Abstract
Introduction
Pathogenic DNA variants in the GLI-Kruppel family member 3 (GLI3) gene are known to cause multiple syndromes: for example, Greig syndrome, preaxial polydactyly-type 4 (PPD4) and Pallister-Hall syndrome. Out of these, Pallister-Hall is a different entity, but the distinction between Greig syndrome and PPD4 is less evident. Using latent class analysis (LCA), our study aimed to investigate the correlation between reported limb anomalies and the reported GLI3 variants in these GLI3-mediated polydactyly syndromes. We identified two subclasses of limb anomalies that relate to the underlying variant.
Methods
Both local and published cases were included for analysis. The presence of individual limb phenotypes was dichotomised and an exploratory LCA was performed. Distribution of phenotypes and genotypes over the classes were explored and subsequently the key predictors of latent class membership were correlated to the different clustered genotypes.
Results
297 cases were identified with 127 different variants in the GLI3 gene. A two-class model was fitted revealing two subgroups of patients with anterior versus posterior anomalies. Posterior anomalies were observed in cases with truncating variants in the activator domain (postaxial polydactyly; hand, OR: 12.7; foot, OR: 33.9). Multivariate analysis supports these results (Beta: 1.467, p=0.013 and Beta: 2.548, p<0.001, respectively). Corpus callosum agenesis was significantly correlated to these variants (OR: 8.8, p<0.001).
Conclusion
There are two distinct phenotypes within the GLI3-mediated polydactyly population: anteriorly and posteriorly orientated. Variants that likely produce haploinsufficiency are associated with anterior phenotypes. Posterior phenotypes are associated with truncating variants in the activator domain. Patients with these truncating variants have a greater risk for corpus callosum anomalies.
Keywords: developmental, molecular genetics, clinical genetics, genetic screening/counselling
Introduction
GLI-Kruppel family member 3 (GLI3) encodes for a zinc finger transcription factor which plays a key role in the sonic hedgehog (SHH) signalling pathway essential in both limb and craniofacial development.1 2 In hand development, SHH is expressed in the zone of polarising activity (ZPA) on the posterior side of the handplate. The ZPA expresses SHH, creating a gradient of SHH from the posterior to the anterior side of the handplate. In the presence of SHH, full length GLI3-protein is produced (GLI3A), whereas absence of SHH causes cleavage of GLI3 into its repressor form (GLI3R).3 4 Abnormal expression of this SHH/GLI3R gradient can cause both preaxial and postaxial polydactyly.2
Concordantly, pathogenic DNA variants in the GLI3 gene are known to cause multiple syndromes with craniofacial and limb involvement, such as: acrocallosal syndrome5 (OMIM: 200990), Greig cephalopolysyndactyly syndrome6 (OMIM: 175700) and Pallister-Hall syndrome7 (OMIM: 146510). Also, in non-syndromic polydactyly, such as preaxial polydactyly-type 4 (PPD4, OMIM: 174700),8 pathogenic variants in GLI3 have been described. Out of these diseases, Pallister-Hall syndrome is the most distinct entity, defined by the presence of central polydactyly and hypothalamic hamartoma.9 The other GLI3 syndromes are defined by the presence of preaxial and/or postaxial polydactyly of the hand and feet with or without syndactyly (Greig syndrome, PPD4). Also, various mild craniofacial features such as hypertelorism and macrocephaly can occur. Pallister-Hall syndrome is caused by truncating variants in the middle third of the GLI3 gene.10–12 The truncation of GLI3 causes an overexpression of GLI3R, which is believed to be the key difference between Pallister-Hall and the GLI3-mediated polydactyly syndromes.9 11 Although multiple attempts have been made, the clinical and genetic distinction between the GLI3-mediated polydactyly syndromes is less evident. This has for example led to the introduction of subGreig and the formulation of an Oro-facial-digital overlap syndrome.10 Other authors, suggested that we should not regard these diseases as separate entities, but as a spectrum of GLI3-mediated polydactyly syndromes.13
Although phenotype/genotype correlation of the different syndromes has been cumbersome, clinical and animal studies do provide evidence that distinct regions within the gene, could be related to the individual anomalies contributing to these syndromes. First, case studies show isolated preaxial polydactyly is caused by both truncating and non-truncating variants throughout the GLI3 gene, whereas in isolated postaxial polydactyly cases truncating variants at the C-terminal side of the gene are observed.12 14 These results suggest two different groups of variants for preaxial and postaxial polydactyly. Second, recent animal studies suggest that posterior malformations in GLI3-mediated polydactyly syndromes are likely related to a dosage effect of GLI3R rather than due to the influence of an altered GLI3A expression.15
Past attempts for phenotype/genotype correlation in GLI3-mediated polydactyly syndromes have directly related the diagnosed syndrome to the observed genotype.10–12 16 Focusing on individual hand phenotypes, such as preaxial and postaxial polydactyly and syndactyly might be more reliable because it prevents misclassification due to inconsistent use of syndrome definition. Subsequently, latent class analysis (LCA) provides the possibility to relate a group of observed variables to a set of latent, or unmeasured, parameters and thereby identifying different subgroups in the obtained dataset.17 As a result, LCA allows us to group different phenotypes within the GLI3-mediated polydactyly syndromes and relate the most important predictors of the grouped phenotypes to the observed GLI3 variants.
The aim of our study was to further investigate the correlation of the individual phenotypes to the genotypes observed in GLI3-mediated polydactyly syndromes, using LCA. Cases were obtained by both literature review and the inclusion of local clinical cases. Subsequently, we identified two subclasses of limb anomalies that relate to the underlying GLI3 variant. We provide evidence for two different phenotypic and genotypic groups with predominantly preaxial and postaxial hand and feet anomalies, and we specify those cases with a higher risk for corpus callosum anomalies.
Methods
Literature review
The Human Gene Mutation Database (HGMD Professional 2019) was reviewed to identify known pathogenic variants in GLI3 and corresponding phenotypes.18 All references were obtained and cases were included when they were diagnosed with either Greig or subGreig syndrome or PPD4.10–12 Pallister-Hall syndrome and acrocallosal syndrome were excluded because both are regarded distinct syndromes and rather defined by the presence of the non-hand anomalies, than the presence of preaxial or postaxial polydactyly.13 19 Isolated preaxial or postaxial polydactyly were excluded for two reasons: the phenotype/genotype correlations are better understood and both anomalies can occur sporadically which could introduce falsely assumed pathogenic GLI3 variants in the analysis. Additionally, cases were excluded when case-specific phenotypic or genotypic information was not reported or if these two could not be related to each other. Families with a combined phenotypic description, not reducible to individual family members, were included as one case in the analysis.
Clinical cases
The Sophia Children’s Hospital Database was reviewed for cases with a GLI3 variant. Within this population, the same inclusion criteria for the phenotype were valid. Relatives of the index patients were also contacted for participation in this study, when they showed comparable hand, foot, or craniofacial malformations or when a GLI3 variant was identified. Phenotypes of the hand, foot and craniofacial anomalies of the patients treated in the Sophia Children's Hospital were collected using patient documentation. Family members were identified and if possible, clinically verified. Alternatively, family members were contacted to verify their phenotypes. If no verification was possible, cases were excluded.
Phenotypes
The phenotypes of both literature cases and local cases were extracted in a similar fashion. The most frequently reported limb and craniofacial phenotypes were dichotomised. The dichotomised hand and foot phenotypes were preaxial polydactyly, postaxial polydactyly and syndactyly. Broad halluces or thumbs were commonly reported by authors and were dichotomised as a presentation of preaxial polydactyly. The extracted dichotomised craniofacial phenotypes were hypertelorism, macrocephaly and corpus callosum agenesis. All other phenotypes were registered, but not dichotomised.
Pathogenic GLI3 variants
All GLI3 variants were extracted and checked using Alamut Visual V.2.14. If indicated, variants were renamed according to standard Human Genome Variation Society nomenclature.20 Variants were grouped in either missense, frameshift, nonsense or splice site variants. In the group of frameshift variants, a subgroup with possible splice site effect were identified for subgroup analysis when indicated. Similarly, nonsense variants prone for nonsense mediated decay (NMD) and nonsense variants with experimentally confirmed NMD were identified.21 Deletions of multiple exons, CNVs and translocations were excluded for analysis. A full list of included mutations is available in the online supplementary materials.
jmedgenet-2020-106948supp001.pdf (152.1KB, pdf)
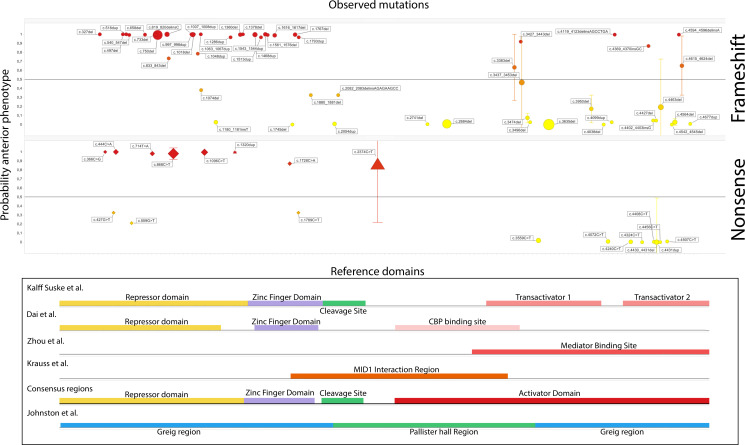
The location of the variant was compared with five known structural domains of the GLI3 gene: (1) repressor domain, (2) zinc finger domain, (3) cleavage site, (4) activator domain, which we defined as a concatenation of the separately identified transactivation zones, the CBP binding domain and the mediator binding domain (MBD) and (5) the MID1 interaction region domain.1 6 22–24 The boundaries of each of the domains were based on available literature (figure 1, exact locations available in the online supplementary materials). The boundaries used by different authors did vary, therefore a consensus was made.
Figure 1.
In this figure the posterior probability of an anterior phenotype is plotted against the location of the variant, stratified for the type of mutation that was observed. For better overview, only variants with a location effect were displayed. The full figure, including all variant types, can be found in the online supplementary figure 1. Each mutation is depicted as a dot, the size of the dot represents the number of observations for that variant. If multiple observations were made, the mean posterior odds and IQR are plotted. For the nonsense variants, variants that were predicted to produce nonsense mediated decay, are depicted using a triangle. Again, the size indicates the number of observations.
jmedgenet-2020-106948supp002.pdf (99.9KB, pdf)
Latent class analysis
To cluster phenotypes and relate those to the genotypes of the patients, an explorative analysis was done using LCA in R (R V.3.6.1 for Mac; polytomous variable LCA, poLCA V.1.4.1.). We used our LCA to detect the number of phenotypic subgroups in the dataset and subsequently predict a class membership for each case in the dataset based on the posterior probabilities.
In order to make a reliable prediction, only phenotypes that were sufficiently reported and/or ruled out were feasible for LCA, limiting the analysis to preaxial polydactyly, postaxial polydactyly and syndactyly of the hands and feet. Only full cases were included. To determine the optimal number of classes, we fitted a series of models ranging from a one-class to a six-class model. The optimal number of classes was based on the conditional Akaike information criterion (cAIC), the non adjusted and the sample-size adjusted Bayesian information criterion (BIC and aBIC) and the obtained entropy.25 The explorative LCA produces both posterior probabilities per case for both classes and predicted class membership. Using the predicted class membership, the phenotypic features per class were determined in a univariate analysis (χ2, SPSS V.25). Using the posterior probabilities on latent class (LC) membership, a scatter plot was created using the location of the variant on the x-axis and the probability of class membership on the y-axis for each of the types of variants (Tibco Spotfire V.7.14). Using these scatter plots, variants that give similar phenotypes were clustered.
Genotype/phenotype correlation
Because an LC has no clinical value, the correlation between genotypes and phenotypes was investigated using the predictor phenotypes and the clustered phenotypes. First, those phenotypes that contribute most to LC membership were identified. Second those phenotypes were directly related to the different types of variants (missense, nonsense, frameshift, splice site) and their clustered locations. Quantification of the relation was performed using a univariate analysis using a χ2 test. Because of our selection criteria, meaning patients at least have two phenotypes, a multivariate using a logistic regression analysis was used to detect the most significant predictors in the overall phenotype (SPSS V.25). Finally, we explored the relation of the clustered genotypes to the presence of corpus callosum agenesis, a rare malformation in GLI3-mediated polydactyly syndromes which cannot be readily diagnosed without additional imaging.
Results
We included 251 patients from the literature and 46 local patients,10–12 16 21 26–43 in total 297 patients from 155 different families with 127 different GLI3 variants, 32 of which were large deletions, CNVs or translocations. In six local cases, the exact variant could not be retrieved by status research.
The distribution of the most frequently observed phenotypes and variants are presented in table 1. Other recurring phenotypes included developmental delay (n=22), broad nasal root (n=23), frontal bossing or prominent forehead (n=16) and craniosynostosis (n=13), camptodactyly (n=8) and a broad first interdigital webspace of the foot (n=6).
Table 1.
Baseline phenotypes and genotypes of selected population
| Phenotypes | Affected/reported cases (n) | |
| Hand | Preaxial polydactyly | 124/294 |
| Postaxial polydactyly | 170/292 | |
| Syndactyly | 124/297 | |
| Foot | Preaxial polydactyly | 238/297 |
| Postaxial polydactyly | 70/295 | |
| Syndactyly | 193/297 | |
| Cranium | Macrocephaly | 85/228 |
| Hypertelorism | 92/237 | |
| Corpus callosum | 16/145 | |
| Genotypes | Cases (n) | |
| Included in analysis | Frameshift | 107 |
| Nonsense | 68 | |
| Missense | 60 | |
| Splice | 24 | |
| Excluded in analysis | CNV | 29 |
| Translocation | 3 | |
| No specific information on mutation | 6 | |
The LCA model was fitted using the six defined hand/foot phenotypes. Model fit indices for the LCA are displayed in table 2. Based on the BIC, a two-class model has the best fit for our data. The four-class model does show a gain in entropy, however with a higher BIC and loss of df. Therefore, based on the majority of performance statistics and the interpretability of the model, a two-class model was chosen. Table 3 displays the distribution of phenotypes and genotypes over the two classes.
Table 2.
Model fit indices for the one-class through six-class model evaluated in our LCA
| Number of classes | Log-likelihood | Residual df | BIC | aBIC | cAIC | Likelihood ratio | Entropy |
| 1 | −1072.0687 | 57 | 2178.316 | 2159.109 | 2184.316 | 299.59038 | – |
| 2 | −966.4844 | 50 | 2006.632 | 1965.407 | 2019.632 | 88.42178 | 0.765 |
| 3 | −949.9799 | 43 | 2013.288 | 1949.865 | 2033.288 | 55.41278 | 0.740 |
| 4 | −942.9999 | 36 | 2038.993 | 1953.372 | 2065.993 | 41.45279 | 0.952 |
| 5 | −937.2077 | 29 | 2067.074 | 1959.255 | 2101.074 | 29.86850 | 0.569 |
| 6 | −933.5159 | 22 | 2099.355 | 1969.338 | 2140.355 | 22.48488 | 0.716 |
BIC, Bayesian information criterion; LCA, latent class analysis.
Table 3.
Distribution of phenotypes and genotypes in the two latent classes (LC)
| LC 1/posterior phenotype | LC 2/anterior phenotype | ||
| Cases in LC (n) | 88 | 201 | |
| Mean probability of class membership | 0.91 (0.88–0.94) | 0.96 (0.95–0.97) | |
| Phenotypes | % of cases in class | ||
| Hand | Preaxial polydactyly | 15.91% | 52.74%* |
| Postaxial polydactyly | 96.59% | 40.80%* | |
| Syndactyly | 12.50% | 53.73%* | |
| Foot | Preaxial polydactyly | 45.45% | 95.52%* |
| Postaxial polydactyly | 69.32% | 1.49%* | |
| Syndactyly | 23.86% | 83.08%* | |
| Cranium | Macrocephaly | 29/60 | 54/162 |
| Hypertelorism | 23/56 | 68/177 | |
| Corpus callosum | 8/44 | 8/98 | |
| Genotypes | Cases (n) | ||
| Total | 85/88 | 173/201 | |
| Included mutations | Frameshift | 52 | 54 |
| Nonsense | 26 | 42 | |
| Missense | 6 | 54* | |
| Splice | 1 | 23* | |
*P<0.00.
Table 1 depicts the baseline phenotypes and genotypes in the obtained population. Note incomplete data especially in the cranium phenotypes. In total 259 valid genotypes were present. In total, 289 cases had complete data for all hand and foot phenotypes (preaxial polydactyly, postaxial polydactyly and syndactyly) and thus were available for LCA. Combined, for phenotype/genotype correlation 258 cases were available with complete genotypes and complete hand and foot phenotypes.
Table 2 depicts the model fit indices for all models that have been fitted to our data.
Table 3 depicts the distribution of phenotypes and genotypes over the two assigned LCs. Hand and foot phenotypes were used as input for the LCA, thus are all complete cases. Malformation of the cranium and genotypes do have missing cases. Note that for the LCA, full case description was required, resulting in eight cases due to incomplete phenotypes. Out of these eight, one also had a genotype that thus needed to be excluded. Missingness of genotypic data was higher in LC2, mostly due to CNVs (table 1).
In 54/60 cases, a missense variant produced a posterior phenotype. Likewise, splice site variants show the same phenotype in 23/24 cases (table 3). For both frameshift and nonsense variants, this relation is not significant (52 anterior vs 54 posterior and 26 anterior vs 42 posterior, respectively). Therefore, only for nonsense and frameshift variants the location of the variant was plotted against the probability for LC2 membership in figure 1. A full scatterplot of all variants is available in online supplementary figure 1.
Figure 1 reveals a pattern for these nonsense and frameshift variants that reveals that variants at the C-terminal of the gene predict anterior phenotypes. When relating the domains of the GLI3 protein to the observed phenotype, we observe that the majority of patients with a nonsense or frameshift variant in the repressor domain, the zinc finger domain or the cleavage site had a high probability of an LC2/anterior phenotype. This group contains all variants that are either experimentally determined to be subject to NMD (triangle marker in figure 1) or predicted to be subject to NMD (diamond marker in figure 1). Frameshift and nonsense variants in the activator domain result in high probability for an LC1/posterior phenotype. These variants will be further referred to as truncating variants in the activator domain.
The univariate relation of the individual phenotypes to these two groups of variants are estimated and presented in table 4. In our multivariate analysis, postaxial polydactyly of the foot and hand are the strongest predictors (Beta: 2.548, p<0001 and Beta: 1.47, p=0.013, respectively) for patients to have a truncating variant in the activator domain. Moreover, the effect sizes of preaxial polydactyly of the hand and feet (Beta: −0.797, p=0123 and −1.772, p=0.001) reveals that especially postaxial polydactyly of the foot is the dominant predictor for the genetic substrate of the observed anomalies.
Table 4.
Univariate and multivariate analysis of the phenotype/genotype correlation
| Univariate analysis | Multivariate analysis | ||||
| OR frameshift/nonsense mutation 5′ side of the zinc finger domain | Beta | P value | |||
| Phenotype | Hand | Preaxial polydactyly | 0.27 (CI: 0.14 – 0.54) | −0.797 | 0.123 |
| Postaxial polydactyly | 12.7 (CI: 5.2 – 31.0) | 1.469 | 0.013 | ||
| Syndactyly | 0.3 (CI: 0.16 – 0.57) | 0.505 | 0.338 | ||
| Foot | Preaxial polydactyly | 0.1 (CI: 0.032 – 0.14) | −1.772 | 0.001 | |
| Postaxial polydactyly | 33.9 (CI: 15.1 – 76.0) | 2.548 | <0.001 | ||
| Syndactyly | 0.1 (CI: 0.054 – 0.19) | −1.773 | <0.001 | ||
| Regression constant | −0.564 | 0.729 | |||
Table 4 shows exploration of the individual phenotypes on the genotype, both univariate and multivariate. The multivariate analysis corrects for the presence of multiple phenotypes in the underlying population.
Although the craniofacial anomalies could not be included in the LCA, the relation between the observed anomalies and the identified genetic substrates can be studied. The prevalence of hypertelorism was equally distributed over the two groups of variants (47/135 vs 21/47 respectively, p<0.229). However for corpus callosum agenesis and macrocephaly, there was a higher prevalence in patients with a truncating variant in the activator domain (3/75 vs 11/41, p<0.001; OR: 8.8, p<0.001) and 42/123 vs 24/48, p<0.05). Noteworthy is the fact that 11/14 cases with corpus callosum agenesis in the dataset had a truncating variant in the activator domain.
Discussion
In this report, we present new insights into the correlation between the phenotype and the genotype in patients with GLI3-mediated polydactyly syndromes. We illustrate that there are two LCs of patients, best predicted by postaxial polydactyly of the hand and foot for LC1, and the preaxial polydactyly of the hand and foot and syndactyly of the foot for LC2. Patients with postaxial phenotypes have a higher risk of having a truncating variant in the activator domain of the GLI3 gene which is also related to a higher risk of corpus callosum agenesis. These results suggest a functional difference between truncating variants on the N-terminal and the C-terminal side of the GLI3 cleavage site.
Previous attempts of phenotype to genotype correlation have not yet provided the clinical confirmation of these assumed mechanisms in the pathophysiology of GLI3-mediated polydactyly syndromes. Johnston et al have successfully determined the Pallister-Hall region in which truncating variants produce a Pallister-Hall phenotype rather than Greig syndrome.11 However, in their latest population study, subtypes of both syndromes were included to explain the full spectrum of observed malformations. In 2015, Demurger et al reported the higher incidence of corpus callosum agenesis in the Greig syndrome population with truncating mutations in the activator domain.12 Al-Qattan in his review summarises the concept of a spectrum of anomalies dependent on haplo-insufficiency (through different mechanisms) and repressor overexpression.13 However, he bases this theory mainly on reviewed experimental data. Our report is the first to provide an extensive clinical review of cases that substantiate the phenotypic difference between the two groups that could fit the suggested mechanisms. We agree with Al-Qattan et al that a variation of anomalies can be observed given any pathogenic variant in the GLI3 gene, but overall two dominant phenotypes are present: a population with predominantly preaxial anomalies and one with postaxial anomalies. The presence of preaxial or postaxial polydactyly and syndactyly is not mutually exclusive for one of these two subclasses; meaning that preaxial polydactyly can co-occur with postaxial polydactyly. However, truncating mutations in the activator domain produce a postaxial phenotype, as can be derived from the risk in table 4. The higher risk of corpus callosum agenesis in this population shows that differentiating between a preaxial phenotype and a postaxial phenotype, instead of between the different GLI3-mediated polydactyly syndromes, might be more relevant regarding diagnostics for corpus callosum agenesis.
We chose to use LCA as an exploratory tool only in our population for two reasons. First of all, LCA can be useful to identify subgroups, but there is no ‘true’ model or number of subgroups you can detect. The best fitting model can only be estimated based on the available measures and approximates the true subgroups that might be present. Second, LC membership assignment is a statistical procedure based on the posterior probability, with concordant errors of the estimation, rather than a clinical value that can be measured or evaluated. Therefore, we decided to use our LCA only in an exploratory tool, and perform our statistics using the actual phenotypes that predict LC membership and the associated genotypes. Overall, this method worked well to differentiate the two subgroups present in our dataset. However, outliers were observed. A qualitative analysis of these outliers is available in the online supplementary data.
The genetic substrate for the two phenotypic clusters can be discussed based on multiple experiments. Overall, we hypothesise two genetic clusters: one that is due to haploinsufficiency and one that is due to abnormal truncation of the activator. The hypothesised cluster of variants that produce haploinsufficiency is mainly based on the experimental data that confirms NMD in two variants and the NMD prediction of other nonsense variants in Alamut. For the frameshift variants, it is also likely that the cleavage of the zinc finger domain results in functional haploinsufficiency either because of a lack of signalling domains or similarly due to NMD. Missense variants could cause haploinsufficiency through the suggested mechanism by Krauss et al who have illustrated that missense variants in the MID1 domain hamper the functional interaction with the MID1-α4-PP2A complex, leading to a subcellular location of GLI3.24 The observed missense variants in our study exceed the region to which Krauss et al have limited the MID-1 interaction domain. An alternative theory is suggested by Zhou et al who have shown that missense variants in the MBD can cause deficiency in the signalling of GLI3A, functionally implicating a relative overexpression of GLI3R.22 However, GLI3R overexpression would likely produce a posterior phenotype, as determined by Hill et al in their fixed homo and hemizygous GLI3R models.15 Therefore, our hypothesis is that all included missense variants have a similar pathogenesis which is more likely in concordance with the mechanism introduced by Krauss et al. To our knowledge, no splice site variants have been functionally described in literature. However, it is noted that the 15 and last exon encompasses the entire activator domain, thus any splice site mutation is by definition located on the 5′ side of the activator. Based on the phenotype, we would suggest that these variants fail to produce a functional protein. We hypothesise that the truncating variants of the activator domain lead to overexpression of GLI3R in SHH rich areas. In normal development, the presence of SHH prevents the processing of full length GLI34 into GLI3R, thus producing the full length activator. In patients with a truncating variant of the activator domain of GLI3, thus these variants likely have the largest effect in SHH rich areas, such as the ZPA located at the posterior side of the hand/footplate. Moreover, the lack of posterior anomalies in the GLI3∆699/- mouse model (hemizygous fixed repressor model) compared with the GLI3∆699/∆699 mouse model (homozygous fixed repressor model), suggesting a dosage effect of GLI3R to be responsible for posterior hand anomalies.15 These findings are supported by Lewandowski et al, who show that the majority of the target genes in GLI signalling are regulated by GLI3R rather than GLI3A.44 Together, these findings suggest a role for the location and type of variant in GLI3-mediated syndromes.
Interestingly, the difference between Pallister-Hall syndrome and GLI3-mediated polydactyly syndromes has also been attributed to the GLI3R overexpression. However, the difference in phenotype observed in the cases with a truncating variant in the activator domain and Pallister-Hall syndrome suggest different functional consequences. When studying figure 1, it is noted that the included truncating variants on the 3′ side of the cleavage site seldomly affect the CBP binding region, which could provide an explanation for the observed differences. This binding region is included in the Pallister-Hall region as defined by Johnston et al and is necessary for the downstream signalling with GLI1.10 11 23 45 Interestingly, recent reports show that pathogenic variants in GLI1 can produce phenotypes concordant with Ellis von Krefeld syndrome, which includes overlapping features with Pallister-Hall syndrome.46 The four truncating variants observed in this study that do affect the CBP but did not result in a Pallister-Hall phenotype are conflicting with this theory. Krauss et al postulate an alternative hypothesis, they state that the MID1-α4-PP2A complex, which is essential for GLI3A signalling, could also be the reason for overlapping features of Opitz syndrome, caused by variants in MID1, and Pallister-Hall syndrome. Further analysis is required to fully appreciate the functional differences between truncating mutations that cause Pallister-Hall syndrome and those that result in GLI3-mediated polydactyly syndromes.
For the clinical evaluation of patients with GLI3-mediated polydactyly syndromes, intracranial anomalies are likely the most important to predict based on the variant. Unfortunately, the presence of corpus callosum agenesis was not routinely investigated or reported thus this feature could not be used as an indicator phenotype for LC membership. Interestingly when using only hand and foot phenotypes, we did notice a higher prevalence of corpus callosum agenesis in patients with posterior phenotypes. The suggested relation between truncating mutations in the activator domain causing these posterior phenotypes and corpus callosum agenesis was statistically confirmed (OR: 8.8, p<0.001). Functionally this relation could be caused by the GLI3-MED12 interaction at the MBD: pathogenic DNA variants in MED12 can cause Opitz-Kaveggia syndrome, a syndrome in which presentation includes corpus callosum agenesis, broad halluces and thumbs.47
In conclusion, there are two distinct phenotypes within the GLI3-mediated polydactyly population: patients with more posteriorly and more anteriorly oriented hand anomalies. Furthermore, this difference is related to the observed variant in GLI3. We hypothesise that variants that cause haploinsufficiency produce anterior anomalies of the hand, whereas variants with abnormal truncation of the activator domain have more posterior anomalies. Furthermore, patients that have a variant that produces abnormal truncation of the activator domain, have a greater risk for corpus callosum agenesis. Thus, we advocate to differentiate preaxial or postaxial oriented GLI3 phenotypes to explain the pathophysiology as well as to get a risk assessment for corpus callosum agenesis.
Footnotes
MB and EBB contributed equally.
Contributors: MB and EBB contributed equally to this study. All authors were involved in the conception, design and acquisition of data and/or in the analysis and interpretation of the data. A detailed description of the contribution of all authors has been submitted to the journal. All authors reviewed and approved the manuscript.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The research protocol was approved by the local ethics board of the Erasmus MC University Medical Center (MEC 2015-679).
References
- 1. Ruppert JM, Vogelstein B, Arheden K, Kinzler KW. Gli3 encodes a 190-kilodalton protein with multiple regions of Gli similarity. Mol Cell Biol 1990;10:5408–15. 10.1128/MCB.10.10.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biesecker LG. What you can learn from one gene: Gli3. J Med Genet 2006;43:465–9. 10.1136/jmg.2004.029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 2000;100:423–34. 10.1016/S0092-8674(00)80678-9 [DOI] [PubMed] [Google Scholar]
- 4. Tickle C, Towers M. Sonic hedgehog signaling in limb development. Front Cell Dev Biol 2017;5. 10.3389/fcell.2017.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Speksnijder L, Cohen-Overbeek TE, Knapen MFCM, Lunshof SM, Hoogeboom AJM, van den Ouwenland AM, de Coo IFM, Lequin MH, Bolz HJ, Bergmann C, Biesecker LG, Willems PJ, Wessels MW. A de novo GLI3 mutation in a patient with acrocallosal syndrome. Am J Med Genet A 2013;161A:1394–400. 10.1002/ajmg.a.35874 [DOI] [PubMed] [Google Scholar]
- 6. Kalff-Suske M, Wild A, Topp J, Wessling M, Jacobsen EM, Bornholdt D, Engel H, Heuer H, Aalfs CM, Ausems MG, Barone R, Herzog A, Heutink P, Homfray T, Gillessen-Kaesbach G, König R, Kunze J, Meinecke P, Müller D, Rizzo R, Strenge S, Superti-Furga A, Grzeschik KH. Point mutations throughout the Gli3 gene cause Greig cephalopolysyndactyly syndrome. Hum Mol Genet 1999;8:1769–77. 10.1093/hmg/8.9.1769 [DOI] [PubMed] [Google Scholar]
- 7. Kang S, Graham JM, Olney AH, Biesecker LG. Gli3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet 1997;15:266–8. 10.1038/ng0397-266 [DOI] [PubMed] [Google Scholar]
- 8. Radhakrishna U, Bornholdt D, Scott HS, Patel UC, Rossier C, Engel H, Bottani A, Chandal D, Blouin JL, Solanki JV, Grzeschik KH, Antonarakis SE. The phenotypic spectrum of Gli3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and Postaxial polydactyly type-A/B; no phenotype prediction from the position of Gli3 mutations. Am J Hum Genet 1999;65:645–55. 10.1086/302557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kalff-Suske M, Paparidis Z, Bornholdt D, Cole T, Kalff-Suske M, Grzeschik K-H. Gene symbol: Gli3. disease: Pallister-Hall syndrome. Hum Genet 2004;114:403. [PubMed] [Google Scholar]
- 10. Johnston JJ, Sapp JC, Turner JT, Amor D, Aftimos S, Aleck KA, Bocian M, Bodurtha JN, Cox GF, Curry CJ, Day R, Donnai D, Field M, Fujiwara I, Gabbett M, Gal M, Graham JM, Hedera P, Hennekam RCM, Hersh JH, Hopkin RJ, Kayserili H, Kidd AMJ, Kimonis V, Lin AE, Lynch SA, Maisenbacher M, Mansour S, McGaughran J, Mehta L, Murphy H, Raygada M, Robin NH, Rope AF, Rosenbaum KN, Schaefer GB, Shealy A, Smith W, Soller M, Sommer A, Stalker HJ, Steiner B, Stephan MJ, Tilstra D, Tomkins S, Trapane P, Tsai AC-H, Van Allen MI, Vasudevan PC, Zabel B, Zunich J, Black GCM, Biesecker LG. Molecular analysis expands the spectrum of phenotypes associated with Gli3 mutations. Hum Mutat 2010;31:1142–54. 10.1002/humu.21328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnston JJ, Olivos-Glander I, Killoran C, Elson E, Turner JT, Peters KF, Abbott MH, Aughton DJ, Aylsworth AS, Bamshad MJ, Booth C, Curry CJ, David A, Dinulos MB, Flannery DB, Fox MA, Graham JM, Grange DK, Guttmacher AE, Hannibal MC, Henn W, Hennekam RCM, Holmes LB, Hoyme HE, Leppig KA, Lin AE, Macleod P, Manchester DK, Marcelis C, Mazzanti L, McCann E, McDonald MT, Mendelsohn NJ, Moeschler JB, Moghaddam B, Neri G, Newbury-Ecob R, Pagon RA, Phillips JA, Sadler LS, Stoler JM, Tilstra D, Walsh Vockley CM, Zackai EH, Zadeh TM, Brueton L, Black GCM, Biesecker LG. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of Gli3 mutations. Am J Hum Genet 2005;76:609–22. 10.1086/429346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Démurger F, Ichkou A, Mougou-Zerelli S, Le Merrer M, Goudefroye G, Delezoide A-L, Quélin C, Manouvrier S, Baujat G, Fradin M, Pasquier L, Megarbané A, Faivre L, Baumann C, Nampoothiri S, Roume J, Isidor B, Lacombe D, Delrue M-A, Mercier S, Philip N, Schaefer E, Holder M, Krause A, Laffargue F, Sinico M, Amram D, André G, Liquier A, Rossi M, Amiel J, Giuliano F, Boute O, Dieux-Coeslier A, Jacquemont M-L, Afenjar A, Van Maldergem L, Lackmy-Port-Lis M, Vincent-Delorme C, Chauvet M-L, Cormier-Daire V, Devisme L, Geneviève D, Munnich A, Viot G, Raoul O, Romana S, Gonzales M, Encha-Razavi F, Odent S, Vekemans M, Attie-Bitach T. New insights into genotype-phenotype correlation for Gli3 mutations. Eur J Hum Genet 2015;23:92–102. 10.1038/ejhg.2014.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Al-Qattan MM, Shamseldin HE, Salih MA, Alkuraya FS. GLI3-related polydactyly: a review. Clin Genet 2017;92:457–66. 10.1111/cge.12952 [DOI] [PubMed] [Google Scholar]
- 14. Radhakrishna U, Wild A, Grzeschik KH, Antonarakis SE. Mutation in Gli3 in Postaxial polydactyly type A. Nat Genet 1997;17:269–71. 10.1038/ng1197-269 [DOI] [PubMed] [Google Scholar]
- 15. Hill P, Götz K, Rüther U. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Dev Biol 2009;328:506–16. 10.1016/j.ydbio.2009.02.017 [DOI] [PubMed] [Google Scholar]
- 16. Jamsheer A, Sowińska A, Trzeciak T, Jamsheer-Bratkowska M, Geppert A, Latos-Bieleńska A. Expanded mutational spectrum of the Gli3 gene substantiates genotype-phenotype correlations. J Appl Genet 2012;53:415–22. 10.1007/s13353-012-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vermunt JK. Latent class modeling with covariates: two improved three-step approaches. Polit. anal. 2010;18:450–69. 10.1093/pan/mpq025 [DOI] [Google Scholar]
- 18. Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NST, Abeysinghe S, Krawczak M, Cooper DN. Human gene mutation database (HGMD): 2003 update. Hum Mutat 2003;21:577–81. 10.1002/humu.10212 [DOI] [PubMed] [Google Scholar]
- 19. Biesecker LG. Pallister-Hall syndrome. Seattle WA: GeneReviews((R)), 1993. [Google Scholar]
- 20. den Dunnen JT. Describing sequence variants using HGVS nomenclature. Methods Mol Biol 2017;1492:243–51. 10.1007/978-1-4939-6442-0_17 [DOI] [PubMed] [Google Scholar]
- 21. Furniss D, Critchley P, Giele H, Wilkie AOM. Nonsense-Mediated decay and the molecular pathogenesis of mutations in Sall1 and Gli3. Am J Med Genet A 2007;143A:3150–60. 10.1002/ajmg.a.32097 [DOI] [PubMed] [Google Scholar]
- 22. Zhou H, Kim S, Ishii S, Boyer TG. Mediator modulates Gli3-dependent sonic hedgehog signaling. Mol Cell Biol 2006;26:8667–82. 10.1128/MCB.00443-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by Gli3. J Biol Chem 1999;274:8143–52. 10.1074/jbc.274.12.8143 [DOI] [PubMed] [Google Scholar]
- 24. Krauss S, So J, Hambrock M, Köhler A, Kunath M, Scharff C, Wessling M, Grzeschik K-H, Schneider R, Schweiger S. Point mutations in Gli3 lead to misregulation of its subcellular localization. PLoS One 2009;4:e7471. 10.1371/journal.pone.0007471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lanza ST, Collins LM, Lemmon DR, Schafer JL. Proc LCA: a SAS procedure for latent class analysis. Struct Equ Modeling 2007;14:671–94. 10.1080/10705510701575602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Al-Qattan MM. A novel frameshift mutation of the Gli3 gene in a family with broad Thumbs with/without big toes, Postaxial polydactyly and variable syndactyly of the hands/feet. Clin Genet 2012;82:502–4. 10.1111/j.1399-0004.2012.01866.x [DOI] [PubMed] [Google Scholar]
- 27. Bilguvar K, Bydon M, Bayrakli F, Ercan-Sencicek AG, Bayri Y, Mason C, DiLuna ML, Seashore M, Bronen R, Lifton RP, State M, Gunel M. A novel syndrome of cerebral cavernous malformation and Greig cephalopolysyndactyly. laboratory investigation. J Neurosurg 2007;107:495–9. 10.3171/PED-07/12/495 [DOI] [PubMed] [Google Scholar]
- 28. Cheng F, Ke X, Lv M, Zhang F, Li C, Zhang X, Zhang Y, Zhao X, Wang X, Liu B, Han J, Li Y, Zeng C, Li S. A novel frame-shift mutation of Gli3 causes non-syndromic and complex digital anomalies in a Chinese family. Clin Chim Acta 2011;412:1012–7. 10.1016/j.cca.2011.02.007 [DOI] [PubMed] [Google Scholar]
- 29. Debeer P, Peeters H, Driess S, De Smet L, Freese K, Matthijs G, Bornholdt D, Devriendt K, Grzeschik K-H, Fryns J-P, Kalff-Suske M. Variable phenotype in Greig cephalopolysyndactyly syndrome: clinical and radiological findings in 4 independent families and 3 sporadic cases with identified Gli3 mutations. Am J Med Genet A 2003;120A:49–58. 10.1002/ajmg.a.20018 [DOI] [PubMed] [Google Scholar]
- 30. Elson E, Perveen R, Donnai D, Wall S, Black GCM. De novo GLI3 mutation in acrocallosal syndrome: broadening the phenotypic spectrum of Gli3 defects and overlap with murine models. J Med Genet 2002;39:804–6. 10.1136/jmg.39.11.804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Furniss D, Kan S-H, Taylor IB, Johnson D, Critchley PS, Giele HP, Wilkie AOM. Genetic screening of 202 individuals with congenital limb malformations and requiring reconstructive surgery. J Med Genet 2009;46:730–5. 10.1136/jmg.2009.066027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hurst JA, Jenkins D, Vasudevan PC, Kirchhoff M, Skovby F, Rieubland C, Gallati S, Rittinger O, Kroisel PM, Johnson D, Biesecker LG, Wilkie AOM. Metopic and sagittal synostosis in Greig cephalopolysyndactyly syndrome: five cases with intragenic mutations or complete deletions of Gli3. Eur J Hum Genet 2011;19:757–62. 10.1038/ejhg.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonald-McGinn DM, Feret H, Nah H-D, Bartlett SP, Whitaker LA, Zackai EH. Metopic craniosynostosis due to mutations in Gli3: a novel association. Am J Med Genet A 2010;152A:1654–60. 10.1002/ajmg.a.33495 [DOI] [PubMed] [Google Scholar]
- 34. Patel R, Tripathi FM, Singh SK, Rani A, Bhattacharya V, Ali A. A novel GLI3c.750delC truncation mutation in a multiplex Greig cephalopolysyndactyly syndrome family with an unusual phenotypic combination in a patient. Meta Gene 2014;2:880–7. 10.1016/j.mgene.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raposo L, Fachada H, Santos Paulo A, Cerveira I, Castedo S, Pereira S. Prenatal diagnosis of Greig cephalopolysyndactyly syndrome: a case report. Prenat Diagn 2015;35:203–5. 10.1002/pd.4506 [DOI] [PubMed] [Google Scholar]
- 36. Sethi SK, Goyal D, Khalil S, Yadav DK. Two Indian families with Greig cephalopolysyndactyly with non-syndromic phenotype. Eur J Pediatr 2013;172:1131–5. 10.1007/s00431-013-1938-2 [DOI] [PubMed] [Google Scholar]
- 37. Tommerup N, Nielsen F. A familial reciprocal translocation t(3;7) (p21.1;p13) associated with the Greig polysyndactyly-craniofacial anomalies syndrome. Am J Med Genet 1983;16:313–21. 10.1002/ajmg.1320160304 [DOI] [PubMed] [Google Scholar]
- 38. Wang Z, Wang J, Li Y, Geng J, Fu Q, Xu Y, Shen Y. Novel frame-shift mutations of Gli3 gene in non-syndromic Postaxial polydactyly patients. Clin Chim Acta 2014;433:195–9. 10.1016/j.cca.2014.03.012 [DOI] [PubMed] [Google Scholar]
- 39. Xiang Y, Wang Z, Bian J, Xu Y, Fu Q. Exome sequencing reveals a novel nonsense mutation of GLI3 in a Chinese family with 'non-syndromic' pre-axial polydactyly. J Hum Genet 2016;61:907–10. 10.1038/jhg.2016.76 [DOI] [PubMed] [Google Scholar]
- 40. Crapster JA, Hudgins L, Chen JK, Gomez-Ospina N. A novel missense variant in the Gli3 zinc finger domain in a family with digital anomalies. Am J Med Genet A 2017;173:3221–5. 10.1002/ajmg.a.38415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ni F, Han G, Guo R, Cui H, Wang B, Li Q. A novel frameshift mutation of Gli3 causes isolated Postaxial polydactyly. Ann Plast Surg 2019;82:570–3. 10.1097/SAP.0000000000001685 [DOI] [PubMed] [Google Scholar]
- 42. Patel R, Singh CB, Bhattacharya V, Singh SK, Ali A. Gli3 mutations in syndromic and non-syndromic polydactyly in two Indian families. Congenit Anom 2016;56:94–7. 10.1111/cga.12139 [DOI] [PubMed] [Google Scholar]
- 43. Rao C, Chen J, Peng Q, Mo Q, Xia X, Lu X. Mutational screening of Gli3, Shh, and Shh ZRS in 78 Chinese children with nonsyndromic polydactyly. Genet Test Mol Biomarkers 2018;22:577–81. 10.1089/gtmb.2018.0096 [DOI] [PubMed] [Google Scholar]
- 44. Lewandowski JP, Du F, Zhang S, Powell MB, Falkenstein KN, Ji H, Vokes SA. Spatiotemporal regulation of Gli target genes in the mammalian limb bud. Dev Biol 2015;406:92–103. 10.1016/j.ydbio.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnston JJ, Olivos-Glander I, Turner J, Aleck K, Bird LM, Mehta L, Schimke RN, Heilstedt H, Spence JE, Blancato J, Biesecker LG. Clinical and molecular delineation of the Greig cephalopolysyndactyly contiguous gene deletion syndrome and its distinction from acrocallosal syndrome. Am J Med Genet A 2003;123A:236–42. 10.1002/ajmg.a.20318 [DOI] [PubMed] [Google Scholar]
- 46. Palencia-Campos A, Ullah A, Nevado J, Yildirim R, Unal E, Ciorraga M, Barruz P, Chico L, Piceci-Sparascio F, Guida V, De Luca A, Kayserili H, Ullah I, Burmeister M, Lapunzina P, Ahmad W, Morales AV, Ruiz-Perez VL. Gli1 inactivation is associated with developmental phenotypes overlapping with Ellis-van Creveld syndrome. Hum Mol Genet 2017;26:4556–71. 10.1093/hmg/ddx335 [DOI] [PubMed] [Google Scholar]
- 47. Graham JM. Schwartz Ce. MED12 related disorders. Am J Med Genet A 2013;161A:2734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2020-106948supp001.pdf (152.1KB, pdf)
jmedgenet-2020-106948supp002.pdf (99.9KB, pdf)
Data Availability Statement
Data are available upon reasonable request.