SUMMARY
Characterizing the proteome composition of organelles and subcellular regions of living cells can facilitate the understanding of cellular organization as well as protein interactome networks. Proximity labeling-based methods coupled with mass spectrometry (MS) offer a high-throughput approach for systematic analysis of spatially-restricted proteomes. Proximity labeling utilizes enzymes that generate reactive radicals to covalently tag neighboring proteins. The tagged endogenous proteins can then be isolated for further analysis by MS. To analyze protein-protein interactions or identify components that localize to discrete subcellular compartments, spatial expression is achieved by fusing the enzyme to specific proteins or signal peptides that target to particular subcellular regions. Although these technologies have only been introduced recently, they have already provided deep insights into a wide range of biological processes. Here, we provide an updated description and comparison of proximity labeling methods, as well as their applications and improvements. As each method has its own unique features, the goal of this review is to describe how different proximity labeling methods can be used to answer different biological questions.
Keywords: Proximity labeling, APEX, BioID, PUP-IT, HRP
INTRODUCTION
Specialized biological processes occur in different organelles and subcellular regions. In addition, protein functions correlate with their subcellular localizations and interactions. Understanding how cellular structures underlie specialized functions requires the comprehensive identification of proteins within spatially-defined cellular domains. Further, identification of interacting proteins is key to elucidating the mechanisms underlying complex cellular processes.
Mass spectrometry (MS) techniques have been used to systematically characterize the proteome of isolated organelles and protein interactors purified by affinity pull-down or following crosslinking. However, these approaches are limited by available purification methods, as it is not possible in many cases to obtain intact organelles of high purity. Moreover, even when purification is possible, contamination that results in false positive identification is common. For example, false positives may be introduced by cellular disruption, as two proteins that normally localize in different subcellular regions may artificially interact when membranes are disrupted. In addition, false negatives often occur due to loss of components caused by disruption of isolated organelles or protein complexes. Additionally, a variety of discrete cellular regions cannot be purified by centrifugation, such as specialized endoplasmic reticulum (ER)-plasma membrane (PM) junctions that are critical for lipid metabolism and Ca2+ signaling1–4. Similarly, transient or weak interactions may be lost during purification of a protein interactome due to stringent washes.
Recently, proximity-dependent labeling methods have been developed and utilized for mapping compartmental proteome and protein interactomes. In this updated review, we compare proximity labeling techniques that utilize different enzymes and describe how they are used to address limitations of traditional methods.
Overview of enzyme-catalyzed proximity labeling for proteomic profiling
In general, proximity labeling relies on enzymes that convert a substrate into a reactive radical that covalently tags neighboring proteins. We will discuss four major enzyme systems utilized for proximity labeling: BioID (proximity-dependent biotin identification), HRP (horseradish peroxidase), APEX (engineered ascorbate peroxidase), and PUP-IT (pupylation-based interaction tagging).
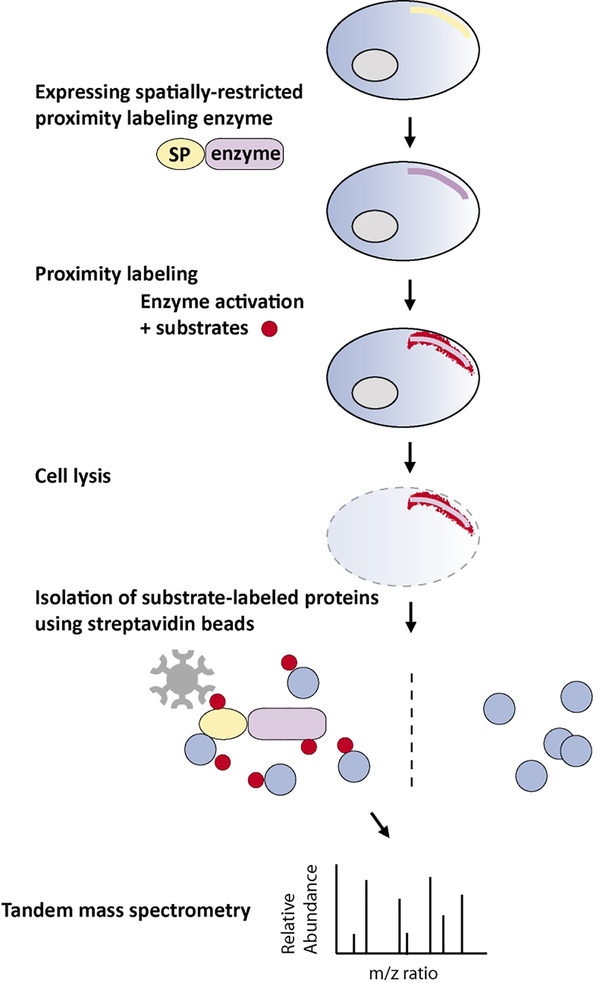
To achieve spatially-restricted labeling, the enzymes are usually fused with a targeting signal peptide, a protein of interest, or antibody. After performing proximity labeling in living cells, cells are then lysed and tagged endogenous proteins are isolated using streptavidin beads. Small peptides from enriched proteins are generated by trypsin digestion and subsequently analyzed by tandem mass spectrometry (aka MS/MS or MS2). The mass-to-charge (m/z) ratio of peptides and their fragment ions are then used to identify the peptide sequence through computational comparison against an established database (Figure 1).
Figure 1. Proximity labeling for proteomic profiling.
To achieve regional protein labeling, the enzymes are usually fused with a targeting signal peptide or a spatially-restricted protein (SP). The enzymes can also be fused with any protein of interest for protein interactome studies. After performing proximity labeling in living cells, the cells are lysed and the tagged endogenous proteins are isolated using steptavidin beads. Small peptides of enriched proteins are generated by trypsin digestion and subsequently ionized for tandem mass spectrometry (MS/MS) analysis. The mass-to-charge (m/z) ratio of each peptide and their fragment ions is then used to identify peptide sequence through computational comparison against established databases.
Importantly, with proximity labeling, cells and tissues remain intact when the proteome or interactome is labeled. Thus, the potential for false-positive identifications is minimized, as artificial interactions caused by disruption of cells and contaminants during purification steps no longer affect the results. Moreover, proximity labeling can be applied to bypass organelle purification steps, offering an alternative approach for systematic proteomic characterization in live cells. As proximity labeling is an emerging method that enables proteomic profiling of organelles, subcellular domains and interactomes, this updated review aims to provide an overview of the different methods to aid planning and execution of future experiments.
BioID-based proximity labeling
BioID-based proximity labeling employs a mutant form of the biotin ligase BirA from E coli5–7. The biotin ligase BirA is a conserved enzyme that mediates the attachment of biotin to target proteins8. In the presence of ATP, BirA biotinylates proteins by catalyzing the conversion of biotin to reactive biotinoyl-5'-AMP, which specifically tags a lysine residue of a subunit of the acetyl-CoA carboxylase5,9. Wild-type BirA has a high affinity to biotinol-5'AMP and keeps it in the active site until the acetyl-CoA carboxylase, or a short acceptor peptide, becomes available10. Since BirA has a high specificity for its target sequence, it has been used to study specific protein-protein interactions 11: BirA is fused to a bait protein and BAP (biotin acceptor peptide) is fused to a prey protein. If the interaction occurs, the prey will be close enough to the bait to become biotinylated.
To achieve promiscuous labeling, the active site of BirA has been mutated, enabling random biotinylation of vicinity proteins without BAP5,6. This method is named proximity-dependent biotin identification (BioID) and the mutated form of BirA for proximity labeling is called BiolD or BirA* to be distinguished from the wild-type and other mutant forms of BirA7 (Figure 2 and Figure 3). When the active site of BirA is mutated (R118G), its affinity to biotin-5'AMP is greatly reduced. The highly-reactive biotinoyl-5'-AMP is released from the active site of BiolD and non-specifically reacts with nearby proteins. Therefore, BioID can covalently tag nearby endogenous proteins on lysine residues. Although the labeling radius of BiolD may vary depending on the local environment, the labeling radius of BioID is estimated to be around 10 nm using the structure of the nuclear pore complex as a "molecular ruler"12.
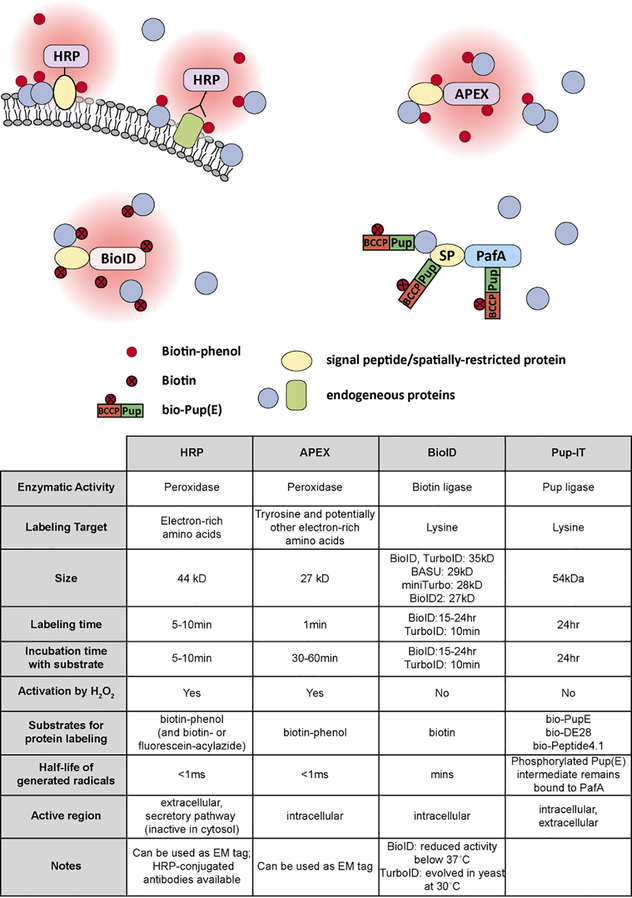
Figure 2. Proximity labeling methods.
HRP and APEX are peroxidases that, when activated by H2O2, are able to turn biotin-phenol substrates into highly-reactive radicals that covalently tag neighboring proteins on electron-rich amino acids. HRP is inactive in a reducing environment, such as the cytosol, but functions extracellularly. BioID, a mutant form of the biotin ligase BirA, can convert biotin into radicals that can covalently tag neighboring proteins on lysine residues. PafA is a ligase that can covalently tag neighboring proteins with the small protein Pup onto lysine residues.
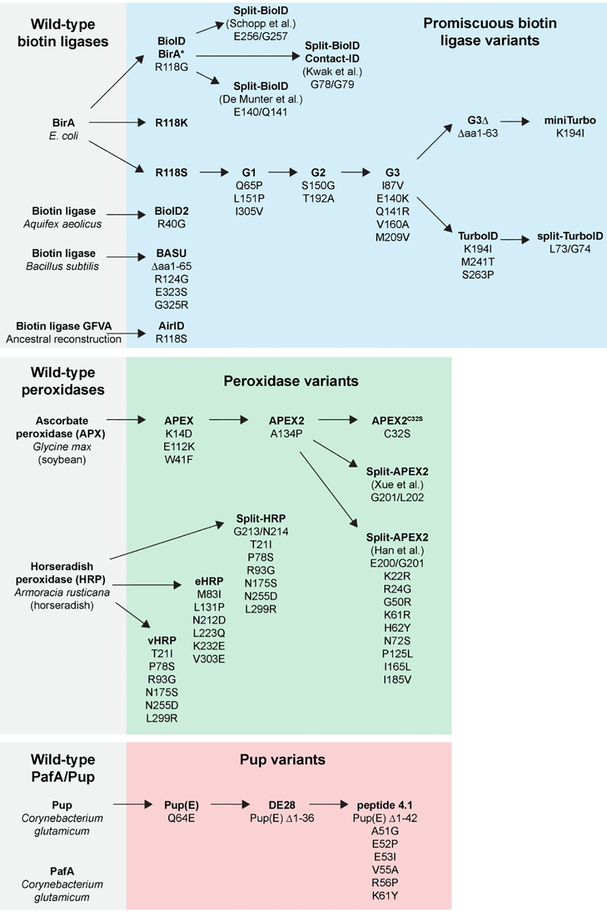
Figure 3. Directed evolution of proximity labeling components.
Proximity labeling enzymes have been modified from their wild-type counterparts by selecting for mutants with promiscuous activity. Directed evolution has been used to isolate enzymes with increased activity, increased stability, smaller molecular weight, and that are split into inactive fragments that reconstitute activity when combined. Smaller Pup substrates have also been identified.
In addition to the E.coli BioID enzyme, promiscuous biotin ligases from other species have been isolated. BioID2 was generated with an R40G mutation in the reactive site of a biotin ligase from Aquifex aeolicus to allow promiscuous labeling13. BioID2 lacks the DNA binding domain at the N-terminus and is thus smaller (233 a.a.) than E.coli BioID (321a.a.), potentially minimizing functional interference with a tagged protein. BioID2 performs similar labeling chemistry as BioID but shows a higher activity and requires less biotin. Similarly, BASU is a promiscuous BirA from Bacillus subtilis with improved biotinylation activity compared to BioID and BioID214. Like BioID2, BASU lacks the N-terminal DNA-binding domain and is smaller than BioID. Finally, ancestral reconstruction of BirA proteins led to the recent isolation of a promiscuous biot inligase called AirID, which exhibits robust biotinlyation in cultured human cells15.
The BioID enzyme has also been engineered for increased activity. TurboID was isolated by directed evolution of BioID for increased biotinlyation activity via yeast display16. In human HEK293T cells, TurboID can label an equivalent amount and diversity of proteins in 10 minutes as BioID, BioID2, or BASU can label in 18 hours. A smaller variant of TurboID called miniTurbo lacks the DNA-binding domain while still retaining robust biotinylation activity. While miniTurbo has ~2x fold less activity than TurboID, it exhibits lower biotinlyation activity in the absence of exogenously added biotin and thus may be more suited to tighter labeling windows. Under extreme conditions (e.g. high expression levels, long labeling times), TurboID expression can be toxic in human cells, flies, and worms, suggesting that the evolution of this enzyme for increased activity may have effectively reached an upper limit. In addition to being useful in spatial proteomics16, TurboID has also proven successful to discover new protein-protein interactions17–20. However, for some bait proteins, TurboID may increase the number of labeled background proteins relative to BioID21, perhaps due to its robust enzymatic activity.
Promiscuous biotin ligases have also been engineered with new functions. By screening all possible mutations at R118 in E. coli BirA, a new promiscuous biotin ligase variant (R118K) was isolated22. While R118K activity was less than R118G (BioID), R118K may be useful for proximity labeling under conditions where exogenous biotin is not added. Three independent studies derived split-BioID proteins, which were identified by screening for inactive fragments of BioID that can reform to restore biotinylation activity when physically brought together23–25. By linking these BioID fragments to two interacting proteins, the split-BioID system can be used to label proximal proteins only associated with this protein-protein pair. Recently, a split-TurboID system has been developed, with more robust labeling upon reconstitution26.
Promiscuous biotin ligase enzymes has been used to map local interactomes, identify transient protein interactions, map organelle components, and thus provide a better understanding of cellular structures as well as interactions occurring during signal transduction. The application and impact of promiscuous biotin ligases have been extensively reviewed27–29. Recent applications include interaction mapping of Ras30, mitochondrial transcription elongation factor31, influenza A virus PA-X32, growth factor independence 1B33, receptor PTPRK34, murine coronavirus replicase transcriptase complex35, PCNA36, NHLRC237, GRPEL1/238, IGF1R39, Toxoplasma gondii conoid proteins40, N-cadherin41, the NuRD complex (BioID2)42, plant N immune receptor17, protein arginine methyltransferase Rmt3 and the RNA exosome subunits, Rrp6 and Dis318, AKAP1843, plant transcription factor FAMA20, and stress granules (SGs) processing bodies (PBs)44, and desmosomes45. In addition, BiolD has recently been used to identify RNA-binding proteins by tethering BiolD to RNA transcripts via MS2 aptamers46, and used in conjunction with traditional affinity purification to improve proteomic coverage and help determine distances between protein complex members47.
HRP-based proximity labeling
HRP is a peroxidase that, when activated by H2O2, is able to convert a substrate into a highly-reactive radical that covalently tags neighboring proteins on electron-rich amino acids48. HRP is inactive in a reducing environment, such as the cytosol, because the structure of HRP, which is maintained with four disulfide bonds and two Ca2+ ion-binding sites, is disrupted in reducing conditions49. This has limited its use for determining intracellular interactomes, and motivated the development of APEX. Nevertheless, HRP is active in oxidizing environments, such as the lumen of the ER or the Golgi and the extracellular region. Thus, HRP has been used for proteomic mapping on the surface of living cells50–53. In addition, HRP can also be used as an electron microscopy (EM) tag54. With H2O2, HRP can catalyze the polymerization of 3,3'-diaminobenzidine (DAB) which precipitates and creates an EM contrast after OsO4 fixation.
Although HRP can catalyze a variety of substrates, for proximity labeling two in particular have been used: 1. the enzyme-mediated activation of radical source (EMARS) method uses fluorescein arylazide or biotin arylazide55–63. Fluorescein arylazide reduces the cytosolic background generated by biotin-aryl azide56, which is membrane permeable during the EMARS reaction and activated by endogenous enzymes55,57; and 2. the selective proteomic proximity labeling assay using tyramide (SPPLAT) method using biotin-tyramide, which is also known as biotin-phenol48,64.
HRP has been used extensively for other applications, such as ELISA and immunochemistry65. Further, antibody-HRP conjugates have been generated that can also be used for proximity labeling. However, this application is limited by the affinity of the antibody. Nevertheless, antibody-HRP conjugates have been successfully used to identify cell surface molecules such as the composition of the B cell receptor cluster, proteins that interact with Thy1, β1 integrin, CD20, and PrPC, and signaling ligands48,51,55–63,66. Antibody-HRP conjugates can also be used to identify proteins in fixed cells67.
New versions of HRP have been isolated with modified functions. A bimolecular complementation version of HRP has recently been reported68. This split HRP has beer generated to characterize intercellular protein-protein interactions and visualize synapses. The two split HRP fragments were fused with neurexin and neuroligin, which bind to each other across the synaptic cleft. When the split fragments are brought together as a result of the neurexin-neuroligin interaction, they reconstitute a functional form of HRP that allows proximity labeling. This binary system offers another level of control to the HRP system, making it useful for finer spatial restriction. In addition, two enhanced versions of HRP have been isolated. vHRP69 was isolated based on stabilizing mutations identified in split-HRP. In parallel, eHRP70 was isolated based on directed evolution. Although split HRP and the enhanced HRP variants have not yet been used for proteomics, their potential use for proteomic mapping of cell-cell interactions is very promising.
APEX-based proximity labeling
APEX, an engineered ascorbate peroxidase derived from plants, uses the same labeling chemistry and rapid kinetics as HRP to convert a substrate into a radical in the presence of H2O271,72. The key advantage of APEX over HRP, however, is that it remains active in the reducing environment of the cellular cytosol. Upon activation by H2O2, APEX catalyzes the conversion of its substrate biotin-phenol into short-lived (<1 ms) and highly-reactive radicals, which can covalently attach to electron-rich amino acids such as tyrosine in nearby endogenous proteins72,73. The labeling reaction can be stopped by the removal of H2O2 and the addition of quenching buffer, and the resulting biotinylated proteins can be subsequently isolated using streptavidin beads and further analyzed by MS. In addition, APEX can catalyze the polymerization and precipitation of DAB creating a contrast after OsO4 fixation71, which can then be used for EM to visualize the structures where APEX is expressed.
Yeast display selection has been performed to screen for mutations that increase APEX activity74. An improved version of APEX, called APEX2, has one additional mutation (A134P) and catalyzes the same chemistry as APEX but with higher activity and sensitivity for promiscuous labeling and EM. APEX2 was further improved with a mutation (C32S) that improved the stability of APEX2-tagged proteins75. Two groups developed a split-APEX2 where inactive fragments of APEX2 can reconstitute and restore enzymatic activity. One group split APEX2 at amino acids 201/20276, whereas a second group split APEX2 at nearly the same site (200/201)77 but used directed evolution of the N-terminal fragment to increase the activity of the reconstituted enzyme.
APEX-mediated proximity labeling was first introduced by Rhee and colleagues to circumvent the limitations of traditional mitochondrial purification and to achieve spatial and temporal specificity of organelle proteome mapping72. As biotin-phenoxyl radicals are not membrane-permeable, APEX is excellent for proteomic profiling of membrane-enclosed subcellular compartments, such as the mitochondria72,73,78 and autophagosomes79. Nevertheless, APEX is not limited to membrane-enclosed organelles, and has been used successfully to map proteins in the cilia80,81, stress granules82, mitochondria-ER contact points83,84, Drosophila ring canals85, mitochondrial nucleoid86, bacterial-host inclusion membrane87, lipid droplets88, and lysosome-RNA granule contact points89. APEX also provides a good tool for identification of protein-protein interactions. For example, APEX fused with bait proteins have revealed interaction networks of VAPB90, OPTN91, Rab proteins92, PAQR393, MIEF1 microprotein94, FGF195, ribosome-associated quality control complex96, and DNA repair factors97. In particular, the fast labeling time of APEX has been leveraged to identify dynamic changes in protein complex composition98,99. APEX has also been used for identification of proteins interacting with specific sequences of RNA14,100,101 and DNA102–104. Finally, we note that APEX has recently been used to directly label and identify RNAs105–107.
PUP-IT proximity labeling
Recently, a new proximity labeling system using the bacterial PafA enzyme was developed called PUP-IT (Pupylation-based interaction tagging)108. Unlike BioID, HRP, and APEX, which tag proteins with biotin (known as biotinylation), PafA tags proteins with a small protein called Pup (known as pupylation). In bacteria, PafA ligates Pup to lysine residues on target proteins, signaling those proteins for degradation. During this reaction, Pup is deaminated at its C-terminus to form Pup(E) (also known as PupGlu), which PafA phosphorylates and conjugates to a lysine residue109. PafA has no consensus binding motif flanking the target lysine, and therefore should ligate Pup to any lysine residue in proximity, making it a potentially useful promiscuous protein-labeling enzyme.
To test the effectiveness of PUP-IT as a proximity labeling system, Liu et al. fused PafA to bait proteins and supplied Pup(E) either as purified protein or via transgenic expression and translation into the cell cytoplasm. This resulted in pupylation of proteins in the close vicinity of the enzyme - PafA itself, the bait protein, and interacting prey proteins - but not distant proteins, which indicates a highly specific proximity-dependent labeling reaction. Pupylated proteins can be detected by molecular weight laddering on protein gels or western blots. In addition, the authors devised a more versatile method for detection of pupylation by fusing a bacterial-derived carboxylase domain (BCCP) to Pup(E). BCCP is biotinylated by endogenous ligases in human cells, allowing “bio-Pup(E)” and pupylated proteins to be detected by western blot using streptavidin-HRP, or purified on streptavidin beads and identified by MS. Using this method, the authors identified known interactors on the intracellular tail of CD28 such as p85. Recently, the PUP-IT system was combined with CRISPR-Cas13a (called CRUIS) to identify RNA-binding proteins110.
Whereas Pup(E) is 64 aa long, two smaller Pup variants were identified called DE28 (28 aa) and Peptide 4.1111 (14 aa). In particular, Peptide 4.1 lacks lysine residues, which may be useful to prevent unwanted branched tags. While these smaller Pup variants may be useful improvements to the PUP-IT system, they have not been tested under conditions of transgenic expression like Pup(E). Finally, like improvements to BiolD, HRP, and APEX, directed evolution of the PafA enzyme may yield increased or modified labeling activity.
Comparison between biotin ligase-based, peroxidase-based, and Pup ligase-based approaches
The major differences between biotin ligase-based, peroxidase-based, and Pup ligase-based (PUP-IT) labeling approaches are the substrates, the targeted amino acid(s), the kinetics, and the working conditions (Figure 2). In addition to differences in proteomic labeling, APEX, like HRP, can be used for EM, thus allowing confirmation of fine subcellular localization. On the other hand, the proper expression and localization of promiscuous biotin ligases and PafA can only be verified by other methods like immunostaining and/or Western blotting to rule out the possibility of false positive from mis-localization of the fusion proteins or slow translation of the fusion protein.
One major difference is the type of substrate used for proteomic analysis. The biotin ligase-based method uses biotin, the peroxidase-based approaches use biotin-phenol, and the PUP-IT method uses biotinylated forms of Pup(E). Delivery of the substrate to the region of interest is a critical factor. Biotin is actively imported into mammalian cells and other organisms though distinct mechanisms112. Even though biotin-phenol can be simply incubated with mammalian cells for cytosolic and mitochondrial protein labeling, a number of studies have shown that biotin-phenol Reviewmaynoteffectivelypenetrate membranes48,64. Moreover, special procedures are required for efficient delivery of biotin-phenol and optimal proximity labeling in yeast113,114. Therefore, optimizing biotin-phenol delivery to a region of interest in a specific cell type may be required to achieve successful protein labeling. Chemically synthesized bio-DE28 and bio-Peptide4.1 can also be incubated with cells but would likely not penetrate the plasma membrane. In contrast, genetically encoded BCCP-PupE is translated into the cytoplasm where it is biotinylated by endogenous ligases. While PupE has the unique advantage of being genetically modifiable with additional domains, this tag is substantially larger than biotin and may interfere with protein function.
The half-life of biotin-5'-AMP radicals generated by promiscuous biotin ligases is on the order of minutes in aqueous solutions115, which is longer than that of APEX-generated biotin-phenoxyl radicals (<1 ms)72,73. The shorter half-life of unstable radicals may result in a smaller labeling radius, which is also determined by other factors, such as local intracellular environments. Unfortunately, the labeling radius of promiscuous biotin ligases and APEX has been estimated by different methods and in different cellular regions. Unlike biotin ligase and peroxidase-based approaches, PafA enzyme does not release the Pup tag, thus ensuring that only proteins in close contact with PafA become labeled. Therefore, PUP-IT labeling will likely not be as useful for spatial proteomics such as organelle mapping. Furthermore, the lack of a diffusible reactive substrate may spatially limit labeling to lysine residues on prey proteins that directly face PafA.
Promiscuous biotin ligases and PafA labels lysine residues of nearby proteins whereas APEX and HRP tag electron-rich tyrosine residues. Generally, the estimated amount of lysine present in proteins is higher than that of tyrosine116,117. Thus, when the number of available tyrosine residues is limited, potential target proteins may not be identified using APEX and HRP.
Promiscuous biotin ligases and PafA overall show slower kinetics than APEX or HRP. The optimal labeling time for APEX (~1 min) is shorter than that for HRP (5–10 min) and much shorter than for BioID (15–24 h) and PafA (24 h). The only exception is TurboID and miniTurbo, which label on timescales closer to APEX and HRP (~10min). Although biotin is not toxic, biotinylation of proteins over a long period may perturb protein function, lead to artificial interactions, and cause cell toxicity, which was confirmed in cultured mammalian cells expressing TurboID longer than 24hrs16. This difference in labeling time will undoubtedly change the specificity of the labeled proteomes. While promiscuous biotin ligases and PUP-IT are useful for capturing entire changes in protein complexes during a longer period of time, APEX is excellent for characterizing rapid dynamic changes in proteomes that can only be achieved with a short labeling window, such as acute responses to drug treatment98,99. The fast labeling times of TurboID suggests it too can be applied in this manner.
Notably, the activity of BioID or BioID2 is greatly reduced at temperatures below 37°C13. For model systems that need to be maintained under 37°C, BioID cannot be easily used. Nevertheless, BioID has been successfully applied to many organisms in addition to mammalian cells, such as single celled organisms (Trypanosoma brucei, Toxoplasma gondii, Dictyostelium discoideum, Plasmodium berghei), invertebrates (Drosophila melanogaster, Caenorhabditis elegans), and plants (Nicotiana benthamiana, Arabidopsis thaliana)118–125. In contrast, TurboID and miniTurbo were evolved in yeast grown at 30°C, perhaps explaining why they perform well in Drosophila and C. elegans, which are grown at 25°C and 20°C, respectively. APEX has been shown to be active in Drosophila cultured cells at 25°C and in yeast cultured at room temperature, in addition to showing good activity in mammalian cells that are cultured at 37 °C. This temperature range allows APEX to be broadly suitable for studies in a variety of model organisms.
Comparison between APEX and HRP-based approaches
Both APEX and HRP catalyze the same proximity labeling chemistry. The key parameter that one should consider for their usage is the environment to which the enzyme will be exposed. As mentioned above, HRP is inactive in the cytosol; however, it is functional when it faces outside the cell on the cell surface and has been successfully used to identify membrane proteins48,50–53,55–63,66,126. Notably, many previous studies used antibody-conjugated HRP48,51,55–63,66,67. A key advantage of the HRP-mediated approach is that many antibody-HRP conjugates are currently available. As noted previously, however, the use of antibody-conjugated HRP in proximity labeling is limited by the affinity of the antibody.
Analysis of proteomic data from proximity labeling approaches
A challenge common to all labeling strategies is to distinguish candidate proteins from background in MS data. Generally, proteins with the highest abundance, and represented by 2 or more independent peptides, are chosen for further study even though low-abundance candidates may potentially be biologically relevant. Researchers have devised additional experimental procedures to help generate a high-confidence and comprehensive list of candidates from MS data: 1. Proximity labeling coupled with quantitative MS can be achieved using metabolic labeling such as SILAC (stable isotope labeling by amino acid in cell culture)127 or done with in vitro chemical labeling, such as iTRAQ (isobaric tags for relative and absolute quantification)128 and TMT (tandem mass tags)129. 2. Additional negative controls can help filter out background labeled proteins. For example, researchers can target a labeling enzyme to a different organelle or protein complex in addition to the primary target73. Furthermore, isogenic cell lines can be used to avoid differences in transgene expression130,131. 3. Background due to non-specific labeling can be reduced by inducibly activating the labeling enzyme84,123 or using endogenous CRISPR/Cas9 tagging of bait proteins to maintain physiological levels of the labeling enzyme132. 4. True positives can be distinguished by identifying peptide biotinylation sites133–135. See additional reviews for detailed considerations for proximity labeling experimental design and data analysis 136–137.
Proximity labeling in developmental systems
Proximity labeling is typically performed in cultured cells due to technical advantages of this system (e.g. easy delivery of labeling reagents, efficient cell lysis of large quantities of cells). However, the application of proximity labeling tools in vivo has specific benefits. For example, in vivo protein labeling allows researchers to identify organelle components or protein interactions from cells in a normal physiological environment, including cell types that would be too difficult grow in culture (e.g. neurons138). Furthermore, by expressing labeling enzymes from transgenes, protein labeling can be restricted to specific cell types or developmental stages. Cells expressing labeling enzymes can also be transplanted into otherwise wild-type host organisms.
Penetration of labeling substrate into target tissues and cells is a significant technical challenge of using proximity labeling tools in vivo. For example, experiments using APEX or HRP require incubating live dissected tissues with biotin-phenol. For some experiments, this dissection step might be too laborious, or make it difficult to collect enough material for pulldown/MS analysis. In contrast, promiscuous biotin ligases can label proteins in intact organisms. This is because biotin is membrane permeable and can be added to an organism’s water/food supply. Temporal labeling experiments may be difficult using this method, as biotin needs to ingested and perfuse to the target tissue. For example, Drosophila adult flies expressing TurboID exhibit significant labeling only after 16hrs of feeding flies biotin16. This problem might be addressed by direct injection of biotin into the organism138, or temporal control of biotin ligase expression. Finally, while Pup-IT has yet to be applied in vivo, the PupE label can be genetically encoded, potentially avoiding tissue penetration entirely.
Many groups have applied proximity labeling tools in developmental systems, such as Arabidopsis20,139,140, C. elegans16,141, Drosophila16,50,78,85,142, and mouse118,143,144. Importantly, some have used proximity-labeling tools to discover new components of developmental processes. For example, APEX was used in Drosophila to identify novel components of the ring canals, which are intercellular pores that transport cellular material from nurse cells to the developing oocyte85. By tagging known ring canal proteins with APEX, and phenotypic screening MS hits by RNAi, they identified eight new proteins important for ring canal morphology. Another study in Drosophila used HRP localized to the cell Forsurface toidentifynewwiring regulators in developing and adult olfactory projection neurons50. RNAi screening of MS hits revealed 20 new developmental regulators of olfactory projection neuron wiring, including the lipoprotein receptor LRP1. Finally, in C. elegans, APEX was expressed under the control of four different tissue-specific enhancer elements, as well as targeted to either the nucleus or cytoplasm141. By comparing MS datasets from each condition, they identified tissue specific and subcellular specific proteins, seven of which were confirmed by GFP-tagging and had no previous such annotation.
Conclusion/Perspectives
Since the recent introduction of proximity labeling, the method has significant contributions to the mapping of local interactomes relevant to a wide range of biological processes. By tagging regional proteomes, proximity labeling overcomes issues associated with traditional approaches of organelle purification and allows proteomic analysis of other types of subcellular regions. A disadvantage that all proximity labeling-based methods have in common is that they cannot distinguish direct binding of two proteins from proximity of two adjacent proteins. Thus, these methods serve as discovery methods that require detailed follow-up studies. Nevertheless, as proximity labeling does not require disruption of cells for complex isolation, these methods not only preserve evidence of weak or transient interactions that are not detectable using traditional approaches but also minimizes false discovery by eliminating false positives generated during lysis or disruption. Importantly, as proximity labeling can be performed inlivingcells, researchers can study protein-protein interactions and proteomic alterations in physiologically-relevant conditions. Proximity labeling has been adapted to several model systems, making this technology available to study diverse biological problems in a wide range of organisms.
Notably, while improved variants of labeling systems are now available (Figure 3), further improvements are likely to be made in the near future. In particular, variants of PafA with faster kinetics and higher activity could be isolated that match the robustness of APEX2 and TurboID enzymes. Furthermore, PafA variants that release diffusible reactive PupE, similar to promiscuous biotin ligase-based and peroxidase-based systems, would make the PUP-IT system more useful for spatial proteomics such as organelle mapping. Furthermore, a split-PafA enzyme would be a valuable addition to existing split labeling tools to fine-tune spatial restrictions.
Importantly, the ease of applying genetically-encoded enzymes will benefit greatly from the powerful genome editing using CRISPR technology145,146, as these enzymes can now be easily fused to any gene of interest via a knock-in approach. In addition, numerous genetic engineering tools already available for organisms such as Drosophila facilitate a wide range of proximity-labeling applications. For example, the existing library of MiMICs, a transposon insertion resource for engineering Drosophila genes, allows for rapid tagging of genes147–148. Altogether, a broad-range of proximity-labeling applications that build on existing tools are now possible and likely to provide deep insights into various biological questions.
Acknowledgments
We thank Alice Ting and members of her laboratory for stimulating discussions of APEX-based methods over the past few years. We are grateful to Raghuvir Viswanatha, Stephanie Mohr, Richard Binari, and Pierre Michel Jean Beltran for helpful comments on the manuscript. J.B. was supported by the Damon Runyon Foundation #2258–16. This work was supported in part by 5R01DK121409 to N.P. N.P is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Carrasco S & Meyer T STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem 80, 973–1000, doi: 10.1146/annurev-biochem-061609-165311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogan PG, Lewis RS & Rao A Molecular basis of calcium signaling in lymphocytes: STIM andO RAI. Annu RevImmunol 28, 491–533, doi: 10.1146/annurev.immunol.021908.132550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefan CJ, Manford AG & Emr SD ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol 25, 434–442, doi: 10.1016/j.ceb.2013.02.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbaz Y & Schuldiner M Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci 36, 616–623, doi: 10.1016/j.tibs.2011.08.004 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Choi-Rhee E, Schulman H & Cronan JE Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci 13, 3043–3050, doi: 10.1110/ps.04911804 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronan JE Targeted and proximity-dependent promiscuous protein biotinylation by a mutant Escherichia coli biotin protein ligase. J Nutr Biochem 16, 416–418, doi: 10.1016/j.jnutbio.2005.03.017 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Roux KJ, Kim DI, Raida M & Burke B A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810, doi: 10.1083/jcb.201112098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravartty V & Cronan JE Altered regulation of Escherichia coli biotin biosynthesis in BirA superrepressor mutant strains. J Bacteriol 194, 1113–1126, doi: 10.1128/JB.06549-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman-Smith A & Cronan JE Jr. Molecular biology of biotin attachment to proteins. J Nutr 129, 477S–484S (1999). [DOI] [PubMed] [Google Scholar]
- 10.Beckett D, Kovaleva E & Schatz PJ A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci 8, 921–929, doi: 10.1110/ps.8.4.921 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Suarez M, Chen TS & Ting AY Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J Am Chem Soc 130, 9251–9253, doi: 10.1021/ja801445p (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DI et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 111, E2453–2461, doi: 10.1073/pnas.1406459111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DI et al. An improved smaller biotin ligase for BiolD proximity labeling. Mol Biol Cell 27, 1188–1196, doi: 10.1091/mbc.E15-12-0844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramanathan M et al. RNA-protein interaction detection in living cells. Nat Methods 15, 207–212, doi: 10.1038/nmeth.4601 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kido K et al. AirID, a novel proximity biotinylation enzyme, for analysis of protein-protein interactions. Elife 9, doi: 10.7554/eLife.54983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branon TC et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880–887, doi: 10.1038/nbt.4201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun 10, 3252, doi: 10.1038/s41467019-11202-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larochelle M, Bergeron D, Arcand B & Bachand F Proximity-dependent biotinylation mediated by TurboID to identify protein-protein interaction networks in yeast. J Cell Sci 132, doi: 10.1242/jcs.232249 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Tachie-Menson T et al. Characterisation of the biochemical and cellular roles of native and pathogenic amelogenesis imperfecta mutants of FAM83H. Cell Signal 72, 109632, doi: 10.1016/j.celIsig.2020.109632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mair A, Xu SL, Branon TC, Ting AY & Bergmann DC Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. Elife 8, doi: 10.7554/eLife.47864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May DG, Scott KL, Campos AR & Roux KJ Comparative Application of BioID and TurboID for Protein-Proximity Biotinylation. Cells 9, doi: 10.3390/cells9051070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oostdyk LT et al. Towards improving proximity labeling by the biotin ligase BirA. Methods 157, 66–79, doi: 10.1016/j.ymeth.2018.11.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Munter S et al. Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett 591, 415–424, doi: 10.1002/1873-3468.12548 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Schopp IM et al. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat Commun 8, 15690, doi: 10.1038/ncommsl5690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak C et al. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.1916584117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho KF et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.1919528117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DI & Roux KJ Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 26, 804–817, doi: 10.1016/j.tcb.2016.09.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varnaite R & MacNeill SA Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 16, 2503–2518, doi: 10.1002/pmic.201600123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li P, Li J, Wang L & Di LJ Proximity Labeling of Interacting Proteins: Application of BiolD as a Discovery Tool. Proteomics 17, doi: 10.1002/pmic.201700002 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Kovalski JR, Shanderson RL & Khavari PA Ras functional proximity proteomics establishes mTORC2 as new direct ras effector. Oncotarget 10, 5126–5135, doi: 10.18632/oncotarget.27025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang S et al. TEFM regulates both transcription elongation and RNA processing in mitochondria. EMBO Rep 20, doi: 10.15252/embr.201948101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaucherand L et al. The Influenza A Virus Endoribonuclease PA-X Usurps Host mRNA Processing Machinery to Limit Host Gene Expression. Cell Rep 27, 776–792 e777, doi: 10.1016/j.celrep.2019.03.063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClellan D et al. Growth Factor Independence IB-Mediated Transcriptional Repression and Lineage Allocation Require Lysine-Specific Demethylase 1-Dependent Recruitment of the BHC Complex. Mol Cell Biol 39, doi: 10.1128/MCB.00020-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fearnley GW et al. The homophilic receptor PTPRK selectively dephosphorylates multiple junctional regulators to promote cell-cell adhesion. Elife 8, doi: 10.7554/eLife.44597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.V'Kovski P et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife 8, doi: 10.7554/eLife.42037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava M et al. Replisome Dynamics and Their Functional Relevance upon DNA Damage through the PCNA Interactome. Cell Rep 25, 3869–3883 e3864, doi: 10.1016/j.celrep.2018.11.099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paakkola T et al. Biallelic mutations in human NHLRC2 enhance myofibroblast differentiation in FINCA disease. Hum Mol Genet 27, 4288–4302, doi: 10.1093/hmg/ddy298 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Konovalova S et al. Redox regulation of GRPEL2 nucleotide exchange factor for mitochondrial HSP70 chaperone. Redox Biol 19, 37–45, doi: 10.1016/j.redox.2018.07.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bareja A, Hodgkinson CP, Soderblom E, Waitt G & Dzau VJ The proximity-labeling technique BiolD identifies sorting nexin 6 as a member of the insulin-like growth factor 1 (IGF1)-IGF1 receptor pathway. J Biol Chem 293, 6449–6459, doi: 10.1074/jbc.RA118.002406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long S, Anthony B, Drewry LL & Sibley LD A conserved ankyrin repeat-containing protein regulates conoid stability, motility and cell invasion in Toxoplasma gondii. Nat Commun 8, 2236, doi: 10.1038/s41467-017-02341-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y et al. The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J Cell Sci 132, doi: 10.1242/jcs.221606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sher F et al. Rational targeting of a NuRD subcomplex guided by comprehensive in situ mutagenesis. Nat Genet 51, 1149–1159, doi: 10.1038/s41588-019-0453-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith FD et al. Single nucleotide polymorphisms alter kinase anchoring and the subcellular targeting of A-kinase anchoring proteins. Proc Natl Acad Sci U S A 115, E11465–E11474, doi: 10.1073/pnas.1816614115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youn JY et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell 69, 517–532e511, doi: 10.1016/j.molcel.2017.12.020 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Badu-Nkansah KA & Lechler T Proteomic analysis of desmosomes reveals novel components required for epidermal integrity. Mol Biol Cell 31, 1140–1153, doi: 10.1091/mbc.E19-09-0542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee J et al. beta-Actin mRNA interactome mapping by proximity biotinylation. Proc Natl Acad Sci U S A 116, 12863–12872, doi: 10.1073/pnas.1820737116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X et al. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat Commun 9, 1188, doi: 10.1038/s41467-018-03523-2(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XW et al. New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J Biol Chem 289, 14434–14447, doi: 10.1074/jbc.M113.529578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hopkins C, Gibson A, Stinchcombe J & Futter C Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol 327, 35–45 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Li J et al. Cell-Surface Proteomic Profiling in the Fly Brain Uncovers Wiring Regulators. Cell 180, 373–386 e315, doi: 10.1016/j.cell.2019.12.029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu G, Nagala M & Crocker PR Identification of lectin counter-receptors on cell membranes by proximity labeling. Glycobiology 27, 800–805, doi: 10.1093/glycob/cwx063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cijsouw T et al. Mapping the Proteome of the Synaptic Cleft through Proximity Labeling Reveals New Cleft Proteins. Proteomes 6, doi: 10.3390/proteomes6040048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loh KH et al. Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 166, 1295–1307 e1221, doi: 10.1016/j.cell.2016.07.041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellisman MH, Deerinck TJ, Shu X & Sosinsky GE Picking faces out of a crowd: genetic labels for identification of proteins in correlated light and electron microscopy imaging. Methods Cell Biol 111, 139–155, doi: 10.1016/B978-0-12-416026-2.00008-X (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honke K & Kotani N Identification of cell-surface molecular interactions under living conditions by using the enzyme-mediated activation of radical sources (EMARS) method. Sensors (Basel) 12, 16037–16045, doi: 10.3390/sl21216037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang S et al. A proteomics approach to the cell-surface interactome using the enzyme-mediated activation of radical sources reaction. Proteomics 12, 54–62, doi: 10.1002/pmic.201100551 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Kotani N et al. Biochemical visualization of cell surface molecular clustering in living cells. Proc Natl Acad Sci U S A 105, 7405–7409, doi: 10.1073/pnas.0710346105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyagawa-Yamaguchi A, Kotani N & Honke K Expressed glycosylphosphatidylinositol-anchored horseradish peroxidase identifies co-clustering molecules in individual lipid raft domains. PLoS One 9, e93054, doi: 10.1371/journal.pone.0093054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyagawa-Yamaguchi A, Kotani N & Honke K Each GPI-anchored protein species forms a specific lipid raft depending on its GPI attachment signal. Glycoconj J 32, 531–540., doi: 10.1007/s10719-015-9595-5 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Iwamaru Y et al. Proximity of SCG10 and prion protein in membrane rafts. J Neurochem, doi: 10.1111/jnc.l3488 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Hashimoto N et al. Proteomic analysis of ganglioside-associated membrane molecules: substantial basis for molecular clustering. Proteomics 12, 3154–3163, doi: 10.1002/pmic.201200279 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Ishiura Y et al. Anomalous expression of Thyl (CD90) in B-cell lymphoma cells and proliferation inhibition by anti-Thy1 antibody treatment. Biochem Biophys Res Commun 396, 329–334, doi: 10.1016/j.bbrc.2010.04.092 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Yamashita R, Kotani N, Ishiura Y, Higashiyama S & Honke K Spatiotemporally-regulated interaction between betal integrin and ErbB4 that is involved in fibronectin-dependent cell migration. J Biochem 149, 347–355, doi: 10.1093/jb/mvql48 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Rees JS, Li XW, Perrett S, Lilley KS & Jackson AP Selective Proteomic Proximity Labeling Assay Using Tyramide (SPPLAT): A Quantitative Method for the Proteomic Analysis of Localized Membrane-Bound Protein Clusters. Curr Protoc Protein Sci 80, 19 27 11–18, doi: 10.1002/0471140864.psl927s80 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Ryan BJ, Carolan N & O'Fagain C Horseradish and soybean peroxidases: comparable tools for alternative niches? Trends Biotechnol 24, 355–363, doi: 10.1016/j.tibtech.2006.06.007 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Chang L et al. Identification of Siglec Ligands Using a Proximity Labeling Method. J Proteome Res 16, 3929–3941, doi: 10.1021/acs.jproteome.7b00625 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Bar DZ et al. Biotinylation by antibody recognition-a method for proximity labeling. Nat Methods 15, 127–133, doi: 10.1038/nmeth.4533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martell JD et al. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat Biotechnol 34, 774–780, doi: 10.1038/nbt.3563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamagata M & Sanes JR Reporter-nanobody fusions (RANbodies) as versatile, small, sensitive immunohistochemical reagents. Proc Natl Acad Sci U S A 115, 2126–2131, doi: 10.1073/pnas.1722491115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cruz-Lopez D, Ramos D, Castilloveitia G & Schikorski T Quintuple labeling in the electron microscope with genetically encoded enhanced horseradish peroxidase. PLoS One 13, e0200693, doi: 10.1371/journal.pone.0200693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martell JD et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30, 1143–1148, doi: 10.1038/nbt.2375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhee HW et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331, doi: 10.1126/science.1230593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hung V et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell 55, 332–341, doi: 10.1016/j.molcel.2014.06.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lam SS et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12, 51–54, doi: 10.1038/nmeth.3179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang MS et al. The cysteine-free single mutant C32S of APEX2 is a highly expressed and active fusion tag for proximity labeling applications. Protein Sci 28, 1703–1712, doi: 10.1002/pro.3685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xue M et al. Optimizing the fragment complementation of APEX2 for detection of specific protein-protein interactions in live cells. Sci Rep 7, 12039, doi: 10.1038/s41598017-12365-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han Y et al. Directed Evolution of Split APEX2 Peroxidase. ACS Chem Biol 14, 619–635, doi: 10.1021/acschembio.8b00919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen CL et al. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci U S A 112, 12093–12098, doi: 10.1073/pnas.1515623112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Guerroue F et al. Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol Cell 68, 786–796 e786, doi: 10.1016/j.molcel.2017.10.029 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Mick DU et al. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell 35, 497–512, doi: 10.1016/j.devcel.2015.10.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohli P et al. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep 18, 1521–1535, doi: 10.15252/embr.201643846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Markmiller S et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513, doi: 10.1016/j.cell.2017.12.032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hung V et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6, doi: 10.7554/eLife.24463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cho IT et al. Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts. J Biol Chem 292, 16382–16392, doi: 10.1074/jbc.M117.795286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mannix KM, Starble RM, Kaufman RS & Cooley L Proximity labeling reveals novel interactomes in live Drosophila tissue. Development 146, doi: 10.1242/dev,176644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han S et al. Proximity Biotinylation as a Method for Mapping Proteins Associated with mtDNA in Living Cells. Cell Chem Biol 24, 404–414, doi: 10.1016/j.chembiol.2017.02.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olson MG et al. Proximity Labeling To Map Host-Pathogen Interactions at the Membrane of a Bacterium-Containing Vacuole in Chlamydia trachomatis-lnfected Human Cells. Infect Immun 87, doi: 10.1128/IAI.00537-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bersuker K et al. A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev Cell 44, 97–112 e117, doi: 10.1016/j.devcel.2017.11.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao YC et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin All as a Molecular Tether. Cell 179, 147–164 e120, doi: 10.1016/j.cell.2019.08.050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.James C et al. Proteomic mapping by rapamycin-dependent targeting of APEX2 identifies binding partners of VAPB at the inner nuclear membrane. J Biol Chem 294, 16241–16254, doi: 10.1074/jbc.RA118.007283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heo JM et al. Integrated proteogenetic analysis reveals the landscape of a mitochondrial-autophagosome synapse during PARK2-dependent mitophagy. Sci Adv 5, eaay4624, doi: 10.1126/sciadv.aay4624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Del Olmo T et al. APEX2-mediated RAB proximity labeling identifies a role for RAB21 in clathrin-independent cargo sorting. EMBO Rep 20, doi: 10.15252/embr.201847192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cao Q et al. PAQR3 Regulates Endoplasmic Reticulum-to-Golgi Trafficking of COPII Vesicle via Interaction with Sec13/Sec31 Coat Proteins. iScience 9, 382–398, doi: 10.1016/j.isci.2018.11.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rathore A et al. MIEF1 Microprotein Regulates Mitochondrial Translation. Biochemistry 57, 5564–5575, doi: 10.1021/acs.biochem.8b00726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhen Y, Haugsten EM, Singh SK & Wesche J Proximity Labeling by a Recombinant APEX2-FGF1 Fusion Protein Reveals Interaction of FGF1 with the Proteoglycans CD44 and CSPG4. Biochemistry 57, 3807–3816, doi: 10.1021/acs.biochem.8b00120 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Zuzow N et al. Mapping the mammalian ribosome quality control complex interactome using proximity labeling approaches. Mol Biol Cell 29, 1258–1269, doi: 10.1091/mbc.E1712-0714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gupta R et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 173, 972–988 e923, doi: 10.1016/j.cell.2018.03.050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lobingier BT et al. An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 169, 350–360 e312, doi: 10.1016/j.cell.2017.03.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paek J et al. Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 169, 338–349 e311, doi: 10.1016/j.cell.2017.03.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaewsapsak P, Shechner DM, Mallard W, Rinn JL & Ting AY Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Elife 6, doi: 10.7554/eLife.29224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu M & Wei W Proximity labeling to detect RNA-protein interactions in live cells. FEBS Open Bio 9, 1860–1868, doi: 10.1002/2211-5463.12706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Myers SA et al. Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat Methods 15, 437–439, doi: 10.1038/s41592-018-0007-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao XD et al. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat Methods 15, 433–436, doi: 10.1038/s41592-018-0006-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu W et al. Determination of local chromatin interactions using a combined CRISPR and peroxidase APEX2 system. Nucleic Acids Res 47, e52, doi: 10.1093/nar/gkz134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou Y et al. Expanding APEX2 Substrates for Proximity-Dependent Labeling of Nucleic Acids and Proteins in Living Cells. Angew Chem Int Ed Engl 58, 11763–11767, doi: 10.1002/anie.201905949 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Fazal FM et al. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 178, 473–490 e426, doi: 10.1016/j.cell.2019.05.027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Padron A, Iwasaki S & Ingolia NT Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol Cell 75, 875–887 e875, doi: 10.1016/j.molcel.2019.07.030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Q et al. A proximity-tagging system to identify membrane protein-protein interactions. Nat Methods 15, 715–722, doi: 10.1038/s41592-018-0100-5 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Iyer LM, Burroughs AM & Aravind L Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct 3, 45, doi: 10.1186/1745-6150-3-45 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Z et al. Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res, doi: 10.1093/nar/gkaa143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun W et al. Identification of a Small Probe That Can Be Conjugated to Proteins by Proximity Labeling. ACS Chem Biol 15, 39–43, doi: 10.1021/acschembio.9b00842 (2020). [DOI] [PubMed] [Google Scholar]
- 112.Azhar A, Booker GW & Polyak SW Mechanisms of Biotin Transport. Biochemistry & Analytical Biochemistry 4, 8 (2015). [Google Scholar]
- 113.Hwang J & Espenshade PJ Proximity-dependent biotin labelling in yeast using the engineered ascorbate peroxidase APEX2. Biochem J 473, 2463–2469, doi: 10.1042/BCJ20160106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singer-Kruger B et al. APEX2-mediated proximity labeling resolves protein networks in Saccharomyces cerevisiae cells. FEBS J 287, 325–344, doi: 10.1111/febs.15007 (2020). [DOI] [PubMed] [Google Scholar]
- 115.Demoss JA, Genuth SM & Novelli GD The Enzymatic Activation of Amino Acids Via Their Acyl-Adenylate Derivatives. Proc Natl Acad Sci U S A 42, 325–332 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Echols N et al. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res 30, 2515–2523 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tourasse NJ & Li WH Selective constraints, amino acid composition, and the rate of protein evolution. Mol Biol Evol 17, 656–664 (2000). [DOI] [PubMed] [Google Scholar]
- 118.Dingar D et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics 118, 95–111, doi: 10.1016/j.jprot.2014.09.029 (2015). [DOI] [PubMed] [Google Scholar]
- 119.Hu H, Zhou Q & Li Z SAS-4 Protein in Trypanosoma brucei Controls Life Cycle Transitions by Modulating the Length of the Flagellum Attachment Zone Filament. J Biol Chem 290, 30453–30463, doi: 10.1074/jbc.M115.694109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McAllaster MR et al. Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol Biol Cell 26, 3013–3029, doi: 10.1091/mbc.E15-04-0219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morriswood B et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell 12, 356–367, doi: 10.1128/EC.00326-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou Q, Hu H & Li Z An EF-hand-containing Protein in Trypanosoma brucei Regulates Cytokinesis Initiation by Maintaining the Stability of the Cytokinesis Initiation Factor CIF1. J Biol Chem 291, 14395–14409, doi: 10.1074/jbc.M116.726133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kehrer J, Frischknecht F & Mair GR Proteomic Analysis of the Plasmodium berghei Gametocyte Egressome and Vesicular biolD of Osmiophilic Body Proteins Identifies Merozoite TRAP-like Protein (MTRAP) as an Essential Factor for Parasite Transmission. Mol Cell Proteomics 15, 2852–2862, doi: 10.1074/mcp.M116.058263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen AL et al. Novel components of the Toxoplasma inner membrane complex revealed by BiolD. MBio 6, e02357–02314, doi: 10.1128/mBio.02357-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Batsios P, Ren X, Baumann O, Larochelle DA & Graf R Srcl is a Protein of the Inner Nuclear Membrane Interacting with the Dictyostelium Lamin NE81. Cells 5, doi: 10.3390/cells5010013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bausch-Fluck D, Milani ES & Wollscheid B Surfaceome nanoscale organization and extracellular interaction networks. Curr Opin Chem Biol 48, 26–33, doi: 10.1016/j.cbpa.2018.09.020 (2019). [DOI] [PubMed] [Google Scholar]
- 127.Ong SE et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1, 376–386 (2002). [DOI] [PubMed] [Google Scholar]
- 128.Ross PL et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3, 1154–1169, doi: 10.1074/mcp.M400129-MCP200 (2004). [DOI] [PubMed] [Google Scholar]
- 129.Thompson A et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75, 1895–1904 (2003). [DOI] [PubMed] [Google Scholar]
- 130.Vandemoortele G et al. A Well-Controlled BiolD Design for Endogenous Bait Proteins. J Proteome Res 18, 95–106, doi: 10.1021/acs.jproteome.8b00367 (2019). [DOI] [PubMed] [Google Scholar]
- 131.Hesketh GG, Youn JY., Samavarchi-Tehrani P., Raught B. & Gingras AC. Parallel Exploration of Interaction Space by BiolD and Affinity Purification Coupled to Mass Spectrometry. Methods Mol Biol 1550, 115–136, doi: 10.1007/978-l-4939-6747-6_10 (2017). [DOI] [PubMed] [Google Scholar]
- 132.Long S, Brown KM & Sibley LD CRISPR-mediated Tagging with BirA Allows Proximity Labeling in Toxoplasma gondii. Bio Protoc 8, doi: 10.21769/BioProtoc.2768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim DI et al. BioSITe: A Method for Direct Detection and Quantitation of Site-Specific Biotinylation. J Proteome Res 17, 759–769, doi: 10.1021/acs.jproteome.7b00775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Udeshi ND et al. Antibodies to biotin enable large-scale detection of biotinylation sites on proteins. Nat Methods 14, 1167–1170, doi: 10.1038/nmeth.4465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee SY et al. Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells. ACS Cent Sci 2, 506–516, doi: 10.1021/acscentsci.6b00137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gingras AC, Abe KT & Raught B Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr Opin Chem Biol 48, 44–54, doi: 10.1016/j.cbpa.2018.10.017 (2019). [DOI] [PubMed] [Google Scholar]
- 137.Samavarchi-Tehrani P, Samson R & Gingras AC Proximity Dependent Biotinylation: Key Enzymes and Adaptation to Proteomics Approaches. Mol Cell Proteomics 19, 757–773, doi: 10.1074/mcp.R120.001941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Han S, Li J & Ting AY Proximity labeling: spatially resolved proteomic mapping for neurobiology. Curr Opin Neurobiol 50, 17–23, doi: 10.1016/j.conb.2017.10.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim T-W et al. Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. doi: 10.1101/636324 (2019). [DOI]
- 140.Khan M, Youn JY, Gingras AC, Subramaniam R & Desveaux D In planta proximity dependent biotin identification (BioID). Sci Rep 8, 9212, doi: 10.1038/s41598-018-27500-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reinke AW, Mak R, Troemel ER & Bennett EJ In vivo mapping of tissue-and subcellular-specific proteomes in Caenorhabditis elegans. Sci Adv 3, e1602426, doi: 10.1126/sciadv.1602426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shinoda N, Hanawa N, Chihara T, Koto A & Miura M Dronc-independent basal executioner caspase activity sustains Drosophila imaginal tissue growth. Proc Natl Acad Sci U S A 116, 20539–20544, doi: 10.1073/pnas.1904647116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Uezu A et al. Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129, doi: 10.1126/science.aag0821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brudvig JJ et al. MARCKS Is Necessary for Netrin-DCC Signaling and Corpus Callosum Formation. Mol Neurobiol 55, 8388–8402, doi: 10.1007/s12035-018-0990-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Housden BE & Perrimon N Cas9-Mediated Genome Engineering in Drosophila melanogaster. Cold Spring Harb Protoc 2016, pdb top086843, doi: 10.1101/pdb.top086843 (2016). [DOI] [PubMed] [Google Scholar]
- 146.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell, doi: 10.1016/j.cell.2016.10.044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Venken KJ et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods 8, 737–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nagarkar-Jaiswal S et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4, doi: 10.7554/eLife.05338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]