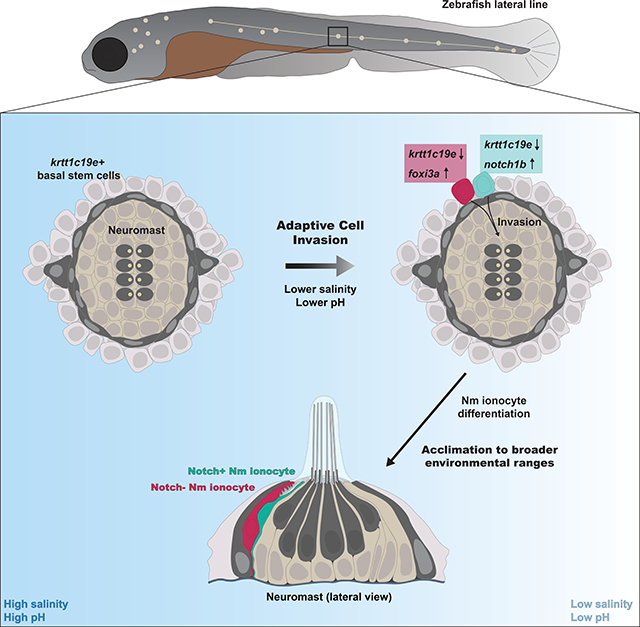
Summary
Mammalian inner ear and fish lateral line sensory hair cells (HCs) detect fluid motion to transduce environmental signals. Actively-maintained ionic homeostasis of the mammalian inner ear endolymph is essential for HC function. In contrast, fish lateral line HCs are exposed to the fluctuating ionic composition of the aqueous environment. Using lineage labeling, in vivo time lapse imaging and scRNA-seq, we discovered highly motile skin-derived cells that invade mature mechanosensory organs of the zebrafish lateral line and differentiate into Neuromast-associated (Nm) ionocytes. This invasion is adaptive as it is triggered by environmental fluctuations. Our discovery of Nm ionocytes challenges the notion of an entirely placodally-derived lateral line and identifies Nm ionocytes as likely regulators of HC function possibly by modulating the ionic microenvironment. Nm ionocytes provide an experimentally accessible in vivo system to study cell invasion and migration, as well as the physiological adaptation of vertebrate organs to changing environmental conditions.
Graphical Abstract
eTOC Blurb:
Peloggia and Münch et al. report the discovery of a new cell type in lateral line neuromasts, Nm ionocytes. Nm ionocytes invade mature sensory organs in a salinity- and pH-dependent manner, a process named Adaptive Cell Invasion. This process allows for physiological adaptation of sensory organs to fluctuating environmental conditions.
Introduction
The vestibular and auditory part of the vertebrate inner ear, as well as the lateral line sensory organ of aquatic vertebrates harbor hair cells immersed in electrogenically-maintained fluid microenvironments essential for sensory transduction. Inner ear hair cells perceive sounds and changes in body position and lateral line hair cells detect water motion and enable the animals to orient themselves, catch prey and avoid predators (Lush and Piotrowski, 2014). Changes in the ionic composition of the fluid environment in the mammalian inner ear leads to hearing loss and vestibular defects (Mittal et al., 2016). Vestibular and auditory systems are known as closed systems because of the specialized K+-rich fluid maintained by these organs, called the endolymph. The lateral line, on the other hand, is referred to as an open system as it is devoid of endolymph and its organs are exposed to the aqueous habitat in which the fishes and amphibians reside. However, lateral line hair cells are covered by a complex gelatinous cupula that provides an electrical and ionic microenvironment that is actively maintained and necessary for the lateral line cells to depolarize when mechanically activated (McGlone et al., 1979; Russell and Sellick, 1976). The mechanisms regulating ionic homeostasis in these open systems are not fully understood, and presently, the cell types maintaining the proper ionic composition within the cupula remain unknown (Baker et al., 2015; Flock, 1967; Flock and Jorgensen, 1974; Flock et al., 1973; Roberts and Ryan, 1971).
Several mechanisms that regulate ionic and osmotic homeostasis have evolved in vertebrates. For example, kidneys, the inner ear, mammalian lungs, as well as gills and embryonic skin of fish possess specialized cells for ion uptake and secretion, some of which are called ionocytes (Hwang and Lee, 2007). While ion-transporting cells in the kidney help maintain body ion homeostasis related to excretion, the loss of several ion channels in the inner ear causes impairment of sensory hair cell function and hearing loss (Mittal et al., 2016). Likewise, both genetic ablation of skin ionocytes in zebrafish or exposure to acidified water cause impaired hair cell function, supporting the notion that lateral line hair cells also need a buffered ionic environment for proper function (Hailey et al., 2012; Lin et al., 2019; Stawicki et al., 2014). In the mammalian inner ear, the mechanisms regulating the ionic composition of the endolymph involve specialized K+, Na+ and Cl− channels and transporters in the epithelial cells neighboring the hair cells (Gillespie and Muller, 2009; Trune, 2010). In the skin and gills of zebrafish, five subtypes of ionocytes are described to fine tune whole-body ionic balance: NaR (Ca2+ uptake), HR (H+ secretion/Na+ uptake/NH4+ excretion), NCC (Na+/Cl− uptake), KS (K+ secretion) and SLC26 (Cl− uptake/HCO3− secretion) (Abbas et al., 2011; Guh et al., 2015; Horng et al., 2007; Liao et al., 2009; Piermarini et al., 2002; Wang et al., 2009).
The zebrafish lateral line has proven to be an excellent model system to study sensory organ development and regeneration in vivo (Aman and Piotrowski, 2011; Lush and Piotrowski, 2014). The superficially located mechanosensory organs, called neuromasts, contain mechanosensory hair cells that are surrounded by several types of support cells. Loss of hair cells triggers a robust regenerative response during which support cells divide and differentiate into new hair cells (Romero-Carvajal et al., 2015). The lateral line system, including the sensory organs, the sensory ganglia, and the axons that innervate the hair cells is derived from sets of ectodermal placodes (Ghysen and Dambly-Chaudiere, 2007; Piotrowski and Baker, 2014). In the trunk, the posterior lateral line placode (primI) migrates toward the tail tip between 24–48 hours post fertilization (hpf) and periodically deposits clusters of cells that differentiate into neuromasts. Starting at 48 hpf, a second placode (primII) migrates along the trunk and deposits neuromasts of a different orientation (Lopez-Schier et al., 2004). Despite different deposition times, neuromasts from all placodes have mature and functional hair cells by 72 hpf (Nicolson et al., 1998; Sidi et al., 2003).
Using a combination of live imaging and single-cell RNA sequencing (scRNA-seq), we report the discovery of an invasive cell type in the zebrafish lateral line possessing ionocyte characteristics, named Neuromast-associated ionocytes (Nm ionocytes). Cre-mediated lineage tracing and time lapse imaging demonstrate that Nm ionocytes are derived from krtt1c19e+ basal keratinocytes (Lee et al., 2014) that surround neuromasts. We find that these cells both migrate extensively and persistently invade the post-embryonic and adult lateral line organs. It remains unclear how prevalent non-pathological cell invasion may be in post-natal and adult tissues and whether or not such events are active contributors to normal organ function. In the lateral line the invasion process is triggered and modulated in response to changing environmental stimuli, such as salinity and pH, a process we refer to as adaptive cell invasion (ACI). The discovery of ACI and Nm ionocytes indicate important and underappreciated roles for post-embryonic developmental processes and environmental stimuli in regulating mechanosensory organs in vertebrates, including humans.
Results
Zebrafish neuromasts contain previously uncharacterized cells that express ionocyte markers
To characterize lateral line cell type heterogeneity in mature 5 dpf neuromasts, we previously performed scRNA-seq analysis with cells isolated via fluorescence activated cell sorting (FACS) from a double transgenic line that labels hair and support cells (Fig. 1A and 1B; Gw57a:EGFP;pou4f3:GFP) (Lush et al., 2019). Using confocal imaging we discovered cells in some neuromasts that were not labeled by lateral line markers (Fig. 1C and 1D, Fig. S1A–C’, asterisks). The cells present in these fluorescence “gaps” were therefore not sorted and included in previously published datasets. The majority of gaps contains a pair of cells, as revealed by the ubiquitous nuclear marker H2B-mCherry (Fig. 1C–E, Fig. S1A” and B”, white dots). Some of the gaps contain only one cell, which is likely due to the partner cell having died. To identify and characterize these cells, we looked for transgenic lines that would label them. We found that the Notch pathway reporters tp1bglobin:mCherry or EGFP (Parsons et al., 2009), label one of the two cells in the pair (Fig. 1F and 1G, white arrowheads, Fig. 1H–J, Fig. S1C”, white dot, Supplementary Video 1). These Notch reporters are mosaic and only label 80% of the observed gaps in fluorescence (Fig. 1J, Fig. S1C, yellow arrowhead). Fluorescence intensity quantification of the lateral line specific she enhancer driving mCherry (she:H2A-mCherry) in Notch reporter-positive (Notch+) cells confirmed that these cells are indeed not labeled by lateral line markers (Fig. S1D–F).
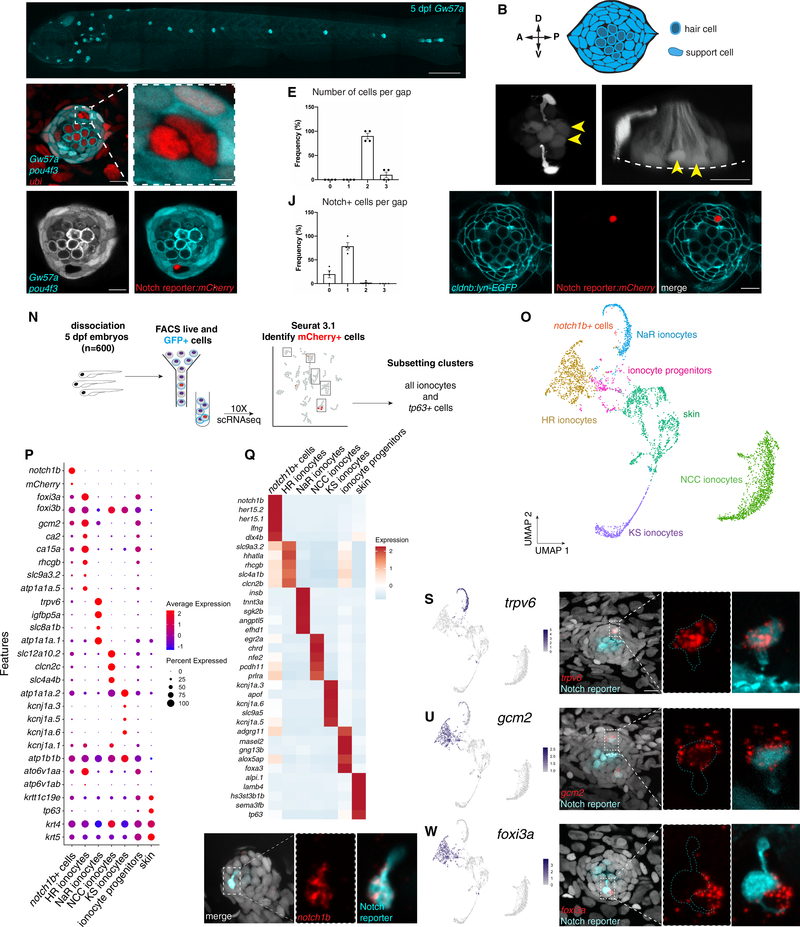
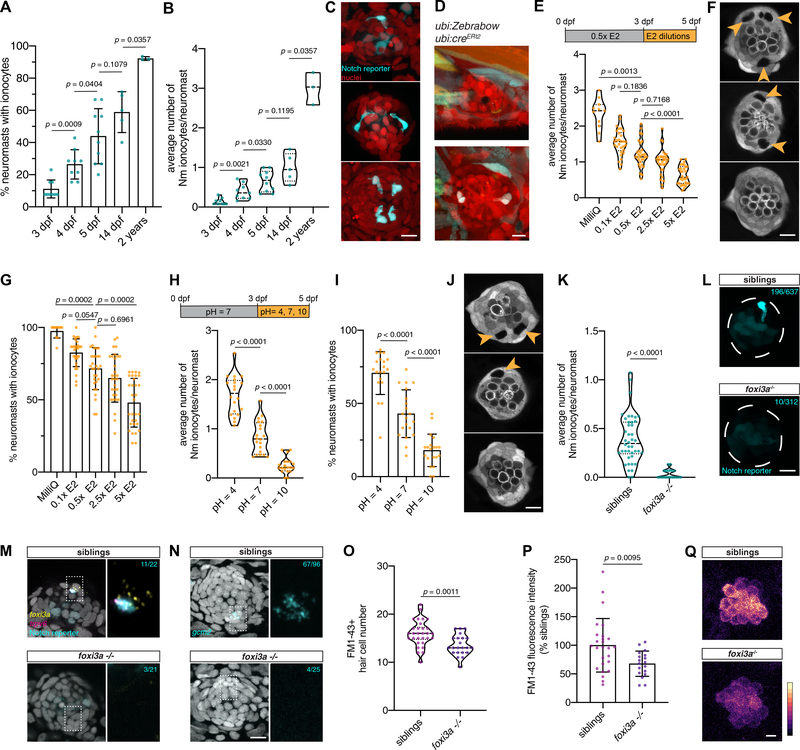
Figure 1: Zebrafish neuromasts contain previously uncharacterized pairs of cells that express ionocyte markers.
(A) Maximum projection of a 5 dpf Gw57a:EGFP;pou4f3:GFP larva. Scale bar = 300 μm. (B) Schematic of neuromast nuclei, dorsal view. (C) ubi:H2A-mCherry;Gw57a:EGFP;pou4f3:GFP larvae labeling nuclei, support cells and hair cells show a pair of cells that does not express neuromast or hair cell markers, dorsal view. (D) Magnification of (C). (E) Quantification of (C) (n = 4 fish, 36 neuromasts). Error bars = standard error of the mean (SEM). (F) The Notch reporter tp1bglobin:EGFP labels one cell of the pair (white arrowheads) and central support cells (yellow arrowheads). (G) 3D projection of a neuromast with a non-lateral line cell (white arrowhead) and central support cells (yellow arrowheads), lateral view (Supplementary Video 1). (H) Z-slice of Gw57a:EGFP;pou4f3:GFP and (I) tp1bglobin:hmgb1:mCherry confirms the Notch+ cell is one of the cells in the gap of (C). (J) Quantification of Notch+ cells per gap (n = 4 fish, 16 gaps). Error bars = SEM. (K-M) cldnb:lyn-GFP;tp1bglobin:mCherry larvae label all neuromasts, skin cells (K), the Notch+ cell (L) and its partner cell. (N) scRNA-seq sorting strategy. (O) UMAP after subsetting ionocytes and tp63+ cells. (P) Dot plot of known ionocyte markers. (Q) Pseudobulk heatmap of cluster markers. (R) notch1b hybridization chain reaction (HCR). (S) Feature plot for trpv6. (T) HCR of trpv6 in the Notch+ cell (n = 6 fish, 12 neuromasts). n-numbers = number of Nm ionocyte pairs analyzed. Magnification of the boxed area. (U) Feature plot for gcm2. (V) gcm2 is expressed in both cells in the pair (n = 4 fish, 11 neuromasts). (W) Feature plot of foxi3a. (X) foxi3a is expressed in the Notch− ionocyte (n = 5 fish, 12 neuromasts). All images, except panels A and G were blurred. Panels A, F and G were non-linear adjusted (gamma). Scale bars = 10 μm, unless otherwise noted. See also Figure Supplementary 1 and Supplementary Video 1.
To determine the identity of the pairs of non-lateral line cells, we performed scRNA-seq of FACS-purified cells from double transgenic cldnb:lyn-GFP;Notch reporter:mCherry fish (Fig. 1K–M). The cldnb promoter labels lateral line neuromasts, skin cells, as well as the Notch+ and Notch− non-lateral line cells, capturing both cells when sorted by GFP expression. In contrast, GFP and mCherry are co-expressed only in Notch+ non-lateral line cells, in neuromast central support cells and rarely in some skin cells (Fig. S1G and S1G’). To capture non-lateral line cells within mature neuromasts, we selected 5 dpf fish, in which around 50% of neuromasts have fluorescence gaps. This allowed us to directly compare our results with previous published neuromast datasets and use mCherry detection in central support cells as a positive control (Lush et al., 2019). We performed mild larvae dissociation to avoid contamination from deeper tissues and sorted for live GFP+ cells (Fig. 1N). After sequencing, alignment and quality controls, we performed dimensionality reduction with UMAP (Fig. S1H, Table 1) (Butler et al., 2018). To identify the Notch+ non-lateral line cells we searched the dataset for the presence of mCherry mRNA-expressing cells. We identified most mCherry transcripts in clusters expressing markers for NCC, NaR and HR ionocytes and, as expected, in the neuromast clusters containing central support cells (Fig. S1I).
To gain a better insight into the identity of mCherry+ cells, we selected for further analysis all detected ionocyte clusters together with tp63+ skin stem cells that contain ionocyte progenitors during embryonic development (Hsiao et al., 2007; Jänicke et al., 2007; Lee et al., 2014). This generated clusters of four known ionocyte subtypes, skin cells and putative ionocyte progenitor populations (Fig. 1O). We annotated ionocyte subtype clusters based on classical markers: trpv6, igfbp5a and slc8a1b are expressed in NaR ionocytes, rhcgb, slc4a1b, foxi3a, ca2 and atp6v1aa in HR ionocytes, clcn2c, slc4a4b and slc12a10.2 in NCC ionocytes and kcnj1a channels in KS ionocytes (Dai et al., 2014; Guh et al., 2015). krtt1c19e and tp63 expressing clusters were combined and identified as “skin” cells, while clusters that form streams between the skin and HR ionocytes are putative ionocyte progenitors, because of their expression of basal keratinocyte (tp63, krtt1c19e, krt4), ionocyte progenitor (foxi3a/b) and ionocyte markers (ca15a, atp1b1b, atp6v1aa) (Guh et al., 2015; Hsiao et al., 2007; Jänicke et al., 2007; Lee et al., 2014). The mCherry+ cells did not naturally fall into a homogenous cluster. To perform differential expression analyses between Notch reporter cells and ionocytes, we selected and clustered all mCherry+ cells that were originally intermingled with ionocyte cells in UMAP space. The most highly expressed Notch receptor in these cells was notch1b (Fig. 1P and 1Q, “notch1b+ cells”). To test if the non-lateral line Notch+ cells express notch1b, we performed hybridization chain reaction (HCR) on Notch reporter:EGFP larvae (Choi et al., 2018). Indeed, notch1b mRNA colocalized with EGFP (Fig. 1R) and Notch+ non-lateral line cells were labeled by anti-Notch1 immunofluorescence (Fig. S1J), suggesting that cells in the notch1b cluster possibly represent one of the cells in the pair of non-lateral line cells.
Our scRNA-seq analysis showed that some traditional ionocyte markers are not as specific for ionocyte subtypes as previously assumed (Fig. 1P) (Guh et al., 2015). We now identified highly specific marker genes for these cell types (Fig. 1Q, Supplementary Table 2). Likewise, notch1b+ cells express specific genes that they do not share with ionocytes, such as her15.1, her 15.2 and lfng (Fig. 1Q). They also express a few NaR and HR ionocyte-associated genes, albeit at a reduced level. Examples are the calcium channel trpv6 expressed in NaR cells and the transcription factor gcm2 involved in both NaR and HR ionocyte specification (Fig. 1P, 1S, 1U, 1W) (Chang et al., 2009; Kumai et al., 2014). On the other hand, the Notch− cell expresses Na+/K+-channels based on immunohistochemistry of Na+/K+-ATPases (NKA), labeling NaR and HR cells, while the Notch+ cell does not (Fig. S1K, S1K’).
We could not distinguish the non-lateral line Notch− cells from skin ionocytes in our scRNA-seq data. Possibly, they failed to form a distinct cluster because we sequenced too few cells, or their transcriptional signature is too similar to skin ionocytes. HCR with candidate genes revealed that the Notch− cells, as well as skin ionocytes strongly express the HR/NaR specification gene gcm2 and the HR ionocyte marker foxi3a (Fig. 1U–X). Additionally, the transcription factor foxi3b and the NaR marker atp1a1a.1 are likely expressed in either of these cells (Fig. S1N–O). Neither the Notch+ nor the Notch− non-lateral line cells express markers for NCC or KS ionocytes (Fig. S1L, S1M). We conclude that the Notch− cell is an ionocyte that expresses both HR and NaR markers. As the Notch+ cell expresses a few ionocyte-associated genes but otherwise does not share the majority of ionocyte marker genes, we hypothesize that the Notch+ cell is likely an accessory cell as has been described for gill ionocytes (Hootman and Philpott, 1980; Katoh et al., 2001; Katoh et al., 2003; Pisam et al., 2000). We termed the pair of cells Neuromast-associated ionocytes (Nm ionocytes).
Nm ionocytes are derived from skin cells surrounding the neuromast
As Nm ionocytes share genes with skin ionocytes but do not express neuromast markers we wondered if they possess a different embryonic origin than the placodally-derived neuromast cells (Fig. 2A). To determine the embryonic origin of these cells, we analyzed Zebrabow fish (ubi:Zebrabow;ubi:creERt2) that allow tracing of cells throughout development (Pan et al., 2013). After Cre-induced recombination of a set of mutually incompatible loxP sites, only one out of three fluorescent proteins per construct (RFP, CFP or YFP) is stochastically expressed in each cell. Fish lines containing more than one insertion produce a combination of colors with clonally related cells having the same color (Fig. 2B). Cre-induced recombination between 4–6 hpf led to mosaically labeled lateral line primordia which then deposited mosaic neuromasts (Fig. 2C–D). When we followed these neuromasts over time we detected two changes: 1) Starting around 3 dpf, some neuromasts suddenly contained new pairs of cells in the poles that had different color hues than the other cells in the neuromast (Fig. 2E, arrowhead) and 2) The vast majority of neuromast cells became clonal in adult fish, highlighting the presence of differently labeled pairs (Fig. 2F, F’, arrowheads). Triple transgenic embryos for ubi:Zebrabow;ubi:creERt2 and a Notch reporter driving the expression of mKate2 revealed that one of the differently colored cells in each pair is mKate2+, demonstrating that the differently colored pairs of cells are Nm ionocytes (Fig. S2A). Thus, Nm ionocytes possess a different embryonic origin than all other placodally-derived neuromast cells.
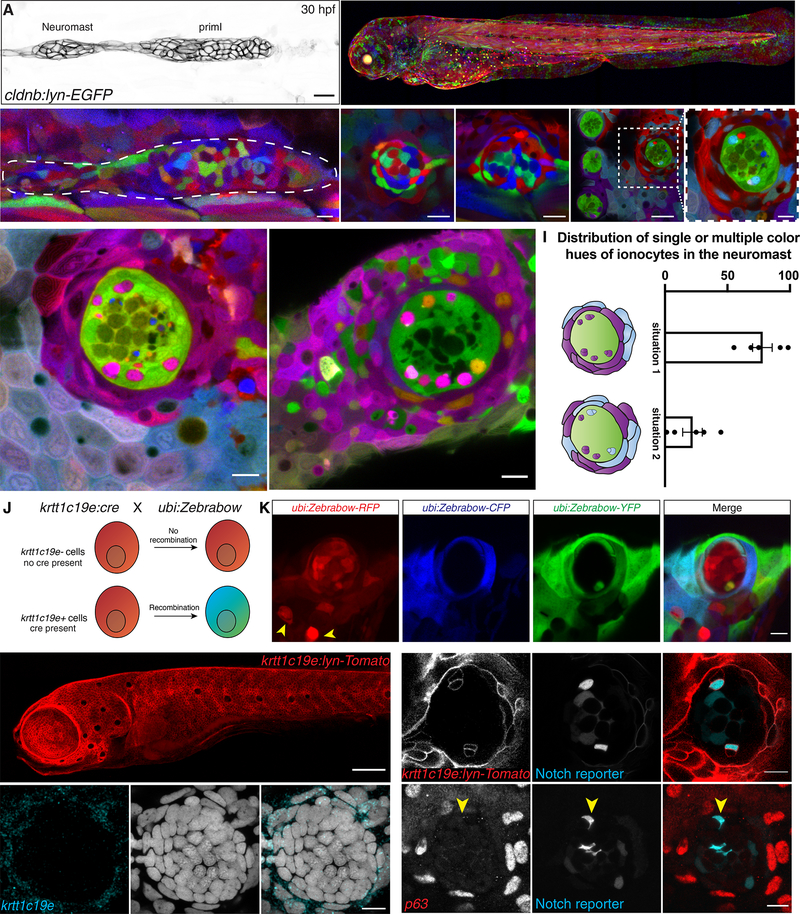
Figure 2: Neuromast-associated ionocytes are derived from skin cells surrounding the neuromast.
(A) 30 hpf cldnb:lyn-GFP+ lateral line primordium and deposited neuromast. (B) Maximum intensity projection of a 3 dpf ubi:Zebrabow larva recombined during shield stage. (C) 24 hpf primordium recombined for 15 min at 4 hpf. Neuromast in (C) is mosaic at 48 hpf (D) and 14 dpf (arrowhead indicates the new cell) (E). (F-F’) Neuromasts in two-year-old fish recombined between 16–24 hpf have become clonal. Scale bar in (F) = 30 μm. (G) Z-slices of 2 year old recombined ubi:Zebrabow;ubi:creERt2 fish show Nm ionocyte pairs of the same color hue as skin cells surrounding the neuromast or (H) Nm ionocyte pairs of two different color hues (white arrows) also shared with surrounding skin cells. (I) Quantification of (G and H) (n = 5 fish, 194 neuromasts). Error bars indicate SEM. (J) Overview of lineage tracing experiments with ubi:Zebrabow and krtt1c19e:cre-MYC. (K) A 5 dpf ubi:Zebrabow;krtt1c19e:cre-MYC neuromast shows a pair of recombined cells (white arrowhead) in between red, unrecombined cells. Skin ionocytes differentiate before Cre expression and are not recombined (yellow arrowheads). (L) 5 dpf krtt1c19e:lyn-Tomato larva labels basal keratinocytes. Scale bar = 100 μm. (M) 5 dpf krtt1c19e:lyn-Tomato;tp1bglobin:EGFP neuromast. krtt1c19e channel was gamma adjusted. (N) krtt1c19e HCR and (O) TP63 immunohistochemistry does not label cells in the neuromast (Notch+ ionocytes indicated by red arrowhead). Scale bars = 10 μm, unless stated otherwise. See also Figure Supplementary 2.
We also observed that Nm ionocytes shared their color hue with skin cells immediately surrounding the neuromasts (Fig. 2G and 2H), suggesting that skin cells might be the source of the Nm ionocytes, or at least share the same origin. In the majority of neuromasts (75%), all ionocytes within a neuromast shared the same color hue, while in the remaining neuromasts (25%) ionocytes had two or more different color hues (Fig. 2I). Irrespective of Nm ionocyte color heterogeneity, we always observed that cells in the surrounding skin expressed the same color hues as Nm ionocytes (n = 194 neuromasts). As Nm ionocytes often exist in pairs of cells we wondered if the two cells are clonally related. While most pairs had cells with the same color (92.4%), a small number of pairs were composed of cells of different colors (7.6%) (Fig. S2B and S2C). However, in the latter case we cannot exclude that they derived from a common progenitor cell if the recombination event occurred after division of this progenitor cell.
To investigate if Nm ionocytes are derived from skin cells, we performed lineage tracing by crossing the ubi:Zebrabow fish to krtt1c19e:cre-MYC, that expresses a constitutively active Cre recombinase with a MYC-tag (Fig. 2J). The krtt1c19e promoter becomes active around 24 hpf, permanently labelling basal keratinocytes and their progeny. Skin ionocytes already differentiated around the 14-somite stage (~16 hpf) and are therefore not recombined (Fig. 2K, yellow arrows). We observed recombined pairs of cells in the neuromasts, suggesting they are derived from Cre-expressing basal keratinocytes (Fig. 2K, white arrow). Likewise, double transgenic larvae for the basal keratinocyte reporter line krtt1c19e:lyn-Tomato (Fig. 2L) and the Notch reporter revealed pairs of Tomato+ cells in the neuromasts, of which one cell expressed the Notch reporter, confirming that Nm ionocytes initially expressed the basal keratinocyte cell marker krtt1c19e (Fig. 2M). Yet, krtt1c19e mRNA and TP63 protein are absent from Nm ionocytes suggesting these markers are downregulated as they differentiate (Fig. 2N and 2O, S2F and S2G). In support of this hypothesis, we found a negative correlation between fluorescence of krtt1c19e and Notch reporter expression (Fig. S2D and S2E), suggesting that the cells stop expressing krtt1c19e as Notch signaling increases. We observed a small number of cells in which H2B-EGFP was not detected. As all ionocytes reported in the literature are derived from tp63+ basal cells (Hsiao et al., 2007; Jänicke et al., 2007; Wu and Tang, 2021), it is possible that the krtt1c19e line is slightly mosaic or that the progenitors of these Nm ionocytes lost their basal stem cell fate before the krtt1c19e promoter was active. Altogether, we conclude from these data that Nm ionocytes are likely derived from basal keratinocytes surrounding the neuromasts and differentiate once inside the sensory organ.
Nm ionocyte progenitors migrate into neuromasts as pairs of cells
Nm ionocytes are not present at the time of neuromast deposition. To investigate the origin and dynamics of Nm ionocytes we took advantage of the superficial location of the lateral line. As we detected the first Nm ionocytes around 3 dpf, we performed time lapse imaging starting at this stage to capture the moment when these cells first appeared. Time lapse recordings revealed that ionocyte progenitors migrate into neuromasts as pairs of cells (Fig. 3A and 3B, colored dots, Supplementary Videos 2 and 3). By tracking individual pairs of Nm ionocytes from their origin outside the neuromast (Fig. 3C) to their final location (Fig. 3D, Supplementary Video 4), we found that after invasion some pairs of cells migrate to the other side of the organ, while others stay closer to their site of entry (Fig. 3C–E). Quantifying the speed of Notch+ cells showed that these cells on average migrate more than seven times the distance from their start to final location (Fig. 3F). Both cells re-arrange extensively between neuromast support cells and hair cells, while remaining closely associated with each other.
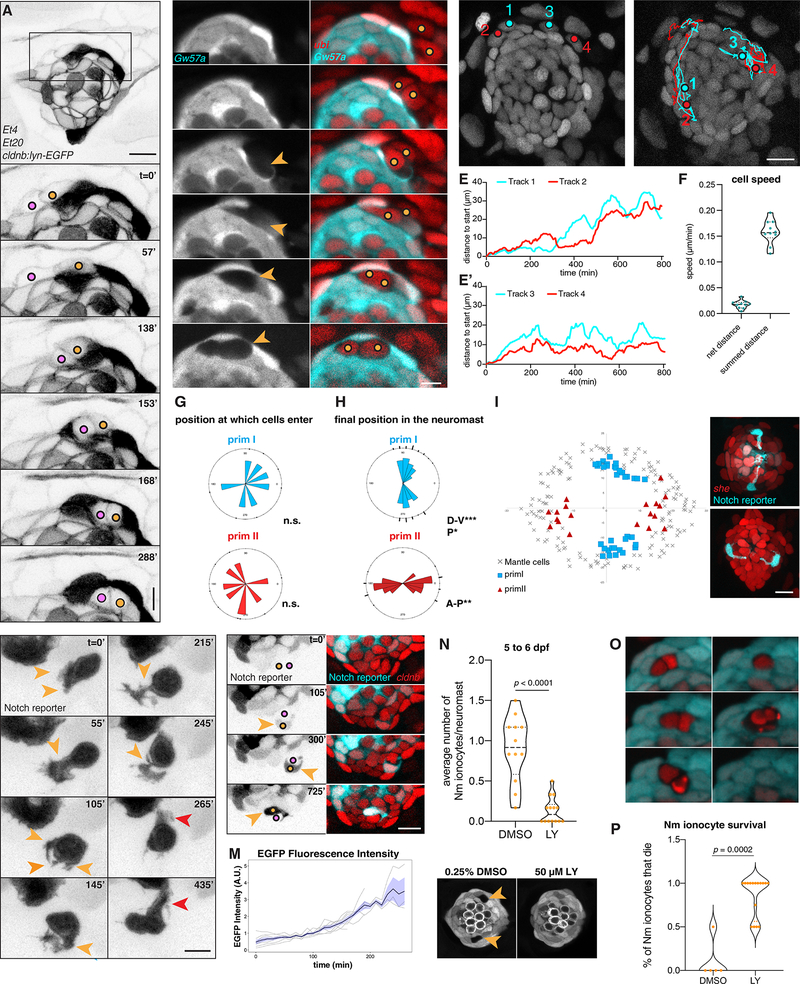
Figure 3: Ionocytes invade neuromasts as rearranging pairs of cells and their migration depends on Notch signaling.
(A) Still images of a time lapse recording of two Nm ionocyte precursors (colored dots) entering a 3 dpf Et4;Et20;cldnb-lynEGFP neuromast (Supplementary Video 2). Magnification of boxed area is shown below. Scale bar = 5 μm, images were gamma-adjusted. (B) Individual z-slices of a time lapse recording of a Gw57a:EGFP;ubi:H2A-mCherry larva at 3 dpf, with Nm ionocyte precursor cells migrating into the neuromast (right panels, colored dots), creating a gap in EGFP fluorescence (left panels, orange arrowheads, Supplementary Video 3). Scale bar = 5 μm. (C-D) Tracks of two Nm ionocyte precursor cell pairs in a 2 dpf cldnb:H2A-mCherry larva (Supplementary Video 4). Red and cyan dots depict the position of the Notch− and the Notch+ cell of each pair at the start (C) and end point of the movie (D). (E-E’) Quantification of the distance from the start position of the cells tracked in C and D over time. (F) Quantification of the cell speed calculated based on the net distance, as well as the summed distance that Notch+ cells traverse (n = 7 neuromasts, 10 cells). (G) Rose plots displaying the angular position at which Nm ionocyte precursors enter neuromasts of different polarities (primI- and primII, blue and red labeling, respectively) (n = 15 neuromasts, 7 fish). Binomial test revealed no directional bias. (H-I) Angular, final position of Nm ionocytes in primI- and primII-derived 5 dpf neuromasts (H), and their spatial distribution in relation to mantle cell position (I) (n = 55 neuromasts). Binomial analysis showed significant bipolar localization with a slight directional bias towards posterior in primI-derived neuromasts. (J-J’) Nm ionocyte localization in primI-derived (J) and primII-derived neuromasts at 5 dpf (J’). Images were gamma-adjusted (K) Still images of a time lapse recording at 3 dpf, depicting protrusions of a Notch+ Nm ionocyte precursor (orange arrowheads) before stabilizing an apical extension (red arrowheads, Supplementary Video 5). Images were gamma-adjusted. Scale bar = 5 μm. (L) Gradual increase in Notch reporter fluorescence (left panel, orange arrowheads; right panel, white arrowheads) as Nm ionocyte precursor cells (left panel, colored dots) re-arrange between neuromast cells (Supplementary Video 6). (M) Quantification of Notch reporter GFP fluorescence over time, normalized by the average of all the curves up to the 160 min time point. (N-N”) Nm ionocyte frequency following inhibition of Notch signaling with 50 μM LY411575 for 24 h (n = 14 larvae, 84 neuromasts) between 5 and 6 dpf compared to DMSO controls (n = 12 larvae, 72 neuromasts; Mann-Whitney test, p < 0.0001). (O) Still images of time lapse from a 4 dpf larva (Gw57a:EGFP; ubi:H2A-mCherry) shows a pair of Nm ionocyte undergoing cell death after Notch inhibition (P) Quantification of time lapses assessing Nm ionocyte death after incubation with 50 μM LY411575 (n = 8 larvae, 18 neuromasts) compared to DMSO controls (n = 3 larvae, 5 neuromasts). Scale bars = 10 μm, unless otherwise noted. Dashed black lines in violin plots indicate the median, while dotted black lines indicate quartiles. See also Figure Supplementary 3 and Supplementary Videos 2–8.
Once inside the neuromast, the Notch+ ionocyte precursor extends highly dynamic protrusions as visualized by the cytoplasmic Notch reporter (Fig. 3K, Supplementary Video 5). While time lapse recordings showed that Nm ionocyte precursors do not enter the organs at predefined positions (Fig. 3G), spatial analyses of Nm ionocytes at 5 dpf demonstrated that they are eventually positioned stereotypically in the D-V and A-P poles of primI- and primII-derived neuromasts, respectively (Fig. 3H–J’). After reaching the poles, Nm ionocytes stop re-arranging and project a stable apical extension towards the cuticular plate of the neuromast (Fig. 3K).
Notch signaling plays a crucial role in skin ionocyte specification via lateral inhibition (Hsiao et al., 2007; Jänicke et al., 2007) and we observed that migration of Nm ionocytes are associated with a progressive increase in Notch signaling in one cell of the pair (Fig. 3L, Fig. 3M, Supplementary Videos 6 and 7). We therefore wondered if Notch plays a role in Nm ionocyte differentiation. Pharmacological inhibition of Notch signaling with the gamma secretase inhibitor LY411575 during neuromast development from 3 to 4 dpf generated an excess of hair cells at the expense of support cells accompanied by a lack of ionocytes (Fig. S3A and S3A”) (Romero-Carvajal et al., 2015). To exclude the possibility that the excess of hair cells spatially prevents ionocyte precursors from invading, we inhibited Notch signaling once all hair cells had differentiated (5 dpf) and some Nm ionocytes were already present in the neuromast. We still observed a significant decrease in Nm ionocytes (Fig. 3N and 3N’). To determine if Nm ionocytes die or migrate out of the neuromasts after Notch inhibition, we performed time lapse imaging of embryos incubated with LY411575. We observed that both cells of the pair died after Notch inhibition, with the Notch+ cell usually dying first (Fig. 3O, 3P, S3B and S3C and Supplementary Videos 8 and 9). The mechanism by which the Notch+ cells maintain the Notch− cell is unknown. We conclude from these data that Notch signaling plays an essential role in Nm ionocyte survival. The role of Notch signaling in the migration and fate determination of the highly protrusive ionocyte progenitors invading mature neuromasts remains to be determined.
3D modeling of Nm ionocytes reveals exquisite morphological characteristics resembling gill and skin ionocytes
Nm ionocyte differentiation is associated with profound changes in cell morphology. Although they commonly exist as pairs, we could only visualize the Notch+ cell due to a dearth of markers. To visualize the structure of both cells, we performed Serial Block Face Scanning Electron Microscopy (SBF-SEM). To identify the ionocyte pair inside the neuromast, we took a correlative approach between SBF-SEM and light microscopy. We imaged ionocyte-containing neuromasts under a confocal microscope with two fluorescent markers: a neuromast nuclei marker (red) and the Notch reporter (cyan) (Fig. 4A). The same embryo was then processed for SBF-SEM. Modeling of the SEM neuromast nuclei resulted in a very similar pattern of nuclei observed under confocal microscopy (Fig. 4B) allowing us to identify the ionocyte pairs in the SBF-SEM dataset (Fig. 4A and 4B). 3D modeling of the cell membranes of the Notch+ and Notch− cells revealed tight association between these cells. The two cells are in close contact with each other along their entire apico-basal axis, their nuclei are located basally and the Notch+ cell (cyan) is smaller than the Notch− cell (white) (Fig. 4C, Supplementary Video 10). Both cells possess extensions that reach the apical surface of the neuromast and frequently contain thin projections (Fig. 4C and 4D, Fig. S4A and S4A’).
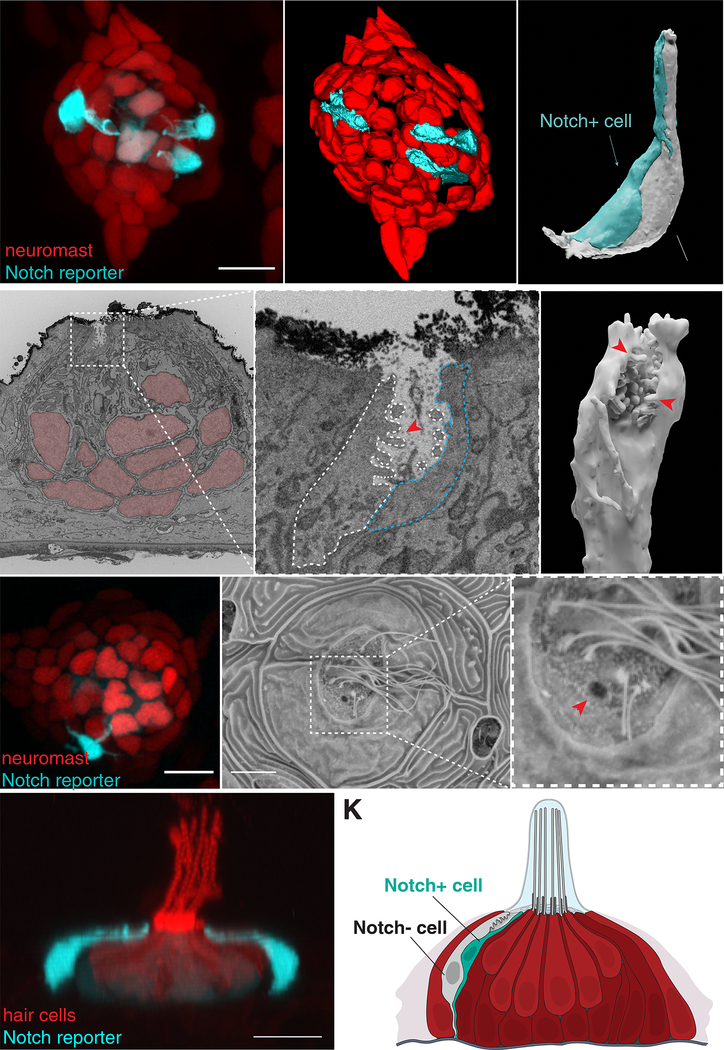
Figure 4: Nm ionocytes share morphological characteristics with skin ionocytes and are exposed to the external environment through an opening to the neuromast cupula.
(A) Maximum intensity projection of a neuromast labeled by she:H2A-mCherry;tp1bglobin:EGFP shows nuclei of the neuromast cells in red and three Notch+ Nm ionocytes in cyan. (B) 3D modeling of the SBF-SEM stack of the same neuromast in (A). (C) 3D modeling of the Nm ionocyte pair (white and cyan). (D) Single image of a transverse section of the SBF-SEM shows the apical part of a pair of Nm ionocytes (white square) and neuromast cells (red nuclei). (E) Magnification of white square in (D) showing microvilli (red arrowhead) and the apical crypt of the ionocyte pair exposed to the outside of the neuromast. (F) 3D modeling of the microvilli (red arrowheads) containing crypt of the Notch− cell. (G) A neuromast labeled by she:H2A-mCherry;tp1bglobin:EGFP. (H) Scanning electron micrograph of the same neuromast in (G). (I) Magnification of the region in white square in (H). Red arrowhead depicts an opening in the neuromast cuticular plate that correlates with the position of the Nm ionocyte pair. (J) Lateral view of a 3D projection of a tp1bglobin:EGFP;Myo6b:Lck-mScarlet-I neuromast (Supplementary Video 11). Nm ionocytes have long apical projections with the apical crypt (white arrowheads) close to the cuticular plate. (K) Model of the neuromast (red) containing a pair of Nm ionocytes (white and cyan). Panels A and G are non-linear adjusted (gamma). See also Figure Supplementary 4 and Supplementary Videos 10 and 11.
The pair forms a crypt at the top of the apical extensions with microvilli projecting only from the Notch− cell into the lumen (Fig. 4E and 4F, Fig. S4B and S4B’). This morphology is reminiscent of gill and skin ionocytes that possess apical crypts exposed to the outside (Katoh et al., 2001; Katoh et al., 2003). To test if Nm ionocytes also open to the external environment, we analyzed correlated SEM and confocal images. Indeed, we observed openings in the neuromast cuticular plate that correlated with presence and location of Nm ionocytes (Fig. 4G–I). Consequently, the crypts are exposed to the gelatinous cupula microenvironment that covers the cuticular plate and hair cell cilia of live animals (Fig. 4J, Supplementary Video 11).
Microvilli-containing ionocytes accompanied by accessory cells have been described mostly in seawater fish (Katoh et al., 2001). We hypothesize that the Notch+ cells act as accessory cells for the Notch− Nm ionocyte. Our results show that the pair of Nm ionocyte precursors differentiates into a microvilli-containing ionocyte and a slightly smaller accessory cell that together form a crypt open to the neuromast cuticular plate. These findings reveal a previously unappreciated cell type diversity in the neuromast (Fig. 4K).
Nm ionocyte frequency is modulated by changes in salinity and pH
The finding that apical openings of Nm ionocytes are in contact with the overlying cupula suggests that they could be involved in controlling the ionic microenvironment surrounding hair cells. However, we observed that not all neuromasts contain ionocytes during the larval stages. To test whether Nm ionocyte frequency increases at later time points, we quantified Nm ionocyte numbers in 14 dpf larvae and 2-year-old adult fish. Indeed, the percentage of ionocyte containing neuromasts (Fig. 5A), as well as the number of Nm ionocytes per neuromast (Fig. 5B and 5C) significantly increases during early larval development (3 dpf to 5 dpf) and from later larval stages (14 dpf) until adult stages (2 years). While we cannot fully rule out the possibility of proliferation of existing Nm ionocytes during these later stages, we observed Nm ionocytes appear in neuromasts without pre-existing ionocytes between 21 dpf and 28 dpf based on Zebrabow analysis (Fig. 5D). This suggests that Nm ionocytes continue to invade neuromasts as animals age.
Figure 5: Nm ionocyte frequency is modulated by environmental changes.
(A) Percentage of neuromasts with Notch+ Nm ionocytes at 3 dpf (n = 9 fish, 117 neuromasts), 4 dpf (n = 9 fish, 125 neuromasts), 5 dpf (n = 10 fish, 162 neuromasts), 14 dpf (n = 5 fish, 49 neuromasts) and in 2-year old adult fish (n = 3 fish, 78 neuromasts, Mann-Whitney test). (B) Average number of Notch+ Nm ionocytes per neuromast at stages quantified in (A), (same n-numbers, Mann-Whitney test). (C) Representative maximum intensity projections of Nm ionocytes (arrowheads) at stages quantified in (A)-(B). (D) Maximum intensity projections of the same ubi:Zebrabow;ubi:creERt2 neuromast at 21 dpf (upper panel) and 28 dpf (lower panel) with newly appeared Nm ionocytes (arrowheads). (E) Nm ionocyte number in larvae incubated in control media (0.5x E2) or different concentrations (MilliQ, n = 11 fish, 154 neuromasts; 0.1x E2, n = 30 fish, 433 neuromasts; 0.5x E2, n = 31 fish, 436 neuromasts; 2.5x E2, n = 31 fish, 449 neuromasts; 5x E2, n = 31 fish, 454 neuromasts; Kruskal-Wallis ANOVA and Dunn’s post hoc test). (F) Representative images of neuromasts and Nm ionocytes (arrowheads) following incubation in E2 media dilutions. (G) Percentage of neuromasts with Nm ionocytes in larvae incubated in different E2 media dilutions (n-numbers as in (E), Kruskal-Wallis ANOVA and Dunn’s post hoc test). (H) Nm ionocyte frequency in acidic (pH = 4; n = 20 fish, 286 neuromasts), alkaline (pH = 10; n = 20 fish, 291 neuromasts) and neutral E2 media (pH = 7, n = 20 fish, 296 neuromasts; unpaired t-test). (I) Percentage of neuromasts with Nm ionocytes following incubation in E2 media of different pH (n-numbers as in (H), Mann-Whitney test). (J) Representative images of Nm ionocytes (arrowheads) as quantified in (I). (K) Average Nm ionocyte frequency in foxi3a−/− fish (n = 20 fish, 312 neuromasts) and their siblings (n = 42 fish, 637 neuromasts; Mann-Whitney test). (L) Representative maximum projections of Nm ionocytes (arrowheads) in foxi3a−/− fish and their siblings. (M) foxi3a and trpv6 HCR in foxi3a−/− fish (lower panel, n = 21 neuromasts, 4 fish) and their siblings (upper panel, n = 22 neuromasts, 7 fish). Cyan n-numbers indicate the number of neuromasts with trpv6+ Nm ionocytes. Magnification of boxed area is shown on the right. (N) gcm2 HCR in foxi3a−/− fish in the Notch reporter background (lower panel, n = 25 neuromasts, 3 fish) and their siblings (upper panel, n = 96 neuromasts, 12 fish). Cyan n-numbers indicate the number of neuromasts with gcm2+ Nm ionocytes. Magnification of boxed area is shown on the right. (O) FM1–43+ hair cell numbers in foxi3a−/− fish (n = 15 fish, 30 neuromasts) and their siblings (n = 11 fish, 22 neuromasts). (P) FM1–43 intensity of hair cells in foxi3a−/− fish (n = 9 fish, 18 neuromasts) compared to their siblings (n = 13 fish, 26 neuromasts; unpaired t-test). Data is shown as fluorescence intensity/background, normalized to the average of the siblings. (Q) FM1–43 dye uptake by hair cells in foxi3a−/− fish and their siblings, as quantified in (P). Scale bars = 5 μm.
All scale bars = 10 μm, unless otherwise noted. Error bars indicate standard deviation. Dashed lines in violin plots indicate the median, dotted lines indicate quartiles. Individual data points in A, B, E, G, H, I and K represent an average of all neuromasts quantified per fish. See also Figure Supplementary 4.
The increase in Nm ionocyte frequency not only correlates with developmental age but also with the transfer of the larvae from ion-rich embryo medium (E2) to more ion-poor water in the housing facility. Since the density of skin and gill ionocytes is modulated in response to different ionic environments (Hwang and Chou, 2013), we wondered whether salinity changes could influence the frequency of Nm ionocyte invasion. To test this hypothesis, we incubated larvae in different dilutions of E2 medium between 3 and 5 dpf and quantified the frequency of Nm ionocytes. Indeed, the number of Nm ionocytes negatively correlated with increasing E2 media salinity (Fig. 5E and 5F). After incubation in MilliQ water, Nm ionocytes displayed frequencies that more closely resembled those observed in adult fish (Fig. 5B and 5E) and 98.7% of neuromasts now contained one or more Nm ionocytes (Fig. 5G). The increase in Nm ionocytes was not caused by a systemic increase in proliferation as the total neuromast cell and hair cell number decreased in MilliQ water (Fig. S5A–C). Similar to Nm ionocytes, the density of NKA+ as well as ca2 and trpv6-expressing skin ionocytes on the yolk increased in MilliQ water (Fig. S5D–G). In contrast, an increase in salinity led to increased Nm ionocyte death (Fig. S5H). These findings provide support for the hypothesis that Nm ionocytes respond to salinity changes and are involved in regulating ion homeostasis.
Since incubation in MilliQ water increased Nm ionocytes but also decreased hair cell numbers, we aimed to test if hair cell death modulates Nm ionocyte numbers. Killing hair cells with the antibiotic neomycin had no effect on Nm ionocyte frequency (Fig. S5I and S5J). Neomycin-induced hair cell death also did not lead to proliferation of the Notch+ Nm ionocyte as revealed by EdU-incorporation experiments (Fig. S5K). These results show that invasion of the neuromast by ionocytes is not triggered by hair cell death.
Lateral line hair cell function is impaired in merovingian mutants that harbor a mutation in gcm2 (Stawicki et al., 2014), a transcription factor involved in HR and NaR ionocyte differentiation (Chang et al., 2009; Kumai et al., 2014). This impairment had been attributed to loss of skin HR ionocytes, as Nm ionocytes had not been discovered (Stawicki et al., 2014). Given the physical proximity to hair cells and the expression of HR ionocyte markers like gcm2 (Fig. 1V), we wondered whether Nm ionocytes could be involved in maintaining the pH in the hair cell microenvironment. We reasoned that Nm ionocyte numbers would increase in acidified environments, as has been described for HR cells (Horng et al., 2009). Indeed, we observed that Nm ionocyte number as well as the percentage of neuromasts with ionocytes increased with decreasing pH (Fig. 5H–J). This finding suggests that Nm ionocytes invade neuromasts not only in response to changes in salinity, but also pH, pointing towards an additional involvement in acid/base secretion (Horng et al., 2009).
Loss of foxi3a reduces Nm ionocyte numbers and impairs hair cell function
To test Nm ionocyte function we sought to identify genes that when knocked out cause Nm ionocyte loss. One candidate is the transcription factor foxi3a that specifies skin ionocytes from epidermal precursors, with foxi3a morphants lacking NaR and HR ionocytes (Hsiao et al., 2007; Jänicke et al., 2007). To determine whether Nm ionocytes are specified by the same transcriptional program, we generated a zebrafish mutant lacking the foxi3a promoter region and part of the first exon by CRISPR/Cas12a-mediated genome editing (Moreno-Mateos et al., 2017) (Fig. S5L). Homozygous foxi3a−/− larvae show a significant reduction of Notch+ Nm ionocytes compared to their siblings (Fig. 5K and 5L). Likewise, HCR revealed a reduction in the number of foxi3a-, trpv6- and gcm2-expressing cells in the neuromasts (Fig. 5M and 5N). These data demonstrate that foxi3a plays a pivotal role in Nm ionocyte development. Interestingly, while foxi3a expression in skin ionocytes on the yolk is nearly absent in foxi3a mutants (Fig. S5M), gcm2-expressing ionocytes are still present (Fig. S5N).
We next wondered whether the absence of Nm ionocytes in foxi3a mutants affects hair cell function. Since uptake of the vital dye FM1–43 depends on functional mechanotransduction (Gale et al., 2001; Meyers et al., 2003; Seiler and Nicolson, 1999), we tested whether hair cells in foxi3a mutants display decreased labeling with this dye. foxi3a mutants show a slight reduction in FM1–43+ hair cell numbers (Fig. 5O) and hair cells display a significant reduction in FM1–43 intensity to 67% of average sibling’s levels (Fig. 5P and 5Q). This suggests that hair cell mechanotransduction is impaired in foxi3a mutants. Although we cannot rule out that skin ionocytes also contribute to the hair cell function defect by modulating global ionic homeostasis, these results suggest that Nm ionocytes might play an important role in regulating the ionic microenvironment surrounding lateral line hair cells.
Discussion
The lateral line system is necessary for schooling behavior, mating, feeding and predator avoidance (Lush and Piotrowski, 2014). Therefore, it is crucial to maintain proper hair cell function, and regulate its microenvironment in the presence of ever-changing environmental factors (Hwang and Lee, 2007). We have made the unexpected discovery that pairs of skin-derived epithelial cells invade mature lateral line sensory organs post-embryonically. These cells are highly motile and, after invasion, rearrange and migrate extensively, before anchoring in stereotypical positions and differentiating into ionocytes.
Cell migration and tissue invasion are essential events driven by diverse signals and involved in processes such as development, regeneration, wound healing, immune system and lateral line formation itself (Kapsimali, 2017). However, these migratory events differ from the invasive processes reported here in two major aspects: 1) they are restricted to developmental stages of organ formation; and 2) cells that contribute to mature organs during regeneration differentiate into sensory organ cells. For example, developing pulmonary neuroendocrine cells are sparsely distributed throughout the lung epithelium and then start to migrate, leading to cell sorting and cluster formation (Kuo and Krasnow, 2015; Steinberg and Takeichi, 1994). Recruitment of cells into mature sensory organs also occurs during taste bud regeneration when basal cells from the lingual epithelium surrounding the sensory organs regenerate taste cells (Barlow and Klein, 2015). But in contrast to these taste bud regenerating basal cells, Nm ionocytes maintain a distinct transcriptome after invading mature sensory organs.
Signals that trigger Adaptive Cell Invasion and determine the Nm ionocyte lineage
The signals that attract epithelial cells to the sensory organs remain poorly understood. Nm ionocyte invasion is not triggered by hair cell death and regeneration signals, instead they robustly respond to environmental changes, such as salinity and pH (Fig. 5E–J). Environmental changes also control ionocyte numbers in the skin via hormones such as cortisol, isotocin and prolactin (Chou et al., 2011; Evans et al., 2005; Flik and Perry, 1989; Guh et al., 2015; Kumai et al., 2015). The scRNA-seq analysis revealed Nm ionocytes indeed express the cortisol receptor nr3c1 (glucocorticoid receptor, GR), a receptor involved in cortisol-induced ionocyte precursor differentiation, as well as the prolactin receptor prlra (Fig. 1Q) (Cruz et al., 2013a; Cruz et al., 2013b). Whether hormones trigger ACI needs to be further tested.
The identity of the signals that specify Nm ionocyte precursors among basal cells surrounding the neuromast is unknown. In Medaka, neuromast border cells (nBCs) are induced by neuromasts (Seleit et al., 2017), however they are unlikely to be Nm ionocyte precursors as they do not enter the neuromasts. Nevertheless, it is likely that neuromasts induce ionocytes in their immediate vicinity. We find individual foxi3a+ cells in contact with neuromasts suggesting that these cells could be pre-invasion Nm ionocytes progenitors (Fig. S1P). foxi3a+ ionocyte progenitors in the skin are specified from epidermal stem cells by Notch-mediated lateral inhibition (Hsiao et al., 2007; Jänicke et al., 2007). Nm ionocytes also upregulate Notch signaling as they invade neuromasts. However, the outcomes of inhibiting Notch are different. In the skin, both DAPT treatment and mindbomb mutants increase the frequency of ionocytes at the expense of epidermal cells (Hsiao et al., 2007; Jänicke et al., 2007). In contrast, Notch inhibition affects Nm ionocyte survival. The Notch pathway has distinct downstream targets depending on ligand-receptor combinations (Nandagopal et al., 2018). As skin ionocytes and Nm ionocytes express different Notch receptors (notch1a/notch3 and notch1b, respectively), Notch signaling possibly has different functions in these two cell types. Future studies will shed light on the divergent roles of Notch signaling in Nm specification and ionocyte invasion, as well as the Notch ligands involved.
Tissue-specific ionocyte function
Ionocytes play crucial roles in the function of many organs. Loss of a rare ionocyte population recently discovered in the lungs of mouse and humans is associated with cystic fibrosis (Montoro et al., 2018; Plasschaert et al., 2018). Different types of ionocytes in fish are functionally and molecularly analogous to ion transporting cells of the kidney of terrestrial vertebrates (Hwang and Chou, 2013). Many kidney ion transporters are also highly expressed in the mammalian inner ear where they regulate the ionic composition of the endolymph (Lang et al., 2007). Consequently, mutations in a H+-ATPase subunit are both associated with distal renal tubular acidosis and progressive sensorineural hearing loss in humans and mouse models (Karet et al., 1999; Lorente-Canovas et al., 2013; Norgett et al., 2012). Similarly, mutations in Pendrin, a HCO3−/Cl− transporter, are associated with hearing loss caused by an acidification of the endolymph (Everett et al., 1997; Nakaya et al., 2007; Wangemann et al., 2007).
In the teleost ear, ionocytes likely regulate the ionic composition of the endolymph (Becerra and Anadon, 1993; Mayer-Gostan et al., 1997). Possible homologs in non-teleost vertebrates include dark cells in the vestibular system and marginal cells of the stria vascularis in the cochlea (Ciuman, 2009; Köppl et al., 2018). Vestibular dark cells are adjacent to light cells, which were proposed to be homologous to ionocyte associate cells (Becerra and Anadon, 1993; Kimura, 1969; Köppl et al., 2018). In contrast to dark cells, light cells lack apical microvilli and send out membrane interdigitations to neighboring cells (Harada et al., 1989; Köppl et al., 2018). While microvilli-containing Nm ionocytes and their accessory cells appear morphologically similar to vestibular dark and light cells, respectively, comparative molecular and functional analysis of these cell types will be necessary to test homology with Nm ionocytes.
Nm ionocyte function
Mutations that affect the development (merovingian) or function (persephone) of HR ionocytes result in mechanotransduction defects (Hailey et al., 2012; Lin et al., 2019; Stawicki et al., 2014). It was proposed that loss of HR skin ionocytes in merovingian/gcm2 mutants leads to a global acidification of the extracellular environment that reduces hair cell mechanotransduction (Horng et al., 2007; Horng et al., 2009; Stawicki et al., 2014). Our discovery of Nm ionocytes that express multiple HR ionocyte genes and respond to acidic environmental conditions provides evidence that they play a role in acid/base regulation in the lateral line. Indeed, the loss of Nm ionocytes in foxi3a mutants correlates with a decrease in mechanotransduction-dependent uptake of FM1–43 (Fig. 5P and 5Q). We propose that Nm ionocytes, located immediately adjacent to hair cells and in direct contact with the hair cell microenvironment, maintain hair cell function throughout different environmental conditions.
Nm ionocyte frequency is regulated by environmental changes and grants plasticity to the system
We have demonstrated that Nm ionocytes are recruited into neuromasts in a salinity- and pH-dependent manner, and we propose this is an adaptive trait to allow for tight control of hair cell microenvironment. This mechanism ensures sensory hair cell function and acclimation in a wider range of environmental conditions. Acclimation is the process of facultative phenotypic changes in response to environmental cues that is reversible and repeatable during the organism’s lifetime (Beaman et al., 2016). Tissue cell number has been observed to be environmentally regulated in the intestine of pythons during cycles of feeding and fasting or in murine adult stem cell compartments during long-term dietary interventions, low temperatures or light/dark cycles (Andrew et al., 2017; Casanova-Acebes et al., 2013; Ma et al., 2020; Mendez-Ferrer et al., 2008; Perry et al., 2019; Shwartz et al., 2020). Acclimation in the case of Nm ionocytes overcomes the need to make genetic changes to control their number and confer higher plasticity within a generation. Plasticity of ionocyte numbers is not restricted to Nm ionocytes and has been observed for most ionocyte subtypes (Sardet et al., 1979; Varsamos et al., 2005). The ability of freshwater fish to respond to osmotic stress correlates with the capacity to modulate their ionocyte composition (Varsamos et al., 2005). It would be interesting to investigate whether euryhaline fish have more pronounced plasticity in their Nm ionocyte frequency compared to stenohaline fish.
ACI allows animals to cope with far from optimal environmental conditions. Even though this particular adaptation occurs within an individual in response to a narrow range of stimuli, it increases the range of environments in which the animal has high fitness. This extension of the geographical distribution that fish can inhabit provides them with a selective advantage and is ultimately a driver of evolution.
Limitations of the Study
Here we report the discovery and characterization of Nm ionocytes. These cells invade mature neuromasts and this invasion is associated with an upregulation of Notch signaling. We identified through scRNAseq notch1b as the main receptor expressed in one Nm ionocyte. While our scRNAseq experimental strategy allowed us to bioinformatically pinpoint the Notch+ cell, we could not identify the Notch− Nm ionocyte. Thus, we were not able to determine the source nor the identity of the Notch ligand in this study and future work will shed light on these features.
Our data also demonstrates that foxi3a mutants have reduced FM1–43 dye uptake, suggesting decreased hair cell mechanotransduction. However, foxi3a is a master regulator of skin ionocyte differentiation (Hsiao et al., 2007; Jänicke et al., 2007), and the distribution of skin ionocytes is also altered in these mutants. It is therefore possible that the hair cell phenotype we observed in foxi3a mutants is caused by a global loss of ion homeostasis. Currently, we lack Nm ionocyte-specific markers that would allow Nm ionocyte specific manipulations and a more precise read-out of their role in the lateral line. Future work will focus on identifying specific markers to allow in-depth molecular characterization of both cells of the Nm ionocyte pair and their function.
The above limitations are discussed in the manuscript.
Conclusions
Even though lateral line organs have been studied since the 19th century, Nm ionocytes have eluded discovery. These newly described cells reported here are skin-derived and capable of invading mature sensory organs in response to environmental stimuli. In vivo time lapse imaging allowed us to characterize the migration of Nm ionocyte precursor cells from the skin into the lateral line. We also observed and quantified the morphological changes these cells display during this highly dynamic process with high resolution, providing a detailed outline of the development of these newly discovered cells. The comprehensive molecular, ultrastructural and functional characterization of Nm ionocytes provides support for the hypothesis that they are involved in maintaining an ionic microenvironment that ensures robust hair cell mechanotransduction. Therefore, the discovery of ACI and Nm ionocytes not only underscores important roles for post-embryonic migration events in regulating the function of vertebrate mechanosensory organs, but also indicates that similar mechanisms may be associated with pathologies afflicting the inner ear.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tatjana Piotrowski (pio@stowers.org).
Materials Availability
Fish lines generated in this study are available from the lead contact.
Data and Code Availability
Original data used for the results reported in this paper may be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1543_2020. The datasets and codes generated and used during this study are available at GEO: GSE168695 and GitHub https://github.com/Piotrowski-Lab/scRNAseq_Nm_ionocytes_analysis_Seuratv3_Peloggia_Muench_et_al_2020.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish lines and husbandry
All experiments were performed following the guidelines from the Stowers Institute IACUC review board. Embryos were raised in 0.5x E2 Medium unless stated otherwise (7.5 mM NaCl, 0.25 mM KCl, 0.5 mM MgSO4, 75 μM KH2PO4, 25 μM Na2HPO4, 0.5 mM CaCl2, 0.5 mg/L NaHCO3, pH = 7.4). The published transgenic lines used were: Tg(pou4f3:GAP-GFP)s356t (Xiao et al., 2005), Tg(−8.0cldnb:lynEGFP)zf106Tg (Haas and Gilmour, 2006), Tg(tp1bglobin:EGFP)um13, Tg(tp1bglobin:hmgb1-mCherry)jh11 (Parsons et al., 2009), ET(krt4:EGFP)SqGw57a (Kondrychyn et al., 2011), Tg(ubi:creERt2)cz1702Tg (Mosimann et al., 2011), Tg(ubi:Zebrabow)a131Tg (Pan et al., 2013), TgBAC(cxcr4b:h2az2a-GFP,cryaa:DsRed)sk84Tg (Kozlovskaja-Gumbriene et al., 2017), Tg(krtt1c19e:lyn-Tomato)sq16Tg (Lee et al., 2014), Tg(−8.0cldnb:H2A-mCherry)psi4Tg (Romero-Carvajal et al., 2015), Tg(krtt1c19e:H2A-mCherry).
For generation of new transgenic lines, fusions of gene and fluorescent protein of interest were cloned into middle entry vectors (pME-MCS) using Gibson assembly (Gibson et al., 2009) and recombined with targeted enhancers (cloned into p5’-Entry) using the tol2 transgenesis kit (Kwan et al., 2007). The following transgenic lines were made Tg(ubi:H2-AmCherry)psi18Tg, Tg(she:H2B-EGFP)psi59Tg, Tg(krtt1c19e:H2B-EGFP)psi63Tg, Tg(she:H2A-mCherry)psi57Tg, Tg(tp1bglobin:lox-mKate2-lox-SpvB)psi64Tg, Tg(krtt1c19e:cre-MYC)psi65Tg, Tg(Myo6:H2B-mScarlet-I)psi66Tg, Tg(Myo6b:Lck-mScarlet-I)psi67Tg and foxi3apsi68.
Generation of foxi3apsi68 mutants
CRISPR/Cas12a-mediated gene editing was performed to delete the foxi3a promoter and a part of the first exon (Moreno-Mateos et al., 2017). Two guide RNAs (gRNAs) targeting sequences 1377 bp upstream and 143 bp downstream of the start codon, respectively, were designed using DeepCpf1 (Luo et al., 2019) and checked for specificity and off-target effects by zebrafish genome BLAST (Altschul et al., 1990) and CRISPRscan (Moreno-Mateos et al., 2017): gRNA 1: 5’-ATATGACAACCTACCGCAGA-3’; gRNA2: 5’-GGAGAATACGCAGGGCAGAC-3’.
Tg(tp1bglobin:EGFP)um13 zebrafish one cell stage embryos were injected with 22.2 μM EnGen® Lba Cas12a (Cpf1) (New England Biolabs) and both gRNAs at a concentration of 13.8 mM each. All GFP+ larvae were raised to adulthood and germline-transmitting founders harboring the promoter deletion were identified by genotyping PCR with the following three primers: foxi3a-F: 5- CGATCAGAAAACGCCTGCAGACTGA-3’, foxi3a-R: 5’-GGGAGGTCTCACGAGTTTCATCAGATC-3’, foxi3a-R2: 5’-GCAACGAATGGAATCAGAATGTACAGTGC-3’. One F0 founder was outcrossed to wildtype fish to generate the F1 generation. F1 fish were raised to adulthood and heterozygous carriers of the deletion were identified by genotyping PCR. Multiple pairs of heterozygous F1 fish were incrossed and F2 larvae were analyzed for all experiments.
METHOD DETAILS
In situ hybridization and Hybridization Chain Reaction
Whole mount in situ hybridization was performed according to http://research.stowers.org/piotrowskilab/in-situ-protocol.html. Probes were cloned and synthesized with the following primer pairs: foxi3b-F: TGACAATGGCAACTTCAGACGC, foxi3b-R: GCCCACAGAACTCAAAGGAG, slc12a10.2-F: CTGAAGCCAAACACACTTGTTTTGG, slc12a10.2-R: CTACATCTGCAGTAATACAATCATTGC, CR936482.1-F: AAACTTCAACTCTTCAAGAC, CR936482.1-R: ATCAAACGAGCTCCTTTATTC, trpv6-F: GGATTTTCTACATGACTCAGGAACC, trpv6-R: CAGTGATGGTGCACACCCTATATATTTG, ca2-F: CAGTTCTCTGGACTACTGGACATACC, ca2-R: GCATGCCTTCTATAGAATGTACGTTCAG. Reverse probes had a T3 polymerase sequence attached 5’ to allow for in vitro synthesis. PCR reaction was conducted with Phusion High Fidelity DNA polymerase (NEB, USA) and the product was purified by gel extraction according to manufacturer’s protocol (Monarch® DNA Gel Extraction Kit). In vitro transcription was carried out with T3 RNA Polymerase and DIG-labeling kit (Roche) for 2h at 37°C, followed by 30 min of DNaseI treatment (Roche). RNA was purified with a LiCl Precipitation Solution (7.5 M) (AM9480, Invitrogen) and stored at −80°C until use.
Hybridization chain reaction (HCR) was performed according to manufacturer’s instructions for notch1b-B2, trpv6-B2, gcm2-B4, foxi3a-B1, krtt1c19e-B1 (Choi et al., 2018) (Molecular Instruments). The amplifiers used were B1–647, B2–546 and B4–488 (Molecular Instruments). HCR fish were subsequently stained with DAPI (5ug/mL) for 30 min at room temperature (RT) in the dark and washed three times with 5x SSCT before imaging.
Immunohistochemistry
Larvae were anesthetized and fixed with 4% PFA overnight at 4°C. Standard antibody staining was performed. Antibodies used were: tp63 (1:100, Santa Cruz, sc-8431), Anti-Notch1 intracellular domain rabbit polyclonal (1:100, Abcam, AB_306525), Monoclonal a5 ATPase, (Na (+) K(+)) alpha subunit (5 ug/mL, Developmental Studies Hybridoma Bank, AB_2166869), and rabbit anti-GFP (1:500, Invitrogen, A11122). Antibody staining was counterstained with DAPI (5ug/mL) for 30 min at RT in the dark and washed three times with PBST before imaging.
Time lapse and confocal imaging
Images were acquired using the confocal microscopes Zeiss LSM700, Zeiss LSM780 or Nikon Ti Eclipse with Yokogawa CSU-W1 spinning disk head equipped with a Hamamatsu Flash 4.0 sCMOS. Objective lenses used were Zeiss Plan-Apochromat 10x/0.45 M27 (air), Plan-Apochromat 20x/0.8 M27 (air) and LD C-Apochromat 40x/1.1 W Korr M27 (water) for the Zeiss Microscopes and a Nikon Plan Apo 40× 1.15NA LWD (water). Temperature was kept constant at 28.5°C using Zeiss 780 standard incubation or a Stage Top Chamber (OkoLab) for the Nikon Microscope.
For live imaging experiments, larvae were immobilized with tricaine (MS-222) up to 150 mg/L and mounted in glass bottom dishes (MatTek) with 0.8% low melting point agarose dissolved in 0.5x E2 or 5x E2 with tricaine (100 mg/L). Time lapse recordings during Notch inhibition were performed after a 30-minute pre-incubation period with LY411575 (50uM). Larvae were exposed to the same concentration throughout the experiment.
Laser lines used on the Zeiss confocal were Diode 405–30, Argon multiline laser (458, 488 and 514 nm), DPSS 561–10, HeNe 594 and 633 nm. A Nikon LUNV solid state laser launch was used for lasers 445, 515, and 561nm for CFP, YFP, and RFP respectively. Emission filters used on the Nikon were 480/30, 535/30, 605/70.
All image acquisition was performed using Zen 2012 SP5 Black (Zeiss) and Nikon Elements AR 4.6 (Nikon) software.
Electron Microscopy
Scanning Electron Microscopy
Zebrafish larvae (5 dpf) raised in 0.5x E2 were anesthetized and Nm ionocytes were mapped using a Zeiss LSM780 with LD C-Apochromat 40x/1.1 W Korr M27 (water) objective. Individually mapped larvae were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in PBS for 1 hour at room temperature on a rocker and stored at 4°C until processing. Secondary fixation and staining with tannic acid, osmium, thiocarbohydrazide, and osmium were carried out as in (Jongebloed et al., 1999). Samples were dehydrated in a graded ethanol series, critical point dried in a Tousimis Samdri 795 critical point dryer, mounted on stubs, and imaged in a Hitachi TM4000 table top SEM at 10 kV.
Serial Block Face Scanning Electron Microscopy
Individual 5 dpf zebrafish larvae were analyzed for the presence of Nm ionocytes using the Notch reporter and then fixed in 2.5% glutaraldehyde, 2% paraformaldehyde, 1mM CaCl2, and 1% sucrose in 50 mM sodium cacodylate buffer for one hour at room temperature on a rocker, and processed for serial block face imaging in one of two ways: 1. (Fig. 4B) As in Dow et al., 2018, except that Hard Plus resin (Electron Microscopy Sciences) was used in place of Durcupan, or 2. (Fig. 4C to 4F) With en bloc staining steps per (Genoud et al., 2018; Hua et al., 2015; Tapia et al., 2012), modified as follows and processed on the ASP-1000 automated sample processing robot (Microscopy Innovations): Buffered reduced osmium with formamide incubation was performed at room temperature for 4 hours, TCH incubation at 60 C for 45 minutes, incubation in 1% UA at room temperature for 4 hours plus an additional 2 hours at 60C, and lead acetate incubation for 2 hours at 60C. Samples were dehydrated in a graded series of acetone with 3– 100% exchanges for 15 minutes each step, and infiltrated with Hard Plus resin at 25% 50% and 75% for 30 minutes each and 3– 100% exchanges for 2 hours each. All samples were polymerized in either flat embed molds (Electron Microscopy Sciences) or using the minimal resin embedding technique as in (Schieber et al., 2017). Once polymerized, samples were mounted on aluminum pins with silver conductive epoxy (Ted Pella, Inc). The minimal resin embedded sample was imaged in the Hitachi TM4000 SEM to verify the neuromast of interest for correlation with fluorescent imaging.
Neuromast image volumes were acquired on a Zeiss Merlin with Gatan 3View 2XP at either 1.5 or 2 kV, with slice thickness either 50nm or 80 nm, 10 nm pixel size and image acquisition area between of 55 μm or 65 μm. Charging was mitigated using focal charge compensation (Zeiss) with nitrogen set between 20% and 45%.
Tracing and Modeling
Tracing and modeling were performed in IMOD 4.9 (Kremer et al., 1996). Final models were exported as wavefront objects and imported into Blender 2.83.4. Shade smooth was added to the objects and rendering was performed using the Blender Cycles Render at 29.97 frames per second. Intel AI Denoiser built-in on Blender was used for animation post-processing and denoising.
Single Cell RNA sequencing
Embryo dissociation and FACS
A total of 600 larvae 5 dpf Tg(−8.0cldnb:lynEGFP);Tg(tp1bglobin:hmgb1-mCherry)jh11 were screened for double positive fluorescence. 300 embryos were dissociated in 4.5 mL of 0.25% trypsin-EDTA (Thermo Fisher Scientific, Waltham, MA. USA) by pipetting them for 2.5 min on ice with 1uM Actinomycin D (Sigma A1410) to inhibit new transcription. Cells were stained with DAPI (5 μg/mL) and Draq5 (1:1000) (Thermo) and sorted for DAPI-negative and Draq5-positive cells for viability control. Cells were sorted into 90% MeOH for fixation. Cell sorting was performed by the Cytometry Facility of the Stowers Institute for Medical Research using the BD Influx Cell Sorter (BD Biosciences, San Jose, CA. USA).
10X Chromium scRNA-seq library construction
MeOH-fixed cells were rehydrated with rehydration buffer (0.5% BSA and 0.5 U/μl RNase-inhibitor in ice-cold DPBS) following manufacturer’s instructions (10X Genomics). Approximately 15.000 cells were loaded into the Chromium Single Cell Controller (10x Genomics). For library preparation, Chromium Next GEM Single Cell 3’ GEM, Library & Gel Bead Kit v3.1 was used. The sample concentration was measured on a Bioanalyzer (Agilent) and sequenced with NextSeq 500 High Output Kit v3 with read length of 28 bp Read 1, 8 bp i7 index and 91 bp Read 2 (150 cycles) (Illumina).
Read Alignment and Quantification and Quality Control
Reads were de-multiplexed and aligned to version Ensembl GRCz11 of the zebrafish genome using the CellRanger (v3.0.0) pipeline. Following analysis of UMI count matrix was performed using Seurat (version 3.1.2) (Butler et al., 2018; Stuart et al., 2019). Initial quality control filtered out all genes expressed in less than three cells, less than 250 genes, more than 30% mitochondrial genes and 30.000 UMIs. Standard Seurat v3.1.2 workflow was performed, following the steps from the pbmc3k online tutorial (https://satijalab.org/seurat/v3.2/pbmc3k_tutorial.html). Dimensional reduction, resolution and the final UMAP were then validated by comparing their biomarkers, differentially expressed genes obtained with the function FindAllMarkers(), to known markers based on literature. Further cluster subset was performed for all detected ionocytes and skin cells (defined as tp63+).
Acclimation of zebrafish larvae to different pH and salinity environments
For the acclimation experiments, 0.1x, 0.5x, 2.5x, 5x E2 dilutions of E2 media were prepared with distilled water from a 20x E2 stock solution (17.5g/L NaCl, 0.75g KCl, 4.9g/L MgSO4•7H2O, 0.41g/L KH2PO4, 0.142g/L Na2HPO4, 2.9g/L CaCl2•2H2O, pH = 6.0–6.1). To prepare pH 4, 7, and 10 media, 0.5x E2 medium was supplemented with 300 μM MES (Chem-Impex), 300 μM MOPS (Sigma), or 300 μM tricine (Sigma), respectively (modified from (Horng et al., 2009). In all experiments, larvae were first raised in regular 0.5x E2 medium to 3 dpf. Between 3 and 5 dpf, larvae were then incubated in diluted E2 media or media with different pH for 48 h. Embryo medium was exchanged daily to ensure stable pH or salinity. No significant differences in larvae survival were observed between the different conditions.
MilliQ water parameters are conductivity of 1.6 μS/cm, salinity of 0.0 ppm, alkalinity of 17.5 ppm (as CaCO3) and general hardness of 7.5 ppm (as CaCO3) and a pH of 7.6. Conductivity and salinity were measured with YSI 30 (YSI incorporated), and alkalinity, general hardness and pH were measure with SpinTouch FF104 (LaMotte).
Quantification of Nm ionocyte frequency
To quantify Nm ionocyte frequency based on the absence Et(Gw57a:EGFP)/Tg(pou4f3:GFP) fluorescence, 5 dpf larvae were embedded in 0.8 % low melting point agarose and gaps in Et(Gw57a) fluorescence were counted using a Zeiss LSM780 confocal. A gap in the fluorescence in the neuromast poles that was fully separated from other fluorescence gaps was considered to be one Nm ionocyte, regardless of the number of cells within the gap, and all of the possible scenarios were considered for further analysis. A total number of 14–16 neuromasts of the anterior lateral line (Ml1, O2, Ml2, IO4, O1; for nomenclature see (Chou et al., 2017), as well as the posterior lateral line (L1-L9, LII.1, LII.2; for nomenclature see (Ghysen and Dambly-Chaudiere, 2007), were quantified per larva, unless otherwise noted.
To quantify Nm ionocyte frequency based on the Notch-reporter (tp1bglob:EGFP) fluorescence, larvae were anesthetized with 150 mg/L tricaine (MS-222) and juvenile and adults were killed using cold-shock protocol. The fish were embedded in 0.8% low melting point agarose and Notch+ cells were counted on a confocal microscope. All cells in the neuromast polar compartments showing strong labeling by the Notch reporter and/or displaying pronounced apical protrusions were included in the analysis. Nm on different body parts were imaged.
Sensory hair cell ablation
To ablate mechanosensory hair cells, 5 dpf tp1bglob:EGFP larvae were incubated in 300 μM neomycin (Sigma-Aldrich) for 30 min (Venero Galanternik et al., 2016). After the antibiotic was washed out, larvae were allowed to recover in 0.5x E2 for 24 h and Nm ionocyte number was quantified based on Notch reporter fluorescence as described above.
Proliferation analysis
5 dpf tp1bglob:EGFP;Et20 larvae were incubated in 3.3 mM EdU (Carbosynth) in 1% DMSO in E2 medium for 20h following Neomycin-induced sensory hair cell ablation (see above). EdU staining, anti-GFP immunohistochemistry and DAPI counterstaining were performed as previously described (Lush et al., 2019).
Manipulation of Notch signaling
Notch signaling was inhibited by incubating larvae in 50 μM of the gamma secretase inhibitor LY411575 (Selleckchem) with 0.25% DMSO in 0.5x E2 media for 24 h. Control larvae were incubated with 0.25% DMSO in 0.5x E2 media only. Nm ionocyte frequency was quantified based on gaps in Et(Gw57a:EGFP)/Tg(pou4f3:GFP) fluorescence as described above. Three neuromasts of both the anterior lateral line (Ml1, O2, Ml2) and the posterior lateral line (L1, L2, L3) were quantified per larva.
FM1–43 dye uptake
To analyze mechanotransduction-dependent uptake of FM1–43FX by hair cells, individual 5 dpf larvae were incubated with 2.25 μM FM1–43FX (Invitrogen, F35355, diluted in 0.5x E2 medium) for 1 min, washed two times with 0.5x E2 containing 200 mg/mL MS-222 for 15s and embedded in a small drop of 0.8% low-melting point agarose supplemented with 140 mg/mL on a glass bottom dish. After letting the agarose solidify for 1 min, the L2 and L3 neuromasts of each larva were imaged as a z-stack of 25 sections (1 μm Z step).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
All statistical tests were performed using GraphPad Prism 8 (version 8.4.3) as indicated in the figure legends. Normal distribution was assessed using Kolmogorov-Smirnov test. When comparing data from more than two groups, statistical significance was calculated using either one-way ANOVA with Tukey’s post hoc test or non-parametric Kruskal-Wallis ANOVA with Dunn’s post hoc test. Data from two groups were compared using two-tailed unpaired t-test or two-tailed Mann–Whitney U test. Binomial analysis was performed in R (Lund et al., 2017; Team, 2013). p-values smaller than 0.05 were considered to be statistically significant. Statistical details of experiments, such as p-values, sample size and dispersion measures are individually specified for each experiment in the figure legends. Plots were made in Microsoft Excel (version 16.39), GraphPad Prism 8 and the R package ggplot2 (Wickham, 2011).
Image Analysis
All image analyses were performed in Fiji (Schindelin et al., 2012) and 3D renderings in Imaris (Bitplane).
Nonlinear operations (gamma)
The she (lateral line) and tp1bglobin (Notch reporter) enhancers drive expression at different ranges in different cells. Therefore, non-linear changes (gamma) were applied to make all features visible. Gamma ranged from 0.5 to 0.8 and was applied whenever stated in figure legends. Image quantification was always performed with the raw file and never with gamma-modified images.
Nm ionocyte position quantification
Cell positions for mantle cells and Notch+ Nm ionocytes were quantified in Fiji or Imaris software, and calculations were performed as previously described (Romero-Carvajal et al., 2015; Venero Galanternik et al., 2016).
Linear unmixing
For proper separations of ubi:Zebrabow and mKate2 fluorescent proteins, linear unmixing was performed. Fluorescent proteins were excited by using 458 laser line for CFP, 514 for YFP and RFP and 594 for mKate2. Z stacks were acquired on a Zeiss 780 confocal set on lambda mode and 8.9nm resolution. Images were unmixed using a collected reference spectrum for mKate2 and by manually detecting CFP, YFP and tdTomato spectra on Zen Black (Zeiss) based on FPBase reference spectra (Lambert, 2019).
Tracking of individual cells of migrating Nm ionocyte cell pairs
To visualize the path of both cells of the migrating Nm ionocyte precursor pair over time, confocal time lapse recordings were analyzed using the mTrackJ plugin (Meijering et al., 2012) in ImageJ and cell nuclei tracked through individual z-planes. Tracks were then visualized on maximum projections of the time lapse recording using the “Movie” function in mTrackJ. The distance to the start point of each track (D2S) was obtained using the “Measure” function in the mTrackJ plugin.
Quantification of migrating Nm ionocyte precursor speed
To quantify migrating Nm ionocyte precursor speed, z-stacks of confocal time lapse recordings were maximum projected and corrected for tissue drift using the StackRegJ plugin (Thevenaz et al., 1998). The nucleus of the Notch+ cell was then tracked based on cldnB:H2A-Cherry or ubi:H2A-Cherry signal and using tp1bglob:EGFP fluorescence as an additional reference. The summed distance of the entire track, as well as the net distance between the first and the last tracking point were calculated using the “Measure” function of the mTrackJ plugin (final track length and final D2S, respectively) and the speed calculated by dividing the distance by the elapsed time.
Quantification of FM1–43 fluorescence intensity
To quantify FM1–43 dye uptake by hair cells, maximum intensity projections of 12 z-planes of the acquired z-stacks were generated, starting just below the cuticular plate and encompassing the entire cell body of the hair cells. A mask was then drawn around the labeled hair cells and the average fluorescence intensity calculated using the ‘Measure’ function. Four identical masks were drawn in multiple regions within close vicinity of the hair cells to calculate the average background intensity. Data is shown as hair cell fluorescence intensity/background, normalized to the average of the siblings and shown as percentage.
Supplementary Material
Supplementary Table 1 - Cluster markers of all obtained clusters, related to Figure 1 and S1: Differentially expressed genes per cluster (cluster markers) obtained for all sequenced cells after quality controls.
Supplementary Table 2 - Cluster markers of skin and ionocyte subsets, related to Figure 1: Differentially expressed genes for the ionocyte and skin subset.
Supplementary Video 1 - 3D rendering of neuromast (confocal), related to Figure 1: 3D animation of a confocal z-stack of a 4 dpf tp1bglobin:EGFP larva, showing a Notch+ non-lateral line cell, as well as central support cells labeled by this reporter line. Scale bar equals 5 μm.
Supplementary Video 2 - Pair of cells migrate into the neuromast, related to Figure 3: Neuromasts in a 3 dpf fish labeled with Et4;Et20;cldnb-lynEGFP shows a pair of cells entering on the dorsal side. Scale bar equals 10 μm.
Supplementary Video 3 - Pair of cells migrate into the neuromast, related to Figure 3: Confocal time lapse recording of a Gw57a:EGFP;ubi:H2A-mCherry larva at 3 dpf showing Nm ionocyte precursor cells migrating into the neuromast as visualized by translocation of the two cell nuclei, as well as the appearance of a gap in Gw57a:EGFP fluorescence. Individual z-slices of a confocal z-stack are shown, intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 4 - Pairs of cells migrate extensively inside the neuromast, related to Figure 3: Maximum intensity projection of confocal time lapse recording of a primII-derived neuromast of a cldnb:H2A-mCherry;tp1bglobin:EGFP larva at 2 dpf, showing two pairs of Nm ionocyte precursors migrating into the organ. The nuclei of the cells were tracked on 3D based on cldnB:H2A-mCherry fluorescence. Future Notch+ cells (based on Notch reporter fluorescence) are labeled with cyan dots, Notch− cells are labeled with red dots. Maximum intensity projection of a confocal z-stack is shown. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 5 - Nm ionocyte develops apical extension, related to Figure 3: Confocal time lapse recording of a neuromast of a tp1bglobin:EGFP transgenic at 3 dpf depicting a Notch+ Nm ionocyte precursor in the ventral pole extending multiple protrusions before developing and stabilizing an apical extension. Maximum intensity projection of a confocal z-stack is shown. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 6 - Notch signaling increases in one Nm ionocytes in a pair, related to Figure 3: Confocal time lapse recording of a neuromast of a cldnb:H2A-mCherry;tp1bglobin:EGFP transgenic at 3 dpf, depicting a pair of Nm ionocyte precursors re-arranging in the organ, with one cell gradually increasing in Notch reporter fluorescence. The nuclei of the cells were tracked based on cldnb:H2A-mCherry fluorescence. Notch+ cells are labeled with cyan dots, Notch− cells are labeled with red dots. An individual z-slice of a confocal z-stack is shown. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 7 - Notch signaling increases in one the Nm ionocytes in a pair, related to Figure 3: ubi:H2A-mCherry;tp1bglobin:EGFP double transgenic shows two pairs of cells entering the neuromast simultaneously while increasing EGFP fluorescence, indicating activation of Notch signaling. Only the Notch+ cell is visible. Scale bar equals 10um.
Supplementary Video 8 – Notch inhibition leads to Nm ionocyte death, related to Figure 3: Gw57a:EGFP; ubi:H2A-mCherry 4 dpf double transgenic shows that both Nm ionocytes of two different pairs die after Notch signaling inhibition with LY411575. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 9 – Notch+ cells frequently die first after Notch inhibition, related to Figure 3 and S3: time lapse of a 3 dpf ubi:H2A-mCherry;tp1bglobin:EGFP shows Notch+ cell death followed by death of an adjacent cell after incubation with the Notch inhibitor LY411575. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 10 - 3D rendering of Nm ionocyte pair (EM), related to Figure 4: Rendering of a 3D reconstruction of a Nm ionocyte pair in the neuromast.
Supplementary Video 11 - 3D rendering of Nm ionocytes and hair cells (confocal), Related to Figure 4: 3D animation of a confocal z-stack of a tp1bglobin:EGFP;myo6b:Lck-mScarlet-I neuromast at 5 dpf, depicting the close association of Notch+ Nm ionocytes and hair cells and the formation of the apical crypt in close proximity to the hair cell bundles.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ΔNp63 (4A4) | Santa Cruz | sc-8431 |
| Rabbit polyclonal anti-Notch1 intracellular domain | Abcam | AB_306525 |
| Mouse monoclonal anti-ATPase, (Na(+) K(+)) alpha subunit, supernatant | Developmental Studies Hybridoma Bank | a5 |
| Rabbit polyclonal anti-GFP | Thermo Fisher Scientific | A11122 |
| Anti-Digoxigenin, Fab fragments | Roche | 11214667001 |
| Alexa Fluor 488 goat anti-rabbit IgG (H+L), Superclonal™ Recombinant | Thermo Fisher Scientific | A27034 |
| Alexa Fluor 594 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | A11005 |
| Alexa Fluor 568 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | A11004 |
| Bacterial and Virus Strains | ||
| E. coli DH5α | New England Biolabs | C2987I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LY411575 | Selleckchem | S2714 |
| EnGen Lba Cas12a (Cpf1) | NEB | M0653T |
| FM1-43FX | Thermo Fisher Scientific | F35355 |
| Neomycin sulfate | Sigma | N6386 |
| EdU | Carbosynth | NE08701 |
| Alexa Fluor-594 Azide | Thermo Fisher Scientific | A10270 |
| DIG labeling mix | Roche | 11277073910 |
| RNAse inhibitor | Roche | 03335402001 |
| DNase I | New England Biolabs | M0303S |
| Proteinase K | Roche | 03115828001 |
| NBT (4-Nitro blue tetrazolium chloride) | Roche | 11383213001 |
| BCIP (5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt) | Roche | 11383221001 |
| Transcription optimized 5x buffer | Promega | P1181 |
| DTT (100mM) | Promega | P1171 |
| T3 RNA polymerase | Promega | P2083 |
| DAPI | Invitrogen | D1306 |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | 25200056 |
| Actinomycin D | Sigma | A1410 |
| DRAQ5™ Fluorescent Probe Solution (5 mM) | Thermo Fisher Scientific | 62251 |
| Critical Commercial Assays | ||
| Hybridization Chain Reaction (HCR) v3.0 | Molecular Instruments | N/A |
| Chromium Next GEM Single Cell 3’ GEM, Library & Gel Bead Kit v3.1 | 10x Genomics | 120267 |
| Deposited Data | ||
| Sequencing data | This paper | GEO accession number GSE168695 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: TU-wildtype | N/A | |
| Tg(pou4f3:GAP-GFP)s356t | Xiao et al., 2005 | s356t; RRID: ZFIN_ZDB-GENO-100820-2 |
| Tg(-8.0cldnb:lynEGFP)zf106Tg | Haas and Gilmour, 2006 | zf106Tg; RRID: ZFIN_ZDB-ALT-060919-2 |
| Tg(tp1bglobin:EGFP)um13Tg | Parsons et al., 2009 | um13; RRID: ZDB-GENO-101201-5 |
| Tg(tp1bglobin:hmgb1-mCherry)jh11Tg | Parsons et al., 2009 | jh11Tg; RRID: ZDB-ALT-101006-1 |
| ET(krt4:EGFP)SqGw57a | Kondrychyn et al., 2011 | SqGw57a |
| Tg(ubi:creERt2)cz1702Tg | Mosimann et al., 2011 | cz1702Tg; RRID: ZDB-GENO-110121-2 |
| Tg(ubi:Zebrabow)a131Tg | Pan et al., 2013 | a131Tg; RRID: ZDB-ALT-130816-2 |
| TgBAC(cxcr4b:h2az2a-GFP,cryaa:DsRed) | Kozlovskaja-Gumbrienė et al., 2017 | sk84Tg; RRID: ZDB-TGCONSTRCT-180201-1 |
| Tg(krtt1c19e:lyn-Tomato)sq16Tg | Lee et al., 2014 | sq16Tg; RRID: ZDB-GENO-140424-5 |
| Tg(-8.0cldnb:H2A-mCherry)psi4Tg | Romero-Carvajal et al., 2015 | psi4Tg; RRID: ZDB-ALT-140623-3 |
| Tg(krtt1c19e:H2A-mCherry)pd309Tg | Han et al., 2019 | pd309Tg |
| Et(krt4:EGFP)sqet20ET | Parinov et al., 2004 | sqet20ET; RRID: ZFIN_ZDB-ALT-070628-20 |
| Tg(ubi:H2A-mCherry)psi18Tg | This paper | N/A |
| Tg(she:H2B-EGFP)psi59Tg | This paper | N/A |
| Tg(krtt1c19e:H2B-EGFP)psi63Tg | This paper | N/A |
| Tg(she:H2A-mCherry)psi57Tg | This paper | N/A |
| Tg(tp1bglobin:lox-mKate2-lox-SpvB)psi64Tg | This paper | N/A |
| Tg(krtt1c19e:cre-MYC)psi65Tg | This paper | N/A |
| Tg(Myo6:H2B-mScarlet-I)psi66Tg | This paper | N/A |
| Tg(Myo6b:Lck-mScarlet-I)psi67Tg | This paper | N/A |
| foxi3apsi68 | This paper | N/A |
| Oligonucleotides | ||
| foxi3a gRNA 1: ATATGACAACCTACCGCAGA | IDT | N/A |
| foxi3a gRNA 2: GGAGAATACGCAGGGCAGAC | IDT | N/A |
| foxi3a F: CGATCAGAAAACGCCTGCAGACTGA | IDT | N/A |
| foxi3a R1: GGGAGGTCTCACGAGTTTCATCAGATC | IDT | N/A |
| foxi3a R2: GCAACGAATGGAATCAGAATGTACAGTGC | IDT | N/A |
| Recombinant DNA | ||
| pDEST-Myo6b:Lck-mScarlet-I | This paper | N/A |
| pDEST-tp1bglobin:lox-mKate2-lox-SpvB | This paper | N/A |
| pDEST-she:H2B-EGFP | This paper | N/A |
| pDEST-krtt1c19e:H2B-EGFP | This paper | N/A |
| pDEST-krtt1c19e:cre-MYC | This paper | N/A |
| pDEST-ubi:H2A-mCherry | This paper | N/A |
| pDEST-she:H2A-mCherry | This paper | N/A |
| pDEST-Myo6:H2B-mScarlet-I | This paper | N/A |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | https://fiji.sc/ |
| Imaris | Bitplane | http://www.bitplane.com/imaris |
| Microsoft Excel | Microsoft | |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| R version 4.0.2 | R Core Team | https://www.r-project.org/ |
| RStudio | RStudio | https://rstudio.com |
| ggplot2 (version: 3.2.1) | Wickham et al., 2016 | https://ggplot2.tidyverse.org/ |
| Cell Ranger 3.0.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat v3.1.2 | Butler et al., 2018; Stuart et al., 2019 | https://satijalab.org/seurat/ |
Highlights:
Mature lateral line sensory organs contain non-lateral line derived cells
Skin-derived Nm ionocyte pairs migrate and invade lateral line sensory organs
Nm ionocytes are recruited in response to environmental fluctuations
Mutants that lack ionocytes possess decreased hair cell mechanotransduction
Acknowledgments
We are grateful to Drs. A. Sánchez Alvarado, A. Schier and N. Denans for critical reading of the manuscript. We also thank Drs. A. Schier, T. Carney, K. Poss, and B. Slaughter for reagents and fish lines; the Stowers Institute Aquatics Facility for fish care; J. Blanck, K. Ferro, S. Baek, Y-Y. Tsai, M. Harwood, S. Nowotarski and A. Peak for experimental assistance; C. Chen, D. Diaz, H. Li and J. Unruh for help with analysis and M. Miller for scientific illustrations. This work was performed to fulfill, in part, requirements for J. Peloggia’s thesis research in the Graduate School of the Stowers Institute. The work was funded by an NIH (NICHD) award 1R01DC015488–01A1 to T.P and by institutional support from the Stowers Institute for Medical Research to T.P.
Footnotes
Declaration of Interests
The authors declare no conflict of interest.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas L, Hajihashemi S, Stead LF, Cooper GJ, Ware TL, Munsey TS, Whitfield TT, and White SJ (2011). Functional and developmental expression of a zebrafish Kir1.1 (ROMK) potassium channel homologue Kcnj1. J Physiol 589, 1489–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. Journal of molecular biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Aman A, and Piotrowski T (2011). Cell-cell signaling interactions coordinate multiple cell behaviors that drive morphogenesis of the lateral line. Cell Adh Migr 5, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AL, Perry BW, Card DC, Schield DR, Ruggiero RP, McGaugh SE, Choudhary A, Secor SM, and Castoe TA (2017). Growth and stress response mechanisms underlying post-feeding regenerative organ growth in the Burmese python. BMC Genomics 18, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker GE, de Grip WJ, Turton M, Wagner HJ, Foster RG, and Douglas RH (2015). Light sensitivity in a vertebrate mechanoreceptor? J Exp Biol 218, 2826–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow LA, and Klein OD (2015). Developing and regenerating a sense of taste, In Curr Top Dev Biol Vol 111, (Elsevier; ), pp 401–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaman JE, White CR, and Seebacher F (2016). Evolution of Plasticity: Mechanistic Link between Development and Reversible Acclimation. Trends in Ecology and Evolution 31, 237–249. [DOI] [PubMed] [Google Scholar]
- Becerra M, and Anadon R (1993). Fine structure and development of ionocyte areas in the labyrinth of the trout (Salmo trutta fario). Journal of Anatomy 183, 463–474. [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Acebes M, Pitaval C, Weiss LA, Nombela-Arrieta C, Chevre R, N ,AG, Kunisaki Y, Zhang D, van Rooijen N, Silberstein LE, et al. (2013). Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WJ, Horng JL, Yan JJ, Hsiao CD, and Hwang PP (2009). The transcription factor, glial cell missing 2, is involved in differentiation and functional regulation of H+-ATPase-rich cells in zebrafish (Danio rerio). American Journal of Physiology - Regulatory Integrative and Comparative Physiology 296, 1192–1201. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Hung JC, Wu LC, Hwang SPL, and Hwang PP (2011). Isotocin controls ion regulation through regulating ionocyte progenitor differentiation and proliferation. Cellular and Molecular Life Sciences 68, 2797–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SW, Chen Z, Zhu S, Davis RW, Hu J, Liu L, Fernando CA, Kindig K, Brown WC, Stepanyan R, et al. (2017). A molecular basis for water motion detection by the mechanosensory lateral line of zebrafish. Nat Commun 8, 2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuman RR (2009). Stria vascularis and vestibular dark cells: characterisation of main structures responsible for inner-ear homeostasis, and their pathophysiological relations. J Laryngol Otol 123, 151–162. [DOI] [PubMed] [Google Scholar]
- Cruz SA, Chao PL, and Hwang PP (2013a). Cortisol promotes differentiation of epidermal ionocytes through Foxi3 transcription factors in zebrafish (Danio rerio). Comparative Biochemistry and Physiology - A Molecular and Integrative Physiology 164, 249–257. [DOI] [PubMed] [Google Scholar]
- Cruz SA, Lin CH, Chao PL, and Hwang PP (2013b). Glucocorticoid receptor, but not mineralocorticoid receptor, mediates cortisol regulation of epidermal ionocyte development and ion transport in zebrafish (Danio rerio). PloS one 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Bai Y, Hebda L, Zhong X, Liu J, Kao J, and Duan C (2014). Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death and Differentiation 21, 568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DH, Piermarini PM, and Choe KP (2005). The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85, 97–177. [DOI] [PubMed] [Google Scholar]
- Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, Adawi F, Hazani E, Nassir E, Baxevanis AD, et al. (1997). Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat Genet 17, 411–422. [DOI] [PubMed] [Google Scholar]
- Flik G, and Perry SF (1989). Cortisol stimulates whole body calcium uptake and the branchial calcium pump in freshwater rainbow trout. J Endocrinol 120, 75–82. [DOI] [PubMed] [Google Scholar]
- Flock A (1967). Lateral line detectors. [Google Scholar]
- Flock A, and Jorgensen JM (1974). The ultrastructure of lateral line sense organs in the juvenile salamander Ambystoma mexicanum. Cell Tissue Res 152, 283–292. [DOI] [PubMed] [Google Scholar]
- Flock A, Jorgensen M, and Russell I (1973). In Basic mechanisms in hearing (New York and London: Academic Press; ). [Google Scholar]
- Gale JE, Marcotti W, Kennedy HJ, Kros CJ, and Richardson GP (2001). FM1–43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J Neurosci 21, 7013–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud C, Titze B, Graff-Meyer A, and Friedrich RW (2018). Fast Homogeneous En Bloc Staining of Large Tissue Samples for Volume Electron Microscopy. Front Neuroanat 12, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A, and Dambly-Chaudiere C (2007). The lateral line microcosmos. Genes Dev 21, 2118–2130. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, and Muller U (2009). Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell 139, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guh YJ, Lin CH, and Hwang PP (2015). Osmoregulation in zebrafish: Ion transport mechanisms and functional regulation. EXCLI Journal 14, 627–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas P, and Gilmour D (2006). Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell 10, 673–680. [DOI] [PubMed] [Google Scholar]
- Hailey DW, Roberts B, Owens KN, Stewart AK, Linbo T, Pujol R, Alper SL, Rubel EW, and Raible DW (2012). Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant persephone. PLoS Genetics 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Tagashira N, and Hirakawa K (1989). Morphological study of the cells in the planum semilunatum. Acta Oto-Laryngologica 108, 17–21. [DOI] [PubMed] [Google Scholar]
- Hootman SR, and Philpott CW (1980). Accessory cells in teleost branchial epithelium. Am J Physiol 238, R199–206. [DOI] [PubMed] [Google Scholar]
- Horng JL, Lin LY, Huang CJ, Katoh F, Kaneko T, and Hwang PP (2007). Knockdown of V-ATPase subunit A (atp6v1a) impairs acid secretion and ion balance in zebrafish (Danio rerio). American Journal of Physiology - Regulatory Integrative and Comparative Physiology 292, 2068–2076. [DOI] [PubMed] [Google Scholar]
- Horng JL, Lin LY, and Hwang PP (2009). Functional regulation of H+-ATPase-rich cells in zebrafish embryos acclimated to an acidic environment. American Journal of Physiology - Cell Physiology 296, 682–692. [DOI] [PubMed] [Google Scholar]
- Hsiao CD, You MS, Guh YJ, Ma M, Jiang YJ, and Hwang PP (2007). A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PLoS ONE 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Laserstein P, and Helmstaedter M (2015). Large-volume en-bloc staining for electron microscopy-based connectomics. Nat Commun 6, 7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PP, and Chou MY (2013). Zebrafish as an animal model to study ion homeostasis. Pflugers Archiv European Journal of Physiology 465, 1233–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PP, and Lee TH (2007). New insights into fish ion regulation and mitochondrion-rich cells. Comparative Biochemistry and Physiology - A Molecular and Integrative Physiology 148, 479–497. [DOI] [PubMed] [Google Scholar]
- Jänicke M, Carney TJ, and Hammerschmidt M (2007). Foxi3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Developmental Biology 307, 258–271. [DOI] [PubMed] [Google Scholar]
- Jongebloed WL, Stokroos I, Van der Want JJ, and Kalicharan D (1999). Non-coating fixation techniques or redundancy of conductive coating, low kV FE-SEM operation and combined SEM/TEM of biological tissues. J Microsc 193, 158–170. [DOI] [PubMed] [Google Scholar]
- Kapsimali M (2017). Epithelial cell behaviours during neurosensory organ formation. Development (Cambridge) 144, 1926–1936. [DOI] [PubMed] [Google Scholar]
- Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez-Soriano J, Santos F, Cremers CW, Di Pietro A, et al. (1999). Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet 21, 84–90. [DOI] [PubMed] [Google Scholar]
- Katoh F, Hasegawa S, Kita J, Takagi Y, and Kaneko T (2001). Distinct seawater and freshwater types of chloride cells in killifish, Fundulus heteroclitus. Canadian Journal of Zoology 79, 822–829. [Google Scholar]
- Katoh F, Hyodo S, and Kaneko T (2003). Vacuolar-type proton pump in the basolateral plasma membrane energizes ion uptake in branchial mitochondria-rich cells of killifish Fundulus heteroclitus, adapted to a low ion environment. Journal of Experimental Biology 206, 793–803. [DOI] [PubMed] [Google Scholar]
- Kimura RS (1969). Distribution, structure, and function of dark cells in the vestibular labyrinth. Ann Otol Rhinol Laryngol 78, 542–561. [DOI] [PubMed] [Google Scholar]
- Kondrychyn I, Teh C, Garcia-Lecea M, Guan Y, Kang A, and Korzh V (2011). Zebrafish Enhancer TRAP transgenic line database ZETRAP 2.0. Zebrafish 8, 181–182. [DOI] [PubMed] [Google Scholar]
- Köppl C, Wilms V, Russell IJ, and Nothwang HG (2018). Evolution of Endolymph Secretion and Endolymphatic Potential Generation in the Vertebrate Inner Ear. Brain, Behavior and Evolution 92, 1–31. [DOI] [PubMed] [Google Scholar]
- Kozlovskaja-Gumbriene A, Yi R, Alexander R, Aman A, Jiskra R, Nagelberg D, Knaut H, McClain M, and Piotrowski T (2017). Proliferation-independent regulation of organ size by Fgf/Notch signaling. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. Journal of structural biology 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Kumai Y, Bernier NJ, and Perry SF (2014). Angiotensin-II promotes Na+ uptake in larval zebrafish, Danio rerio, in acidic and ion-poor water. J Endocrinol 220, 195–205. [DOI] [PubMed] [Google Scholar]
- Kumai Y, Kwong RWM, and Perry SF (2015). A role for transcription factor glial cell missing 2 in Ca2+ homeostasis in zebrafish, Danio rerio. Pflugers Archiv European Journal of Physiology 467, 753–765. [DOI] [PubMed] [Google Scholar]
- Kuo CS, and Krasnow MA (2015). Formation of a Neurosensory Organ by Epithelial Cell Slithering. Cell 163, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Lambert TJ (2019). FPbase: a community-editable fluorescent protein database. Nat Methods 16, 277–278. [DOI] [PubMed] [Google Scholar]
- Lang T, Hansson GC, and Samuelsson T (2007). Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci U S A 104, 16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RT, Asharani PV, and Carney TJ (2014). Basal keratinocytes contribute to all strata of the adult zebrafish epidermis. PLoS One 9, e84858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BK, Chen RD, and Hwang PP (2009). Expression regulation of Na+-K+-ATPase α1-subunit subtypes in zebrafish gill ionocytes. American Journal of Physiology - Regulatory Integrative and Comparative Physiology 296, 1897–1906. [DOI] [PubMed] [Google Scholar]
- Lin LY, Hung GY, Yeh YH, Chen SW, and Horng JL (2019). Acidified water impairs the lateral line system of zebrafish embryos. Aquatic Toxicology 217. [DOI] [PubMed] [Google Scholar]
- Lopez-Schier H, Starr CJ, Kappler JA, Kollmar R, and Hudspeth AJ (2004). Directional cell migration establishes the axes of planar polarity in the posterior lateral-line organ of the zebrafish. Dev Cell 7, 401–412. [DOI] [PubMed] [Google Scholar]
- Lorente-Canovas B, Ingham N, Norgett EE, Golder ZJ, Karet Frankl FE, and Steel KP (2013). Mice deficient in H+-ATPase a4 subunit have severe hearing impairment associated with enlarged endolymphatic compartments within the inner ear. Dis Model Mech 6, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund U, Agostinelli C, and Agostinelli MC (2017). Package ‘circular’. Repository CRAN. [Google Scholar]
- Luo J, Chen W, Xue L, and Tang B (2019). Prediction of activity and specificity of CRISPR-Cpf1 using convolutional deep learning neural networks. BMC Bioinformatics 20, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush ME, Diaz DC, Koenecke N, Baek S, Boldt H, St Peter MK, Gaitan-Escudero T, Romero-Carvajal A, Busch-Nentwich EM, Perera AG, et al. (2019). scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush ME, and Piotrowski T (2014). Sensory hair cell regeneration in the zebrafish lateral line. Dev Dyn 243, 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Sun S, Geng L, Song M, Wang W, Ye Y, Ji Q, Zou Z, Wang S, He X, et al. (2020). Caloric Restriction Reprograms the Single-Cell Transcriptional Landscape of Rattus norvegicus Aging. Cell 180, 984–1001 e1022. [DOI] [PubMed] [Google Scholar]
- Mayer-Gostan N, Kossmann H, Watrin A, Payan P, and Boeuf G (1997). Distribution of ionocytes in the saccular epithelium of the inner ear of two teleosts (Oncorhynchus mykiss and Scophthalmus maximus). Cell and Tissue Research 289, 53–61. [DOI] [PubMed] [Google Scholar]
- McGlone FP, Russell IJ, and Sand O (1979). Measurement of calcium ion concentrations in the lateral line cupulae of Xenopus laevis. J Exp Biol 83, 123–130. [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, and Smal I (2012). Methods for cell and particle tracking. Methods Enzymol 504, 183–200. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Lucas D, Battista M, and Frenette PS (2008). Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447. [DOI] [PubMed] [Google Scholar]
- Meyers JR, MacDonald RB, Duggan A, Lenzi D, Standaert DG, Corwin JT, and Corey DP (2003). Lighting up the senses: FM1–43 loading of sensory cells through nonselective ion channels. J Neurosci 23, 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Chan B, Grati M, Mittal J, Patel K, Debs LH, Patel AP, Yan D, Chapagain P, and Liu XZ (2016). Molecular Structure and Regulation of P2X Receptors With a Special Emphasis on the Role of P2X2 in the Auditory System. J Cell Physiol 231, 1656–1670. [DOI] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, Khokha MK, Doudna JA, and Giraldez AJ (2017). CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun 8, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, and Zon LI (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya K, Harbidge DG, Wangemann P, Schultz BD, Green ED, Wall SM, and Marcus DC (2007). Lack of pendrin HCO3- transport elevates vestibular endolymphatic [Ca2+] by inhibition of acid-sensitive TRPV5 and TRPV6 channels. Am J Physiol Renal Physiol 292, F1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, and Elowitz MB (2018). Dynamic Ligand Discrimination in the Notch Signaling Pathway. Cell 172, 869–880 e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson T, Rusch A, Friedrich RW, Granato M, Ruppersberg JP, and Nusslein-Volhard C (1998). Genetic analysis of vertebrate sensory hair cell mechanosensation: the zebrafish circler mutants. Neuron 20, 271–283. [DOI] [PubMed] [Google Scholar]
- Norgett EE, Golder ZJ, Lorente-Canovas B, Ingham N, Steel KP, and Karet Frankl FE (2012). Atp6v0a4 knockout mouse is a model of distal renal tubular acidosis with hearing loss, with additional extrarenal phenotype. Proc Natl Acad Sci U S A 109, 13775–13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, and Schier AF (2013). Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, and Leach SD (2009). Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of Development 126, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BW, Andrew AL, Mostafa Kamal AH, Card DC, Schield DR, Pasquesi GIM, Pellegrino MW, Mackessy SP, Chowdhury SM, Secor SM, et al. (2019). Multi-species comparisons of snakes identify coordinated signalling networks underlying post-feeding intestinal regeneration. Proc Biol Sci 286, 20190910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piermarini PM, Verlander JW, Royaux IE, and Evans DH (2002). Pendrin immunoreactivity in the gill epithelium of a euryhaline elasmobranch. Am J Physiol Regul Integr Comp Physiol 283, R983–992. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, and Baker CV (2014). The development of lateral line placodes: taking a broader view. Dev Biol 389, 68–81. [DOI] [PubMed] [Google Scholar]
- Pisam M, Massa F, Jammet C, and Prunet P (2000). Chronology of the appearance of β, A, and α mitochondria-rich cells in the gill epithelium during ontogenesis of the brown trout (Salmo trutta). Anatomical Record 259, 301–311. [DOI] [PubMed] [Google Scholar]
- Plasschaert LW, Žilionis R, Choo-wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B, and Ryan K (1971). The fine structure of the lateral-line sense organs of dogfish. Proceedings of the Royal Society of London Series B Biological Sciences 179, 157–169. [Google Scholar]
- Romero-Carvajal A, Navajas Acedo J, Jiang L, Kozlovskaja-Gumbriene A, Alexander R, Li H, and Piotrowski T (2015). Regeneration of Sensory Hair Cells Requires Localized Interactions between the Notch and Wnt Pathways. Dev Cell 34, 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell IJ, and Sellick PM (1976). Measurement of potassium and chloride ion concentrations in the cupulae of the lateral lines of Xenopus laevis. The Journal of Physiology 257, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardet C, Pisam M, and Maetz J (1979). The surface epithelium of teleostean gish gills. Journal of Cell Biology 80, 96–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber NL, Machado P, Markert SM, Stigloher C, Schwab Y, and Steyer AM (2017). Minimal resin embedding of multicellular specimens for targeted FIB-SEM imaging. Methods Cell Biol 140, 69–83. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C, and Nicolson T (1999). Defective calmodulin-dependent rapid apical endocytosis in zebrafish sensory hair cell mutants. J Neurobiol 41, 424–434. [PubMed] [Google Scholar]
- Seleit A, Kramer I, Riebesehl BF, Ambrosio EM, Stolper JS, Lischik CQ, Dross N, and Centanin L (2017). Neural stem cells induce the formation of their physical niche during organogenesis. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz Y, Gonzalez-Celeiro M, Chen CL, Pasolli HA, Sheu SH, Fan SM, Shamsi F, Assaad S, Lin ET, Zhang B, et al. (2020). Cell Types Promoting Goosebumps Form a Niche to Regulate Hair Follicle Stem Cells. Cell 182, 578–593 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidi S, Friedrich RW, and Nicolson T (2003). NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science 301, 96–99. [DOI] [PubMed] [Google Scholar]
- Stawicki TM, Owens KN, Linbo T, Reinhart KE, Rubel EW, and Raible DW (2014). The zebrafish merovingian mutant reveals a role for pH regulation in hair cell toxicity and function. DMM Disease Models and Mechanisms 7, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MS, and Takeichi M (1994). Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci U S A 91, 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia JC, Kasthuri N, Hayworth KJ, Schalek R, Lichtman JW, Smith SJ, and Buchanan J (2012). High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nat Protoc 7, 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.C. (2013). R: A language and environment for statistical computing (Vienna, Austria: ). [Google Scholar]
- Thevenaz P, Ruttimann UE, and Unser M (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7, 27–41. [DOI] [PubMed] [Google Scholar]
- Trune DR (2010). Ion homeostasis in the ear: mechanisms, maladies, and management. Curr Opin Otolaryngol Head Neck Surg 18, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsamos S, Nebel C, and Charmantier G (2005). Ontogeny of osmoregulation in postembryonic fish: A review. Comparative Biochemistry and Physiology - A Molecular and Integrative Physiology 141, 401–429. [DOI] [PubMed] [Google Scholar]
- Venero Galanternik M, Navajas Acedo J, Romero-Carvajal A, and Piotrowski T (2016). Imaging collective cell migration and hair cell regeneration in the sensory lateral line. Methods Cell Biol 134, 211–256. [DOI] [PubMed] [Google Scholar]
- Wang YF, Tseng YC, Yan JJ, Hiroi J, and Hwang PP (2009). Role of SLC12A10.2, a Na-Cl cotransporter-like protein, in a Cl uptake mechanism in zebrafish (Danio rerio). American Journal of Physiology - Regulatory Integrative and Comparative Physiology 296, 1650–1660. [DOI] [PubMed] [Google Scholar]
- Wangemann P, Nakaya K, Wu T, Maganti RJ, Itza EM, Sanneman JD, Harbidge DG, Billings S, and Marcus DC (2007). Loss of cochlear HCO3- secretion causes deafness via endolymphatic acidification and inhibition of Ca2+ reabsorption in a Pendred syndrome mouse model. Am J Physiol Renal Physiol 292, F1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2011). ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics 3, 180–185. [Google Scholar]
- Wu H, and Tang N (2021). Stem cells in pulmonary alveolar regeneration. Development 148. [DOI] [PubMed] [Google Scholar]
- Xiao T, Roeser T, Staub W, and Baier H (2005). A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development 132, 2955–2967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 - Cluster markers of all obtained clusters, related to Figure 1 and S1: Differentially expressed genes per cluster (cluster markers) obtained for all sequenced cells after quality controls.
Supplementary Table 2 - Cluster markers of skin and ionocyte subsets, related to Figure 1: Differentially expressed genes for the ionocyte and skin subset.
Supplementary Video 1 - 3D rendering of neuromast (confocal), related to Figure 1: 3D animation of a confocal z-stack of a 4 dpf tp1bglobin:EGFP larva, showing a Notch+ non-lateral line cell, as well as central support cells labeled by this reporter line. Scale bar equals 5 μm.
Supplementary Video 2 - Pair of cells migrate into the neuromast, related to Figure 3: Neuromasts in a 3 dpf fish labeled with Et4;Et20;cldnb-lynEGFP shows a pair of cells entering on the dorsal side. Scale bar equals 10 μm.
Supplementary Video 3 - Pair of cells migrate into the neuromast, related to Figure 3: Confocal time lapse recording of a Gw57a:EGFP;ubi:H2A-mCherry larva at 3 dpf showing Nm ionocyte precursor cells migrating into the neuromast as visualized by translocation of the two cell nuclei, as well as the appearance of a gap in Gw57a:EGFP fluorescence. Individual z-slices of a confocal z-stack are shown, intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 4 - Pairs of cells migrate extensively inside the neuromast, related to Figure 3: Maximum intensity projection of confocal time lapse recording of a primII-derived neuromast of a cldnb:H2A-mCherry;tp1bglobin:EGFP larva at 2 dpf, showing two pairs of Nm ionocyte precursors migrating into the organ. The nuclei of the cells were tracked on 3D based on cldnB:H2A-mCherry fluorescence. Future Notch+ cells (based on Notch reporter fluorescence) are labeled with cyan dots, Notch− cells are labeled with red dots. Maximum intensity projection of a confocal z-stack is shown. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 5 - Nm ionocyte develops apical extension, related to Figure 3: Confocal time lapse recording of a neuromast of a tp1bglobin:EGFP transgenic at 3 dpf depicting a Notch+ Nm ionocyte precursor in the ventral pole extending multiple protrusions before developing and stabilizing an apical extension. Maximum intensity projection of a confocal z-stack is shown. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 6 - Notch signaling increases in one Nm ionocytes in a pair, related to Figure 3: Confocal time lapse recording of a neuromast of a cldnb:H2A-mCherry;tp1bglobin:EGFP transgenic at 3 dpf, depicting a pair of Nm ionocyte precursors re-arranging in the organ, with one cell gradually increasing in Notch reporter fluorescence. The nuclei of the cells were tracked based on cldnb:H2A-mCherry fluorescence. Notch+ cells are labeled with cyan dots, Notch− cells are labeled with red dots. An individual z-slice of a confocal z-stack is shown. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 7 - Notch signaling increases in one the Nm ionocytes in a pair, related to Figure 3: ubi:H2A-mCherry;tp1bglobin:EGFP double transgenic shows two pairs of cells entering the neuromast simultaneously while increasing EGFP fluorescence, indicating activation of Notch signaling. Only the Notch+ cell is visible. Scale bar equals 10um.
Supplementary Video 8 – Notch inhibition leads to Nm ionocyte death, related to Figure 3: Gw57a:EGFP; ubi:H2A-mCherry 4 dpf double transgenic shows that both Nm ionocytes of two different pairs die after Notch signaling inhibition with LY411575. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 9 – Notch+ cells frequently die first after Notch inhibition, related to Figure 3 and S3: time lapse of a 3 dpf ubi:H2A-mCherry;tp1bglobin:EGFP shows Notch+ cell death followed by death of an adjacent cell after incubation with the Notch inhibitor LY411575. Intervals between frames are 5 min, scale bar equals 10 μm.
Supplementary Video 10 - 3D rendering of Nm ionocyte pair (EM), related to Figure 4: Rendering of a 3D reconstruction of a Nm ionocyte pair in the neuromast.
Supplementary Video 11 - 3D rendering of Nm ionocytes and hair cells (confocal), Related to Figure 4: 3D animation of a confocal z-stack of a tp1bglobin:EGFP;myo6b:Lck-mScarlet-I neuromast at 5 dpf, depicting the close association of Notch+ Nm ionocytes and hair cells and the formation of the apical crypt in close proximity to the hair cell bundles.
Data Availability Statement
Original data used for the results reported in this paper may be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1543_2020. The datasets and codes generated and used during this study are available at GEO: GSE168695 and GitHub https://github.com/Piotrowski-Lab/scRNAseq_Nm_ionocytes_analysis_Seuratv3_Peloggia_Muench_et_al_2020.