Abstract
Rationale and Objectives:
We investigated the feasibility of utilizing convolutional neural network (CNN) for predicting patients with pure Ductal Carcinoma In Situ (DCIS) versus DCIS with invasion using mammographic images.
Materials and Methods:
An IRB-approved retrospective study was performed. 246 unique images from 123 patients were used for our CNN algorithm. In total, 164 images in 82 patients diagnosed with DCIS by stereotactic-guided biopsy of calcifications without any upgrade at the time of surgical excision (pure DCIS group). A total of 82 images in 41 patients with mammographic calcifications yielding occult invasive carcinoma as the final upgraded diagnosis on surgery (occult invasive group). Two standard mammographic magnification views (CC and ML/LM) of the calcifications were used for analysis. Calcifications were segmented using an open source software platform 3D Slicer and resized to fit a 128 × 128 pixel bounding box. A 15 hidden layer topology was used to implement the neural network. The network architecture contained five residual layers and dropout of 0.25 after each convolution. Five-fold cross validation was performed using training set (80%) and validation set (20%). Code was implemented in open source software Keras with TensorFlow on a Linux workstation with NVIDIA GTX 1070 Pascal GPU.
Results:
Our CNN algorithm for predicting patients with pure DCIS achieved an overall diagnostic accuracy of 74.6% (95% CI, ±5) with area under the ROC curve of 0.71 (95% CI, ±0.04), specificity of 91.6% (95% CI, ±5%) and sensitivity of 49.4% (95% CI, ±6%).
Conclusion:
It’s feasible to apply CNN to distinguish pure DCIS from DCIS with invasion with high specificity using mammographic images.
Keywords: DCIS, CNN, Calcifications
INTRODUCTION
Ductal Carcinoma In Situ (DCIS) is considered the earliest form of breast cancer, defined by the presence of abnormal cells limited to the mammary ducts within the breast, without extension beyond the basement membrane. DCIS may be subdivided into three pathologic subtypes: high, intermediate, and low. Although pathologic grade at time of diagnosis is a factor in patient management, no conclusive data regarding progression of DCIS to invasive carcinoma has been clearly established. Surgical management remains the standard of care for patients with DCIS, regardless of grade (1).
Although the incidence of DCIS has continued to increase with more patients undergoing screening mammography, there has been no subsequent decline in the rate of invasive carcinoma (1). The risk of overdiagnosis and treatment of DCIS must be weighed against the risk to observation alone in this patient population. Currently, there are two large clinical trials, in the US and Europe aimed at identifying potential low risk DCIS and comparing the necessity of surgical management versus observation (1,2). Recent studies have shown that there is still approximately 20% risk of upgrade upon excision in women diagnosed with "low risk" DCIS based upon the eligibility criteria of each of these clinical trials, respectively (1,2). In order to safely manage patients with DCIS by observation alone, alternate methodology is needed for more appropriate patient selection.
Mammography is used as the primary screening modality in the detection of breast cancer and microcalcification is the commonest mammographic feature of DCIS, visualized in approximately 80%—90% of cases (3). Little research has been done to evaluate the use of mammography in the detection of intratumor heterogeneity, (4) however there has been growing interest in radiomics in recent years with the advances in machine learning. Recently a subset of machine learning named Convolutional Neural Networks (CNN) has made great strides in medical imaging analysis. Compared to traditional machine learning, which primarily relies on human extracted feature analysis, neural networks depend on the input of raw data and allow the computer to automatically construct predictive statistical models through increasingly complex layers and self-optimization processes (5).
The purpose of this study is to determine the feasibility of utilizing CNN for predicting which patients have pure DCIS versus DCIS with invasion using mammographic images.
MATERIALS AND METHODS
An IRB-approved retrospective query was performed on patients at our institution from January 2015 to January 2018. Inclusion criteria include all patients who underwent stereotactic guided biopsy and subsequent surgical excision after identification of calcifications on a diagnostic mammogram with two standard magnification views (craniocaudal [CC] and mediolateral/lateromedial [ML/LM]). In total, 123 patients were identified, representing 246 unique images mammographic images. Of these, 164 images in 82 patients represented pure DCIS group (DCIS by stereotactic-guided biopsy of calcifications without any upgrade at the time of surgical excision). A total of 82 images in 41 patients represented occult invasive group (occult invasive carcinoma as the final upgraded diagnosis on surgery).
Mammograms at our institution were performed on dedicated mammography units (Senographe Essential, GE Healthcare). The views obtained consisted of the standard mediolateral oblique and CC views. Additional magnification views were obtained of the calcifications in CC and ML/LM projections. All statistical analyses were performed using a statistical software program (SPSS Statistics for Windows, Version 24. Chicago: SPSS Inc.). A two-sided p value of ≤0.05 was considered significant.
All of the patients in this study underwent stereotactic-guided core needle biopsy with a nine-gauge needle. Clinical and pathologic data were collected including patient’s age, size of the calcifications’ extent, and pathology result. All statistical analyses were performed using a statistical software program (SPSS Statistics for Windows, Version 24. Chicago: SPSS Inc.). A two-sided p value of ≤0.05 was considered significant.
DATA AUGMENTATION AND SEGREGATION
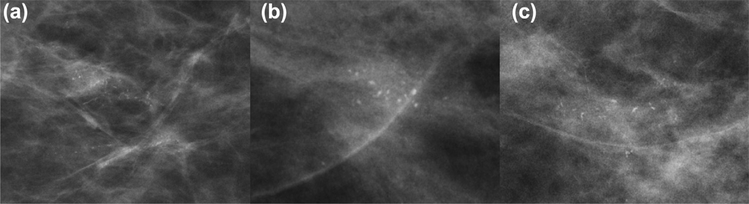
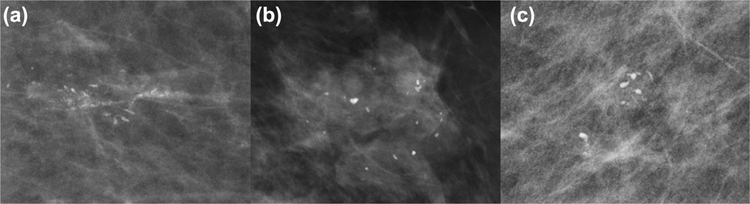
The raw magnification views (CC and ML/LM) of each patient’s mammogram was loaded into a segmentation program (3D Slicer) (18). Segmentations were manually extracted encompassing the regions of the magnification view which contained calcifications. Each image was scaled in size based on the radius of the segmentations and resized to fit a 128 × 128 pixel bounding box. The pathology report was used as the ground truth, from which patients were split into pure DCIS and occult invasive group (Fig. 1 and 2).
Figure 1.
Example mammographic cases of pure ductal carcinoma in situ before data augmentation.
Figure 2.
Example mammographic cases of ductal carcinoma in situ with invasion before data augmentation.
The entire image batch was normalized by dividing the nonair pixel intensity values by the standard deviation and subtracting by the mean. To perform data augmentation, queued images were randomly flipped vertically and/or horizontally, rotated by a random angle between +0.52 and −0.52 radians, and randomly cropped to a box 80% of their initial size. Due to the nature of obtaining compression for mammograms, a small amount of random shear was artificially applied do each input batch in order to simulate differing compression force on mammograms. The degree of affine warping, including shear was visually inspected on 1000 images to ensure that realistic augmentations were obtained. Finally, to simulate the effect of differing radiographic acquisition parameters due to slightly different kVp and mA, a random gaussian noise matrix was added to each input batch.
NETWORK ARCHITECTURE
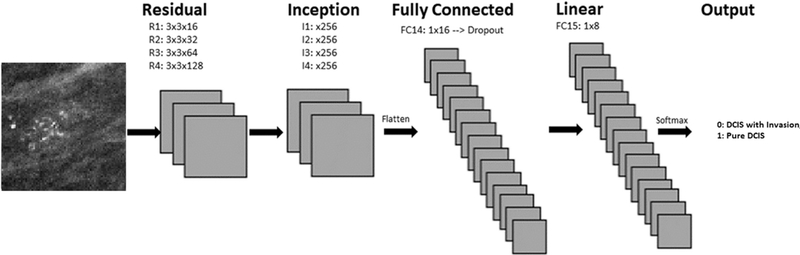
A novel 15 hidden layer customized CNN architecture (Fig 3) was designed to create a network architecture that would balance the most current strategies while keeping the overall number of trainable parameters as small as possible given the relatively small datasets seen in medical imaging. The network was trained from random weight initializations for evaluation of calcification types. Originally described by LeCun et al. in 1998 CNNs involve applying a series of convolution matrices to a vectorized input image that iteratively separates the input to a target vector space (6).
Figure 3.
Convolutional neural network architecture for two classification model.
After an initial standard convolutional layer, a series of residual layers are utilized in the first portion of the network. Originally described by He et al., residual neural networks are able to stabilize gradients during back propagation, leading to improved optimization, and facilitating greater network depth (7).
Beginning with the 10th hidden layer, inception V2 style layers are utilized. The Inception layer architecture, initially described in 2015 by Szegedy et al., introduced a computationally efficient method of allowing a network to selectively determine the appropriate filter architectures to an input feature map, leading to improved learning rates (8). A fully connected layer with 16 neurons was implemented after the 13th hidden layer followed by a linear layer with eight neurons. A final Softmax output layer with two classes was inserted as the last layer.
Training was implemented using the Adam optimizer four combined with the Nesterov accelerated gradient five described by Dozat (9—11). Parameters were initialized using the heuristic described by Glorot et al. (12). L2 regularization was implemented to prevent overfitting of data by limiting the squared magnitude of the kernel weights. Dropout was also employed to prevent overfitting by limiting unit coadaptation (13). Batch normalization was utilized to improve network training speed and regularization performance by reducing internal covariate shift (14).
Software code for this study was written utilizing the Python TensorFlow v1.5 library. Experiments and network training was performed on an Ubuntu 16.04 workstation with an NVIDIA TITAN X Pascal GPU. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
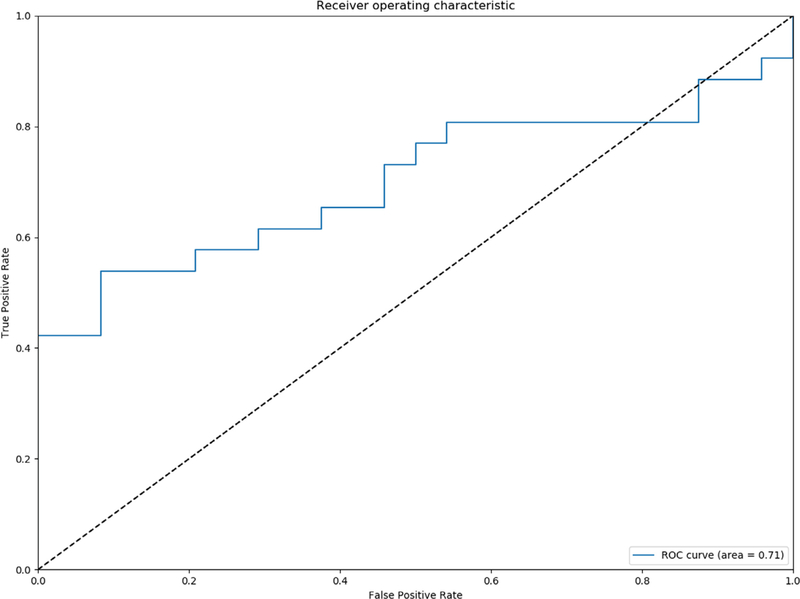
Softmax with cross entropy hinge loss was utilized as the primary objective function of the network to provide a more intuitive output of normalized class probabilities. A class sensitive cost function penalizing incorrect classification of the underrepresented class was utilized. Raw logits from the CC and ML/ LM view were summed to and a Softmax function was applied to arrive at the predicted class. Area under curve (AUC) was employed as the primary performance metric (Fig 4). Sensitivity, specificity, and accuracy were also calculated as secondary performance metrics.
Figure 4.
Receiver operating curve analysis for two class prediction model.
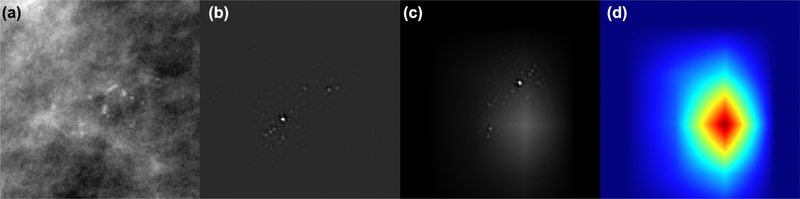
Five-fold cross validation was performed using training set (80%) and validation set (20%). In cross validation, the data set is split into five different equal pieces where one of the five pieces is used to test the performance of the network and the remaining pieces are used as a training set. The piece used for testing is then changed to a different piece and the process is repeated until all the pieces have been used as a testing set. Visualization of network predictions was performed using the gradient-weighted class activation mapping (Grad-CAM) techniques described by Ramprasaath et al. (15) (Fig 5).
Figure 5.
Results of class activation mapping on a patient with pure DCIS. Input image showing heterogeneous calcifications (a). Guided backpropagation (b), highlighting every pixel used in making the decision for each class, whether that pixel was a negative or positive predictor. In GRAD-CAM, the areas that are highlighted are the regions that were a positive factor in predicting the specific class (c). In Guided Grad-CAM (d), previous methods are combined to highlight pixels the network pays attention to in order to provide positive inputs, which include the region of calcifications as well as intervening breast parenchyma.
This study was IRB approved by our institution’s review board on July 18, 17.
RESULTS
The average patient age was 62.1 years (SD, 11.3 years) in the pure DCIS group and 61.0 years (SD, 11.8 years) in the DCIS with invasion group. The difference in age between the two groups was not statistically significant (p = 0.6). The average size of the mammographic calcifications ‘extent was larger (2.1 cm, SD, 1.66 cm) in the DCIS group with invasion compared to the pure DCIS group (1.54 cm, SD, 1.36 cm). But the difference between the two groups was no significant (p = 0.06). The average number of core samples obtained per biopsy was 9.6 cores (SD3.5 cores) in the pure DCIS group and 8.9 cores (SD 3.6 cores) in the DCIS with invasion group. The number of cores between the two groups was not significantly different (p = 0.2).
Based on the initial biopsy pathology result, 65.9% (54/82) of cases were high grade in the pure DCIS group. In the DCIS with invasion group, 63.4% (26/41) of cases were high grade. The frequency of high-grade cases between the two groups was not significant (p = 0.8). Comedonecrosis was present in 41.5% (17/41) of the cases in the DCIS with invasion group and 47.6% (39/82) of the cases in the pure DCIS group. The frequency of Comedonecrosis cases between the two groups was not significant (p = 0.6).
In total, 246 unique images from 123 patients were used for our CNN algorithm. A total of 164 images in82 patients diagnosed with DCIS by stereotactic-guided biopsy of calcifications without any upgrade at the time of surgical excision (pure DCIS group). In total, 82 images in 41 patients with mammographic calcifications yielding occult invasive carcinoma as the final upgraded diagnosis on surgery (occult invasive group). The network was trained for 300 epochs. The CNN algorithm for predicting patients with pure DCIS versus DCIS with invasion achieved an overall diagnostic accuracy of 74.6% (95% CI, ±5) with area under the ROC curve of 0.71 (95% CI, ±0.04), specificity of 91.6% (95% CI, ±5%), and sensitivity of 49.4% (95% CI, ±6%). The "positive class" was defined as pure DCIS group in this study and thus specificity represents minimizing the amount of falsely labeled pure DCIS cases.
Generated Grad-CAM maps indicated salient regions to include calcifications and the intervening breast parenchyma. Example is shown in Figure 5. The method of visualizing the pixels from the input image (Fig 5a) that the network pays attention to is the “guided backpropagation” (Fig 4b) showing heterogeneous calcifications. This highlights every pixel used in making the decision for each class, whether that pixel was a negative or positive predictor. In GRAD-CAM, the areas that are highlighted are the regions that were a positive factor in predicting the specific class (Fig 5c). In Guided Grad-CAM (Fig 5d), previous methods are combined to highlight pixels the network pays attention to in order to provide positive inputs, which include the region of calcifications as well as intervening breast parenchyma.
DISCUSSION
In our study, we demonstrated a feasibility of predicting patients with pure DCIS using mammography dataset utilizing a novel CNN developed at our institution. For potential use in selecting patients for DCIS observation trials, our CNN model was designed to maximize specificity in order to limit cases with occult invasive cancers. Our model was able to achieve very high specificity of 91.6%.
COMET and LORIS are two large clinical trials aimed at determining patients with DCIS diagnosis on core biopsy that may undergo observation rather than surgery. Despite using extensive inclusion and exclusion criteria in selecting patient, the accuracy of these trials in predicting pure DCIS patients is approximately 40% and 39%, respectively (2). In comparison, our CNN algorithm solely utilizing the mammographic image data yielded significantly higher accuracy of 74.6%. Potential of combining clinical information used in these clinical trials and the results of our CNN algorithm in order to further improve overall prediction model is under investigation.
Only few prior studies have evaluated mammographic image data to predict potential occult invasive cancers in patients diagnosed with DCIS. A study by Shi et al. in 2017 demonstrated that the traditional "handcrafted" computer vision mammographic features could be used to predict DCIS upstaging with performance comparable to a radiologist (16). In their study of 99 patients (74 pure DCIS; 25 DCIS with occult invasion), the manually extracted mammographic features, was able to distinguish DCIS with occult invasion from pure DCIS, with an area under the curve for ROC (AUC-ROC) equal to 0.70 (95% CI: 0.59 0.81). Major drawback of this traditional machine leaning approach is that the process of feature engineering is subjective and time consuming and likely not captures all the relevant image information.
Due to these limitations above, the same group (Shi et al.) utilized the VGG-16 model pretrained on ImageNet (nonmedical images such as animals, plants, instruments) as the feature extractor and compared it to their original study (17). In this study, the deep features were able to distinguish DCIS with occult invasion from pure DCIS, with an area under the ROC curve (AUC-ROC) equal to 0.70 (95% CI: 0.68 0.73). This performance was reported comparable to the handcrafted CV features (AUC-ROC = 0.68, 95% CI: 0.66—0.71). They concluded that the VGG-16 model pretrained on ImageNet might not be the optimal one to extract the off-the shelf deep features from digital mammograms, although it is well recognized as one of the most generalizable models for many different tasks.
In contrast to traditional algorithms that utilize handcrafted features based on human extracted patterns, neural networks allow the computer to automatically construct predictive statistical models, tailored to solve a specific problem subset (5). The laborious task of human engineers inputting specific patterns to be recognized could be replaced by inputting curated data and allowing this technology to self-optimize and discriminate through increasingly complex layers. In our study, we developed a novel CNN to distinguish pure DCIS from DCIS with invasion. We tailored the algorithm to increase specificity during training in order to minimize DCIS cases with invasion miscategorized as pure DCIS cases. To our knowledge, 246 unique images from 123 patients in our study is the largest study to date yielding a reasonable diagnostic accuracy of 74.6% and high specificity of 91.6%.
Our study is limited given the small sample size and the retrospective nature of the study performed at a single institution. The performance of CNN has been shown to increase logarithmically with larger datasets (16). Larger MRI datasets are likely to significantly improve our prediction model. In addition, mammography-based imaging data are more robust to data augmentation than are other medical images, because of the typical variance in normal biologic breast tissue compression rates and shear rates during image acquisition. This enabled us to use more-extensive image warps for data augmentation than typically is available, mitigating our small dataset size. Furthermore, potential of combining clinical information used in clinical trials (COMET and LORIS) and the results of our CNN algorithm in order to further improve overall prediction model is under investigation. In our study, ground truth was determined on the basis of pathologic findings, which has intrinsic limitations secondary to interobserver variability. We did not conduct interpretation by multiple pathologists. Although our algorithm has not been externally validated, a prospective validation study is planned in the near future.
Lastly, because training a CNN is an end-to-end process, it does not clearly reveal the reasoning behind the final result in a deterministic manner. Many methods have been developed to improve human understanding and intuition behind the predictions of a neural network; however, this is an ongoing area of research.
It’s feasible to apply CNN to distinguish pure DCIS from DCIS with invasion with high specificity using mammographic images. This can potentially aid in appropriate patient selection for observation trial in patients diagnosed with DCIS on core biopsy.
Acknowledgments
Disclosure: Richard Ha MD, MS. NVIDIA GPU provided by the GPU Grant Program. NVIDIA Corporation.
Footnotes
Work originated from Columbia University Medical Center. This work was presented at RSNA in 2018: SSM02–04.
Contributor Information
Simukayi Mutasa, Department of Radiology, New York, New York.
Peter Chang, Division of Neuroradiology, Center for Artificial Intelligence in Diagnostic Medicine (CAIDM), UCI Health, Department of Radiological Sciences, Orange, California.
Eduardo P. Van Sant, Department of Radiology, New York, New York.
John Nemer, Department of Radiology, New York, New York.
Michael Liu, Department of Radiology, New York, New York.
Jenika Karcich, Department of Radiology, New York, New York.
Gita Patel, Department of Radiology, New York, New York.
Sachin Jambawalikar, Department of Medical Physics and Radiology, Columbia University Medical Center, New York, New York.
Richard Ha, Breast Imaging Section, 622 West 168th Street, PB-1-301, New York, NY 10032.
REFERENCES
- 1.Pilewskie M, Olcese C, Patil S, et al. Women with low-risk DCIS eligible for the LORIS trial after complete surgical excision: how low is their risk after standard therapy? Ann Surg Oncol 2016; 23: 4253–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel G, Van Sant EP, Taback B, et al. Patient selection for ductal carcinoma in situ observation trials: are the lesions truly low risk? Am J Roentgenol 2018; 211(3):712–713. [DOI] [PubMed] [Google Scholar]
- 3.Evans A. The diagnoses and management of preinvasive breast disease: radiological diagnosis. Breast Cancer Res 2003; 5(5):250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song JL, Chen C, Yuan JP, et al. Progress in the clinical detection of heterogeneity in breast cancer. Cancer Med 2016; 5:3475–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeChun Y, Bengio T, Hinton G. Deep learning. Nature 2015; 521:436–444. [DOI] [PubMed] [Google Scholar]
- 6.LeCun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proc IEEE 1998; 86(11):2278–2324. [Google Scholar]
- 7.He K, Zhang S, Ren S, et al. Deep residual learning for image recognition. Proc IEEE Conf Comput Vis Pattern Recognit 2016.
- 8.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. Proc IEEE Conf Comput Vis Pattern Recognit 2015.
- 9.Kingma D, and Ba J. "Adam: a method for stochastic optimization." arXiv preprint arXiv:1412.6980 (2014).
- 10.Dozat T. "Incorporating nesterov momentum into adam." (2016).
- 11.Nesterov Y. "Gradient methods for minimizing composite objective function." (2007).
- 12.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics; 2010. [Google Scholar]
- 13.Srivastava N, Hinton G, Krizhevsky A, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014; 15 (1):1929–1958. [Google Scholar]
- 14.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Int Conf Mach Learn 2015: 4. [Google Scholar]
- 15.Ramprasaath RS. "Grad-cam: Why did you say that? visual explanations from deep networks via gradient-based localization." CVPR 2016. (2016).
- 16.Shi B, Grimm LJ, Mazurowski M, et al. Can occult invasive disease in ductal carcinoma in situ be predicted using computer-extracted mammographic features? Acad Radiol 2017; 24(9):1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi B, Grimm LJ, Mazurowski MA, et al. Prediction of occult invasive disease in ductal carcinoma in situ using deep learning features. J Am Coll Radiol 2018; 15(3 Pt B):527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.3D Slicer. https://www.slicer.org/