The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory disease coronavirus 2 (SARS-CoV-2), has led to millions of confirmed cases and deaths worldwide. Efficient diagnostic tools are in high demand, as rapid and large-scale testing plays a pivotal role in patient management and decelerating disease spread.
KEYWORDS: COVID-19, SARS-CoV-2, 2019-nCoV, NAAT, PCR, serology, antigen, coronavirus, biomarkers, next-generation sequencing
SUMMARY
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory disease coronavirus 2 (SARS-CoV-2), has led to millions of confirmed cases and deaths worldwide. Efficient diagnostic tools are in high demand, as rapid and large-scale testing plays a pivotal role in patient management and decelerating disease spread. This paper reviews current technologies used to detect SARS-CoV-2 in clinical laboratories as well as advances made for molecular, antigen-based, and immunological point-of-care testing, including recent developments in sensor and biosensor devices. The importance of the timing and type of specimen collection is discussed, along with factors such as disease prevalence, setting, and methods. Details of the mechanisms of action of the various methodologies are presented, along with their application span and known performance characteristics. Diagnostic imaging techniques and biomarkers are also covered, with an emphasis on their use for assessing COVID-19 or monitoring disease severity or complications. While the SARS-CoV-2 literature is rapidly evolving, this review highlights topics of interest that have occurred during the pandemic and the lessons learned throughout. Exploring a broad armamentarium of techniques for detecting SARS-CoV-2 will ensure continued diagnostic support for clinicians, public health, and infection prevention and control for this pandemic and provide advice for future pandemic preparedness.
INTRODUCTION
While coronavirus disease 2019 (COVID-19) is not the first pandemic of the 21st century (1), it has generated unprecedented global concern and responses. COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is thought to have emerged from a zoonotic source (2) and spread rapidly in humans through respiratory droplets and contact. There is some concern for airborne transmission, but the role of this transmission route outside the potential aerosolizing procedures in health care settings is unclear (3–5). With an estimated reproductive number, R naught (R0), of between 1.4 and 5.6, SARS-CoV-2 rapidly spread worldwide (6–9). Since the first cases reported in December 2019 (10–12), there have been over 106 million confirmed cases and 2.3 million deaths reported worldwide (as of 9 February 2021) (13).
From a disease manifestation perspective, SARS-CoV-2 infection can be asymptomatic (14), and COVID-19 spans from a mild influenza-like illness (ILI) to life-threatening complications (15, 16). SARS-CoV-2 not only affects the respiratory tract, resulting in pneumonia, but also can affect gastrointestinal (GI), neurological, or cardiovascular systems. Atypical presentations of COVID-19 include cutaneous manifestations such as a Kawasaki-like disease in children and ophthalmic/gustatory dysfunction (i.e., anosmia and ageusia, which are the loss of smell and taste, respectively), which may have been underestimated in initial reports (17–20).
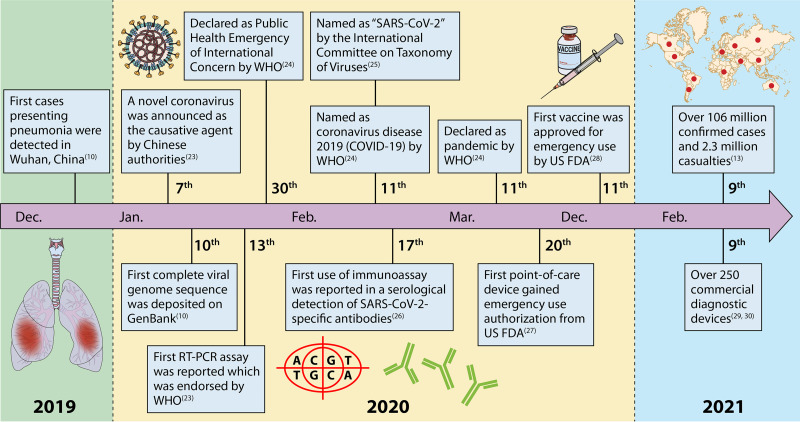
Despite numerous therapeutic options being explored (e.g., convalescent-phase plasma), no large-scale treatments are available. Public health interventions have evolved over time to limit viral spread (Fig. 1) and have included the use of personal protective equipment (PPE) like masks, handwashing, and containment measures such as city lockdowns, travel restrictions, and physical distancing (21–30). Although these strategies have been essential to reduce the virus’s spread, they have had significant adverse socioeconomic impacts, and adherence to these prevention strategies is challenging to sustain (22). Currently, cases of COVID-19 have declined following a first pandemic wave in some areas, whereas other areas are experiencing subsequent waves of activity. Fortunately, many vaccine candidates are under development and undergoing regulatory approval processes (31–35). Recently, COVID-19 mRNA vaccines have been the first licensed for use and are rapidly being administered as supplies are provided (28, 36). However, given the time required for adequate immunization coverage in the population at large, subsequent pandemic waves are anticipated (31, 37–39). Therefore, detection methods for SARS-CoV-2 remain a crucial part of containment and mitigation strategies, and lessons learned from this pandemic may help prepare against future pandemics.
FIG 1.
Timeline of COVID-19 spread and the global response to it (10, 13, 23–30). Of note, while SARS-CoV-2 was initially thought to have emerged from China in December 2020, there are data to suggest that it may have circulated more broadly earlier than initially recorded in other countries, and further studies are under way to investigate this possibility in other areas (571–574).
In terms of testing, real-time reverse transcription-PCR (RT-PCR) remains the most common method used to identify SARS-CoV-2 (40). While common in diagnostic laboratories worldwide, many laboratories remain faced with supply chain shortages for real-time RT-PCR reagents and consumables, all while being asked to increase testing capacity. As such, delays were common for test results, prompting the exploration of alternative testing options such as specimen pooling or laboratory testing using methods other than RT-PCR. Methods that could enhance testing capacity, streamline testing (i.e., automation), or provide more rapid results in easy-to-use formats that are amenable to point-of-care (POC) applications without complex instrumentation (e.g., isothermal technologies) were all desired (41–47). Rigorous research escalated quickly from the academic to industry partners, and this research is ongoing to develop testing alternatives or complements to existing technologies.
While recent reviews have been published on the management of SARS-CoV-2 (41, 47–55), recent advancements in novel diagnostic methods justify the need for a more comprehensive synthesis of the current literature. In this review, first, the biological characteristics of SARS-CoV-2 are described in order to fully understand the molecular and immunological methods for its detection. Following a brief discussion on the COVID-19 manifestations, compatible signs and symptoms, and disease biomarkers, diagnostic imaging techniques are described in relation to COVID-19 lower respiratory tract involvement, including applications such as monitoring disease severity, the progression of the illness, or complications. Next, a comprehensive review of current and recent advances in molecular, antigen (Ag), and serological immunodiagnostic methods is covered, including rapid diagnostic tests (RDTs) used in the laboratory setting and POC applications. Overall, this review expands our knowledge of current and exploratory avenues for detecting SARS-CoV-2 and COVID-19.
It should be noted that some of the references used in this review were preprints that have not been peer reviewed, and recognizing that data on the detection of SARS-CoV-2 or COVID-19 are rapidly evolving, some details on testing options and guidelines may no longer be recent and should thus be reviewed in the context of recent findings and recommendations. Nonetheless, this review provides a comprehensive synthesis of the most current data available to date, along with current recommendations for the detection of SARS-CoV-2 or the diagnosis of COVID-19.
SARS-CoV-2 GENOME AND STRUCTURE
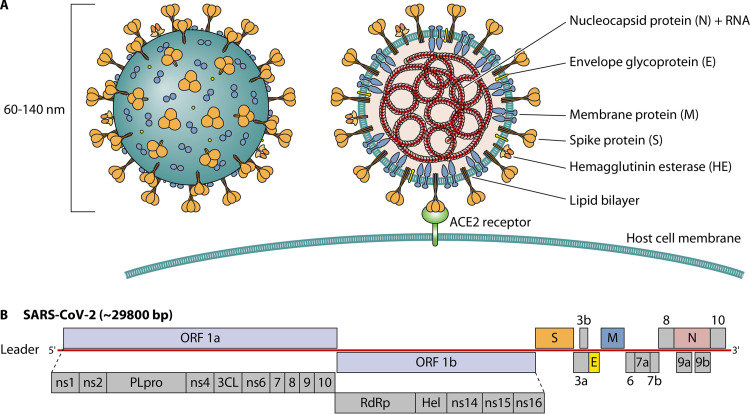
Understanding the genetic and structural properties of SARS-CoV-2 is a prerequisite to developing effective diagnostic tools. SARS-CoV-2 was first isolated and sequenced in China in January 2020 (10–12). Transmission electron microscopy revealed that SARS-CoV-2 has a diameter in the range of 60 to 140 nm, and its morphology was consistent with those of other members of the Coronaviridae family (Fig. 2A) (12, 25). SARS-CoV-2 is an enveloped, positive-strand RNA virus, and on the genetic level, it shares 96%, 80%, and 50% sequence identities with bat coronavirus (RaTG13), SARS-CoV-1, and Middle East respiratory syndrome coronavirus (MERS-CoV), respectively (11, 56). Based on these analyses, the International Committee on Taxonomy of Viruses named the virus SARS-CoV-2, which was formerly referred to as the 2019 novel coronavirus (2019-nCoV) or human coronavirus 2019 (hCoV-19) (25).
FIG 2.
Physical and genome structure of SARS-CoV-2. (A) Diagram of the SARS-CoV-2 virion. (B) Genome organization and proteins with known or unknown functions.
Our understanding of SARS-CoV-2 structure and function has been largely derived from research on SARS-CoV-1, MERS-CoV, and seasonal coronaviruses. SARS-CoV-2 has a single-stranded positive-sense RNA genome of between 26 and 35 kb, encoding approximately 27 proteins with similarity to proteins of known functions, while others are unclear/unknown or putative (Fig. 2B) (21, 37, 53, 57, 58). The first open reading frame (ORF1a/b) on the 5′ end of the viral genome occupies ∼71% of the entire genome and produces two polyproteins (pp’s), pp1a and pp1ab. These two polyproteins are processed by the viral proteases into 15 nonstructural proteins (nsp’s), and these proteins are collectively involved in polyprotein processing, viral RNA replication, and mRNA synthesis (53, 57). The remaining proteins, including the structural and accessory proteins, are expressed from several nested subgenomic mRNAs produced through a process known as discontinuous transcription by the viral RNA-dependent RNA polymerase (RdRp).
The structural proteins include the small envelope (E) protein, membrane (M) protein (also known as the matrix protein), nucleocapsid (N) protein, hemagglutinin-esterase (HE) protein, and spike (S) glycoprotein (Fig. 2A) (57, 59). The E and M proteins are primarily involved in viral assembly, budding, and virion morphogenesis (60–62), while the N protein complexes with the viral genomic RNA to generate the nucleocapsid (63). The S protein is the major surface glycoprotein on SARS-CoV-2, forming approximately 40 trimers that play an important role in both receptor binding and membrane fusion through the two functional subunits S1 and S2 (37, 64). The S protein trimers contain a stable stalk separated from the globular heads by three flexible hinges, allowing for orientation freedom to interact with host cell receptors (65). The S1 subunit contains the receptor-binding domain (RBD) that directly interacts with the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface, whereas the S2 subunit contains a structural loop responsible for fusion events between the viral and host cell membranes, resulting in the release of the viral genomic RNA into the cytoplasm (66, 67). Of note, along with engaging the ACE2 host cell receptor, the cellular serine protease TMPRSS2 is engaged for S protein priming, and this cofactor has been investigated as a possible antiviral target using viral entry inhibitors (68, 69).
Overall, having knowledge of SARS-CoV-2 pathogenesis can help in understanding disease manifestations and help guide the development of molecular and immunological tools for the identification of this virus.
CLINICAL MANIFESTATIONS OF COVID-19
The spectrum of SARS-CoV-2 infection can vary from asymptomatic infection to life-threatening complications of COVID-19 (37). Using modeling, it was estimated that over 59% of transmissions arise from asymptomatic individuals, with 35% from individuals in presymptomatic stages of infection and 24% from individuals who never develop symptoms (70). These estimates are concerning but emphasize the need for the wide use of vaccines and maintaining key public health interventions like mask wearing, hand hygiene, and social distancing.
In most symptomatic cases, COVID-19 presents as a mild to moderate upper respiratory illness, with signs and symptoms compatible with those of other respiratory viruses (71). As such, the diagnostic accuracy of any individual sign or symptom is very poor, and neither the presence nor the absence of any sign or symptom can be used to rule in or out COVID-19 (71). With the possibility of other pathogens that could present like SARS-CoV-2 infection, case definitions based on clinical presentation are not sufficiently specific but can help support the investigation of suspect COVID-19 cases. On the other hand, given that the list of possible presentations and atypical manifestations of COVID-19 could mirror those of other diseases, identifying the etiology of illness as SARS-CoV-2 requires laboratory testing.
In a recent Cochrane review, a summary of 16 studies (7,706 patients) was presented (71). Only six of the possible signs and symptoms of COVID-19 had sensitivities of >50%, and results were highly variable between studies and settings. The most common signs and symptoms (and their performances) are summarized as follows: cough (with sensitivity and specificity from 43 to 71% and 14 to 54%, respectively), sore throat (5 to 71% and 55 to 80%), fever (7 to 91% and 16% to 94%), musculoskeletal symptoms (e.g., arthralgias or myalgias) (19 to 86% and 45 to 91%), fatigue (10 to 57% and 60 to 94%), and headache (3 to 71% and 78 to 98%) (71). It was noted that possible confounders were present, and the high heterogeneity between data suggested that signs and symptoms are variable between individuals (71). Other less common clinical presentations have been documented, including alterations in smell or taste (i.e., anosmia or dysgeusia) as well as neurological or cutaneous manifestations (17–19, 72–76). It is noteworthy that in the early stages of the pandemic, some of these symptoms may have been missed or underreported, but knowledge on possible clinical presentations of COVID-19 have evolved over time.
In some cases of COVID-19, progression to lower respiratory tract illness (e.g., pneumonia) can occur and may require hospitalization, intensive care unit (ICU) support, and mechanical ventilation, and complications can arise, which include acute respiratory distress, multiorgan dysfunction, and death (71, 77–85). In general, adverse outcomes and deaths are more common with increasing age or in individuals with underlying medical comorbidities such as respiratory system disease, cardiovascular disease, and diabetes (78–80). Fatality rates vary among studies and countries but are generally high in the hospital setting (e.g., 4 to 11%) compared to the overall case fatality rates (e.g., 2 to 3%) in the general population (80, 82, 85, 86). In terms of recovery, the median duration of hospital stay is 10 to 14 days, and resolution generally occurs within 2 to 3 weeks (85). There is a lack of evidence on whether some symptoms can persist after recovery. In one study, patients were monitored up to 60 days after recovery, with 87.4% reporting at least one symptom (86). The most common symptoms were fatigue (53.1%), dyspnea (43.4%), joint pain (27.3%), and chest pain (21.7%).
Overall, while some signs or symptoms may be compatible with COVID-19, none are specific, and laboratory testing is required to confirm the diagnosis. Further studies are required to help identify the frequency of atypical clinical presentations, and additional studies looking at known clinical presentations of COVID-19 should consider possible confounders such as the possibility of other etiologies, host factors (e.g., comorbidities), disease severity, and the times from infection and symptom onset.
BIOMARKERS FOR COVID-19 AND ROUTINE LABORATORY INVESTIGATIONS
Apart from laboratory tests specific for detecting SARS-CoV-2 discussed throughout this review, routine laboratory testing spanning hematological, biochemical, and chemical markers is used to assess a patient’s health or identify possible clues to a disease state (87–90). Such routine laboratory workup of individuals is used to refine a medical differential diagnosis, thereby supporting or refuting potential causes of the clinical presentation based on typical outcomes of these investigations for a defined disease. Many of these investigations can evolve through the clinical course of illness, and additional testing can be ordered by physicians based on the clinical presentation. These can include tests such as white blood cell (WBC) counts, markers for inflammatory conditions (C-reactive protein [CRP], procalcitonin [PCT], or interleukin 6 [IL-6]), tests for anticoagulation, and indicators of tissue damage (alanine aminotransferase [ALT], aspartate aminotransferase [AST], lactate dehydrogenase [LDH], and creatine kinase [CK]). While biomarkers for COVID-19 have been the subject of much investigation during the current pandemic, none of these tests are sensitive or specific for COVID-19. In a Cochrane review analyzing 67 laboratory tests from 21 studies encompassing 14,126 COVID-19 cases and 56,585 non-COVID-19 cases, only three markers showed sensitivity and specificity values of >50%: a decrease in the lymphocyte count and increases in the inflammatory markers CRP and IL-6 (90). Overall, no individual biomarker can be used reliably to rule COVID-19 in or out, and laboratory testing should be performed. However, it should be noted that some laboratory markers have value for patient management as they can help assess the severity of the disease or progression of the illness or even act as risk factors for death. In the most recent Centers for Disease Control and Prevention (CDC) guidance documents for clinicians caring for patients with COVID-19, a summary of important laboratory tests is described, with lymphopenia being the most common laboratory finding in patients hospitalized with COVID-19 (87). Laboratory markers associated with increased illness severity include lymphopenia, neutropenia, and elevated serum ALT, AST, LDH, CRP, and ferritin (87, 88). Patients with critical illness have high plasma levels of inflammatory makers, and elevated levels of d-dimer and lymphopenia have been associated with an increased risk of death.
Of note, this section is not intended to be a comprehensive review of all biomarkers used in routine or exploratory investigations for COVID-19. We recognize the availability of guidelines for clinicians caring for patients with suspected or confirmed infection with SARS-CoV-2 (87, 88) as well as the expertise of medical staff in ordering laboratory tests to help guide evolving differential diagnoses throughout the clinical course of illness. However, this section also recognizes the ongoing efforts of researchers who are dedicated to understanding the role of existing or novel biomarkers. Overall, no laboratory marker to date is diagnostic for COVID-19, but they have value in patient management over time, regardless of SARS-CoV-2 infection status. Biomarkers for COVID-19 severity or prognosis remain an active area of research that may not only lead to new diagnostic approaches but also help us understand disease progression and host responses to COVID-19 (91–94).
DIAGNOSTIC IMAGING FOR COVID-19
While testing of specimens collected from the upper respiratory tract is common for diagnosing SARS-CoV-2 infection, the progression of the disease may involve the lower respiratory tract (e.g., pneumonia), with or without detectable SARS-CoV-2 in the upper respiratory tract (55, 95–103). Testing of specimens from the lower respiratory tract (e.g., bronchoalveolar lavage [BAL] fluid) is possible using nucleic acid amplification tests (NAATs) like RT-PCR, but obtaining lower respiratory tract specimens is not always possible (104–107). Along with laboratory testing, diagnostic imaging can complement investigations of COVID-19 to assess the involvement of disease in the lower respiratory tract or other anatomical sites. Diagnostic imaging techniques include chest radiography (or chest X ray [CXR]), computed tomography (CT) scan, ultrasound, magnetic resonance imaging (MRI), and positron emission tomography-CT (PET/CT) (108–116). Among these, CT scans are the most frequently used methods for diagnosis of lower tract involvement or follow-up of COVID-19 cases (110–112). CT scans produce cross-sectional images at different angles, thereby providing a three-dimensional (3D) look at the targeted anatomy. Chest CT scan images can be assembled and assessed by radiologists to check for possible abnormalities suggestive of lower tract disease such as viral pneumonia (53, 112, 117). Typical features of a chest CT image in COVID-19 are ground-glass or reticular opacities (GGOs) with or without consolidations that present bilaterally, peripherally, or in posterior distributions (113).
The utility of diagnostic imaging for routine screening for COVID-19 has been a subject of debate and has not been recommended by most radiology societies (113, 114, 118–121). On the other hand, due to the shortage of RT-PCR supplies during the early days of the pandemic and the possibility of false-negative RT-PCR results from sampling the upper respiratory tract, some hospitals in the Hubei province of China included CT scans in the diagnosis of SARS-CoV-2 infection (53, 117, 122, 123). While diagnostic imaging techniques like CT have merits to help assess lower respiratory tract disease involvement, to monitor disease progression, or to investigate other complications of COVID-19, it should be noted that diagnostic imaging methods are less sensitive than sampling the lower respiratory tract and testing using molecular methods, and specificity is low, given that typical features of COVID-19 are common to other respiratory viruses or illnesses (113–116, 124–128). Initial reports of the utility of CT scans in the diagnosis of COVID-19 suggested an increased sensitivity of CT over real-time RT-PCR, but others have suggested explanations for the disparities between RT-PCR results and diagnostic imaging assessments, including poor sampling techniques, differences in the performances of testing methods, the anatomical site of RT-PCR testing (upper versus lower tract), and disease prevalence (111, 124, 126, 129–134). High sensitivities (i.e., >90%) have been reported for CT scans in high-prevalence populations, while low sensitivities (<60%) were reported in studies with low-prevalence populations (112–114, 118–121, 123). In a Cochrane review for confirmed cases of COVID-19, the pooled sensitivities were 93.1% (95% confidence interval [CI], 90.2% to 95.0%) for chest CT and 82.1% (95% CI, 62.5% to 92.7%) for CXR, but heterogeneity between studies was considerable (121). Specificity for diagnostic imaging is low, at 18.1% (95% CI, 3.7 to 55.8%) (121). In other words, approximately 80% of individuals would have received a diagnosis of COVID-19 in the absence of disease. As such, the use of diagnostic imaging techniques should be accompanied by careful consideration of factors such as disease prevalence in the study population, severity of the illness, performance and context of the methods used, differences in radiologist opinions, and possible confounding diagnoses (112–114, 118–121, 123, 126, 132–135). On the other hand, it is also important to recognize that diagnostic imaging is a useful tool for patient management with or without a confirmed etiology through laboratory testing, as it can be used to monitor the severity of illness and disease progression and assess possible complications (136–141).
Understanding the benefits and limitations of diagnostic imaging for COVID-19 is an active area of research, along with applications of artificial intelligence (AI) (also known as machine learning) (142–144). AI-based methods can be used in diagnostic imaging to help recognize abnormal features in images and classify them into defined categories, thus increasing accuracy, standardization, and speed of analyses by radiologists (50, 109, 142–146). AI approaches can be categorized into three main groups: approaches that analyze CT scan images, methods based on X ray, and those that realize diagnosis through jointly analyzing CT scan and X-ray images (147–151). While AI-based applications have shown benefits for diagnostic imaging methodologies (50, 109, 145, 146), more clinical investigations are needed to evaluate their possible incorporation into routine procedures for investigations of suspected cases of COVID-19, and laboratory testing is required to confirm the disease etiology. Furthermore, acquiring a reliable AI-based system requires access to a comprehensive training data set that includes all variations of COVID-19 as well as other lung diseases; providing such an all-inclusive data set is difficult and labor-intensive.
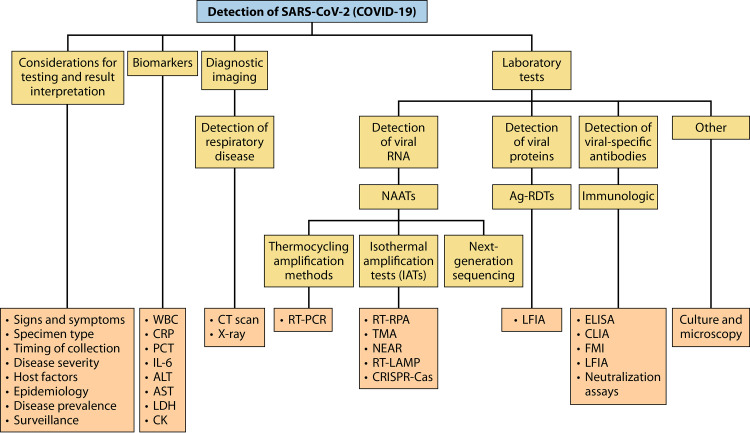
LABORATORY METHODS FOR THE DETECTION OF SARS-CoV-2
Diagnosis of COVID-19 can be performed using molecular detection of SARS-CoV-2 RNA, which is now widely available. Immunodiagnostic methods for identifying viral antigens and serology to recognize an immune response to the virus are also available. The following sections describe the commonly used and potential methods for the laboratory detection of SARS-CoV-2, with important consideration for factors like specimen type and timing of specimen collection.
Specimen Types
Prior to describing methods for SARS-CoV-2 detection, it should be recognized that accurate detection of any infectious disease requires adequate specimen collection at the anatomical site of infection, at a time when the pathogen of interest should be present (152–154). While the virus has been detected in a variety of specimen types using molecular methods (26, 96, 155–161), few have been widely adopted due to unreliable detection or a lack of sensitivity. The use of blood, serum, or plasma for SARS-CoV-2-specific serology and other immunodiagnostic tests is discussed in later sections of this review.
For respiratory viruses like SARS-CoV-2, specimens collected from the upper respiratory tract using a flocked nasopharyngeal (NP) swab that is placed in universal or viral transport medium (UTM or VTM, respectively) are the gold standards (162–164). In contrast to other swabs (e.g., cotton swabs on wooden sticks), specimen collection using flocked NP swabs that are coated with multilength fibers allows for enhanced recovery of respiratory viruses and bacteria, and the UTM or VTM allows a stable medium for transport to the laboratory (133). Other than NP swab specimens, alternative specimens and collection methods have been validated and gained interest, including the use of nasal midturbinate swabs, sampling of the anterior nares (Na), oropharyngeal (OP) swabs, or washes/aspirates from the nasopharynx, nose, or throat (96, 97, 154, 165–172). Specimen combinations can also be used. For example, paired collection using an OP swab along with sampling of the anterior nares was shown to be equivalent to NP swab collection for the detection of SARS-CoV-2, and different options are available for OP/Na collection (133, 154, 167, 168, 173). It is worth mentioning that during the COVID-19 pandemic, procurement of NP swabs and UTM (or VTM) was challenged by global supply chain shortages. Several groups have developed and validated the possibility of using 3D-printed swabs as alternatives to commercial NP or nasal swabs, but while some have been clinically validated for detecting SARS-CoV-2 RNA, further investigations are required for their applicability in SARS-CoV-2 antigen detection (174–178). For media used for swab transport to laboratories, other than the typical UTM or VTM, alternatives have been investigated for use for SARS-CoV-2 testing, including Amies transport medium, sterile normal saline, phosphate-buffered saline (PBS), M4 medium, and minimal essential medium (MEM), and stability analyses have assessed ideal transport and storage conditions (168, 169, 179).
While NP swabs are considered the gold standard for respiratory virus sampling of the upper respiratory tract, hospitalized adults with progression of COVID-19 to lower tract disease may require additional specimen types (170). When lower tract infection is suspected through clinical presentation or with the aid of diagnostic imaging, specimens such as BAL fluid, endotracheal secretions, or sputum should be considered (55, 95–102, 180).
Recently, the use of noninvasive collection methods like saliva and throat gargles has gained much interest, as these samples are amenable to self-collection and have the potential for large-scale population-based surveillance (181–186). While some studies have demonstrated that the performance of saliva for the detection of SARS-CoV-2 was comparable to that of NP or nasal swab collections (183, 184, 187–190), others challenged the performance of saliva for SARS-CoV-2 detection (191). The variability in saliva collection or differences in the patient populations tested might explain these inconsistencies, but further analyses are required (191, 192). Also, although not used routinely in many laboratories, detecting SARS-CoV-2 RNA from stool is possible in the presence or absence of gastrointestinal symptoms (193). The possibility of culturing SARS-CoV-2 from stool opens discussions regarding the possibility of fecal-oral transmission and human health or ecological risks (194, 195) and also opens the opportunity for research into community-based surveillance in low-prevalence settings using wastewater (193, 196).
In postmortem examinations, the extent of investigations will be dependent on several factors, but NP swabs, swabs from the lungs, and tissue samples can be used for diagnostic testing for SARS-CoV-2 (197–200). Specimens in 10% buffered neutral saline or formalin-fixed paraffin-embedded (FFPE) specimens commonly used for histopathological examinations can also be used, but these pose challenges for NAATs like real-time RT-PCR as RNA can be degraded by formalin, and sensitivity for the detection of SARS-CoV-2 RNA by real-time RT-PCR could be compromised (201). The CDC recommends that these media be used in limited settings (197). Immunohistochemical (IHC) and in situ hybridization (ISH) assays for the detection of SARS-CoV-2 have now been developed, but limited data are available on their performance (202).
Timing of Specimen Collection
SARS-CoV-2 has been identified in various clinical specimen types (26, 96, 154–161), but the timing of detection differs between methods and the specimen types collected for testing. SARS-CoV-2 RNA can be detected early in the presymptomatic stage of the disease and later on, even after recovery. However, the timing of specimen collection is critical, as testing too early or too late following exposure can potentially lead to false-negative results (203). It was shown that real-time RT-PCR false-negative rates could be minimized by testing 2 to 3 days after symptom onset, with an average time of symptom onset of 5 days postexposure (204–210). Repeat testing can be considered for individuals with an initial negative test result but for whom there is a high level of clinical suspicion (134). Of note, viral shedding studies are often performed using RNA detection alone and less often in combination with virus culture; however, the absence of cultivable virus does not preclude the potential for SARS-CoV-2 transmission, and laboratory detection of SARS-CoV-2 using molecular methods does not imply infectious virus (208, 211–217). For the purpose of this section, viral shedding is described in the context of RNA detection without implying the potential for viral transmission. A discussion regarding the association of SARS-CoV-2 RNA detection with potential infectivity is covered later in this section as well as in the real-time RT-PCR section below.
The magnitude of the viral load and duration of shedding depend on the specimen type, the anatomical site of illness, the severity of illness, and, likely, the host immune response to infection (170, 208, 218–220). The average duration of SARS-CoV-2 RNA detection in the upper respiratory tract of patients with mild disease ranged from 7.9 to 20 days after symptom onset and from 6 to 30.8 days in cases with moderate to severe illness. The detection of SARS-CoV-2 in the lower respiratory tract ranged from 8 to 38.4 days for mild cases of COVID-19 and spanned between 6 and 26.9 days for moderate to severe illness (221). In a systematic review and meta-analysis, the pooled estimates of the mean duration of SARS-CoV-2 RNA detection from symptom onset in mild adult cases were 12.1 days (95% CI, 10.1 to 14.1 days) in the upper respiratory tract and 24.1 days (95% CI, 10.0 to 38.2 days) in the lower respiratory tract. For moderate to severe cases, the pooled estimates for the duration of SARS-CoV-2 RNA positivity in the upper respiratory tract were 15.8 days (95% CI, 11.1 to 20.6 days) and 23.2 days (95% CI, 21.5 to 25.0 days) in the lower respiratory tract (221). In a systematic review and meta-analysis, the temporal dynamics of SARS-CoV-2 viral loads were stratified by COVID-19 severity and sampling site. In cases of mild adult disease, SARS-CoV-2 RNA in the upper respiratory tract was maximal on day 4, at approximately 6.6 × 108 copies/ml, whereas lower tract viral loads peaked at approximately 2.7 × 108 copies/ml on day 6 after symptom onset (221). In cases of moderate to severe adult disease, maximal SARS-CoV-2 RNA detected in the upper respiratory tract occurred on day 8, at 4.6 × 109 copies/ml, and on day 11, at approximately 3.5 × 108 copies/ml, in the lower respiratory tract (221). Regarding the differences in viral loads and durations of shedding between symptomatic and asymptomatic patients, the literature is inconsistent. Some publications observed little to no difference in viral loads between the two groups (222–226), while others suggested significantly higher viral loads in symptomatic patients (224). Whether these differences are attributed to differences in disease severity, variations in the performances of methods used, or host factors remains to be determined.
As highlighted above, the median duration of viral shedding is variable between individuals and likely dependent on disease severity and several host factors such as age, immunocompromising conditions, or medical comorbidities (208, 212–217, 227–229). While most individuals with mild disease clear the virus within 10 to 20 days, in some cases with severe COVID-19, the duration of shedding can be prolonged (217, 230). The longest durations of viral RNA shedding reported to date were 83 and 111 days after symptom onset (231, 232); however, the persistence of RNA suggestive of low viral loads may be of little clinical significance, as the detection of SARS-CoV-2 RNA does not necessarily imply infectivity (170, 208, 212–220, 227–229). Moreover, many factors can affect the detection of SARS-CoV-2 RNA, such as the quality of sample collection, transport, and variables in laboratory processing; RNA positivity can be intermittent and inaccurate at the later stages of illness (233). Therefore, the CDC recommends that the discontinuation of transmission-based precautions for patients with confirmed SARS-CoV-2 infection should be based on the resolution of symptoms and not based on testing (230). While some countries have similar recommendations for discharge from quarantine, there is some heterogeneity in approaches, and these often vary based on the severity of illness and the presence or absence of symptoms (234).
While not used routinely in many laboratories, detecting SARS-CoV-2 RNA from stool is possible in the presence or absence of gastrointestinal (GI) symptoms (193, 235). However, only 1% of patients had detectable RNA in their stool in the absence of positive respiratory specimens (193, 235). For some patients, viral shedding in stool can occur for a longer period than in the respiratory samples and could help diagnose infection if upper and lower respiratory tract specimens are negative but there is a high suspicion of disease (156–160, 193, 236). Of individuals who test positive with GI specimens, the median duration of RNA shedding in the GI tract is 12.5 days following negative respiratory tract specimens (193, 235). Less frequently, shedding in stool can be prolonged and has been documented up to 70 days after symptom onset or 33 days following clearance from the respiratory tract (236, 237). As for respiratory tract specimens, RNA detection does not necessarily imply that infectious virions are produced, but SARS-CoV-2 has been cultured from stool specimens in some studies (236, 237).
Like molecular methods, antigen testing can be used to detect SARS-CoV-2 proteins in the acute stages of the disease following the incubation period in upper respiratory tract specimens such as NP swabs, nasal swabs, and possibly saliva. Antigen detection using immunoassays like lateral flow rapid diagnostic tests (RDTs) is often less sensitive than molecular methods (203, 238), but these tests can detect SARS-CoV-2 antigen reliably when the viral load is high in the clinical specimens (i.e., typically from 1 to 3 days before the onset of symptoms to 5 to 7 days after symptom onset), whereas the likelihood of SARS-CoV-2 detection decreases in the second week after symptom onset (238).
In contrast to RNA and antigen detection, immunological responses take longer to appear, with antibodies typically beginning to appear 6 days after symptom onset, as viral RNA levels begin to decline (207). Typically, the first detectible antibody in human blood is immunoglobulin M (IgM), followed by immunoglobulin G (IgG). However, concomitant increases of the IgM and IgG immunoglobulin classes as well as IgG first seroconversion have also been observed (239). Few data are also available for immunoglobulin A (IgA) detection, a marker of mucosal immune responses, but it is evident that both IgA and IgM decline rapidly over the course of infection (240). The median seroconversion times for total antibody, IgM, and IgG were 9, 10, and 12 days after symptom onset (or 15, 18, and 20 days after exposure), respectively (240). It is unclear how long IgG responses last or whether they confer protection against subsequent SARS-CoV-2 reinfection (241). The longest study on the antibody dynamics tracked IgG up to 115 days after symptom onset in sera and saliva (242). Immune responses may vary depending on disease severity and host factors such as immunocompromising conditions or other medical comorbidities, and the value of immune responses will be dependent on the ability to provide neutralizing antibodies (nAbs) or cellular immunity capable of viral clearance. The applications and limitations of serology and other immunodiagnostics are discussed in more detail in later sections of this paper.
Specimen Preprocessing Requirements
While detection of SARS-CoV-2 from respiratory specimens can be performed using RNA or antigen detection, they sometimes require a preprocessing step like heat lysis or inactivation using guanidinium salts before nucleic acid extraction and amplification or testing, to ensure safe handling conditions, depending on local biosafety risk assessments (133, 154, 173, 180). Specimen types such as sputum may require mucolytic agents such as dithiothreitol (DTT), N-acetyl-l-cysteine (NALC), or proteinase K (PK) to reduce specimen viscosity prior to testing (243). Other preprocessing steps would include centrifugation for specimens like stool (236), PK digests for tissues (e.g., lung biopsy specimens), and specimen aliquoting into compatible tubes for testing (if testing from primary specimen containers is not possible). With any manipulation of the primary specimen (i.e., preprocessing steps), careful consideration should be undertaken to ensure that there are no potential impacts on downstream testing (e.g., RNA or antigen stability). Of note, some preprocessing steps, like specimen lysis, can be done in conjunction with nucleic acid extraction using automated instrumentation (discussed below in the real-time RT-PCR section of this review).
Overall, the choice of the specimen and timing of collection are crucial for the accurate detection of SARS-CoV-2, as are factors such as the severity of illness. Given that the performance characteristics of diagnostic methods depend on numerous variables as well as the method(s) used as a comparator and disease prevalence, a comprehensive synthesis of all method performances falls outside the scope of this review. However, general concepts for performance characteristics, important considerations, and a description of the technologies used for SARS-CoV-2 detection in the clinical setting or in development are presented in the following sections.
Molecular Methods for Viral RNA Detection
While no true reference standard exists for detecting SARS-CoV-2, nucleic acid amplification tests (NAATs) such as real-time RT-PCR are the methods of choice for SARS-CoV-2 diagnostic testing (40, 41, 48, 53, 54, 244). Following sequencing of its genome (10), laboratory-developed tests (LDTs) for the detection of SARS-CoV-2 were quickly developed, and protocols were circulated broadly by health care regulatory bodies such as the World Health Organization (WHO) and the U.S. Centers for Disease Control and Prevention (CDC) (23, 40, 245–247). Many commercial kits have since become available and were authorized for use through emergency use authorization (EUA) by entities such as the Food and Drug Administration (FDA) and Health Canada. Lists of authorized medical devices related to COVID-19 in Canada and the United States are regularly updated online (29, 30), and examples of them are summarized in Table 1.
TABLE 1.
Examples of the NAATs approved for emergency use by the U.S. FDA for detection of SARS-CoV-2 RNAa
| Device/assay (manufacturer) | Method | Target gene(s) | Specimen type(s) | Authorized setting(s) | Time/throughput | LoDb | Reference |
|---|---|---|---|---|---|---|---|
| cobas 6800/cobas SARS-CoV-2 (Roche Molecular Systems, USA) | RT-PCR | ORF1ab + E | NS, NPS, OPS | H, M, H-pooling | 3 h for the first-run results but 90 min per run in continuous mode/864 samples per 8 h | 46 copies/ml | 575 |
| Abbott m2000/RealTime SARS-CoV-2 (Abbott Diagnostics, USA) | RT-PCR | RdRp + N | NS, NPS, OPS, BAL fluid | H | 7 h per run/470 samples per 24 h | 100 copies/ml | 576 |
| NeuMoDx 288/NeuMoDx SARS-CoV-2 (NeuMoDx Molecular, USA) | RT-PCR | Nsp2 + N | NS, NPS, OPS, BAL fluid, saliva | H, M | 1.3 h per run/288 samples per 8 h | 150 copies/ml | 577 |
| Panther Fusion/Aptima SARS-CoV-2 (Hologic, USA) | TMA | ORF1ab | NS, NPS, OPS, MTS, NPW, NPA, NA | H, pooling | 2.4 h per run/500 samples per 8 h | 0.026 TCID50/ml | 578 |
| Liaison MDX/Simplexa COVID-19 Direct (DiaSorin Molecular, Italy) | RT-PCR | ORF1ab + S | NS, NPS, NW, NA, BAL fluid | H, M | 1 h per run/8 samples per run | 500 copies/ml | 318 |
| FilmArray/BioFire Respiratory Panel 2.1 (BioFire Diagnostics, USA) | RT-PCR | S + M | NPS | H, M | 2-min hands-on time/1 h per run | 160 copies/ml | 579 |
| ePlex/ePlex SARS-CoV-2 (GeneMark Diagnostics, USA) | RT-PCR | N | NPS | H, M | 2-min hands-on time/1.5 h per run | 750 copies/ml | 319 |
| GeneXpert Xpress/Xpert Xpress SARS-CoV-2 (Cepheid, USA) | RT-PCR | E + N2 | NS, NPS, OPS, MTS, NW, NA | H, M, W | 1-min hands-on time/45 min per run | 0.02 PFU/ml | 580 |
| Accula Dock/Accula SARS-CoV-2 (Mesa Biotech, USA) | RT-PCR | N | NS, MTS | H, M, W | 5-min hands-on time/30 min per run | 150 copies/reaction | 320 |
| ID Now/ID Now COVID-19 (Abbott Diagnostics, USA) | NEAR | RdRp | NS, NPS, OPS | H, M, W | 2-min hands-on time/13 min per run | 125 copies/ml | 581 |
| SHERLOCK CRISPR SARS-CoV-2 kit (Sherlock Biosciences, USA) | RT-LAMP, CRISPR-Cas13 | ORF1ab + N | NS, NPS, OPS, NPW, NPA, NA, BAL fluid | H | 1 h per run | 6,750 copies/ml | 582 |
| SARS-CoV-2 DETECTR reagent kit (Mammoth Biosciences, USA) | RT-LAMP, CRISPR-Cas12 | N | NPS, OPS, MTS, ANS, NPW, NPA, NA | H | 45 min per run | 20,000 copies/ml | 583 |
| NovaSeq 6000/Illumina COVIDSeq test (Illumina Inc., USA) | Next-generation sequencing | 98 targets on the virus | NPS, OPS, MTS, ANS, NPW, NPA, NA, BAL fluid | H | 3,072 samples per 12 h | 500 copies/ml | 584 |
The full list is available in reference 29. Abbreviations: RT-PCR, reverse transcription-PCR; TMA, transcription-mediated amplification; NEAR, nicking enzyme amplification reaction; RT-LAMP, reverse transcription–loop-mediated isothermal amplification; NS, nasal swab; NPS, nasopharyngeal swab; OPS, oropharyngeal (throat) swab; BAL, bronchoalveolar lavage; MTS, midturbinate nasal swab; NPW, nasopharyngeal wash; NPA, nasopharyngeal aspirate; NA, nasal aspirate; NW, nasal wash; ANS, anterior nasal swab; H, laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high-complexity tests; M, laboratories certified under CLIA, 42 U.S.C. §263a, that meet requirements to perform moderate-complexity tests; W, patient care settings operating under a CLIA certificate of waiver; TCID50, median tissue culture infectious dose.
The LoD (limit of detection) of each assay is the lowest LoD reported in the instructions for use for that assay, regardless of the specimen types.
Real-time RT-PCR.
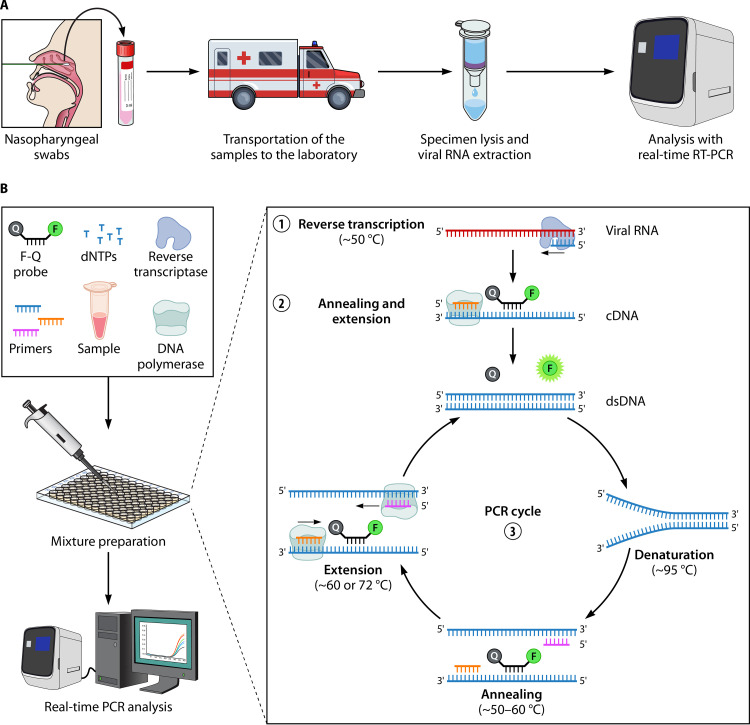
Among NAATs, real-time RT-PCR is the most widely used method for the detection of SARS-CoV-2. As shown in Fig. 3A, the sample workflow for SARS-CoV-2 real-time RT-PCR includes specimen collection, transportation of the samples to the laboratory, specimen lysis, purification of viral RNA through nucleic acid extraction, and real-time RT-PCR amplification, detection, and analysis. Prior to real-time RT-PCR amplification, specimens are lysed to provide access to the SARS-CoV-2 RNA, and nucleic acid extraction is performed to remove potential inhibitors that could impede the amplification of the target. Both lysis/extraction and RT-PCR amplification can be performed sequentially through manual processing on individual instruments, or the entire process can be automated.
FIG 3.
Real-time RT-PCR analysis. (A) Typical steps required for the detection of SARS-CoV-2 with real-time RT-PCR. (B) Principle of real-time RT-PCR. (1) During reverse transcription, reverse transcriptase (RT) creates a cDNA from the viral RNA template, with the aid of the reverse primer (or random oligonucleotides). The RNase H activity of the RT digests the initial RNA template. (2) The DNA polymerase activity of RT (or of the Taq polymerase) completes the second DNA strand guided by the forward primer and cDNA. (3) The newly formed double-stranded DNA (dsDNA) is used as a template for the PCR portion of the assay. At the annealing stage, the reverse primer binds to the sense strand of dsDNA in a sequence-specific manner, and the forward primer and a dually labeled probe bind to the antisense strand of the DNA. In this stage, the fluorophore (F) present on the probe is masked by the quencher (Q). During the extension step, the DNA polymerase extends the forward primer and, in the process, hydrolyzes the probe, resulting in the release of the fluorophore. Next, following excitation, fluorescence emission can be captured by the real-time thermocycler. With each round of PCR amplification, the dsDNA amplicon is multiplied by a 2-fold factor, with a proportional increase in the overall fluorescence signal. After 30 to 40 cycles of amplification, the RT-PCR is complete. The PCR cycle at which the fluorescence signal crosses the threshold for positivity is called the threshold cycle (CT), and CT values are inversely proportional to the quantity of the target present in the reaction mixture.
(i) Specimen lysis and RNA purification.
To release viral RNA from host cells and virions, specimen lysis can be performed using physical (e.g., heat, sonication, or homogenization), chemical (e.g., organic solvents, detergents, chelating agents, or chaotropic agents), or enzymatic (e.g., proteases) methods (54, 243, 248, 249). Lysis steps based on enzymatic digestion (e.g., proteinase K digestion) are common for nucleic acid extraction in clinical laboratories. Following specimen lysis, extraction of viral RNA is performed to remove cellular debris and contaminants that could potentially inhibit the RT-PCR and purify the nucleic acids (250–252). In many automated instruments, silica-coated magnetic microbeads are used to capture nucleic acids, which can be sequentially transferred into different wash solutions by a robotic pipetting instrument with a magnetic head (248–252). The efficiencies of several extraction methods have been compared for detecting SARS-CoV-2, and the results favor commercial kits over manual methods like organic extractions containing guanidinium thiocyanate-phenol-chloroform (253).
It should be noted that while nucleic acid extraction is essential to achieve optimal sensitivity in molecular assays, recent studies have described extraction-free protocols for molecular testing for SARS-CoV-2 to circumvent the potential bottleneck of extraction if the supply of extraction reagents or consumables is limited (or to provide options for low-income environments) (254–262). However, without a nucleic acid extraction step to remove PCR inhibitors in clinical specimens, there is a notable reduction in sensitivity, but the extent is dependent on the method and target used for SARS-CoV-2 detection, the type and duration of the lysis/inactivation method (heat or chemical), the input volume, the specimen type, the transport media, and the viral load in the specimen (253, 257, 263–265). For example, the sensitivity of a 60-min heat inactivation alone reached 100% for specimens with moderate to high viral loads (threshold cycle [CT] values of between 20 and 30) but declined to 54% in specimens with CT values of >30 (264).
(ii) Target amplification and detection.
Amplification in real-time RT-PCR involves two main steps. First, an enzyme called reverse transcriptase creates a cDNA from the viral RNA. The cDNA is then used as a template in a real-time PCR amplification step where fluorescence is produced as DNA amplification occurs (41, 266). The PCR portion of real-time RT-PCR contains a fluorescent probe or dye to generate fluorescence (e.g., dually labeled hydrolysis probe or intercalating dyes that bind to double-stranded DNA [dsDNA], like SYBR green) (267–272). Figure 3B illustrates the principle of a typical real-time RT-PCR using a dually labeled hydrolysis probe. Overall, if amplification of the target genes occurs during cycling through the denaturation, annealing, and extension stages, a fluorescent signal is produced that can be captured by the real-time thermocycler (23, 41, 273). If the fluorescence crosses a defined threshold, the cycle in which it occurred is termed the threshold cycle (CT). CT values help interpret results as positive, negative, or indeterminate (or equivocal), and each real-time RT-PCR method must validate its cutoff values, as they may differ among methods and instruments (40, 53, 274).
The performance of real-time RT-PCR depends on a number of factors, including the specimen type, the timing of collection, the quality and quantity of viral RNA, the primers and probes designed and their viral RNA target, the reagents used for the RT-PCR(s), the instrument and its operational parameters, and the signal/cutoffs used for result interpretation (23, 40, 53). Typically, real-time RT-PCR assays demonstrate high sensitivity and specificity for SARS-CoV-2. In a systematic review and meta-analysis by Mustafa Hellou et al., the pooled sensitivity for SARS-CoV-2 detection from 29 studies was 96.2% (95% CI, 91.0% to 98.4%), and the pooled specificity was 98.1% (95% CI, 95.9% to 99.2%) (106). Sensitivity is of the utmost importance to ensure that cases in a population are identified, and specificity is important to ensure that no false-positive results occur. SARS-CoV-2 NAATs like real-time RT-PCR do not cross-react with other respiratory viruses, including human coronaviruses.
(iii) SARS-CoV-2 targets and data interpretation.
Various targets have been used for SARS-CoV-2 real-time RT-PCR, but the genes encoding E, N, S, and ORF1ab are used widely (40, 41, 53, 275–279). Despite limited access to control materials early in the pandemic, LeBlanc et al. assessed the analytical performances of various LDTs and commercial RT-PCR assays and found that most of them had a high sensitivity with similar limits of detection (LoDs) in the range of 3.4 to 4.5 log10 copies/ml (11.2 to 141 copies/reaction) (40). Similar proficiency testing across laboratories has been performed by others, demonstrating high sensitivity and specificity across different NAAT methods and instruments (280–282).
In the initial stages of the pandemic, dual- or multigene detection strategies were adopted for real-time RT-PCRs to ensure assay specificity (40). As the pandemic progressed and the disease prevalence increased, some laboratories implemented single-target detection of SARS-CoV-2 in LDTs to streamline the workflow; however, many commercial methods continue to rely on detection using two or more targets (283). In duplex or multiplex real-time RT-PCR assays, if the identification of any target is considered a positive result, sensitivity would be enhanced compared to the requirement of at least two targets to be positive for a SARS-CoV-2 result to be released as such (40). However, this strategy comes at the risk of decreasing the specificity and potentially increasing the false-positivity rate (40). Regardless of the approach for testing, each target should have a validated range of values that define a positive, negative, or indeterminate result as well as conditions that would trigger an invalid result (e.g., processing or quality failures) (40, 284). CT values can be used in real-time RT-PCR to define these cutoffs, and indeterminate (or equivocal) results arise for values falling between the CT cutoff values of negative results and the reproducible CT value cutoff for positivity. This is sometimes termed the diagnostic gray zone for result interpretation (40, 285).
Low CT values suggest that more viral RNA was present in the specimen, whereas high CT values represent specimens with lower virus burdens, as more cycles were required to amplify the viral target. Therefore, CT values are sometimes used as a surrogate for viral load. While low viral loads (indicated by high CT values) could represent early or late disease, they could also be explained by nonspecific reactions (i.e., false-positive reactions), poor collection techniques, specimen integrity issues during storage or transport, or a problem occurring during laboratory processing. As discussed in the section on the timing of specimen collection above, some studies have evaluated the correlation between CT values and infectivity (208, 211–217). There are some data to suggest that specimens that have SARS-CoV-2 RT-PCR results with CT values of >24 cannot be effectively grown in tissue culture (212), yet other data have shown that SARS-CoV-2 can be recovered from cultured specimens with a CT value of >35, at a lower frequency (213, 214). While methodologies may have differed between studies to explain these differences (e.g., fresh versus frozen specimens), it should be noted that the infectious dose required for human infection with SARS-CoV-2 is unknown and influenced by many biological and environmental variables. Given the variability that can occur in specimen collection, transport, and processing, there are no biological correlates accurately linking CT values to infectivity or the potential for transmission. Detectable virus by NAATs does not imply infectious virus. Of note, even if such a correlate existed, CT value cutoffs cannot be applied universally to all NAATs, as they are method, reagent, and target specific, and to date, there is no international standard that can be used for calibration. Of note, other studies have investigated the role of CT values in predicting the clinical course of COVID-19 or prognosis (286, 287); however, given the number of factors that could influence CT values, along with the inability to standardize respiratory specimens, the role of CT values in accurately predicting clinical outcomes would likely be inconsistent, and further research is needed. While the interpretation of CT values requires careful consideration, it is clear that CT values vary based on the viral burden, which itself varies throughout SARS-CoV-2 infection. Staging infection can provide epidemiological clues and can help with patient management. For example, low viral loads (i.e., high CT values) are seen during presymptomatic, early, or late stages of infection, whereas low CT values are seen between the early and late stages. If clinically indicated, patients with high CT values should undergo repeat testing within 24 to 48 h to determine if the CT value is stable, rising, or declining to help stage potential exposures in contact tracing (167, 168, 230, 288). However, CT value interpretation is complicated in asymptomatic infections, where the time of infection onset may be unknown. Therefore, to rule out potential false-positive results, repeat testing is recommended for patients with high CT values suggestive of low viral loads (167, 168, 230, 288).
(iv) Automation.
LDTs and commercial assays for moderate- to high-throughput testing for SARS-CoV-2 require relatively expensive equipment and experienced personnel to obtain accurate and robust data, and the turnaround time for results can take several hours. Real-time RT-PCR assays are constantly being improved to increase specimen throughput, provide rapid specimen turnaround times, reduce the hands-on time, and facilitate result interpretation and reporting. Automated high-throughput instruments are capable of performing over 1,000 tests daily, with performance characteristics greater than or equivalent to those of LDTs (167, 168, 251, 289–295). LDTs typically require separate nucleic acid extraction and amplification steps, but these processes can occur simultaneously with high-throughput instruments, along with full traceability, and results can be directly reported through interfacing with the laboratory information system. One of the first high-throughput instruments with a commercially available SARS-CoV-2 detection assay was the cobas 6800 instrument (Roche Molecular Systems, USA), but other highly automated instruments relying on NAAT technology are now available, with similar performances, testing capacities, and workflow benefits. These include the Abbott RealTime SARS-CoV-2 assay on the m2000 instrument (Abbott Molecular, USA), the Hologic Panther SARS-CoV-2 assay (Hologic, USA), the NeuMoDx SARS-CoV-2 assay (NeuMoDx Molecular, USA), and BD Max reagents (Becton, Dickinson, USA) (296–302). Advances have also been made for LDTs for SARS-CoV-2 testing using semiautomated robotics to streamline specimen processing, nucleic acid extraction, RT-PCR setup and amplification, data interpretation, and interfacing for data reporting (295). LDTs for SARS-CoV-2 have also been adapted for other instruments, such as droplet digital PCR (ddPCR). In ddPCR, water-oil emulsions are used to partition nucleic acid samples into thousands of nanoliter-sized droplets, and PCR amplification is carried out within each droplet (303–306). To date, the performance of ddPCR has been shown to be equivalent to or slightly more sensitive than LDT comparators, but limited data are available for its use in clinical laboratories (303–306).
(v) Specimen pooling.
Regardless of the NAAT used for SARS-CoV-2 detection, manufacturers of nucleic acid purification kits or RT-PCR reagents and consumables have been challenged with the rapid increase in testing demands that came with the global spread of SARS-CoV-2. With challenges to meet testing resources and limitations in the supply chain, research into alternative testing strategies has been explored (254). A possible strategy to increase testing capacity and gain laboratory efficiencies is group testing (i.e., specimen pooling) (307). While many pooling permutations are possible, its simplest application involves combining patient samples before testing and retesting individual specimens following the identification of a positive pool (308–312). The optimal number of specimens within pools (i.e., pool depth) varies with disease prevalence and assay performance (307, 311, 312). While larger pool depths may achieve higher efficiency, particularly for high-throughput instruments, the trade-off is the accompanying reduced sensitivity, with the potential generation of false-negative results (307). When prevalence is low, typically only a subset of specimens with low viral loads pass undetected, while the testing capacity is increased and the cost of testing is reduced (307, 311, 312). In settings of high disease prevalence, the merits of pooling are lost given the high number of pools that need to be resolved. Other challenges for pooling include the increased human resource requirements for specimen registration and processing, but robotics and pooling software can help mitigate some of these issues (311). For lower-throughput analyzers like NAAT-based RDTs, pooling can also be considered (312). Overall, while thorough validation and careful consideration of potential impacts of pooling should be considered before implementation on any instrument, pooling can offer an opportunity for clinical laboratories to increase testing capacity, reduce costs, and mitigate the supply chain limitations of laboratory testing (311).
(vi) RT-PCR-based rapid diagnostic tests.
Unlike high-throughput automated instruments that are focused on large specimen volumes, rapid diagnostic tests (RDTs), as their name implies, are focused on speed. While this is acceptable for routine testing, RDTs have been developed to provide rapid results with easy-to-use testing, with minimal hands-on processing steps to facilitate training and testing. The first RDT based on real-time RT-PCR that obtained EUA from the FDA and Canada was the Xpert Xpress SARS-CoV-2 assay on the Cepheid GeneXpert platform (Cepheid Inc., USA), which provides results in about 45 min, with a <5-min hands-on specimen processing time (27). This NAAT-based RDT showed analytical and clinical performance characteristics often greater than those of LDTs and other commercial NAATs (99, 167, 168, 290–292, 296, 313, 314). It should be noted that while Xpert Xpress is often referred to as a point-of-care (POC) test, this testing is not typically performed at the time and place of patient assessment and is more commonly performed in a laboratory setting; therefore, the term RDT would be more appropriate. The most current version of the Xpert SARS-CoV-2 assay is multiplexed with influenza A and B viruses as well as respiratory syncytial virus (RSV), which can present with similar respiratory symptoms (315). More highly multiplexed assays like BioFire Respiratory Panel 2.1 with SARS-CoV-2 (BioFire Diagnostics, USA) are also available, which allow a syndromic approach with the simultaneous detection of SARS-CoV-2 and several other respiratory viruses (316, 317). While syndromic testing is also being developed for larger instruments, such assays on RDTs are particularly useful for remote communities or resource-limited settings or for testing of populations where rapid diagnosis would be of benefit (e.g., patients admitted to the ICU).
Other devices with a focus on potential POC applications have integrated RT-PCR with rapid (5- to 30-min) technologies such as digital microfluidics, visual lateral flow readouts, and portable instruments (318–325). All these assays have the advantage of speed and simplicity but are prone to limitations such as low sensitivity, low specimen throughput, and minimal scalability (288). For developing countries or other resource-limited settings where instrumentation is lacking, other cost-sparing testing alternatives are being explored. Arumugam et al. demonstrated a proof of principle of an RT-PCR that could be conducted in 12 min using a setup consisting of thin-walled PCR tubes, sous vide immersion heaters/circulators, and an endpoint readout performed with a light-emitting diode (LED) gel-viewing box (326). Such creative and innovative solutions from industry and academic settings help meet the global needs for SARS-CoV-2 laboratory testing, besides other NAATs rapidly being developed and validated.
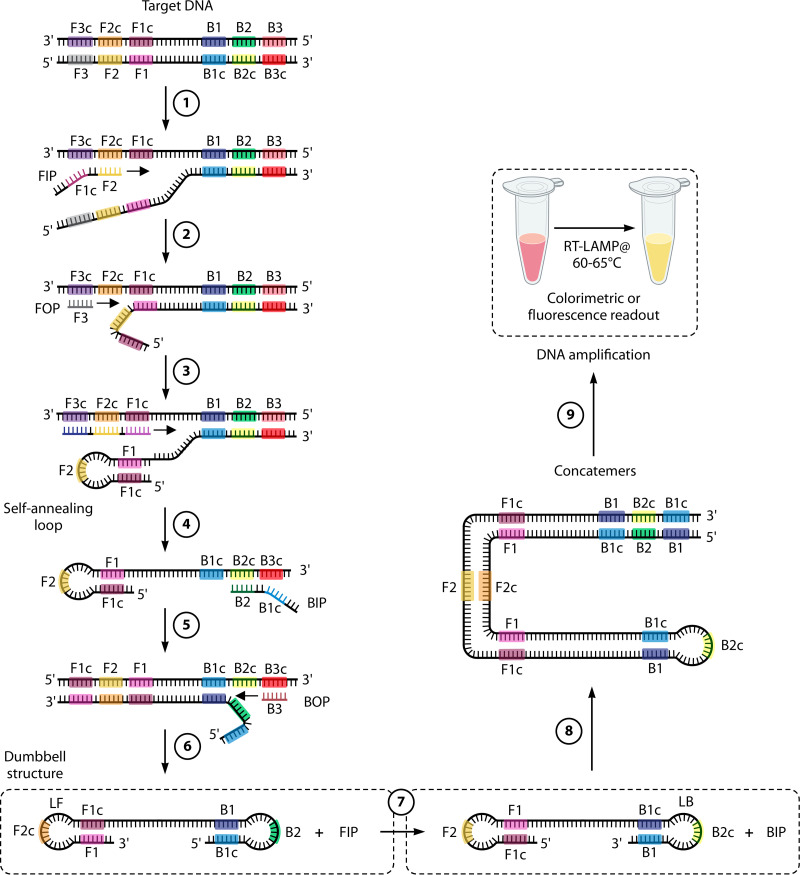
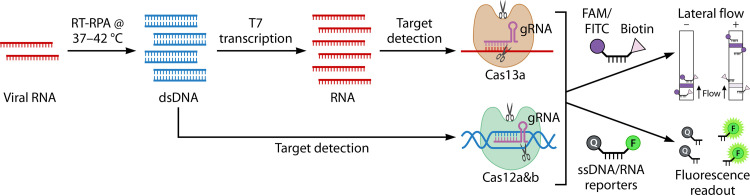
Isothermal amplification technologies.
In efforts to develop portable and rapid diagnostic tests for SARS-CoV-2, NAATs other than RT-PCR have been investigated. Isothermal amplification technologies (IATs) are conducted at a constant temperature, eliminating the need for expensive equipment such as thermocyclers. The principles behind IATs rely on thermal or enzymatic denaturation of nucleic acids, followed by nucleic acid amplification reactions, and have been reviewed in detail elsewhere (244). Isothermal NAAT technologies include transcription-mediated amplification (TMA), nicking enzyme-assisted reaction (NEAR), loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), and systems using clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated (Cas) (CRISPR-Cas) systems. While most IAT methods have been applied to DNA, they can often be adapted to RNA amplification by adding an RT step (e.g., RT-LAMP and RT-RPA) (244, 271, 327, 328). Other IATs were designed for the intent of RNA amplification (e.g., TMA). Only TMA has been commercialized on a high-throughput instrument, but other IATs have been explored for uses as RDTs or for potential POC applications (e.g., RT-RPA, RT-LAMP, NEAR, and CRISPR-Cas) (244). The following sections describe examples of IATs and current and potential applications.
(i) Reverse transcription-recombinase polymerase amplification.
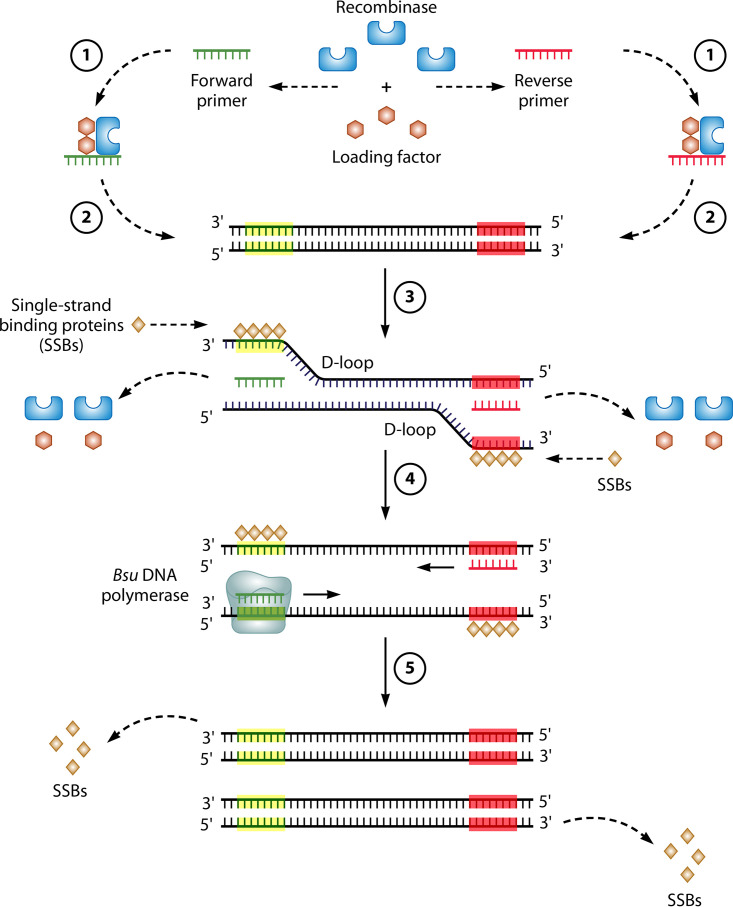
As shown in Fig. 4, the mechanism of RPA relies on homologous recombination (329). RT-RPA uses a DNA polymerase to extend forward and reverse primers and make copies of each DNA strand (like PCR). However, to unwind and copy the DNA strands generated from reverse transcription, RPA requires the ATP-dependent targeting activity of a recombinase complex as well as the polymerase activity of a strand displacement DNA polymerase (e.g., Bsu) (329, 330).
FIG 4.
Mechanism of RT-RPA. The RT-RPA reactions typically occur at between 37°C and 42°C in the following steps. (1) The reaction is initiated by the binding of a recombinase (e.g., T4 UvsX) and a loading factor (e.g., T4 UvsY) to each of the forward and reverse primers. (2) These recombinase/loading factor/oligonucleotide complexes search for homologous sequences in dsDNA, formed in the RT reaction from viral RNA (not depicted). (3) Once sequence homology is found, the recombinase complex invades the duplex DNA, forming a structure called a D-loop in an ATP-dependent reaction, where there is the unwinding of dsDNA and binding of the primer to its complementary sequence. Access to the primer-binding sequence is possible due to the stabilization of the opposite strand by SSBs (e.g., T4 gp32). Subsequently, the recombinase and loading factor disassemble and are released to initiate other rounds of target recognition. (4) Following the binding of the forward and reverse primers, these primers are extended at their 3′ ends using a strand displacement DNA polymerase (e.g., Bsu), and during the elongation process, there is a further separation of the two strands. (5) Eventually, SSBs are displaced, and the replication of both strands is complete.
While the mechanisms for RT-RPA are relatively simple, the reaction components are fairly complex. Single-tube RT-RPA reactions include forward and reverse primers, core enzymes (e.g., reverse transcriptase, recombinase, recombinase loading factor, and a strand displacement DNA polymerase), proteins like single-stranded binding protein (SSB), and a number of ancillary components such as deoxynucleoside triphosphates (dNTPs), salts, buffers, cofactors, crowding agents, ATP, and an enzymatic system to generate additional ATP (phosphocreatine and creatine kinase [CK]). Once added, magnesium (Mg2+) initiates the RPA reaction. Fortunately, various kits are now commercially available for RPA (e.g., TwistDx, United Kingdom), with variations for the probe used in the detection step (329). For example, the TwistAmp exonuclease (exo) probes are used for fluorescence detection through a mechanism involving exonuclease III, whereas a detection system designed for a lateral flow assay (LFA) output can be incorporated using endonuclease IV (nfo) probes (329). Alternative fluorescence technologies have also been used, such as fluorescence resonance energy transfer (FRET) probes or CRISPR-Cas technology (271, 327).
Unlike real-time RT-PCR, RT-RPA does not require sophisticated instrumentation like thermocyclers, thereby simplifying the testing process. The ease of use of this isothermal technology makes RT-RPA an attractive candidate for point-of-care molecular tests. RT-RPA technology has been applied to the detection of other RNA viruses like Ebola virus (329); however, to date, data presenting its use for the detection of SARS-CoV-2 are scarce (271, 327, 331). Kim et al. used a modified version of RT-RPA to detect SARS-CoV-2 and achieved a sensitivity of approximately 4 copies/reaction in a 10-min reaction that used a lateral flow immunoassay (LFIA) readout. Their RT-RPA correctly identified all 18 contrived specimens generated by spiking heat-inactivated virus into NP swabs or saliva (327). A second publication by Xia and Chen described another modified single-tube version of RT-RPA introduced by GenDx called reverse transcription-enzymatic recombinase amplification (RT-ERA) as well as the whole-course encapsulated procedure for exponential amplification from RNA (WEPEAR) protocol (271). The WEPEAR protocol contains all the reaction components necessary for RT-ERA, except the activator Mg2+, which is loaded into the microtube’s lid. Following the RT reaction, the tube can be spun and mixed to initiate the modified RPA reaction. Using FRET probes for a fluorescence output or nfo probes for LFIA-based detection, the WEPEAR protocol achieved high sensitivity in the range of a single copy per reaction. Unfortunately, this method was attempted on only a single clinical specimen and would require further validation. Other applications of the RT-RPA for the detection of SARS-CoV-2 involve the use of CRISPR-Cas technology, which is covered in a later section [see “Isothermal amplification technologies. (v) CRISPR-Cas technology,” below].
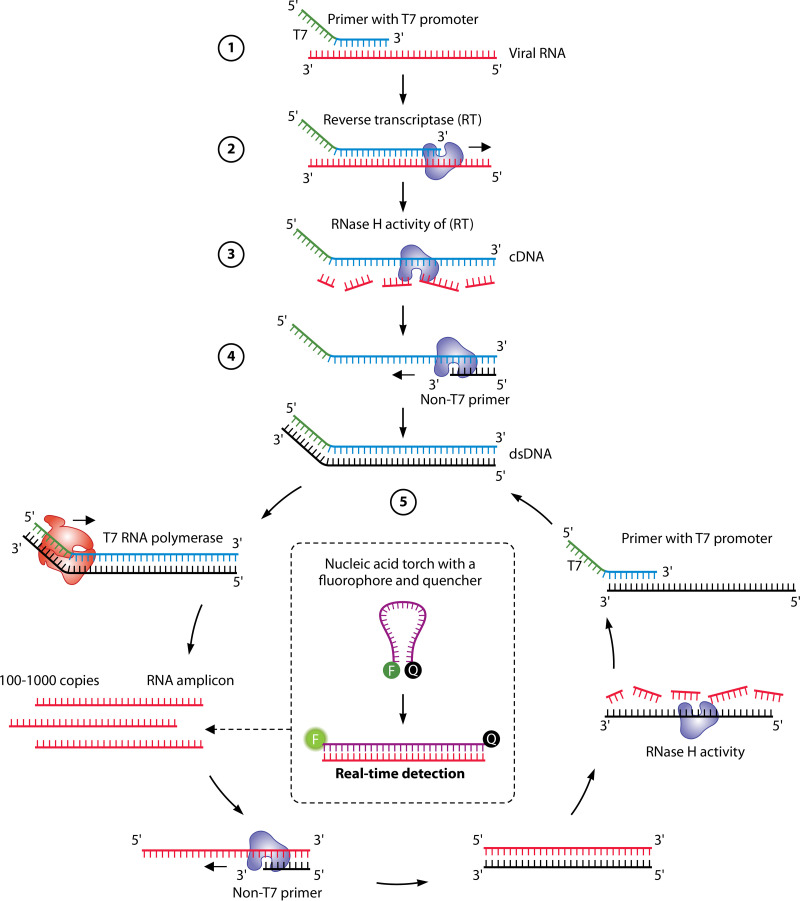
(ii) Transcription-mediated amplification.
Transcription-mediated amplification (TMA) is an IAT that amplifies RNA from an RNA template (41, 332–334), and this technology has been applied to SARS-CoV-2 diagnostics on high-throughput analyzers (296, 322, 335, 336). Figure 5 illustrates the principle of TMA.
FIG 5.
Principle of TMA. (1) The reactions use a reverse primer that is complementary to the sequence of the RNA template, but the reverse primer also contains an overhang with a promoter sequence for T7 RNA polymerase at its 5′ end. (2) Reverse transcription is conducted by the RT; the newly transcribed cDNA includes both the target sequence and the T7 promoter. (3) The RNA template is digested by the RNase H activity of the RT. (4) dsDNA is produced by the DNA polymerase activity of the RT. (5) The produced dsDNA is used as the template for transcription mediated by the T7 RNA polymerase. RNA is thereby amplified severalfold and, through the activity of the same enzyme(s), can serve as the template for a new TMA reaction. As the cycle progress, exponential amplification ensues. Detection of the amplified RNA is usually accomplished using sequence-specific molecular beacons (“torch”) or hybridization probes targeting the single-stranded RNA (ssRNA).
The Aptima SARS-CoV-2 assay is performed on the Hologic Panther instrument, a highly automated instrument capable of processing over 1,000 specimens daily (41). Its principle combines a purification step using target capture, TMA for RNA amplification, and chemiluminescent probes for RNA detection. In the target capture step, SARS-CoV-2 RNA is isolated from specimens using magnetic microparticles coupled to oligomers containing sequences complementary to specific regions of the target RNA molecules as well as polydeoxyadenosine residues. By modifying the temperature, sequential hybridization can occur between the RNA target and the sequence-specific portion of the capture oligomers, and a hybridization step then occurs between the polydeoxyadenosine region of the capture oligomer and the polydeoxythymidine sequence that is covalently bound to the magnetic microparticles (337). After the purification step, TMA reactions occur, while detection is achieved through the hybridization of sequence-specific single-stranded oligonucleotide probes labeled with acridinium ester. A reagent is applied to generate a chemiluminescence signal that can distinguish between free and bound probes. A luminometer captures the resulting light emitted from bound probes, expressed as relative light units (RLU).
The Hologic Aptima SARS-CoV-2 assay has only recently been authorized by the FDA and Health Canada, but data on its performance are scarce. The Aptima SARS-CoV-2 assay showed higher analytical sensitivity than some LDTs using real-time RT-PCR, and the performance against other high-throughput analyzers was equivalent (296, 335, 336). Compared to the Hologic Panther Fusion SARS-CoV-2 assay (i.e., real-time RT-PCR on a highly automated instrument), the Aptima SARS-CoV-2 assay showed similar analytical sensitivity, with LoDs ranging between 62.5 and 125 copies/ml, and the clinical performance was equivalent (322). Given the widespread use of Panther instruments in clinical laboratories for other pathogens (e.g., Chlamydia trachomatis and Neisseria gonorrhoeae) (338), the SARS-CoV-2 assay on this high-throughput instrument was highly anticipated.
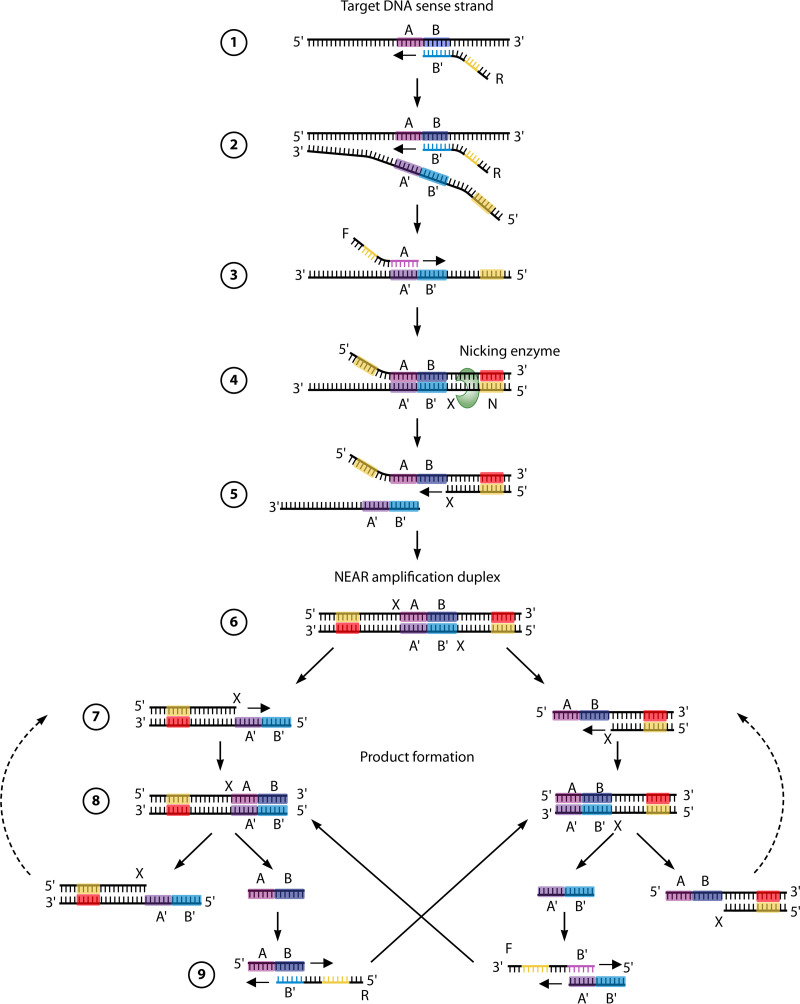
(iii) Nicking enzyme-assisted reaction.
The ID Now COVID-19 (IDNCOV) assay performed on the ID Now instrument (Abbott Diagnostics Inc., USA) is an IAT that uses nicking enzyme-assisted reaction (NEAR) technology, and this RDT was recently authorized for use for POC testing in the United States and Canada. NEARs are typically coupled to fluorescence detection following exponential amplification of DNA but can be used to detect an RNA template with the addition of a reverse transcription step (339, 340). NEARs occur under isothermal conditions (at 60°C) and in several steps mediated by two main enzymes: (i) a nicking endonuclease that recognizes specific restriction endonuclease sites in DNA (e.g., Nt.BstNBI [5′-GAGTCNNNN^N-3′]) but nicks only one strand and (ii) a strand-displacing DNA polymerase like Bst that can displace downstream DNA during synthesis at temperatures of around 65°C (Fig. 6). Strand displacement is possible due to the fact that the Bst DNA polymerase lacks 5′→3′ exonuclease activity common to other DNA polymerases (e.g., Taq polymerase).
FIG 6.
Principle of NEAR technology. The NEAR amplification reactions occur at 60°C and can be broken down into two milestones: NEAR amplification duplex formation and product formation. (1) The target recognition region (B′) of the reverse primer (R) binds to the complementary sequence (B) of the target DNA sense strand and is fully extended by the strand displacement DNA polymerase. (2) A second R primer binds to the template DNA and, during extension, displaces the elongated product of the first R primer extension. (3) The recognition region (A) of the forward primer (F) binds to its complementary sequence (A′) in the R extension product, and F is extended to create a double-stranded nicking enzyme recognition site (N). (4) The nicking enzyme recognizes N and cleaves a single strand of DNA in a sequence-specific manner at the cut site (X). (5) This releases a fragment of the R extension product. The remaining fragment serves as a primer and is extended at its 3′ end. (6) This extension completes the double-stranded complex, termed the NEAR amplification duplex, which is the starting point for product formation. (7) Nicking enzymes bind to the nicking enzyme recognition sites on both ends of the NEAR amplification duplex and cleave at X. (8) The resulting single-strand nicks create two complexes, each consisting of a single-stranded target region flanked by a nicking enzyme recognition region. (9) Repeated nicking, polymerization, and strand displacement activities result in the amplification of the AB and A′B′ target products. Cleaved complexes are regenerated, while the AB and A′B′ products can anneal to R and F primers, respectively. In turn, the bidirectional extension of the primer and product each creates duplexes that lead to the generation of the opposite product upon cleavage. Product amplification continues until reagents or enzymes are depleted.
While the mechanism for nucleic acid amplification with NEAR may be complex, IDNCOV testing is simple and rapid. The assay has processing times as low as 5 min for positive results with high viral loads and 15 min for specimens with lower viral loads or negative results. Compared to LDTs or commercial NAATs, many recent studies have demonstrated an excellent specificity/negative percent agreement (NPA) near 100% but relatively poor sensitivity/positive percent agreement (PPA) of between 48% and 70% for the detection of SARS-CoV-2, while other studies showed a high specificity/NPA (∼100%) as well as high sensitivity/PPA values above 90% (290, 301, 341–347).
The reasons for the disparities in sensitivity/PPA between studies are likely multifold and include differences in the patient population (setting, host factors, and the presence or not of compatible symptoms), the specimen type, the timing between collection and testing, the transport conditions used (dry swabs or transport media), the quality of specimens (prospective versus retrospective), the spectrum of viral loads in the specimens evaluated (proportion of specimens with low viral loads), or differences in performance characteristics of the comparator method(s) (290, 301, 341–347). For example, the swab type affected IDNCOV performance, where NP swabs showed a PPA of 64%, compared to 48% with nasal swabs (348). Using residual positive and negative NP swabs collected in VTM, Mitchell and St. George compared IDNCOV to the CDC real-time RT-PCR, and IDNCOV showed sensitivity and specificity of 71.7% and 100%, respectively (341). All false-negative results corresponded to specimens for which CT values were between 35 and 40, suggesting low viral loads. Smithgall et al. used residual NP swabs tested with the Roche cobas assay and found NPAs of 100% and 92.0% for INDCOV and Cepheid Xpert Xpress and overall PPAs of 73.9% and 98.9%, respectively (290). However, they also noted that the PPA varied with viral loads. When specimens were categorized by CT values, both INDCOV and Xpert showed 100% PPA for specimens with medium to high viral loads (CT values of <30), but at low viral loads (CT values of >30), the PPA for IDNCOV was 34.3%, versus 97.1% for Xpert (290). In a recent study, Stokes et al. compared IDNCOV to an LDT and showed an excellent PPA of 89.1% (95% CI, 82.0% to 94.1%) for IDNCOV (347). Notably, the PPA increased to 98.2% by following the manufacturer’s recommendations for testing under EUA for symptomatic individuals tested ≤7 days after symptom onset and within an hour of collection using the appropriate swab (347). Overall, these studies not only demonstrate that the performance characteristics of a test are dependent on numerous factors but also reflect the need for validations or verification of these factors in the settings and conditions where NAAT-based RDTs are applied (274, 288).
(iv) Reverse transcription–loop-mediated isothermal amplification.
Reverse transcription–loop-mediated isothermal amplification (RT-LAMP) is an IAT that is gaining interest for potential POC applications and is also being explored for routine diagnostic testing (41, 53, 54, 244, 349–351). Like RT-PCR, RT-LAMP begins with reverse transcription of the target RNA into cDNA by reverse transcriptase, which is done either in a separate reaction or in the same tube as the LAMP reaction. The LAMP reaction can take place in a single tube at 60°C to 65°C (260, 352, 353), and the process can be performed in as little as 20 to 60 min (353, 354). LAMP reactions consist of a strand displacement DNA polymerase (e.g., Bst polymerase); a DNA template; dNTPs, typically from 4 to 6 primers; and, depending on the LAMP permutation for signal detection, either a pH-sensitive colorimetric dye or a fluorescent dye (41, 54, 244, 350, 351, 355, 356). The mechanism for DNA amplification using LAMP is summarized in Fig. 7.
FIG 7.
Amplification of nucleic acids using RT-LAMP. Overall, there are four core primers that mediate all the processes in a LAMP reaction by recognizing six distinct regions of the target DNA through several steps. (1) After the conversion of the template RNA into dsDNA via reverse transcription (not shown), the LAMP reaction starts from strand invasion by the forward inner primer (FIP), which hybridizes through its F2 region to the F2c region of the target DNA. This initiates complementary-strand synthesis using a strand displacement DNA polymerase. (2) The forward outer primer (FOP) (also termed the F3 primer) then hybridizes to the F3c region of the target DNA and, during extension, displaces the newly elongated strand from the FIP. (3) Given that the FIP also contains an F1c sequence, the strand displacement triggered by the DNA polymerase and the FOP leads to the formation of a self-annealing loop in the 5′ end of the FIP-linked strand (regions F1 and F1c). (4) This single-stranded DNA with a stem-loop at its 5′ end then serves as a template for the backward inner primer (BIP), which hybridizes to the B2c region of the template DNA through its B2 sequence. (5) During elongation, the complementary strand opens the 5′-end stem-loop. Next, the backward outer primer (BOP) (also termed the B3 primer) hybridizes to the B3c region of the target DNA, and its elongation displaces the BIP-linked complementary strand. (6) The displacement of the BIP-linked strand results in self-hybridization on both the 5′ and 3′ ends, leading to two stem-loops and the formation of a dumbbell-shaped DNA. (7 to 9) The amplification of the dumbbell structure with the FIP leads to a concatemer and the formation of a second dumbbell structure that can be amplified with the BIP. Amplification can occur from the 3′ end of each dumbbell structure or with the annealing of primers such as the FIP and BIP. Additional loop primers (i.e., loop F [LF] and loop B [LB] primers) can also be used to increase the speed and sensitivity (41, 350, 351). Visualization of LAMP amplification is typically done by using pH-sensitive colorimetric or intercalating fluorescent dyes.
Variations of LAMP have been developed for potential POC applications with reactions that are monitored in one of three ways. (i) Turbidity can be measured with a spectrophotometer at an optical density (OD) of 400 nm, as magnesium pyrophosphate precipitates in the solution as a by-product of the LAMP reaction (41, 357–359). (ii) Colorimetric detection can be performed using pH-sensitive dyes (e.g., cresol red or phenol red) that change color from the incorporation of dNTPs during amplification or using a metal indicator (e.g., hydroxynaphthol blue) that would assess the concentration of Mg2+, used as a cofactor for dNTP incorporation during DNA synthesis (349, 360–362). Color changes can be read by the naked eye or spectrophotometry (359, 362). (iii) Fluorescence detection can be performed if an intercalating dye (e.g., SYBR green) is used in the LAMP reaction. When complexed with dsDNA, intercalating dyes can be excited to emit fluorescence, which can then be captured in real time with a fluorometer or a compatible thermocycler (354, 360). Alternative detection systems for RT-LAMP include CRISPR-Cas technology, and these are covered in a later section of this review.
Fluorescence detection tends to be the most sensitive of the visualization methods, which makes the LAMP technology amenable to real-time monitoring and high-throughput testing (349, 354). However, to our knowledge, no high-throughput instruments have adopted this technology to date. On the other hand, a commercial kit for real-time SARS-CoV-2 RT-LAMP (Variplex; Amplex Diagnostics, Germany) has been developed and compared to real-time RT-PCR. The commercial RT-LAMP kit showed moderate agreement with real-time RT-PCR, with a clinical sensitivity of 76.3% (363, 364). Given its simplicity, RT-LAMP technology has also been used to develop rapid POC products (349). Results are ready in 30 min using RapiPrep COVID-19 (MicrosensDX, England), which integrates magnetic bead-based RNA extraction with LAMP technology. However, a relatively low sensitivity of 80% and a specificity of 73% were observed when tested on 21 nasal swabs compared to real-time RT-PCR (365). The authors of that study suggested that while the sensitivity and specificity were poor, the assay still had merit in some clinical applications such as algorithms using repeat testing over time. Other studies that used RT-LAMP demonstrated varying performance compared to LDTs based on real-time RT-PCR for commonly used specimen types (357, 366). The analytical sensitivity of most RT-LAMP assays was found to be in the range of 100 to 200 copies per reaction (360, 367), while others have reported analytical sensitivities of as low as 10 copies/reaction (354, 368). Altogether, this range is consistent with those of some LDTs and commercial real-time RT-PCR assays (e.g., cobas SARS-CoV-2 test) (40, 278, 368, 369). The variability in the performance of RT-LAMP assays could be attributed to differences in processing steps, specimen types, the quality of the nucleic acid extraction, LAMP reagents, viral targets, detection methods (manual versus automated), the methodology (e.g., measuring turbidity, using colorimetry, or using fluorescence), viral loads, patient populations tested, numbers of specimens evaluated, methods used as a comparator, or other undefined or uncharacterized factors like inhibition rates (244, 260–262, 354, 357, 367, 370–372). Overall, RT-LAMP technology shows promise for large-scale testing (373) and POC testing (260, 278, 368, 369, 374, 375), but further optimization is still required.
(v) CRISPR-Cas technology.
CRISPR and its Cas proteins are derived from prokaryotic defense systems against foreign nucleic acids (376–381). When activated, Cas proteins can exhibit local DNase or RNase activity resulting in local cleavage (cis-cleavage) of the target DNA or RNA as well as collateral damage (trans-cleavage) to neighboring single-stranded DNA (ssDNA) or RNA. A number of different Cas proteins have been identified that differ in nucleotide specificity for their cis-cleavage targets and in their ability to cause collateral damage to nearby nucleic acids (331, 382–384). The high degree of collateral damage caused by Cas12a or Cas13 can break down neighboring RNA or ssDNA, which can be exploited for detection. For example, dually labeled ssDNA or RNA probes with a fluorophore-quencher combination can be used, and when the Cas12a or Cas13 system binds to the target DNA or RNA, the Cas proteins are activated to cleave the target and the probe. With a blue-light generator, fluorescence is generated and visualized or captured by a fluorometer (385–387). Alternatively, the RNA or ssDNA probe can be labeled with biotin, and the cleavage reaction can be observed with the aid of a specific immunochromatographic device (e.g., LFIA) and colorimetric detection (385–387). Understanding the mechanism and permutations of CRISPR-Cas systems led to many technological advances in genome editing (388, 389) and diagnostic applications such as the detection of RNA viruses (Fig. 8) (382–384, 386, 387, 390, 391).
FIG 8.
Principle of CRISPR-Cas technology for viral RNA detection. First, the viral RNA is subjected to reverse transcription and amplification, e.g., in an RT-RPA reaction at 37°C to 42°C, to generate dsDNA. The dsDNA can be targeted by guide RNAs (gRNAs) directly in a CRISPR-Cas12 detection system, whereas RNA detection using the CRISPR-Cas13 system requires an additional T7 transcription step. When Cas12 or Cas13 is activated by the recognition of gRNA, there will be cleavage of the target as well as nonspecific cleavage of dually labeled oligonucleotide probes. The probes are ssDNA or ssRNA for the CRISPR-Cas12 or CRISPR-Cas13 systems, respectively. The readout for either method can be colorimetric by the incorporation of fluorescein amidite (FAM)/fluorescein isothiocyanate (FITC)-biotin probes and the use of lateral flow dipsticks, or fluorometric readouts can be used by the incorporation of dually labeled fluorophore (F)-quencher (Q) probes. Upon collateral cleavage, the unquenched fluorophore can be excited with blue light, and the resulting emission of fluorescence can be visualized or captured with a fluorometer.
The use of CRISPR-Cas as a diagnostic tool was proposed by two laboratories, which founded the Cas12a-based system named DETECTR (DNA endonuclease-targeted CRISPR trans-reporter) (392) and the Cas13-based system termed specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) (393, 394) (Fig. 8). Recently, a SHERLOCK-based COVID-19 test received EUA from the FDA, which is the first EUA for any CRISPR technology. However, many variations of CRISPR-Cas12 and CRISPR-Cas13 systems have emerged for the detection of SARS-CoV-2 in a variety of clinical specimens, in both potential POC and high-throughput testing formats (395). These include the SHERLOCK testing in one pot Covid test (STOPCovid), all-in-one dual CRISPR (AIOD-CRISPR), Cas12b-mediated detection (CDtection), CRISPR-assisted detection (CASdetect), and Cas13-based, rugged, equitable, scalable testing (CREST) (387, 392, 395–399).
The composition and conditions of each stage of a CRISPR-Cas detection system can affect its speed and performance. For SARS-CoV-2 detection, the initial step in the CRISPR-Cas system can be the lysis of the specimens, with or without an RNA extraction step; however, purified RNA showed higher sensitivity (392, 395, 396). For example, Broughton et al. demonstrated a DETECTR system that had an LoD of approximately 10 copies/μl using purified RNA, but removing the nucleic acid extraction step decreased the LoDs to 15,000 and 500 copies/μl for contrived specimens consisting of spiked SARS-CoV-2 in ≥10% UTM and ≥20% PBS, respectively (392). As such, the purification of RNA was recommended. Next, the time to results for CRISPR-Cas-based methods was low compared to real-time RT-PCR, at 20 min (395), 40 min (386, 392, 397), 50 min (331), 60 min (387), or 70 min (396), but varied between methods and detection systems. All CRISPR-Cas methods described here require the conversion of SARS-CoV-2 RNA into dsDNA, which can be accomplished by RT-PCR, RT-LAMP, or RT-RPA. Huang et al. compared the CDC real-time RT-PCR to a DETECTR assay using either RT-PCR or RT-RPA with a fluorescence readout and achieved low analytical sensitivity (2 copies per sample, regardless of the method used for amplification) (331). Cas12-based CRISPR systems are typically faster than Cas13-based methods, as Cas12 directly detects dsDNA, but the latter requires an additional transcription step to RNA (Fig. 8).
The methods used for SARS-CoV-2 result readout can also affect the speed and performance of the assays. While initially, CRISPR-Cas assays used visual readouts on commercial lateral flow assays, coupling the CRISPR-Cas technology to fluorescent reporters instead leads to more rapid results and sensitive detection (392, 393, 400). For example, Joung et al. validated a SHERLOCK-based method that returned results in 70 min using the LFIA readout, while fluorescence testing was completed in 40 min (396). Guo et al. demonstrated that the analytical sensitivity of CASdetec, a CRISPR-Cas12b assay based on reverse transcription recombinase-aided amplification (RT-RAA) and fluorescence output, was 103 copies/ml, whereas the sensitivity of DETECTR and SHERLOCK ranged between 104 and 105 copies/ml using an LFIA for detection (387). Also, CRISPR-Cas systems have been adapted to one-tube reactions using fluorescence rather than amplification followed by detection using lateral flow methods, not only to reduce processing steps but also to decrease the potential for contamination (395–397). To optimize the reaction in single-tube formats, the temperature for the IAT must be compatible from the RT step to Cas-based detection. Ding et al. described a one-pot reaction at 37°C combining RT-RPA and a CRISPR-Cas12a system and could achieve detection of 1.3 copies of SARS-CoV-2 RNA (397). With the high temperatures required for RT-LAMP (55°C to 65°C), CRISPR-Cas12b systems have been developed with a thermostable Cas12b protein derived from Alicyclobacillus acidiphilus that could maintain activity at higher temperatures (396, 401). A similar approach was used by Ali et al., who developed a single-tube RT-LAMP- and CRISPR-Cas12a-based system able to reach an analytical sensitivity (10 copies/reaction) comparable that of to the CDC real-time RT-PCR (5 copies/reaction) (395). Given the many variables for CRISPR-Cas technology, optimization of this methodology for SARS-CoV-2 and other diagnostic testing is an active area of research.
While the analytical performances of CRISPR-Cas technology showed some promise for detecting SARS-CoV-2, validation on clinical specimens has been scarce to date. Hou et al. showed that an RT-RPA CRISPR-Cas13a system with fluorescence detection was able to correctly identify 52 positive specimens for SARS-CoV-2, whereas an undefined real-time RT-PCR failed to identify 5 specimens with low viral loads (402). Huang et al. compared the CDC real-time RT-PCR to a DETECTR assay using either RT-PCR or RT-RPA with fluorescence detection on 19 positive clinical specimens and identified all of them; however, 3 additional positive specimens were detected (331). These could represent false-negative results for the real-time RT-PCR or false-positive results for the CRISPR-Cas12 assay (i.e., specificity would be 71.4% for the latter). The LoD for the CRISPR-based method was 2 copies/reaction, compared to 5 copies/reaction for the CDC real-time RT-PCR, suggesting that CRISPR-based detection may be able to identify SARS-CoV-2 at lower viral loads. Broughton et al. compared the CDC real-time RT-PCR to an RT-LAMP DETECTR system with LFIA or fluorescence readouts using 83 clinical specimens (41 positive and 42 negative specimens) and demonstrated 95% positive agreement (392). Of the 21 specimens positive for SARS-CoV-2 identified by the CDC real-time RT-PCR, Ali et al., using an RT-LAMP CRISPR-Cas12 system, were able to identify 18 (85.7%) using the fluorescence readout, but weak or absent signals were noted with LFIA detection (395). Overall, these data show variability in performance for CRISPR-based assays, ranging from 80 to 100% sensitivity.
While CRISPR-based methods are showing promise for SARS-CoV-2 detection, research into this technology is evolving. Recently, an RT-LAMP-based CRISPR-Cas12 system was developed using an electric field gradient on a microfluidic device (403). This allowed for on-chip, automated separation of nucleic acids from nasopharyngeal swab samples in 30 min, followed by CRISPR-based detection (403). While further validation is required to fully understand the benefits of CRISPR-Cas technology, it has much potential for applications for POC devices or high-throughput testing platforms (331, 395).
SARS-CoV-2 next-generation sequencing.
Understanding the genomic sequence of SARS-CoV-2 obtained from clinical specimens can help identify the COVID-19 pandemic origins, delineate transmission events, unravel clues to pathogenesis, and monitor viral evolution over time (404–408). Over the last few decades, DNA sequencing technologies have relied on modifications of Sanger sequencing, which was developed in the 1970s (409). While Sanger sequencing technology is still used for small sequences (∼0.5 to 1 kb) such as single-gene targets, next-generation sequencing (NGS) technologies have allowed sequencing to be performed as massively paralleled reactions, allowing rapid access to complete genomes at a scale and cost that are feasible for many laboratories (404–406, 410–412). For SARS-CoV-2 genomes of <30 kb, high-quality sequences can readily be obtained with NGS directly from clinical specimens using strategies like amplicon enrichment or bait capture techniques to favor the sequencing of the viral targets (413). NGS involves technologies such as sequencing by synthesis, sequencing by ligation, and ion semiconductor sequencing (e.g., nanopore sequencing), each with its own advantages and limitations (404–406, 410–412). The principle of each NGS technology has been reviewed elsewhere (404–408), and some of them are illustrated in Fig. 9.
FIG 9.
Sequencing techniques for identification of SARS-CoV-2. (A) Sanger sequencing. First, SARS-CoV-2 RNA is often amplified by RT-PCR (not depicted). Sanger sequencing reactions can be undertaken to analyze either of the DNA strands, but only one strand per reaction can be assessed. The extension of the primer annealing to the template DNA occurs in the presence of DNA polymerase, buffer, cofactors, deoxynucleotide triphosphates (dNTPs), and fluorescently labeled dideoxynucleotide triphosphates (ddNTPs). The binding of the ddNTPs to the oligonucleotide strands will cease the extension, resulting in various DNA structures with different lengths. Next, the extended DNAs undergo capillary gel electrophoresis in which the shorter DNA strands move faster, resulting in the detection of the fluorescently labeled nucleotides in the order of the size of the DNA strands. Finally, as DNA fragments are resolved and nucleotide-specific fluorescence signals are captured by a detector, a chromatogram is assembled to reveal the sequence of the template. (B) Next-generation sequencing (NGS) by synthesis. First, a library of millions of DNA fragments is created from the template (or enhanced by multiplex RT-PCR for SARS-CoV-2). Adapters are bound to the two ends of each DNA fragment. The adapters consist of a universal primer-binding site and a unique sequence (i.e., barcode) that can be hybridized to a specific sequence on the support (e.g., flow cell). Following hybridization, with complementary sequences of the adapters, bridge amplification is used to amplify each DNA fragment at a defined physical position. In the sequencing and detection steps, fluorescently labeled nucleotides are bound to the forward strands in the presence of a primer and a polymerase, which results in the generation of fluorescent light that is detected by an analyzer in real time. Many other NGS technologies are also available. (C) For example, NGS by nanopore technology is presented. After creating a library of DNA fragments by multiplex RT-PCR and barcoding, the library is loaded onto a membrane containing nanopores. The nanopores are proteins that open the DNA double strand, and as each nucleotide is passed through the membrane, it causes a specific change in the ionic current that can then be translated into the nucleotide sequence of the templates.
To date, a single commercial kit (Illumina Inc., USA) for NGS has been approved as a clinical diagnostic test under FDA EUA guidelines for COVID-19, which is based on sequencing by synthesis (414). However, no data are available to date to describe its performance, advantages, or limitations compared to commonly used detection methods like real-time RT-PCR. Also, only a limited number of studies that have explored the use of NGS for SARS-CoV-2 detection for the purpose of diagnostic testing are available (413, 415–417). For example, using a laboratory-developed protocol for NGS, Bhoyar et al. compared NGS and real-time RT-PCR on 752 clinical specimens processed in duplicate on a single flow cell (417). They demonstrated high concordance between the methods and a diagnostic increase in the positivity of 5.7% with NGS (with the detection of 6 cases that tested negative by PCR and 21 cases where PCR results were inconclusive). This study demonstrates the feasibility of processing 1,536 specimens in a total of 17 h (11 h for sequencing and 6 h for analysis) (417). In another study, a low-cost NGS approach was shown to achieve high sensitivity for the detection of SARS-CoV-2 (84 genome units/ml), which is equal to or higher than those of some RT-PCR methods; however, this study tested only 10 specimens (5 positive and 5 negative specimens) (415). It is unclear whether high sensitivity would still occur if the throughput would be increased to their proposed workflow of 192 specimens in 8 h. Bloom et al. showed 100% concordance between NGS and RT-PCR with a limited number of specimens (31 positive and 33 negative NP swabs) (416). These authors propose NGS as a tool for population-based surveillance rather than individualized testing for medical decisions. While postulated to be able to achieve screening of thousands of samples on high-throughput NGS platforms, no data were provided to support the feasibility of this approach or the impact of such a high level of specimen pooling. Overall, some data support the potential of NGS as a diagnostic tool for SARS-CoV-2, yet further analyses are required to understand its benefits and limitations.
Despite the potential for NGS as a diagnostic tool, the limitations of SARS-CoV-2 genome sequencing using NGS technologies should also be recognized. For example, NGS technologies are challenged with specimens with low viral loads (413, 418), as insufficient data or poor-quality results are obtained for subsequent analyses. Efforts to increase sensitivity and quality are under way using target enrichment processes with NGS techniques such as multiplex PCR amplicon-based sequencing, hybrid capture-based sequencing, and ultrahigh-throughput metatranscriptomic sequencing (413, 419). Another limitation of NGS technologies is cost, which may be prohibitive for many diagnostic laboratories. Furthermore, the complexity of the NGS workflow and requirement for sophisticated instrumentation and bioinformatics expertise may pose significant barriers to NGS access in many laboratories. If resources are limited, specimens could be prioritized and sent to referral laboratories to help inform public health responses and global surveillance initiatives.
While not routinely used for SARS-CoV-2 diagnostic testing in clinical laboratories, genome sequencing of SARS-CoV-2-positive specimens with NGS technologies has paved the way for numerous applications, including investigations of disease pathogenesis, epidemiology, virus phylogenetics, SARS-CoV-2 evolution, and the impact of viral evolution on diagnostic testing or interventions like therapeutics and vaccines (420–424). For example, Meredith et al. used SARS-CoV-2 nanopore sequencing on PCR-positive specimens combined with epidemiological data to help identify nosocomial transmission events and inform infection control interventions (418). They demonstrated the feasibility of rapid NGS in a health care setting by providing sample-to-sequence information in less than 24 h. From the discovery of SARS-CoV-2, NGS has been used to understand its origins and transmission. Initial phylogenetic analysis of the genomes of SARS-CoV-2 showed that it was closely similar to human SARS-CoV and potentially used the same cell entry receptor (i.e., ACE2) and helped postulate the probable zoonotic origins of the virus (i.e., bats) (10, 425–429). Next, the first SARS-CoV-2 genomes were made available less than a month from the first recognition of disease reported from Wuhan, Hubei, China (10). With rapid access to SARS-CoV-2 genome data, molecular methods like real-time RT-PCR were rapidly developed at the early stages of the COVID-19 pandemic and became the method of choice for SARS-CoV-2 detection worldwide. Through remarkable efforts from public health agencies and researchers, SARS-CoV-2 genome sequences have been made available in public data repositories such as the Global Initiative on Sharing All Influenza Data (GISAID) (https://www.gisaid.org/) and the National Center for Biotechnology Information (NCBI) GenBank database (https://www.ncbi.nlm.nih.gov/). These data help provide a snapshot of global diversity and data that could be used for epidemiological investigations. In the initial stages of the pandemic, the high diversity of SARS-CoV-2 genomes was attributable to multiple independent importations of SARS-CoV-2 by travel overseas in countries of initial virus activity, and transmission routes could be investigated (418, 430–434). Following global spread and closure of international borders, sequence diversity was more limited, and sequencing of SARS-CoV-2 genomes was used in outbreak investigations and became particularly useful for cases in areas of unknown community transmission (418, 430–434). However, with natural evolution in the human host, or selective pressures from the recent introduction of SARS-CoV-2 vaccines or exploratory therapeutics (i.e., antivirals or convalescent-phase sera), there are increasing chances for SARS-CoV-2 to further diversify and acquire mutations (435–439).
The genetic diversity of SARS-CoV-2 stems from naturally occurring mutations in its genome, which is common at higher frequencies in RNA viruses (440). Some mutations might have no impact on SARS-CoV-2 protein sequences (i.e., synonymous substitutions), but these may affect the performance of diagnostic tests using NAATs if they occur in the target region for the assays (40, 441–443). From the various genomes of SARS-CoV-2 that have been sequenced since its discovery, the M, E, and RdRp genes were shown to be fairly conserved compared to the high divergence observed in some regions of the S genes (420, 421, 439, 444–450). Mutations in the genome of SARS-CoV-2 could also occur from point mutations, insertions, deletions, or recombination events that could affect protein sequence, structure, and function (i.e., nonsynonymous substitutions). Both synonymous and nonsynonymous mutations can be useful for epidemiological investigations, but it should be noted that nonsynonymous mutations are of significant interest as they could have impacts in terms of disease transmissibility and severity or could help the virus escape from therapeutic (i.e., convalescent-phase sera or antivirals) or preventative (i.e., vaccines) interventions. Mutations in S are of particular concern as S glycoprotein epitopes are major targets for current and exploratory vaccines (35, 451, 452). Multiple SARS-CoV-2 strain variants are circulating globally, but few are variants of concern (VOCs) (439). In the United Kingdom, a novel variant of SARS-CoV-2 (0B/501Y.V1, VOC 202012/01, or B.1.1.7 lineage) emerged, with an unusually large number of mutations. This VOC has since been reported in several countries, including Canada and the United States. It contains a number of mutations in the S gene (e.g., N501Y, 69/70 deletion, and P681H), and while these mutations have yet to show any impact on disease severity or vaccine effectiveness, some preliminary epidemiological data suggest that this variant is associated with increased transmissibility (439). In Brazil, a SARS-CoV-2 variant from lineage B.1.1.248 was reported, with an E484K S gene mutation associated with reduced neutralization capability by convalescent-phase plasma (453–455). Such mutations are concerning due to potential failures of therapeutic options or prevention strategies like vaccines that target similar viral protein epitopes. Furthermore, a novel VOC from lineage B.1.1.248 called P1 was identified, with 12 mutations in the S gene, including both E484K and N501Y, suggesting the potential for increased transmissibility and immune escape (438, 439). Ongoing surveillance should be encouraged to identify novel SARS-CoV-2 variants and characterize the potential impacts of VOCs.
Overall, sequence-based surveillance of SARS-CoV-2 is important to ensure that diagnostic tests accurately identify the virus and that there are no changes with novel variants in disease spread, disease severity, or the effectiveness of vaccines or antivirals. Whether NGS will eventually become a common diagnostic tool remains to be determined.
SARS-CoV-2 Antigen Detection
Antigen detection methods, like NAATs, are used to detect active replicating viruses in the early stages of SARS-CoV-2 infection. Unlike NAATs that rely on the detection of viral RNA, antigen detection is based on the identification of SARS-CoV-2 proteins. The two main antigens in SARS-CoV-2 detection assays are S and N proteins. Antigen detection assays use technologies similar to those of serological methods. Like serology, high-throughput antigen-based testing can be performed on semiautomated or automated instruments using enzyme immunoassay (EIA) technologies like enzyme-linked immunosorbent assays (ELISAs) or chemiluminescence immunoassays (CLIAs). However, most antigen detections to date have aimed for methods allowing easy-to-use and rapid testing using portable devices, like LFIAs (also termed lateral flow immunochromatographic assays or lateral flow assays [LFAs]). The mechanism for LFIAs is shown in Fig. 10A, and other technologies (e.g., ELISA and CLIA) are covered in the subsequent serology sections. This section summarizes the current knowledge on the clinical performance of antigen-based detection assays and their potential applications to support COVID-19 responses.
FIG 10.
Antigen testing for the detection of SARS-CoV-2. (A) Principle of a lateral flow immunochromatographic assay (LFIA). The design of the LFIA for antigen detection is a qualitative immunological reaction confined to a small portable device (e.g., cassette or dipstick) that can be performed in the laboratory or a POC setting. Briefly, antigens in specimens (e.g., nasal swabs, nasopharyngeal swabs, and saliva) are placed in a well with a sample pad, and the fluid containing the antigen flows through the device via capillary action. The bottom of the well where the specimen is inoculated contains a sample pad, which is in contact with the conjugate pad used as a support for SARS-CoV-2-specific monoclonal antibodies (mAbs) that are labeled with colloidal gold nanoparticles (AuNPs) or other tags. If present, SARS-CoV-2 antigen (usually S or N protein) forms a complex with the mAbs bound to the AuNPs, and the entire complex migrates via capillary action until it is captured by other SARS-CoV-2 antigen-specific mAbs immobilized on the nitrocellulose membrane (i.e., the test line). As antigen-antibody complexes are trapped at this location, they form a line that can be visualized by the naked eye or with the aid of a detector. Also, mAbs-AuNPs, whether conjugated with antigens or not, continue to migrate until captured by an isotype-specific antibody directed against the fragment crystallizable (Fc) portion of the mAb at the control line. This ensures proper liquid flow through the device and test validity. (B) Point-of-care detection of SARS-CoV-2 antigen using a FET-based sensor. Upon binding of spike (S) proteins to the anti-S monoclonal antibodies immobilized on the graphene sheet via the PBASE linker, a change in the voltage-ampere diagram reveals the presence of the virus. (Panel B is adapted from reference 465 with permission of the American Chemical Society.)
Hundreds of companies have been developing antigen rapid diagnostic tests (Ag-RDTs) for SARS-CoV-2 detection (238). Ag-RDTs (e.g., LFIAs) are often promoted as POC devices for rapid testing and immediate management of patients in settings such as physicians’ offices or clinics; however, these tests are not always performed in these settings. RDT is a more appropriate term if performed in a laboratory setting. At the time of this review, few antigen-based detection assays have received EUA approval in the United States and Canada, but many more applications have been submitted as laboratory and/or POC tests (29, 30, 456, 457). Examples of SARS-CoV-2 Ag-RDTs that have received EUA for testing in laboratory or POC settings in the United States or Canada include technologies relying on a colorimetric LFIA with a visual readout, instrument-based antigen detection using a fluorescence-based LFIA, a microfluidic immunofluorescence assay, and a chromatographic digital immunoassay (Table 2).
TABLE 2.
Examples of antigen tests approved for emergency use by the U.S. FDA for detection of SARS-CoV-2a
| Device or assay (manufacturer) | Methods | Target antigen protein | Specimen type(s) | Authorized detection window (dpo) | Authorized settings | Time (min) | LoD (TCID50/ml)b | Reference |
|---|---|---|---|---|---|---|---|---|
| BinaxNOW COVID-19 Ag card home testc (Abbott Diagnostics, USA) | LFIA, visual readout | N | NS, ANS | 7 | Home, H, M, W | 15 | 140.6 | 585 |
| CareStart COVID-19 antigen test (Access Bio, USA) | LFIA, visual readout | N | NPS | 5 | H, M, W | 10 | 800 | 586 |
| QuickVue SARS antigen testc (Quidel Corporation, USA) | LFIA, visual readout | N | ANS | 5 | H, M, W | 10 | 7,570 | 587 |
| Veritor system for rapid detection of SARS-CoV-2 (BD, USA) | LFIA, instrument readout | N | NS | 5 | H, M, W | 15 | 140 | 588 |
| Sofia 2 Flu + SARS antigen FIAc (Quidel Corporation, USA) | LFIA, IFA, instrument readout | N | NS, NPS | 5 | H, M, W | 15 | 91.7 | 589 |
| LumiraDx SARS-CoV-2 Ag testc (LumiraDx Ltd., UK) | Microfluidics, IFA, instrument readout | N | NS | 12 | H, M, W | 12 | 32 | 590 |
| Clip COVID rapid antigen testc (Luminostics Inc., USA) | LFIA, ILA, instrument readout | N | ANS | 5 | H, M, W | 30 | 88 | 591 |
| Ellume COVID-19 home testc (Ellume Limited, Australia) | LFIA, IFA, smartphone readout | N | MTS | NA | Home, H, M, W | 15 | 6,309 | 592 |
The full list is available in reference 29. Abbreviations: LFIA, lateral flow immunoassay; IFA, immunofluorescence assay; ILA, immunoluminescence assay; BD, Becton, Dickinson and Company; dpo, days post-symptom onset; NS, nasal swab; ANS, anterior nasal (naris) swab; NPS, nasopharyngeal swab; MTS, midturbinate nasal swab; H, laboratories certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a, that meet requirements to perform high-complexity tests; M, laboratories certified under the CLIA, 42 U.S.C. §263a, that meet requirements to perform moderate-complexity tests; W, patient care settings operating under a CLIA certificate of waiver; TCID50, median tissue culture infectious dose.
The limit of detection (LoD) of each assay is the lowest LoD reported in the instructions for use of that assay, regardless of the specimen types.
The assay does not differentiate between SARS-CoV-2 and SARS-CoV.
While EUA was granted for some Ag-RDTs, at the time of this review, limited data have been published on their clinical performance characteristics. The performance of antigen- and molecular-based POC tests for detecting SARS-CoV-2 was the subject of a recent Cochrane review (203), but limited data were available to be summarized (369, 458–463). Briefly, the review summarized data from five studies, representing 943 samples. The average sensitivity of Ag-RDTs was found to be 56.2% (95% CI, 29.5% to 79.8%), and the average specificity was 99.5% (95% CI, 98.1% to 99.9%). In comparison, NAAT-based RDTs had an average sensitivity of 95.2% (95% CI, 86.7% to 98.3%) and an average specificity of 98.9% (95% CI, 97.3% to 99.5%). High specificity was observed for both molecular and antigen-based detection methods; however, low sensitivity was noted for Ag-RDTs, but a high level of heterogeneity was noted between studies. This variability could be explained by factors including the methods used for SARS-CoV-2 detection, the comparator methods during the evaluation, the patient populations assessed, SARS-CoV-2 prevalences, the timing of specimen collection, the specimen types used, and antigen stability during analyses.
Consistent with the low clinical sensitivity of Ag-RDTs, the analytical sensitivity was defined in some studies, which described LoDs of Ag-RDTs that were approximately 1,000-fold lower than those of culture-based detection of SARS-CoV-2 and 10,000-fold lower than those of NAATs (463). Mertens et al. noted that when specimens with high viral loads were evaluated (defined as specimens with real-time RT-PCR CT values of <25), the sensitivity of Ag-RDTs increased to 74.8%, compared to an overall sensitivity of 57.6%, when all specimens were considered (459). Scohy et al. observed an overall sensitivity of Ag-RDTs of 30.2% (461). When specimens were further characterized by real-time RT-PCR CT values, the impact of the poor sensitivity of Ag-RDTs was clear. With specimens with CT values of <25 (1.8 × 105 copies/ml), CT values of <30 (9.4 × 103 copies/ml), and CT values of <35 (4.9 × 102 copies/ml), the sensitivities of Ag-RDTs were shown to be 100%, 70.6%, and 46.9%, respectively. Linares et al. showed that an Ag-RDT achieved 86.5% sensitivity (95% CI, 75.0% to 97.0%) if symptomatic patients with high or moderate viral loads were tested within 7 days of symptom onset (457). As seen with all other SARS-CoV-2 tests, the timing of testing likely plays an important role in assay performance. Recognizing the relatively poor sensitivity compared to NAATs but the potential role for Ag-RDTs, WHO interim guidance described situations where such tests could be considered (238). The WHO supports the use of Ag-RDTs for conditions such as (i) testing in areas where NAATs are not available (e.g., remote areas) or when the result turnaround times using NAATs are long; (ii) in outbreak investigations, but the frequency of testing in these settings remains unclear; (iii) in areas where the prevalence is high; and (iv) for testing of asymptomatic contacts of positive cases.
Given the poor sensitivity of Ag-RDTs for detecting SARS-CoV-2, research is being performed to improve sensitivity using novel sensor and biosensor technologies (464). So far, a few studies have leveraged the power of electronic and electrochemical methods to create fast and sensitive diagnostic devices for SARS-CoV-2 detection. For example, Seo et al. described a field-effect transistor (FET)-based biosensing device that detected SARS-CoV-2 at concentrations of 2.42 × 102 copies/ml in clinical specimens, without sample pretreatment, in approximately 3 min (465). The FET sensor used SARS-CoV-2 monoclonal antibodies (mAbs) to spike proteins, which were coupled to graphene sheets through a 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE) linker (Fig. 10B). With the high conductivity and other properties of graphene, real-time dose-dependent detection of SARS-CoV-2 antigen can be achieved down to 1 fg/ml. While further optimization is needed to commercialize this technology, it has the potential to help increase the sensitivity of antigen detection. In another study, Mahari et al. developed an electrochemical device using a screen-printing technique. They immobilized antibodies against SARS-CoV-2 spike proteins on screen-printed carbon electrodes (SPCEs) to detect the virus and developed an integrated in-house-built portable device that could measure the changes in electrical conductivity upon the reaction between antigens and antibodies. The device was able to detect spiked SARS-CoV-2 antigens in saliva with an LoD of 90 fM within 10 to 30 s (466).
While FETs and electrochemical sensors show promise to increase the sensitivity of antigen detection, the desire to achieve the highest sensitivity possible with RDTs has been the subject of recent debate (288, 299, 301, 467, 468). Some authors have proposed that with repeat testing, the chance of identifying SARS-CoV-2-infected individuals increases (288, 467–470). This may be true on a population level, where testing may otherwise not have been performed (e.g., asymptomatic testing), and there would be fewer consequences if detection of the virus is missed. However, repeat testing and the possibility of false-negative results in an acute-care setting pose many more challenges, particularly when decisions have to be made at the time of or shortly after the time of presentation. For existing or novel technologies, studies with more robust data are needed to better define the utility of Ag-RDTs in various settings. These should include prospective analyses that consider factors such as the timing of collection, time of symptom onset, symptomatic and asymptomatic patient populations, disease prevalence, stability, and following the manufacturer’s recommendations for stability and transport (i.e., direct testing of specimens and not using specimens in transport media).
Serological Immunological Methods for SARS-CoV-2 Detection
Serological assays detect antibodies specific to SARS-CoV-2 in blood sources like serum, plasma, or whole blood (including fingerstick and heel pricks sometimes used in POC testing), and the possibility of antibody detection in other body fluids such as saliva is being explored (471–473). Given that the typical time required to detect immune responses to SARS-CoV-2 is around 1 to 2 weeks, serological tests have limited utility for SARS-CoV-2 diagnostics in the acute stages of the illness but could have value once immune responses have had time to occur (241, 474–478). Many serological assays for detecting SARS-CoV-2 antibodies have been developed and commercialized, and a list of authorized medical devices related to COVID-19 in the United States and Canada is regularly updated online (29, 30, 472, 479). Despite access to serological assays, the interpretation of their results is further complicated in the postvaccine era for SARS-CoV-2, and the relevance of antibody detection must be considered in the context in which it is used and its limitations. Following a section describing serological methods, a discussion on the relevance and possible applications is provided.
Serological assays often use recombinant antigens to capture SARS-CoV-2-specific antibodies, with the N protein and the receptor-binding domain (RBD) of the S1 subunit of the S glycoprotein being the most commonly used antigens. Serological methods can target one or more immunoglobulin isotypes (i.e., IgA, IgM, or IgG) or total antibody, and theoretically, any of these isotypes could provide neutralizing activity against SARS-CoV-2 (480). IgM and IgG are commonly used immunoglobulins in serological assays for SARS-CoV-2, while IgA detection is less commonly used (479). IgA and IgM have shown some merits for the detection of early immune responses to SARS-CoV-2, but there has been some concern regarding the rapid decay of IgA and IgM in serum and saliva compared to IgG (242, 481, 482). While more data are needed to fully understand the serological responses in different settings or patient populations, it is clear that IgA, IgM, and IgG all have the potential to generate neutralizing responses to SARS-CoV-2 (242, 481, 482).
The most common methods used are ELISAs in a 96-well plate format and CLIAs for automated higher-throughput instruments (49, 479). LFIAs are also available to provide rapid results. It is unclear whether detectable antibodies in commercial serology methods are associated with immune protection, as large population-level studies and quantitative immunoglobulin analyses are required to assess if detection of SARS-CoV-2-specific immunoglobulins correlates with immune protection. However, studies are under way to investigate the extent of neutralizing antibodies produced over time (242, 481, 482).
The performance of immunological assays remains unclear as there is a lack of a gold standard for method comparison. Methods are often compared among themselves from a consensus standard, but the true performance and relevance of antibody responses, and the optimal testing algorithm, require further investigation (483). For now, the applications of serology include a role as an adjunct to molecular testing to support the identification of SARS-CoV-2 in a patient suspected of having COVID-19 (i.e., persistent or progressing symptoms) but with the absence of testing or repeated negative (or indeterminate) results obtained by NAATs. It should be noted that some patients may be antibody negative at the time of testing, but that does not preclude memory B cell activity or functional T cell responses with subsequent exposures to SARS-CoV-2. Some studies argue that immunological methods could also be used in seroprevalence studies to aid in ongoing outbreak investigations (to identify cases beyond the detectable window of NAATs), to determine past exposures of populations to SARS-CoV-2 (e.g., health care workers or the general population), or to assess attack rates in defined populations or geographical areas. However, correlations to neutralizing antibody titers would be required to fully understand serological data, along with the impact of prior infection with SARS-CoV-2 and other human coronaviruses as well as the impact of vaccines. High sensitivity and specificity are desired, but with these many confounding factors, result interpretation is complicated (49, 484). The following sections describe common methods used for SARS-CoV-2 serology, including ELISAs, CLIAs, and LFIAs, along with a discussion of the potential value and limitations of immunological methods.
Enzyme-linked immunosorbent assay.
While many permutations exist, enzyme-linked immunosorbent assays (ELISAs) can be categorized into four main types: sandwich, direct, indirect, and competitive (485). For the detection of SARS-CoV-2, indirect (210, 486–489), modified indirect (210), and double-antigen sandwich (206) ELISAs are the most commonly used methods (Fig. 11A).
FIG 11.
Common serological immunoassays for the detection of SARS-CoV-2-specific antibodies. (A) Common designs of ELISA methods, including indirect, modified indirect, and double-antigen sandwich assays. (B) Magnetic bead-based CLIA.
In the indirect ELISA method, a solid support, such as wells of a 96-well microplate, is coated with SARS-CoV-2 recombinant antigens (210, 486–489). Patient serum or plasma is added, and if present, SARS-CoV-2-specific antibodies will bind to the immobilized antigens. Following washing steps, a secondary antibody is then added that is specific to the isotype targeted (i.e., anti-human IgM, anti-human IgG, or anti-human IgA). This secondary antibody (or conjugate) is linked to a fluorophore that can generate fluorescence or conjugated to an enzyme for colorimetric or chemiluminescent signal generation following the addition of a substrate. Usually, colorimetric methods are used in which the optical density (OD) (i.e., absorbance) of the solution can be measured using a spectrophotometer and correlated with the concentration of the target antibodies. Other permutations of ELISAs are also possible, but while these techniques all share similarities in terms of their signal generation mechanisms, each can vary in performance. Typically, the time to result for ELISAs is 1 to 5 h (41), and more recently, POC applications of ELISAs for the detection of anti-SARS-CoV-2 antibodies have been explored using microfluidics technology (490). The performance characteristics and limitations of SARS-CoV-2 serology are discussed in a later section.
Chemiluminescence immunoassay.
The chemiluminescence assay (CLIA) is an immunoassay that is commonly used in highly automated serological instruments and is known for its high sensitivity compared to other serological assays (491). Similar to ELISAs, variations of CLIAs have been applied for the detection of SARS-CoV-2 antibodies, such as indirect (492, 493) and sandwich (239) methods.
The CLIA has two main differences from colorimetric ELISAs. First, the final reaction produces light, which is detected by a luminometer in relative light units (RLU) rather than the OD, which is an absolute value. To produce light, the conjugate uses an enzyme-substrate reaction such as the alkaline phosphatase (AP) and Lumigen APS-5 substrates. Second, magnetic microspheres in a liquid-phase reaction rather than multiwell plates are coated with the antigenic materials, which allows the easy separation of bound and unbound molecules by a magnet and faster reactions due to providing a large surface area and allowing the reactions to occur entirely in suspension (494–496) (Fig. 11B). Some CLIAs have been commercialized and used on high-throughput automatic analyzers. For example, the Liaison assay (DiaSorin, Italy) uses magnetic beads coated with SARS-CoV-2 S1 and S2 antigens to detect IgG antibodies. This assay is capable of providing up to 170 results/h in a fully automated manner.
Fluorescent microparticle immunoassays.
Recently, Norman et al. described a fluorescent nanoparticle immunoassay (FMI)-based single-molecule array assay that consisted of a mixture of four types of dye-encoded beads coupled to S, S1, S2, and N proteins and used for the detection and differentiation of SARS-CoV-2 IgM, IgG, and IgA antibodies (497). In this technique, an excess number of beads is utilized in comparison to the number of antibody molecules in the samples in a way that either zero or one antibody molecule binds to each bead. After the reactions occur between the coated beads and the antibodies and other reagents, the beads are loaded into an array of 216,000 femtoliter-sized wells for imaging, which allows the detection of antibodies with single-molecule resolution. When tested on a set of 81 plasma samples, the technique showed 86% sensitivity and 100% specificity for the samples during the first week after symptom onset and 100% sensitivity and specificity for the samples taken after the first week after symptom onset (497).
Similarly, to facilitate the simultaneous detection of the IgG, IgM, and IgA isotypes of SARS-CoV-2-specific immunoglobulins, a multiplex SARS-CoV-2 antibody immunoassay based on Luminex technology was developed for detection on the Bioplex automated flow cytometer (Bio-Rad Laboratories) (473). This multiplex immunoassay was used to demonstrate SARS-CoV-2 antigen-specific antibody responses in 33 saliva and 206 serum samples from participants with RT-PCR-confirmed SARS-CoV-2 infection. Overall, this technology could detect prior SARS-CoV-2 infection with high sensitivity and specificity at ≥10 days after symptom onset, but the performance varied with the targets (473). FMI technology allows robust antibody profiles to be simultaneously Tdetected in a single reaction. A commercial assay for the Bioplex 2200 system has recently received FDA approval, but no data on this assay have been published to date.
Lateral flow immunoassays.
As described in the antigen detection section above, lateral flow immunoassays (LFIAs) can be used as an RDT or POC test to detect antibodies against SARS-CoV-2 in blood or serum samples. The most frequently used antigens are recombinant S or N proteins to capture total antibody or detect and sometimes differentiate IgM and IgG antibodies (479). The LFIA devices are low cost, portable, and rapid (∼15 min) and require only a few microliters of samples, making them suitable for fingerstick tests (498–500). Figure 12A illustrates an example of an LFIA used for the simultaneous colorimetric detection of SARS-CoV-2-specific IgM and IgG. Many different types of LFIAs have been developed to detect SARS-CoV-2, with variations in antigens used for antibody capture, immunoglobulin isotypes detected, and overall performances (479, 501).
FIG 12.
Serological lateral flow immunoassays for the detection of SARS-CoV-2-specific antibodies. (A) Schematic of a lateral flow immunoassay device for the simultaneous detection of IgM and IgG antibodies. Upon the addition of the sample, the liquid moves toward the preimmobilized reagents through capillary action and reacts with them. If IgM/IgG antibodies are present in the sample, they bind and form a complex with the recombinant SARS-CoV-2 antigen conjugated with colloidal gold nanoparticles (AuNPs). The complex is then captured in one of the test lines by anti-human IgM/IgG antibodies, resulting in a pink color due to the accumulation of the AuNPs. There is also an AuNP-rabbit IgG conjugate that will be captured in the control line, indicating proper liquid flow through the device. The results will be observable in ∼15 min. (B) Variations of results of the lateral flow assay device in a cassette format. (This figure was inspired by the work in reference 498.)
Performance of serological immunological methods for SARS-CoV-2.
TIn a systematic review and meta-analysis, the results of 40 studies up to 30 April 2020 were summarized, and differences in sensitivity between serological methods used for SARS-CoV-2 antibody detection as well as antibody classes were presented (49). Data for IgA were scarce, and no conclusions could be drawn. For IgM detection, the pooled sensitivity (95% CI) for LFIAs was the lowest, at 61.8% (50.8% to 71.8%), whereas that for ELISAs was higher, at 81.7% (71.8% to 88.5%), and that for CLIAs was the highest, at 84.3% (70.7% to 93.0%). Similarly, the pooled sensitivities for IgG detection were 64.9% (53.8% to 75.4%), 80.6% (71.9% to 87.9%), and 93.5% (84.9% to 98.1%) for the LFIA, ELISA, and CLIA, respectively. The pooled specificities were fairly similar among the LFIA, ELISA, and CLIA methods, at 96.6% (93.8% to 98.4%), 99.7% (99.0% to 100.0%), and 96.6% (84.7% to 99.5%) for IgM detection and 97.6% (96.2% to 98.8%), 98.9% (96.7% to 99.8%), and 97.8% (62.9% to 99.9%) for IgG detection, respectively. Other meta-analyses and systematic reviews yielded consistent conclusions for sensitivity and specificity for IgM, IgG, or total antibodies (472, 502, 503).
One important issue affecting the diagnostic performance of serological assays is the time after symptom onset when the samples are taken for serological analyses, as the antibody profiles change over the course of the disease. In a meta-analysis and systematic review (49), the pooled sensitivity (95% CI) for IgM detection was low in the first week after symptom onset, at 25.3% (16.3% to 31.1%), for the LFIA but increased to 51.8% (30.3% to 69.6%) after 2 weeks and to 69.9% (58.4% to 79.9%) after 3 weeks. ELISA detection of IgM showed similar values for weeks 1, 2, and 3 after symptom onset, at 26.7% (15.6% to 35.6%), 57.6% (15.9% to 88.2%), and 78.4% (54.1% to 91.9%), respectively. The CLIA followed the same trend but with higher values, at 50.3% (10.9% to 81.2%), 74.3% (16.1% to 99.4%), and 90.6% (51.8% to 99.4%), respectively. Next, the pooled sensitivities for IgG at weeks 1, 2, and 3 after symptom onset were 13.4% (4.7 to 29.6%), 50.1% (24.8% to 77.0%), and 79.7% (71.4% to 86.9%) for the LFIA; 23.7% (12.7% to 38.1%), 65.3% (46.3% to 79.4%), and 82.1% (76.4% to 89.0%) for the ELISA; and 53.2% (28.7% to 67.6%), 85.4% (48.1% to 98.1%), and 98.9% (86.9% to 100.0%) for the CLIA, respectively (49).
Overall, the performances of serology assays varied between methods and were dependent on the timing of collection, and the applicability of these methods is limited to date. It is important to recognize that other factors such as antigens and the inherent characteristics of a test or instrument used for antibody detection can also impact the performance of immunological assays. Also, data summaries in recent meta-analyses and systematic reviews so far have shown high levels of heterogeneity and a risk of bias and applicability between studies (472, 502, 503). Thus, careful attention should be paid to the design of the studies, their limitations, and whether the conclusions derived from these evaluations are justified. It remains unclear whether antibodies will provide durable protective responses against subsequent SARS-CoV-2 infections or what will be the impact of prior infection with seasonal human coronaviruses (474, 475). It is possible that prior infection with other coronaviruses or other conditions (e.g., pregnancy or chronic illnesses) may cross-react with SARS-CoV-2 serology assays, and robust evaluation of methodologies should include specificity analyses against related viruses and possible interfering substances (504, 505). The recent introduction of vaccine programs will further complicate the use of immunological methods as diagnostic tests but may expand their use to determine whether individuals are immune once exposed to SARS-CoV-2 through natural infection or following immunization. This remains an active area of research that requires comparisons to neutralizing antibody titers and cell-mediated immunity.
Neutralization assays and research areas of interest for serology.
The efforts of the scientific community to help understand the host responses to SARS-CoV-2 are fundamental in understanding COVID-19 pathophysiology and the interpretation of SARS-CoV-2 laboratory diagnostics (506–509). There has been an extraordinary effort to rapidly develop serological methods in various formulations, and while some were licensed under EUA in the United States and Canada, their use to make recommendations on a patient’s current or future susceptibility to SARS-CoV-2 infection remains unclear (29, 30). Given the timing of the immune responses to SARS-CoV-2 infection, it is clear that serology would not be useful for diagnosis in acute phases of illness; antibodies would not be present in substantial quantities in this time frame to allow public health interventions (133). In fact, positive serology does not preclude viral shedding (208, 510), and the absence of antibodies does not preclude the possibility of immunity. To date, recommendations for the use of serology include clinical research (e.g., seroprevalence studies) to help inform public health strategies or policies on a population level (102, 133, 241). For example, seroepidemiological studies can support ongoing outbreak investigations or retrospectively assess infection rates in certain populations (133, 511). On an individual level, serology may have considerations for individual patients with multisystem inflammatory syndrome or in situations where suspect cases repeatedly test negative by NAATs, but symptoms persist (and the timing fits into a period of at least 2 weeks after symptom onset) (102, 133, 241). Future research is required to further understand the full value of SARS-CoV-2 serology, especially in the context of immunization with COVID-19 vaccines.
Key areas of research interest should include the relevance and duration of antibody-based immune responses to better understand whether antibody responses in commercial tests or LDTs correlate with protective immune responses against subsequent SARS-CoV-2 infection as well as the magnitude and duration of these responses. Understanding this correlation can be an important function to assess immunity on an individual or population level or even to qualify blood donors desiring to become donors of convalescent-phase plasma that could subsequently be used in clinical trials (512, 513). This would require a quantitative analysis of seroconversion (detection of measurable antibodies following infection), which would not be possible with qualitative assays such as LFIAs but may be possible using semiquantitative ELISAs (where antibody detection relies on a signal value that is normalized to a calibrator to establish a signal-to-cutoff [S/CO] ratio) or, ideally, using quantitative ELISAs. Quantitative detection of SARS-CoV-2 antibodies over time could be used to demonstrate seroconversion, and quantitative ELISAs have become commercially available (e.g., the liquid-based double-antigen sandwich ELISA from Roche [Elecsys anti-SARS-CoV-2 S]). Quantitative antibody analyses, paired with a fundamental understanding of how well serological methods correlate with neutralizing antibodies (nAbs), may help establish a threshold where populations are protected from SARS-CoV-2 reinfection (known as a correlate of protection).
The establishment of a correlate of protection first requires an in-depth understanding of whether nAbs are produced, the extent to which they are produced, whether they provide protection, and the duration of these responses and then analyzing these data in large population studies to correlate a level of nAbs that may confer protection against subsequent infection (514). The quantification of nAbs in serum or plasma is performed using target- and immunoglobulin isotype-specific neutralization assays 9like plaque reduction neutralization tests (PRNTs) (41, 241, 424, 515). Briefly, PRNTs rely on the ability of nAbs to effectively prevent SARS-CoV-2 from infecting cultured cells that would otherwise cause cytopathic effects (i.e., cell death/lysis) (41, 241, 511, 515–517).
Nonetheless, PRNTs are known to be time-consuming and laborious. To increase specimen throughput, microneutralization (MN) tests have been developed to detect nAbs to SARS-CoV-2 that rely on a principle similar to that of PRNTs but are performed in 96-well formats and without the need for semisolid overlays (511, 517). To further automate the detection of nAbs, a focus reduction neutralization test (FRNT) can be used where patient sera/plasma dilutions can be assessed for their ability to reduce viral plaque formation, foci, or individually infected cells using immunocolorimetric staining and visualization using digital imaging (480). Despite MN, FRNTs are amenable to moderate-throughput testing, and alternatives have been developed using fluorescence detection and luminescence to increase sensitivity. On the other hand, MN tests and FRNTs share a challenge similar to that with PRNTs: they all rely on SARS-CoV-2 propagation in cell culture, which requires laboratory facilities with higher levels of biological containment (i.e., biosafety level 3 [BSL3] in Canada or physical containment level 3 [PC3] as defined by the WHO) (424). These practices are not common outside reference laboratories (241). To circumvent these biosafety requirements, pseudovirus-based neutralization assays (PBNAs) have been developed, where SARS-CoV-2 proteins are expressed on a surrogate virus backbone such as a lentivirus, retrovirus, or vesicular stomatitis virus (VSV) (516, 518, 519). Other alternatives include the use of a protein-based surrogate neutralization ELISA (snELISA), which is based on the competition between ACE2 and nAbs for binding to recombinant antigens (520–526).
Few studies have looked at nAbs, and data on the extent and duration of nAbs over the course of SARS-CoV-2 infection are scarce (508, 527–530). Suthar et al. used an FRNT to demonstrate that nAb binding to the SARS-CoV-2 S protein RBD is detectable within 6 days after PCR confirmation (480). While days postonset would have better defined the antibody profiles over time, these authors showed that there is rapid isotype switching in nAbs from IgA and IgM to IgG1 and IgG3. In a limited data set from Long et al., it was noted that there was a decline in nAbs over time in both asymptomatic and symptomatic individuals (239). Similarly, Seow et al. performed a longitudinal study using sequential serum sampling and demonstrated that seroconversion was observed in 95% of cases, with nAbs present 8 days after the onset of symptoms (531). They also noticed that the magnitude of nAb responses depended on disease severity and waned over time, which was consistent with the results of others (206, 239, 532–535). In some cases, nAbs persisted up to 60 days, while others approached the limit of detection in this period (531). In a more recent study by Isho et al., the duration of antibody responses was evaluated in both serum and saliva (242). Antibodies to S protein were detected in 90% of cases approximately 10 days after symptom onset, and longitudinal studies showed the persistence of nAbs up to 105 days. Similarly, nAbs (defined using snELISAs) reached a maximum from days 30 to 45. Interestingly, the sensitivities of anti-S or anti-RBD IgG in saliva were 89% and 85%, showing promise for noninvasive methods for seroepidemiological studies and vaccine trials. The sensitivities of IgA, on the other hand, were 51% and 30%, and those for IgM were 57% and 33%, respectively. It should be noted that the limited number of longitudinal studies seem to suggest that SARS-CoV-2-specific antibodies decrease over a 2- or 3-month period; however, the longevity of humoral immunity is not yet fully understood for SARS-CoV-2. It is possible that memory T cells may still be able to mount effective responses upon reexposure to SARS-CoV-2 (509, 536–538). Research into the role of cell-mediated immune responses is needed to help unravel knowledge gaps for SARS-CoV-2 immune responses (508, 509).
Overall, whether derived from vaccines or natural infection, quantitative analyses of nAbs and establishing a correlate of protection would help inform public health and health care policies. Using neutralization assays with targets outside those used in vaccines could still be of benefit to understand seroepidemiology to help better define populations at risk, which would be important for vaccine trials or evaluations of novel SARS-CoV-2-specific therapies like the use of hyper-IgG preparations or convalescent-phase sera (520, 523, 539–541). Like any other method for SARS-CoV-2 detection, serology research should consider antibody and cellular immune responses over time, have a population- and individual-level testing perspective, as well as account for factors like the presence or absence of symptoms, the timing from symptom onset (if present), the severity of illness, host factors (e.g., age and medical comorbidities), and any collection, transport, or processing steps that may affect the reliability of the analyte profiles over time. Additional research is still needed to meet these goals.
CONCLUSIONS AND OUTLOOK
Despite the recent availability of vaccines, with the ongoing and rapid spread of SARS-CoV-2 worldwide, laboratory testing remains the cornerstone of public health containment and mitigation strategies. Given the thoroughness of the data on methodologies in the body of this review, only key findings, some considerations for testing, and areas for improvement and successes are discussed below; however, a summary of testing modalities is presented in Fig. 13.
FIG 13.
Methods used for SARS-CoV-2 detection or identification of COVID-19. Of note, cell culture and microscopy are not used for clinical diagnosis but are used for research purposes. Abbreviations: WBC, white blood cell; CRP, C-reactive protein; PCT, procalcitonin; IL-6, interleukin 6; ALT, alanine aminotransferase; AST, aspartate aminotransferase; LDH, lactate dehydrogenase; CK, creatine kinase; NAAT, nucleic acid amplification test; RT-PCR, reverse transcription-PCR; TMA, transcription-mediated amplification; RT-LAMP, reverse transcription–loop-mediated isothermal amplification; RT-RPA, reverse transcription-recombinase polymerase amplification; CRISPR-Cas, clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR associated (Cas); NEAR, nicking enzyme-assisted reaction; Ag-RDT, antigen rapid diagnostic test; LFIA, lateral flow immunoassay; ELISA, enzyme-linked immunosorbent assay; CLIA, chemiluminescence immunoassay; FMI, fluorescent microparticle immunoassay; CT scan, computed tomography scan.
While compatible signs and symptoms, routine laboratory testing for biomarkers of health or disease, and diagnostic imaging have roles to play in diagnostic investigations, none alone are sufficient for diagnosing SARS-CoV-2, and the reliance on specific laboratory testing is paramount. Many technological advances have been made to detect SARS-CoV-2, which include NAATs to detect viral RNA and immunoassays to detect viral antigens or virus-specific antibodies generated by the immune system in response to SARS-CoV-2. There is a growing list of commercially available methods for testing for SARS-CoV-2 (29, 30), and while many have been validated, the results of any test should be interpreted with consideration of the context in which they are used and based on the sum of all diagnostic evidence. When using any method to rule in or out SARS-CoV-2 infection, many factors should be considered, such as the timing and type of specimen collection, the anatomical site of sampling, the method and its expected performance characteristics, host factors like compatible signs and symptoms (versus asymptomatic testing), risk factors for serious outcomes, and disease prevalence in the population (82, 542–544).
NAATs like real-time RT-PCR quickly became the gold standard for diagnostic testing. However, apart from those listed above, other considerations for NAATs are inherent to the methodologies that are all based on the detection of SARS-CoV-2 RNA. For example, it is important to recognize that the performance of any molecular diagnostic method can be influenced by sequence mismatches between the method’s targets and the different genome permutations of circulating SARS-CoV-2 lineages and variants. A possible strategy to reduce the chance of false-negative results that could occur by target mismatches is the simultaneous use of molecular methods targeting more than one gene, as failure to detect a signal in one target gene may not preclude detection in another (40, 164, 443). Alternative strategies could include the use of degenerate primers and probes, a strategy used with other RNA viruses that are prone to mutation (443, 545, 546). Ongoing molecular surveillance using sequencing technologies should also be encouraged to monitor changes in SARS-CoV-2 genome sequences and COVID-19 epidemiology, particularly with an emphasis on variants linked to failures in diagnostic testing, with increased transmissibility, increased severity, or decreased susceptibility to convalescent-phase sera or responses to vaccines.
The primary reliance on NAATs for SARS-CoV-2 detection during the pandemic came with many challenges globally. With human resource strains and supply chain shortages, providing NAATs during the pandemic was challenged by factors such as PPE, human resource strains for sample collection and testing, swabs, and test reagent availability for NAATs. From a clinical perspective, alternatives to NP swabs traditionally used for respiratory specimens were rapidly validated, as was the use of various transport media or swab-free options that are amenable to self-collection (e.g., saliva or saline gargles). The application of these specimens in the laboratory added complexity to the laboratory workflow and required rapid validations to ensure compatibility with new or existing instrumentation.
To help meet capacity demands in the laboratory, laboratories were faced with the need to rapidly procure large quantities of supplies, acquire instrumentation, train additional personnel, and validate specimens, reagents, and equipment. Despite this, supply chain challenges and rapid escalation of testing demands led to the need for resource-sparing strategies for NAATs, including specimen pooling and extraction-free NAAT protocols. Such strategies come at the cost of a relatively reduced sensitivity, which is associated with the potential risk of missing detection of SARS-CoV-2 in specimens with low viral loads. It could be argued that small reductions in sensitivity may have little impact on the detection of most cases. Remnant SARS-CoV-2 at the outset of illness can persist for weeks and is unlikely to represent a period of communicability. On the other hand, missing detection of SARS-CoV-2 in a specimen with a low viral load at the early stages of illness may potentially lead to further virus spread. These testing strategies should be carefully considered prior to implementation.
SARS-CoV-2-specific NAATs evolved over time to facilitate testing and streamline the laboratory workflow. For example, to further streamline testing for respiratory viruses, some methods have now been multiplexed to simultaneously detect SARS-CoV-2, influenza A and B viruses, and other respiratory viruses like respiratory syncytial virus (RSV). In nonpandemic years, influenza and other viral etiologies of respiratory tract infections represented a leading cause of death in North America, particularly among hospitalized patients with community-acquired pneumonia (547–553). Interestingly, while cocirculation and coinfection with other respiratory viruses were reported, there was little activity for non-SARS-CoV-2 respiratory viruses (554). It is unclear whether public health interventions (e.g., travel restrictions, social distancing, handwashing, or PPE like masks) resulted in this decline or whether other factors inadvertently contributed to biases in data, such as fewer individuals seeking routine medical attention or the lack of testing for influenza and other respiratory viruses due to competition for resources used for SARS-CoV-2 testing (554–558). Concomitant diagnostic testing using multiplex technologies could provide an option for syndromic testing that would ensure surveillance for SARS-CoV-2 and other important respiratory viruses like influenza virus and appropriate interventions as needed (e.g., antivirals).
The public and political pressure for laboratory testing evolved with public health indications, and laboratory testing continues to guide public health policies as restrictions ease or escalate throughout the ongoing pandemic (47, 484). One area of significant advancement in NAAT methodologies includes the use of automation to minimize hands-on processing time and increase specimen throughput. With the high demand for laboratory testing, automation is an important consideration to avoid the possibility of staff repetitive-stress injuries. Over the course of the COVID-19 pandemic, the scale and demand for laboratory testing have been unmatched by other pandemics. Concerns over SARS-CoV-2 prompted the testing of both symptomatic and asymptomatic individuals, in the context of public health case contact tracing and surveillance purposes. This includes testing for SARS-CoV-2 in both health care settings where patients are at increased risk (e.g., hospitals and long-term-care facilities) and situations where testing would not otherwise have been performed (i.e., professional sports teams, public events, prior to or after travel, and various workplaces). Thus, innovative, dynamic, and adaptable approaches were required to meet the testing demands, which are covered extensively in this review.
Other areas are recognized as being crucial to increase laboratory testing capacity but are not covered in this review, such as supply procurement, distribution, and management and the coordination of training or recruitment of increased human resources for specimen collection, testing, processing, and registration. Also, rapid validation of laboratory tests and collection devices to meet regulatory requirements despite availability through EUA from regulatory bodies as well as the development of an information technology infrastructure to continuously improve the laboratory information management system (LIMS) in the laboratory testing workflow and dissemination of near-real-time laboratory data to various stakeholders from local to national levels and dissemination to the public through various media formats are of great value. While digitization was not an absolute requirement, the interconnectivity of data ensured transparency and up-to-date information as decision support tools for recommendations, policies, and guidelines (53, 559–561). All of these have been identified by the WHO as key factors to control the COVID-19 pandemic (562).
To further enhance testing capacity and provide access for rapid SARS-CoV-2 testing options, both antigen- and NAAT-based RDTs have been developed and are now readily available (29, 30). These can support rapid laboratory and POC applications, screening of large patient populations, or rapid deployment for assessments of target areas. NAAT-based RDTs rely on real-time RT-PCR or isothermal amplification methods (e.g., RT-RPA, RT-LAMP, and NEAR) and, like antigen-based RDTs, can provide rapid results without complex instrumentation. The ease of use of these portable devices allows access to SARS-CoV-2 diagnostics in settings that may have been prohibitive for traditional laboratory NAATs. Throughout the literature, the main limitation noted for RDTs is their reduced sensitivity compared to traditional NAATs, but the evaluations of their performance do not always reflect conditions in which the RDT was licensed under EUA, and the comparator method and distribution of expected specimen results can have a large impact on assay performance (274, 290, 341, 342, 457). Even if these issues are not considered, and all SARS-CoV-2 detection results were considered of value, a possible strategy to mitigate the relatively reduced sensitivity of antigen- or NAAT-based RDTs is to increase the frequency of testing in the patient population over time, to increase the chances of identifying individuals who fall in a period of high viral shedding (which would be less likely to be impacted by reduced sensitivity) (288, 299, 301, 467, 468). On the other hand, the implementation of such strategies has challenges of its own due to the limited scalability of RDTs, and the balance between sensitivity and testing frequency to achieve optimal SARS-CoV-2 detection in the target population would need to be defined (288). Moreover, if, for example, EUA defined the use of RDTs as within 7 days of symptom onset, this excluded their use for testing of asymptomatic individuals. While much development is ongoing to enhance existing RDT technologies or explore novel methodologies (53, 465, 466, 563–565), how RDTs can effectively be used in practice is the subject of ongoing debate, and further research is needed to understand what setting they would best be of benefit.
For serology, currently available commercial assays are based on ELISAs, CLIAs, and LFIAs and are designed to detect SARS-CoV-2 antibodies. However, the performance of serological assays varies across the different technologies, the timing from disease onset, and comparator methods. Clinical validation of serological methods is ongoing, but recent meta-analyses and systematic reviews so far have shown high levels of heterogeneity and a risk of bias and applicability (472, 502, 503). For example, with the time required to mount anti-SARS-CoV-2 antibodies during seroconversion (or possibly the absence of seroconversion in mild disease), serology has limited value in identifying SARS-CoV-2 in the early stages of illness. Serology has value for seroepidemiological studies to help with ongoing outbreak investigations, can aid in the diagnosis of suspect cases for whom NAATs were persistently negative or not performed, or can help with conditions in children and adolescents like multisystem inflammatory syndrome where NAAT results may be negative (102, 133, 241, 566–570). Much research is needed to expand the knowledge on the use of immunological methods, particularly in the context of the recent availability of vaccines.
Key areas of research interest for serology should include the relevance, magnitude, and duration of protective antibody responses, particularly faced with a population that may have been exposed to SARS-CoV-2 or other human coronaviruses or vaccinated against SARS-CoV-2. Detection of different antibody isotypes is possible with commercial or laboratory-developed serological assays, and recent quantitative methods have now become commercially available; however, much is yet to be learned on how these methods correlate with protective immune responses. Establishing correlates of protection will require longitudinal studies with parallel assessments of quantitative levels of nAbs and cell-mediated immune responses. Various neutralization assays have been established to assess and quantify antibodies against SARS-CoV-2 in serum or plasma (e.g., PRNTs, MN tests, or surrogate assays like PBNAs and snELISAs), but data are scarce when it comes to longitudinal studies undertaken to fully understand the differences or similarities between commercial assays for the qualitative detection of antibodies, quantitative antibody analyses over time, and relevance to the level and duration of protective nAb titers. While the longevity of humoral immunity is not yet fully understood for SARS-CoV-2, some data suggest that antibody levels against SARS-CoV-2 wane over 3 months. Notwithstanding these observations, it is possible that memory T cells may still be able to mount effective responses upon reexposure to SARS-CoV-2, and the role of cell-mediated immune responses may also provide additional benefits (508, 509). Further research is needed, as a greater understanding of the immune response to SARS-CoV-2 is fundamental in making informed recommendations for the use of immunological methods to assess current or future protection against the virus, to determine meaningful endpoints in the development and evaluation of effective vaccines, or to assess effectiveness following immunization with existing vaccines.
Overall, methods based on RNA, antigen, or antibody detection as well as diagnostic imaging all have a place in our response to SARS-CoV-2, but ongoing research is crucial to further optimize and apply all these testing modalities. The emergence of the SARS-CoV-2 pandemic has created a wave of innovative and creative thinking, and more and more creative methods and platforms are being introduced with goals to increase the armamentarium of diagnostic methods for SARS-CoV-2 detection. An understanding of the advantages and limitations of each method used for SARS-CoV-2 detection as well as the development of novel methods will help us unravel the unknowns of disease pathogenesis, epidemiology, and transmissibility and help us develop interventions to mitigate and contain its spread. There will always be a need for laboratory testing and collaboration between clinical laboratories, public health, infection prevention and control, and many others who contribute to the efforts to contain the spread of COVID-19. On the other hand, it is also recognized that maintaining large investments in the rapid deployment of translational research and such a high degree of laboratory testing for COVID-19 will likely not be sustainable from an economic perspective, and justifications for such investments will be more difficult if cases decline significantly with vaccines. However, the lessons learned from SARS-CoV-2 could potentially be used in the preparedness for potential future pandemic threats, thus strengthening global health and surveillance systems.
ACKNOWLEDGMENTS
This research received no specific financial support. S.H.S.T., Z.S., O.D.O., and S.J.-A. receive funding from the Natural Sciences and Engineering Research Council of Canada (NSERC) through a discovery grant and the Concordia FRDP grant for development of point-of-care analytical devices for medical diagnostic and environmental application. The funders had no role in data collection, interpretation, or the decision to submit the work for publication.
We thank Seyed Hossein Safiabadi-Tali and Pegah Hadavi for their kind scientific support during the writing of this paper. We also recognize all the efforts of frontline health care providers, including medical laboratory personnel; researchers in academic, industry, and health care settings who are instrumental in developing novel methods for the identification of SARS-CoV-2; as well as the ongoing efforts of infection control and prevention programs, public health staff and epidemiologists, and governmental agencies who have provided unwavering support throughout this pandemic.
Biographies

Seyed Hamid Safiabadi Tali is a Ph.D. candidate in the Anbuhi Research Group in the Chemical and Materials Engineering Department at Concordia University, which he joined in June 2019. He obtained his B.Sc. and M.Sc. in chemical engineering with a specialty in biotechnology from the University of Tehran, Iran, in 2015 and 2018, respectively. His main area of research is the development of low-cost and portable sensor and biosensor devices for the detection of pathogens and biomarkers for different applications, from food and environmental monitoring to disease diagnostics.

Jason J. LeBlanc, Ph.D., FCCM, d(ABMM), is a Professor in the Department of Pathology, Faculty of Medicine, at Dalhousie University, with cross-appointments in Medicine (infectious diseases) and the Department of Microbiology and Immunology in the same institution. He is the Director of Virology, Immunology, and Molecular Microbiology in the Division of Microbiology, Department of Pathology and Laboratory Medicine, Nova Scotia Health, Halifax, Nova Scotia, Canada, and holds credentials as a Fellow of the Canadian College of Microbiologists (FCCM) and Diplomat of the American Board of Medical Microbiologists [d(ABMM)]. Dr. LeBlanc has a long-standing research interest in molecular diagnostics, molecular epidemiology, and molecular pathogenesis, with application to the fields of vaccine-preventable diseases and emerging respiratory pathogens like SARS-CoV-2.

Zubi Sadiq, Ph.D., is a researcher at Concordia University, Canada, and is interested in the detection of pathogens in food, water, and the environment. She has received her B.S. (Hons) and Ph.D. in Chemistry from Lahore College for Women University, Pakistan. She received a gold medal and academic roll of honor in her M.S. studies. She served as lecturer at the Preparatory Year Deanship, King Faisal University, Saudi Arabia. Her area of expertise also includes the development of sustainable protocols for heterocyclic molecules and evaluation of different bioactivities against infectious microbes.

Oyejide Damilola Oyewunmi obtained his B.Sc. in Chemical Engineering at the University of Lagos, Nigeria. He is currently a graduate student at the Department of Chemical and Materials Engineering at Concordia University, Montreal, Canada. His primary research field is the investigation into paper-based sensors and biosensors for environmental monitoring and biological diagnostics at the Anbuhi Research Group.

Carolina Camargo, Ph.D., is a postdoctoral fellow in the Sagan laboratory in the Department of Microbiology and Immunology at McGill University. Dr. Camargo received her Ph.D. from the University of Missouri—Columbia. Dr. Camargo has been working in virology since her graduate studies using pseudoviruses to study different envelope glycoproteins such as those of HIV, Ebola virus, and Marburg virus. Currently, she is working on negative-strand RNA viruses, mainly focusing on respiratory syncytial virus.

Bahareh Nikpour received her B.Sc. and M.Sc. in Electrical and Computer Engineering from Shahid Bahonar University of Kerman in 2010 and 2016, respectively. She is currently a Ph.D. student at the Department of Electrical and Computer Engineering, McGill University, Montreal, Canada. Her current research interests include machine learning, reinforcement learning, and their applications.

Narges Armanfard received her Ph.D. in Electrical and Computer Engineering from McMaster University, Hamilton, ON, Canada, in 2016. She completed her postdoctoral studies at the University of Toronto in 2018. She is currently an Assistant Professor at the Department of Electrical and Computer Engineering, McGill University, and Mila-Quebec AI Institute, Montreal, QC, Canada. She performs fundamental and applied research in machine learning.

Selena M. Sagan, Ph.D., is an Associate Professor in the Department of Microbiology and Immunology and the Department of Biochemistry, McGill University. Dr. Sagan received her Ph.D. from the University of Ottawa and completed her postdoctoral fellowship training in the Department of Microbiology and Immunology at Stanford University. Dr. Sagan is a Canada Research Chair in RNA Biology and Viral Infections, and her laboratory studies positive-strand RNA viruses of the Flaviviridae family (including hepatitis C virus, dengue virus, Zika virus, and coronaviruses) as well as negative-strand RNA viruses (including respiratory syncytial virus). The main focus of her research program is RNA-RNA and protein-RNA interactions at the host-virus interface.

Sana Jahanshahi-Anbuhi, Ph.D., is currently an Assistant Professor in the Chemical and Materials Engineering Department at Concordia University, which she joined in summer 2018. She earned her Ph.D. in 2015, after which she stayed on at McMaster University and continued her research as a postdoctoral research fellow until joining Concordia University. Her primary field of research focuses on developing diagnostic tools, bioactive analytical devices, microfluidic chips, and paper-based sensors for the detection of biomarkers, point-of-care testing, and environmental monitoring. She has been the principal investigator of many research projects on thermal stabilization of viral vaccines as well as the development of ultrastable and low-cost biosensors and monitoring tools.
REFERENCES
- 1.LeBlanc JJ, Li Y, Bastien N, Forward KR, Davidson RJ, Hatchette TF. 2009. Switching gears for an influenza pandemic: validation of a duplex reverse transcriptase PCR assay for simultaneous detection and confirmatory identification of pandemic (H1N1) 2009 influenza virus. J Clin Microbiol 47:3805–3813. 10.1128/JCM.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding H, Broom A, Broom J. 2020. Aerosol-generating procedures and infective risk to healthcare workers from SARS-CoV-2: the limits of the evidence. J Hosp Infect 105:717–725. 10.1016/j.jhin.2020.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang S, Mao Y, Jones RM, Tan Q, Ji JS, Li N, Shen J, Lv Y, Pan L, Ding P, Wang X, Wang Y, MacIntyre CR, Shi X. 2020. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ Int 144:106039. 10.1016/j.envint.2020.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvis MC. 2020. Aerosol transmission of SARS-CoV-2: physical principles and implications. Front Public Health 8:590041. 10.3389/fpubh.2020.590041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai AN, Patel P. 2020. Stopping the spread of COVID-19. JAMA 323:1516. 10.1001/jama.2020.4269. [DOI] [PubMed] [Google Scholar]
- 7.Chen J. 2020. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microbes Infect 22:69–71. 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. 2020. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1470–1477. 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temime L, Gustin M-P, Duval A, Buetti N, Crépey P, Guillemot D, Thiébaut R, Vanhems P, Zahar J-R, Smith DRM, Opatowski L. 2021. A conceptual discussion about the basic reproduction number of severe acute respiratory syndrome coronavirus 2 in healthcare settings. Clin Infect Dis 72:141–143. 10.1093/cid/ciaa682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. 2021. WHO coronavirus disease (COVID-19) dashboard. World Health Organization, Geneva, Switzerland: https://covid19.who.int/?gclid=Cj0KCQjwz4z3BRCgARIsAES_OVezBT1BH_I8YhZousdOX0PeMERwgm-YmKNco1F1bpTPcArm6HIgwM0aAigBEALw_wcB. Accessed 9 February 2021. [Google Scholar]
- 14.Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, Ma H, Chen W, Lin Y, Zheng Y, Wang J, Hu Z, Yi Y, Shen H. 2020. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 63:706–711. 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamil S, Mark N, Carlos G, Cruz CSD, Gross JE, Pasnick S. 2020. Diagnosis and management of COVID-19 disease. Am J Respir Crit Care Med 201:P19–P20. 10.1164/rccm.2020C1. [DOI] [PubMed] [Google Scholar]
- 16.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen KY, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang JL, Liang Z, Peng Y, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N, China Medical Treatment Expert Group for Covid-19 . 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baj J, Karakuła-Juchnowicz H, Teresiński G, Buszewicz G, Ciesielka M, Sitarz E, Forma A, Karakuła K, Flieger W, Portincasa P, Maciejewski R. 2020. COVID-19: specific and non-specific clinical manifestations and symptoms: the current state of knowledge. J Clin Med 9:1753. 10.3390/jcm9061753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akca UK, Kesici S, Ozsurekci Y, Aykan HH, Batu ED, Atalay E, Demir S, Sag E, Vuralli D, Bayrakci B, Bilginer Y, Ozen S. 2020. Kawasaki-like disease in children with COVID-19. Rheumatol Int 40:2105–2115. 10.1007/s00296-020-04701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abobaker A, Raba AA, Alzwi A. 2020. Extrapulmonary and atypical clinical presentations of COVID‐19. J Med Virol 92:2458–2464. 10.1002/jmv.26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohindra R, Sainath KG, Kanta P, Singh M, Goyal K, Lakshmi PM, Bhalla A, Suri V. 2020. Anosmia and ageusia as presenting complaints of coronavirus disease (COVID-19) infection. J Family Med Prim Care 9:4406–4408. 10.4103/jfmpc.jfmpc_685_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, Singh KP, Chaicumpa W, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. Coronavirus disease 2019—COVID-19. Clin Microbiol Rev 33:e00028-20. 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, Agha M, Agha R. 2020. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg 78:185–193. 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization. 2020. Timeline of WHO’s response to COVID-19. World Health Organization, Geneva, Switzerland. https://www.who.int/news-room/detail/29-06-2020-covidtimeline. Accessed 21 January 2021. [Google Scholar]
- 25.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Du R-H, Li B, Zheng X-S, Yang X-L, Hu B, Wang Y-Y, Xiao G-F, Yan B, Shi Z-L, Zhou P. 2020. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 9:386–389. 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.US Food and Drug Administration. 2020. Coronavirus (COVID-19) update: FDA issues first emergency use authorization for point of care diagnostic. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-first-emergency-use-authorization-point-care-diagnostic#:∼:text=Today%2C%20the%20U.S.%20Food%20and,SARS-CoV-2%20test. Accessed 21 July 2020. [Google Scholar]
- 28.US Food and Drug Administration. 2020. FDA takes key action in fight against COVID-19 by issuing emergency use authorization for first COVID-19 vaccine. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/news-events/press-announcements/fda-takes-key-action-fight-against-covid-19-issuing-emergency-use-authorization-first-covid-19. Accessed 20 January 2021. [Google Scholar]
- 29.US Food and Drug Administration. 2021. In vitro diagnostics EUAs. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas. Accessed 11 April 2021. [Google Scholar]
- 30.Health Canada. 2021. Authorized medical devices for uses related to COVID-19: list of authorized testing devices. Health Canada, Ottawa, ON, Canada. https://www.canada.ca/en/health-canada/services/drugs-health-products/covid19-industry/medical-devices/authorized/list.html. Accessed 21 January 2021. [Google Scholar]
- 31.Sharma O, Sultan AA, Ding H, Triggle CR. 2020. A review of the progress and challenges of developing a vaccine for COVID-19. Front Immunol 11:585354. 10.3389/fimmu.2020.585354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong W-P, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, et al. 2020. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586:567–571. 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKay PF, Hu K, Blakney AK, Samnuan K, Brown JC, Penn R, Zhou J, Bouton CR, Rogers P, Polra K, Lin PJC, Barbosa C, Tam YK, Barclay WS, Shattock RJ. 2020. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun 11:3523. 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T. 2021. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384:403–416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group . 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.US Food and Drug Administration. 2020. Moderna COVID-19 vaccine EUA letter of authorization. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/media/144636/download. Accessed 2 February 2021. [Google Scholar]
- 37.Ortiz-Prado E, Simbaña-Rivera K, Gómez-Barreno L, Rubio-Neira M, Guaman LP, Kyriakidis NC, Muslin C, Jaramillo AMG, Barba-Ostria C, Cevallos-Robalino D, Sanches-SanMiguel H, Unigarro L, Zalakeviciute R, Gadian N, López-Cortés A. 2020. Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the coronavirus disease 2019 (COVID-19), a comprehensive literature review. Diagn Microbiol Infect Dis 98:115094. 10.1016/j.diagmicrobio.2020.115094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warne DJ, Ebert A, Drovandi C, Hu W, Mira A, Mengersen K. 2020. Hindsight is 2020 vision: a characterisation of the global response to the COVID-19 pandemic. BMC Public Health 20:1868. 10.1186/s12889-020-09972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaxiras E, Neofotistos G. 2020. Multiple epidemic wave model of the COVID-19 pandemic: modeling study. J Med Internet Res 22:e20912. 10.2196/20912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeBlanc JJ, Gubbay JB, Li Y, Needle R, Arneson SR, Marcino D, Charest H, Desnoyers G, Dust K, Fattouh R, Garceau R, German G, Hatchette TF, Kozak RA, Krajden M, Kuschak T, Lang ALS, Levett P, Mazzulli T, McDonald R, Mubareka S, Prystajecky N, Rutherford C, Smieja M, Yu Y, Zahariadis G, Zelyas N, Bastien N, COVID-19 Pandemic Diagnostics Investigation Team of the Canadian Public Health Laboratory Network (CPHLN) Respiratory Virus Working Group . 2020. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J Clin Virol 128:104433. 10.1016/j.jcv.2020.104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso JM, Gregg AC, Soares DJ, Beskid TR, Jervey SR, Liu C. 2020. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci 6:591–605. 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jahanshahi-Anbuhi S, Kannan B, Pennings K, Monsur Ali M, Leung V, Giang K, Wang J, White D, Li Y, Pelton RH, Brennan JD, Filipe CDM. 2017. Automating multi-step paper-based assays using integrated layering of reagents. Lab Chip 17:943–950. 10.1039/c6lc01485b. [DOI] [PubMed] [Google Scholar]
- 43.Ali MM, Brown CL, Jahanshahi-Anbuhi S, Kannan B, Li Y, Filipe CDM, Brennan JD. 2017. A printed multicomponent paper sensor for bacterial detection. Sci Rep 7:12335. 10.1038/s41598-017-12549-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahanshahi-Anbuhi S, Kannan B, Leung V, Pennings K, Liu M, Carrasquilla C, White D, Li Y, Pelton RH, Brennan JD, Filipe CDM. 2016. Simple and ultrastable all-inclusive pullulan tablets for challenging bioassays. Chem Sci 7:2342–2346. 10.1039/c5sc04184h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M, Hui CY, Zhang Q, Gu J, Kannan B, Jahanshahi-Anbuhi S, Filipe CDM, Brennan JD, Li Y. 2016. Target-induced and equipment-free DNA amplification with a simple paper device. Angew Chem Int Ed Engl 55:2709–2713. 10.1002/ange.201509389. [DOI] [PubMed] [Google Scholar]
- 46.Jahanshahi-Anbuhi S, Pennings K, Leung V, Liu M, Carrasquilla C, Kannan B, Li Y, Pelton R, Brennan JD, Filipe CDM. 2014. Pullulan encapsulation of labile biomolecules to give stable bioassay tablets. Angew Chem Int Ed Engl 53:6155–6158. 10.1002/anie.201403222. [DOI] [PubMed] [Google Scholar]
- 47.Morales-Narváez E, Dincer C. 2020. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens Bioelectron 163:112274. 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu CY, Chan KG, Yean CY, Ang GY. 2021. Nucleic acid-based diagnostic tests for the detection SARS-CoV-2: an update. Diagnostics 11:53. 10.3390/diagnostics11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui L-P, Johnston JC, Lan Z, Law S, MacLean E, Trajman A, Menzies D, Benedetti A, Ahmad Khan F. 2020. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ 370:m2516. 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D. 2021. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev Biomed Eng 14:4–15. 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 51.Zhu H, Zhang H, Ni S, Korabečná M, Yobas L, Neuzil P. 2020. The vision of point-of-care PCR tests for the COVID-19 pandemic and beyond. Trends Analyt Chem 130:115984. 10.1016/j.trac.2020.115984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pokhrel P, Hu C, Mao H. 2020. Detecting the coronavirus (COVID-19). ACS Sens 5:2283–2296. 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. 2020. Diagnosing COVID-19: the disease and tools for detection. ACS Nano 14:3822–3835. 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 54.Qin Z, Peng R, Baravik IK, Liu X. 2020. Fighting COVID-19: integrated micro- and nanosystems for viral infection diagnostics. Matter 3:628–651. 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kowalik MM, Trzonkowski P, Łasińska-Kowara M, Mital A, Smiatacz T, Jaguszewski M. 2020. COVID-19—toward a comprehensive understanding of the disease. Cardiol J 27:99–114. 10.5603/CJ.a2020.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. 2020. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 7:1012–1023. 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Lee J-Y, Yang J-S, Kim JW, Kim VN, Chang H. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181:914–921.e10. 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kandimalla R, John A, Abburi C, Vallamkondu J, Reddy PH. 2020. Current status of multiple drug molecules, and vaccines: an update in SARS-CoV-2 therapeutics. Mol Neurobiol 57:4106–4116. 10.1007/s12035-020-02022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ. 2011. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol 174:11–22. 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoeman D, Fielding BC. 2019. Coronavirus envelope protein: current knowledge. Virol J 16:69. 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saxena SK (ed). 2020. Coronavirus disease 2019 (COVID-19): epidemiology, pathogenesis, diagnosis, and therapeutics. Springer Singapore, Singapore. [Google Scholar]
- 63.Zeng W, Liu G, Ma H, Zhao D, Yang Y, Liu M, Mohammed A, Zhao C, Yang Y, Xie J, Ding C, Ma X, Weng J, Gao Y, He H, Jin T. 2020. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun 527:618–623. 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang Y, Yang C, Xu X, Xu W, Liu S. 2020. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 41:1141–1149. 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turoňová B, Sikora M, Schürmann C, Hagen WJH, Welsch S, Blanc FEC, von Bülow S, Gecht M, Bagola K, Hörner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Mühlebach MD, Hummer G, Krijnse Locker J, Beck M. 2020. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370:203–208. 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R. 2020. SARS‐CoV‐2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J 39:e105114. 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansson MA, Quandelacy TM, Kada S, Prasad PV, Steele M, Brooks JT, Slayton RB, Biggerstaff M, Butler JC. 2021. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open 4:e2035057. 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Struyf T, Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R, Hooft L, Emperador D, Dittrich S, Domen J, Horn SRA, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group . 2020. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19 disease. Cochrane Database Syst Rev 7:CD013665. 10.1002/14651858.CD013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spinato G, Fabbris C, Polesel J, Cazzador D, Borsetto D, Hopkins C, Boscolo-Rizzo P. 2020. Alterations in smell or taste in mildly symptomatic outpatients with SARS-CoV-2 infection. JAMA 323:2089–2090. 10.1001/jama.2020.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moein ST, Hashemian SM, Mansourafshar B, Khorram‐Tousi A, Tabarsi P, Doty RL. 2020. Smell dysfunction: a biomarker for COVID‐19. Int Forum Allergy Rhinol 10:944–950. 10.1002/alr.22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seirafianpour F, Sodagar S, Pour Mohammad A, Panahi P, Mozafarpoor S, Almasi S, Goodarzi A. 2020. Cutaneous manifestations and considerations in COVID-19 pandemic: a systematic review. Dermatol Ther 33:e13986. 10.1111/dth.13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rose-Sauld S, Dua A. 2020. COVID toes and other cutaneous manifestations of COVID-19. J Wound Care 29:486–487. 10.12968/jowc.2020.29.9.486. [DOI] [PubMed] [Google Scholar]
- 76.Almqvist J, Granberg T, Tzortzakakis A, Klironomos S, Kollia E, Öhberg C, Martin R, Piehl F, Ouellette R, Ineichen BV. 2020. Neurological manifestations of coronavirus infections—a systematic review. Ann Clin Transl Neurol 7:2057–2071. 10.1002/acn3.51166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang G, Hu C, Luo L, Fang F, Chen Y, Li J, Peng Z, Pan H. 2020. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J Clin Virol 127:104364. 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. 2020. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience 42:505–514. 10.1007/s11357-020-00186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu X, Zhang L, Du H, Zhang J, Li YY, Qu J, Zhang W, Wang Y, Bao S, Li Y, Wu C, Liu H, Liu D, Shao J, Peng X, Yang Y, Liu Z, Xiang Y, Zhang F, Silva RM, Pinkerton KE, Shen K, Xiao H, Xu S, Wong GWK, Chinese Pediatric Novel Coronavirus Study Team . 2020. SARS-CoV-2 infection in children. N Engl J Med 382:1663–1665. 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, Ji R, Wang H, Wang Y, Zhou Y. 2020. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis 94:91–95. 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim G-U, Kim M-J, Ra SH, Lee J, Bae S, Jung J, Kim S-H. 2020. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin Microbiol Infect 26:948.e1–948.e3. 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L-Q, Huang T, Wang Y-Q, Wang Z-P, Liang Y, Huang T-B, Zhang H-Y, Sun W, Wang Y. 2020. COVID‐19 patients’ clinical characteristics, discharge rate, and fatality rate of meta‐analysis. J Med Virol 92:577–583. 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li R, Tian J, Yang F, Lv L, Yu J, Sun G, Ma Y, Yang X, Ding J. 2020. Clinical characteristics of 225 patients with COVID-19 in a tertiary hospital near Wuhan, China. J Clin Virol 127:104363. 10.1016/j.jcv.2020.104363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lombardi A, Consonni D, Carugno M, Bozzi G, Mangioni D, Muscatello A, Castelli V, Palomba E, Cantù AP, Ceriotti F, Tiso B, Pesatori AC, Riboldi L, Bandera A, Lunghi G, Gori A. 2020. Characteristics of 1573 healthcare workers who underwent nasopharyngeal swab testing for SARS-CoV-2 in Milan, Lombardy, Italy. Clin Microbiol Infect 26:1413.e9–1413.e13. 10.1016/j.cmi.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singhal T. 2020. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr 87:281–286. 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carfì A, Bernabei R, Landi F, Gemelli Against COVID-19 Post-Acute Care Study Group . 2020. Persistent symptoms in patients after acute COVID-19. JAMA 324:603–605. 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.US Centers for Disease Control and Prevention. 2020. Interim clinical guidance for management of patients with confirmed coronavirus disease (COVID-19), updated December 8, 2020. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html. Accessed 15 January 2021. [Google Scholar]
- 88.Bhimraj A, Morgan RL, Shumaker AH, Lavergne V, Baden L, Cheng VC-C, Edwards KM, Gandhi R, Muller WJ, O’Horo JC, Shoham S, Murad MH, Mustafa RA, Sultan S, Falck-Ytter Y. 27 April 2020. Infectious Diseases Society of America guidelines on the treatment and management of patients with COVID-19. Clin Infect Dis 10.1093/cid/ciaa478. [DOI] [PMC free article] [PubMed]
- 89.O’Shea PM, Lee GR, Griffin TP, Tormey V, Hayat A, Costelloe SJ, Griffin DG, Srinivasan S, O’Kane M, Burke CM, Faul J, Thompson CJ, Curley G, Tormey WP. 2020. COVID-19 in adults: test menu for hospital blood science laboratories. Ir J Med Sci 189:1147–1152. 10.1007/s11845-020-02252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stegeman I, Ochodo EA, Guleid F, Holtman GA, Yang B, Davenport C, Deeks JJ, Dinnes J, Dittrich S, Emperador D, Hooft L, Spijker R, Takwoingi Y, Van den Bruel A, Wang J, Langendam M, Verbakel JY, Leeflang MM, Cochrane COVID-19 Diagnostic Test Accuracy Group . 2020. Routine laboratory testing to determine if a patient has COVID-19. Cochrane Database Syst Rev 11:CD013787. 10.1002/14651858.CD013787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng Y, Zhang Y, Chi H, Chen S, Peng M, Luo L, Chen L, Li J, Shen B, Wang D. 2020. The hemocyte counts as a potential biomarker for predicting disease progression in COVID-19: a retrospective study. Clin Chem Lab Med 58:1106–1115. 10.1515/cclm-2020-0377. [DOI] [PubMed] [Google Scholar]
- 92.Zeng F, Huang Y, Guo Y, Yin M, Chen X, Xiao L, Deng G. 2020. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis 96:467–474. 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fajnzylber J, Regan J, Coxen K, Corry H, Wong C, Rosenthal A, Worrall D, Giguel F, Piechocka-Trocha A, Atyeo C, Fischinger S, Chan A, Flaherty KT, Hall K, Dougan M, Ryan ET, Gillespie E, Chishti R, Li Y, Jilg N, Hanidziar D, Baron RM, Baden L, Tsibris AM, Armstrong KA, Kuritzkes DR, Alter G, Walker BD, Yu X, Li JZ, Massachusetts Consortium for Pathogen Readiness . 2020. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 11:5493. 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Guo H. 2020. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv Biomark Sci Technol 2:1–23. 10.1016/j.abst.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joob B, Wiwanitkit V. 2020. Bronchoalveolar specimen can help detect COVID-19 in suspicious case with negative PCR for nasopharyngeal specimen test. Lung India 37:286–287. 10.4103/lungindia.lungindia_126_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. 2020. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323:1843–1844. 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang Y, Yang M, Shen C, Wang F, Yuan J, Li J, Zhang M, Wang Z, Xing L, Wei J, Peng L, Wong G, Zheng H, Liao M, Feng K, Li J, Yang Q, Zhao J, Zhang Z, Liu L, Liu Y. 2020. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 10.1101/2020.02.11.20021493. [DOI]
- 98.COVID-19 Investigation Team. 2020. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 26:861–868. 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- 99.Wong RC-W, Wong AH, Ho YI-I, Leung EC-M, Lai RW-M. 2020. Evaluation on testing of deep throat saliva and lower respiratory tract specimens with Xpert Xpress SARS-CoV-2 assay. J Clin Virol 131:104593. 10.1016/j.jcv.2020.104593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bwire GM, Majigo MV, Njiro BJ, Mawazo A. 2021. Detection profile of SARS‐CoV‐2 using RT‐PCR in different types of clinical specimens: a systematic review and meta‐analysis. J Med Virol 93:719–725. 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mohammadi A, Esmaeilzadeh E, Li Y, Bosch RJ, Li JZ. 2020. SARS-CoV-2 detection in different respiratory sites: a systematic review and meta-analysis. EBioMedicine 59:102903. 10.1016/j.ebiom.2020.102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanson KE, Caliendo AM, Arias CA, Englund JA, Hayden MK, Lee MJ, Loeb M, Patel R, Altayar O, El Alayli A, Sultan S, Falck-Ytter Y, Lavergne V, Morgan RL, Murad MH, Bhimraj A, Mustafa RA. 12 September 2020. Infectious Diseases Society of America guidelines on the diagnosis of COVID-19: serologic testing. Clin Infect Dis 10.1093/cid/ciaa1343. [DOI] [PMC free article] [PubMed]
- 103.Qanadli SD, Beigelman-Aubry C, Rotzinger DC. 2020. Vascular changes detected with thoracic CT in coronavirus disease (COVID-19) might be significant determinants for accurate diagnosis and optimal patient management. AJR Am J Roentgenol 215:W15. 10.2214/AJR.20.23185. [DOI] [PubMed] [Google Scholar]
- 104.Murphy K. 2020. SARS CoV-2 detection from upper and lower respiratory tract specimens. Chest 158:1804–1805. 10.1016/j.chest.2020.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang K, Zhang X, Sun J, Ye J, Wang F, Hua J, Zhang H, Shi T, Li Q, Wu X. 2020. Differences of severe acute respiratory syndrome coronavirus 2 shedding duration in sputum and nasopharyngeal swab specimens among adult inpatients with coronavirus disease 2019. Chest 158:1876–1884. 10.1016/j.chest.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mustafa Hellou M, Górska A, Mazzaferri F, Cremonini E, Gentilotti E, De Nardo P, Poran I, Leeflang MM, Tacconelli E, Paul M. 2021. Nucleic acid amplification tests on respiratory samples for the diagnosis of coronavirus infections: a systematic review and meta-analysis. Clin Microbiol Infect 27:341–351. 10.1016/j.cmi.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mallett S, Allen AJ, Graziadio S, Taylor SA, Sakai NS, Green K, Suklan J, Hyde C, Shinkins B, Zhelev Z, Peters J, Turner PJ, Roberts NW, di Ruffano LF, Wolff R, Whiting P, Winter A, Bhatnagar G, Nicholson BD, Halligan S. 2020. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med 18:346. 10.1186/s12916-020-01810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schiaffino S, Tritella S, Cozzi A, Carriero S, Blandi L, Ferraris L, Sardanelli F. 2020. Diagnostic performance of chest X-ray for COVID-19 pneumonia during the SARS-CoV-2 pandemic in Lombardy, Italy. J Thorac Imaging 35:W105–W106. 10.1097/RTI.0000000000000533. [DOI] [PubMed] [Google Scholar]
- 109.Dong D, Tang Z, Wang S, Hui H, Gong L, Lu Y, Xue Z, Liao H, Chen F, Yang F, Jin R, Wang K, Liu Z, Wei J, Mu W, Zhang H, Jiang J, Tian J, Li H. 2021. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng 14:16–29. 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 110.Wang K, Kang S, Tian R, Zhang X, Zhang X, Wang Y. 2020. Imaging manifestations and diagnostic value of chest CT of coronavirus disease 2019 (COVID-19) in the Xiaogan area. Clin Radiol 75:341–347. 10.1016/j.crad.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hope MD, Raptis CA, Henry TS. 2020. Chest computed tomography for detection of coronavirus disease 2019 (COVID-19): don’t rush the science. Ann Intern Med 173:147–148. 10.7326/M20-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Esposito A, Palmisano A, Scotti GM, Morelli MJ, Vignale D, De Cobelli F, Tonon G, Tacchetti C. 2020. Why is chest CT important for early diagnosis of COVID-19? Prevalence matters. medRxiv 10.1101/2020.03.30.20047985. [DOI]
- 113.Fields BKK, Demirjian NL, Dadgar H, Gholamrezanezhad A. 30 November 2020. Imaging of COVID-19: CT, MRI, and PET. Semin Nucl Med 10.1053/j.semnuclmed.2020.11.003. [DOI] [PMC free article] [PubMed]
- 114.Fields BKK, Demirjian NL, Gholamrezanezhad A. 2020. Coronavirus disease 2019 (COVID-19) diagnostic technologies: a country-based retrospective analysis of screening and containment procedures during the first wave of the pandemic. Clin Imaging 67:219–225. 10.1016/j.clinimag.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Larici AR, Cicchetti G, Marano R, Merlino B, Elia L, Calandriello L, del Ciello A, Farchione A, Savino G, Infante A, Larosa L, Colosimo C, Manfredi R, Natale L. 2020. Multimodality imaging of COVID-19 pneumonia: from diagnosis to follow-up. A comprehensive review. Eur J Radiol 131:109217. 10.1016/j.ejrad.2020.109217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao Y, Liu X, Xiong L, Cai K. 2020. Imaging and clinical features of patients with 2019 novel coronavirus SARS‐CoV‐2: a systematic review and meta‐analysis. J Med Virol 92:1449–1459. 10.1002/jmv.25822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Whiting P, Singatullina N, Rosser J. 2015. Computed tomography of the chest: I. Basic principles. BJA Educ 15:299–304. 10.1093/bjaceaccp/mku063. [DOI] [Google Scholar]
- 118.Litmanovich DE, Chung M, Kirkbride RR, Kicska G, Kanne JP. 9 June 2020. Review of chest radiograph findings of COVID-19 pneumonia and suggested reporting language. J Thorac Imaging 10.1097/RTI.0000000000000541. [DOI] [PubMed]
- 119.Simpson S, Kay FU, Abbara S, Bhalla S, Chung JH, Chung M, Henry TS, Kanne JP, Kligerman S, Ko JP, Litt H. 2020. Radiological Society of North America expert consensus statement on reporting chest CT findings related to COVID-19. endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA—secondary publication. J Thorac Imaging 35:219–227. 10.1097/RTI.0000000000000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Akl EA, Blazic I, Yaacoub S, Frija G, Chou R, Appiah JA, Fatehi M, Flor N, Hitti E, Jafri H, Jin Z-Y, Kauczor HU, Kawooya M, Kazerooni EA, Ko JP, Mahfouz R, Muglia V, Nyabanda R, Sanchez M, Shete PB, Ulla M, Zheng C, van Deventer E, Perez MDR. 2021. Use of chest imaging in the diagnosis and management of COVID-19: a WHO rapid advice guide. Radiology 298:E63–E69. 10.1148/radiol.2020203173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salameh J-P, Leeflang MM, Hooft L, Islam N, McGrath TA, van der Pol CB, Frank RA, Prager R, Hare SS, Dennie C, Spijker R, Deeks JJ, Dinnes J, Jenniskens K, Korevaar DA, Cohen JF, Van den Bruel A, Takwoingi Y, van de Wijgert J, Damen JA, Wang J, Cochrane COVID-19 Diagnostic Test Accuracy Group, McInnes MDF. 2020. Thoracic imaging tests for the diagnosis of COVID-19. Cochrane Database Syst Rev 9:CD013639. 10.1002/14651858.CD013639.pub2. [DOI] [PubMed] [Google Scholar]
- 122.Yang W, Sirajuddin A, Zhang X, Liu G, Teng Z, Zhao S, Lu M. 2020. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19). Eur Radiol 30:4874–4882. 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu B, Xing Y, Peng J, Zheng Z, Tang W, Sun Y, Xu C, Peng F. 2020. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol 30:5720–5727. 10.1007/s00330-020-06934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Waller JV, Kaur P, Tucker A, Lin KK, Diaz MJ, Henry TS, Hope M. 2020. Diagnostic tools for coronavirus disease (COVID-19): comparing CT and RT-PCR viral nucleic acid testing. AJR Am J Roentgenol 215:834–838. 10.2214/AJR.20.23418. [DOI] [PubMed] [Google Scholar]
- 125.Zhang J-J, Dong X, Cao Y-Y, Yuan Y-D, Yang Y-B, Yan Y-Q, Akdis CA, Gao Y-D. 2020. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy 75:1730–1741. 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 126.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. 2020. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 296:E32–E40. 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y, Xia L. 2020. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol 214:1280–1286. 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 128.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. 2020. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology 296:E15–E25. 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. 2020. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 296:E115–E117. 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C, Zeng B, Li Z, Li X, Li H. 2020. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol 126:108961. 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang P, Liu T, Huang L, Liu H, Lei M, Xu W, Hu X, Chen J, Liu B. 2020. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology 295:22–23. 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He J-L, Luo L, Luo Z-D, Lyu J-X, Ng M-Y, Shen X-P, Wen Z. 2020. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir Med 168:105980. 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.World Health Organization. 2020. Diagnostic testing for SARS-CoV-2: interim guidance, 11 September 2020. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2. Accessed 15 October 2020. [Google Scholar]
- 134.Hanson KE, Caliendo AM, Arias CA, Englund JA, Lee MJ, Loeb M, Patel R, El Alayli A, Kalot MA, Falck-Ytter Y, Lavergne V, Morgan RL, Murad MH, Sultan S, Bhimraj A, Mustafa RA. 16 June 2020. Infectious Diseases Society of America guidelines on the diagnosis of coronavirus disease 2019. Clin Infect Dis 10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed]
- 135.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, Pan I, Shi L-B, Wang D-C, Mei J, Jiang X-L, Zeng Q-H, Egglin TK, Hu P-F, Agarwal S, Xie F, Li S, Healey T, Atalay MK, Liao W-H. 2020. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology 296:E46–E54. 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. 2020. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol 215:87–93. 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 137.Jin Y-H, Cai L, Cheng Z-S, Cheng H, Deng T, Fan Y-P, Fang C, Huang D, Huang L-Q, Huang Q, Han Y, Hu B, Hu F, Li B-H, Li Y-R, Liang K, Lin L-K, Luo L-S, Ma J, Ma L-L, Peng Z-Y, Pan Y-B, Pan Z-Y, Ren X-Q, Sun H-M, Wang Y, Wang Y-Y, Weng H, Wei C-J, Wu D-F, Xia J, Xiong Y, Xu H-B, Yao X-M, Yuan Y-F, Ye T-S, Zhang X-C, Zhang Y-W, Zhang Y-G, Zhang H-M, Zhao Y, Zhao M-J, Zi H, Zeng X-T, Wang Y-Y, Wang X-H, for the Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team, Evidence-Based Medicine Chapter of China International Exchange and Promotive Association for Medical and Health Care (CPAM) . 2020. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res 7:4. 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. 2020. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology 295:715–721. 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pan Y, Guan H, Zhou S, Wang Y, Li Q, Zhu T, Hu Q, Xia L. 2020. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol 30:3306–3309. 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li K, Fang Y, Li W, Pan C, Qin P, Zhong Y, Liu X, Huang M, Liao Y, Li S. 2020. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur Radiol 30:4407–4416. 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu K-C, Xu P, Lv W-F, Qiu X-H, Yao J-L, Gu J-F, Wei W. 2020. CT manifestations of coronavirus disease-2019: a retrospective analysis of 73 cases by disease severity. Eur J Radiol 126:108941. 10.1016/j.ejrad.2020.108941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, Geis JR, Pandharipande PV, Brink JA, Dreyer KJ. 2018. Current applications and future impact of machine learning in radiology. Radiology 288:318–328. 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. 2018. Artificial intelligence in radiology. Nat Rev Cancer 18:500–510. 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Alom MZ, Rahman MMS, Nasrin MS, Taha TM, Asari VK. 2020. COVID_MTNet: COVID-19 detection with multi-task deep learning approaches. arXiv http://arxiv.org/abs/2004.03747.
- 145.Burlacu A, Crisan-Dabija R, Popa IV, Artene B, Birzu V, Pricop M, Plesoianu C, Generali D. 2020. Curbing the AI-induced enthusiasm in diagnosing COVID-19 on chest X-rays: the present and the near-future. medRxiv 10.1101/2020.04.28.20082776. [DOI]
- 146.Shoeibi A, Khodatars M, Alizadehsani R, Ghassemi N, Jafari M, Moridian P, Khadem A, Sadeghi D, Hussain S, Zare A, Sani ZA, Bazeli J, Khozeimeh F, Khosravi A, Nahavandi S, Acharya UR, Shi P. 2020. Automated detection and forecasting of COVID-19 using deep learning techniques: a review. arXiv http://arxiv.org/abs/2007.10785.
- 147.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J. 2020. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 296:E65–E71. 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kang H, Xia L, Yan F, Wan Z, Shi F, Yuan H, Jiang H, Wu D, Sui H, Zhang C, Shen D. 2020. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans Med Imaging 39:2606–2614. 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 149.He X, Yang X, Zhang S, Zhao J, Zhang Y, Xing E, Xie P. 2020. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. medRxiv 10.1101/2020.04.13.20063941. [DOI]
- 150.Wang L, Wong A. 2020. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv http://arxiv.org/abs/2003.09871. [DOI] [PMC free article] [PubMed]
- 151.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK. 2020. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv http://arxiv.org/abs/2004.00038.
- 152.Lippi G, Simundic A-M, Plebani M. 2020. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med 58:1070–1076. 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 153.De Virgilio A, Costantino A, Mercante G, Spriano G. 2020. How to increase the SARS-CoV-2 detection rate through the nasopharyngeal swab? Oral Oncol 106:104802. 10.1016/j.oraloncology.2020.104802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.US Centers for Disease Control and Prevention. 2021. Interim guidelines for collecting, handling, and testing clinical specimens for COVID-19, updated January 6, 2021. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed 15 January 2021. [Google Scholar]
- 155.Wu J, Liu J, Li S, Peng Z, Xiao Z, Wang X, Yan R, Luo J. 2020. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis 37:101673. 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tang A, Tong Z, Wang H, Dai Y, Li K, Liu J, Wu W, Yuan C, Yu M, Li P, Yan J. 2020. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg Infect Dis 26:1337–1339. 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu J, Xiao Y, Shen Y, Shi C, Chen Y, Shi P, Gao Y, Wang Y, Lu B. 2020. Detection of SARS‐CoV‐2 by RT‐PCR in anal from patients who have recovered from coronavirus disease 2019. J Med Virol 92:1769–1771. 10.1002/jmv.25875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kipkorir V, Cheruiyot I, Ngure B, Misiani M, Munguti J. 2020. Prolonged SARS‐CoV‐2 RNA detection in anal/rectal swabs and stool specimens in COVID‐19 patients after negative conversion in nasopharyngeal RT‐PCR test. J Med Virol 92:2328–2331. 10.1002/jmv.26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. 2020. Asymptomatic SARS‐CoV‐2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol 92:1807–1809. 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang J, Wang S, Xue Y. 2020. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol 92:680–682. 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Trypsteen W, Van Cleemput J, van Snippenberg W, Gerlo S, Vandekerckhove L. 2020. On the whereabouts of SARS-CoV-2 in the human body: a systematic review. PLoS Pathog 16:e1009037. 10.1371/journal.ppat.1009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Charlton CL, Babady E, Ginocchio CC, Hatchette TF, Jerris RC, Li Y, Loeffelholz M, McCarter YS, Miller MB, Novak-Weekley S, Schuetz AN, Tang Y-W, Widen R, Drews SJ. 2019. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin Microbiol Rev 32:e00042-18. 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schleihauf E, Fathima S, Pettipas J, LeBlanc JJ, Hatchette TF, Drews SJ, Haldane D. 2015. Respiratory outbreak investigations: how many specimens should be tested? Infect Control Hosp Epidemiol 36:1344–1347. 10.1017/ice.2015.171. [DOI] [PubMed] [Google Scholar]
- 164.LeBlanc JJ, ElSherif M, Mulpuru S, Warhuus M, Ambrose A, Andrew M, Boivin G, Bowie W, Chit A, Dos Santos G, Green K, Halperin SA, Hatchette TF, Ibarguchi B, Johnstone J, Katz K, Langley JM, Lagacé-Wiens P, Loeb M, Lund A, MacKinnon-Cameron D, McCarthy A, McElhaney JE, McGeer A, Poirier A, Powis J, Richardson D, Semret M, Shinde V, Smyth D, Trottier S, Valiquette L, Webster D, Ye L, McNeil SA. 2020. Validation of the Seegene RV15 multiplex PCR for the detection of influenza A subtypes and influenza B lineages during national influenza surveillance in hospitalized adults. J Med Microbiol 69:256–264. 10.1099/jmm.0.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.To KK-W, Tsang OT-Y, Yip CC-Y, Chan K-H, Wu T-C, Chan JM-C, Leung W-S, Chik TS-H, Choi CY-C, Kandamby DH, Lung DC, Tam AR, Poon RW-S, Fung AY-F, Hung IF-N, Cheng VC-C, Chan JF-W, Yuen K-Y. 2020. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 71:841–843. 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, Klausner JD. 2020. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. medRxiv 10.1101/2020.04.11.20062372. [DOI] [PMC free article] [PubMed]
- 167.LeBlanc JJ, Heinstein C, MacDonald J, Pettipas J, Hatchette TF, Patriquin G. 2020. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J Clin Virol 128:104442. 10.1016/j.jcv.2020.104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Patriquin G, Davis I, Heinstein C, MacDonald J, Hatchette TF, LeBlanc JJ. 2020. Exploring alternative swabs for use in SARS-CoV-2 detection from the oropharynx and anterior nares. J Virol Methods 285:113948. 10.1016/j.jviromet.2020.113948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Garnett L, Bello A, Tran KN, Audet J, Leung A, Schiffman Z, Griffin BD, Tailor N, Kobasa D, Strong JE. 2020. Comparison analysis of different swabs and transport mediums suitable for SARS-CoV-2 testing following shortages. J Virol Methods 285:113947. 10.1016/j.jviromet.2020.113947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. 2020. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol 58:e00512-20. 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guo W-L, Jiang Q, Ye F, Li S-Q, Hong C, Chen L-Y, Li S-Y. 2020. Effect of throat washings on detection of 2019 novel coronavirus. Clin Infect Dis 71:1980–1981. 10.1093/cid/ciaa416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vlek ALM, Wesselius TS, Achterberg R, Thijsen SFT. 2021. Combined throat/nasal swab sampling for SARS-CoV-2 is equivalent to nasopharyngeal sampling. Eur J Clin Microbiol Infect Dis 40:193–195. 10.1007/s10096-020-03972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.US Centers for Disease Control and Prevention. 2021. Interim laboratory biosafety guidelines for handling and processing specimens associated with coronavirus disease 2019 (COVID-19), updated January 6, 2021. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html. Accessed 11 January 2021. [Google Scholar]
- 174.Alghounaim M, Almazeedi S, Al Youha S, Papenburg J, Alowaish O, AbdulHussain G, Al-Shemali R, Albuloushi A, Alzabin S, Al-Wogayan K, Al-Mutawa Y, Al-Sabah S. 2020. Low-cost polyester-tipped three-dimensionally printed nasopharyngeal swab for the detection of severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2). J Clin Microbiol 58:e01668-20. 10.1128/JCM.01668-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Callahan CJ, Lee R, Zulauf KE, Tamburello L, Smith KP, Previtera J, Cheng A, Green A, Abdul Azim A, Yano A, Doraiswami N, Kirby JE, Arnaout RA. 2020. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. J Clin Microbiol 58:e00876-20. 10.1128/JCM.00876-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Oland G, Garner O, de St Maurice A. 2021. Prospective clinical validation of 3D printed nasopharyngeal swabs for diagnosis of COVID-19. Diagn Microbiol Infect Dis 99:115257. 10.1016/j.diagmicrobio.2020.115257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Decker SJ, Goldstein TA, Ford JM, Teng MN, Pugliese RS, Berry GJ, Pettengill M, Silbert S, Hazelton TR, Wilson JW, Shine K, Wang Z-X, Hutchinson M, Castagnaro J, Bloom OE, Breining DA, Goldsmith BM, Sinnott JT, O’Donnell DG, Crawford JM, Lockwood CJ, Kim K. 10 September 2020. 3D printed alternative to the standard synthetic flocked nasopharyngeal swabs used for coronavirus disease 2019 testing. Clin Infect Dis 10.1093/cid/ciaa1366. [DOI] [PMC free article] [PubMed]
- 178.Williams E, Bond K, Isles N, Chong B, Johnson D, Druce J, Hoang T, Ballard SA, Hall V, Muhi S, Buising KL, Lim S, Strugnell D, Catton M, Irving LB, Howden BP, Bert E, Williamson DA. 2020. Pandemic printing: a novel 3D‐printed swab for detecting SARS‐CoV‐2. Med J Aust 213:276–279. 10.5694/mja2.50726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rogers AA, Baumann RE, Borillo GA, Kagan RM, Batterman HJ, Galdzicka MM, Marlowe EM. 2020. Evaluation of transport media and specimen transport conditions for the detection of SARS-CoV-2 by use of real-time reverse transcription-PCR. J Clin Microbiol 58:e00708-20. 10.1128/JCM.00708-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nörz D, Frontzek A, Eigner U, Oestereich L, Wichmann D, Kluge S, Fischer N, Aepfelbacher M, Pfefferle S, Lütgehetmann M. 2020. Pushing beyond specifications: evaluation of linearity and clinical performance of the cobas 6800/8800 SARS-CoV-2 RT-PCR assay for reliable quantification in blood and other materials outside recommendations. J Clin Virol 132:104650. 10.1016/j.jcv.2020.104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhu J, Guo J, Xu Y, Chen X. 2020. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect 81:e48–e50. 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guest JL, Sullivan PS, Valentine-Graves M, Valencia R, Adam E, Luisi N, Nakano M, Guarner J, Del Rio C, Sailey C, Goedecke Z, Siegler AJ, Sanchez TH. 2020. Suitability and sufficiency of telehealth clinician-observed, participant-collected samples for SARS-CoV2 testing: the iCollect cohort pilot study. JMIR Public Health Surveill 6:e19731. 10.2196/19731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Liu AX, Paterson A, Anceva-Sami S, Barati S, Crowl G, Faheem A, Farooqi L, Khan S, Prost K, Poutanen S, Yip L, Zhong Z, McGeer AJ, Mubareka S, for the Toronto Invasive Bacterial Diseases Network COVID-19 Investigators . 2020. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). medRxiv 10.1101/2020.05.01.20081026. [DOI]
- 184.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Warren JL, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Venkataraman A, Lu-Culligan A, Klein J, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman O-E, Wong P, Yang Y, Bermejo S, Odio CD, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. 2020. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 383:1283–1286. 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bennett S, Davidson RS, Gunson RN. 2017. Comparison of gargle samples and throat swab samples for the detection of respiratory pathogens. J Virol Methods 248:83–86. 10.1016/j.jviromet.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 186.Malecki M, Lüsebrink J, Teves S, Wendel AF. 2021. Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect Control Hosp Epidemiol 42:248–279. 10.1017/ice.2020.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Khurshid Z, Zohaib S, Joshi C, Moin SF, Zafar MS, Speicher DJ. 2020. Saliva as a non-invasive sample for the detection of SARS-CoV-2: a systematic review. medRxiv https://www.medrxiv.org/content/10.1101/2020.05.09.20096354v1.
- 188.Iwasaki S, Fujisawa S, Nakakubo S, Kamada K, Yamashita Y, Fukumoto T, Sato K, Oguri S, Taki K, Senjo H, Sugita J, Hayasaka K, Konno S, Nishida M, Teshima T. 2020. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect 81:e145–e147. 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zheng S, Yu F, Fan J, Zou Q, Xie G, Yang X, Chen W, Wang Q, Zhang D, Wang R, Feng B, Lin S, Gong R, Ma Z, Lu S, Wang X, Yu L, Xu K, Sheng J, Chen Y. 2020. Saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. SSRN 10.2139/ssrn.3543605. [DOI] [PMC free article] [PubMed]
- 190.Rutgers Clinical Genomics Laboratory. 2020. Rutgers Clinical Genomics Laboratory TaqPath SARS-CoV-2 assay: instructions for use. Rutgers Clinical Genomics Laboratory, Newark, NJ. https://www.fda.gov/media/136875/download. Accessed 15 October 2020. [Google Scholar]
- 191.Becker D, Sandoval E, Amin A, De Hoff P, Diets A, Leonetti N, Lim YW, Elliott C, Laurent L, Grzymski J, Lu JT. 2020. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv 10.1101/2020.05.11.20092338. [DOI]
- 192.Xu R, Cui B, Duan X, Zhang P, Zhou X, Yuan Q. 2020. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci 12:11. 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.van Doorn AS, Meijer B, Frampton CMA, Barclay ML, de Boer NKH. 2020. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment Pharmacol Ther 52:1276–1288. 10.1111/apt.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Foladori P, Cutrupi F, Segata N, Manara S, Pinto F, Malpei F, Bruni L, La Rosa G. 2020. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci Total Environ 743:140444. 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kitajima M, Ahmed W, Bibby K, Carducci A, Gerba CP, Hamilton KA, Haramoto E, Rose JB. 2020. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci Total Environ 739:139076. 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mathavarajah S, Stoddart AK, Gagnon GA, Dellaire G. 2021. Pandemic danger to the deep: the risk of marine mammals contracting SARS-CoV-2 from wastewater. Sci Total Environ 760:143346. 10.1016/j.scitotenv.2020.143346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.US Centers for Disease Control and Prevention. 2020. Collection and submission of postmortem specimens from deceased persons with confirmed or suspected COVID-19. Postmortem guidance. Updated December 2, 2020. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/hcp/guidance-postmortem-specimens.html#:∼:text=Medical%20examiners%2C%20coroners%2C%20and%20pathologists%20should%20work%20with,choice%20for%20upper%20respiratory%20tract%20swab-based%20SARS-CoV-2%20testing. Accessed 11 January 2021. [Google Scholar]
- 198.Sekulic M, Harper H, Nezami BG, Shen DL, Sekulic SP, Koeth AT, Harding CV, Gilmore H, Sadri N. 2020. Molecular detection of SARS-CoV-2 infection in FFPE samples and histopathologic findings in fatal SARS-CoV-2 cases. Am J Clin Pathol 154:190–200. 10.1093/ajcp/aqaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Brouwer AF, Myers JL, Martin ET, Konopka KE, Lauring AS, Eisenberg MC, Lephart PR, Nguyen T, Jaworski A, Schmidt CJ. 2 September 2020. SARS-CoV-2 surveillance in decedents in a large, urban medical examiner’s office. Clin Infect Dis 10.1093/cid/ciaa1312. [DOI] [PMC free article] [PubMed]
- 200.Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T, Yarid N, Marshall DA. 2020. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 396:320–332. 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. 2007. Determinants of RNA quality from FFPE samples. PLoS One 2:e1261. 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Best Rocha A, Stroberg E, Barton LM, Duval EJ, Mukhopadhyay S, Yarid N, Caza T, Wilson JD, Kenan DJ, Kuperman M, Sharma SG, Larsen CP. 2020. Detection of SARS-CoV-2 in formalin-fixed paraffin-embedded tissue sections using commercially available reagents. Lab Invest 100:1485–1489. 10.1038/s41374-020-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, Spijker R, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group . 2020. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 8:CD013705. 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Danis K, Epaulard O, Bénet T, Gaymard A, Campoy S, Botelho-Nevers E, Bouscambert-Duchamp M, Spaccaferri G, Ader F, Mailles A, Boudalaa Z, Tolsma V, Berra J, Vaux S, Forestier E, Landelle C, Fougere E, Thabuis A, Berthelot P, Veil R, Levy-Bruhl D, Chidiac C, Lina B, Coignard B, Saura C, Investigation Team . 2020. Cluster of coronavirus disease 2019 (COVID-19) in the French Alps, February 2020. Clin Infect Dis 71:825–832. 10.1093/cid/ciaa424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kim ES, Chin BS, Kang CK, Kim NJ, Kang YM, Choi J-P, Oh DH, Kim J-H, Koh B, Kim SE, Yun NR, Lee J-H, Kim JY, Kim Y, Bang JH, Song K-H, Kim HB, Chung KH, Oh MD, Korea National Committee for Clinical Management of COVID-19 . 2020. Clinical course and outcomes of patients with severe acute respiratory syndrome coronavirus 2 infection: a preliminary report of the first 28 patients from the Korean Cohort Study on COVID-19. J Korean Med Sci 35:e142. 10.3346/jkms.2020.35.e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 2020. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 71:2027–2034. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lou B, Li T-D, Zheng S-F, Su Y-Y, Li Z-Y, Liu W, Yu F, Ge S-X, Zou Q-D, Yuan Q, Lin S, Hong C-M, Yao X-Y, Zhang X-J, Wu D-H, Zhou G-L, Hou W-H, Li T-T, Zhang Y-L, Zhang S-Y, Fan J, Zhang J, Xia N-S, Chen Y. 2020. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J 56:2000763. 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 209.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang Q-W, Xu S-Y, Zhu H-D, Xu Y-C, Jin Q, Sharma L, Wang L, Wang J. 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 71:778–785. 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu L, Liu W, Zheng Y, Jiang X, Kou G, Ding J, Wang Q, Huang Q, Ding Y, Ni W, Wu W, Tang S, Tan L, Hu Z, Xu W, Zhang Y, Zhang B, Tang Z, Zhang X, Li H, Rao Z, Jiang H, Ren X, Wang S, Zheng S. 2020. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect 22:206–211. 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Salvatore PP, Dawson P, Wadhwa A, Rabold EM, Buono S, Dietrich EA, Reses HE, Vuong J, Pawloski L, Dasu T, Bhattacharyya S, Pevzner E, Hall AJ, Tate JE, Kirking HL. 28 September 2020. Epidemiological correlates of PCR cycle threshold values in the detection of SARS-CoV-2. Clin Infect Dis 10.1093/cid/ciaa1469. [DOI] [PMC free article] [PubMed]
- 212.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 2020. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 71:2663–2666. 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Basile K, McPhie K, Carter I, Alderson S, Rahman H, Donovan L, Kumar S, Tran T, Ko D, Sivaruban T, Ngo C, Toi C, O’Sullivan MV, Sintchenko V, Chen SC-A, Maddocks S, Dwyer DE, Kok J. 24 October 2020. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis 10.1093/cid/ciaa1579. [DOI] [PMC free article] [PubMed]
- 214.Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill 25:2001483. 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gombar S, Chang M, Hogan CA, Zehnder J, Boyd S, Pinsky BA, Shah NH. 2020. Persistent detection of SARS-CoV-2 RNA in patients and healthcare workers with COVID-19. J Clin Virol 129:104477. 10.1016/j.jcv.2020.104477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, Gautret P, Raoult D. 2020. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 39:1059–1061. 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Widders A, Broom A, Broom J. 2020. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health 25:210–215. 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26:672–675. 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 219.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, Ng O-T, Marimuthu K, Ang LW, Mak TM, Lau SK, Anderson DE, Chan KS, Tan TY, Ng TY, Cui L, Said Z, Kurupatham L, Chen MI-C, Chan M, Vasoo S, Wang L-F, Tan BH, Lin RTP, Lee VJM, Leo Y-S, Lye DC, Singapore 2019 Novel Coronavirus Outbreak Research Team . 2020. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323:1488–1494. 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, Li Y, Ni Q, Zou R, Li X, Xu M, Zhang Y, Zhao H, Zhang X, Yu L, Su J, Lang G, Liu J, Wu X, Guo Y, Tao J, Shi D, Yu L, Cao Q, Ruan B, Liu L, Wang Z, Xu Y, Liu Y, Sheng J, Li L. 2020. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19). Clin Infect Dis 71:799–806. 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Weiss A, Jellingsø M, Sommer MOA. 2020. Spatial and temporal dynamics of SARS-CoV-2 in COVID-19 patients: a systematic review and meta-analysis. EBioMedicine 58:102916. 10.1016/j.ebiom.2020.102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, Ahern S, Carty PG, O’Brien KK, O’Murchu E, O’Neill M, Smith SM, Ryan M, Harrington P. 2020. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect 81:357–371. 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Corman VM, Rabenau HF, Adams O, Oberle D, Funk MB, Keller‐Stanislawski B, Timm J, Drosten C, Ciesek S. 2020. SARS-CoV-2 asymptomatic and symptomatic patients and risk for transfusion transmission. Transfusion 60:1119–1122. 10.1111/trf.15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhou R, Li F, Chen F, Liu H, Zheng J, Lei C, Wu X. 2020. Viral dynamics in asymptomatic patients with COVID-19. Int J Infect Dis 96:288–290. 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Long Q-X, Tang X-J, Shi Q-L, Li Q, Deng H-J, Yuan J, Hu J-L, Xu W, Zhang Y, Lv F-J, Su K, Zhang F, Gong J, Wu B, Liu X-M, Li J-J, Qiu J-F, Chen J, Huang A-L. 2020. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 26:1200–1204. 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 226.Lu Y, Li Y, Deng W, Liu M, He Y, Huang L, Lv M, Li J, Du H. 2020. Symptomatic infection is associated with prolonged duration of viral shedding in mild coronavirus disease 2019: a retrospective study of 110 children in Wuhan. Pediatr Infect Dis J 39:e95–e99. 10.1097/INF.0000000000002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zheng Y, Liu X, Le W, Xie L, Li H, Wen W, Wang S, Ma S, Huang Z, Ye J, Shi W, Ye Y, Liu Z, Song M, Zhang W, Han J-DJ, Belmonte JCI, Xiao C, Qu J, Wang H, Liu G-H, Su W. 2020. A human circulating immune cell landscape in aging and COVID-19. Protein Cell 11:740–770. 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Andrew M, Searle SD, McElhaney JE, McNeil SA, Clarke B, Rockwood K, Kelvin DJ. 2020. COVID-19, frailty and long-term care: implications for policy and practice. J Infect Dev Ctries 14:428–432. 10.3855/jidc.13003. [DOI] [PubMed] [Google Scholar]
- 229.Vellas C, Delobel P, De Souto Barreto P, Izopet J. 2020. COVID-19, virology and geroscience: a perspective. J Nutr Health Aging 24:685–691. 10.1007/s12603-020-1416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.US Centers for Disease Control and Prevention. 2020. Discontinuation of transmission-based precautions and disposition of patients with COVID-19 in healthcare settings (interim guidance), updated August 10, 2020. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html. Accessed 11 January 2021. [Google Scholar]
- 231.Zhou C, Zhang T, Ren H, Sun S, Yu X, Sheng J, Shi Y, Zhao H. 2020. Impact of age on duration of viral RNA shedding in patients with COVID-19. Aging (Albany NY) 12:22399–22404. 10.18632/aging.104114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Li N, Wang X, Lv T. 2020. Prolonged SARS‐CoV‐2 RNA shedding: not a rare phenomenon. J Med Virol 92:2286–2287. 10.1002/jmv.25952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.AlJishi JM, Al-Tawfiq JA. 2021. Intermittent viral shedding in respiratory samples of patients with SARS-CoV-2: observational analysis with infection control implications. J Hosp Infect 107:98–100. 10.1016/j.jhin.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sze S, Pan D, Williams CML, Barker J, Minhas JS, Miller CJ, Tang JW, Squire IB, Pareek M. 2021. The need for improved discharge criteria for hospitalised patients with COVID-19—implications for patients in long-term care facilities. Age Ageing 50:16–20. 10.1093/ageing/afaa206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. 2020. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 5:434–435. 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kim J-M, Kim HM, Lee EJ, Jo HJ, Yoon Y, Lee N-J, Son J, Lee Y-J, Kim MS, Lee Y-P, Chae S-J, Park KR, Cho S-R, Park S, Kim SJ, Wang E, Woo S, Lim A, Park S-J, Jang J, Chung Y-S, Chin BS, Lee J-S, Lim D, Han M-G, Yoo CK. 2020. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res Perspect 11:112–117. 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xiao F, Sun J, Xu Y, Li F, Huang X, Li H, Zhao J, Huang J, Zhao J. 2020. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis 26:1920–1922. 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.World Health Organization. 2020. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays interim guidance, 11 September 2020. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays. Accessed 1 December 2020. [Google Scholar]
- 239.Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, Liao P, Qiu J-F, Lin Y, Cai X-F, Wang D-Q, Hu Y, Ren J-H, Tang N, Xu Y-Y, Yu L-H, Mo Z, Gong F, Zhang X-L, Tian W-G, Hu L, Zhang X-X, Xiang J-L, Du H-X, Liu H-W, Lang C-H, Luo X-H, Wu S-B, Cui X-P, Zhou Z, Zhu M-M, Wang J, Xue C-J, Li X-F, Wang L, Li Z-J, Wang K, Niu C-C, Yang Q-J, Tang X-J, Zhang Y, Liu X-M, Li J-J, Zhang D-C, Zhang F, Liu P, Yuan J, Li Q, Hu J-L, Chen J, et al. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 240.Moncunill G, Mayor A, Santano R, Jiménez A, Vidal M, Tortajada M, Sanz S, Méndez S, Llupià A, Aguilar R, Alonso S, Barrios D, Carolis C, Cisteró P, Chóliz E, Cruz A, Fochs S, Jairoce C, Hecht J, Lamoglia M, Martínez MJ, Moreno J, Mitchell RA, Ortega N, Pey N, Puyol L, Ribes M, Rosell N, Figueroa-Romero A, Sotomayor P, Torres S, Williams S, Barroso S, Vilella A, Trilla A, Varela P, Dobaño C, Garcia-Basteiro AL. 2021. SARS-CoV-2 seroprevalence and antibody kinetics among health care workers in a Spanish hospital after 3 months of follow-up. J Infect Dis 223:62–71. 10.1093/infdis/jiaa696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Van Caeseele P, Bailey D, Forgie SE, Dingle TC, Krajden M, for the Canadian Public Health Laboratory Network . 2020. SARS-CoV-2 (COVID-19) serology: implications for clinical practice, laboratory medicine and public health. CMAJ 192:E973–E979. 10.1503/cmaj.201588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Isho B, Abe KT, Zuo M, Jamal AJ, Rathod B, Wang JH, Li Z, Chao G, Rojas OL, Bang YM, Pu A, Christie-Holmes N, Gervais C, Ceccarelli D, Samavarchi-Tehrani P, Guvenc F, Budylowski P, Li A, Paterson A, Yue FY, Marin LM, Caldwell L, Wrana JL, Colwill K, Sicheri F, Mubareka S, Gray-Owen SD, Drews SJ, Siqueira WL, Barrios-Rodiles M, Ostrowski M, Rini JM, Durocher Y, McGeer AJ, Gommerman JL, Gingras A-C. 2020. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 5:eabe5511. 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Peng J, Lu Y, Song J, Vallance BA, Jacobson K, Yu HB, Sun Z. 2020. Direct clinical evidence recommending the use of proteinase K or dithiothreitol to pretreat sputum for detection of SARS-CoV-2. Front Med (Lausanne) 7:549860. 10.3389/fmed.2020.549860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.James A, Alawneh J. 2020. COVID-19 infection diagnosis: potential impact of isothermal amplification technology to reduce community transmission of SARS-CoV-2. Diagnostics 10:399. 10.3390/diagnostics10060399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Corman V, Bleicker T, Brunink S, Drosten C. 2020. Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR. World Health Organization, Geneva, Switzerland. https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf?sfvrsn=d381fc88_2. [Google Scholar]
- 246.US Food and Drug Administration. 2020. CDC 2019-nCoV real-time RT-PCR diagnostic panel emergency use authorization. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/media/134919/download. Accessed 15 December 2020. [Google Scholar]
- 247.US Centers for Disease Control and Prevention. 2020. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel: instructions for use. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.fda.gov/media/134922/download. Accessed 15 October 2020. [Google Scholar]
- 248.Beall SG, Cantera J, Diaz MH, Winchell JM, Lillis L, White H, Kalnoky M, Gallarda J, Boyle DS. 2019. Performance and workflow assessment of six nucleic acid extraction technologies for use in resource limited settings. PLoS One 14:e0215753. 10.1371/journal.pone.0215753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tan SC, Yiap BC. 2009. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol 2009:574398. 10.1155/2009/574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wozniak A, Cerda A, Ibarra-Henriquez C, Sebastian V, Armijo G, Lamig L, Miranda C, Lagos M, Solari S, Guzmán AM, Quiroga T, Hitschfeld S, Riveras E, Ferres M, Gutiérrez RA, García P. 2020. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. bioRxiv 10.1101/2020.05.07.083048. [DOI] [PMC free article] [PubMed]
- 251.Poljak M, Korva M, Knap Gašper N, Fujs Komloš K, Sagadin M, Uršič T, Avšič Županc T, Petrovec M. 2020. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol 58:e00599-20. 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ali N, Rampazzo RDCP, Costa ADT, Krieger MA. 2017. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. Biomed Res Int 2017:9306564. 10.1155/2017/9306564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ambrosi C, Prezioso C, Checconi P, Scribano D, Sarshar M, Capannari M, Tomino C, Fini M, Garaci E, Palamara AT, De Chiara G, Limongi D. 2021. SARS-CoV-2: comparative analysis of different RNA extraction methods. J Virol Methods 287:114008. 10.1016/j.jviromet.2020.114008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fomsgaard AS, Rosenstierne MW. 2020. An alternative workflow for molecular detection of SARS-CoV-2—escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill 25:2000398. 10.2807/1560-7917.ES.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bruce EA, Huang M, Perchetti GA, Tighe S, Laaguiby P, Hoffman JJ, Gerrard DL, Nalla AK, Wei Y, Greninger AL, Diehl SA, Shirley DJ, Leonard DGB, Huston CD, Beth D, Dragon JA, Crothers JW, Jerome KR, Botten JW. 2020. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. bioRxiv 10.1101/2020.03.20.001008. [DOI] [PMC free article] [PubMed]
- 256.Hasan MR, Mirza F, Al-Hail H, Sundararaju S, Xaba T, Iqbal M, Alhussain H, Yassine HM, Perez-Lopez A, Tang P. 2020. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS One 15:e0236564. 10.1371/journal.pone.0236564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lübke N, Senff T, Scherger S, Hauka S, Andrée M, Adams O, Timm J, Walker A. 2020. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J Clin Virol 130:104579. 10.1016/j.jcv.2020.104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Srivatsan S, Han P, van Raay K, Wolf C, McCulloch D, Kim A, Brandstetter E, Martin B, Gehring J, Chen W, Kosuri S, Konnick E, Lockwood C, Rieder M, Nickerson D, Chu H, Shendure J, Starita L. 2020. Preliminary support for a “dry swab, extraction free” protocol for SARS-CoV-2 testing via RT-qPCR. bioRxiv 10.1101/2020.04.22.056283. [DOI]
- 259.Lalli MA, Langmade SJ, Chen X, Fronick CC, Sawyer CS, Burcea LC, Wilkinson MN, Fulton RS, Heinz M, Buchser WJ, Head RD, Mitra RD, Milbrandt J. 2021. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem 67:415–424. 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lee JYH, Best N, McAuley J, Porter JL, Seemann T, Schultz MB, Sait M, Orlando N, Mercoulia K, Ballard SA, Druce J, Tran T, Catton MG, Pryor MJ, Cui HL, Luttick A, McDonald S, Greenhalgh A, Kwong JC, Sherry NL, Graham M, Hoang T, Herisse M, Pidot SJ, Williamson DA, Howden BP, Monk IR, Stinear TP. 2020. Validation of a single-step, single-tube reverse transcription-loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. bioRxiv 10.1101/2020.04.28.067363. [DOI] [PMC free article] [PubMed]
- 261.Ben-Assa N, Naddaf R, Gefen T, Capucha T, Hajjo H, Mandelbaum N, Elbaum L, Rogov P, King DA, Kaplan S, Rotem A, Chowers M, Szwarcwort-Cohen M, Paul M, Geva-Zatorsky N. 2020. SARS-CoV-2 on-the-spot virus detection directly from patients. medRxiv 10.1101/2020.04.22.20072389. [DOI] [PMC free article] [PubMed]
- 262.Rabe BA, Cepko C. 2020. SARS-CoV-2 detection using an isothermal amplification reaction and a rapid, inexpensive protocol for sample inactivation and purification. medRxiv 10.1101/2020.04.23.20076877. [DOI] [PMC free article] [PubMed]
- 263.Smyrlaki I, Ekman M, Lentini A, Rufino de Sousa N, Papanicolaou N, Vondracek M, Aarum J, Safari H, Muradrasoli S, Rothfuchs AG, Albert J, Högberg B, Reinius B. 2020. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun 11:4812. 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mokhtar KM. 2020. Improved COVID-19 testing by extraction-free SARS-CoV-2 RT-PCR. EJIFCC 31:362–368. [PMC free article] [PubMed] [Google Scholar]
- 265.Beltrán-Pavez C, Alonso-Palomares LA, Valiente-Echeverría F, Gaggero A, Soto-Rifo R, Barriga GP. 2021. Accuracy of a RT-qPCR SARS-CoV-2 detection assay without prior RNA extraction. J Virol Methods 287:113969. 10.1016/j.jviromet.2020.113969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wong ML, Medrano JF. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39:75–85. 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 267.Dorlass EG, Monteiro CO, Viana AO, Soares CP, Machado RRG, Thomazelli LM, Araujo DB, Leal FB, Candido ED, Telezynski BL, Valério CA, Chalup VN, Mello R, Almeida FJ, Aguiar AS, Barrientos ACM, Sucupira C, De Paulis M, Sáfadi MAP, Silva DGBP, Sodré JJM, Soledade MP, Matos SF, Ferreira SR, Pinez CMN, Buonafine CP, Pieroni LNF, Malta FM, Santana RAF, Souza EC, Fock RA, Pinho JRR, Ferreira LCS, Botosso VF, Durigon EL, Oliveira DBL. 2020. Lower cost alternatives for molecular diagnosis of COVID-19: conventional RT-PCR and SYBR green-based RT-qPCR. Braz J Microbiol 51:1117–1123. 10.1007/s42770-020-00347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Won J, Lee S, Park M, Kim TY, Park MG, Choi BY, Kim D, Chang H, Kim VN, Lee CJ. 2020. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19). Exp Neurobiol 29:107–119. 10.5607/en20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Meza-Robles C, Barajas-Saucedo CE, Tiburcio-Jimenez D, Mokay-Ramírez KA, Melnikov V, Rodriguez-Sanchez IP, Martinez-Fierro ML, Garza-Veloz I, Zaizar-Fregoso SA, Guzman-Esquivel J, Ramirez-Flores M, Newton-Sanchez OA, Espinoza-Gómez F, Delgado-Enciso OG, Centeno-Ramirez AS, Delgado-Enciso I. 2020. One-step nested RT-PCR for COVID-19 detection: a flexible, locally developed test for SARS-CoV2 nucleic acid detection. J Infect Dev Ctries 14:679–684. 10.3855/jidc.12726. [DOI] [PubMed] [Google Scholar]
- 270.González-González E, Trujillo-de Santiago G, Lara-Mayorga IM, Martínez-Chapa SO, Alvarez MM. 2020. Portable and accurate diagnostics for COVID-19: combined use of the miniPCR thermocycler and a well-plate reader for SARS-CoV-2 virus detection. PLoS One 15:e0237418. 10.1371/journal.pone.0237418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Xia S, Chen X. 2020. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov 6:37. 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hirotsu Y, Mochizuki H, Omata M. 2020. Double-quencher probes improve detection sensitivity toward severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods 284:113926. 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.VanGuilder HD, Vrana KE, Freeman WM. 2008. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44:619–626. 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 274.Mitchell SL, St George K, Rhoads DD, Butler-Wu SM, Dharmarha V, McNult P, Miller MB. 2020. Understanding, verifying, and implementing emergency use authorization molecular diagnostics for the detection of SARS-CoV-2 RNA. J Clin Microbiol 58:e00796-20. 10.1128/JCM.00796-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Nalla AK, Casto AM, Huang M-LW, Perchetti GA, Sampoleo R, Shrestha L, Wei Y, Zhu H, Jerome KR, Greninger AL. 2020. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol 58:e00557-20. 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Chen C-J, Hsieh L-L, Lin S-K, Wang C-F, Huang Y-H, Lin S-Y, Lu P-L. 2020. Optimization of the CDC protocol of molecular diagnosis of COVID-19 for timely diagnosis. Diagnostics 10:333. 10.3390/diagnostics10050333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. 2020. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66:549–555. 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bulterys PL, Garamani N, Stevens B, Sahoo MK, Huang C, Hogan CA, Zehnder J, Pinsky BA. 2020. Comparison of a laboratory-developed test targeting the envelope gene with three nucleic acid amplification tests for detection of SARS-CoV-2. J Clin Virol 129:104427. 10.1016/j.jcv.2020.104427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Toptan T, Hoehl S, Westhaus S, Bojkova D, Berger A, Rotter B, Hoffmeier K, Ciesek S, Widera M. 2020. Optimized qRT-PCR approach for the detection of intra- and extra-cellular SARS-CoV-2 RNAs. bioRxiv 10.1101/2020.04.20.052258. [DOI] [PMC free article] [PubMed]
- 280.Matheeussen V, Corman VM, Donoso Mantke O, McCulloch E, Lammens C, Goossens H, Niemeyer D, Wallace PS, Klapper P, Niesters HGM, Drosten C, Ieven M, RECOVER Project and Collaborating Networks . 2020. International external quality assessment for SARS-CoV-2 molecular detection and survey on clinical laboratory preparedness during the COVID-19 pandemic, April/May 2020. Euro Surveill 25:2001223. 10.2807/1560-7917.ES.2020.25.27.2001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sung H, Han M-G, Yoo C-K, Lee S-W, Chung Y-S, Park J-S, Kim M-N, Lee H, Hong KH, Seong M-W, Lee K, Chun S, Lee WG, Kwon G-C, Min W-K. 2020. Nationwide external quality assessment of SARS-CoV-2 molecular testing, South Korea. Emerg Infect Dis 26:2353–2360. 10.3201/eid2610.202551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Görzer I, Buchta C, Chiba P, Benka B, Camp JV, Holzmann H, Puchhammer-Stöckl E, Aberle SW. 2020. First results of a national external quality assessment scheme for the detection of SARS-CoV-2 genome sequences. J Clin Virol 129:104537. 10.1016/j.jcv.2020.104537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Government of Canada. 2020. Interim national case definition: coronavirus disease (COVID-19), updated April 2, 2020. Government of Canada, Ottawa, Ontario, Canada. https://www.canada.ca/en/public-health/services/diseases/2019-novel-coronavirus-infection/health-professionals/national-case-definition.html. Accessed 15 October 2020. [Google Scholar]
- 284.Li C, Debruyne DN, Spencer J, Kapoor V, Liu LY, Zhou B, Lee L, Feigelman R, Burdon G, Liu J, Oliva A, Borcherding A, Tan H, Urban AE, Liu G, Liu Z, Zhang B, Lee L, Feigelman R, Burdon G, Liu J, Oliva A, Borcherding A, Xu J, Urban AE, Liu G, Liu Z. 2020. High sensitivity detection of SARS-CoV-2 using multiplex PCR and a multiplex-PCR-based metagenomic method. bioRxiv 10.1101/2020.03.12.988246. [DOI]
- 285.Lang ALS, Roberts C, Mazzulli T, Hatchette TF, LeBlanc JJ. 2014. Detection and differentiation of herpes simplex viruses by use of the Viper platform: advantages, limitations, and concerns. J Clin Microbiol 52:2186–2188. 10.1128/JCM.03636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Rao SN, Manissero D, Steele VR, Pareja J. 2020. A narrative systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther 9:573–586. 10.1007/s40121-020-00324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Gaston DC, Malinis M, Osborn R, Peaper DR, Landry M, Juthani‐Mehta M, Azar MM. 2021. Clinical implications of SARS‐CoV‐2 cycle threshold values in solid organ transplant recipients. Am J Transplant 21:1304–1311. 10.1111/ajt.16357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Patriquin G, LeBlanc JJ. 2021. SARS-CoV-2 sensitivity limbo—how low can we go? Int J Infect Dis 103:23–24. 10.1016/j.ijid.2020.11.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. 2020. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill 25:2000152. 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Smithgall MC, Scherberkova I, Whittier S, Green DA. 2020. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the rapid detection of SARS-CoV-2. J Clin Virol 128:104428. 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lowe CF, Matic N, Ritchie G, Lawson T, Stefanovic A, Champagne S, Leung V, Romney MG. 2020. Detection of low levels of SARS-CoV-2 RNA from nasopharyngeal swabs using three commercial molecular assays. J Clin Virol 128:104387. 10.1016/j.jcv.2020.104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Broder K, Babiker A, Myers C, White T, Jones H, Cardella J, Burd EM, Hill CE, Kraft CS. 2020. Test agreement between Roche cobas 6800 and Cepheid GeneXpert Xpress SARS-CoV-2 assays at high cycle threshold ranges. J Clin Microbiol 58:e01187-20. 10.1128/JCM.01187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Pujadas E, Ibeh N, Hernandez MM, Waluszko A, Sidorenko T, Flores V, Shiffrin B, Chiu N, Young-Francois A, Nowak MD, Paniz‐Mondolfi AE, Sordillo EM, Cordon‐Cardo C, Houldsworth J, Gitman MR. 2020. Comparison of SARS‐CoV‐2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS‐CoV‐2 test and a laboratory‐developed real‐time RT‐PCR test. J Med Virol 92:1695–1698. 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lieberman JA, Pepper G, Naccache SN, Huang M-L, Jerome KR, Greninger AL. 2020. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol 58:e00821-20. 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Nörz D, Fischer N, Schultze A, Kluge S, Mayer-Runge U, Aepfelbacher M, Pfefferle S, Lütgehetmann M. 2020. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J Clin Virol 128:104390. 10.1016/j.jcv.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Mostafa HH, Hardick J, Morehead E, Miller JA, Gaydos CA, Manabe YC. 2020. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J Clin Virol 130:104578. 10.1016/j.jcv.2020.104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Craney AR, Velu PD, Satlin MJ, Fauntleroy KA, Callan K, Robertson A, La Spina M, Lei B, Chen A, Alston T, Rozman A, Loda M, Rennert H, Cushing M, Westblade LF. 2020. Comparison of two high-throughput reverse transcription-PCR systems for the detection of severe acute respiratory syndrome coronavirus 2. J Clin Microbiol 58:e00890-20. 10.1128/JCM.00890-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hogan CA, Sahoo MK, Huang C, Garamani N, Stevens B, Zehnder J, Pinsky BA. 2020. Comparison of the Panther Fusion and a laboratory-developed test targeting the envelope gene for detection of SARS-CoV-2. J Clin Virol 127:104383. 10.1016/j.jcv.2020.104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Fung B, Gopez A, Servellita V, Arevalo S, Ho C, Deucher A, Thornborrow E, Chiu C, Miller S. 2020. Direct comparison of SARS-CoV-2 analytical limits of detection across seven molecular assays. J Clin Microbiol 58:e01535-20. 10.1128/JCM.01535-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Degli-Angeli E, Dragavon J, Huang M-L, Lucic D, Cloherty G, Jerome KR, Greninger AL, Coombs RW. 2020. Validation and verification of the Abbott RealTime SARS-CoV-2 assay analytical and clinical performance. J Clin Virol 129:104474. 10.1016/j.jcv.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Harrington A, Cox B, Snowdon J, Bakst J, Ley E, Grajales P, Maggiore J, Kahn S. 2020. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol 58:e00798-20. 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Perng C-L, Jian M-J, Chang C-K, Lin J-C, Yeh K-M, Chen C-W, Chiu S-K, Chung H-Y, Wang Y-H, Liao S-J, Li S-Y, Hsieh S-S, Tsai S-H, Chang F-Y, Shang H-S. 2020. Novel rapid identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by real-time RT-PCR using BD Max Open system in Taiwan. PeerJ 8:e9318. 10.7717/peerj.9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Liu X, Feng J, Zhang Q, Guo D, Zhang L, Suo T, Hu W, Guo M, Wang X, Huang Z, Xiong Y, Chen G, Chen Y, Lan K. 2020. Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg Microbes Infect 9:1175–1179. 10.1080/22221751.2020.1772679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Alteri C, Cento V, Antonello M, Colagrossi L, Merli M, Ughi N, Renica S, Matarazzo E, Di Ruscio F, Tartaglione L, Colombo J, Grimaldi C, Carta S, Nava A, Costabile V, Baiguera C, Campisi D, Fanti D, Vismara C, Fumagalli R, Scaglione F, Epis OM, Puoti M, Perno CF. 2020. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One 15:e0236311. 10.1371/journal.pone.0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Suo T, Liu X, Feng J, Guo M, Hu W, Guo D, Ullah H, Yang Y, Zhang Q, Wang X, Sajid M, Huang Z, Deng L, Chen T, Liu F, Xu K, Liu Y, Zhang Q, Liu Y, Xiong Y, Chen G, Lan K, Chen Y. 2020. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect 9:1259–1268. 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Falzone L, Musso N, Gattuso G, Bongiorno D, Palermo C, Scalia G, Libra M, Stefani S. 2020. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med 46:957–964. 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.US Centers for Disease Control and Prevention. 2020. Interim guidance for use of pooling procedures in SARS-CoV-2 diagnostic, screening, and surveillance testing, updated October 23, 2020. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/pooling-procedures.html. Accessed 11 January 2021. [Google Scholar]
- 308.Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, Iwen PC. 2020. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol 153:715–718. 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Ben-Ami R, Klochendler A, Seidel M, Sido T, Gurel-Gurevich O, Yassour M, Meshorer E, Benedek G, Fogel I, Oiknine-Djian E, Gertler A, Rotstein Z, Lavi B, Dor Y, Wolf DG, Salton M, Drier Y, Hebrew University-Hadassah COVID-19 Diagnosis Team . 2020. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect 26:1248–1253. 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Hogan CA, Sahoo MK, Pinsky BA. 2020. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 323:1967–1969. 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.LeBlanc JJ, Patriquin G, Pettipas J, Warhuus M, Sarty D, Jackson C, Heinstein C, MacDonald J, Haldane D, Hatchette TF. 2020. Look before diving into pooling of SARS-CoV-2 samples on high throughput analyzers. medRxiv 10.1101/2020.08.17.20176982. [DOI]
- 312.Becker MG, Taylor T, Kiazyk S, Cabiles DR, Meyers AFA, Sandstrom PA. 2020. Recommendations for sample pooling on the Cepheid GeneXpert system using the Cepheid Xpert Xpress SARS-CoV-2 assay. PLoS One 15:e0241959. 10.1371/journal.pone.0241959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Wolters F, van de Bovenkamp J, van den Bosch B, van den Brink S, Broeders M, Chung NH, Favié B, Goderski G, Kuijpers J, Overdevest I, Rahamat-Langedoen J, Wijsman L, Melchers WJ, Meijer A. 2020. Multi-center evaluation of Cepheid Xpert Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol 128:104426. 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Loeffelholz MJ, Alland D, Butler-Wu SM, Pandey U, Perno CF, Nava A, Carroll KC, Mostafa H, Davies E, McEwan A, Rakeman JL, Fowler RC, Pawlotsky J-M, Fourati S, Banik S, Banada PP, Swaminathan S, Chakravorty S, Kwiatkowski RW, Chu VC, Kop J, Gaur R, Sin MLY, Nguyen D, Singh S, Zhang N, Persing DH. 2020. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 test. J Clin Microbiol 58:e00926-20. 10.1128/JCM.00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Mostafa HH, Carroll KC, Hicken R, Berry GJ, Manji R, Smith E, Rakeman JL, Fowler RC, Leelawong M, Butler-Wu SM, Quintero D, Umali-Wilcox M, Kwiatkowski RW, Persing DH, Weir F, Loeffelholz MJ. 2021. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV test. J Clin Microbiol 59:e02955-20. 10.1128/JCM.02955-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Creager HM, Cabrera B, Schnaubelt A, Cox JL, Cushman-Vokoun AM, Shakir SM, Tardif KD, Huang M-L, Jerome KR, Greninger AL, Drobysheva D, Spaulding U, Rogatcheva M, Bourzac KM, Hinrichs SH, Broadhurst MJ, Fey PD. 2020. Clinical evaluation of the BioFire Respiratory Panel 2.1 and detection of SARS-CoV-2. J Clin Virol 129:104538. 10.1016/j.jcv.2020.104538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Eckbo EJ, Locher K, Caza M, Li L, Lavergne V, Charles M. 2021. Evaluation of the BioFire COVID-19 test and Respiratory Panel 2.1 for rapid identification of SARS-CoV-2 in nasopharyngeal swab samples. Diagn Microbiol Infect Dis 99:115260. 10.1016/j.diagmicrobio.2020.115260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.DiaSorin Molecular. 2020. Simplexa COVID-19 direct: instructions for use. DiaSorin Molecular, Cypress, CA. https://www.fda.gov/media/136286/download. Accessed 22 January 2021. [Google Scholar]
- 319.GeneMark Diagnostics. 2020. ePlex SARS-CoV-2 test: instructions for use. GeneMark Diagnostics, Carlsbad, CA. https://www.fda.gov/media/136282/download. Accessed 22 September 2020. [Google Scholar]
- 320.Mesa Biotech. 2021. Accula SARS-CoV-2 test: instructions for use. Mesa Biotech, San Diego, CA. https://www.fda.gov/media/136355/download. Accessed 22 January 2021. [Google Scholar]
- 321.Sakai J, Tarumoto N, Orihara Y, Kawamura R, Kodana M, Matsuzaki N, Matsumura R, Ogane K, Kawamura T, Takeuchi S, Imai K, Murakami T, Maesaki S, Maeda T. 2020. Evaluation of a high-speed but low-throughput RT-qPCR system for detection of SARS-CoV-2. J Hosp Infect 105:615–618. 10.1016/j.jhin.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Smith E, Zhen W, Manji R, Schron D, Duong S, Berry GJ. 2020. Analytical and clinical comparison of three nucleic acid amplification tests for SARS-CoV-2 detection. J Clin Microbiol 58:e01134-20. 10.1128/JCM.01134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Liotti FM, Menchinelli G, Marchetti S, Morandotti GA, Sanguinetti M, Posteraro B, Cattani P. 2020. Evaluating the newly developed BioFire COVID-19 test for SARS-CoV-2 molecular detection. Clin Microbiol Infect 26:1699–1700. 10.1016/j.cmi.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Tanida K, Koste L, Koenig C, Wenzel W, Fritsch A, Frickmann H. 2020. Evaluation of the automated cartridge-based ARIES SARS-CoV-2 assay (RUO) against automated Cepheid Xpert Xpress SARS-CoV-2 PCR as gold standard. Eur J Microbiol Immunol 10:156–164. 10.1556/1886.2020.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Green K, Graziadio S, Turner P, Fanshawe T, Allen J. 2020. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. University of Oxford, Oxford, United Kingdom. https://www.cebm.net/covid-19/molecular-and-antibody-point-of-care-tests-to-support-the-screening-diagnosis-and-monitoring-of-covid-19/. [Google Scholar]
- 326.Arumugam A, Faron ML, Yu P, Markham C, Wong S. 2020. A rapid COVID-19 RT-PCR detection assay for low resource settings. bioRxiv 10.1101/2020.04.29.069591. [DOI] [PMC free article] [PubMed]
- 327.Kim Y, Yaseen AB, Kishi JY, Hong F, Saka SK, Sheng K, Gopalkrishnan N, Schaus TE, Yin P. 2020. Single-strand RPA for rapid and sensitive detection of SARS-CoV-2 RNA. medRxiv 10.1101/2020.08.17.20177006. [DOI]
- 328.Wang J, Cai K, He X, Shen X, Wang J, Liu J, Xu J, Qiu F, Lei W, Cui L, Ge Y, Wu T, Zhang Y, Yan H, Chen Y, Yu J, Ma X, Shi H, Zhang R, Li X, Gao Y, Niu P, Tan W, Wu G, Jiang Y, Xu W, Ma X. 2020. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin Microbiol Infect 26:1076–1081. 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Li J, Macdonald J, von Stetten F. 2019. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 144:31–67. 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- 330.Piepenburg O, Williams CH, Stemple DL, Armes NA. 2006. DNA detection using recombination proteins. PLoS Biol 4:e204. 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Huang Z, Tian D, Liu Y, Lin Z, Lyon CJ, Lai W, Fusco D, Drouin A, Yin X, Hu T, Ning B. 2020. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens Bioelectron 164:112316. 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Buchan BW, Ledeboer NA. 2014. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev 27:783–822. 10.1128/CMR.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Giachetti C, Linnen JM, Kolk DP, Dockter J, Gillotte-Taylor K, Park M, Ho-Sing-Loy M, McCormick MK, Mimms LT, McDonough SH. 2002. Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J Clin Microbiol 40:2408–2419. 10.1128/jcm.40.7.2408-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Shen C-H. 2019. Diagnostic molecular biology, p 215–247. Elsevier, London, United Kingdom. [Google Scholar]
- 335.Gorzalski AJ, Tian H, Laverdure C, Morzunov S, Verma SC, VanHooser S, Pandori MW. 2020. High-throughput transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J Clin Virol 129:104501. 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Trémeaux P, Lhomme S, Abravanel F, Raymond S, Mengelle C, Mansuy J-M, Izopet J. 2020. Evaluation of the Aptima transcription-mediated amplification assay (Hologic) for detecting SARS-CoV-2 in clinical specimens. J Clin Virol 129:104541. 10.1016/j.jcv.2020.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Ginocchio CC. 2004. Life beyond PCR: alternative target amplification technologies for the diagnosis of infectious diseases, part II. Clin Microbiol Newsl 26:129–136. 10.1016/j.clinmicnews.2004.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Frontzek A, Aretzweiler G, Winkens D, Duncan D, Marlowe EM. 2019. High-volume workflow and performance comparisons for Chlamydia trachomatis and Neisseria gonorrhoeae testing using automated molecular platforms. BMC Infect Dis 19:797. 10.1186/s12879-019-4442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Qian C, Wang R, Wu H, Ji F, Wu J. 2019. Nicking enzyme-assisted amplification (NEAA) technology and its applications: a review. Anal Chim Acta 1050:1–15. 10.1016/j.aca.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 340.Nie S, Roth RB, Stiles J, Mikhlina A, Lu X, Tang Y-W, Babady NE. 2014. Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J Clin Microbiol 52:3339–3344. 10.1128/JCM.01132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Mitchell SL, St George K. 2020. Evaluation of the COVID19 ID NOW EUA assay. J Clin Virol 128:104429. 10.1016/j.jcv.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Basu A, Zinger T, Inglima K, Woo K, Atie O, Yurasits L, See B, Aguero-Rosenfeld ME. 2020. Performance of Abbott ID Now COVID-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J Clin Microbiol 58:e01136-20. 10.1128/JCM.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. 2020. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA emergency use authorization methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol 58:e00760-20. 10.1128/JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Moore NM, Li H, Schejbal D, Lindsley J, Hayden MK. 2020. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV reverse transcriptase PCR assay for the detection of SARS-CoV-2. J Clin Microbiol 58:e00938-20. 10.1128/JCM.00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zhen W, Smith E, Manji R, Schron D, Berry GJ. 2020. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J Clin Microbiol 58:e00783-20. 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Thwe PM, Ren P. 2020. How many are we missing with ID NOW COVID-19 assay using direct nasopharyngeal swabs? Findings from a mid-sized academic hospital clinical microbiology laboratory. Diagn Microbiol Infect Dis 98:115123. 10.1016/j.diagmicrobio.2020.115123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Stokes W, Berenger BM, Singh T, Adeghe I, Schneider A, Portnoy D, King T, Scott B, Pabbaraju K, Shokoples S, Wong AA, Gill K, Turnbull L, Hu J, Tipples G. 2020. Acceptable performance of the Abbott ID NOW among symptomatic individuals with confirmed COVID-19. medRxiv 10.1101/2020.12.24.20248786. [DOI] [PMC free article] [PubMed]
- 348.Lephart PR, Bachman MA, LeBar W, McClellan S, Barron K, Schroeder L, Newton DW. 2021. Comparative study of four SARS-CoV-2 nucleic acid amplification test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type. Diagn Microbiol Infect Dis 99:115200. 10.1016/j.diagmicrobio.2020.115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Tröger V, Niemann K, Gärtig C, Kuhlmeier D. 2015. Isothermal amplification and quantification of nucleic acids and its use in microsystems. J Nanomed Nanotechnol 6:3. 10.4172/2157-7439.1000282. [DOI] [Google Scholar]
- 350.Dunbar S, Das S. 2019. Amplification chemistries in clinical virology. J Clin Virol 115:18–31. 10.1016/j.jcv.2019.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. 2020. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 26:771–783. 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Zhang H, Xu Y, Fohlerova Z, Chang H, Iliescu C, Neuzil P. 2019. LAMP-on-a-chip: revising microfluidic platforms for loop-mediated DNA amplification. Trends Analyt Chem 113:44–53. 10.1016/j.trac.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Asiello PJ, Baeumner AJ. 2011. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 11:1420–1430. 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- 354.Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, Chen W-H, Yin X. 2020. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem 66:975–977. 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Notomi T, Mori Y, Tomita N, Kanda H. 2015. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 53:1–5. 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 356.Li J-J, Xiong C, Liu Y, Liang J-S, Zhou X-W. 2016. Loop-mediated isothermal amplification (LAMP): emergence as an alternative technology for herbal medicine identification. Front Plant Sci 7:1956. 10.3389/fpls.2016.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Yan C, Cui J, Huang L, Du B, Chen L, Xue G, Li S, Zhang W, Zhao L, Sun Y, Yao H, Li N, Zhao H, Feng Y, Liu S, Zhang Q, Liu D, Yuan J. 2020. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 26:773–779. 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Mori Y, Notomi T. 2009. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother 15:62–69. 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, Freistaedter A, Papagiannidis D, Galmozzi C, Stanifer ML, Boulant S, Klein S, Chlanda P, Khalid D, Barreto Miranda I, Schnitzler P, Kräusslich H-G, Knop M, Anders S. 2020. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med 12:eabc7075. 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. 2020. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci 21:2826. 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Tomita N, Mori Y, Kanda H, Notomi T. 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3:877–882. 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 362.Goto M, Honda E, Ogura A, Nomoto A, Hanaki K-I. 2009. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167–172. 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 363.Rödel J, Egerer R, Suleyman A, Sommer-Schmid B, Baier M, Henke A, Edel B, Löffler B. 2020. Use of the variplex SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J Clin Virol 132:104616. 10.1016/j.jcv.2020.104616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Eckel F, Küsters F, Drossel B, Konert M, Mattes H, Schopf S. 2020. Variplex test system fails to reliably detect SARS-CoV-2 directly from respiratory samples without RNA extraction. Eur J Clin Microbiol Infect Dis 39:2373–2377. 10.1007/s10096-020-03983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Österdahl MF, Lee KA, Lochlainn MN, Wilson S, Douthwaite S, Horsfall R, Sheedy A, Goldenberg SD, Stanley CJ, Spector TD, Steves CJ. 2020. Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect Dis 20:783. 10.1186/s12879-020-05484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Baek YH, Um J, Antigua KJC, Park J-H, Kim Y, Oh S, Kim Y-I, Choi W-S, Kim S-G, Jeong JH, Chin BS, Nicolas HDG, Ahn J-Y, Shin KS, Choi YK, Park J-S, Song M-S. 2021. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect 9:998–1007. 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Park G-S, Ku K, Baek S-H, Kim S-J, Kim SI, Kim B-T, Maeng J-S. 2020. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J Mol Diagn 22:729–735. 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Kitagawa Y, Orihara Y, Kawamura R, Imai K, Sakai J, Tarumoto N, Matsuoka M, Takeuchi S, Maesaki S, Maeda T. 2020. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol 129:104446. 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, Horiuchi M, Kato K, Imoto Y, Iwata M, Mimura S, Ito T, Tamura K, Kato Y. 2020. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription–loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol 58:e01438-20. 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Yang W, Dang X, Wang Q, Xu M, Zhao Q, Zhou Y, Zhao H, Wang L, Xu Y, Wang J, Han S, Wang M, Li H, Pei F, Wang Y. 2020. Rapid and accurate detection of three genes in SARS-CoV-2 using RT-LAMP method. Res Sq 10.21203/rs.3.rs-28070/v1. [DOI]
- 371.Yang W, Dang X, Wang Q, Xu M, Zhao Q, Zhou Y, Zhao H, Wang L, Xu Y, Wang J, Han S, Wang M, Pei F, Wan Y. 2020. Rapid detection of SARS-CoV-2 using reverse transcription RT-LAMP method. medRxiv 10.1101/2020.03.02.20030130. [DOI]
- 372.Hu X, Deng Q, Li J, Chen J, Wang Z, Zhang X, Fang Z, Li H, Zhao Y, Yu P, Li W, Wang X, Li S, Zhang L, Hou T. 2020. Development and clinical application of a rapid and sensitive loop-mediated isothermal amplification test for SARS-CoV-2 infection. mSphere 5:e00808-20. 10.1128/mSphere.00808-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser A-M, Glass B, Meurer M, Morales I, Schillak A, Freistaedter A, Ambiel I, Winter SL, Zimmermann L, Naumoska T, Bubeck F, Kirrmaier D, Ullrich S, Barreto Miranda I, Anders S, Grimm D, Schnitzler P, Knop M, Kräusslich H-G, Dao Thi VL, Börner K, Chlanda P. 2020. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses 12:863. 10.3390/v12080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Ganguli A, Mostafa A, Berger J, Aydin MY, Sun F, Stewart de Ramirez SA, Valera E, Cunningham BT, King WP, Bashir R. 2020. Rapid isothermal amplification and portable detection system for SARS-CoV-2. bioRxiv 10.1101/2020.05.21.108381. [DOI] [PMC free article] [PubMed]
- 375.Augustine R, Hasan A, Das S, Ahmed R, Mori Y, Notomi T, Kevadiya BD, Thakor AS. 2020. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel) 9:182. 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E. 2018. The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259. 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 377.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology (Reading) 151:653–663. 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 378.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182. 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 379.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading) 151:2551–2561. 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 380.Wright AV, Nuñez JK, Doudna JA. 2016. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164:29–44. 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 381.Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355. 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Li Y, Li S, Wang J, Liu G. 2019. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol 37:730–743. 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 383.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356:438–442. 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. 2018. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360:436–439. 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, Sappakhaw K, Leelahakorn N, Ruenkam T, Wongsatit T, Athipanyasilp N, Eiamthong B, Lakkanasirorat B, Phoodokmai T, Niljianskul N, Pakotiprapha D, Chanarat S, Homchan A, Tinikul R, Kamutira P, Phiwkaow K, Soithongcharoen S, Kantiwiriyawanitch C, Pongsupasa V, Trisrivirat D, Jaroensuk J, Wongnate T, Maenpuen S, Chaiyen P, Kamnerdnakta S, Swangsri J, Chuthapisith S, Sirivatanauksorn Y, Chaimayo C, Sutthent R, Kantakamalakul W, Joung J, Ladha A, Jin X, Gootenberg JS, Abudayyeh OO, Zhang F, Horthongkham N, Uttamapinant C. 2020. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng 4:1140–1149. 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 386.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, Wu J, Liao Y, Gou X, Li Y, Wang X, Su H, Wang J, Gu B, Xu T, Wang J, Xu T. 2020. Development and evaluation of a CRISPR-based diagnostic for 2019-novel coronavirus. medRxiv 10.1101/2020.02.22.20025460. [DOI] [PMC free article] [PubMed]
- 387.Guo L, Sun X, Wang X, Liang C, Jiang H, Gao Q, Dai M, Qu B, Fang S, Mao Y, Chen Y, Feng G, Gu Q, Wang RR, Zhou Q, Li W. 2020. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov 6:34. 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, Jin S. 2020. CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off‐target evaluation, and strategies to mitigate off‐target effects. Adv Sci (Weinh) 7:1902312. 10.1002/advs.201902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Kim H, Kim J-S. 2014. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15:321–334. 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 390.Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78. 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O’Connor DH, Gehrke L, Collins JJ. 2016. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165:1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 392.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan C, Guevara H, Wadford DA, Chen JS, Chiu CY. 2020. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 38:870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Zhang F, Abudayyeh OO, Gootenberg JS. 2020. A protocol for detection of COVID-19 using CRISPR diagnostics. Broad Institute, MIT, Cambridge, MA. https://go.idtdna.com/rs/400-UEU-432/images/Zhang%20et%20al.%2C%202020%20COVID-19%20detection%20%28updated%29.pdf.
- 394.LaManna CM, Pyhtila B, Barrangou R. 2020. Sharing the CRISPR toolbox with an expanding community. Crispr J 3:248–252. 10.1089/crispr.2020.0075. [DOI] [PubMed] [Google Scholar]
- 395.Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, Pain A, Alofi FS, Alsomali A, Hashem AM, Khogeer A, Almontashiri NAM, Abedalthagafi M, Hassan N, Mahfouz MM. 2020. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res 288:198129. 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang M-LW, Kim N-G, Yu X, Li J, Walker BD, Greninger AL, Jerome KR, Gootenberg JS, Abudayyeh OO, Zhang F. 2020. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv 10.1101/2020.05.04.20091231. [DOI]
- 397.Ding X, Yin K, Li Z, Liu C. 2020. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv 10.1101/2020.03.19.998724. [DOI] [PMC free article] [PubMed]
- 398.Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, Pande T, Endy D, La Russa MF, Lewis DB, Qi LS. 2020. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 181:865–876.e12. 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Teng F, Guo L, Cui T, Wang X-G, Xu K, Gao Q, Zhou Q, Li W. 2019. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol 20:132. 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Milenia Biotec. 2019. Lateral flow readout for CRISPR/Cas-based detection strategies. Milenia Biotec, Giessen, Germany. https://www.milenia-biotec.com/uploads/2019/07/Confusion-T-and-C-Line_final.pdf. Accessed 21 July 2020. [Google Scholar]
- 401.Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, Li T, Li J, Zhou Q, Li W. 2018. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov 4:63. 10.1038/s41421-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, Wu J, Liao Y, Gou X, Li Y, Wang X, Su H, Gu B, Wang J, Xu T. 2020. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog 16:e1008705. 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Ramachandran A, Huyke DA, Sharma E, Sahoo MK, Banaei N, Pinsky BA, Santiago JG. 2020. Electric-field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. bioRxiv 10.1101/2020.05.21.109637. [DOI] [PMC free article] [PubMed]
- 404.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 405.Hess JF, Kohl TA, Kotrová M, Rönsch K, Paprotka T, Mohr V, Hutzenlaub T, Brüggemann M, Zengerle R, Niemann S, Paust N. 2020. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol Adv 41:107537. 10.1016/j.biotechadv.2020.107537. [DOI] [PubMed] [Google Scholar]
- 406.Muzzey D, Evans EA, Lieber C. 2015. Understanding the basics of NGS: from mechanism to variant calling. Curr Genet Med Rep 3:158–165. 10.1007/s40142-015-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Kumar KR, Cowley MJ, Davis RL. 2019. Next-generation sequencing and emerging technologies. Semin Thromb Hemost 45:661–673. 10.1055/s-0039-1688446. [DOI] [PubMed] [Google Scholar]
- 408.Vincent AT, Derome N, Boyle B, Culley AI, Charette SJ. 2017. Next-generation sequencing (NGS) in the microbiological world: how to make the most of your money. J Microbiol Methods 138:60–71. 10.1016/j.mimet.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 409.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Ronaghi M. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363–365. 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 411.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309:1728–1732. 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 412.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 413.Nasir JA, Kozak RA, Aftanas P, Raphenya AR, Smith KM, Maguire F, Maan H, Alruwaili M, Banerjee A, Mbareche H, Alcock BP, Knox NC, Mossman K, Wang B, Hiscox JA, McArthur AG, Mubareka S. 2020. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses 12:895. 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Nature Publishing Group. 2020. First NGS-based COVID-19 diagnostic. Nat Biotechnol 38:777. 10.1038/s41587-020-0608-y. [DOI] [PubMed] [Google Scholar]
- 415.Hilaire BGS, Durand NC, Mitra N, Pulido SG, Mahajan R, Blackburn A, Colaric ZL, Theisen JWM, Weisz D, Dudchenko O, Gnirke A, Rao SSP, Kaur P, Aiden EL, Aiden AP. 2020. A rapid, low cost, and highly sensitive SARS-CoV-2 diagnostic based on whole genome sequencing. bioRxiv 10.1101/2020.04.25.061499. [DOI] [PMC free article] [PubMed]
- 416.Bloom JS, Jones EM, Gasperini M, Lubock NB, Sathe L, Munugala C, Booeshaghi AS, Brandenberg OF, Guo L, Boocock J, Simpkins SW, Lin I, LaPierre N, Hong D, Zhang Y, Oland G, Choe BJ, Chandrasekaran S, Hilt EE, Butte MJ, Damoiseaux R, Cooper AR, Yin Y, Pachter L, Garner OB, Flint J, Eskin E, Luo C, Kosuri S, Kruglyak L, Arboleda VA. 2020. Swab-Seq: a high-throughput platform for massively scaled up SARS-CoV-2 testing. medRxiv 10.1101/2020.08.04.20167874. [DOI]
- 417.Bhoyar RC, Jain A, Sehgal P, Divakar MK, Sharma D, Imran M, Jolly B, Ranjan G, Rophina M, Sharma S, Siwach S, Pandhare K, Sahoo S, Sahoo M, Nayak A, Mohanty JN, Das J, Bhandari S, Mathur SK, Kumar A, Sahlot R, Rojarani P, Lakshmi JV, Surekha A, Sekhar PC, Mahajan S, Masih S, Singh P, Kumar V, Jose B, Mahajan V, Gupta V, Gupta R, Arumugam P, Singh A, Nandy A, Raghavendran PV, Jha RM, Kumari A, Gandotra S, Rao V, Faruq M, Kumar S, Reshma GB, Varma GN, Roy SS, Sengupta A, Chattopadhyay S, Singhal K, Pradhan S, et al. 2020. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next generation sequencing. bioRxiv 10.1101/2020.08.10.242677. [DOI] [PMC free article] [PubMed]
- 418.Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, Curran MD, Parmar S, Caller LG, Caddy SL, Khokhar FA, Yakovleva A, Hall G, Feltwell T, Forrest S, Sridhar S, Weekes MP, Baker S, Brown N, Moore E, Popay A, Roddick I, Reacher M, Gouliouris T, Peacock SJ, Dougan G, Török ME, Goodfellow I. 2020. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis 20:1263–1271. 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Xiao M, Liu X, Ji J, Li M, Li J, Yang L, Sun W, Ren P, Yang G, Zhao J, Liang T, Ren H, Chen T, Zhong H, Song W, Wang Y, Deng Z, Zhao Y, Ou Z, Wang D, Cai J, Cheng X, Feng T, Wu H, Gong Y, Yang H, Wang J, Xu X, Zhu S, Chen F, Zhang Y, Chen W, Li Y, Li J. 2020. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med 12:57. 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Jary A, Leducq V, Malet I, Marot S, Klement-Frutos E, Teyssou E, Soulié C, Abdi B, Wirden M, Pourcher V, Caumes E, Calvez V, Burrel S, Marcelin A-G, Boutolleau D. 2020. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin Microbiol Infect 26:1560.e1–1560.e4. 10.1016/j.cmi.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Karamitros T, Papadopoulou G, Bousali M, Mexias A, Tsiodras S, Mentis A. 2020. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J Clin Virol 131:104585. 10.1016/j.jcv.2020.104585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Rueca M, Bartolini B, Gruber CEM, Piralla A, Baldanti F, Giombini E, Messina F, Marchioni L, Ippolito G, Di Caro A, Capobianchi MR. 2020. Compartmentalized replication of SARS-Cov-2 in upper vs. lower respiratory tract assessed by whole genome quasispecies analysis. Microorganisms 8:1302. 10.3390/microorganisms8091302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, Rambaut A, Robertson DL. 2020. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 5:1408–1417. 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 424.Gulholm T, Basile K, Kok J, Chen SC-A, Rawlinson W. 2020. Laboratory diagnosis of severe acute respiratory syndrome coronavirus 2. Pathology 52:745–753. 10.1016/j.pathol.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Wang H, Li X, Li T, Zhang S, Wang L, Wu X, Liu J. 2020. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis 39:1629–1635. 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Zhang T, Wu Q, Zhang Z. 2020. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol 30:1346–1351.e2. 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Liu P, Chen W, Chen J-P. 2019. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses 11:979. 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Banerjee A, Doxey AC, Mossman K, Irving AT. 2021. Unraveling the zoonotic origin and transmission of SARS-CoV-2. Trends Ecol Evol 36:180–184. 10.1016/j.tree.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Chan JF-W, Kok K-H, Zhu Z, Chu H, To KK-W, Yuan S, Yuen K-Y. 2020. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9:221–236. 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Rahimi A, Mirzazadeh A, Tavakolpour S. 2021. Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 113:1221–1232. 10.1016/j.ygeno.2020.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Zhang W, Govindavari JP, Davis BD, Chen SS, Kim JT, Song J, Lopategui J, Plummer JT, Vail E. 2020. Analysis of genomic characteristics and transmission routes of patients with confirmed SARS-CoV-2 in Southern California during the early stage of the US COVID-19 pandemic. JAMA Netw Open 3:e2024191. 10.1001/jamanetworkopen.2020.24191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Oude Munnink BB, Nieuwenhuijse DF, Stein M, O’Toole Á, Haverkate M, Mollers M, Kamga SK, Schapendonk C, Pronk M, Lexmond P, van der Linden A, Bestebroer T, Chestakova I, Overmars RJ, van Nieuwkoop S, Molenkamp R, van der Eijk AA, GeurtsvanKessel C, Vennema H, Meijer A, Rambaut A, van Dissel J, Sikkema RS, Timen A, Koopmans M, Dutch-Covid-19 Response Team . 2020. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med 26:1405–1410. 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 433.Stefanelli P, Faggioni G, Lo Presti A, Fiore S, Marchi A, Benedetti E, Fabiani C, Anselmo A, Ciammaruconi A, Fortunato A, De Santis R, Fillo S, Capobianchi MR, Gismondo MR, Ciervo A, Rezza G, Castrucci MR, Lista F, on behalf of the ISS COVID-19 Study Group . 2020. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill 25:2000305. 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Rockett RJ, Arnott A, Lam C, Sadsad R, Timms V, Gray K-A, Eden J-S, Chang S, Gall M, Draper J, Sim EM, Bachmann NL, Carter I, Basile K, Byun R, O’Sullivan MV, Chen SC-A, Maddocks S, Sorrell TC, Dwyer DE, Holmes EC, Kok J, Prokopenko M, Sintchenko V. 2020. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med 26:1398–1404. 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 435.Woo H-J, Reifman J. 2012. A quantitative quasispecies theory-based model of virus escape mutation under immune selection. Proc Natl Acad Sci U S A 109:12980–12985. 10.1073/pnas.1117201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Petrova VN, Russell CA. 2018. The evolution of seasonal influenza viruses. Nat Rev Microbiol 16:47–60. 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 437.Volkov I, Pepin KM, Lloyd-Smith JO, Banavar JR, Grenfell BT. 2010. Synthesizing within-host and population-level selective pressures on viral populations: the impact of adaptive immunity on viral immune escape. J R Soc Interface 7:1311–1318. 10.1098/rsif.2009.0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.World Health Organization. 2020. SARS-CoV-2 variants, updated December 31, 2020. World Health Organization, Geneva, Switzerland. https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/. Accessed 8 February 2021. [Google Scholar]
- 439.US Centers for Disease Control and Prevention. 2021. Emerging SARS-CoV-2 variants, updated January 28, 2021. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html. Accessed 2 February 2021. [Google Scholar]
- 440.Sanjuán R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Hasan MR, Sundararaju S, Manickam C, Mirza F, Al-Hail H, Lorenz S, Tang P. 2021. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by reverse transcription-quantitative PCR. J Clin Microbiol 59:e03278-20. 10.1128/JCM.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Chan WS, Au CH, Lam HY, Wang CLN, Ho DN-Y, Lam YM, Chu DKW, Poon LLM, Chan TL, Zee JS-T, Ma ESK, Tang BSF. 2020. Evaluation on the use of Nanopore sequencing for direct characterization of coronaviruses from respiratory specimens, and a study on emerging missense mutations in partial RdRP gene of SARS-CoV-2. Virol J 17:183. 10.1186/s12985-020-01454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Rooney B-L, Pettipas J, Grudeski E, Mykytczuk O, Pang X-L, Booth TF, Hatchette TF, LeBlanc JJ. 2014. Detection of circulating norovirus genotypes: hitting a moving target. Virol J 11:129. 10.1186/1743-422X-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Franco-Muñoz C, Álvarez-Díaz DA, Laiton-Donato K, Wiesner M, Escandón P, Usme-Ciro JA, Franco-Sierra ND, Flórez-Sánchez AC, Gómez-Rangel S, Rodríguez-Calderon LD, Barbosa-Ramirez J, Ospitia-Baez E, Walteros DM, Ospina-Martinez ML, Mercado-Reyes M. 2020. Substitutions in spike and nucleocapsid proteins of SARS-CoV-2 circulating in South America. Infect Genet Evol 85:104557. 10.1016/j.meegid.2020.104557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Charre C, Ginevra C, Sabatier M, Regue H, Destras G, Brun S, Burfin G, Scholtes C, Morfin F, Valette M, Lina B, Bal A, Josset L. 2020. Evaluation of NGS-based approaches for SARS-CoV-2 whole genome characterisation. Virus Evol 6:veaa075. 10.1093/ve/veaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Wang C, Liu Z, Chen Z, Huang X, Xu M, He T, Zhang Z. 2020. The establishment of reference sequence for SARS‐CoV‐2 and variation analysis. J Med Virol 92:667–674. 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Zuckerman NS, Pando R, Bucris E, Drori Y, Lustig Y, Erster O, Mor O, Mendelson E, Mandelboim M. 2020. Comprehensive analyses of SARS-CoV-2 transmission in a public health virology laboratory. Viruses 12:854. 10.3390/v12080854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Pillay S, Giandhari J, Tegally H, Wilkinson E, Chimukangara B, Lessells R, Moosa Y, Mattison S, Gazy I, Fish M, Singh L, Khanyile KS, San JE, Fonseca V, Giovanetti M, Alcantara LC, Jr, de Oliveira T. 2020. Whole genome sequencing of SARS-CoV-2: adapting Illumina protocols for quick and accurate outbreak investigation during a pandemic. Genes (Basel) 11:949. 10.3390/genes11080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Alm E, Broberg EK, Connor T, Hodcroft EB, Komissarov AB, Maurer-Stroh S, Melidou A, Neher RA, O’Toole Á, Pereyaslov D, WHO European Region Sequencing Laboratories and GISAID EpiCoV Group . 2020. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Euro Surveill 25:2001410. 10.2807/1560-7917.ES.2020.25.32.2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Krammer F. 2020. SARS-CoV-2 vaccines in development. Nature 586:516–527. 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 452.Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. 2020. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther 5:237. 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, Monego SD, Pantano E, Manganaro N, Manenti A, Manna R, Casa E, Hyseni I, Benincasa L, Montomoli E, Amaro RE, McLellan JS, Rappuoli R. 2020. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed]
- 454.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. 2021. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv 10.1101/2020.12.31.425021. [DOI] [PMC free article] [PubMed]
- 455.Nonaka CKV, Franco MM, Gräf T, Mendes AVA, de Aguiar RS, Giovanetti M, Souza BSDF. 2021. Genomic evidence of a Sars-Cov-2 reinfection case with E484K spike mutation in Brazil. Preprints 10.20944/preprints202101.0132.v1. [DOI] [PMC free article] [PubMed]
- 456.Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, Griego-Fullbright C, Burgard C, Fernandez C, Eckert K, Andrews JC, Ren H, Allen J, Ackerman R, Cooper CK. 2021. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS antigen point-of-care test. J Clin Microbiol 59:e02338-20. 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, Arroyo T, Cuadros J. 2020. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol 133:104659. 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso A-M, Le Goff J, Delaugerre C. 2020. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol 58:e00977-20. 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, Cuypers L, Van den Wijngaert S, Monteil V, Melin P, Stoffels K, Yin N, Mileto D, Delaunoy S, Magein H, Lagrou K, Bouzet J, Serrano G, Wautier M, Leclipteux T, Van Ranst M, Vandenberg O, LHUB-ULB SARS-CoV-2 Working Diagnostic Group . 2020. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front Med (Lausanne) 7:225. 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, Pizarro G, Vial P, Iruretagoyena M, Dittrich S, Weitzel T. 2020. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis 99:328–333. 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. 2020. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol 129:104455. 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, Chen J, Pan Y, Chen L, Dan Y, Wang J, Chen Y, Deng G, Zhou H, Wu Y. 2020. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv 10.1101/2020.03.07.20032524. [DOI]
- 463.Mak GC, Cheng PK, Lau SS, Wong KK, Lau C, Lam ET, Chan RC, Tsang DN. 2020. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol 129:104500. 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Cui F, Zhou HS. 2020. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens Bioelectron 165:112349. 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG, Kim S-J, Lee J-O, Kim BT, Park EC, Kim SI. 2020. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14:5135–5142. 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 466.Mahari S, Roberts A, Shahdeo D, Gandhi S. 2020. eCovSens—ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19. bioRxiv 10.1101/2020.04.24.059204. [DOI]
- 467.Mina MJ, Parker R, Larremore DB. 2020. Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med 383:e120. 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 468.Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, Tambe M, Mina MJ, Parker R. 2020. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. medRxiv 10.1101/2020.06.22.20136309. [DOI] [PMC free article] [PubMed]
- 469.Paltiel AD, Zheng A, Walensky RP. 2020. Assessment of SARS-CoV-2 screening strategies to permit the safe reopening of college campuses in the United States. JAMA Netw Open 3:e2016818. 10.1001/jamanetworkopen.2020.16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Chin ET, Huynh BQ, Murrill M, Basu S, Lo NC. 2020. Frequency of routine testing for COVID-19 in high-risk environments to reduce workplace outbreaks. medRxiv 10.1101/2020.04.30.20087015. [DOI] [PMC free article] [PubMed]
- 471.Krammer F, Simon V. 2020. Serology assays to manage COVID-19. Science 368:1060–1061. 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 472.Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. 2020. Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics 10:319. 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Pisanic N, Randad PR, Kruczynski K, Manabe YC, Thomas DL, Pekosz A, Klein SL, Betenbaugh MJ, Clarke WA, Laeyendecker O, Caturegli PP, Larman HB, Detrick B, Fairley JK, Sherman AC, Rouphael N, Edupuganti S, Granger DA, Granger SW, Collins MH, Heaney CD. 2021. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. J Clin Microbiol 59:e02204-20. 10.1128/JCM.02204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Sun B, Feng Y, Mo X, Zheng P, Wang Q, Li P, Peng P, Liu X, Chen Z, Huang H, Zhang F, Luo W, Niu X, Hu P, Wang L, Peng H, Huang Z, Feng L, Li F, Zhang F, Li F, Zhong N, Chen L. 2020. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect 9:940–948. 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Zhang G, Nie S, Zhang Z, Zhang Z. 2020. Longitudinal change of severe acute respiratory syndrome coronavirus 2 antibodies in patients with coronavirus disease 2019. J Infect Dis 222:183–188. 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Kofler N, Baylis F. 2020. Ten reasons why immunity passports are a bad idea. Nature 581:379–381. 10.1038/d41586-020-01451-0. [DOI] [PubMed] [Google Scholar]
- 477.Weinstein MC, Freedberg KA, Hyle EP, Paltiel AD. 2020. Waiting for certainty on Covid-19 antibody tests—at what cost? N Engl J Med 383:e37. 10.1056/NEJMp2017739. [DOI] [PubMed] [Google Scholar]
- 478.Jiang S, Hillyer C, Du L. 2020. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 41:355–359. 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Espejo AP, Akgun Y, Al Mana AF, Tjendra Y, Millan NC, Gomez-Fernandez C, Cray C. 2020. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol 154:293–304. 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, Vanderheiden A, Nyhoff L, Davis CW, Adekunle O, Affer M, Sherman M, Reynolds S, Verkerke HP, Alter DN, Guarner J, Bryksin J, Horwath MC, Arthur CM, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson EJ, Menachery VD, Rouphael N, Mehta AK, Stephens DS, Ahmed R, Roback JD, Wrammert J. 2020. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med 1:100040. 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Sterlin D, Mathian A, Miyara M, Mohr A, Anna F, Claër L, Quentric P, Fadlallah J, Devilliers H, Ghillani P, Gunn C, Hockett R, Mudumba S, Guihot A, Luyt C-E, Mayaux J, Beurton A, Fourati S, Bruel T, Schwartz O, Lacorte J-M, Yssel H, Parizot C, Dorgham K, Charneau P, Amoura Z, Gorochov G. 2021. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med 13:eabd2223. 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Peterhoff D, Glück V, Vogel M, Schuster P, Schütz A, Neubert P, Albert V, Frisch S, Kiessling M, Pervan P, Werner M, Ritter N, Babl L, Deichner M, Hanses F, Lubnow M, Müller T, Lunz D, Hitzenbichler F, Audebert F, Hähnel V, Offner R, Müller M, Schmid S, Burkhardt R, Glück T, Koller M, Niller HH, Graf B, Salzberger B, Wenzel JJ, Jantsch J, Gessner A, Schmidt B, Wagner R. 2021. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection 49:75–82. 10.1007/s15010-020-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Charlton CL, Kanji JN, Johal K, Bailey A, Plitt SS, MacDonald C, Kunst A, Buss E, Burnes LE, Fonseca K, Berenger BM, Schnabl K, Hu J, Stokes W, Zelyas N, Tipples G. 2020. Evaluation of six commercial mid- to high-volume antibody and six point-of-care lateral flow assays for detection of SARS-CoV-2 antibodies. J Clin Microbiol 58:e01361-20. 10.1128/JCM.01361-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Metcalf CJE, Farrar J, Cutts FT, Basta NE, Graham AL, Lessler J, Ferguson NM, Burke DS, Grenfell BT. 2016. Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet 388:728–730. 10.1016/S0140-6736(16)30164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Hosseini S, Vázquez-Villegas P, Rito-Palomares M, Martinez-Chapa SO. 2018. Enzyme-linked immunosorbent assay (ELISA): from A to Z. Springer Singapore, Singapore. [Google Scholar]
- 486.Zhao R, Li M, Song H, Chen J, Ren W, Feng Y, Gao GF, Song J, Peng Y, Su B, Guo X, Wang Y, Chen J, Li J, Sun H, Bai Z, Cao W, Zhu J, Zhang Q, Sun Y, Sun S, Mao X, Su J, Chen X, He A, Gao W, Jin R, Jiang Y, Sun L. 2020. Early detection of SARS-CoV-2 antibodies in COVID-19 patients as a serologic marker of infection. Clin Infect Dis 71:2066–2072. 10.1093/cid/ciaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma W-L. 2020. Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin Infect Dis 71:1930–1934. 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch B-J, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Tan X, Krel M, Dolgov E, Park S, Li X, Wu W, Sun Y-L, Zhang J, Khaing Oo MK, Perlin DS, Fan X. 2020. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens Bioelectron 169:112572. 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Cinquanta L, Fontana DE, Bizzaro N. 2017. Chemiluminescent immunoassay technology: what does it change in autoantibody detection? Auto Immun Highlights 8:9. 10.1007/s13317-017-0097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Cai X-F, Chen J, Hu J-L, Long Q-X, Deng H-J, Liu P, Fan K, Liao P, Liu B-Z, Wu G-C, Chen Y-K, Li Z-J, Wang K, Zhang X-L, Tian W-G, Xiang J-L, Du H-X, Wang J, Hu Y, Tang N, Lin Y, Ren J-H, Huang L-Y, Wei J, Gan C-Y, Chen Y-M, Gao Q-Z, Chen A-M, He C-L, Wang D-X, Hu P, Zhou F-C, Huang A-L, Wang D-Q. 2020. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J Infect Dis 222:189–193. 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J, Dai Y, Xu Y, Cai Y, Chen X, Huang K, Zhang Z. 2020. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur J Clin Microbiol Infect Dis 39:2271–2277. 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Zhang QY, Chen H, Lin Z, Lin JM. 2012. Comparison of chemiluminescence enzyme immunoassay based on magnetic microparticles with traditional colorimetric ELISA for the detection of serum α-fetoprotein. J Pharm Anal 2:130–135. 10.1016/j.jpha.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Wang X, Lin J-M, Ying X. 2007. Evaluation of carbohydrate antigen 50 in human serum using magnetic particle-based chemiluminescence enzyme immunoassay. Anal Chim Acta 598:261–267. 10.1016/j.aca.2007.07.052. [DOI] [PubMed] [Google Scholar]
- 496.Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT, Cohen JI. 2020. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. medRxiv 10.1101/2020.04.20.20071423. [DOI] [PMC free article] [PubMed]
- 497.Norman M, Gilboa T, Ogata AF, Maley AM, Cohen L, Cai Y, Zhang J, Feldman JE, Hauser BM, Caradonna TM, Chen B, Schmidt AG, Alter G, Charles RC, Ryan ET, Walt DR. 2020. Ultra-sensitive high-resolution profiling of anti-SARS-CoV-2 antibodies for detecting early seroconversion in COVID-19 patients. medRxiv 10.1101/2020.04.28.20083691. [DOI]
- 498.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. 2020. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 92:1518–1524. 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Koczula KM, Gallotta A. 2016. Lateral flow assays. Essays Biochem 60:111–120. 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Yoon J-Y. 2016. Introduction to biosensors: from electric circuits to immunosensors, 2nd ed. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 501.Montesinos I, Gruson D, Kabamba B, Dahma H, Van den Wijngaert S, Reza S, Carbone V, Vandenberg O, Gulbis B, Wolff F, Rodriguez-Villalobos H. 2020. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol 128:104413. 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Caini S, Bellerba F, Corso F, Díaz-Basabe A, Natoli G, Paget J, Facciotti F, De Angelis SP, Raimondi S, Palli D, Mazzarella L, Pelicci PG, Vineis P, Gandini S. 2020. Meta-analysis of diagnostic performance of serological tests for SARS-CoV-2 antibodies up to 25 April 2020 and public health implications. Euro Surveill 25:2000980. 10.2807/1560-7917.ES.2020.25.23.2000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Chen X, Chen Z, Azman AS, Deng X, Chen X, Lu W, Zhao Z, Yang J, Viboud C, Ajelli M, Leung DT, Yu H. 2020. Serological evidence of human infection with SARS-CoV-2: a systematic review and meta-analysis. medRxiv 10.1101/2020.09.11.20192773. [DOI] [PMC free article] [PubMed]
- 504.Lv H, Wu NC, Tsang OT-Y, Yuan M, Perera RAPM, Leung WS, So RTY, Chan JMC, Yip GK, Chik TSH, Wang Y, Choi CYC, Lin Y, Ng WW, Zhao J, Poon LLM, Peiris JSM, Wilson IA, Mok CKP. 2020. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep 31:107725. 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Lipsitch M, Grad YH, Sette A, Crotty S. 2020. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol 20:709–713. 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Agrawal M, Sinai Immunology Review Project . 2020. Immunology of COVID-19: current state of the science. Immunity 52:910–941. 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Zohar T, Alter G. 2020. Dissecting antibody-mediated protection against SARS-CoV-2. Nat Rev Immunol 20:392–394. 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Ni L, Ye F, Cheng M-L, Feng Y, Deng Y-Q, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin C, Chen F, Dong C. 2020. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52:971–977.e3. 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. 2020. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol 11:1949. 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Atkinson B, Petersen E. 2020. SARS-CoV-2 shedding and infectivity. Lancet 395:1339–1340. 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Perera RAPM, Mok CK, Tsang OTY, Lv H, Ko RLW, Wu NC, Yuan M, Leung WS, Chan JMC, Chik TSH, Choi CYC, Leung K, Chan KH, Chan KCK, Li K-C, Wu JT, Wilson IA, Monto AS, Poon LLM, Peiris M. 2020. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill 25:2000421. 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, van Buskirk C, Grossman BJ, Joyner M, Henderson JP, Pekosz A, Lau B, Wesolowski A, Katz L, Shan H, Auwaerter PG, Thomas D, Sullivan DJ, Paneth N, Gehrie E, Spitalnik S, Hod EA, Pollack L, Nicholson WT, Pirofski L, Bailey JA, Tobian AAR. 2020. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest 130:2757–2765. 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, Peng C, Yuan M, Huang J, Wang Z, Yu J, Gao X, Wang D, Yu X, Li L, Zhang J, Wu X, Li B, Xu Y, Chen W, Peng Y, Hu Y, Lin L, Liu X, Huang S, Zhou Z, Zhang L, Wang Y, Zhang Z, Deng K, Xia Z, Gong Q, Zhang W, Zheng X, Liu Y, Yang H, Zhou D, Yu D, Hou J, Shi Z, Chen S, Chen Z, Zhang X, Yang X. 2020. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A 117:9490–9496. 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Kellam P, Barclay W. 2020. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol 101:791–797. 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.D’Cruz RJ, Currier AW, Sampson VB. 2020. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Front Cell Dev Biol 8:468. 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, Zhang L, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Fan C, Huang W, Xu M, Wang Y. 2020. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9:680–686. 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Manenti A, Maggetti M, Casa E, Martinuzzi D, Torelli A, Trombetta CM, Marchi S, Montomoli E. 2020. Evaluation of SARS‐CoV‐2 neutralizing antibodies using a CPE‐based colorimetric live virus micro‐neutralization assay in human serum samples. J Med Virol 92:2096–2104. 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Schmidt F, Weisblum Y, Muecksch F, Hoffmann H-H, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, Bieniasz PD. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217:e20201181. 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12:513. 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, Gong Y, Xiao H, Fan Z, Tan S, Wu G, Tan W, Lu X, Fan C, Wang Q, Liu Y, Zhang C, Qi J, Gao GF, Gao F, Liu L. 2020. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368:1274–1278. 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JF-W, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen K-Y, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD. 2020. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584:450–456. 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 522.Byrnes JR, Zhou XX, Lui I, Elledge SK, Glasgow JE, Lim SA, Loudermilk RP, Chiu CY, Wang TT, Wilson MR, Leung KK, Wells JA. 2020. Competitive SARS-CoV-2 serology reveals most antibodies targeting the spike receptor-binding domain compete for ACE2 binding. mSphere 5:e00802-20. 10.1128/mSphere.00802-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, Hong W, Yang Y, Zhao Y, Ye F, Lin S, Deng W, Chen H, Lei H, Zhang Z, Luo M, Gao H, Zheng Y, Gong Y, Jiang X, Xu Y, Lv Q, Li D, Wang M, Li F, Wang S, Wang G, Yu P, Qu Y, Yang L, Deng H, Tong A, Li J, Wang Z, Yang J, Shen G, Zhao Z, Li Y, Luo J, Liu H, Yu W, Yang M, Xu J, Wang J, Li H, Wang H, et al. 2020. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586:572–577. 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 524.Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, Budylowski P, Dupuis AP, II, Girardin RC, Rathod B, Wang JH, Barrios-Rodiles M, Colwill K, McGeer AJ, Mubareka S, Gommerman JL, Durocher Y, Ostrowski M, McDonough KA, Drebot MA, Drews SJ, Rini JM, Gingras A-C. 2020. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight 5:e142362. 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Barnes CO, West AP, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB, Gaebler C, Muecksch F, Lorenzi JCC, Finkin S, Hägglöf T, Hurley A, Millard KG, Weisblum Y, Schmidt F, Hatziioannou T, Bieniasz PD, Caskey M, Robbiani DF, Nussenzweig MC, Bjorkman PJ. 2020. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182:828–842.e16. 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Tan CW, Chia WN, Qin X, Liu P, Chen MI-C, Tiu C, Hu Z, Chen VC-W, Young BE, Sia WR, Tan Y-J, Foo R, Yi Y, Lye DC, Anderson DE, Wang L-F. 2020. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol 38:1073–1078. 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 527.Tang MS, Case JB, Franks CE, Chen RE, Anderson NW, Henderson JP, Diamond MS, Gronowski AM, Farnsworth CW. 2020. Association between SARS-CoV-2 neutralizing antibodies and commercial serological assays. Clin Chem 66:1538–1547. 10.1093/clinchem/hvaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Meschi S, Colavita F, Bordi L, Matusali G, Lapa D, Amendola A, Vairo F, Ippolito G, Capobianchi MR, Castilletti C, INMICovid-19 Laboratory Team . 2020. Performance evaluation of Abbott ARCHITECT SARS-CoV-2 IgG immunoassay in comparison with indirect immunofluorescence and virus microneutralization test. J Clin Virol 129:104539. 10.1016/j.jcv.2020.104539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. 2020. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol 129:104480. 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Jääskeläinen A, Kuivanen S, Kekäläinen E, Ahava M, Loginov R, Kallio-Kokko H, Vapalahti O, Jarva H, Kurkela S, Lappalainen M. 2020. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 129:104512. 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O’Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeño JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O’Connell L, O’Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, Doores KJ. 2020. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 5:1598–1607. 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Yongchen Z, Shen H, Wang X, Shi X, Li Y, Yan J, Chen Y, Gu B. 2020. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect 9:833–836. 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, Adriano A, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Dittrich S, Emperador D, Hooft L, Leeflang MMG, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group . 2020. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev 6:CD013652. 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Mizumoto K, Kagaya K, Zarebski A, Chowell G. 2020. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill 25:2000180. 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Nishiura H, Kobayashi T, Miyama T, Suzuki A, Jung S, Hayashi K, Kinoshita R, Yang Y, Yuan B, Akhmetzhanov AR, Linton NM. 2020. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int J Infect Dis 94:154–155. 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, López-Camacho C, Slon-Campos J, Zhao Y, Stuart DI, Paesen GC, Grimes JM, Antson AA, Bayfield OW, Hawkins DEDP, Ker D-S, Wang B, Turtle L, Subramaniam K, Thomson P, Zhang P, Dold C, Ratcliff J, Simmonds P, de Silva T, Sopp P, Wellington D, Rajapaksa U, Chen Y-L, Salio M, Napolitani G, Paes W, Borrow P, Kessler BM, Fry JW, Schwabe NF, Semple MG, Baillie JK, Moore SC, Openshaw PJM, Ansari MA, Dunachie S, Barnes E, Frater J, Kerr G, Goulder P, Lockett T, et al. 2020. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol 21:1336–1345. 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Altmann DM, Boyton RJ. 2020. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci Immunol 5:eabd6160. 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- 538.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI-C, Wang L-F, Ooi EE, Kalimuddin S, Tambyah PA, Low JG-H, Tan Y-J, Bertoletti A. 2020. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584:457–462. 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 539.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, Wei J, Xiao H, Yang Y, Qu J, Qing L, Chen L, Xu Z, Peng L, Li Y, Zheng H, Chen F, Huang K, Jiang Y, Liu D, Zhang Z, Liu Y, Liu L. 2020. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA 323:1582–1589. 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Amanat F, Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity 52:583–589. 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Dagotto G, Yu J, Barouch DH. 2020. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe 28:364–370. 10.1016/j.chom.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, Li Q, Jiang C, Zhou Y, Liu S, Ye C, Zhang P, Xing Y, Guo H, Tang W. 2020. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect 81:e16–e25. 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Tian W, Jiang W, Yao J, Nicholson CJ, Li RH, Sigurslid HH, Wooster L, Rotter JI, Guo X, Malhotra R. 2020. Predictors of mortality in hospitalized COVID‐19 patients: a systematic review and meta‐analysis. J Med Virol 92:1875–1883. 10.1002/jmv.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Xu L, Mao Y, Chen G. 2020. Risk factors for 2019 novel coronavirus disease (COVID-19) patients progressing to critical illness: a systematic review and meta-analysis. Aging (Albany NY) 12:12410–12421. 10.18632/aging.103383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Binkhamis K, Gillis H, Lafreniere JD, Hiebert J, Mendoza L, Pettipas J, Severini A, Hatchette TF, LeBlanc JJ. 2017. Comparison of monoplex and duplex RT-PCR assays for the detection of measles virus. J Virol Methods 239:58–60. 10.1016/j.jviromet.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 546.Leblanc JJ, Pettipas J, Davidson RJ, Tipples GA, Hiebert J, Hatchette TF. 2008. Detection of mumps virus RNA by real-time one-step reverse transcriptase PCR using the LightCycler platform. J Clin Microbiol 46:4049–4051. 10.1128/JCM.01446-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Andrew MK, McElhaney JE, McGeer AA, Hatchette TF, Leblanc J, Webster D, Bowie W, Poirier A, Nichols MK, McNeil SA, Canadian Immunization Research Network (CIRN) Serious Outcomes Surveillance Network Investigators . 2020. Influenza surveillance case definitions miss a substantial proportion of older adults hospitalized with laboratory-confirmed influenza: a report from the Canadian Immunization Research Network (CIRN) Serious Outcomes Surveillance (SOS) Network. Infect Control Hosp Epidemiol 41:499–504. 10.1017/ice.2020.22. [DOI] [PubMed] [Google Scholar]
- 548.Andrew MK, MacDonald S, Godin J, McElhaney JE, LeBlanc J, Hatchette TF, Bowie W, Katz K, McGeer A, Semret M, McNeil SA. 2021. Persistent functional decline following hospitalization with influenza or acute respiratory illness. J Am Geriatr Soc 69:696–703. 10.1111/jgs.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Nichols MK, Andrew MK, Ye L, Hatchette TF, Ambrose A, Boivin G, Bowie W, Dos Santos G, Elsherif M, Green K, Haguinet F, Katz K, Leblanc J, Loeb M, MacKinnon-Cameron D, McCarthy A, McElhaney JE, McGeer A, Powis J, Richardson D, Semret M, Sharma R, Shinde V, Smyth D, Trottier S, Valiquette L, Webster D, McNeil SA, Serious Outcomes Surveillance (SOS) Network of the Canadian Immunization Research Network (CIRN) . 2019. The impact of prior season vaccination on subsequent influenza vaccine effectiveness to prevent influenza-related hospitalizations over 4 influenza seasons in Canada. Clin Infect Dis 69:970–979. 10.1093/cid/ciy1009. [DOI] [PubMed] [Google Scholar]
- 550.McNeil SA, Andrew MK, Ye L, Haguinet F, Hatchette TF, ElSherif M, LeBlanc J, Ambrose A, McGeer A, McElhaney JE, Loeb M, MacKinnon-Cameron D, Sharma R, Dos Santos G, Shinde V, on behalf of the Investigators of the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network . 2015. Interim estimates of 2014/15 influenza vaccine effectiveness in preventing laboratory-confirmed influenza-related hospitalisation from the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network, January 2015. Euro Surveill 20:21024. 10.2807/1560-7917.ES2015.20.5.21024. [DOI] [PubMed] [Google Scholar]
- 551.McNeil SA, Shinde V, Andrew M, Hatchette TF, LeBlanc J, Ambrose A, Boivin G, Bowie WR, Diaz-Mitoma F, ElSherif M, Green K, Haguinet F, Halperin S, Ibarguchi B, Katz K, Langley JM, Lagacé-Wiens P, Light B, Loeb M, McElhaney JE, MacKinnon-Cameron D, McCarthy AE, Poirier M, Powis J, Richardson D, Semret M, Smith S, Smyth D, Stiver G, Trottier S, Valiquette L, Webster D, Ye L, McGeer A, on behalf of the Public Health Agency of Canada/Canadian Institutes of Health Research Influenza Research Network Serious Outcomes Surveillance Network, Toronto Invasive Bacterial Diseases Network . 2014. Interim estimates of 2013/14 influenza clinical severity and vaccine effectiveness in the prevention of laboratory-confirmed influenza-related hospitalisation, Canada, February 2014. Euro Surveill 19:20729. 10.2807/1560-7917.ES2014.19.9.20729. [DOI] [PubMed] [Google Scholar]
- 552.Ng C, Ye L, Noorduyn SG, Hux M, Thommes E, Goeree R, Ambrose A, Andrew MK, Hatchette T, Boivin G, Bowie W, ElSherif M, Green K, Johnstone J, Katz K, Leblanc J, Loeb M, MacKinnon-Cameron D, McCarthy A, McElhaney J, McGeer A, Poirier A, Powis J, Richardson D, Sharma R, Semret M, Smith S, Smyth D, Stiver G, Trottier S, Valiquette L, Webster D, McNeil SA, Serious Outcomes Surveillance Network of the Canadian Immunization Research Network (CIRN) Investigators, Toronto Invasive Bacterial Diseases Network (TIBDN) Investigators . 2018. Resource utilization and cost of influenza requiring hospitalization in Canadian adults: a study from the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network. Influenza Other Respir Viruses 12:232–240. 10.1111/irv.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Mulpuru S, Li L, Ye L, Hatchette T, Andrew MK, Ambrose A, Boivin G, Bowie W, Chit A, Dos Santos G, ElSherif M, Green K, Haguinet F, Halperin SA, Ibarguchi B, Johnstone J, Katz K, Langley JM, LeBlanc J, Loeb M, MacKinnon-Cameron D, McCarthy A, McElhaney JE, McGeer A, Powis J, Richardson D, Semret M, Shinde V, Smyth D, Trottier S, Valiquette L, Webster D, McNeil SA, Lagace-Wiens P, Light B, Poirier A, Smith S, Taylor G. 2019. Effectiveness of influenza vaccination on hospitalizations and risk factors for severe outcomes in hospitalized patients with COPD. Chest 155:69–78. 10.1016/j.chest.2018.10.044. [DOI] [PubMed] [Google Scholar]
- 554.Olsen SJ, Azziz-Baumgartner E, Budd AP, Brammer L, Sullivan S, Pineda RF, Cohen C, Fry AM. 2020. Decreased influenza activity during the COVID-19 pandemic—United States, Australia, Chile, and South Africa, 2020. MMWR Morb Mortal Wkly Rep 69:1305–1309. 10.15585/mmwr.mm6937a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Singer BD. 2020. COVID-19 and the next influenza season. Sci Adv 6:eabd0086. 10.1126/sciadv.abd0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Zhang K, Misra A, Kim PJ, Moghadas SM, Langley JM, Smieja M. 2020. Rapid disappearance of influenza following the implementation of COVID-19 mitigation measures in Hamilton, Ontario. medRxiv 10.1101/2020.11.27.20240036. [DOI] [PMC free article] [PubMed]
- 557.Konala VM, Adapa S, Naramala S, Chenna A, Lamichhane S, Garlapati PR, Balla M, Gayam V. 2020. A case series of patients coinfected with influenza and COVID-19. J Invest Med High Impact Case Rep 8:232470962093467. 10.1177/2324709620934674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Nowak MD, Sordillo EM, Gitman MR, Paniz Mondolfi AE. 2020. Coinfection in SARS‐CoV‐2 infected patients: where are influenza virus and rhinovirus/enterovirus? J Med Virol 92:1699–1700. 10.1002/jmv.25953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.US Centers for Disease Control and Prevention. 2021. How to report COVID-19 laboratory data, updated January 5, 2021. US Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/reporting-lab-data.html#what-to-include. Accessed 11 January 2021. [Google Scholar]
- 560.Baumgart DC. 2020. Digital advantage in the COVID-19 response: perspective from Canada’s largest integrated digitalized healthcare system. NPJ Digit Med 3:114. 10.1038/s41746-020-00326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Cheng MP, Papenburg J, Desjardins M, Kanjilal S, Quach C, Libman M, Dittrich S, Yansouni CP. 2020. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2. Ann Intern Med 172:726–734. 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Bedford J, Enria D, Giesecke J, Heymann DL, Ihekweazu C, Kobinger G, Lane HC, Memish Z, Oh M-D, Sall AA, Schuchat A, Ungchusak K, Wieler LH, WHO Strategic and Technical Advisory Group for Infectious Hazards . 2020. COVID-19: towards controlling of a pandemic. Lancet 395:1015–1018. 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Wood CS, Thomas MR, Budd J, Mashamba-Thompson TP, Herbst K, Pillay D, Peeling RW, Johnson AM, McKendry RA, Stevens MM. 2019. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature 566:467–474. 10.1038/s41586-019-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Nayak S, Blumenfeld NR, Laksanasopin T, Sia SK. 2017. Point-of-care diagnostics: recent developments in a connected age. Anal Chem 89:102–123. 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Sheridan C. 2020. COVID-19 spurs wave of innovative diagnostics. Nat Biotechnol 38:769–772. 10.1038/s41587-020-0597-x. [DOI] [PubMed] [Google Scholar]
- 566.Rowley AH. 2020. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat Rev Immunol 20:453–454. 10.1038/s41577-020-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Kaushik A, Gupta S, Sood M, Sharma S, Verma S. 2020. A systematic review of multisystem inflammatory syndrome in children associated with SARS-CoV-2 infection. Pediatr Infect Dis J 39:e340–e346. 10.1097/INF.0000000000002888. [DOI] [PubMed] [Google Scholar]
- 568.Whittaker E, Bamford A, Kenny J, Kaforou M, Jones CE, Shah P, Ramnarayan P, Fraisse A, Miller O, Davies P, Kucera F, Brierley J, McDougall M, Carter M, Tremoulet A, Shimizu C, Herberg J, Burns JC, Lyall H, Levin M, PIMS-TS Study Group and EUCLIDS and PERFORM Consortia . 2020. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA 324:259–269. 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Feldstein LR, Rose EB, Horwitz SM, Collins JP, Newhams MM, Son MBF, Newburger JW, Kleinman LC, Heidemann SM, Martin AA, Singh AR, Li S, Tarquinio KM, Jaggi P, Oster ME, Zackai SP, Gillen J, Ratner AJ, Walsh RF, Fitzgerald JC, Keenaghan MA, Alharash H, Doymaz S, Clouser KN, Giuliano JS, Jr, Gupta A, Parker RM, Maddux AB, Havalad V, Ramsingh S, Bukulmez H, Bradford TT, Smith LS, Tenforde MW, Carroll CL, Riggs BJ, Gertz SJ, Daube A, Lansell A, Coronado Munoz A, Hobbs CV, Marohn KL, Halasa NB, Patel MM, Randolph AG, Overcoming COVID-19 Investigators, CDC COVID-19 Response Team . 2020. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med 383:334–346. 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Radia T, Williams N, Agrawal P, Harman K, Weale J, Cook J, Gupta A. 11 August 2020. Multi-system inflammatory syndrome in children & adolescents (MIS-C): a systematic review of clinical features and presentation. Paediatr Respir Rev 10.1016/j.prrv.2020.08.001. [DOI] [PMC free article] [PubMed]
- 571.Gámbaro F, Behillil S, Baidaliuk A, Donati F, Albert M, Alexandru A, Vanpeene M, Bizard M, Brisebarre A, Barbet M, Derrar F, van der Werf S, Enouf V, Simon-Loriere E. 2020. Introductions and early spread of SARS-CoV-2 in France, 24 January to 23 March 2020. Euro Surveill 25:2001200. 10.2807/1560-7917.ES.2020.25.26.2001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Amendola A, Bianchi S, Gori M, Colzani D, Canuti M, Borghi E, Raviglione MC, Zuccotti GV, Tanzi E. 2021. Evidence of SARS-CoV-2 RNA in an oropharyngeal swab specimen, Milan, Italy, early December 2019. Emerg Infect Dis 27:648–650. 10.3201/eid2702.204632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Hogan CA, Garamani N, Sahoo MK, Huang C, Zehnder J, Pinsky BA. 2020. Retrospective screening for SARS-CoV-2 RNA in California, USA, late 2019. Emerg Infect Dis 26:2487–2488. 10.3201/eid2610.202296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Kanji JN, Diggle M, Bulman D, Hume S, Taylor S, Kelln R, Haase S, Tomaszewski R, Walker C, Pabbaraju K, Li V, Croxen M, Zelyas N, Hinshaw D, Tipples G. 15 March 2021. Retrospective testing of respiratory specimens for COVID-19 to assess for earlier SARS-CoV-2 infections in Alberta, Canada. Off J Assoc Med Microbiol Infect Dis Can 10.3138/jammi-2020-0035. [DOI] [PMC free article] [PubMed]
- 575.Roche Molecular Systems. 2020. cobas SARS-CoV-2: instructions for use. Roche Molecular Systems, Branchburg, NJ. https://www.fda.gov/media/136049/download. Accessed 21 January 2021. [Google Scholar]
- 576.Abbott Diagnostics. 2020. Abbott RealTime SARS-CoV-2: instructions for use. Abbott Molecular Inc, Des Plaines, IL. https://www.fda.gov/media/136258/download. Accessed 21 September 2020. [Google Scholar]
- 577.NeuMoDx Molecular. 2021. NeuMoDx SARS-CoV-2 assay: instructions for use. NeuMoDx Molecular, Ann Arbor, MI. https://www.fda.gov/media/136565/download. Accessed 24 January 2021. [Google Scholar]
- 578.Hologic. 2021. Aptima SARS-CoV-2 assay (Panther system): instructions for use. Hologic, San Diego, CA. https://www.fda.gov/media/138096/download. Accessed 24 January 2021. [Google Scholar]
- 579.BioFire Diagnostics. 2020. BioFire Respiratory Panel 2.1 (RP2.1): instructions for use. BioFire Diagnostics, Salt Lake City, UT. https://www.fda.gov/media/137583/download. Accessed 21 January 2021. [Google Scholar]
- 580.Cepheid. 2021. Xpert Xpress SARS-CoV-2: instructions for use. Cepheid, Sunnyvale, CA. https://www.fda.gov/media/136314/download. Accessed 22 January 2021. [Google Scholar]
- 581.Abbott Diagnostics. 2020. ID NOW COVID-19: instructions for use. Abbott Diagnostics, Scarborough, ME. https://www.fda.gov/media/136525/download. Accessed 22 September 2020. [Google Scholar]
- 582.Sherlock Biosciences. 2021. Sherlock CRISPR SARS-CoV-2 kit: instructions for use. Sherlock Biosciences, Boston, MA. https://www.fda.gov/media/137746/download. Accessed 27 January 2021. [Google Scholar]
- 583.Mammoth Biosciences. 2020. SARS-CoV-2 DETECTR reagent kit: instructions for use. Mammoth Biosciences, Brisbane, CA. https://www.fda.gov/media/141765/download. Accessed 20 January 2021. [Google Scholar]
- 584.Illumina Inc. 2020. Illumina CovidSeq test: instructions for use. Illumina Inc, San Diego, CA. https://www.fda.gov/media/138776/download. Accessed 22 January 2021. [Google Scholar]
- 585.Abbott Diagnostics. 2020. BinaxNOW COVID-19 Ag card home test: instructions for use. Abbott Diagnostics, Scarborough, ME. https://www.fda.gov/media/144574/download. Accessed 11 January 2021. [Google Scholar]
- 586.Access Bio. 2020. CareStart COVID-19 antigen: instructions for use. Access Bio, Somerset, NJ. https://www.fda.gov/media/142919/download. Accessed 1 December 2020. [Google Scholar]
- 587.Quidel Corporation. 2020. QuickVue SARS antigen test: instructions for use. Quidel Corporation, San Diego, CA. https://www.fda.gov/media/144668/download. Accessed 27 January 2021. [Google Scholar]
- 588.Becton, Dickinson and Company. 2021. BD Veritor system: instructions for use. Becton, Dickinson and Company, Sparks, MD. https://www.fda.gov/media/139755/download. Accessed 22 January 2021. [Google Scholar]
- 589.Quidel Corporation. 2020. Sofia 2 Flu + SARS antigen FIA: instructions for use. Quidel Corporation, San Diego, CA. https://www.fda.gov/media/142704/download. Accessed 1 December 2020. [Google Scholar]
- 590.LumiraDx. 2020. LumiraDx SARS-CoV-2 Ag test: instruction for use. LumiraDx, Waltham, MA. https://www.fda.gov/media/141304/download. Accessed 1 December 2020. [Google Scholar]
- 591.Luminostics Inc. 2020. Clip COVID rapid antigen test: instructions for use. Luminostics Inc, Milpitas, CA. https://www.fda.gov/media/144256/download. Accessed 22 January 2021. [Google Scholar]
- 592.Ellume Limited. 2020. Ellume COVID-19 home test: instructions for use. Ellume Limited, Brisbane, Queensland, Australia. https://www.fda.gov/media/144592/download. Accessed 21 January 2021. [Google Scholar]