Abstract
Endosomal trafficking plays an integral role in various eukaryotic cellular activities and is vital for higher-order functions in multicellular organisms. RAB GTPases are important proteins that influence various aspects of membrane traffic, which consequently influence many cellular functions and responses. Compared to yeast and mammals, plants have evolved a unique set of plant-specific RABs that play a significant role in their development. RABs form the largest family of small guanosine triphosphate (GTP)-binding proteins, and are divided into eight sub-families named RAB1, RAB2, RAB5, RAB6, RAB7, RAB8, RAB11 and RAB18. Recent studies on different species suggest that RAB proteins play crucial roles in intracellular trafficking and cytokinesis, in autophagy, plant microbe interactions and in biotic and abiotic stress responses. This review recaptures and summarizes the roles of RABs in plant cell functions and in enhancing plant survival under stress conditions.
Keywords: Abiotic stress, biotic stress, GTP binding protein, RAB, Guanosine triphosphate, vesicle trafficking
1. INTRODUCTION
Endocytic trafficking in eukaryotes is vital for normal growth and development as well as in stress signaling. The major players involved in the process include RAS, ARF, RAN and SNAREs (soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptors). Vesicular trafficking targets the newly synthesized membrane or soluble cargo proteins either for secretion via the plasma membrane (PM) or for degradation in lysosomes\vacuoles via the endoplasmic reticulum (ER) and trans-Golgi network (TGN) pathway.
In animals and lower eukaryotes, RAS family members function as signaling molecules that are activated by extracellular stimuli and regulate intracellular signaling, which, in turn, controls gene transcription, thus influencing cell growth and differentiation. Presently, no homologue of the mammalian RAS has been identified in plants [1, 2]. In contrast, plant RAB, ARF, RHO and RAN GTPase proteins are well characterized. It seems that only the RAS and RHO GTPases are involved in signaling networks, whereas RAB and ARF family of proteins are involved in the regulation of vesicular trafficking. RAN proteins are abundant in cells and are involved in nuclear transport [3-5]. Most of these RAS family members signal through a wide range of effectors, such as coat complexes (COP, AP-1 and AP-3) and lipid- modifying enzymes (PLD1, phosphatidylinositol 4,5-kinase, and phosphatidylinositol 4-kinase). RAB GTPase regulates intracellular vesicular transport and the trafficking of proteins between different organelles via endocytotic and secretory pathways in both plants and animals [6-8]. They are more diverse in plants and mammals than in yeast [9]. Multiple copies of RABs have been reported in plants and other organisms; 52 in Oryza sativa [10], 57 in Arabidopsis thal- iana [11], 67 in Populus trichocarpa [12], 11 in Saccha- romyces cerevisiae [13, 14], 7 in Schizosaccharomyces pom- be [13], 31 in Caenorhabditis elegans [15], 33 in Drosophila melanogaster [10] and 65 in Homo sapiens [16].
Phylogenetic analyses of the amino acid sequences of RAB GTPases suggest their segregation into different sub- families on the basis of their localization and/or function in membrane trafficking [17]. Plant RABs are divided into eight sub-families: RAB1, RAB2, RAB5, RAB6, RAB7, RAB8, RAB11 and RAB18. In silico analyses revealed that monocots and dicots have divergent profiles, probably to execute unique cellular functions between the two groups [10]. Plant RAB proteins appear to have followed a different evolutionary pathway than animal or fungal RABs, as revealed by recent functional information on plant RAB proteins and discussed more in detail later in this review.
Plant and animal cells have similar core elements for G protein-coupled signaling, however, they differ from animal cells only in network architecture and intrinsic properties of G protein elements. In comparison to animal G proteins, plant G proteins are self-activating, and therefore the regulation of their activation occurs at the deactivation step [18]. Molecular and biochemical analyses have shown that plants have both heterotrimeric and small G proteins (small GTPases), both of which work as protein switches that are turned on and off by the nucleotide GTP. When GTP is bound, the G proteins and GTPases are “on” and activate downstream signals [8, 19-21].
In recent years, remarkable progress has been made in understanding the endomembrane system in plants. However it is still not clear how small GTPases participate in the signaling steps that regulate developmental processes, control hormone biosynthesis or activity, and mediate biotic and abiotic stress tolerance. In this review, we will focus mainly on the structural and functional characterization of RAB proteins in plants. However, the eukaryotic trafficking system has to be summarized briefly so as to provide a better understanding of how plant RABs are compared to those of other organisms.
2. EUKARYOTIC PROTEIN TRAFFICKING PATHWAYS
In eukaryotic organisms, each organelle membrane has a specific arrangement of proteins and lipids and operates an accurate and highly co-ordinated vesicular trafficking pathway for the establishment and maintenance of compartmentalization. This intracellular compartment creates specialized environments for various chemical reactions important for cellular functions [22]. The trafficking process begins with the budding of transport vesicles from donor membranes and ends with their fusion to target organelles. A single round in the transport process from vesicle budding to its fusion involves two classes of RAS-like, small GTPase family proteins. These are SAR (Secretion-associated Ras-related)/ARF, and RAB. The SAR/ARF act during vesicle budding, and RAB functions in targeting and/or tethering transport vesicles to specific acceptor compartments. The SAR/ARF proteins are highly conserved among species, from yeast to mammals as well as in plants. In any one species, the Rab gene number is typically larger than the number of Sar/Arf genes. For example, in S. cerevisiae there are 11 RAB/Ypt (yeast protein 2), one SAR and three ARF GTPases [23, 24].
3. SMALL GTP BINDING PROTEINS IN PLANTS
Small GTP-binding proteins are monomeric G proteins with molecular masses of 20-30 kDa, and they are related to the α-subunit of heterotrimeric G proteins. The small GTPase super-family is structurally divided into five families, including RAS, RHO, RAB, RAN and ARF [17, 25, 26]. The RAB, RAN and ARF families are conserved in eukaryotes and are involved in different cellular processes. The genome of Arabidopsis has 11 Rho, 57 Rab, 4 Ran and 21 Arf genes [11]. In fact, RAB proteins constitute the largest family of the small GTPases [27, 28].
RAS and RHO are signaling proteins mainly responsible for transmitting extracellular signals in yeast and animals. RAS homologs are absent in Arabidopsis [1]. The RHO family in animals is composed of conserved sub-families, CDC42, RAC and RHO, while in plants it belongs to a unique sub-family, ROP [4]. The RHO-related GTPases have higher similarity with human RAC GTPases and therefore are called RAC GTPases [29].
The ROP GTPases control the assembly of filamentous actin structures and the development of cell polarity in eukaryotes. ROPs also regulate multiple responses of plants to abscisic acid (ABA) and auxin, including changes in gene expression, pollen tube growth, root hair development, cell cycle progression and H2O2 production [30].
RAN proteins regulate trafficking of RNA and protein molecules through nuclei by regulating nucleocytoplasmic transport during the G1, S, and G2 phases of the cell cycle. They also play a role in the organization of microtubules during the M phase [26, 27, 31]. In contrast to other signaling proteins like RAS and RHO, RAN itself is not a membrane-bound protein.
ARF family members regulate intracellular vesicle trafficking. They recruit cytosolic coat proteins (COP-I, COP-II and clathrin coats) to transport vesicles [28, 32].
4. A BRIEF OVERVIEW OF THE STRUCTURE AND FUNCTION (S) OF RABS
The RAB GTPases make up the largest subfamily of the RAS super-family, and their role has been analysed in many different plant species. A comparison of different RAB’s shows that they share 30-55% similarity at the primary sequence level. RAB proteins have several conserved structural domains (Fig. 1), including four guanine nucleotide-binding domains (G1, G3, G4 and G5), an effector-binding domain (G2), a membrane attachment domain (C-residues) and YRG domain [33]. The G1 (GDSGVGKT) domain is involved in Mg+ and phosphate binding. The G3 (WDTAGQ) domain is the site where DTAG sequence interacts with the γ-phosphate of GTP. The G4 (GNKXD) domain is the region that determines guanine specificity, and the G5 (ETSAK) domain is the site where ETSA sequence interacts with the D residue in the NKXD sequence. The effector region G2 (YKATIGADF) specifies RAB function, and its TIGADF motif interacts with specific GTPase-activating proteins GAPs [17]. The role of YRG is not well established.
Fig. (1).
Schematic representation of the domain architecture of a typical RAB protein. Coloured rectangle represents different conserved motifs of GTP binding and hydrolysis. G1, G3, G4 and G5 are guanine nucleotide-binding domains and G2 is an effector-binding domain. YRG domain function is not yet known. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
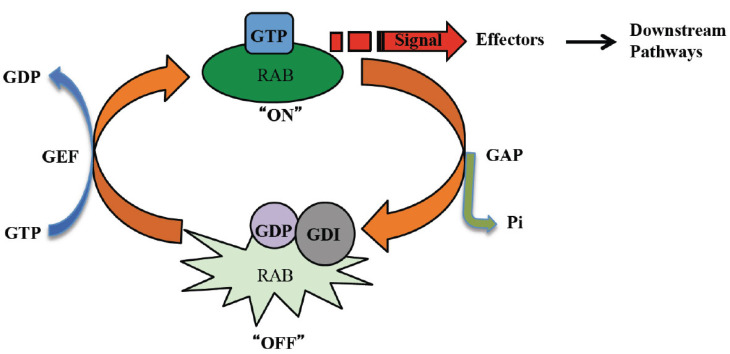
RABs are the first RAS proteins involved in the regulation of vesicle trafficking. Like other small GTPases, they function as molecular switches that cycle between a GTP, membrane-bound, active form and a GDP, cytosolic, inactive form. In the GTP-bound activated form, the RAB transmits signals to downstream effectors. In the inactive GDP-bound form, RAB detaches itself from the membrane (Fig. 2) [8].
Fig. (2).
The RAB GTPase regulatory cycle. RAB-GTPases cycle between an active GTP-bound and an inactive GDP-bound state. GEFs activate RAB, which, in turn, interact with specific effectors to mediate downstream pathways. GAPs stimulate the GTPase activity of RAB proteins, catalyse the inactivation of their regulatory activity. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
RAB proteins have to recruit GTPase activating protein (GAP) to efficiently hydrolyse GTP because their intrinsic GTPase activity is low. Once GTP is hydrolyzed, the GDP-bound RAB binds to the GDP dissociation inhibitor (GDI), which masks the geranylgeranyl moiety at the C-terminus of RAB, resulting in membrane association. Thus, GDI solubilizes RAB GTPases in the cytosol for the next round of GTPase cycle. Activation of RAB is catalyzed by the guanine nucleotide exchange factor (GEF), which replaces GDP with GTP on RAB proteins. For signal execution, RAB GTPases must detach from RAB GDI and move to the target organelle membranes. This step is facilitated by the GDI displacement factor (GDF) [34], which comprises a group of small membrane proteins that belong to the PRA/Yip family [35]. Active RABs bound to GTP associate with RAB effectors to activate downstream reactions. Through genetic and biochemical approaches, a large number of RAB effectors have been identified [36-38] and can be categorized as (1) tethers, (2) regulatory factors (i.e., GEF or GAP), or (3) others, like APPL (RAB5 effectors), a protein that directly links RAB5 to endosomal signaling [39-41]. Tethers are RAB effector proteins that function by attaching transport vesicles to the target membranes and are divided into two groups (1) long fibrous proteins and (2) large multiprotein complexes [22]. GM130, giantin, p115, and EEA1 are examples of long fibrous RAB effector tether proteins, while TRAPP, COG, exocyst, CORVET, and HOPS are RAB effector tether multiprotein complexes.
5. FAMILY OF RAB PROTEINS IN PLANTS
The complete genome sequencing of A. thaliana, O. sativa, S. cerevisiae, S. pombe, C. elegans, D. melanogaster and H. sapiens has uncovered divergent organizations of RABs encoded in different genomes. Plant RABs are more diverse in comparison with those of other eukaryotes.
The genome of A. thaliana contains 57 RAB GTPases, which are grouped into eight clades (RAB1/RABD, RAB2/RABB, RAB5/RABF, RAB6/RABH, RAB7/RABG, RAB8/RABE, RAB11/RABA, and RAB18/RABC). Out of eight clades, six clades (RAB1, RAB5, RAB6, RAB7, RAB8 and RAB11) are conserved among plants, yeasts and animals. The other two clades are not found in yeast but have high similarity with the mammalian RAB2 and RAB18 [23].
The gene organization of each sub-family is conserved in the genomes of Arabidopsis, maize and rice. In these genomes, RAB1 and RAB8 genes have eight exons; RAB2, RAB6, and RAB18 have six exons, and RAB5 (except OsRAB5D1) and RAB18 clade and RAB7 have seven exons, indicating the common ancestral origin of the genes in a sub-family. However, RAB11 members have a different origin, as evidenced by their highly diverged gene organization with exon numbers varying between one and four [10].
6. ROLE OF RAB’S IN INTRACELLULAR TRAFFICKING
On the basis of localization of RAB’s in different organelles, their role in trafficking can be categorised as discussed in the following sections.
6.1. ER to Golgi Trafficking
The RAB1/RABD group regulates traffic between the ER and the Golgi in yeast and mammalian cells and is widely conserved among eukaryotes. Plant homologs of RAB1 complemented a mutation in the yeast RAB1 ortholog, YPT1, indicating that its function is conserved in plants [42, 43]. Transient co-expression of mutant AtRABD2a and fluorescent markers in tobacco cells showed there are five different A. thaliana RAB1 homologs involved in ER-Golgi trafficking [44].
The RAB2/RABB homolog group is conserved in plants and animals but it is not present in budding yeast [23]. In mammalian cells, RAB2 localizes on cis-Golgi membranes, interacts with Golgi matrix proteins, and acts in ER-Golgi trafficking. RAB2 homologs are well conserved among plants. Some RAB2 group members in A. thaliana and Nicotiana tabacum are expressed predominantly in pollen, and there is evidence they are important for pollen function. Dominant-negative mutants of NtRAB2 showed slow growth of pollen tubes and reduced transport between the ER and the Golgi [45]. Fluorescent protein-tagging and immunoelectron microscopy studies showed that NtRAB2 is localized to the Golgi apparatus. Thus, as in mammals, RAB2 proteins in plants could be an important regulator of ER-Golgi trafficking.
RAB6/RABH are involved in retrograde trafficking pathways from the Golgi to the ER. An A. thaliana RAB6 homolog, AtRABH1b could replace the function of yeast YPT6 [46]. Additionally, A. thaliana RAB6 proteins can physically interact with Golgi proteins that act as putative tethers [47].
6.2. Secretion
The RAB8/RABE in mammals and SEC4, its yeast counterpart, also participate in retrograde membrane trafficking. Transgenic A. thaliana constitutively expressing GFP-RABE1d revealed these to be associated with the Golgi apparatus and PM in leaf. These localization studies support their proposed role in mediating the traffic of secretory vesicles from the Golgi to the PM [48]. A study in A. thaliana demonstrated that the RAB8 group regulates vesicular trafficking in the secretory pathway, either at or after the Golgi [49]. When a dominant-negative AtRABE1 was co-expressed with secGFP, the secretion of secGFP was disturbed resulting in its accumulation in cells and getting misdirected to the vacuolar transport pathway. In tobacco cells, the localization of YFP-AtRABE1 on the Golgi also suggests that it may function in the late secretory pathway. This result further supported the conclusion that RABE1 acts downstream of RABD2.
The RAB11/RABA group is the largest sub-clade in plant RABs. In A. thaliana, the RAB11 group comprises 26 members, out of the total of 57 RABs [50]. Similarly, 17 RAB11 group members are present in rice out of 52 RABs [10]. However, in H. sapiens only 3 RAB11 members are classified as 66 RAB GTPases and 2 RAB11 group members out of 11 RAB GTPases in S. cerevisiae [16, 51]. In non-polarized mammalian cells, RAB11a localizes to the membrane of recycling endosomes and regulates transport from the sorting endosomes to the recycling compartments. In polarized epithelial cells, RAB11a, RAB11b and RAB25 localize to the apical recycling endosomes. Though the RAB11 and RAB25 sub-classes appear to co-localize, yet it seems these control distinct transport routes of the recycling endosomes and the Golgi to the PM. The S. cerevisiae RAB11 homolog, Ypt31/32, is implicated in endocytic recycling and vesicular export from the late Golgi compartment to the PVC or the PM [52].
In plants, the RAB11 group is divided into six sub- groups (RABA1 to RABA6). Out of the six sub-groups, RABA1 is the largest subgroup in A. thaliana, which has nine members (RABA1a-RABA1i). RABA1 homologs are absent in yeast or mammals. In A. thaliana, RABA1 sub- group, RABA1a, RABA1b, RABA1c and RABA1d, expression is abundant in all tissues and these RABA1 proteins localize to a compartment adjacent to the TGN. Several lines of evidence further indicate that RABA1 regulates transport between the TGN and the PM and plays a role in salinity stress tolerance in plants [52]. The N. tabacum RABA1 protein, NtRAB11b, localizes to the apical zone of the elongating pollen tubes and its proper functioning is essential for the secretion and endocytic recycling at the tips of pollen tubes [53]. The RABA2 sub-group shows high similarity to mammalian RAB11 and yeast Ypt31/32, suggesting it may function in post-Golgi or endocytic pathways. The RABA3, A4, A5, and A6 have no homologs in yeast and mammals. A recent study demonstrated that A. thaliana RABA2 and RABA3 co-localize on a new post-Golgi membrane domain in root tip cells [54]. The RABA2/A3 compartment was distinct from, but often close to, Golgi stacks and the PVC, and partly overlapped with the VHA-a1-positive TGN. Intriguingly, RABA2/A3 localized on cell plates in dividing cells [54]. This finding suggests that these sub-groups might play crucial roles in cytokinesis via the regulation of polarized secretion.
Experimental evidence suggests that even the RABA4 subgroup in A. thaliana functions in polarized secretion as shown in root hair cells. Enhanced yellow fluorescent protein (EYFP)-tagged RABA4b specifically localized in the tips of growing root hair cells but was absent in mature root hair cells that had stopped expansion [55]. The tip-localized fluorescence was disrupted by treatment with latrunculin B, which disrupts the actin cytoskeleton of cells, indicating that the proper localization of RABA4b required actin polymerization. Interestingly, most of this protein was found in a unique membrane compartment that did not co-fractionate with the TGN SNAREs, SYP41 or SYP51 [55]. Nevertheless, RABA4b was detected on the TGN by immunoelectron microscopy [56]. These results suggest that there are at least two distinct TGN-related compartments with different protein contents. Sub-cellular localization of P. sativum GFP tagged RABA3 and RABA4 proteins expressed in tobacco BY-2 cells showed PRA2 on Golgi stacks and endosomes predominantly, while PRA3 was present in the TGN and/or the pre-vacuolar compartment, suggesting their differential localization/function in the trafficking pathway [57].
At present, there are very few reports on the functions of RABA5 and RABA6 proteins. Immunoelectron microscopy of pollen indicated that one member of the A. thaliana RABA5 group, ARA4 (RABA5c), is localized on the Golgi and adjacent vesicular structures [58]. The yeast two-hybrid system was used to screen molecules that interacted with ARA4 [59]. However, the physiological function of RABA5/A6 is still not clear.
Overall the plant RAB11/RABA group of RAB proteins appear to reside on post-Golgi organelles, including the TGN, and mediate both exocytic and endocytic trafficking. Thus, they are likely to play an especially important role in vesicle trafficking in polarized cells, including cell plate formation during cytokinesis and the tip growth of pollen tubes or root hairs.
6.3. Vacuolar and PVC/endosomal Trafficking
The RAB7/RABG and Ypt7p function in lysosomal and vacuolar biogenesis in mammals and yeast, respectively. Transient expression of GFP-fused OsRAB7 showed that it localizes to vacuoles in Arabidopsis protoplasts, which suggested it could have a role in vacuole biogenesis [60]. Transgenic plants constitutively overexpressing AtRAB7 in Arabidopsis had accelerated endocytosis in roots, leaves and protoplasts, which could be due to faster vesicle trafficking during endosome-vacuole fusion [61]. Other RAB7-related proteins, like AtRABG3b/AtRAB75 were also shown to be localized on the vacuolar membrane [62]. A proteomic analysis of vacuoles isolated from rosette leaves of A. thaliana revealed the presence of AtRABG3b and AtRABG3d/AtRAB72 [63], and the vacuolar membrane fraction showed RABD2a/ARA5, RABE1c/ARA3, RABB1a and RABD2b. Some of these RAB7-related proteins have been reported to function in ER-Golgi trafficking, which may suggest a direct trafficking route from the ER to vacuoles in plants [64]. The physiological function of the RAB7 protein group is not yet clear. However, overexpression of AtRABG3e in A. thaliana conferred tolerance to both salt and osmotic stress [61]. Similarly, the constitutive overexpression of PgRAB7 in tobacco and rice plants also enhanced tolerance to salt and osmotic stress [65, 66]. These reports suggest that the RAB7/RABG group plays important roles in stress physiology by regulating vacuolar functions.
In yeast and mammalian systems, the Yip/PRA1 family of proteins facilitates the delivery of RAB GTPases to the membrane by dissociating the RAB-GDI complex during vesicle trafficking. As evaluated by the yeast two-hybrid assay, OsPRA1 (a putative prenylated rice RAB acceptor) is an interacting partner of OsRAB7 along with other RAB GTPases that are involved in vacuolar trafficking. Studies using GFP as a tag found that OsPRA1 was localized to the pre- vacuolar compartment, and a Northern blot analysis revealed its high expression in rice shoots and mature stems. These results suggest that OsPRA1 may function as a RAB effector for vacuolar trafficking in plants [67]. A trafficking assay using Arabidopsis protoplasts showed that the point mutant of OsPRA1 (Y94A) strongly inhibits the vacuolar trafficking of cargo proteins but has no inhibitory effect on the PM trafficking of H+-ATPase-GFP, suggesting its specific involvement in vacuolar trafficking. OsPRA1 also interacted with OsVAMP3, implying its involvement in vesicle fusion. A study using a yeast expression system showed that OsPRA1 counteracts OsGDI2 activity and facilitates the delivery of OsRAB7 to the target membrane [68]. This result is consistent with the conclusion that OsPRA1 targets OsRAB7 to the tonoplast during vacuolar trafficking [68].
The RAB5/RABF is a very well characterized RAB GTPase in mammals with respect to its localization and functions. It localizes on the PM and early endosomes and is shown to be involved in a wide range of endocytic events, like biogenesis of clathrin coated vesicles, a homotypic fusion between early endosomes, endosomal motility, compartmentalization of endosomal membrane domains, modification of lipid moieties in endosomal membranes and endosomal signaling [23]. All green plants have a mammalian RAB5 homolog in addition to a group of plant-specific RAB5s with unique structural features. This group lacks a C-terminal variable region and the Cys motif, structural features essential for localization, membrane binding, interaction with GDI and the function of RAB GTPases. These plant RAB5s have an extra amino acid sequence present at their N-terminus that is acylated and helps in membrane anchorage.
In plants, RAB5 was first identified in Lotus japonicas [50], and later in Mesembryanthemum crystallinum [69] and A. thaliana [70]. TBC2 functions as a GAP to cycle RAB5 from an active GTP-bound to an inactive GDP-bound state, which is required for maintaining the dynamic changes of RAB5 on phagosomes and serves as a switch for the progression of phagosome maturation [71]. There are three RAB5 homologs in the A. thaliana genome, ARA7/RABF2b, RHA1/RABF2a and ARA6/RABF1. ARA7 and RHA1 are conventional RAB5s and ARA6 is a unique RAB5 in plants. All these RAB5 proteins are involved in the endocytic pathway [70-74]. The endocytic nature of the ARA7-compartments was futher confirmed by using BOR1, a boron transporter that promotes endocytosis when induced by high concentration of boron [75]. During endocytosis, BOR1 passed through ARA7 endosomes before reaching the vacuoles. Functional analysis with a dominant-negative ARA7 also indicated its involvement in endocytosis. When the dominant-negative ARA7 was overexpressed in A. thaliana plants and tobacco BY-2 cells, endocytosis was severely suppressed [76].
Several lines of evidence have suggested that plant RAB5s also function in the vacuolar transport pathway. The overexpression of dominant negative mutants of ARA7 and RHA1 disturbed the transport of soluble vacuolar proteins to the vacuole in N. tabacum leaf cells and A. thaliana protoplasts, respectively [77, 78]. These results indicate that conventional RAB5s play their role where endocytic and biosynthetic pathways are closely associated or merged. Recently, using immunoelectron microscopy it was demonstrated that all RAB5s are localized on multivesicular bodies [79], suggesting that at least some portion of the RAB5-bearing endosomes corresponded to the organelle previously identified as the pre-vacuolar compartment. This evidence is also consistent with the observation that RAB5 proteins mediate both endocytic and vacuolar transport. However, transient expression analysis showed contradictory results about the participation of plant unique RAB5 proteins in vacuolar transport. The overexpression of dominant-negative ARA6 did not perturb the transport of sporamin-GFP to the vacuole [78] while a dominant negative construct of m-RABmc, a M. crystallinum ARA6 homolog, inhibited the trafficking of aleurain-GFP to the vacuole [69]. This discrepancy can be explained by the fact that sporamin is a storage protein in tubers, while aleurain is sorted to the lytic vacuole. Further physiological and genetic analyses with loss-of-function mutants may clarify the differential effect of targeting by these proteins.
In mammalian cells, the RAB18/RABC protein plays a role in multiple cellular functions, including endocytic transport [80], lipid droplet localization [81] and secretion [82]; however, its precise function is still unclear. Members of the RAB sub-family C in Arabidopsis have homologs called RAB18 in mammals, but not in yeast. Mammalian RAB18s are expressed in the polar epithelial cells and control endocytosis [6, 83]. Thus substantial variation in the amino acid sequences of plant and animal RAB18 proteins in the conserved domains defines sub-class specificity and hence their different roles.
6.4. Cytokinesis
Cytokinesis is the final stage of eukaryotic cell division leading to the partitioning of cytoplasm between daughter cells. Recent evidence from different organisms suggests that some RAB proteins may play a crucial role during cytokinesis. When tagged with YFP, RAB11/RABA proteins relocate from punctate cytoplasmic structures to the cell plate during cytokinesis, which suggests a role for RABA GTPase in co-ordinating membrane traffic to or from the cell plate [84]. A functional analysis of small RAB GTPases in Arabidopsis revealed that RABA1c has a role in cytokinesis [84, 85]. Similarly, RABA2 and RABA3 were found on a novel domain of the post-Golgi membrane and co-localized with KNOLLE/AtSYP111 on cell plates during mitotic phase [54]. In particular, these proteins localized to the growing edges of the cell plates, where VHA-a1, GNOM and PVC residents were excluded. Conditional expression of a dominant-negative RABA2a construct resulted in enlarged polynucleate cells with cell wall stubs. As these stubs are localized on the growing edges of cell plates, this result could indicate that RABA2/A3 plays a role in cell plate formation, possibly by regulating secretion or endocytosis associated with developing cell plates.
7. ROLE OF RABs IN PLANT
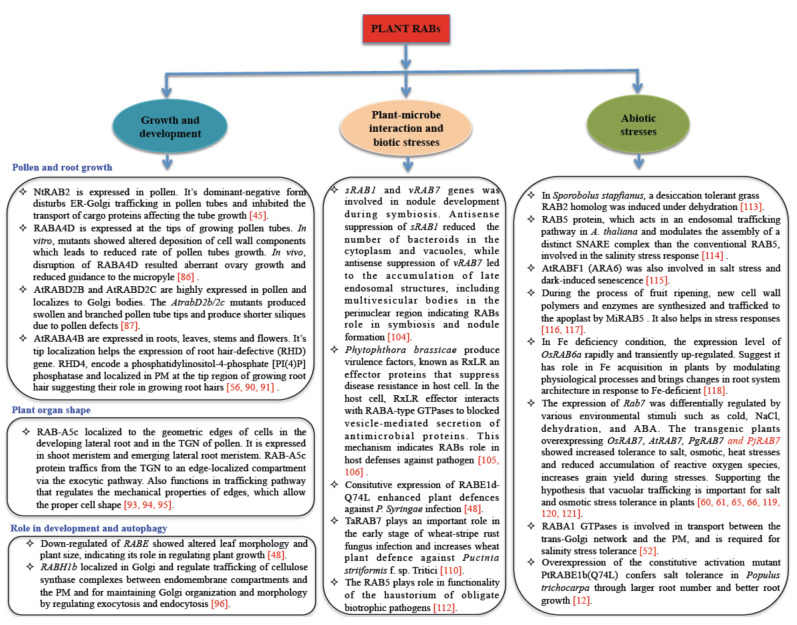
RABs have been extensively studied in animals, but not much work has been carried out in plants. The available literature suggests RABs were involved in plant growth and development, autophagy, abiotic stress, plant-microbe interaction and biotic stresses (Fig. 3). The RAB GTPase and its functions are summarized in Table 1.
Fig. (3).
Role of RAB GTPase in plants. Role of RAB GTPase are mainly devided into three parts in plants: (1) growth and development, (2) plant-microbe interaction and biotic stresses, and (3) abiotic stress. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Table 1.
Summary shows the RAB GTPase different functions with references.
| S. No. | Functions | Gene Names | ID (GenBank Number) | References |
|---|---|---|---|---|
| 1. | Pollen and root growth | NtRAB2AtRABA4DAtRABD2B and AtRABD2C AtSEC22AtRABA4BRAB-A5C | AF397451.1OAP01381.1AED95483.1 and AEE83907.1AEE28809.1AEE87150.1BAH19889.1 | [45] [86] [87] [89] [56, 90, 91] [95] |
| 2. | Development and autophagy | AtRABE/RAB8OsRAB5AAtRABG3E/AtRAB7AtRABG3BRAB3GAP,AtRABD2A | NP_001078278.1CAC19792.1AEE32414.1OAP16282.1NP_848805.2OAP18309.1 | [48] [98] [100] [100, 103] [21] [21] |
| 3. | Plant-microbe interaction and biotic stresses | sRAB1, sRAB7, vRAB7MtRab7MtRAB7A1MtRAB7A2AtRABE1DAtPEN1TaRAB7AoRAB-7A and AoRAB-2AtRAB5 | L14929.1; L14930.2; L14928.1-TC101145TC94423AED90617.1OAO99188.1AFC93432.1XP_011120064.1;XP_011119978.1 AEE79298.1 | [104] [107, 108] [48] [109] [110] [111] [112] |
| 4. | Abiotic stresses | SsRAB2AtARA6AtRABF1 (ARA6)MiRAB5OsRAB6AOsRAB7AtRAB7/ AtRABG3EPgRAB7PjRAB7AtRABA5E/AtCPRABA5eAtRABA1AtRABA1AAtRABA1BAtRABA1CAtRABA1DPtRABE1BSlRABGAPSlRABGAP18SlRaABGAP22 | AAD30658.1NP_567008.1NP_001327724.1AHF81484.1LOC_Os03g09140.1AAO67728.1Q9XI98.1AY829438.1EF591762.1NM_100462.5-NM_100520.4NM_101553.3NM_123942.2NM_117996.6Potri.008G051700-Solyc01g101090Solyc03g082590 | [113] [114] [115] [116] [118] [60, 87] [120, 121] [61] [65, 66] [119] [122] [52] [12] [21] |
7.1. Role of RABs in Plant Growth and Development
7.1.1. Pollen and Root Growth
RAB proteins are crucial regulators of vesicular trafficking in tip growth both in pollen tubes and root hairs [85]. During reproduction in flowering plants, the germinating pollen tubes grow in a directional manner towards the female gametophyte to fertilize the egg cell. This polarized pollen tube growth at the tip is facilitated by the transport of newly synthesized membrane and cell wall components. Two tobacco RAB GTPases NtRAB2 and NtRAB11b are important in this process. NtRAB2, a Golgi-localized RAB2 protein, is expressed predominantly in pollen [45]. The expression of a dominant-negative form of NtRAB2 was shown to disturb ER-Golgi trafficking in pollen tubes and inhibited the transport of cargo proteins affecting the tube growth. However, when expressed in leaf cells, NtRAB2 was not properly recruited to the Golgi apparatus. This result suggested that pollen tubes may possess a specialized mechanism for localizing NtRAB2 to the Golgi. For the correct localization, these vesicles require an intact actin cytoskeleton and functional GTPase cycle [45].
In Arabidopsis, there are also several RAB proteins associated with pollen tube growth. For example, RABA4D is expressed at the tips of growing pollen tubes. Mutant pollen with a disrupted RABA4D gene showed bulges with a reduced rate of growth in vitro and altered deposition of cell wall components. The wild type phenotypes could be restored only with the EYFP-RABA4d expression and not with EYFP-RABA4b in raba4d mutant pollen tubes. In vivo, disruption of RABA4D also resulted in a male-specific transmission defect, which was aberrant growth in the ovary and reduced guidance to the micropyle, suggesting its important role in pollen tube growth [86].
Two other RABs associated with pollen in Arabidopsis are AtRABD2b and AtRABD2c. These two have different but overlapping expression patterns, but both are highly expressed in pollen. These proteins localize to Golgi bodies and play important roles in pollen development, germination and tube elongation. Both single as well as double mutants of AtrabD2b and AtrabD2c did not show any morphological changes in comparison to the wild-type plants during vegetative growth, except that the siliques were shorter than the wild type in the double mutants. Compared with wild-type plants, AtrabD2b/2c mutants produced swollen and branched pollen tube tips, and their shorter siliques could also be due to pollen defects [87].
Tube growth depends on vesicle trafficking that transports phospholipid and pectin to the tube tip. There are 328 vesicle trafficking genes, of which 14 are up-regulated by the gibberellin signaling pathway during pollen development, including RABA4D and SNARE genes [88]. Loss of AtSEC22 function affects gametophyte development. Atsec22 mutant pollen becomes abnormal during the bi-cellular stage, giving rise to degenerated pollen grains. In many mutants, embryo sacs failed to support embryogenesis and displayed unfused polar nuclei in their central cell [89].
A role for RABs has also been demonstrated in root hair growth. RAB11/RABA, (AtRABA4b) are expressed in roots, leaves, stems and flowers. Tip localization of AtRABA4b-domains are required for the expression of root hair-defective (RHD) gene products, which were found mislocalized in rhd mutants. Recently, RHD4, a mutated rhd gene, was found to encode a phosphatidylinositol-4-phosphate [PI(4)P] phosphatase. The product of the enzyme PI(4)P was
localized in the PM at the tip region of growing root hair suggesting that PI(4)P, or one of its derivatives (e.g., PI(4,5)P, generated by phosphatidylinositol-phosphate 5-kinase), may act as a positional cue in this region [90, 91]. This is an important finding linking phosphoinositide metabolism to RAB11/RABA functions. In further support of this link, a study on the AtRABA4b effector protein revealed that it interacted with the plant phosphoinositide kinase (Pl-4Kβ1, phosphatidylinositol 4-OH kinase) and that both proteins co-localized to the tip-localized membranes, and played a role in controlling the growth of root hairs [56].
7.1.2. Plant Organ Shape
Plants acquired diverse organ shapes that are generated from the geometries of their constituent cells. While animal cells achieve shape through the internal support of their cytoskeleton, in plant rigid exterior walls provide structural support along with the turgor pressure, which determines cell shape. The plant cell wall resists or responds to the changes in tension, compression, and shear that result from cell division and expansion as organs take shape during development. Previous studies have suggested that the junctions of two planar cell walls (the geometric edges) have special mechanical properties to deal with these dynamic forces [92]. However, little is known about the trafficking mechanisms that could regulate plant cell geometry.
Recent reports provide some insight on how RAB-A5c, a RAB-GTPase could help regulate cell shape. This RAB is preferentially expressed in the shoot meristem and emerging lateral root meristem, which rapidly attains a conical organ shape that pushes through the outer layers of the parent root.
A quantitative analysis of the distribution of RAB-A5c showed its preferential localization to the geometric edges of cells in the developing lateral root and that it traffics from the TGN to an edge-localized compartment via the exocytic pathway [93]. A series of localization and perturbation experiments showed that RAB-A5c also functions in the trafficking pathway that regulates the mechanical properties of edges, which would allow the lateral root cells to attain proper shape [93-95].
7.1.3. Role in Development and Autophagy
RAB proteins have also been implicated in growth control. When RABE expression was down-regulated in Arabidopsis, the transgenic plants showed altered leaf morphology and reduced plant size, indicating that RABE-GTPases play an important role in regulating plant growth [48]. During plant growth and development, deposition of cellulose microfibrils in cell-wall is essential. Cellulose synthase complexes located in the PM are the enzymes responsible for cellulose synthesis. Trafficking of these complexes between endomembrane compartments (EC) and the PM is vital for cellulose biosynthesis. In A. thaliana, RAB-H1b, a Golgi-localized small GTPase, participates in the trafficking of CELLULOSE SYNTHASE 6 (CESA6) to PM. Loss of function of RAB-H1b resulted in altered distribution and motility of CESA6 in the PM and reduced cellulose content. Seedlings with this defect exhibited short, fragile etiolated hypocotyls. This may be due to the fact that exocytosis of CESA6 was impaired in rab-h1b cells, and endocytosis in mutant cells was also compromised. Impaired exocytosis and endocytosis of CESA6 lead to the accumulation of vesicles around an abnormal Golgi apparatus, which has an increased number of cisternae in rab-h1b cells, suggesting a defect in cisternal homeostasis caused by RAB-H1b loss function. This finding is consistent with the hypothesis that RAB GTPases play a key role in cellulose biosynthesis during hypocotyl growth and suggests that RAB-H1b is crucial for modulating the trafficking of cellulose synthase complexes between endomembrane compartments and the PM and for maintaining Golgi organization and morphology [96].
Several lines of evidence have suggested a relationship between vacuolar/endosomal trafficking and autophagy. In mammalian cells, RAB7 is needed for the maturation of autophagosomes and, thus, for the progression of autophagy. The RabG3b gene is also implicated in autophagy [97, 98]. In Arabidopsis, accumulation of autophagosome-like structures was observed in the embryos of mutants that have impairments in VCL1, a VPS16 homolog, and in VPS9a, which encodes a GEF for RAB5/RABF proteins [99, 100]. Both these mutants exhibited embryonic lethality; but otherwise, they had distinct phenotypes. This suggested that VCL1 and VPS9a act in distinct trafficking pathways and that they might not play a direct role in autophagy. Recent report found two new members of subclass VI with a RAB3GAP domain whose functional roles in plants unknown [21]. However, in animal homolog, genes participate in the deactivation of RAB3 (a homolog of plant RABD), interaction with ATG8 in autophagy processes, or activation of a RAB GTPase of subfamily C, acting as a GEF in degradative pathways and macro-autophagy. Intriguingly, a RABD of plants (RABD2A) localizes with EHD1 or SYP41 proteins associated with salt-stress tolerance.
The phenotypes of transgenic plants overexpressing wild type (RABG3bOX), constitutively active (RABG3bCA), and dominant negative (RABG3bDN) forms of RABG3b were indistinguishable from wild-type plants under normal growth conditions. However, both RABG3bOX and RABG3bCA plants displayed unrestricted hypersensitive programmed cell death when exposed to a fungal toxin, Fumonisin B1, or a fungal pathogen Alternaria brassicicola, whereas no major difference was observed between wild- type and RABG3bDN plants. In addition, RABG3bOX and RABG3bCA plants underwent accelerated leaf senescence compared to wild type and RABG3b DN plants. These results suggest that RABG3b is a modulator for cell death progression during pathogen responses and senescence processes in plants [101].
Roles for RABs in development have also been studied in crop plants. In peach 14 of the 24 genes encoding small G-proteins are RABs. Some of these play key roles in fruit development and maturation [102]. In rice, OsRAB5a plays an essential role in the trafficking of storage protein to PBII, as part of its function in organizing the endomembrane system in developing endosperm cells [103].
7.2. Plant-microbe Interaction and Biotic Stresses
A number of studies have confirmed the role of vesicular trafficking during symbiosis and nodule formation in legumes. The peri-bacteroid membrane (PBM) surrounding rhizobium in nodules originates from the fusion and expansion of newly synthesized vesicles with the PM. To explore the function of RAB GTPases in PBM biogenesis and nodule development, several genes encoding RAB GTPases were isolated from legumes, including sRAB1 and sRAB7 from Glycine max and vRAB7 from Vigna aconitifolia [104]. Elevations in the transcript abundance of sRAB1 and vRAB7 genes were observed during nodule formation, and antisense suppression of these genes affected different steps of nodule development. Antisense suppression of sRAB1 reduced the number of bacteroids in the cytoplasm and vacuoles, while antisense suppression of vRAB7 led to the accumulation of late endosomal structures, including multivesicular bodies in the perinuclear region. These results indicate RABs play critical roles in symbiosis and nodule formation.
Rhizobium bacteria from N2-fixing organelles, called symbiosomes, in the root nodules of legumes. The endocytic-like entry of rhizobia into nodule cells resembles the phagocytosis of bacteria into animal cells. Using membrane identity markers such as regulatory small GTPases and SNARE proteins in Medicago truncatula, rhizobial symbiosomes of Sinorhizobium meliloti were found to contain the syntaxin MtSYP132 from the initiation of symbiosome till their complete development and contained endosomal MtRAB7 once bacteria stopped dividing inside the host cells [105, 106].
Roles for RABs in mediating biotic stress responses have also been documented. Phytophthora brassicae, a plant-damaging oomycete that causes significant amount of crop losses worldwide, produces virulence factors, known as RxLR effector proteins, that are transferred into host cells to suppress disease resistance. In the host cell, the conserved RxLR effector interacts with host RABA-type GTPases to block vesicle-mediated secretion of antimicrobial proteins such as PATHOGENESIS RELATED PROTEIN 1 (PR-1) and DEFENSIN (PDF1.2), which function to block pathogen growth in the extracellular space. This important discovery reveals a key role for RABs in plant-pathogen interactions during host defenses [107, 108].
Other studies also point to the importance of RABs in plants’ responses to biotic stress. Although plant susceptibility to pathogenic Pseudomonas syringae bacteria was not affected by the down regulation of RABE, constitutive expression of RABE1d-Q74L enhanced plant defences, conferring resistance to P. syringae infection [48]. When elicitor molecules of microbial origin called Microbe Associated Molecular Patterns (MAMPs) act to trigger basal defence responses in plants, RAB-SNARE interactions may help mediate these responses. The Arabidopsis syntaxin PEN1, a member of the SNARE superfamily, is needed for the elicitor-mediated induction of lipopolysaccharides (LPS) as part of plant defenses, which would indicate it has a role in vesicle trafficking during the signaling process [109]. In wheat, knocking down TaRAB7 (Wheat RAB7 homologue) expression by virus-induced gene silencing enhanced the susceptibility of wheat cv. Suwon 11 to an avirulent race CYR23 [110]. These results indicate that TaRAB7 plays an important role in the early stage of wheat-stripe rust fungus interaction and increases defense of wheat plant against the fungus Pucinia striiformis f. sp. tritici. It was recently reported that in fungi Arthrobotrys oligospora, two RAB GTPases (AoRAB-7A and AoRAB-2) are involved in the regulation of mycelial growth, conidiation, trap formation, stress resistance, and pathogenicity in the nematode [111].
Localization studies also implicate RABs in the biotic defense responses of plants. Plant RAB5 from the ARA6 group, which regulates trafficking events distinct from RAB5 GTPases, was found to accumulate at the interface between host plants and biotrophic fungal and oomycete pathogens. Recent findings suggest that A. thaliana ARA6/RABF1, a plant-specific RAB5, is localized to the specialized membrane that surrounds the haustorium (infection hyphae), the extrahaustorial membrane (EHM), formed by the A. thaliana-adapted powdery mildew fungus Golovinomyces orontii. The conventional RAB5, ARA7/RABF2b was also localized to the EHM, endosomal SNARE whereas the RAB5-activating proteins were not, which suggests that the EHM has modified endosomal characteristic. The recruitment of host RAB5 to the EHM was a property shared by the barley- adapted powdery mildew fungus Blumeria graminis f.sp. hordei and the oomycete Hyaloperonospora arabidopsidis, but the extrahyphal membrane surrounding the hypha of the hemibiotrophic fungus Colletotrichum higginsianum at the biotrophic stage was devoid of RAB5. The localization of RAB5 to the EHM appears a link with the functionality of the haustorium. This finding sheds light on the relationship between plant RAB5 and obligate biotrophic pathogens [112].
7.3. Abiotic Stress
Several reports reveal an important role of RAB proteins in abiotic stress responses. A RAB2 homolog was induced under dehydration in a desiccation-tolerant grass, Sporobolus stapfianus [113]. Similarly, the enhanced expression of RABF1 (ARA6), a plant-unique RAB5 protein, which regulates an endosomal trafficking pathway in A. thaliana [114], confers greater tolerance to salinity stress [115], and its absence in rabF1 plants accelerates the senescence induced by dark treatments [115]. Recently quantitative RT-PCR analysis of MiRAB5, a RAB5 gene that is ubiquitously expressed in various tissues of the mango tree Mangifera indica, demonstrated that it is up-regulated in response to various abiotic stress conditions such as cold, salinity, and PEG treatments, indicating that it participates in the signaling response to these stresses [116].
Vesicle trafficking is shown to play a key role in facilitating the synthesis and modification of cell walls in fruits. During the process of fruit ripening, new cell wall polymers and enzymes are synthesized and trafficked to the apoplast. Recent review supports the finding that plant RABs (GTPases) play an important role in fruit development and ripening through vesicle trafficking to cell wall [117]. They also help to mediate the Fe acquisition needed for plant growth and development. The expression of a gene encoding the rice small GTPase, OsRAB6a was rapidly and transiently up-regulated by Fe deficiency. In Fe-sufficient medium, there were no differences in growth and development among the OsRAB6a-overexpression, OsRAB6a-RNAi (RNA interference) and wild-type plants, but when grown in Fe-deficient medium, plants overexpressing OsRAB6a showed greater tolerance to the deficiency in comparison with OsRab6a-RNAi and wild- type plants, exhibiting larger root systems than wild-type and RNAi plants. In both wild-type and transgenic rice plants, exposure to Fe-deficient medium led to up-regulation of OsIRO2, OsIRT1, OsNAS1 and OsNAS2 correlated with the expression pattern of OsRAB6a. These findings suggest that OsRAB6a plays an important role in the regulation of Fe acquisition in rice plants both by modulating physiological processes involved in Fe acquisition and by promoting an expanded root system architecture in response to Fe-deficient medium [118].
The expression of a rice RAB7 gene was differentially regulated by various environmental stimuli such as cold, NaCl, dehydration, and ABA [60]. The transgenic plants overexpressing AtRAB7 showed increased tolerance to salt and osmotic stresses and reduced accumulation of reactive oxygen species during salt stress [61]. Transgenic tobacco constitutively overexpressing PgRAB7 (from Pennisetum gluacum) also showed tolerance to both hyperosmotic and hyperionic stress [65] and those overexpressing a RAB7 from Prosopis juliflora (PjRAB7) showed enhanced salt stress tolerance [119]. When RAB7 from O. sativa was ectopically expressed in Escherichia coli it conferred tolerance to heat, cold and salt stress [87]. Overexpression of OsRAB7 enhanced tolerance to salt stress in transgenic rice plants, further supporting the hypothesis that vacuolar trafficking is important for salt tolerance in plants [120]. Consistent with this result, transgenic O. sativa plants constitutively overexpressing PgRAB7 also showed tolerance to both drought and salinity stress. Rice plants overexpressing PgRab7 under NaCl stress showed higher seed germination, survival and better growth as compared to wild-type plants [66]. A recent report found that overexpression of the OsRAB7 gene that improves drought and heat tolerance also increases grain yield in transgenic rice by modulating osmolytes, antioxidants and the expression of abiotic stress-responsive genes [121]. This finding adds even stronger support for the importance of RAB7 in abiotic stress tolerance.
Studies on a novel RAB GTPase protein in Arabidopsis, CPRABA5e, which is chloroplast localized, suggest that it plays a role in development, stress responses, and photosynthesis related processes. Infact, it has been demonstrated to function in transport to and from thylakoids, similar to cytosolic RAB proteins which function in vesicle transport [122]. A study in Arabidopsis indicates that RABA1 GTPases (a RAB11 member) is involved in transport between the trans-Golgi network and the PM, and is required for salinity stress tolerance [52]. Overexpression of the constitutive activation mutant PtRABE1b (Q74L) confers salt tolerance in Populus trichocarpa through larger root number and better root growth. In PtRABE1b (Q74L) overexpression transgenic lines the transcript level of trafficking-related genes (Got1-like, KEU, p24, PHF1, SEC14, SKD1 and SYP61), a stress-related TF (bZIP60), an autophagy-related gene (ATG18a-1) and development-related genes (EPC1) were up-regulated [12]. A comprehensive study of the transcriptional profiles of different tissues and developmental stages of S. lycopersicum, in response to heat, drought and salt stress conditions, revealed that RABGAP genes associated with endocytic and pre-vacuolar trafficking are up-regulated in roots subjected to salt stress, suggesting that these genes play an important role during salt stress in tomato [21]. Overall these findings suggest that RAB proteins that are involved in transporting vesicles play an important role in plant tolerance to salinity, osmotic, cold and heat stress.
CONCLUSION
In this review, we present reports showing that endosomal trafficking plays an integral role in plants as it does in other eukaryotic organisms. An understanding of the importance of endosomal trafficking in plants is rapidly growing, but the molecular mechanism of this process is mostly unknown. In eukaryotes, endocytic trafficking is basically mediated by five gene families of the RAS superfamily of which, the RAB family is the largest. In plants, the RAB family is divided into eight subfamilies, and members of these families perform a wide variety of cellular functions, including protein trafficking from ER to Golgi and vacuolar and PVC/endosomal trafficking, which are important processes required for secretion and cytokinesis. Recently several studies have begun to functionally characterize the plant RABs and their role in diverse cellular processes. These studies show that RABs are important components of membrane trafficking involved in pollen and root tip growth, autophagy, and also in protecting plants from damage induced by biotic and abiotic stress conditions. However, the complete mechanism(s) by which RAB overexpression provides stress tolerance remains to be better understood. More detailed molecular and functional characterization of RABs will likely reveal the basis for their role in plant growth and development and in biotic and abiotic stress tolerance.
ACKNOWLEDGEMENTS
We thank Dr. Stanley J. Roux and Dr. Greg Clark, Department of Molecular Bioscience, The University of Texas at Austin, USA, for critical reading and correcting the manuscript.
AUTHORS’ CONTRIBUTIONS
All authors (MKT, RD, SS) have made a substantial, direct and intellectual contribution to the work and approved it for publication.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
MKT acknowledges support from CSIR, India in the form of Senior Research Associateship (Scientist Pool Scheme) Fellowship. SKS acknowledges support from SERB, India in the form of Distinguished Fellowship.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Bos J.L. Ras. In: Hall A., editor. GTPases. Oxford University Press. Oxford: 2000. pp. 67–88. [Google Scholar]
- 2.Rojas A.M., Fuentes G., Rausell A., Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012;196(2):189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore M.S. Ran and nuclear transport. J. Biol. Chem. 1998;273(36):22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Z.L., Yang Z. The Rop GTPase: an emerging signaling switch in plants. Plant Mol. Biol. 2000;44(1):1–9. doi: 10.1023/A:1006402628948. [DOI] [PubMed] [Google Scholar]
- 5.Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. J. Cell Sci. 2005;118(Pt 5):843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 6.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 7.Grosshans B.L., Ortiz D., Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen E., Cheung A.Y., Ueda T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 2008;147(4):1516–1526. doi: 10.1104/pp.108.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutherford S., Moore I. The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 2002;5(6):518–528. doi: 10.1016/S1369-5266(02)00307-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Hill D.R., Sylvester A.W. Diversification of the RAB guanosine triphosphatase family in dicots and monocots. J. Integr. Plant Biol. 2007;49:1129–1141. doi: 10.1111/j.1672-9072.2007.00520.x. [DOI] [Google Scholar]
- 11.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Li Y., Liu B., Wang L., Zhang L., Hu J., Chen J., Zheng H., Lu M. Characterization of the Populus Rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 2018;18(1):124. doi: 10.1186/s12870-018-1342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazar T., Götte M., Gallwitz D. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem. Sci. 1997;22(12):468–472. doi: 10.1016/S0968-0004(97)01150-X. [DOI] [PubMed] [Google Scholar]
- 14.Calero M., Collins R.N. Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem. Biophys. Res. Commun. 2002;290(2):676–681. doi: 10.1006/bbrc.2001.6242. [DOI] [PubMed] [Google Scholar]
- 15.Gallegos M.E., Balakrishnan S., Chandramouli P., Arora S., Azameera A., Babushekar A., Bargoma E., Bokhari A., Chava S.K., Das P., Desai M., Decena D., Saramma S.D., Dey B., Doss A.L., Gor N., Gudiputi L., Guo C., Hande S., Jensen M., Jones S., Jones N., Jorgens D., Karamchedu P., Kamrani K., Kolora L.D., Kristensen L., Kwan K., Lau H., Maharaj P., Mander N., Mangipudi K., Menakuru H., Mody V., Mohanty S., Mukkamala S., Mundra S.A., Nagaraju S., Narayanaswamy R., Ndungu-Case C., Noorbakhsh M., Patel J., Patel P., Pendem S.V., Ponakala A., Rath M., Robles M.C., Rokkam D., Roth C., Sasidharan P., Shah S., Tandon S., Suprai J., Truong T.Q., Uthayaruban R., Varma A., Ved U., Wang Z., Yu Z. The C. elegans rab family: identification, classification and toolkit construction. PLoS One. 2012;7(11):e49387. doi: 10.1371/journal.pone.0049387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira-Leal J.B., Seabra M.C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001;313(4):889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal P., Reddy M.K., Sopory S.K., Agarwal P.K. Plant Rabs: Characterization, functional diversity, and role in stress tolerance. Plant Mol. Biol. Report. 2009;27:417–430. doi: 10.1007/s11105-009-0100-9. [DOI] [Google Scholar]
- 18.Urano D., Jones A.M. Heterotrimeric G protein-coupled signaling in plants. Annu. Rev. Plant Biol. 2014;65:365–384. doi: 10.1146/annurev-arplant-050213-040133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rehman R.U., Di Sansebastiano G.P. Plant Rab GTPases in membrane trafficking and signaling. In: Hakeem K., Rehman R., Tahir I., editors. Plant Signaling: Understanding the Molecular Crosstalk. New Delhi, India: Springer; 2014. pp. 51–73. [DOI] [Google Scholar]
- 20.Ku Y.S., Sintaha M., Cheung M.Y., Lam H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018;19(10):E3206. doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madrid-Espinoza J., Salinas-Cornejo J., Ruiz-Lara S. The RabGAP gene family in tomato (Solanum lycopersicum) and wild relatives: Identification, interaction networks, and transcriptional analysis during plant development and in response to salt stress. Genes (Basel) 2019;10(9):E638. doi: 10.3390/genes10090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren G., Mellman I. “Protein trafficking between membranes”. In: Lewin B., editor. Cell. Sudbury: Jones & Bartlett; 2006. pp. 153–204. [Google Scholar]
- 23.Saito C., Ueda T. Chapter 4: functions of RAB and SNARE proteins in plant life. Int. Rev. Cell Mol. Biol. 2009;274:183–233. doi: 10.1016/S1937-6448(08)02004-2. [DOI] [PubMed] [Google Scholar]
- 24.Yorimitsu T., Sato K., Takeuchi M. Molecular mechanisms of Sar/Arf GTPases in vesicular trafficking in yeast and plants. Front. Plant Sci. 2014;5:411. doi: 10.3389/fpls.2014.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischoff F., Molendijk A., Rajendrakumar C.S., Palme K. GTP-binding proteins in plants. Cell. Mol. Life Sci. 1999;55(2):233–256. doi: 10.1007/s000180050287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81(1):153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z. Small GTPases: versatile signaling switches in plants. Plant Cell. 2002;14(Suppl.):S375–S388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernoud V., Horton A.C., Yang Z., Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131(3):1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemichez E., Wu Y., Sanchez J.P., Mettouchi A., Mathur J., Chua N.H. Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 2001;15(14):1808–1816. doi: 10.1101/gad.900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feiguelman G., Fu Y., Yalovsky S. ROP GTPases structure-function and signaling pathways. Plant Physiol. 2018;176(1):57–79. doi: 10.1104/pp.17.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke P.R., Zhang C. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 2001;11(9):366–371. doi: 10.1016/S0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- 32.Matozaki T., Nakanishi H., Takai Y. Small G-protein networks: their crosstalk and signal cascades. Cell. Signal. 2000;12(8):515–524. doi: 10.1016/S0898-6568(00)00102-9. [DOI] [PubMed] [Google Scholar]
- 33.Tripathy M.K., Reddy M.K., Deswal R., Sopory S.K. Muralidharan K and Siddiq EA, eds. 2013. International dialogue on perception and prospects of designer rice. Society for advancement of rice research, Directorate of rice research, Hyderabad 500030, India; 2013. Towards developing transgenic rice for salinity and drought tolerance: role of Rab7. pp. 228–237. [Google Scholar]
- 34.Dirac-Svejstrup A.B., Sumizawa T., Pfeffer S.R. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J. 1997;16(3):465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivars U., Aivazian D., Pfeffer S.R. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425(6960):856–859. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- 36.Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12(5):671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Markgraf D.F., Peplowska K., Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581(11):2125–2130. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 38.Novick P., Medkova M., Dong G., Hutagalung A., Reinisch K., Grosshans B. Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem. Soc. Trans. 2006;34(Pt 5):683–686. doi: 10.1042/BST0340683. [DOI] [PubMed] [Google Scholar]
- 39.Benmerah A. Endocytosis: signaling from endocytic membranes to the nucleus. Curr. Biol. 2004;14(8):R314–R316. doi: 10.1016/j.cub.2004.03.053. [DOI] [PubMed] [Google Scholar]
- 40.Horazdovsky B. Endosomal protein traffic meets nuclear signal transduction head on. Dev. Cell. 2004;6(2):161–162. doi: 10.1016/S1534-5807(04)00035-8. [DOI] [PubMed] [Google Scholar]
- 41.Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler-Joseph S., Habermann B., Wilm M., Parton R.G., Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116(3):445–456. doi: 10.1016/S0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 42.Park Y.S., Song O., Kwak J.M., Hong S.W., Lee H.H., Nam H.G. Functional complementation of a yeast vesicular transport mutation ypt1-1 by a Brassica napus cDNA clone encoding a small GTP-binding protein. Plant Mol. Biol. 1994;26(6):1725–1735. doi: 10.1007/BF00019487. [DOI] [PubMed] [Google Scholar]
- 43.Kim W.Y., Cheong N.E., Lee D.C., Lee K.O., Je D.Y., Bahk J.D., Cho M.J., Lee S.Y. Isolation of an additional soybean cDNA encoding Ypt/Rab-related small GTP-binding protein and its functional comparison to Sypt using a yeast ypt1-1 mutant. Plant Mol. Biol. 1996;31(4):783–792. doi: 10.1007/BF00019466. [DOI] [PubMed] [Google Scholar]
- 44.Batoko H., Zheng H.Q., Hawes C., Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell. 2000;12(11):2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheung A.Y., Chen C.Y., Glaven R.H., de Graaf B.H., Vidali L., Hepler P.K., Wu H.M. Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell. 2002;14(4):945–962. doi: 10.1105/tpc.000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bednarek S.Y., Reynolds T.L., Schroeder M., Grabowski R., Hengst L., Gallwitz D., Raikhel N.V. A small GTP-binding protein from Arabidopsis thaliana functionally complements the yeast YPT6 null mutant. Plant Physiol. 1994;104(2):591–596. doi: 10.1104/pp.104.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latijnhouwers M., Gillespie T., Boevink P., Kriechbaumer V., Hawes C., Carvalho C.M. Localization and domain characterization of Arabidopsis golgin candidates. J. Exp. Bot. 2007;58(15-16):4373–4386. doi: 10.1093/jxb/erm304. [DOI] [PubMed] [Google Scholar]
- 48.Speth E.B., Imboden L., Hauck P., He S.Y. Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol. 2009;149(4):1824–1837. doi: 10.1104/pp.108.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng H., Camacho L., Wee E., Batoko H., Legen J., Leaver C.J., Malhó R., Hussey P.J., Moore I. A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell. 2005;17(7):2020–2036. doi: 10.1105/tpc.105.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borg S., Brandstrup B., Jensen T.J., Poulsen C. Identification of new protein species among 33 different small GTP-binding proteins encoded by cDNAs from Lotus japonicus, and expression of corresponding mRNAs in developing root nodules. Plant J. 1997;11(2):237–250. doi: 10.1046/j.1365-313X.1997.11020237.x. [DOI] [PubMed] [Google Scholar]
- 51.Stenmark H., Olkkonen V.M. The Rab GTPase family. Genome Biol. 2001;2(5):S3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asaoka R., Uemura T., Ito J., Fujimoto M., Ito E., Ueda T., Nakano A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013;73(2):240–249. doi: 10.1111/tpj.12023. [DOI] [PubMed] [Google Scholar]
- 53.de Graaf B.H., Cheung A.Y., Andreyeva T., Levasseur K., Kieliszewski M., Wu H.M. Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell. 2005;17(9):2564–2579. doi: 10.1105/tpc.105.033183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chow C.M., Neto H., Foucart C., Moore I. Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell. 2008;20(1):101–123. doi: 10.1105/tpc.107.052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preuss M.L., Serna J., Falbel T.G., Bednarek S.Y., Nielsen E. The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell. 2004;16(6):1589–1603. doi: 10.1105/tpc.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preuss M.L., Schmitz A.J., Thole J.M., Bonner H.K., Otegui M.S., Nielsen E. A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J. Cell Biol. 2006;172(7):991–998. doi: 10.1083/jcb.200508116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inaba T., Nagano Y., Nagasaki T., Sasaki Y. Distinct localization of two closely related Ypt3/Rab11 proteins on the trafficking pathway in higher plants. J. Biol. Chem. 2002;277(11):9183–9188. doi: 10.1074/jbc.M111491200. [DOI] [PubMed] [Google Scholar]
- 58.Ueda T., Anai T., Tsukaya H., Hirata A., Uchimiya H. Characterization and subcellular localization of a small GTP-binding protein (Ara-4) from Arabidopsis: conditional expression under control of the promoter of the gene for heat-shock protein HSP81-1. Mol. Gen. Genet. 1996;250(5):533–539. doi: 10.1007/BF02174441. a. [DOI] [PubMed] [Google Scholar]
- 59.Ueda T., Matsuda N., Anai T., Tsukaya H., Uchimiya H., Nakano A. An Arabidopsis gene isolated by a novel method for detecting genetic interaction in yeast encodes the GDP dissociation inhibitor of Ara4 GTPase. Plant Cell. 1996;8(11):2079–2091. doi: 10.1105/tpc.8.11.2079. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nahm M.Y., Kim S.W., Yun D., Lee S.Y., Cho M.J., Bahk J.D. Molecular and biochemical analyses of OsRab7, a rice Rab7 homolog. Plant Cell Physiol. 2003;44(12):1341–1349. doi: 10.1093/pcp/pcg163. [DOI] [PubMed] [Google Scholar]
- 61.Mazel A., Leshem Y., Tiwari B.S., Levine A. Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol. 2004;134(1):118–128. doi: 10.1104/pp.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saito C., Ueda T., Abe H., Wada Y., Kuroiwa T., Hisada A., Furuya M., Nakano A. A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 2002;29(3):245–255. doi: 10.1046/j.0960-7412.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 63.Carter C., Pan S., Zouhar J., Avila E.L., Girke T., Raikhel N.V. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;16(12):3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hara-Nishimura I., Shimada T., Hatano K., Takeuchi Y., Nishimura M. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell. 1998;10(5):825–836. doi: 10.1105/tpc.10.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agarwal P.K., Agarwal P., Jain P., Jha B., Reddy M.K., Sopory S.K. Constitutive overexpression of a stress-inducible small GTP-binding protein PgRab7 from Pennisetum glaucum enhances abiotic stress tolerance in transgenic tobacco. Plant Cell Rep. 2008;27(1):105–115. doi: 10.1007/s00299-007-0446-0. [DOI] [PubMed] [Google Scholar]
- 66.Tripathy M.K., Tiwari B.S., Reddy M.K., Deswal R., Sopory S.K. Ectopic expression of PgRab7 in rice plants (Oryza sativa L.) results in differential tolerance at the vegetative and seed setting stage during salinity and drought stress. Protoplasma. 2017;254(1):109–124. doi: 10.1007/s00709-015-0914-2. [DOI] [PubMed] [Google Scholar]
- 67.Rho S.H., Heo J.B., Bang W.Y., Hwang S.M., Nahm M.Y., Kwon H.J., et al. The role of OsPRA1 in vacuolar trafficking by OsRab GTPases in plant system. Plant Sci. 2009;177:411–417. doi: 10.1016/j.plantsci.2009.07.003. [DOI] [Google Scholar]
- 68.Heo J.B., Bang W.Y., Kim S.W., Hwang S.M., Son Y.S., Im C.H., Acharya B.R., Kim C.W., Kim S.W., Lee B.H., Bahk J.D. OsPRA1 plays a significant role in targeting of OsRab7 into the tonoplast via the prevacuolar compartment during vacuolar trafficking in plant cells. Planta. 2010;232(4):861–871. doi: 10.1007/s00425-010-1226-6. [DOI] [PubMed] [Google Scholar]
- 69.Bolte S., Schiene K., Dietz K.J. Characterization of a small GTP-binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol. Biol. 2000;42(6):923–936. doi: 10.1023/A:1006449715236. [DOI] [PubMed] [Google Scholar]
- 70.Ueda T., Yamaguchi M., Uchimiya H., Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 2001;20(17):4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W., Zou W., Zhao D., Yan J., Zhu Z., Lu J., Wang X. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 2009;136(14):2445–2455. doi: 10.1242/dev.035949. [DOI] [PubMed] [Google Scholar]
- 72.Ueda T., Uemura T., Sato M.H., Nakano A. Functional differentiation of endosomes in Arabidopsis cells. Plant J. 2004;40(5):783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- 73.Ito E., Ebine K., Choi S.W., Ichinose S., Uemura T., Nakano A., Ueda T. Integration of two RAB5 groups during endosomal transport in plants. eLife. 2018;7:7. doi: 10.7554/eLife.34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minamino N., Ueda T. RAB GTPases and their effectors in plant endosomal transport. Curr. Opin. Plant Biol. 2019;52:61–68. doi: 10.1016/j.pbi.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 75.Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA. 2005;102(34):12276–12281. doi: 10.1073/pnas.0502060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dhonukshe P., Baluska F., Schlicht M., Hlavacka A., Samaj J., Friml J., Gadella T.W., Jr Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev. Cell. 2006;10(1):137–150. doi: 10.1016/j.devcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Kotzer A.M., Brandizzi F., Neumann U., Paris N., Moore I., Hawes C. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 2004;117(Pt 26):6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- 78.Sohn E.J., Kim E.S., Zhao M., Kim S.J., Kim H., Kim Y.W., Lee Y.J., Hillmer S., Sohn U., Jiang L., Hwang I. Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell. 2003;15(5):1057–1070. doi: 10.1105/tpc.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haas T.J., Sliwinski M.K., Martínez D.E., Preuss M., Ebine K., Ueda T., Nielsen E., Odorizzi G., Otegui M.S. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell. 2007;19(4):1295–1312. doi: 10.1105/tpc.106.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lütcke A., Parton R.G., Murphy C., Olkkonen V.M., Dupree P., Valencia A., Simons K., Zerial M. Cloning and subcellular localization of novel rab proteins reveals polarized and cell type-specific expression. J. Cell Sci. 1994;107(Pt 12):3437–3448. doi: 10.1242/jcs.107.12.3437. [DOI] [PubMed] [Google Scholar]
- 81.Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 2005;118(Pt 12):2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 82.Vazquez-Martinez R., Cruz-Garcia D., Duran-Prado M., Peinado J.R., Castaño J.P., Malagon M.M. Rab18 inhibits secretory activity in neuroendocrine cells by interacting with secretory granules. Traffic. 2007;8(7):867–882. doi: 10.1111/j.1600-0854.2007.00570.x. [DOI] [PubMed] [Google Scholar]
- 83.Segev N. Ypt/rab gtpases: regulators of protein trafficking. Sci. STKE. 2001;2001(100):re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 84.Qi X., Zheng H. Functional analysis of small Rab GTPases in cytokinesis in Arabidopsis thaliana. Methods Mol. Biol. 2013;1043:103–112. doi: 10.1007/978-1-62703-532-3_11. [DOI] [PubMed] [Google Scholar]
- 85.Yao H.Y., Xue H.W. Signals and mechanisms affecting vesicular trafficking during root growth. Curr. Opin. Plant Biol. 2011;14(5):571–579. doi: 10.1016/j.pbi.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 86.Szumlanski A.L., Nielsen E. The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell. 2009;21(2):526–544. doi: 10.1105/tpc.108.060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng J., Ilarslan H., Wurtele E.S., Bassham D.C. AtRabD2b and AtRabD2c have overlapping functions in pollen development and pollen tube growth. BMC Plant Biol. 2011;11:25. doi: 10.1186/1471-2229-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kato N., He H., Steger A.P. A systems model of vesicle trafficking in Arabidopsis pollen tubes. Plant Physiol. 2010;152(2):590–601. doi: 10.1104/pp.109.148700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Kasmi F., Pacher T., Strompen G., Stierhof Y.D., Müller L.M., Koncz C., Mayer U., Jürgens G. Arabidopsis SNARE protein SEC22 is essential for gametophyte development and maintenance of Golgi-stack integrity. Plant J. 2011;66(2):268–279. doi: 10.1111/j.1365-313X.2011.04487.x. [DOI] [PubMed] [Google Scholar]
- 90.Kusano H., Testerink C., Vermeer J.E., Tsuge T., Shimada H., Oka A., Munnik T., Aoyama T. The Arabidopsis Phosphatidylinositol Phosphate 5-Kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell. 2008;20(2):367–380. doi: 10.1105/tpc.107.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thole J.M., Vermeer J.E., Zhang Y., Gadella T.W., Jr, Nielsen E. Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell. 2008;20(2):381–395. doi: 10.1105/tpc.107.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Routier-Kierzkowska A.L., Weber A., Kochova P., Felekis D., Nelson B.J., Kuhlemeier C., Smith R.S. Cellular force microscopy for in vivo measurements of plant tissue mechanics. Plant Physiol. 2012;158(4):1514–1522. doi: 10.1104/pp.111.191460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kirchhelle C., Chow C.M., Foucart C., Neto H., Stierhof Y.D., Kalde M., Walton C., Fricker M., Smith R.S., Jérusalem A., Irani N., Moore I. The specification of geometric edges by a plant Rab GTPase is an essential cell-patterning principle during organogenesis in Arabidopsis. Dev. Cell. 2016;36(4):386–400. doi: 10.1016/j.devcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ambrose C., Allard J.F., Cytrynbaum E.N., Wasteneys G.O. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2011;2:430. doi: 10.1038/ncomms1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rahni R., Birnbaum K.D. Plant cell shape: Trafficking gets edgy. Dev. Cell. 2016;36(4):353–354. doi: 10.1016/j.devcel.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 96.He M., Lan M., Zhang B., Zhou Y., Wang Y., Zhu L., Yuan M., Fu Y. Rab-H1b is essential for trafficking of cellulose synthase and for hypocotyl growth in Arabidopsis thaliana. J. Integr. Plant Biol. 2018;60(11):1051–1069. doi: 10.1111/jipb.12694. [DOI] [PubMed] [Google Scholar]
- 97.Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 98.Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E.L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117(Pt 20):4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 99.Rojo E., Gillmor C.S., Kovaleva V., Somerville C.R., Raikhel N.V. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell. 2001;1(2):303–310. doi: 10.1016/S1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- 100.Goh T., Uchida W., Arakawa S., Ito E., Dainobu T., Ebine K., Takeuchi M., Sato K., Ueda T., Nakano A. VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell. 2007;19(11):3504–3515. doi: 10.1105/tpc.107.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwon S.I., Cho J.H., Bae K., Jung H.J., Jin H.C., Park O.K. Role of an Arabidopsis Rab GTPase RabG3b in pathogen response and leaf senescence. J. Plant Biol. 2009;52:79–87. doi: 10.1007/s12374-009-9011-4. [DOI] [Google Scholar]
- 102.Falchi R., Cipriani G., Marrazzo T., Nonis A., Vizzotto G., Ruperti B. Identification and differential expression dynamics of peach small GTPases encoding genes during fruit development and ripening. J. Exp. Bot. 2010;61(10):2829–2842. doi: 10.1093/jxb/erq116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y., Ren Y., Liu X., Jiang L., Chen L., Han X., Jin M., Liu S., Liu F., Lv J., Zhou K., Su N., Bao Y., Wan J. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J. 2010;64(5):812–824. doi: 10.1111/j.1365-313X.2010.04370.x. [DOI] [PubMed] [Google Scholar]
- 104.Cheon C.I., Lee N.G., Siddique A.B., Bal A.K., Verma D.P. Roles of plant homologs of Rab1p and Rab7p in the biogenesis of the peribacteroid membrane, a subcellular compartment formed de novo during root nodule symbiosis. EMBO J. 1993;12(11):4125–4135. doi: 10.1002/j.1460-2075.1993.tb06096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Catalano C.M., Czymmek K.J., Gann J.G., Sherrier D.J. Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta. 2007;225(3):541–550. doi: 10.1007/s00425-006-0369-y. [DOI] [PubMed] [Google Scholar]
- 106.Limpens E., Ivanov S., van Esse W., Voets G., Fedorova E., Bisseling T. Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell. 2009;21(9):2811–2828. doi: 10.1105/tpc.108.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tomczynska I., Stumpe M., Mauch F. A conserved RxLR effector interacts with host RABA-type GTPases to inhibit vesicle-mediated secretion of antimicrobial proteins. Plant J. 2018;95(2):187–203. doi: 10.1111/tpj.13928. [DOI] [PubMed] [Google Scholar]
- 108.Brandizzi F. Rabs: targets in the plant-pathogen battlefield. Plant J. 2018;95(2):185–186. doi: 10.1111/tpj.13997. [DOI] [PubMed] [Google Scholar]
- 109.Erbs G., Molinaro A., Dow J.M., Newman M.A. Lipopolysaccharides and plant innate immunity. Subcell. Biochem. 2010;53:387–403. doi: 10.1007/978-90-481-9078-2_17. [DOI] [PubMed] [Google Scholar]
- 110.Liu F., Guo J., Bai P., Duan Y., Wang X., Cheng Y., Feng H., Huang L., Kang Z. Wheat TaRab7 GTPase is part of the signaling pathway in responses to stripe rust and abiotic stimuli. PLoS One. 2012;7(5):e37146. doi: 10.1371/journal.pone.0037146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang X., Ma N., Yang L., Zheng Y., Zhen Z., Li Q., Xie M., Li J., Zhang K.Q., Yang J. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2018;102(10):4601–4613. doi: 10.1007/s00253-018-8929-1. [DOI] [PubMed] [Google Scholar]
- 112.Inada N., Betsuyaku S., Shimada T.L., Ebine K., Ito E., Kutsuna N., Hasezawa S., Takano Y., Fukuda H., Nakano A., Ueda T. Modulation of Plant RAB GTPase-Mediated Membrane Trafficking Pathway at the Interface Between Plants and Obligate Biotrophic Pathogens. Plant Cell Physiol. 2016;57(9):1854–1864. doi: 10.1093/pcp/pcw107. [DOI] [PubMed] [Google Scholar]
- 113.O’Mahony P.J., Oliver M.J. Characterization of a desiccation-responsive small GTP-binding protein (Rab2) from the desiccation- tolerant grass Sporobolus stapfianus. Plant Mol. Biol. 1999;39(4):809–821. doi: 10.1023/A:1006183431854. [DOI] [PubMed] [Google Scholar]
- 114.Ebine K., Fujimoto M., Okatani Y., Nishiyama T., Goh T., Ito E., Dainobu T., Nishitani A., Uemura T., Sato M.H., Thordal-Christensen H., Tsutsumi N., Nakano A., Ueda T. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 2011;13(7):853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 115.Yin C., Karim S., Zhang H., Aronsson H. Arabidopsis RabF1 (ARA6) is involved in salt stress and dark-induced senescence (DIS). Int. J. Mol. Sci. 2017 doi: 10.3390/ijms18020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Z.L., Luo C., Dong L., Van Toan C., Wei P.X., He X.H. Molecular characterization and expression analysis of a GTP-binding protein (MiRab5) in Mangifera indica. Gene. 2014;540(1):86–91. doi: 10.1016/j.gene.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 117.Lawson T., Mayes S., Lycett G.W., Chin C.F. Plant Rabs and the role in fruit ripening. Biotechnol. Genet. Eng. Rev. 2018;34(2):181–197. doi: 10.1080/02648725.2018.1482092. [DOI] [PubMed] [Google Scholar]
- 118.Yang A., Zhang W.H. A small GTPase, OsRab6a, is involved in the regulation of iron homeostasis in rice. Plant Cell Physiol. 2016;57(6):1271–1280. doi: 10.1093/pcp/pcw073. [DOI] [PubMed] [Google Scholar]
- 119.George S., Parida A. Over-expression of a Rab family GTPase from phreatophyte Prosopis juliflora confers tolerance to salt stress on transgenic tobacco. Mol. Biol. Rep. 2011;38(3):1669–1674. doi: 10.1007/s11033-010-0278-9. [DOI] [PubMed] [Google Scholar]
- 120.Peng X., Ding X., Chang T., Wang Z., Liu R., Zeng X., Cai Y., Zhu Y. Overexpression of a Vesicle Trafficking Gene, OsRab7, enhances salt tolerance in rice. Scientific World J. 2014;2014:483526. doi: 10.1155/2014/483526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.El-Esawi M.A., Alayafi A.A. Overexpression of rice Rab7 gene improves drought and heat tolerance and increases grain yield in rice (Oryza sativa L.). Genes (Basel) 2019;10(1):E56. doi: 10.3390/genes10010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karim S., Alezzawi M., Garcia-Petit C., Solymosi K., Khan N.Z., Lindquist E., Dahl P., Hohmann S., Aronsson H. A novel chloroplast localized Rab GTPase protein CPRabA5e is involved in stress, development, thylakoid biogenesis and vesicle transport in Arabidopsis. Plant Mol. Biol. 2014;84(6):675–692. doi: 10.1007/s11103-013-0161-x. [DOI] [PubMed] [Google Scholar]