Abstract
Objective
MicroRNAs (miRNAs) with functional relevance have not been previously identified in mantle cell lymphoma (MCL). Here, we aimed to evaluate the relationships between miR-34a and miR-155-5p and MCL clinicopathology and prognosis.
Methods
Seventy-five paraffin-embedded tissue samples from patients with MCL who completed at least four cycles of chemotherapy from January 2006 to October 2016, and 27 samples from control patients with reactive lymphoid hyperplasia (RLH), were collected. MiRNA expression levels were measured by qRT-PCR.
Results
The miR-155-5p levels were significantly higher in patients with MCL than in the controls. The Eastern Cooperative Oncology Group (ECOG) ≥ 2 and Sex-Determining Region Y-Box transcription factor 11 (SOX11) < median value (M) groups presented lower miR-34a expression than the ECOG < 2 and SOX11 ≥ M groups, respectively. MiR-155-5p expression differed between low, intermediate, and high MCL International Prognostic Index risk groups. The AUCs of miR-34a and miR-155-5p were 0.5819 and 0.7784, respectively. The median survival times of the miR-34a ≤ 0.2150 and miR-155-5p > 2.11 groups were shorter than those of the miR-34a > 0.2150 and miR-155-5p ≤ 2.11 groups, respectively.
Conclusions
Low miR-34a and elevated miR-155-5p levels may be correlated with poor prognosis in MCL.
Keywords: Mantle cell lymphoma, miR-155-5p, miR-34a, prognostic marker, survival, SOX11
Introduction
Mantle cell lymphoma (MCL) is a subtype of mature B-cell lymphoma that accounts for 5% to 10% of non-Hodgkin's lymphoma cases and has a median patient age of 60 to 65 years. It originates from B cells in the mantle zone of the lymph node, and extranodal invasion is common. The median survival time of patients with MCL is significantly lower than that of patients with other subtypes of B-cell lymphoma.1,2 There has been no substantial improvement in patient survival with the common treatment protocols for MCL, including the use of high-dose Ara-c and rituximab, a CD20 antibody, although autologous peripheral blood stem cell transplantation (APBSCT) improves the long-term remission rate.3
The characteristic cytogenetics of MCL include a translocation between chromosomes 11 and 14, that is, t (11;14), which leads to the overexpression of cyclin D1.1,4 In addition to this major genetic alteration, most MCL cases have several recurrent secondary mutations with important roles in disease pathogenesis.5 These mutations occur in coding genes involved in various cell functions, including cell cycle regulation, post-DNA damage responses, and signaling pathways. Recent studies indicate that genes in these pathways may be downregulated by microRNAs (miRNAs), and these changes in gene expression may be associated with clinicopathological features.6,7 Interestingly, studies have suggested an association between miRNAs and MCL prognosis.7,8
MiRNAs are small non-coding single-stranded RNA molecules and are typically 19 to 25 nucleotides in length. They play a direct role in the post-transcriptional regulation of many genes, some of which are involved in cell differentiation, proliferation, and apoptosis.9,10 Abnormal expression of multiple miRNAs has been detected in several solid tumors and hematologic neoplasms, including lymphoma.11–13 In 2002, Calin et al. demonstrated that a deletion at 13q14.3 leads to a decrease in miR-15/miR-16 expression, thereby affecting the development of chronic lymphocytic leukemia (CLL).14 Subsequent studies have identified various miRNAs that play important roles in lymphoma development and prognosis by regulating the expression of target genes.12,15 For example, abnormalities in the expression of miR-17–92, miR-34a, miR-150, and miR-155-5p are common events in B-cell tumorigenesis.6 MiR-34a is involved in the regulation of the tumor suppressor gene TP53 via forkhead box protein P1 (FOXP1) and B-cell lymphoma 2 (BCL2), thus affecting the differentiation and apoptosis of B cells. The decreased expression levels of miR-34a in lymphomas, such as diffuse large B-cell lymphoma (DLBCL), are associated with prognosis.6,16 The causal role of miR-155-5p in lymphoma has been established in two mouse model studies. Functional studies of miR-155-5p in anaplastic large cell lymphoma have demonstrated its association with lymphocyte differentiation and immunity.17 Furthermore, elevated expression of miR-155-5p is associated with the transformation of monoclonal B lymphocytosis (MBL) to CLL, a poor therapeutic response, and short survival in patients with CLL.18
Only a few studies have evaluated the role of miRNAs in MCL. Some studies have reported SOX11 as a critical gene with diagnostic value,19,20 but it may also have prognostic value.21 Furthermore, the biological information databases TargetScan and miRdb predict that miR-155-5p may participate in the regulation of Sex-Determining Region Y-Box transcription factor 11 (SOX11), as indicated by a systematic search of miRNAs targeting SOX11, a definitive diagnostic indicator of MCL.22 In the present study, we identified miR-34a and miR-155 as candidate markers, evaluated their expression, and assessed their prognostic value in MCL.
Patients and methods
Patients
We obtained paraffin-embedded MCL tissue samples (excluding biopsy specimens) from the Department of Pathology, Shanxi Tumor Hospital, and Shanxi Provincial People’s Hospital from January 2006 to October 2016. Patients with detailed clinical data who had completed at least four cycles of chemotherapy including the CHOP/R-CHOP or HyperCAVD/R-HyperCAVD regimen3 were selected and subjected to follow-up. Subsequently, 75 samples from patients with MCL and 27 samples from patients with reactive lymphoid hyperplasia (RLH) (controls) were obtained. The baseline characteristics of the cases were the same as those previously reported by He et al.23 The study was approved by the Ethics Committee of Shanxi Tumor Hospital (Taiyuan, China; Date of approval: 29 July 2019; Approval number: 201935) and written informed consent was obtained from all participants. It was performed in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The reporting of this study conforms to the STROBE statement.24
Exemption
The study was exempted from ethical committee review owing to its retrospective nature.
Quantitative real-time-polymerase chain reaction (qRT-PCR)
The Recover All Total Acid Isolation Kit (ABI AM1975; Thermo Fisher Scientific, Waltham, MA, USA) was used to extract total RNA from paraffin-embedded tissues according to the manufacturer’s instructions. The RNA concentrations were measured by spectrophotometry. The TaqMan microRNA RT Kit (ABI 4366597; Thermo Fisher Scientific) was used to generate cDNA. The final reaction mixture comprised 5 µL of total RNA and 3 µL of RT primer. Quantitative real time-polymerase chain reaction (qRT-PCR) was conducted to measure miRNA expression using a fluorescence quantitative PCR amplifier (AB71700 ABI; Thermo Fisher Scientific). The total volume of the reaction mixture was 20 µL, and it contained 1.33 μL of cDNA, 1 μL of primer and probe mixture (20×, miR-34a-specific primer, ABI 4427975; miR-155-5p-specific primer, ABI 4427975; U6 nuclear RNA, ABI 4427975), 10 μL of TaqMan general mixture (2×, ABI), and 7.67 μL of nuclease-free water. The amplification conditions were as follows: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. MiRNA expression levels were calculated using the 2−ΔCt method, where ΔCt (cycle threshold) = (Ct miRNA − Ct U6) for the experimental group − (Ct miRNA − Ct U6) for the control group.
Statistical analyses
SPSS 22.0 (IBM Corp., Armonk, NY, USA) was used to analyze the data. The median and quartile values were used for descriptive statistics. Wilcoxon rank-sum test and Kruskal–Wallis H test was used for intergroup and intragroup comparisons, respectively, and the Nemenyi method was used for multiple comparisons. The Kaplan–Meier method was used to analyze patient prognosis and survival. The log-rank test was used for univariate analyses and the Cox proportional hazards model was used for multivariate analyses. Additionally, the prognostic values of miR-34a and miR-155-5p for patients with MCL were analyzed by receiver operating characteristic (ROC) curve. Results with P < 0.05 were considered statistically significant.
Results
Expression of miR-34a and miR-155-5p in patients with MCL
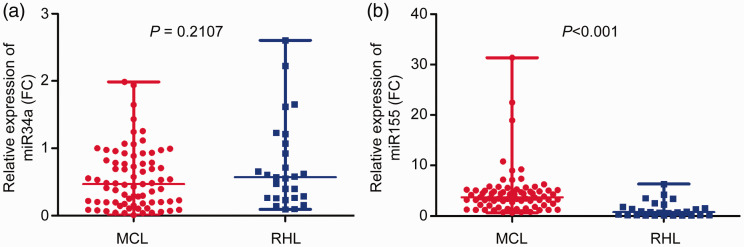
The expression of miR-34a was 0.478 (0.202–0.821) and 0.572 (0.265–1.072) in patients with MCL and RLH, respectively, with no significant difference between the groups (P = 0.2107) (Figure 1a). The expression of miR-155-5p was 3.482 (2.136–5.048) and 0.774 (0.083–1.834) in patients with MCL and RLH, respectively, with significantly higher expression in patients with MCL than in patients with RLH (P < 0.001) (Figure 1b).
Figure 1.
Expression levels of miR-34a and miR155-5p in the mantle cell lymphoma (MCL) and reactive lymphoid hyperplasia (RLH) groups. (a) No significant difference in expression between patients with MCL and RLH (P = 0.2107). The three horizontal lines represent the maximum, median, and minimum values of miR-34a in MCL (0.821, 0.478, and 0.202) and RLH (1.072, 0.572, and 0.265), respectively. (b) Expression of miR-155-5p in patients with MCL was significantly higher than that in patients with RLH (P < 0.001). The three horizontal lines represent the maximum, median, and minimum values of miR-155-5p in MCL (5.048, 3.482, and 2.136) and RLH (1.834, 0.774, and 0.083), respectively.
Correlations between miR-34a or miR-155-5p levels and clinicopathological features
As indicated in Table 1, miR-34a expression was significantly lower in patients with ECOG ≥ 2 than in those with ECOG < 2 (0.385 vs. 0.768, P < 0.05). Similarly, a significantly lower expression was detected in patients with SOX11 < M (median value) than in patients with SOX11 ≥ M (0.162 vs. 0.503, P < 0.05) by Kruskal–Wallis rank test (Table 1).
Table 1.
Correlation between miR-34a and miR-155-5p levels and clinicopathological features in mantle cell lymphoma (MCL).
| Variable | n | miR-34a M (Q1, Q3) | P-value | miR-155-5p M (Q1, Q3) | P-value |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 58 | 0.494 (0.203, 0.927) | 0.4144 | 3.393 (2.085, 4.74) | 0.1226 |
| Female | 17 | 0.47 (0.198, 0.576) | 4.84 (3.031, 5.618) | ||
| Age (y) | |||||
| ≥60 | 46 | 0.54 (0.203, 0.933) | 0.125 | 3.423 (1.828, 4.874) | 0.4761 |
| <60 | 29 | 0.362 (0.202, 0.553) | 3.643 (3.031, 5.223) | ||
| Morphology | |||||
| Classic morphology | 69 | 0.485 (0.204, 0.821) | 0.3337 | 3.458 (2.085, 4.874) | 0.4065 |
| Blastoid morphology | 6 | 0.337 (0.066, 0.707) | 4.512 (2.704, 5.408) | ||
| Ki67 | |||||
| ≥30% | 22 | 0.718 (0.195, 1) | 0.1827 | 4.164 (2.274, 5.098) | 0.9258 |
| <30% | 53 | 0.429 (0.203, 0.693) | 3.458 (2.136, 5.011) | ||
| ECOG | |||||
| ≥2 | 48 | 0.385 (0.184, 0.69) | 0.0214* | 4.37 (2.489, 5.352) | 0.0738 |
| <2 | 27 | 0.768 (0.228, 0.976) | 3.193 (1.51, 4.469) | ||
| WBC | |||||
| ≥10 × 109/L | 11 | 0.693 (0.107, 1.061) | 0.3649 | 4.469 (2.136, 5.046) | 0.4722 |
| <10 × 109/L | 64 | 0.45 (90.203, 0.786) | 3.422 (2.144, 5.054) | ||
| LDH | |||||
| ≥240 IU/L | 17 | 0.771 (0.36, 1.061) | 0.0611 | 3.891 (2.274, 4.74) | 0.9093 |
| <240 IU/L | 58 | 0.42 (0.202, 0.73) | 3.464 (2.085, 5.134) | ||
| β2-MG | |||||
| ≥3 mg/L | 30 | 0.36 (0.173, 0.771) | 0.4021 | 3.891 (2.274, 5.046) | 0.8268 |
| <3 mg/L | 45 | 0.494 (0.283, 0.821) | 3.452 (2.136, 5.011) | ||
| p53 | |||||
| Positive | 16 | 0.466 (0.196, 0.906) | 0.7269 | 4.025 (2.733, 5.513) | 0.2889 |
| Negative | 59 | 0.478 (0.202, 0.787) | 3.458 (1.51, 5.011) | ||
| Ann Arbor stage | |||||
| I–II | 19 | 0.429 (0.204, 0.821) | 0.9272 | 3.643 (1.404, 5.098) | 0.817 |
| III–IV | 56 | 0.491 (0.2, 0.835) | 3.47 (2.359, 5.028) | ||
| B symptoms | |||||
| No | 62 | 0.444 (0.203, 0.886) | 0.7742 | 3.393 (2.136, 5.134) | 0.5242 |
| Yes | 13 | 0.576 (0.195, 0.73) | 4.469 (3.482, 4.84) | ||
| Bone marrow | |||||
| Involvement | 22 | 0.531 (0.107, 0.771) | 0.8706 | 4.454 (3.204, 5.223) | 0.0513 |
| Normal | 53 | 0.459 (0.204, 0.821) | 3.283 (1.51, 4.84) | ||
| MIPI | |||||
| Low-risk | 21 | 0.54 (0.228, 0.959) | 0.1559 | 3.031 (1.27, 3.643) | 0.0432* |
| Intermediate-risk | 31 | 0.693 (0.195, 0.933) | 4.438 (2.704, 5.134) | ||
| High-risk | 23 | 0.379 (0.173, 0.553) | 4.438 (2.99, 5.296) | ||
| SOX11 mRNA | |||||
| ≥M | 31 | 0.503 (0.219, 0.886) | 0.0309* | 3.458 (2.085, 4.84) | 0.1352 |
| <M | 44 | 0.162 (0.107, 0.362) | 5.117 (3.182, 5.598) | ||
| Chemotherapy regimen | |||||
| CHOP/R-CHOP | 36 | 0.481 (0.118, 0.964) | 0.687 | 3.423 (1.587, 5.125) | 0.574 |
| yperCAVD/R-HyperCAVD | 39 | 0.429 (0.204, 0.771) | 3.891 (3.031, 5.046) | ||
Bone marrow involvement = lymphoma cells ≥ 5%; B symptoms = fever, night sweat, and weight loss.
*P < 0.05 indicates statistical significance.
LDH, lactate dehydrogenase; WBC, white blood cell; β 2-MG, β 2-microglobulin; MIPI, MCL International Prognostic Index; ECOG, Eastern Cooperative Oncology Group; SOX11, Sex-Determining Region Y-Box transcription factor 11; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone; R-hyper-CVAD, rituximab, fractionated cyclophosphamide, vincristine, adriamycin, and dexamethasone.
The expression of miR-155-5p was 3.031 (1.27–3.643) in the MCL International Prognostic Index (MIPI) low-risk subgroup, 4.438 (2.704–5.134) in the medium-risk subgroup, and 4.438 (2.99–5.296) in the high-risk subgroup. A Kruskal–Wallis rank-sum test demonstrated statistical differences among the subgroups (P = 0.0432) (Table 1). The Nemenyi method was used for multiple comparisons25 (Table 2). The expression of miR-155-5p in the low-risk subgroup was lower than that in the medium-risk and high-risk subgroups, with no statistical difference in the miR-155-5p expression between the high- and medium-risk subgroups.
Table 2.
Multiple comparisons of miR-155-5p levels among the three Mantle Cell Lymphoma International Prognostic Index (MIPI) subgroups.
| MIPI subgroup | Difference | 95%CI |
|---|---|---|
| High-risk vs. low-risk | 14.552 | 0.208 to 28.896*** |
| Intermediate-risk vs. low-risk | 13.627 | 0.196 to 27.059*** |
| High-risk vs. intermediate-risk | 4.529 | −8.963 to 19.527 |
*** indicates P < 0.05.
MIPI, MCL International Prognostic Index; CI, confidence interval.
Evaluation of the prognostic value of miR-34a and miR-155-5p in MCL
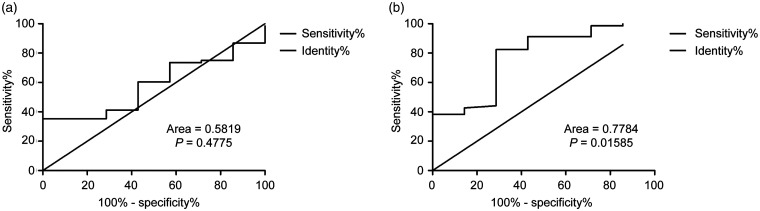
The ROC curve analysis showed that the area under the curve (AUC) of miR-34a and miR-155-5p expression was 0.5819 (95%CI = 0.4111–0.7527, P = 0.477) and 0.7784 (95%CI = 0.5912–0.9655, P = 0.015), and the cut-off values were 0.2150 and 2.110, respectively (Figure 2a and 2b).
Figure 2.
Evaluation of the prognostic value of miRNA-34a and miRNA-155-5p in mantle cell lymphoma (MCL). The receiver operating characteristic (ROC) curves of miR-34a and miR-155-5p are shown. (a) The area under the curve (AUC) of miR-34a was 0.5819 (95%CI = 0.4111–0.7527, P = 0.4775) and the cut-off value was 0.2150. (b) The AUC of miR-155-5p was 0.7784 (95%CI = 0.5912–0.9655, P = 0.01585) and the cut-off value was 2.110.
Survival analysis using miR-34a and miR-155-5p expression levels in MCL
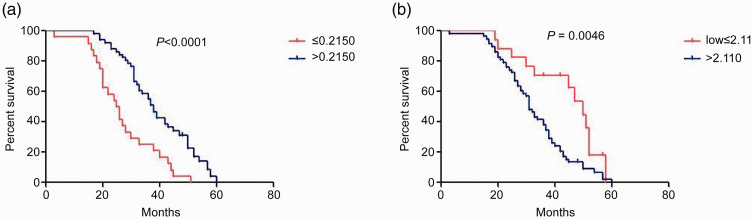
To determine the prognostic values of miR-34a and miR-155-5p in MCL, we divided the patients into two subgroups according to miRNA expression levels. Using the cut-off value of the ROC curve as the threshold for dividing patients, patient survival was evaluated using a Kaplan–Meier curve. The median survival time of patients with miR-34a expression ≤0.2150 was 25 months, whereas that of patients with miR-34a expression >0.2150 was 37 months. The median survival time of patients with miR-34a expression ≤0.2150 was significantly lower than that of patients with miR-34a expression >0.2150 (P < 0.0001) using the log-rank test (Figure 3a). The median survival time of patients in the miR-155-5p expression >2.11 subgroup was significantly shorter than that of patients in the miR-155-5p expression ≤2.11 subgroup (32 vs. 48 months), as indicated by analysis using the log-rank test (P = 0.0046) (Figure 3b).
Figure 3.
Survival analysis of miR-34a and miR-155-5p expression in patients with mantle cell lymphoma (MCL). (a) The median survival time of patients with miR-34a expression ≤ 0.2150 was significantly shorter than that of patients with miR-34a expression > 0.2150 (25 vs. 37 months; P < 0.0001). (b) The median survival time of patients in the miR-155-5p expression > 2.11 subgroup was significantly shorter than that of patients in the miR-155-5p expression ≤ 2.11 subgroup (32 vs. 48 months; P = 0.0046).
Multivariate analysis of the main predictors in MCL
We performed a multivariate analysis of the main predictors, including International Prognostic Index (IPI), P53, miR-155-5p, and miR-34a. The results suggested that only IPI and miR-34a were associated with MCL prognosis. MiR-34a was a negative independent factor for OS. Survival time of patients with low miR-34a expression was 0.343 times that of patients with high miR-34a expression (P < 0.001). MiR-155-5p was also an independent factor for OS. Survival time of patients with high miR-155-5p expression was 1.734 times that of patients with low miR-155-5p expression (P < 0.001) (Table 3).
Table 3.
Multivariate analysis of the main predictors in mantle cell lymphoma (MCL).
| Variable | B | SE | Wald | df | Sig. | Exp(B) | 95%CI for Exp(B) |
|
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| miR-34a | −1.069 | 0.269 | 15.827 | 1 | 0.000 | 0.343 | 0.203 | 0.581 |
| IPI | 0.550 | 0.166 | 11.046 | 1 | 0.001 | 1.734 | 1.253 | 2.398 |
IPI, International Prognostic Index; CI, confidence interval; SE, standard error; B, partial regression coefficient; Wald, Wald test; Sig., significance; df, degree of freedom.
Discussion
MCL is a B-cell non-Hodgkin's lymphoma subtype with a poor prognosis. Although the recent treatment regimen containing a high cytarabine dose, combined with rituximab, and followed by APBSCT has significantly improved the remission rate and survival time of patients with MCL, there is no complete cure for this disease.26 Therefore, it is important to identify new prognostic factors for targeted treatment to improve the remission rate and survival time of patients, especially in relapsed and refractory cases. Recent gene expression analyses have suggested the importance of miRNAs in the pathogenesis of hematological tumors.7,14,27
miR-155-5p is located on non-coding B cell integration cluster (BIC) transcripts on chromosome 21 and has a regulatory relationship with BIC transcript levels. It was the first miRNA identified to be involved in tumorigenesis and cancer development.28 Previous studies indicate that miR-155-5p is highly expressed in most hematological tumors, including DLBCL, CLL, natural killer (NK)/T-cell lymphoma, and MCL.28,29 Interestingly, studies on CLL indicate the association of high miR-155-5p expression with poor prognosis.29–31
In the present study, we found that miR-155-5p expression in patients with MCL was significantly higher than that in the control group, indicating that miR-155-5p may play a role in the development of the disease. A subgroup analysis revealed that miR-155-5p expression in the low-risk subgroup was significantly lower than that in the medium- and high-risk subgroups. Additionally, survival analysis confirmed that lower miR-155-5p expression is associated with an increased survival time in patients with MCL. These results suggest the possibility that miR-155-5p expression is related to the prognosis of MCL, and a high level of miR-155-5p expression is possibly indicative of poor prognosis. Limited by the small cohort of our study, future studies with a higher number of patients are needed to confirm the findings.
MiR-34a is a member of the miRNA-34 family along with miR-34b and miR-34c, with the expression of miR-34a higher than that of other miRNAs in most tissues.32,33 Recent studies have shown that 1p36, which includes the miR-34a site, is deleted in several tumors, resulting in low expression of miR-34a and high expression of its target gene TP53. This thereby affects cell differentiation, apoptosis, and senescence.32,34,35 He et al.16 demonstrated that significantly decreased expression of miR-34a, combined with high levels of Bcl-2 and p53, is observed in patients with gastric mucosa-associated lymphoma and DLBCL, with miR-34a serving as an independent poor prognostic factor.
In this study, the expression levels of miR-34a in patients with newly diagnosed MCL demonstrated no significant difference compared with those of the control group. This could be explained by the relatively small number of cases in our study or confounding factors. However, the subgroup analysis demonstrated that the expression of miR-34a in patients with ECOG ≥ 2 was lower than that in patients with ECOG < 2. In addition, the survival analysis indicated that low miR-34a expression is related to a shorter survival time than high miR-34a expression, suggesting that low miR-34a expression may be a factor for poor prognosis. In this study, an association between miR-34a and its target gene TP53 was not found. This could be explained by the complex interacting mechanisms and involvement of more than one biological pathway. Although this study showed a correlation between prognosis and both miR-155-5p and miR-34a, the multivariate analysis suggested that miR-34a, rather than miR-155-5p, is an independent prognostic factor. This finding is not consistent with the previous prediction and may be related to the small sample size, retrospective nature of the study, or sample bias.
In conclusion, the expression levels of miR-34a and miR-155-5p may be correlated with prognosis in patients with MCL, and this could possibly help differentiate patients with poor prognosis. However, further studies with a higher number of patients are warranted.
Acknowledgements
We thank our colleagues in the Department of Pathology for technical assistance. We would like to thank Editage (www.editage.cn) for their assistance with English language editing.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This study was supported by Key Science and Technology Projects of Shanxi Province [20140313011-10].
ORCID iD: Jianxia He https://orcid.org/0000-0003-0138-0570
References
- 1.Vardiman JW. The World Health Organization (WHO) classification of tumors of the hematopoietic and lymphoid tissues: an overview with emphasis on the myeloid neoplasms. Chem Biol Interact 2010; 184: 16–20. [DOI] [PubMed] [Google Scholar]
- 2.Pérez-Galán P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 2011; 117: 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortelazzo S, Ponzoni M, Ferreri AJ, et al. Mantle cell lymphoma. Crit Rev Oncol Hematol 2012; 82: 78–101. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 2003; 3: 185–197. [DOI] [PubMed] [Google Scholar]
- 5.Royo C, Salaverria I, Hartmann EM, et al. The complex landscape of genetic alterations in mantle cell lymphoma. Semin Cancer Biol 2011; 21: 322–334. [DOI] [PubMed] [Google Scholar]
- 6.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia 2015; 29: 1004–1017. [DOI] [PubMed] [Google Scholar]
- 7.Di LL, Gómez-López G, Sánchez-Beato M, et al. Mantle cell lymphoma: transcriptional regulation by microRNAs. Leukemia 2010; 24: 1335–1342. [DOI] [PubMed] [Google Scholar]
- 8.Zhao JJ, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010; 115: 2630–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooi JY, Bernardo BC, Singla S, et al. Identification of miR-34 regulatory networks in settings of disease and antimiR-therapy: Implications for treating cardiac pathology and other diseases. RNA Biol 2016; 14: 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med 2013; 5: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabbri M, Croce CM. Role of microRNAs in lymphoid biology and disease. Curr Opin Hematol 2011; 18: 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iqbal J, Shen Y, Liu Y, et al. Genome-wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood 2012; 119: 4939–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenquist R, Beà S, Du MQ, et al. Genetic landscape and deregulated pathways in B-cell lymphoid malignancies. J Intern Med 2017; 282: 371–394. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci 2002; 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi Y, Li J, Zan L, et al. Micro-RNA-16 expression in paraffin-embedded specimen correlates with overall survival of T-lymphoblastic lymphoma/leukemia. Hum Pathol 2013; 44: 1011–1016. [DOI] [PubMed] [Google Scholar]
- 16.He M, Gao L, Zhang S, et al. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL. Gastric Cancer 2014; 17: 431–441. [DOI] [PubMed] [Google Scholar]
- 17.O'Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 2008; 205: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrajoli A, Shanafelt TD, Ivan C, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013; 122: 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica 2009; 94: 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meggendorfer M, Kern W, Haferlach C, et al. SOX11 overexpression is a specific marker for mantle cell lymphoma and correlates with t(11;14) translocation, CCND1 expression and an adverse prognosis. Leukemia 2013; 27: 2388–2391. [DOI] [PubMed] [Google Scholar]
- 21.Nordström L, Sernbo S, Eden P, et al. SOX11 and TP53 add prognostic information to MIPI in a homogenously treated cohort of mantle cell lymphoma–a Nordic Lymphoma Group study. Br J Haematol 2014; 166: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrando AA. SOX11 is a mantle cell lymphoma oncogene. Blood 2013; 121: 2169–2170. [DOI] [PubMed] [Google Scholar]
- 23.He JX, Xi YF, Su LP, et al . Association of SOX11 gene expression with clinical features and prognosis of mantle cell lymphoma. Eur Rev Med Pharmacol Sci 2018; 22: 2556–2563. [DOI] [PubMed] [Google Scholar]
- 24.Von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007; 147: 573–577. [DOI] [PubMed] [Google Scholar]
- 25.Elliott AC, Hynan LS. A SAS(®) macro implementation of a multiple comparison post hoc test for a Kruskal-Wallis analysis. Comput Methods Programs Biomed 2011; 102: 75–80. [DOI] [PubMed] [Google Scholar]
- 26.Cohen JB, Zain JM, Kahl BS. Current approaches to mantle cell lymphoma: Diagnosis, prognosis, and therapies. Am Soc Clin Oncol Educ Book 2017; 37: 512–525. [DOI] [PubMed] [Google Scholar]
- 27.Arakawa F, Kimura Y, Yoshida N, et al. Identification of miR-15b as a transformation-related factor in mantle cell lymphoma. Int J Oncol 2016; 48: 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji WG, Zhang XD, Sun XD, et al. miRNA-155 modulates the malignant biological characteristics of NK/T-cell lymphoma cells by targeting FOXO3a gene. J Huazhong Univ Sci Technolog Med Sci 2014; 34: 882–888. [DOI] [PubMed] [Google Scholar]
- 29.Due H, Svendsen P, Bødker JS, et al. miR-155 as a Biomarker in B-Cell Malignancies. Biomed Res Int 2016; 2016: 9513037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui B, Chen L, Zhang S, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014; 124: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caivano A, La Rocca F, Simeon V, et al. MicroRNA-155 in serum-derived extracellular vesicles as a potential biomarker for hematologic malignancies - a short report. Cell Oncol 2017; 40: 97–103. [DOI] [PubMed] [Google Scholar]
- 32.Okada N, Lin CP, Ribeiro MC, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 2014; 28: 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmid G, Notaro S, Reimer D, et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer 2016; 16: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacombe J, Zenhausern F. Emergence of miR-34a in radiation therapy. Crit Rev Oncol Hematol 2017; 109: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louie DC, Offit K, Jaslow R, et al. p53 overexpression as a marker of poor prognosis in mantle cell lymphomas with t(11;14)(q13;q32). Blood 1995; 86: 2892–2899. [PubMed] [Google Scholar]