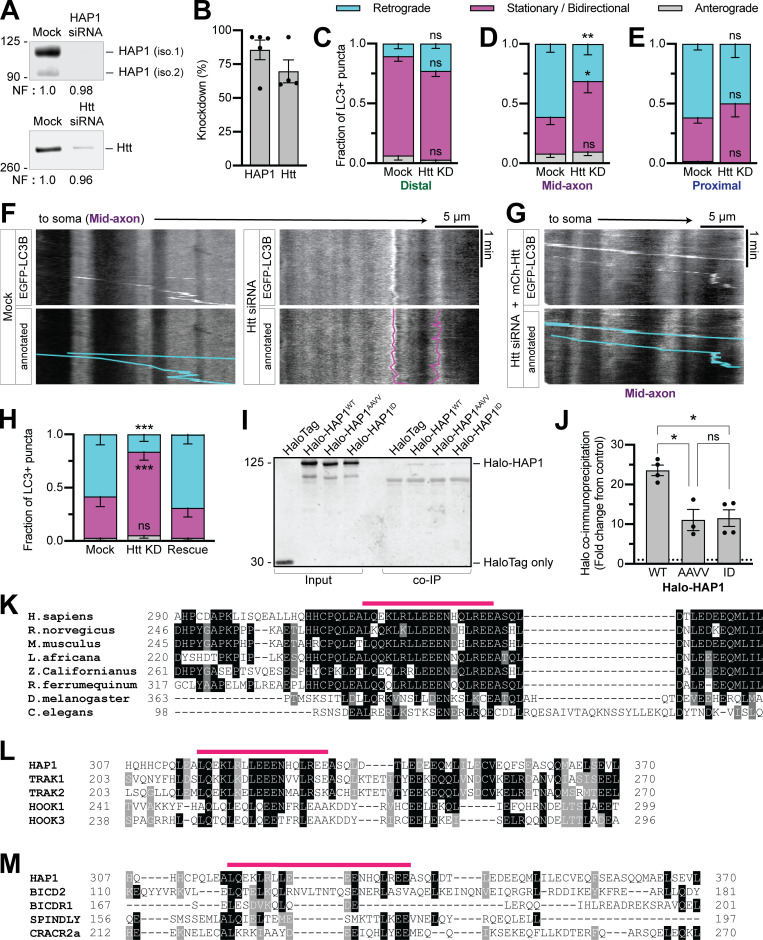
To test the function of HAP1 in axonal autophagosome transport, we used siRNA to knock down (KD) HAP1 (∼85% KD efficiency in PC12 cells; Fig. S2, A and B) in neurons transduced with LC3B-GFP. As expected, the majority (61% ± 7% in the mid-axon) of LC3+ puncta moved retrograde toward the soma under control conditions (Fig. 2 A). HAP1 depletion decreased retrograde autophagosomal motility (30% ± 7%) in the mid-axon, where the majority of LC3+ puncta exhibited stationary or bidirectional motility, defined as events moving <10 µm during the video (Fig. 2, B and D). Interestingly, the effect was limited to the mid-axon with no change in the distal (Fig. 2 C) or proximal (Fig. 2 E) axon. To confirm specificity of KD, we expressed siRNA-resistant Halo-HAP1 (HAP1WT) in KD neurons, which rescued motility to control levels (Fig. 2, F and G).
Figure S2.
Htt regulates autophagosomal motility in the mid-axon and HAP1 contains conserved dynein–dynactin binding sites. (A and B) Immunoblotting and quantification of PC12 cell lysates show KD efficiency of HAP1 and Htt siRNAs. Normalization factor (NF) determined using Revert Total Protein Stain. n = 4–5 repeats. (C–E) Quantification of LC3+ puncta motile behavior in the distal, mid-, and proximal axonal regions. n = 6–21 neurons. Two-way ANOVA with Sidak’s multiple comparisons test (mid retrograde mock vs. Htt KD, P = 0.0076; mid stationary/bidirectional [Stat/Bidir] mock vs. Htt KD, P = 0.0132). (F) Example kymographs from the mid-axon of a mock-transfected (control) neuron and a neuron transfected with Htt siRNA. (G and H) Example kymograph and quantification from the mid-axon of a neuron transfected with Htt siRNA and siRNA-resistant mCh-Htt. Two-way ANOVA with Tukey’s multiple comparisons test (retrograde mock vs. Htt KD, P = 0.0005 = 4; Stat/Bidir mock vs. Htt KD, P = 0.0009). Symbols indicate comparison to mock. (I and J) Immunoblot and quantification of purified LIC1 pulldown by HAP1 constructs (COS-7 lysate). n = 3–4 repeats; results normalized to HaloTag only pulldown (negative control; dotted black line); mixed-effects model (fixed effect, P = 0.0233) with Tukey’s multiple comparisons test (WT vs. AAVV, P = 0.0234; WT vs. ID, P = 0.0373; AAVV vs. ID, P = 0.9936). (K) Sequence alignment of HAP1 across species. (L and M) Alignment of human HAP1 with known and putative dynein-activating adaptors (human canonical sequences). Alignments run with T-Coffee (version 11.00.d625267) default settings. Prepared for visualization by BoxShade (ExPAy, Swiss Institute of Bioinformatics) with RTF_new output. Pink lines indicate Glued motif. Black boxes indicate aa identical in ≥60% of sequences. Gray boxes indicate aa similar in ≥ 60% of sequences. Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 2.
HAP1 regulates autophagosomal motility specifically in the mid-axon. (A and B) Example kymographs and micrographs from the mid-axon of a mock-transfected (control) neuron and a neuron transfected with HAP1 siRNA. Kymographs depict distance on the x-axis and time on the y-axis. Annotated kymographs mirror the above kymographs with the LC3+ puncta paths pseudo-colored for visualization. (C–E) Quantification of LC3+ puncta motility in the distal, mid-, and proximal axon. n = 8–21 neurons; two-way ANOVA with Sidak’s multiple comparisons test (mid retrograde mock vs. HAP1 KD, P = 0.0033; mid stationary/bidirectional [Stat/Bidir] mock vs. HAP1 KD, P = 0.0009). (F) Example kymograph and micrograph from the mid-axon of a neuron transfected with HAP1 siRNA and siRNA-resistant Halo-HAP1WT. (G) Quantification of LC3+ puncta motility in the mid-axon. Two-way ANOVA with Tukey’s multiple comparisons test (retrograde mock vs. HAP1 KD, P = 0.0005; Stat/Bidir mock vs. HAP1 KD, P = 0.0020). Bars throughout show mean ± SEM. **, P < 0.01; ***, P < 0.001 compared to mock. Scale bar, 5 µm.
Htt KD (Fig. S2, A and B) likewise decreased the retrograde fraction (31% ± 9%) in the mid-axon with no effect in the distal or proximal axon (Fig. S2, C–F). Expression of siRNA-resistant mCh–Htt rescued motility (Fig. S2, G and H). Together, our data show that the HAP1–Htt complex is required for autophagosome motility exclusively in the mid-axon.
HAP1 binds dynein–dynactin via activating adaptor motifs
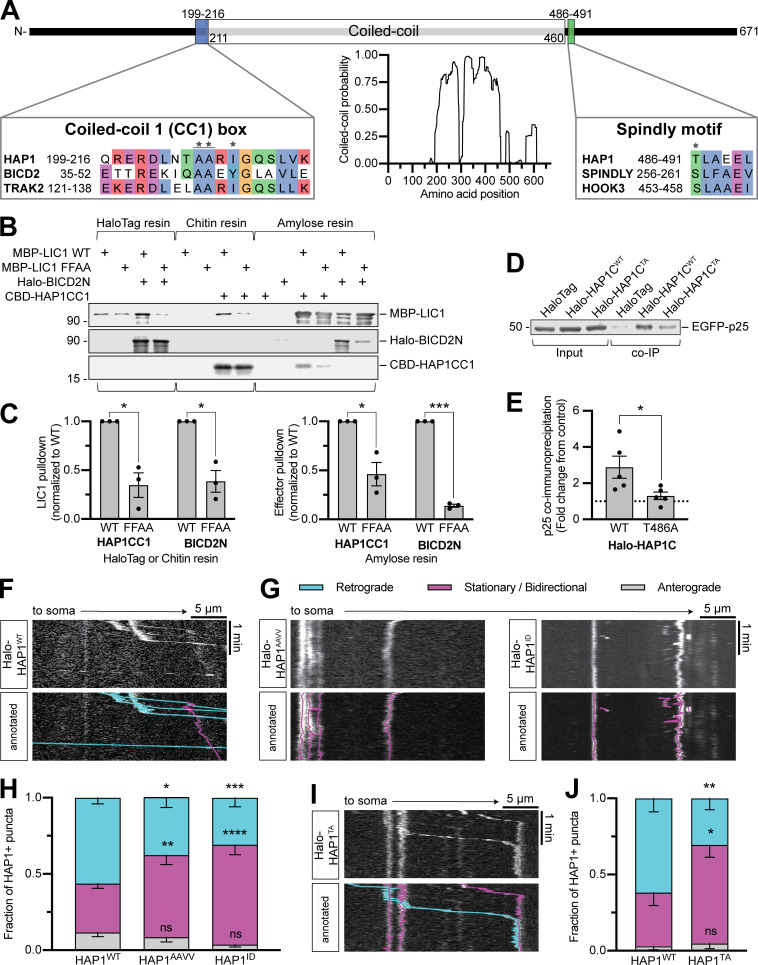
We next queried the mechanism by which HAP1 regulates autophagosome motility. Sequence analysis of HAP1 using coiled-coil prediction programs (Lupas et al., 1991; Trigg et al., 2011; Vincent et al., 2013; Ludwiczak et al., 2019) identified an extended coiled-coil segment (aa 211–460; Fig. 3 A). Many dynein-activating adaptor proteins dock along dynactin’s filament of actin-related protein 1 (Arp1) via a similar length coiled coil (Hodgkinson et al., 2005; Urnavicius et al., 2015, 2018); assuming 3.5 residues per turn (Truebestein and Leonard, 2016), the predicted coiled coil in HAP1 extends 35 nm, the appropriate length to dock onto the 37-nm Arp1 filament (Schafer et al., 1994).
Figure 3.
HAP1 binds dynein–dynactin via activating adaptor motifs. (A) Schematic illustrating the domain architecture of HAP1. Coiled-coil probability was calculated using predictions from four different programs. Residues in the sequence alignments are colored using the clustal color scheme where conserved. Stars indicate point mutants. (B and C) Immunoblot and quantification of purified HAP1CC1 (aa 168–261) or BICD2N (aa 1–572) pulldown by dynein LIC1WT or mutant LIC1FFAA and vice versa. Results were normalized to the LIC1WT condition; n = 3 pulldowns from n = 2 independent purifications; paired two-tailed t test; LIC1 pulldown (HAP1CC1 WT vs. FFAA, P = 0.0353; BICD2N WT vs. FFAA, P = 0.0051); effector pulldown (HAP1CC1 WT vs. FFAA, P = 0.0109; BICD2N WT vs. FFAA, P < 0.0001). (D and E) Example immunoblot and quantification of EGFP-p25 coimmunoprecipitation by HAP1CWT (aa 470–671) or mutant HAP1CTA (coexpressed in COS-7 cells). Results were normalized to p25 pulled down by HaloTag only (negative control; dotted black line); n = 5 repeats; paired t test (two-tailed; P = 0.0450). (F–J) Example kymographs and quantification of motile behavior of punctate HAP1WT (F), HAP1 CC1 box (G and H), and HAP1 Spindly (I and J) mutants. n = 11–16 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test; AAVV (retrograde, P = 0.0333; stationary/bidirectional [Stat/Bidir], P = 0.0099); ID (retrograde, P = 0.0009; Stat/Bidir, P < 0.0001); TA (retrograde, P = 0.0063; Stat/Bidir, P = 0.0109). Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 compared to WT.
The coiled-coil region in HAP1 is flanked by conserved sequence motifs found in dynein activating adaptors (Fig. 3 A). The coiled-coil 1 (CC1) box motif preceding the coiled-coil region is predicted to bind a conserved helix in the C terminus of dynein LIC1 (Lee et al., 2018; Celestino et al., 2019; Lee et al., 2020). To test the predicted CC1 box in HAP1, we performed pull-down experiments using purified recombinant LIC1 and HAP1 spanning residues 168–261 (HAP1CC1). The well-characterized dynein activator bicaudal-D 2 (BICD2N) was used as a positive control. HAP1CC1 and BICD2N pulled down LIC1, and vice versa (Fig. 3, B and C), while an LIC1 mutant known to disrupt LIC1-activator binding (F447A, F448A; termed FFAA; Lee et al., 2018) showed markedly less pulldown (Fig. 3, B and C). We also generated two HAP1 mutant constructs expected to inhibit LIC1 binding (Fig. 3 A): HAP1AAVV altered conserved alanine residues (A207V, A208V; termed AAVV) required for LIC1 binding in other dynein activators (Schlager et al., 2014; Gama et al., 2017; Lee et al., 2020), and HAP1ID introduced a single isoleucine-to-aspartic acid mutation (I210D; termed ID) expected to disrupt the hydrophobic groove that accommodates LIC1 (Lee et al., 2020). When expressed in COS-7 cell lysate, both mutants pulled down significantly less purified LIC1WT than HAP1WT (Fig. S2, I and J). HAP1 thus binds dynein using the CC1 box motif.
The Spindly motif, first described in activating adaptor Spindly, follows the coiled coil in dynein activators and mediates an interaction with the pointed-end capping complex of dynactin’s Arp1 filament (Hodgkinson et al., 2005; Gama et al., 2017). To test the motif in HAP1, we generated a HAP1 C-terminal construct (aa 470–671) and coexpressed it in COS-7 cells with the dynactin pointed-end complex protein EGFP-p25. HAP1CWT coimmunoprecipitated p25, while a threonine-to-alanine point mutant (T486A; TA) predicted to deleteriously impact dynactin binding (Fig. 3 A; Gassmann et al., 2010; Gama et al., 2017) coimmunoprecipitated significantly less p25 (Fig. 3, D and E). HAP1 thus contains a functional Spindly motif for binding dynactin.
Finally, to test the function of these motifs in cells, we overexpressed Halo-HAP1 in primary neurons and imaged along the axon. Overexpressed Halo-HAP1WT formed puncta in the axon that mostly moved retrograde (59% ± 6%), indicating dynein activity (Fig. 3 F). Halo-HAP1AAVV (38% ± 7%), HAP1ID (31% ± 6%), and the full-length Spindly motif mutant HAP1TA (31% ± 7%) all showed significantly less retrograde motility than HAP1WT (Fig. 3, G–J). Hence, both the dynein and dynactin binding regions in HAP1 are required for dynein-driven HAP1 transport in cells.
HAP1 contains a novel conserved p150Glued binding site
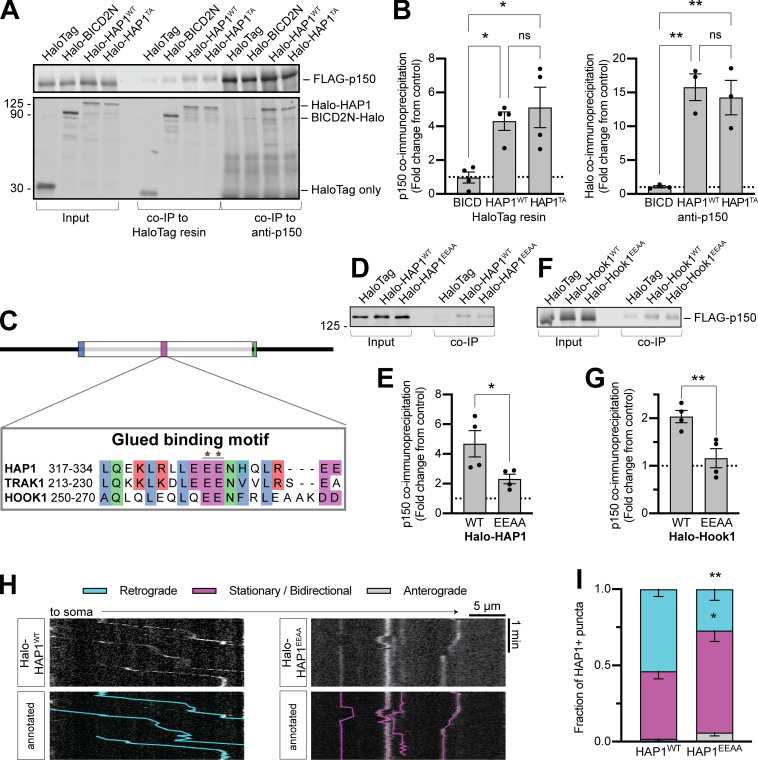
A yeast two-hybrid screen (Engelender et al., 1997) identified an interaction between dynactin subunit p150Glued (p150) and HAP1 aa 280–445, a region well conserved across species (Fig. S2 K). We cotransfected COS-7 cells with FLAG-p150 and either Halo-HAP1WT, Halo-HAP1TA, or BICD2N-Halo and found that, while BICD2N was incapable of coimmunoprecipitating p150 above negative control (HaloTag only) levels, both HAP1WT and HAP1TA could coimmunoprecipitate p150 and vice versa (Fig. 4, A and B). Thus, p150 binding and pointed-end complex binding are independent.
Figure 4.
HAP1 contains a novel conserved p150Glued binding site. (A and B) Example immunoblot and quantification of p150 coimmunoprecipitation by HAP1 or BICD2N (coexpressed in COS-7 cells) and vice versa. n = 3–4 repeats; results were normalized to HaloTag only (negative control; dotted black line); one-way ANOVA of the three conditions with Tukey’s multiple comparisons test (left: BICD2N vs. WT, P = 0.0349; BICD2N vs. TA, P = 0.0114; WT vs. TA, P = 0.7518; right: BICD2N vs. WT, P = 0.0034; BICD2N vs. TA, P = 0.0059; WT vs. TA, P = 0.8347). (C) Schematic showing sequence alignment for the Glued binding motif. Stars indicate point mutant. (D–G) Immunoblotting and quantification show HAP1 (D and E) and Hook1 (F and G) Glued motif mutants (EEAA) coimmunoprecipitation of FLAG-p150 (coexpressed in COS-7 cells). n = 4 repeats; results were normalized to HaloTag only (negative control; dotted black line); paired t test (HAP1, P = 0.0268; Hook1, P = 0.0037). (H and I) Example kymographs and quantification of HAP1+ puncta motile behavior in the mid-axon. n = 13–15 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test (Retrograde WT vs. EEAA, P = 0.0019; stationary/bidirectional WT vs. EEAA, P = 0.0103). Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01.
We identified a highly conserved motif within this region (aa 317–334) that is also conserved within the evolutionarily related TRAK family and the HOOK family of dynein activating adaptors (Fig. S2 L; Lumsden et al., 2016), but not in other dynein activators, including Spindly and the BICD protein family (Fig. S2 M). We engineered HAP1 (E326-327A; HAP1EEAA) and Hook1 (E259-260A; Hook1EEAA) mutant constructs expected to affect p150 binding (Fig. 4 C) and coexpressed them in COS-7 cells with FLAG-p150. Both showed reduced coimmunoprecipitation of FLAG-p150 compared with WT (Fig. 4, D–G). To test the function of the motif in cells, we transfected HAP1EEAA into primary neurons where it displayed significantly less retrograde motility (27% ± 7%) than HAP1WT (54% ± 5%), indicating the importance of this binding site for a motile dynein complex (Fig. 4, H and I). We therefore conclude that both the HAP/TRAK family and the HOOK family possess a novel Glued binding motif that is independent of pointed-end complex binding.
HAP1 activates dynein motility
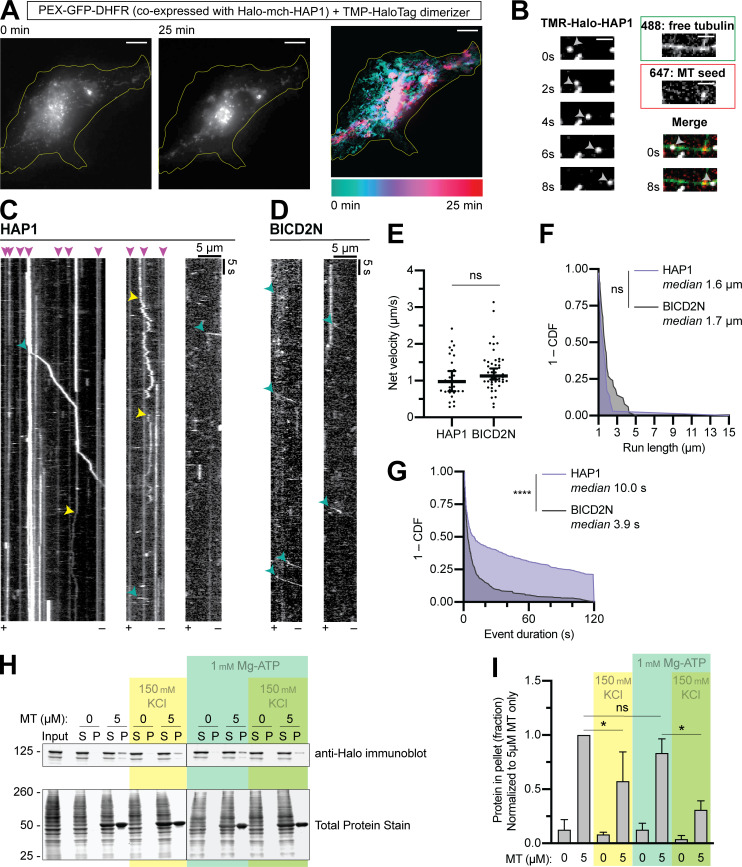
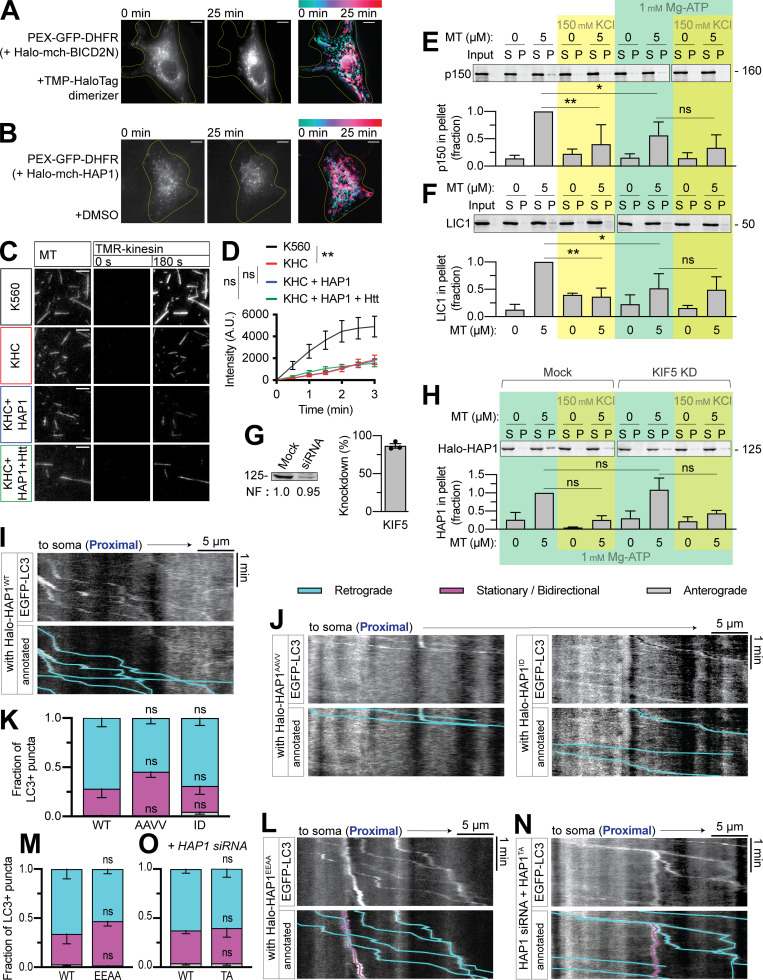
Based on our biochemical data, we hypothesized that HAP1 functions as a dynein activating adaptor. We assayed the ability of HAP1 to redistribute organelles in live COS-7 cells using an induced dimerization assay. Our cell-permeant dimerizer irreversibly binds a dihydrofolate reductase (DHFR)–tagged protein and a Halo-tagged protein (Ballister et al., 2015; Olenick et al., 2016). We assayed peroxisomes because they are generally immotile and uniformly distributed under control conditions (Smith and Aitchison, 2013). The addition of dimerizer to cells expressing peroxisome marker PEX3-GFP-Halo and either HAP1-mCh-DHFR (Fig. 5 A) or BICD2N-mCh-DHFR (Fig. S3 A) induced retrograde motility of peroxisomes and eventual clustering in the perinuclear region; perinuclear clustering was not seen in vehicle-treated cells (Fig. S3 B). HAP1 thus induces minus-end–directed motility in a cellular assay.
Figure 5.
HAP1 activates dynein motility. (A) Peroxisome recruitment assay wherein peroxisomes (PEX-GFP-DHFR) were tethered to Halo-mCh-HAP1 via dimerizer and imaged for 25 min to show relocalization to the cell center. Left: Initial (0 min) and concluding (25 min) stills of peroxisomes. Right: Maximum time projection pseudo-colored by frame (scale below). Yellow line indicates cell outline. Scale bar, 10 µm. (B) Example micrographs from single-molecule motility TIRF assay showing TMR-labeled Halo-HAP1 traversing an MT from the dynamic plus end to the minus end, indicated by the 647-labeled MT seed. Single stills from both tubulin channels are shown (488 and 647) as well as the first and last frames merged (all three channels). Scale bar, 2 µm. (C and ;D) Example kymographs showing dynein-directed motility of HAP1 and BICD2N puncta in the lysate-based motility assay. Kymographs span the length of each MT with the plus end (+) and minus end (−) labeled at the bottom. Arrowheads represent motile minus end–directed events (teal), diffusive events (yellow), and sustained MT binding events (pink). (E–G) Quantifications of single-molecule motility assay showing the average velocity (E), run length (F), and stationary event duration (G). n = 30–51 runs (E and F), 457–471 events (G) in three independent trials on 57 total MTs. Welch’s two-tailed t test (E: P = 0.1969), Mann-Whitney U test (F: P = 0.0568; G: P < 0.0001). Y-axes in F and G represent 1 - the cumulative distribution function (CDF); x-axis in F begins at 1 µm because only runs ≥1 µm were analyzed; x-axis in G ends at 120 s, the length of videos acquired. (H and I) Blotting and quantification of cell extract MT pelleting assay. n = 3 independent assays; one-way ANOVA followed by Tukey’s multiple comparisons test (ATP addition, P = 0.6691; KCl without ATP, P = 0.0075; KCl with ATP, P = 0.0011). Bars throughout show mean ± SEM. *, P < 0.05; ****, P < 0.0001.
Figure S3.
HAP1 binds MTs independently of motors and is not required for autophagosome motility in the proximal axon. (A and B) Peroxisome recruitment assay showing peroxisome (PEX-GFP-DHFR) localization for 25 min following addition of dimerizer or DMSO. Left: Initial (0 min) and concluding (25 min) stills of peroxisomes. Right: Maximum time projection pseudo-colored by frame (scale above). Yellow line indicates cell outline. Scale bar, 10 µm. (C and D) Quantification and example micrographs from kinesin MT recruitment TIRF assay wherein cell extracts expressing constitutively active N-terminal kinesin construct K560 (positive control) or full-length kinesin-1 heavy chain (KHC) were added to Taxol-stabilized MT in a TIRF chamber with nonhydrolyzable ATP homologue AMPPNP. TMR-labeled kinesin intensity is measured across a 3-min video. n = 20 MT from four independent trials; one-way ANOVA with Tukey’s multiple comparisons test (KHC vs. K560, P = 0.0019; KHC vs. KHC + HAP1, P = 0.9992; KHC vs. KHC + HAP1 + Htt, P = 0.9833). Scale bar, 5 µm. (E and F) Blotting and quantification of cell extract MT pelleting assay for dynactin p150 (E) and dynein LIC1 (F) normalized to 5 µM MT only condition. n = 3–4 independent assays; one-way ANOVA with Tukey’s multiple comparisons test. p150 (ATP addition, P = 0.0463; KCl without ATP, P = 0.0025; KCl with ATP, P = 0.6624); DIC (ATP addition, P = 0.0264; KCl without ATP, P = 0.0025; KCl with ATP, P > 0.9999). (G) Immunoblot and quantification of KIF5 KD. Normalization factor (NF) determined using Revert Total Protein Stain). (H) Blotting and quantification of cell extract MT pelleting assay showing HAP1 binds MT independently of KIF5. Normalized to mock 5 µM MT with 1 mM Mg-ATP and no added salt; n = 3 independent assays; one-way ANOVA with Tukey’s multiple comparisons test (KD, P > 0.9999; KCl in mock, P = 0.0849; KCl in KD, P = 1662). (I) Example kymograph of LC3 in the proximal with Halo-HAP1WT. (J–M) Example kymographs and quantification showing no effect of HAP1 CC1 box or Glued motif mutants on LC3+ puncta motility in the proximal axon. n = 9–13 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test (retrograde: AAVV, P = 0.1337; ID, P > 0.9999; EEAA, P = 0.5408; stationary/bidirectional [Stat/Bidir]: AAVV, P = 0.0900; ID, P > 0.9999; EEAA, P = 0.4316.) (N and O) Example kymograph and quantification showing no effect of HAP Spindly mutant in the proximal axon of cells transfected with HAP1 siRNA. n = 10–12 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test (retrograde, P > 0.9999; Stat/Bidir, P > 0.9999). Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01 compared to WT unless otherwise indicated.
We next performed lysate-based in vitro single molecule motility assays using total internal reflection fluorescence (TIRF) microscopy (Ayloo et al., 2014). We bound MT seeds (labeled with HiLyte 647 tubulin) to coverslips and then flowed in GTP and free tubulin (labeled with HiLyte 488 tubulin) and imaged MT dynamics. Plus ends were identified by their higher rates of both growth and catastrophe. We then introduced COS-7 cell lysates containing tetramethylrhodamine (TMR)-labeled HAP1 (Fig. 5, B and C) or BICD2N (Fig. 5 D), both of which induced minus end–directed runs (teal arrowheads) with similar average velocities (1.1 µms−1) and run lengths (1.6 µm) to previous reports of dynein motility (Fig. 5, E and F; Olenick et al., 2016; Lee et al., 2020).
BICD2N exhibited almost exclusively (90%) minus end–directed runs, while HAP1 displayed ∼30% plus end–directed runs, not surprising given the interaction between HAP1 and kinesin; however, in both primary axons and in vitro, HAP1 demonstrated majority minus end–directed motility. We assessed HAP1 activation of kinesin by performing a kinesin MT recruitment assay in TIRF with full-length kinesin-1 (KHC; Fig. S3, C and D). Although HAP1 has been shown to activate kinesin in coordination with another effector, GRIP1 (Twelvetrees et al., 2019), we found that HAP1 could not activate kinesin independently or with Htt in this assay.
In addition to processive runs, HAP1 complexes displayed diffusive events (yellow arrowheads; Fig. 5 C) and frequent long immotile MT binding events, often lasting the entire length of the video (pink arrowheads; Fig. 5, C and G). These sustained MT binding events and diffusive runs, not seen in the BICD2N condition, are similar to those observed with purified TRAK1 (Henrichs et al., 2020). TRAK1 directly binds MTs (Henrichs et al., 2020), leading us to hypothesize that the frequent nonprocessive events observed in our assay were due to HAP1 MT binding. We performed a cell extract–based MT pelleting assay in which COS-7 lysate expressing Halo-HAP1 was incubated with 5 µM MT under varying buffer conditions. HAP1 robustly pelleted with MT, and this pelleting was unaffected by the addition of Mg-ATP (1 mM; Fig. 5, H and I), which caused the dynein–dynactin complex to disassociate from the MT (Fig. S3, E and F). Likewise, HAP1 pelleting was unaffected by kinesin-1 KD (Fig. S3, G and H). Increased ionic strength (150 mM KCl) decreased but did not eliminate HAP1 pelleting (Fig. 5, H and I; and Fig. S3 H), suggesting that the interaction is partially dependent on electrostatic interactions. Together, these observations indicate HAP1 can bind MTs and activate dynein motility.
HAP1 drives autophagosomal transport by binding to dynein and dynactin
Based on the above results, we hypothesized HAP1 activates dynein on autophagosomes. We coexpressed EGFP-LC3 with each Halo-HAP1 construct and imaged the mid-axon, where all the HAP1 constructs colocalized with LC3 (Fig. 6, A and D). LC3+ puncta in neurons expressing HAP1WT moved primarily retrograde (77% ± 5%; Fig. 6 E), while LC3+ puncta in neurons expressing CC1 box mutants HAP1AAVV or HAP1ID (Fig. 6, F and G) or the Glued motif mutant HAP1EEAA (Fig. 6, H and I) showed less retrograde motility (48% ± 7%, 54% ± 9%, or 31% ± 7% respectively), indicating a dominant-negative effect. Like HAP1 KD, this effect was limited to the mid-axon (Fig. S3, I–M).
Figure 6.
HAP1 drives autophagosomal transport by binding to dynein and dynactin. (A–D) Representative micrographs showing HAP1WT and the four HAP1 point mutants colocalize with LC3+ puncta in the axon. Scale bars, 5 µm. (E) Example kymograph illustrates the typical motility of EGFP-LC3+ puncta in the presence of overexpressed Halo-HAP1WT. (F–I) Example kymographs and quantification showing dominant-negative effect of HAP1 CC1 box mutants (HAP1AAVV and HAP1ID) and HAP1 Glued motif mutant (HAP1EEAA) on LC3+ puncta motility. n = 8–15 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test (retrograde: AAVV, P = 0.0002; ID, P = 0.0013; EEAA, P < 0.0001; stationary/bidirectional [Stat/Bidir]: AAVV, P = 0.0009; ID, P = 0.0091; EEAA, P = 0.0002). (J and K) Example kymograph and quantification showing the HAP1 Spindly mutant (HAP1TA) has no effect on LC3+ puncta motile behavior in the mid-axon. n = 16 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test (retrograde, P = 0.4635; Stat/Bidir, P = 0.3500). (L and M) Example kymograph and quantification of LC3+ puncta in cells transfected with both HAP1 siRNA and HAP1WT or HAP1TA. n = 16 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test (retrograde, P = 0.0071; Stat/Bidir, P = 0.0075). Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared to WT. All data are from the mid-axon.
The Spindly mutant HAP1TA (Fig. 6, J and K) did not affect LC3 motility, perhaps due to interaction with endogenous HAP1. We therefore cotransfected neurons with HAP1 siRNA and Halo-HAP1TA or Halo-HAP1WT. In the absence of endogenous HAP1, HAP1WT induced retrograde motility of LC3+ puncta (69% ± 4%), but HAP1TA induced significantly less (43% ± 7%; Fig. 6, L and M). Motility in the proximal axon still appeared normal (Fig. S3, N and O). These findings confirm that HAP1 functions mechanistically in autophagosomal transport by binding and activating the dynein–dynactin complex.
JIP3 regulates autophagosomal motility in the mid- and proximal axon
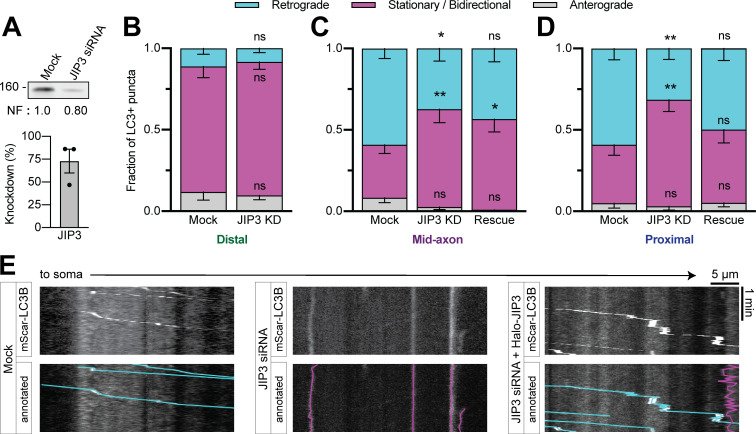
We next queried the role of JIP3 in autophagosomal transport. We used siRNA to deplete endogenous JIP3 (∼70% KD efficiency in PC12 cells; Fig. 7 A) in neurons transfected with mScarlet-LC3. JIP3 KD decreased retrograde autophagosomal motility in the mid- (37% ± 8%) and proximal (32% ± 7%) axon with a concomitant increase in the nonmotile fraction, but did not affect motility in the distal axon (Fig. 7, B–E). Expression of siRNA-resistant Halo-JIP3 could rescue motility fully in the proximal axon (Fig. 7, D and E) and partially in the mid-axon (Fig. 7 C), confirming the specificity of KD (Fig. 7, D and E). Thus, JIP3 regulates the activity of dynein on autophagosomes closer to the soma.
Figure 7.
JIP3 regulates autophagosomal motility in the mid- and proximal axon. (A) Immunoblotting and quantification of PC12 cell lysates demonstrate KD efficiency of JIP3 siRNA. n = 3 repeats. Normalization factor (NF) determined using Revert Total Protein Stain. (B–D) Quantification of LC3+ puncta motile behavior in the distal, mid-, and proximal axon. n = 12–19 neurons. Two-way ANOVA for each with Tukey’s multiple comparisons test; mid (retrograde: mock vs. KD, P = 0.0212; mock vs. rescue, P = 0.2045; KD vs. rescue, P = 0.7854; stationary/bidirectional [Stat/Bidir]: mock vs. KD, P = 0.0025; mock vs. rescue, P = 0.0347; KD vs. rescue, P = 0.8816); proximal (retrograde: mock vs. KD, P = 0.0044; mock vs. rescue, P = 0.5203; KD vs. rescue, P = 0.0858; Stat/Bidir: mock vs. KD, P = 0.0021; mock vs. rescue, P = 0.5410; KD vs. rescue, P = 0.0469). (E) Example kymographs showing proximal axon. Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01 compared to mock.
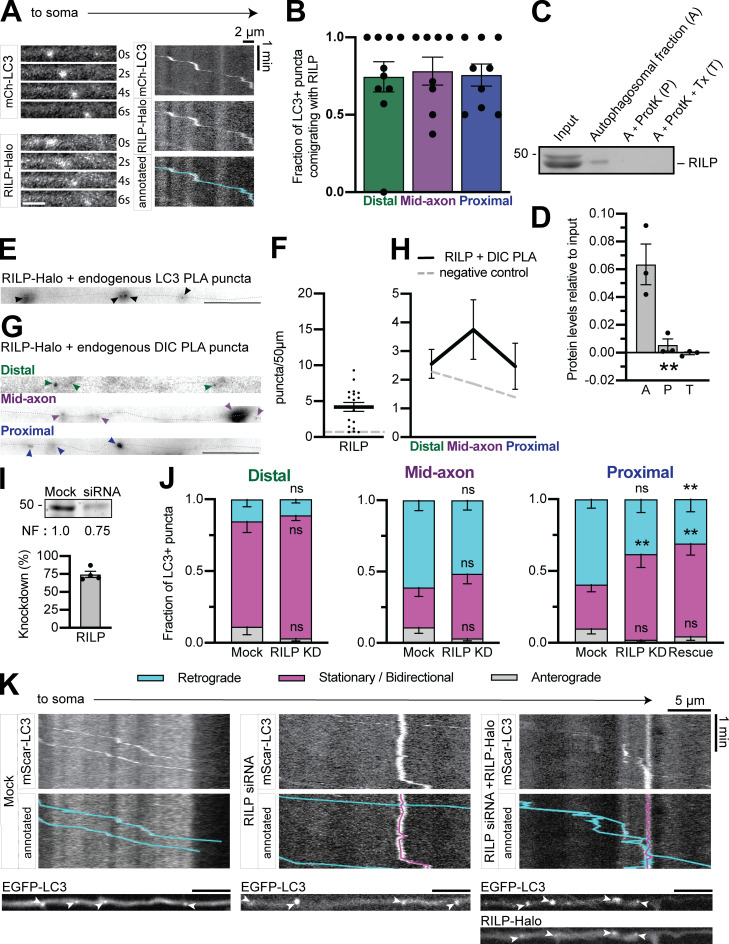
In both the JIP3 (Fig. 7) and HAP1 (Fig. 2) KD experiments, a subpopulation (∼25%) of autophagosomes continued moving retrograde. This sustained motility could be due to incomplete KD or an additional dynein effector. Rab-interacting lysosomal protein (RILP) was recently shown to be involved in autophagosomal motility in primary rat cortical neurons (Khobrekar et al., 2020). We found RILP comigrating with autophagosomes in the axon (Fig. S4, A and B), enriched on the autophagosomal membrane (Fig. S4, C and D), and within 40 nm of dynein and LC3 via PLA (Fig. S4, E–H). However, RILP KD using siRNA (∼80% efficiency in PC12 cells) induced no effect on motility in the distal or mid-axon, and only a minor effect in the proximal axon (Fig. S4, I–K). The minor phenotype was not rescued by expression of siRNA-resistant Halo-RILP, despite the colocalization between Halo-RILP and LC3 (Fig. S4, J and K). We therefore conclude that RILP likely plays a minor or indirect role in autophagosomal transport in hippocampal axons.
Figure S4.
RILP may play a minor role in axonal autophagosome transport. (A) Time series and kymograph from separate LC3+ autophagosomes demonstrating comigration with RILP. Scale bars, 2 µm. (B) Quantification of LC3+ puncta comigrating with RILP in different subaxonal regions. n = 8–10 neurons; one-way ANOVA (P = 0.9577). (C and D) Example immunoblot and quantification of autophagosomal isolation illustrating enrichment of RILP on the outer membrane. n = 3 preps; one-way ANOVA (P = 0.0039). (E–H) Representative micrographs and quantifications of PLA puncta for Halo-tagged RILP and endogenous LC3 (n = 18 neurons) or endogenous DIC (n = 8 per region) along the axon (dotted gray line). Arrowheads indicate PLA puncta. Scale bar, 10 µm. Dashed gray line indicates negative control (missing primary antibody). One-way ANOVA. (I) Immunoblotting and quantification of PC12 cell lysates show KD efficiency of RILP siRNA. n = 4 repeats. Normalization factor (NF) determined using Revert Total Protein Stain. (J) Quantification of LC3+ puncta motile behavior in the distal, mid-, and proximal axon. n = 10–13 neurons. Two-way ANOVA with Tukey’s multiple comparisons test; proximal (retrograde: mock vs. KD, P = 0.0650; mock vs. rescue, P = 0.0094; KD vs. rescue, P = 0.7205; stationary/bidirectional: mock vs. KD, P = 0.0067; mock vs. rescue, P = 0.0015; KD vs. rescue, P = 0.8641). Symbols indicate comparison to mock. (K) Example kymographs from the proximal axon. Corresponding micrographs below. Scale bar, 5 µm. Bars throughout show mean ± SEM. **, P < 0.01.
Dynein effectors exhibit preference for autophagosome maturity
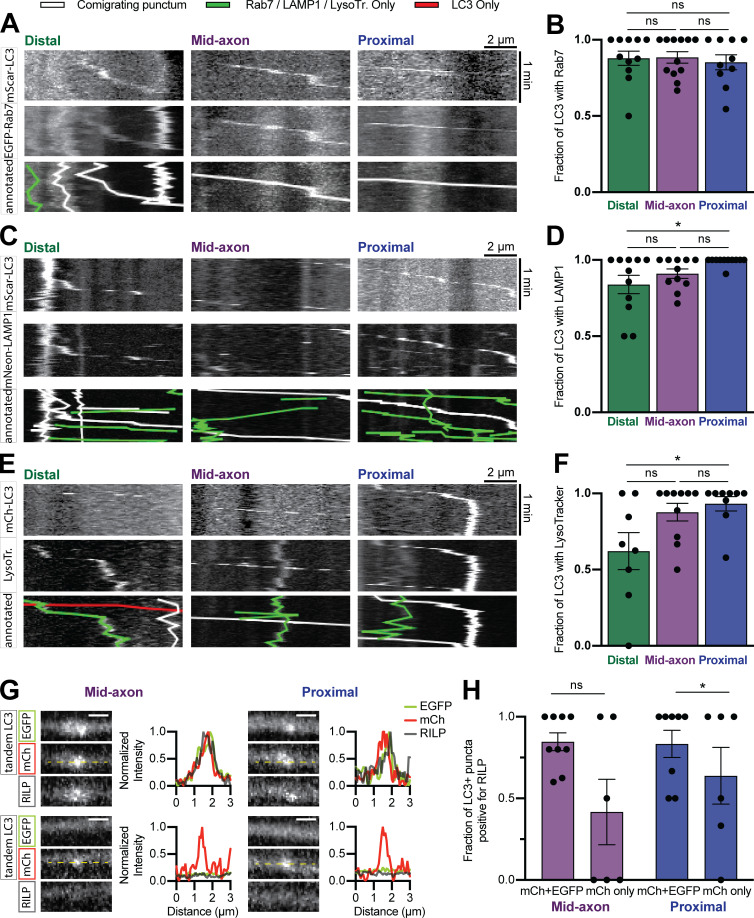
JIP3, HAP1, and JIP1 are active on autophagosomes in different axonal subregions. Autophagosomes mature via lysosomal fusion en route to the soma (Maday et al., 2012), so we asked whether dynein effectors associate with autophagosomes based on maturation state. The majority of LC3+ puncta in the distal axon comigrated with Rab7 (88% ± 5%) and LAMP1 (84% ± 6%) and were positive for LysoTracker DeepRed (62% ± 12%), a fluorescent dye that labels acidic organelles, suggesting that autophagosomes fuse with endolysosomes before exit from the distal tip (Fig. S5, A–F), consistent with observations in DRG axons (Maday et al., 2012). As an alternate way to test acidification, we transfected cells with the dual-color LC3 reporter mCh-EGFP-LC3; EGFP quenches in acidic environments, resulting in a shift from dually labeled green/red puncta to red only (Pankiv et al., 2007). 27% ± 7% of autophagosomes in the proximal region of hippocampal axons were mCh only (Fig. 8, A and B) compared with ∼70% in proximal DRG axons (Maday et al., 2012). Thus, despite early acquisition of endolysosomal membrane markers, autophagosomes in hippocampal axons mature slowly.
Figure S5.
Autophagosomes in hippocampal axons mature slowly. (A–F) Example kymographs and quantification of LC3 comigrating with Rab7 (A and B), LAMP1 (C and D), or LysoTracker (LysoTr) Deep Red dye (E and F) in different subaxonal regions. n = 8–12 neurons. Two-way ANOVA with Tukey’s multiple comparisons test (distal vs. mid: Rab7, P = 0.9956; LAMP1, P = 0.4166; LysoTracker, P = 0.0709; distal vs. proximal: Rab7, P = 0.9088; LAMP1, P = 0.0264; LysoTracker, P = 0.0280; mid vs. proximal: Rab7, P = 0.8655; LAMP1, P = 0.3205; LysoTracker, P = 0.8623.) (G and H) Example micrographs with line scans and quantification demonstrating RILP association with autophagosomes of differing maturity. n = 8–9 neurons. Two-way ANOVA with Sidak’s multiple comparisons test (mid, P = 0.4894; proximal, P = 0.0403). Dashed yellow lines indicate line scan. Scale bar, 1 µm. Bars throughout show mean ± SEM. *, P < 0.05.
Figure 8.
Dynein effectors exhibit preference for autophagosome maturity. (A and B) Example kymographs and quantification of mCh-EGFP-LC3 in different subaxonal regions. n = 11–12 neurons. Two-way ANOVA with Tukey’s multiple comparisons test (distal vs. mid, P = 0.4452; distal vs. proximal, P = 0.0195; mid vs. proximal, P = 0.2608). (C–H) Example micrographs, line scans, and quantifications showing the colocalization of JIP1 (C and D); HAP1 (E and F); and JIP3 (G and H) with mCh-EGFP-LC3. n = 10–18 neurons. Two-way ANOVA with Sidak’s multiple comparisons test (JIP1: mid, P = 0.0069; proximal, P = 0.0092; HAP1: mid, P = 0.0007; proximal, P = 0.0007; JIP3: mid, P = 0.6893; proximal, P = 0.1700.) Dashed yellow lines represent line scan. Scale bar, 1 µm. Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We coexpressed mCh-EGFP-LC3 with each Halo-tagged effector and then assayed comigration in the axon. The distal axon was excluded due to minimal mCh-only autolysosomes in that region (Fig. 8 B). JIP1 (Fig. 8, C and D), HAP1 (Fig. 8, E and F), and RILP (Fig. S5, G and H) primarily associated with immature autophagosomes. In contrast, JIP3 displayed no maturation-based preference (Fig. 8, G and H), consistent with its lack of location-based preference (Fig. 1). Thus, some motor regulators associate with autophagosomes based on maturity.
Maturity regulates the association and function of dynein effectors
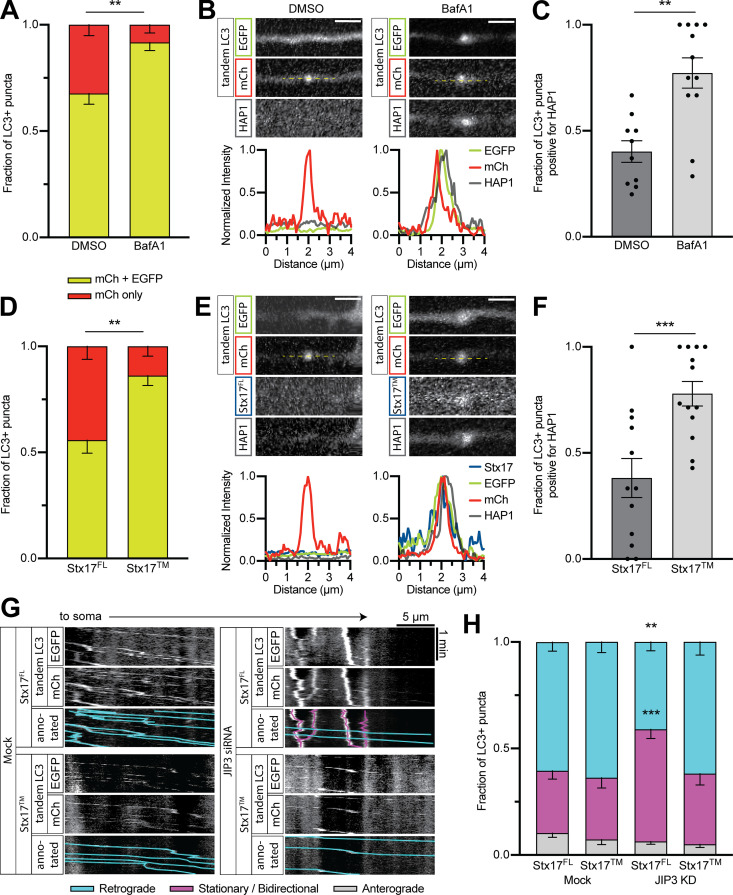
We next investigated whether autophagosomal dynein scaffolding was regulated by the maturation process. The drug bafilomycin A1 (BafA1) blocks acidification by interrupting autophagosome–lysosome fusion and inhibiting the vacuolar ATPase (Mauvezin et al., 2015). We treated neurons coexpressing mCh-EGFP-LC3 and Halo-HAP1 with 100 nM BafA1 or equal volume of vehicle (DMSO) for 2 h and imaged autophagosomes in the proximal axon. BafA1 treatment increased the fraction of EGFP+ autophagosomes, indicating more immature autophagosomes (Fig. 9 A). Accordingly, we saw an increase in the number of HAP1+ autophagosomes in the proximal axon (Fig. 9, B and C).
Figure 9.
Maturity regulates the association and function of dynein effectors. (A) Quantification of autophagosomal maturity under BafA1 or vehicle only (DMSO) treatment. n = 11–14 neurons. Mann-Whitney U test of fraction with EGFP (P = 0.0050). (B and C) Example line scans and quantification showing HAP1 colocalization with LC3 in the proximal axon after BafA1 treatment. n = 10–12 neurons. Mann-Whitney U test (P = 0.0007). Scale bar, 2 µm. (D) Quantification of autophagosomal maturity under SNAP-Stx17FL or dominant-negative mutant (Stx17TM) expressing conditions. n = 12–13 neurons. Mann-Whitney U test of fraction with EGFP (P = 0.0013). (E and F) Example line scans and quantification showing HAP1 colocalization with LC3 in the proximal axon with Stx17 expression. n = 12–13 neurons. Mann-Whitney U test (P = 0.0020). Scale bar, 2 µm. (G and H) Example kymographs and quantification of LC3+ puncta motility in the proximal axon following JIP3 siRNA transfection and/or Stx17 expression. n = 19–20 neurons. Two-way ANOVA with Tukey’s multiple comparisons test for all LC3+ puncta (retrograde: mock Stx17FL vs. mock Stx17TM, P = 0.9372; mock Stx17FL vs. KD Stx17FL, P = 0.0037; mock Stx17FL vs. KD Stx17TM, P = 0.9954; KD Stx17FL vs. KD Stx17TM, P = 0.0019; stationary/bidirectional: mock Stx17FL vs. mock Stx17TM, P > 0.9999; mock Stx17FL vs. KD Stx17FL, P = 0.0003; mock Stx17FL vs. KD Stx17TM, P = 0.9018; KD Stx17FL vs. KD Stx17TM, P = 0.0044.) Bars throughout show mean ± SEM. **, P < 0.01; ***, P < 0.001 compared to mock.
SNARE protein Syntaxin 17 (Stx17FL) interacts with SNAP29 and VAMP8 to induce autophagosome–lysosome fusion; expression of a transmembrane-only (Stx17TM) version of the protein has a dominant-negative effect on fusion (Itakura et al., 2012). We coexpressed mCh-EGFP-LC3, Halo-HAP1, and either SNAP-Stx17FL or SNAP-Stx17TM and then imaged in the proximal axon. Like BafA1, expression of Stx17TM increased the fraction of EGFP+ autophagosomes (Fig. 9 D) and resulted in more HAP1+ autophagosomes (Fig. 9, E and F).
Although JIP3 is present on autophagosomes throughout the axon, JIP3 depletion only affected autophagosomal motility in the mid- and proximal axon (Fig. 7) where autophagosomes are more mature (Fig. 8). We therefore asked whether the function of JIP3 on autophagosomes is maturation dependent. We cotransfected neurons with JIP3 siRNA, mCh-EGFP-LC3, and either SNAP-Stx17FL or SNAP-Stx17TM and then measured motility in the proximal axon. Autophagosomes in cells without siRNA (Mock) moved retrograde regardless of Stx17 construct (63% ± 5%; Fig. 9, G and H). Autophagosomes in cells cotransfected with JIP3 siRNA and Stx17FL displayed less motility (41% ± 4%; Fig. 9, G and H), as seen previously with JIP3 KD; however, expression of Stx17TM in JIP3 KD cells rescued normal autophagosomal motility (62% ± 6%; Fig. 9, G and H). This restored autophagosomal motility is likely facilitated by HAP1 based on HAP1’s increased colocalization with LC3 upon Stx17TM expression (Fig. 9, E and F). JIP3 thus drives the transport of mature autolysosomes, despite its comigration with both mature and immature autophagosomes, while HAP1 associates specifically with immature autophagosomes.
Discussion
We find that the motor effector proteins JIP1, HAP1, and JIP3 associate with axonal autophagosomes and regulate their motility (Fig. 10). Motile organelles, including autophagosomes, associate simultaneously with plus-end–directed and minus-end–directed motors (Maday et al., 2012; Hendricks et al., 2010). These opposing motors compete and/or coordinate to direct motility (Kural et al., 2005; Müller et al., 2008; Hendricks et al., 2010; Kunwar et al., 2011; Hancock, 2014; Fu and Holzbaur, 2014). Regulatory proteins that modulate motor function are essential to determine the net direction of motion (Hancock, 2014; Elshenawy et al., 2019, 2020; Feng et al., 2020). For axonal autophagosomes, which demonstrate highly processive unidirectional motility essential for their function (Maday et al., 2012; Wong and Holzbaur, 2014; Fu et al., 2014), this regulation is critical.
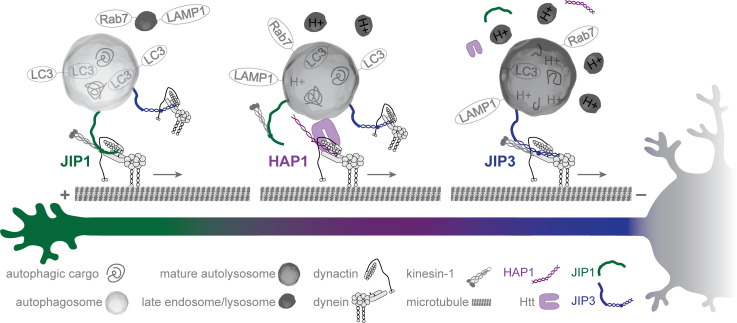
Figure 10.
Dynein scaffolding on axonal autophagosomes is regulated by maturation. Graphic summary of the model. Autophagosomes form in the distal tip (left) where they fuse with endolysosomes and bind JIP1, promoting the initiation of retrograde motility. In the mid-axon (middle), HAP1 binds autophagosomes via Htt and activates their dynein-driven motility. As the autophagosome matures, HAP1 and JIP1 disassociate and JIP3 promotes the continued retrograde motility in the mid- and proximal axon (right). Finally, degradation and recycling are completed in the soma.
Previous work has implicated JIP1, HAP1, and JIP3 in autophagosome transport (Fu et al., 2014; Wong and Holzbaur, 2014; Hill et al., 2019), but our study now uncovers the interplay between these effectors. This handoff between dynein regulators on a single cargo is unique, but may represent a more general paradigm relevant to other organelles, especially those that mature (e.g., phagosomes) or traverse long distances (e.g., signaling endosomes). For example, previous work found the dynein effector Hook1 activates motility of signaling endosomes only in the distal axon (Olenick et al., 2019), suggesting another effector may take over more proximally.
We find JIP1 localizes specifically to autophagosomes in the distal and mid-axon (Fig. 1), consistent with previous work showing JIP1 is required for exit from the distal tip and sustained motility in the mid-axon (Fu et al., 2014). JIP1 interacts with kinesin and dynein, with phosphorylation of residue S421 acting as a switch between anterograde and retrograde transport (Fu and Holzbaur, 2013; Fu et al., 2014). JIP1-LC3 binding blocks KHC activation in motility assays (Fu et al., 2014), but it is unclear whether JIP1 directly activates dynein.
Our finding that HAP1 and Htt drive autophagosome transport in the mid-axon (Fig. 2) is consistent with previous work in DRG axons (Wong and Holzbaur, 2014). We identify conserved dynein- and dynactin-binding sites within HAP1 (Figs. 3 and 4) that are necessary for autophagosome motility (Fig. 6). The CC1 box motif forms a hydrophobic pocket, found in multiple unrelated dynein effector families, that is necessary for the binding of dynein subunit LIC1 (Gama et al., 2017; Lee et al., 2018, 2020; Celestino et al., 2019). Our work shows the CC1 box in HAP1 is required for dynein activation on autophagosomes in neurons (Fig. 6). Likewise, a mutation in the CC1 box of BICD causes a hypomorphic loss-of-function phenotype in Drosophila (Oh et al., 2000). The well-conserved Spindly motif binds the pointed-end complex of dynactin’s Arp1 filament (Gama et al., 2017) and is required for autophagosomal transport in cells (Fig. 6) and robust dynein–dynactin complex motility in vitro (Schroeder and Vale, 2016). A popular hypothesis suggests the pointed-end complex protein p25 contains a mutually exclusive binding site that either binds a Spindly motif or the tail of autoinhibited p150; therefore, activator binding to p25 releases p150 from autoinhibition (Cianfrocco et al., 2015; Schroeder and Vale, 2016; Qiu et al., 2018; Lau et al., 2021).
We identify a third conserved binding site through which dynein-activating adaptors can bind dynactin, independent of the Spindly motif. This p150Glued binding site is conserved and functional among the HAP/TRAK and HOOK families of dynein effectors but is not found in all dynein effectors (Figs. 4, 6, and S3). A yeast two-hybrid assay showed that this region in HAP1 binds the second coiled-coil region of p150 (Engelender et al., 1997), which has been suggested to regulate membrane binding by dynactin (Kumar et al., 2001). Because this motif falls within a coiled-coil region, structural studies are essential to determine whether the mutations affect coiled-coil formation and to better understand the interaction.
Additionally, HAP1 likely binds MTs (Fig. 5, H and I) independently of dynein and kinesin (Fig. S3, E–H), providing an additional anchor to the MT that may help the motor complex better navigate crowded MT surfaces during the long trip through the axon (Henrichs et al., 2020). Together with HAP1’s ability to induce dynein activity in single-molecule motility assays (Fig. 5), these findings introduce HAP1 to the growing family of dynein-activating adaptors (Reck-Peterson et al., 2018; Olenick and Holzbaur, 2019).
We find that Htt is required in complex with HAP1 to direct mid-axon autophagosomal motility (Fig. S2). We hypothesize that the primary role of Htt in autophagosome transport is as a scaffold to bring the dynein–dynactin–HAP1 complex into contact with the autophagosome. Htt interacts with HAP1, dynein, dynactin, and multiple autophagosomal membrane proteins, including LC3B and GABARAPL1, making it the perfect scaffold to link the HAP1–motor complex to autophagosomes (Li et al., 1996; Engelender et al., 1997; Li et al., 1998; Caviston et al., 2007; Ochaba et al., 2014; Saudou and Humbert, 2016).
While JIP3 is present on autophagosomes throughout the axon, it primarily drives the motility of autolysosomes (Figs. 7 and 9), consistent with its canonical role in endolysosomal transport (Drerup and Nechiporuk, 2013). JIP3 interacts with kinesin and dynein–dynactin (Cockburn et al., 2018; Cavalli et al., 2005; Vilela et al., 2019) and regulates axonal autophagosome transport in C. elegans (Hill et al., 2019). JIP3 may interact with the autophagosome via its binding partner, Arf6, which is important for autophagosomal trafficking in zebrafish (Montagnac et al., 2009; George et al., 2016).
In HAP1 (Fig. 2) and JIP3 (Fig. 7) KD experiments, we observed low levels of persistent retrograde transport. RILP was recently shown to mediate autophagosomal transport in rat cortical neurons (Khobrekar et al., 2020), but RILP KD did not induce a robust effect in rat hippocampal neurons (Fig. S4). We propose that RILP may be playing an indirect role in autophagosomal transport, perhaps via v-ATPase regulation (De Luca et al., 2014), since autophagosomal acidification mediates autophagosome transport (Fig. 9).
Maturation regulates the association and function of dynein effectors on the autophagosome. HAP1 is aberrantly retained on proximal autophagosomes following pharmacological or genetic inhibition of autophagosome maturation, indicating that maturation induces dissociation (Fig. 9). JIP3 interacts indiscriminately with autophagosomes but regulates the transport of autolysosomes (Figs. 7, 8, and 9). Maturation could affect motor scaffolding through a number of mechanisms. The dissociation of immature effectors (HAP1 and JIP1) may be caused by loss of binding partners (e.g., LC3) on the fused membrane, or a membrane protein acquired during fusion could promote dissociation. Membrane proteins acquired during fusion could also activate mature dynein scaffolds (JIP3), or acidification could trigger an inside-out signaling cascade causing dissociation and/or activation of scaffolding proteins. While there is a clear genetic interaction between autophagosomal maturation and transport (Hill et al., 2019), more work is essential.
Htt, HAP1, JIP1, and JIP3 have all been implicated in neurodegenerative disease (Helbecque et al., 2003; Gunawardena et al., 2003; Beeler et al., 2009; Weiss and Littleton, 2016; Choudhary et al., 2017; Gowrishankar et al., 2017; Liu et al., 2020). Since both axonal transport and autophagy are strongly associated with neuronal health and neurodegeneration, future studies on the regulation of axonal autophagosome transport are likely to provide valuable insights into human disease.
Materials and methods
Plasmids and reagents
Constructs, all of which were verified by DNA sequencing, are included in Table 1.
Table 1. Plasmids and reagents.
| Construct | Source |
|---|---|
| pEGFP-LC3B | Gift from T. Yoshimori, Osaka University, Osaka, Japan (rat) |
| mScarlet-LC3B | Subcloned from above with mScarlet (Addgene 85054) |
| mCh-EGFP-LC3 | Gift from T. Johansen, University of Tromsø, Tromsø, Norway |
| BFP-JIP1 | Human myc-JIP1 (gift from D. Holtzmann, Washington University in St. Louis, St. Louis, MO) subcloned into pBI-BFP plasmid |
| Halo-JIP1 | Subcloned from above with HaloTag on the N terminus of JIP1 in pEGFP vector backbone |
| Halo-HAP1 | HAP1 isoform 2 (gift from X. J. Li, Emory University, Atlanta, GA) subcloned into pHTN HaloTag vector (Promega, G7721) was lengthened into isoform 1 using transOMIC. The initial HAP1 isoform 2 differed from the National Center for Biotechnology Information database sequence by two codons and one amino acid; those differences were propagated to the final construct but had no effect on protein function or expression. HAP1 truncated and mutant constructs were all generated from this final construct. |
| HAP1-mCh-DHFR | mCherry, HAP1, and DHFR domains fused in pHTN vector backbone |
| Halo-JIP3 | HaloTag fused to the N terminus of hJIP3 from cDNA Clone (GE: MGC9053013) fused in pEGFP vector backbone |
| mCh-Htt | pARIS, Q23 (Pardo et al., 2010); gift from F. Saudou, Institut Curie, Paris, France |
| EGFP | pEGFP-N1 vector (Clontech, 6085-1) alone (for GFP fill) |
| HaloTag | pHTN vector with STOP codon following the HaloTag in order to express tag (for negative control) |
| PEX3-GFP-Halo | N-terminal 42 aa of human PEX3 for peroxisome targeting (Kapitein et al., 2010) generated in laboratory via PCR for a previous paper (Olenick et al., 2016) |
| BICD2N-Halo | Full-length mouse BICD2 in the pEGFP vector (gift from A. Akhmanova, Utrecht University, Utrecht, the Netherlands) was used to generate a truncated construct spanning residues 1–572 fused to the HaloTag and cloned into pcDNA3.1 |
| BICD2N-mCh-DHFR | Gift from E. Ballister, University of Pennsylvania, Philadelphia, PA |
| KHC-Halo | Full-length mouse kinesin-1 heavy chain (KIF5B) in pRK5 myc plasmid (gift from J. Kittler, University of Surrey, Guildford, UK) with HaloTag fused to the C terminus |
| K560-Halo | First 560 aa of human KIF5B from pET17: K560 GFP ST (gift from R. Vale, University of California, San Francisco, San Francisco, CA) subcloned into pHTC-HaloTag CMVneo vector (Promega) |
| EGFP-p25 | Gift from T. Schroer, Johns Hopkins University, Baltimore, MD |
| FLAG-p150 | Gift from T. Schwarz, Boston Children’s Hospital, Boston, MA |
| CBD-HAP1CC1 | Chitin binding domain (CBD) and HAP1 CC1 (residues 168-261) cloned in-frame into pRSF-Duet1 (EMD Millipore) |
| MBP-LIC1 (WT and FFAA) | Created in Lee et al. (2018) |
| LAMP1-EGFP | Subcloned from Addgene 1817, replacing RFP with EGFP |
| EGFP-Rab7 | Addgene 12605 |
| SNAP-Stx17 (FL and TM) | Addgene 45909-45910 subcloned into pSNAPf vector (New England Biolabs) |
| RILP-Halo | RILP (gift from J. Neefjes, Leiden University, Leiden, the Netherlands) subcloned into pHTC vector |
| Halo-Hook1 | WT and mutant constructs generated from human Hook1 sequence with HaloTag from the pHTN Halo tag CMV-neo vector (Promega; Olenick et al., 2016) |
As an alternative to transfecting an LC3 plasmid, some experiments used the Premo Autophagy Sensor BacMam 2.0 LC3B-GFP (P36235; Thermo Fischer Scientific), which introduces DNA via insect Baculovirus with a Mammalian promoter. Individual ON-TARGETplus siRNA to HAP1 (5′-GAAGUAUGUCCUCCAGCAAUU-3′), Htt (5′-GCAGCUUGUCCAGGUUUAUUU-3′), JIP3 (5′-CAGCUGGCUUUAGCCAGCGUCGCAAUU-3′), and RILP (5′-CGGUGAACAUCUUGGUCUG-3′), plus an individual siGENOME to KIF5B (5′-GCAGUCAGGUCAAAGAAUA-3′), were obtained from Dharmacon (Horizon Discovery). Chemical compounds used include LysoTracker Deep Red (L12492; Thermo Fisher Scientific) and BafilomycinA1 (B1793; Sigma-Aldrich). HaloTag constructs were labeled with Janelia Fluor 646 (GA1120; Promega), Janelia Fluor 549 (GA1110; Promega), or TMR (G8251; Promega). SNAP-Tag constructs were labeled with SNAP-Cell 430 (S9109S; New England BioLabs) or SNAP-Cell 647-SiR (S9102S; New England BioLabs), or Janelia Fluor 646 (provided by Luke Lavis [Janelia, Ashburn, VA]).
Antibodies used are included in Table 2.
Table 2. . Antibodies.
| Antibody | Source | Application/dilution | |
|---|---|---|---|
| Anti-LC3 | ab48394 (Abcam) | WB @ 1:1,000; PLA @ 1:250 | |
| Anti-GM130 | 610823 (BD Biosciences) | WB @ 1:500 | |
| Anti-dynein light intermediate chain 1 | NBP2-14935 (Novus) | WB @ 1:250 | |
| Anti-dynein light intermediate chain 1 | ab157468 (Abcam) | WB @ 1:500 | |
| Anti-JIP1 | AF4366 (Novus) | WB @ 1:400 | |
| Anti-HAP1 | NB110-74569 (Novus) | WB @ 1:500 | |
| Anti-HAP1 | MA1-46412 (Thermo Fisher Scientific) | WB @ 1:500 | |
| Anti-JIP3 | ab196761 (Abcam) | WB @ 1:200 | |
| Anti-JIP3 | SC-46663 (Santa Cruz Biotechnology) | WB @ 1:50 | |
| Anti-JIP3 | KIAA1066 (MyBioSource) | WB @ 1:300 | |
| Anti-Htt | MAB2166 (EMD Millipore) | WB @ 1:500 | |
| Anti-Htt | ab109115 (Abcam) | WB @ 1:2,000 | |
| Anti-Halo (polyclonal) | G9281 (Promega) | WB @ 1:500; PLA @ 1:400 | |
| Anti-Halo (monoclonal) | G9211 (Promega) | WB @ 1:500; PLA @ 1:250 | |
| Anti-dynein intermediate chain | MAB1618 (EMD Millipore) | WB @ 1:1,000; PLA @ 1:200 | |
| Anti-p150 | 610474 (BD Biosciences) | WB @ 1:10,000 | |
| Anti-GFP | GFP-1020 (Aves Labs) | WB @ 1:500 | |
| Anti-CBD | E80345 (New England Biolabs) | WB @ 1:800 | |
| Anti-tubulin | T5201 (Sigma-Aldrich) | TIRF @ 1:40 | |
| Anti-RILP | ab140188 (Abcam) | WB @ 1:250 | |
WB, Western blot.
Primary hippocampal culture
Sprague-Dawley rat hippocampal neurons at embryonic day 18 were obtained from the Neurons R Us Culture Service Center at the University of Pennsylvania. Cells (proximity ligation assay, 40,000 cells on 7-mm glass; live imaging, 200,000 cells on 20-mm glass) were plated in glass-bottom 35-mm dishes (MatTek) that were precoated with 0.5 mg/ml poly-L-lysine (Sigma-Aldrich). Cells were initially plated in attachment media (MEM supplemented with 10% horse serum, 33 mM D-glucose, and 1 mM sodium pyruvate) that was replaced with maintenance media (Neurobasal [Gibco] supplemented with 33 mM D-glucose, 2 mM GlutaMAX [Invitrogen], 100 U/ml penicillin, 100 mg/ml streptomycin, and 2% B-27 [Thermo Fisher Scientific]) after 5–20 h. Neurons were maintained at 37°C in a 5% CO2 incubator. Cytosine arabinoside (final concentration, 1 µM) was added the day after plating to prevent glia cell proliferation. For DNA only or DNA plus siRNA transfections, neurons (4–6 DIV) were transfected with 0.35–1.5 µg of total plasmid DNA and optional 45 pmol siRNA using Lipofectamine 2000 Transfection Reagent (11668030; Thermo Fisher Scientific) and incubated for 18–48 h. For siRNA only transfections (Figs. 2 and S2), neurons were transfected with 45 pmol siRNA using Lipofectamine RNAiMAX (13778030; Thermo Fisher Scientific) and incubated for 36–48 h. To label LC3 in those experiments, ∼20 multiplicity of infection BacMam LC3 (40 μl) was added immediately following transfection.
Live-cell neuron imaging and analysis
One hour before imaging, HaloTag ligands and/or most SNAP-tag ligands were applied for 15 min at a final concentration of 100 nM, followed by a 30–45-min washout. SNAP-tag ligand Blue 430 was applied for 30 min at a final concentration of 2 µM, followed by a 30-min washout. In applicable experiments, neurons were incubated with Lysotracker (25 nM) for 15–30 min, which was then removed for imaging. In applicable experiments, BafilomycinA1 (100 nM) or DMSO was added 2 h before imaging, and then neurons were imaged in the third hour of continued treatment. Neurons were imaged in Imaging Media (HibernateE [Brain Bits] supplemented with 2% B27 and 33 mM D-glucose). Autophagosome behavior was monitored in the proximal (<100 µm from the soma), distal (<100 µm from the distal tip), or mid-axon of 6–8 DIV neurons imaged at a rate of 1 time point/s for 2–3 min. Neurons were imaged in an environmental chamber at 37°C on a PerkinElmer UltraView Vox spinning-disk confocal on a Nikon Eclipse Ti Microscope with an Apochromat 100 × 1.49 NA oil-immersion objective and a Hamamatsu EMCCD C9100-50 camera driven by Volocity (PerkinElmer). Only cells expressing moderate levels of fluorescent proteins were imaged to avoid overexpression artifacts or aggregation. It should be noted that the quality of the primary neuron dissections affected autophagosomal motility, but compared conditions were always collected from the same dissections and imaging sessions.
Kymographs were generated in ImageJ (https://imagej.net/ImageJ2) using the MultiKymograph plugin (line width, 5) and analyzed either in ImageJ or using the MatLab program KymoSuite (J. Nirschl, University of Pennsylvania, Philadelphia, PA). Puncta were classified as either anterograde (moving ≥10 µm toward the axon tip), retrograde (moving ≥10 µm toward the soma), or stationary/bidirectional (net movement <10 µm during the video). Because fluorescent LC3 is cytosolic (as well as punctate) and neurites occasionally crossed in culture, raw videos were referenced throughout kymograph analysis to ensure only real puncta (≥1.5 SD from the axon mean) were included in analyses. All comigration analyses were performed using kymographs. Line scans were generated for presentation purposes from raw video stills and normalized either within that line (for positive channels) or to the local region (for negative channels; surrounding ∼10-µm area).
Autophagosome fractionation
Enriched autophagosome fractions were isolated from mouse brain via sequential ultracentrifugation, adding Gly-Phe-β-naphthylamide to inactivate and deplete lysosomal vesicles and thus enhance the integrity of autophagosome-associated proteins (Strømhaug et al., 1998). Detailed protocols and validations can be found in Goldsmith et al. (unpublished data). Briefly, brains were collected from WT mice on C57BL/6J background (Ref 14699058) and homogenized in a tissue grinder in ice-cold buffered 10 mM Hepes, 1 mM EDTA, and 250 mM sucrose solution, and then subjected to three differential centrifugations through Nycodenz and Percoll discontinuous gradients to isolate vesicles of the appropriate size and density. The autophagosome-enriched fraction was then divided and either immediately lysed for the identification of all internal and externally associated proteins on autophagosomes (A fraction), treated with 10 µg proteinase K for 45 min at 37°C to degrade externally associated proteins and enrich for membrane-protected autophagosome cargo (P fraction), or membrane permeabilized by addition of 0.2% Triton X-100 before proteinase K treatment to confirm proteinase K efficacy (T fraction). The lysis buffer used contained a final concentration of 0.5% NP-40 with 1× protease and phosphatase inhibitors, PMSF and Pepstatin A. Protein concentration was measured by Bradford assay and equal amounts of protein in denaturing buffer were run on SDS-PAGE gels.
Immunoblotting
For fluorescence Western blotting, samples were analyzed by SDS-PAGE and transferred onto polyvinylidene difluoride Immobilon FL (Millipore). Membranes were dried for at least 1 h, rehydrated in methanol, and stained for total protein (LI-COR REVERT Total Protein Stain). Following imaging of the total protein, membranes were destained, blocked for 1 h in TrueBlack Blocking Buffer (23013; Biotium), and incubated overnight at 4°C with primary antibodies diluted in TrueBlack Antibody Dilutent (23013; Biotium) with 0.2% Tween-20. Membranes were washed four times for 5 min in 1×TBS washing solution (50 mM Tris-HCl [pH 7.4], 274 mM NaCl, 9 mM KCl, 0.1% Tween-20), incubated in secondary antibodies diluted in TrueBlack Antibody Dilutent (23013; Biotium) with 0.2% Tween-20 and 0.01% SDS for 1 h, and again washed four times for 5 min in washing solution. Membranes were immediately imaged using an Odyssey CLx Infrared Imaging System (LI-COR). In a limited number of cases, the membrane was stripped using NewBlot IR Stripping Buffer (928–40028; LI-COR) according to manufacturer’s instructions. Band intensity was measured in the LI-COR Image Studio application.
PLA
Neurons were transfected (Lipofectamine 2000) with 0.3 µg EGFP plasmid (for GFP fill) and 0.5 µg Halo-tagged effector following above protocol and then 24 h later (DIV 7–8) fixed in PBS containing 4% paraformaldehyde and 4% sucrose for 8 min. Duolink In Situ PLA Mouse/Rabbit kit with red detection reagents (DUO92101-1KT; Sigma-Aldrich) was used according to manufacturer’s protocol. A Halo antibody (either mouse G9211 or rabbit G9281) was used in every experiment along with either an LC3 antibody (rabbit ab48394), a dynein intermediate chain antibody (mouse MAB1618), or no second 1° antibody (negative control). Both 2° antibodies (mouse and rabbit) were added for all experiments, including negative control. Z-stacks (0.25-µm steps) were acquired on an inverted epifluorescence microscope (DMI6000B; Leica) with an Apochromat 63 × 1.4 NA oil immersion objective and a charge-coupled device camera (ORCA-R2; Hamamatsu Photonics) using LAS-AF software (Leica). Puncta were counted manually using ImageJ.
Cell line culture
COS-7 cells (American Type Culture Collection [ATCC]) were maintained in DMEM (Corning) supplemented with 1% GlutaMAX and 10% FBS. PC12 cells (ATCC) were maintained in DMEM supplemented with 1% GlutaMAX, 5% FBS, and 5% horse serum. Cells were maintained at 37°C in a 5% CO2 incubator. For peroxisome recruitment assay experiments, COS-7 cells were plated on 35-mm glass-bottom plates. 24 h before imaging, cells were cotransfected with human PEX3-GFP-Halo and either human HAP1-mCherry-eDHFR or mouse BICD2N-mCherry-eDHFR using FuGENE 6 (1 µg total DNA; Promega), and 48 h before imaging, with control siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific). For motility assays and coimmunoprecipitation experiments, COS-7 cells were plated on 10-cm plates and transfected 24 h before lysis using FuGENE 6 (6–12 µg total DNA; Promega). For siRNA tests, PC12 cells were plated in 6-well dishes and transfected 48 h before lysis with 45 pmol siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific). Cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) and protease inhibitors (1 mM PMSF, 0.01 mg/ml Nα-p-tosyl-L-arginine methyl ester [TAME], 0.01 mg/ml leupeptin, 0.001 mg/ml pepstatin A, 1 mM DTT) for 30 min at 4°C followed by centrifugation at 4°C for 10 min at 17,000 × g and a BCA assay of the supernatant to determine total protein concentration. Cells were routinely tested for mycoplasma using a MycoAlert detection kit (LT07; Lonza). COS-7 and PC12 cells were authenticated by ATCC.
Protein purification
BICD2N-Halo and MBP-LIC1 (WT and FFAA) were purified as described previously (Lee et al., 2018). In short, proteins were expressed in Escherichia coli BL21 (DE3) cells (Invitrogen), grown in Terrific Broth medium at 37°C, followed by 16 h at 19°C in the presence of 0.25 mM IPTG. Cells were collected by centrifugation, resuspended in 20 mM Tris (pH 7.0), 100 mM NaCl, 4 mM benzamidine hydrochloride, 1 mM PMSF, and 1 mM DTT, and then lysed using a Microfluidizer large-scale homogenizer (Microfluidics). The proteins were purified through an amylose affinity column according to the manufacturer’s protocol (New England Biolabs). The proteins were then loaded onto a StrepTactin Sepharose column (IBA Lifesciences) and eluted after extensive washing with 3 mM desthiobiotin, 20 mM Tris (pH 7.0), 100 mM NaCl, and 1 mM DTT.
CBD-HAP1CC1 was expressed in ArcticExpress(DE3) RIL cells (Agilent Technologies), grown in Terrific Broth medium for 6 h at 37°C to a density of ∼1.5 to 2 OD at 600 nm (OD600), followed by 24 h at 10°C in the presence of 0.4 mM IPTG. Cells were harvested by centrifugation, resuspended in 20 mM Hepes (pH 7.5), 200 mM NaCl, 10 mM imidazole, and 2 mM PMSF and then lysed using a microfluidizer (Microfluidics). The protein was first purified using Ni-NTA resin according to the manufacturer’s protocol and eluted using the same buffer supplemented with 300 mM imidazole. A final step of purification was performed on a gel filtration SD200HL 16/60 column (GE Healthcare) in 20 mM Hepes (pH 7.5) and 100 mM NaCl.
Coimmunoprecipitation and pull-down experiments
COS-7 cells were lysed 24 h after transfection in 50 mM Hepes (pH 7.4), 1 mM EDTA, 1 mM MgCl2, and 25 mM NaCl, with 0.5% Triton X-100 and protease inhibitors (1 mM PMSF, 0.01 mg/ml TAME, 0.01 mg/ml leupeptin, 0.001 mg/ml pepstatin A, 1 mM DTT), and then clarified at 17 × g at 4°C for 10 min. Dynabeads Protein G (incubated with anti-p150 antibody [BD 610474] for 15 min before addition of lysate; 10007D; Thermo Fisher Scientific) or Magne HaloTag (Promega G7281) beads were then washed with lysis buffer and incubated with lysate for 15 min at 25°C. Following incubation, beads were washed 3 × 300 µl in lysis buffer and then resuspended in 60 µl denaturing buffer and boiled to release the bound proteins. Coimmunoprecipitation was analyzed by Western blot.
Pull-down experiments with exclusively purified protein (Fig. 3, B and C) were performed in 50 mM Hepes (pH 7.5) with 100 mM NaCl, 1 mM DTT, and 0.1% Tween20 (PD buffer). MBP-LIC1 (50 pmol) and either BICD2N-Halo (50 pmol) or CBD-HAP1CC1 (100 pmol) was incubated with resin (Magne HaloTag; Chitin, New England BioLabs S6651; or Amylose, New England Biolabs E8021) for 1 h at 4°C. For pull-down experiments mixing purified protein and cell lysate (Fig. S2, I and J), COS-7 cells were lysed 24 h after transfection in 50 mM Hepes (pH 7.4), 1 mM EDTA, 1 mM MgCl2, and 25 mM NaCl, with 0.1% Tween 20 and protease inhibitors and then clarified at 10 × g at 4°C for 10 min. Lysate from three 10-cm plates was incubated with MBP-LIC1 (50 pmol) and Amylose resin for 30 min at 25°C. In both cases, following incubation, resin was washed 3 × 500 µl in PD buffer and then resuspended in 100 µl denaturing buffer and boiled to release the bound proteins. Protein pulldown was analyzed by Western blot.
Peroxisome recruitment assay
The uncaged dimerizer TMP-Htag (Ballister et al., 2015) was dissolved in DMSO to 10 mM concentration and stored at −20°C. After capturing stills and a 2-min predimerizer video, the dimerizer (or DMSO) was applied at a working concentration of 10 μM dropwise directly to the cells followed by a 25-min video of the dimerization (0.5 frames/s) followed by postdimerizer stills. Imaging medium was L-15 medium (Thermo Fisher Scientific) supplemented with 10% FBS. Videos were acquired at 37° on an inverted epifluorescence microscope (DMI6000B; Leica) with an Apochromat 63 × 1.4 NA oil immersion objective and a charge-coupled device camera (ORCA-R2; Hamamatsu Photonics) using LAS-AF software (Leica). Color-coded time projections were generated in ImageJ using a publicly available plugin (K. Miura, EMBL Heidelberg, Heidelberg, Germany; https://imagej.net/Temporal-Color_Code).
Motility assay
The movement of HAP1- or BICD2N-containing complexes from cell extracts was tracked using TIRF microscopy. Motility assays were performed in flow chambers constructed with a glass slide and a coverslip silanized with PlusOne Repel-Silane ES (GE Healthcare), held together with vacuum grease to form a ∼10-µl chamber. Rigor kinesin-1E236A (0.5 µM) was nonspecifically absorbed to the coverslip (Wagenbach et al., 2008), and the chamber was then blocked with 5% pluronic F-127 (Sigma-Aldrich). 250 nM GMPCPP MT seeds, labeled at a 1:40 ratio with HiLyte Fluor 647 tubulin (Cytoskeleton), were flowed into the chamber and immobilized by interaction with rigor kinesin-1E236A. 11.25 µM free tubulin (labeled at a 1:20 ratio with HiLyte Fluor 488 tubulin) was added with the lysate to grow dynamic MTs from the seeds. COS-7 cells grown in 10-cm plates to 70% to 80% confluence expressing full-length Halo-tagged HAP1, BICD2N, or HaloTag alone were labeled with TMR 18–24 h after transfection and then lysed in 100 µl lysis buffer (40 mM Hepes [pH 7.4], 120 mM NaCl, 1 mM EDTA, 1 mM ATP, 0.1% Triton X-100, 1 mM PMSF, 0.01 mg⋅ml−1 TAME, 0.01 mg⋅ml−1 leupeptin, and 1 µg⋅ml−1 pepstatin-A). Cell lysates were clarified by centrifugation (17,000 × g) and diluted in P12 motility buffer (12 mM Pipes [pH 6.8], 1 mM EGTA, and 2 mM MgCl2) supplemented with 1 mM Mg-ATP, 1 mM GTP, 0.08 mg⋅ml−1 casein, 0.08 mg⋅ml−1 BSA, 2.55 mM DTT, 0.05% methylcellulose, and an oxygen-scavenging system (0.5 mg⋅ml−1 glucose oxidase, 470 U⋅ml−1 catalase, and 3.8 mg⋅ml−1 glucose). All videos (2 min, 4 frames⋅s−1) were acquired at 37°C using a Nikon TIRF microscopy system (PerkinElmer) on an inverted Ti microscope equipped with a 100× objective and an ImageEM C9100-13 camera (Hamamatsu Photonics) with a pixel size of 0.158 µm and controlled with the program Volocity (Improvision). At least five MTs per video were analyzed by generating kymographs using the MultiKymograph plugin of ImageJ and analyzed in Excel (Microsoft). During acquisition, seeds (647) were imaged at a rate of 1 frame⋅min−1 and the free tubulin (488) at 12 frames⋅min−1 and only nonbundled MTs with a clear plus end (one end clearly growing faster and longer away from the seed) were analyzed. At least 15 MTs were analyzed per replicate; three biological and technical replicates were performed for a final n = 57 MTs per condition. Processive runs were defined as events (≥1 SD above the background) ≥1 µm in length and lasting ≥1 s. Runs ending within 0.5 µm of the MT minus end were excluded from the run-length analysis.
Kinesin MT binding assay
COS-7 cells grown in 10-cm plates to 70% to 80% confluence expressing Halo-tagged K560 or KIF5B (KHC) were labeled with TMR 18–24 h after transfection and then lysed in P12 buffer (as described above) supplemented with 20 µM Taxol, 1 mM Mg-ATP, 0.1% Triton, and 10 µM DTT supplemented with protease inhibitors (described above). Separate plates expressing Halo-HAP1 or Halo-HAP1 plus mCh-Htt were likewise lysed but not labeled. All lysates were clarified by a low speed (17,000 × g) and a high speed (287,582 × g) centrifugation. Flow chambers were assembled as described above. A 1:40 dilution of monoclonal anti-tubulin antibody was flowed in and incubated for 5 min. The chamber was then blocked with two 5-min incubations of 5% pluronic F-127 (Sigma-Aldrich). Labeled (labeling ratio of 1:40; HiLyte 488; Cytoskeleton) Taxol-stabilized MTs were then flowed into the chamber and immobilized on the antibody. Finally, diluted cell lysates were flowed in with P12 buffer containing 10 mM AMPPNP, 20 µM Taxol, 0.3 mg/ml BSA, 0.3 mg/ml casein, 10 mM DTT, and an oxygen-scavenging system (described above). One 3-min video was acquired for each chamber at 0.067 frames/s. The mean fluorescence intensity for five unbundled MTs per video was collected at 30-s intervals in ImageJ.
MT pelleting assay
Unlabeled tubulin was polymerized at 5 mg/ml in BRB80 (80 mM Pipes, 1 mM EGTA, and 1 mM MgCl2 [pH 6.8]) with 1 mM GMPCPP. To generate cell lysate, COS-7 cells transfected with Halo-HAP1 (for 18–20 h) were lysed in BRB80 buffer with 0.5% Triton X-100 and protease inhibitors (as described above) and clarified with two centrifugation steps (at 17,000 and 27,000 × g). 0 or 5 µM MTs were incubated with equal volumes of cell lysate at 37°C for 20 min, with 1 mM Mg-ATP or 150 mM KCl where appropriate. Then samples were centrifuged at 10,000 × g at 25°C for 20 min. The supernatant and the pellet were then separated, denatured, and analyzed by SDS-PAGE. Protein in the pellet and supernatant fractions was analyzed by Western blot.
Statistics
All statistical analyses were performed in Prism (GraphPad). All error bars represent SEM and n indicates the number of events or cells pooled across at least three trials per experiment. Parametric or nonparametric tests were used where appropriate, but formal normality testing was not performed. Statistical measures are described in the legends.
Online supplemental material
Fig. S1 shows candidate effectors closely apposed to dynein and autophagosomes in axons. Fig. S2 compares Htt KD in the different axonal subregions and illustrates the conservation of the novel Glued binding motif. Fig. S3 shows that HAP1 binds MTs independently of motors and is not required for autophagosome motility in the proximal axon. Fig. S4 illustrates RILP associates with axonal autophagosomes but plays only a minor role in their transport. Fig. S5 compares the autophagosomal maturation state in the axonal subregions.
Acknowledgments
We thank Mariko Tokito for technical assistance; Chanat Aonbangkhen and David Chenoweth (University of Pennsylvania, Philadelphia, PA) for the dimerizer reagent; somersault 18:24 for cartoons; and Andrea Stavoe, Chantell Evans, Alex Boecker, and Adam Fenton for insights and discussions.
This research was supported by National Institutes of Health grants R35 GM126950 (E.L.F. Holzbaur) and RM1 GM136511 (R. Dominguez and E.L.F. Holzbaur), National Institutes of Health T32 grant AR053461 (P.J. Carman), and National Science Foundation Graduate Research Fellowship DGE-1845298 (S.E. Cason).
The authors declare no competing financial interests.
Author contributions: S.E. Cason: conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, project administration, and writing – original draft and review/editing; P.J. Carman: methodology, resources, and writing – review/editing; C. Van Duyne: investigation, formal analysis, and writing – review/editing; J. Goldsmith: resources, validation, writing – review/editing; R. Dominguez: conceptualization, supervision, and writing – review/editing; E.L.F. Holzbaur: conceptualization, supervision, funding acquisition, project administration, and writing – original draft and review/editing.
References
- Alam, M.S. 2018. Proximity Ligation Assay (PLA). Curr. Protoc. Immunol. 123:e58. 10.1002/cpim.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto, M., Koushika S.P., Choudhary B.C., Li C., Matsumoto K., and Hisamoto N.. 2011. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31:2216–2224. 10.1523/JNEUROSCI.2653-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayloo, S., Lazarus J.E., Dodda A., Tokito M., Ostap E.M., and Holzbaur E.L.F.. 2014. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat. Commun. 5:4807. 10.1038/ncomms5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister, E.R., Ayloo S., Chenoweth D.M., Lampson M.A., and Holzbaur E.L.F.. 2015. Optogenetic control of organelle transport using a photocaged chemical inducer of dimerization. Curr. Biol. 25:R407–R408. 10.1016/j.cub.2015.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler, N., Riederer B.M., Waeber G., and Abderrahmani A.. 2009. Role of the JNK-interacting protein 1/islet brain 1 in cell degeneration in Alzheimer disease and diabetes. Brain Res. Bull. 80:274–281. 10.1016/j.brainresbull.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Boecker, C.A., Goldsmith J., Dou D., Cajka G.G., and Holzbaur E.L.F.. 2021. Increased LRRK2 kinase activity alters neuronal autophagy by disrupting the axonal transport of autophagosomes. Curr. Biol. 10.1016/j.cub.2021.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli, V., Kujala P., Klumperman J., and Goldstein L.S.B.. 2005. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 168:775–787. 10.1083/jcb.200410136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J.P., Ross J.L., Antony S.M., Tokito M., and Holzbaur E.L.. 2007. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA. 104:10045–10050. 10.1073/pnas.0610628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino, R., Henen M.A., Gama J.B., Carvalho C., McCabe M., Barbosa D.J., Born A., Nichols P.J., Carvalho A.X., Gassmann R., and Vögeli B.. 2019. A transient helix in the disordered region of dynein light intermediate chain links the motor to structurally diverse adaptors for cargo transport. PLoS Biol. 17:e3000100. 10.1371/journal.pbio.3000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, B., Kamak M., Ratnakaran N., Kumar J., Awasthi A., Li C., Nguyen K., Matsumoto K., Hisamoto N., and Koushika S.P.. 2017. UNC-16/JIP3 regulates early events in synaptic vesicle protein trafficking via LRK-1/LRRK2 and AP complexes. PLoS Genet. 13:e1007100. 10.1371/journal.pgen.1007100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco, M.A., DeSantis M.E., Leschziner A.E., and Reck-Peterson S.L.. 2015. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol. 31:83–108. 10.1146/annurev-cellbio-100814-125438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn, J.J.B., Hesketh S.J., Mulhair P., Thomsen M., O’Connell M.J., and Way M.. 2018. Insights into Kinesin-1 Activation from the Crystal Structure of KLC2 Bound to JIP3. Structure. 26:1486–1498.e6. 10.1016/j.str.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca, M., Cogli L., Progida C., Nisi V., Pascolutti R., Sigismund S., Di Fiore P.P., and Bucci C.. 2014. RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J. Cell Sci. 127:2697–2708. 10.1242/jcs.142604 [DOI] [PubMed] [Google Scholar]
- Drerup, C.M., and Nechiporuk A.V.. 2013. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 9:e1003303. 10.1371/journal.pgen.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshenawy, M.M., Canty J.T., Oster L., Ferro L.S., Zhou Z., Blanchard S.C., and Yildiz A.. 2019. Cargo adaptors regulate stepping and force generation of mammalian dynein-dynactin. Nat. Chem. Biol. 15:1093–1101. 10.1038/s41589-019-0352-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshenawy, M.M., Kusakci E., Volz S., Baumbach J., Bullock S.L., and Yildiz A.. 2020. Lis1 activates dynein motility by modulating its pairing with dynactin. Nat. Cell Biol. 22:570–578. 10.1038/s41556-020-0501-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender, S., Sharp A.H., Colomer V., Tokito M.K., Lanahan A., Worley P., Holzbaur E.L., and Ross C.A.. 1997. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 6:2205–2212. 10.1093/hmg/6.13.2205 [DOI] [PubMed] [Google Scholar]
- Feng, Q., Gicking A.M., and Hancock W.O.. 2020. Dynactin p150 promotes processive motility of DDB complexes by minimizing diffusional behavior of dynein. Mol. Biol. Cell. 31:782–792. 10.1091/mbc.E19-09-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, M.M., and Holzbaur E.L.F.. 2013. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol. 202:495–508. 10.1083/jcb.201302078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, M.M., and Holzbaur E.L.F.. 2014. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 24:564–574. 10.1016/j.tcb.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, M.M., Nirschl J.J., and Holzbaur E.L.F.. 2014. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev. Cell. 29:577–590. 10.1016/j.devcel.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama, J.B., Pereira C., Simões P.A., Celestino R., Reis R.M., Barbosa D.J., Pires H.R., Carvalho C., Amorim J., Carvalho A.X., et al. 2017. Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly. J. Cell Biol. 216:943–960. 10.1083/jcb.201610108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, R., Holland A.J., Varma D., Wan X., Çivril F., Cleveland D.W., Oegema K., Salmon E.D., and Desai A.. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971. 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, A.A., Hayden S., Stanton G.R., and Brockerhoff S.E.. 2016. Arf6 and the 5'phosphatase of synaptojanin 1 regulate autophagy in cone photoreceptors. BioEssays. 38(Suppl 1):S119–S135. 10.1002/bies.201670913 [DOI] [PubMed] [Google Scholar]
- Gill, S.R., Schroer T.A., Szilak I., Steuer E.R., Sheetz M.P., and Cleveland D.W.. 1991. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol. 115:1639–1650. 10.1083/jcb.115.6.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar, S., Wu Y., and Ferguson S.M.. 2017. Impaired JIP3-dependent axonal lysosome transport promotes amyloid plaque pathology. J. Cell Biol. 216:3291–3305. 10.1083/jcb.201612148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena, S., Her L.-S., Brusch R.G., Laymon R.A., Niesman I.R., Gordesky-Gold B., Sintasath L., Bonini N.M., and Goldstein L.S.B.. 2003. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 40:25–40. 10.1016/S0896-6273(03)00594-4 [DOI] [PubMed] [Google Scholar]
- Hancock, W.O. 2014. Bidirectional cargo transport: moving beyond tug of war. Nat. Rev. Mol. Cell Biol. 15:615–628. 10.1038/nrm3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Heidemann, S.R., Landers J.M., and Hamborg M.A.. 1981. Polarity orientation of axonal microtubules. J. Cell Biol. 91:661–665. 10.1083/jcb.91.3.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbecque, N., Abderrahamani A., Meylan L., Riederer B., Mooser V., Miklossy J., Delplanque J., Boutin P., Nicod P., Haefliger J.-A., et al. 2003. Islet-brain1/C-Jun N-terminal kinase interacting protein-1 (IB1/JIP-1) promoter variant is associated with Alzheimer’s disease. Mol. Psychiatry. 8:413–422: 363. 10.1038/sj.mp.4001344 [DOI] [PubMed] [Google Scholar]
- Hendricks, A.G., Perlson E., Ross J.L., Schroeder H.W. III, Tokito M., and Holzbaur E.L.F.. 2010. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20:697–702. 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichs, V., Grycova L., Barinka C., Nahacka Z., Neuzil J., Diez S., Rohlena J., Braun M., and Lansky Z.. 2020. Mitochondria-adaptor TRAK1 promotes kinesin-1 driven transport in crowded environments. Nat. Commun. 11:3123. 10.1038/s41467-020-16972-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, S.E., Kauffman K.J., Krout M., Richmond J.E., Melia T.J., and Colón-Ramos D.A.. 2019. Maturation and Clearance of Autophagosomes in Neurons Depends on a Specific Cysteine Protease Isoform, ATG-4.2. Dev. Cell. 49:251–266.e8. 10.1016/j.devcel.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson, J.L., Peters C., Kuznetsov S.A., and Steffen W.. 2005. Three-dimensional reconstruction of the dynactin complex by single-particle image analysis. Proc. Natl. Acad. Sci. USA. 102:3667–3672. 10.1073/pnas.0409506102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura, E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Schlager M.A., van der Zwan W.A., Wulf P.S., Keijzer N., and Hoogenraad C.C.. 2010. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J. 99:2143–2152. 10.1016/j.bpj.2010.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobrekar, N.V., Quintremil S., Dantas T.J., and Vallee R.B.. 2020. The Dynein Adaptor RILP Controls Neuronal Autophagosome Biogenesis, Transport, and Clearance. Dev. Cell. 53:141–153.e4. 10.1016/j.devcel.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., and Tanaka K.. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Kulkarni, A., Chen J., and Maday S.. 2018. Neuronal autophagy and intercellular regulation of homeostasis in the brain. Curr. Opin. Neurobiol. 51:29–36. 10.1016/j.conb.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Kumar, S., Zhou Y., and Plamann M.. 2001. Dynactin-membrane interaction is regulated by the C-terminal domains of p150(Glued). EMBO Rep. 2:939–944. 10.1093/embo-reports/kve202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar, A., Tripathy S.K., Xu J., Mattson M.K., Anand P., Sigua R., Vershinin M., McKenney R.J., Yu C.C., Mogilner A., and Gross S.P.. 2011. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. Proc. Natl. Acad. Sci. USA. 108:18960–18965. 10.1073/pnas.1107841108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural, C., Kim H., Syed S., Goshima G., Gelfand V.I., and Selvin P.R.. 2005. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 308:1469–1472. 10.1126/science.1108408 [DOI] [PubMed] [Google Scholar]
- Lau, C.K., O’Reilly F.J., Santhanam B., Lacey S.E., Rappsilber J., and Carter A.P.. 2021. Cryo-EM reveals the complex architecture of dynactin’s shoulder region and pointed end. EMBO J. 40(8):e106164. 10.15252/embj.2020106164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I.-G., Olenick M.A., Boczkowska M., Franzini-Armstrong C., Holzbaur E.L.F., and Dominguez R.. 2018. A conserved interaction of the dynein light intermediate chain with dynein-dynactin effectors necessary for processivity. Nat. Commun. 9:986. 10.1038/s41467-018-03412-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I.-G., Cason S.E., Alqassim S.S., Holzbaur E.L.F., and Dominguez R.. 2020. A tunable LIC1-adaptor interaction modulates dynein activity in a cargo-specific manner. Nat. Commun. 11:5695. 10.1038/s41467-020-19538-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.J., Sharp A.H., Li S.H., Dawson T.M., Snyder S.H., and Ross C.A.. 1996. Huntingtin-associated protein (HAP1): discrete neuronal localizations in the brain resemble those of neuronal nitric oxide synthase. Proc. Natl. Acad. Sci. USA. 93:4839–4844. 10.1073/pnas.93.10.4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.-H., Gutekunst C.-A., Hersch S.M., and Li X.-J.. 1998. Interaction of huntingtin-associated protein with dynactin P150Glued. J. Neurosci. 18:1261–1269. 10.1523/JNEUROSCI.18-04-01261.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Cheng S., Yang H., Zhu L., Pan Y., Jing L., Tang B., Li S., and Li X.-J.. 2020. Loss of Hap1 selectively promotes striatal degeneration in Huntington disease mice. Proc. Natl. Acad. Sci. USA. 117:20265–20273. 10.1073/pnas.2002283117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwiczak, J., Winski A., Szczepaniak K., Alva V., and Dunin-Horkawicz S.. 2019. DeepCoil-a fast and accurate prediction of coiled-coil domains in protein sequences. Bioinformatics. 35:2790–2795. 10.1093/bioinformatics/bty1062 [DOI] [PubMed] [Google Scholar]
- Lumsden, A.L., Young R.L., Pezos N., and Keating D.J.. 2016. Huntingtin-associated protein 1: Eutherian adaptation from a TRAK-like protein, conserved gene promoter elements, and localization in the human intestine. BMC Evol. Biol. 16:214. 10.1186/s12862-016-0780-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke M., and Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Maday, S., Wallace K.E., and Holzbaur E.L.F.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417. 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis, T., Akhmanova A., Wulf P., Del Nery E., Weide T., Stepanova T., Galjart N., Grosveld F., Goud B., De Zeeuw C.I., et al. 2002. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4:986–992. 10.1038/ncb891 [DOI] [PubMed] [Google Scholar]
- Mauvezin, C., Nagy P., Juhász G., and Neufeld T.P.. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6:7007. 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire, J.R., Rong J., Li S.-H., and Li X.-J.. 2006. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J. Biol. Chem. 281:3552–3559. 10.1074/jbc.M509806200 [DOI] [PubMed] [Google Scholar]
- McKenney, R.J., Huynh W., Tanenbaum M.E., Bhabha G., and Vale R.D.. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 345:337–341. 10.1126/science.1254198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac, G., Sibarita J.-B., Loubéry S., Daviet L., Romao M., Raposo G., and Chavrier P.. 2009. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19:184–195. 10.1016/j.cub.2008.12.043 [DOI] [PubMed] [Google Scholar]
- Müller, M.J.I., Klumpp S., and Lipowsky R.. 2008. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. USA. 105:4609–4614. 10.1073/pnas.0706825105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch, A.L., Neufeld T.P., and Hays T.S.. 2017. A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J. Cell Biol. 216:441–461. 10.1083/jcb.201606082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba, J., Lukacsovich T., Csikos G., Zheng S., Margulis J., Salazar L., Mao K., Lau A.L., Yeung S.Y., Humbert S., et al. 2014. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. USA. 111:16889–16894. 10.1073/pnas.1420103111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J., Baksa K., and Steward R.. 2000. Functional domains of the Drosophila bicaudal-D protein. Genetics. 154:713–724. 10.1093/genetics/154.2.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenick, M.A., and Holzbaur E.L.F.. 2019. Dynein activators and adaptors at a glance. J. Cell Sci. 132:jcs227132. 10.1242/jcs.227132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenick, M.A., Tokito M., Boczkowska M., Dominguez R., and Holzbaur E.L.F.. 2016. Hook Adaptors Induce Unidirectional Processive Motility by Enhancing the Dynein-Dynactin Interaction. J. Biol. Chem. 291:18239–18251. 10.1074/jbc.M116.738211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenick, M.A., Dominguez R., and Holzbaur E.L.F.. 2019. Dynein activator Hook1 is required for trafficking of BDNF-signaling endosomes in neurons. J. Cell Biol. 218:220–233. 10.1083/jcb.201805016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv, S., Clausen T.H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Pardo, R., Molina-Calavita M., Poizat G., Keryer G., Humbert S., and Saudou F.. 2010. pARIS-htt: an optimised expression platform to study huntingtin reveals functional domains required for vesicular trafficking. Mol. Brain. 3:17. 10.1186/1756-6606-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, R., Zhang J., and Xiang X.. 2018. p25 of the dynactin complex plays a dual role in cargo binding and dynactin regulation. J. Biol. Chem. 293:15606–15619. 10.1074/jbc.RA118.004000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Redwine W.B., Vale R.D., and Carter A.P.. 2018. The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol. 19:382–398. 10.1038/s41580-018-0004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou, F., and Humbert S.. 2016. The Biology of Huntingtin. Neuron. 89:910–926. 10.1016/j.neuron.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Schafer, D.A., Gill S.R., Cooper J.A., Heuser J.E., and Schroer T.A.. 1994. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol. 126:403–412. 10.1083/jcb.126.2.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager, M.A., Serra-Marques A., Grigoriev I., Gumy L.F., Esteves da Silva M., Wulf P.S., Akhmanova A., and Hoogenraad C.C.. 2014. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell Rep. 8:1248–1256. 10.1016/j.celrep.2014.07.052 [DOI] [PubMed] [Google Scholar]
- Schroeder, C.M., and Vale R.D.. 2016. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J. Cell Biol. 214:309–318. 10.1083/jcb.201604002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., and Aitchison J.D.. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817. 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe, A.K.H., Hill S.E., Hall D.H., and Colón-Ramos D.A.. 2016. KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev. Cell. 38:171–185. 10.1016/j.devcel.2016.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug, P.E., Berg T.O., Fengsrud M., and Seglen P.O.. 1998. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 335:217–224. 10.1042/bj3350217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg, J., Gutwin K., Keating A.E., and Berger B.. 2011. Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone. PLoS One. 6:e23519. 10.1371/journal.pone.0023519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truebestein, L., and Leonard T.A.. 2016. Coiled-coils: The long and short of it. BioEssays. 38:903–916. 10.1002/bies.201600062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees, A.E., Lesept F., Holzbaur E.L.F., and Kittler J.T.. 2019. The adaptor proteins HAP1a and GRIP1 collaborate to activate the kinesin-1 isoform KIF5C. J. Cell Sci. 132:jcs215822. 10.1242/jcs.215822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius, L., Zhang K., Diamant A.G., Motz C., Schlager M.A., Yu M., Patel N.A., Robinson C.V., and Carter A.P.. 2015. The structure of the dynactin complex and its interaction with dynein. Science. 347:1441–1446. 10.1126/science.aaa4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius, L., Lau C.K., Elshenawy M.M., Morales-Rios E., Motz C., Yildiz A., and Carter A.P.. 2018. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature. 554:202–206. 10.1038/nature25462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela, F., Velours C., Chenon M., Aumont-Nicaise M., Campanacci V., Thureau A., Pylypenko O., Andreani J., Llinas P., and Ménétrey J.. 2019. Structural characterization of the RH1-LZI tandem of JIP3/4 highlights RH1 domains as a cytoskeletal motor-binding motif. Sci. Rep. 9:16036. 10.1038/s41598-019-52537-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, T.L., Green P.J., and Woolfson D.N.. 2013. LOGICOIL--multi-state prediction of coiled-coil oligomeric state. Bioinformatics. 29:69–76. 10.1093/bioinformatics/bts648 [DOI] [PubMed] [Google Scholar]
- Wagenbach, M., Domnitz S., Wordeman L., and Cooper J.. 2008. A kinesin-13 mutant catalytically depolymerizes microtubules in ADP. J. Cell Biol. 183:617–623. 10.1083/jcb.200805145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, K.R., and Littleton J.T.. 2016. Characterization of axonal transport defects in Drosophila Huntingtin mutants. J. Neurogenet. 30:212–221. 10.1080/01677063.2016.1202950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.C., and Holzbaur E.L.F.. 2014. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 34:1293–1305. 10.1523/JNEUROSCI.1870-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.C., and Holzbaur E.L.F.. 2015. Autophagosome dynamics in neurodegeneration at a glance. J. Cell Sci. 128:1259–1267. 10.1242/jcs.161216 [DOI] [PMC free article] [PubMed] [Google Scholar]