Abstract
Mosquitoes are the greatest animal threat to human health, causing hundreds of millions of infections and around 1 million deaths each year. All mosquito-borne pathogens must traverse the salivary glands (SGs) to be transmitted to the next host, making this organ an ideal target for interventions. The adult SG develops from precursor cells located in the larval SG duct bud. Characterization of the larval SG has been limited. We sought to better understand larval SG architecture, secretion and gene expression. We developed an optimized method for larval SG staining and surveyed hundreds of larval stage 4 (L4) SGs using fluorescence confocal microscopy. Remarkable variation in SG cell and chromatin organization differed among individuals and across the L4 stage. Lumen formation occurred during L4 stage through secretion likely involving a coincident cellular apical lipid enrichment and extracellular vesicle-like structures. Meta-analysis of microarray data showed that larval SG gene expression is divergent from adult SGs, more similar to larval gastric cecae, but different from other larval gut compartments. This work highlights the variable cell architecture of larval Anopheles gambiae SGs and provides candidate targets for genetic strategies aiming to disrupt SGs and transmission of mosquito-borne pathogens.
Keywords: Anopheles, larvae, salivary gland, brush border-like, vesicles
Introduction
Half of the world’s population is at risk of mosquito-borne illness and 700 million people are infected annually, leading to around one million deaths each year (Riveron et al.,, 2018; World Health Organization, 2019). Current vector control measures (insecticide-coated bed nets or insecticides sprayed indoors and/or outdoors) have resulted in a marked decrease in deaths since 2000; however, this decrease in death toll has stopped since 2015, largely due to rising rates of resistance (Alout et al., 2017; Riveron et al. 2018). New control strategies and targets are sorely needed. Recent interruptions in malaria control necessitated by the COVID-19 pandemic are predicted to increase the burden caused by malaria (Thornton, 2020).
Despite the broad diversity of mosquito-borne pathogens (Müller et al., 2019), all must invade and traverse the mosquito salivary glands (SGs) to be passed to a new host during a subsequent blood meal (Mueller et al., 2010). Many disease vectors (including mosquitoes, ticks and tsetse flies) require pathogen traversal of the SGs for transmission (Mueller et al., 2010). Sandfly transmission of leishmaniasis is an exception, involving only development of the pathogen in the gut, not the SGs (Lestinova et al., 2017). However, sandfly saliva has been shown to enhance transmission of leishmania (Norsworthy et al., 2004; Rohousova and Volf, 2006; Oliveira et al., 2013; Lestinova et al., 2017). This traversal requirement for transmission makes the SGs an ideal target for intervention strategies (James, 2003).
Mosquito and SG development are understudied topics of critical human health importance. The mosquito life cycle involves aquatic eggs that hatch into aquatic larvae, which progress (via increases in cell size) through four larval stages, until pupation (also aquatic) occurs, leading to terrestrial/aerial adulthood (World Health Organization, 2019). The length of each stage varies by genera, and sometimes species, but the overall mosquito lifespan is around 25–30 days (Styer et al., 2007; Papadopoulos et al., 2016; World Health Organization, 2019). All mosquito larvae are filter feeders. Plant nectar is used as a food source by all adults, while blood meals that contribute proteins to oocyte maturation are taken only by adult females. Larval food quantity, quality and the microbiome have been linked to adult vectorial capacity in numerous ways, including morphometry, pathogen load, microbiome composition and immunity, longevity and gene expression (Capone et al., 2013; Gimonneau et al., 2014; Price et al., 2015; Shapiro et al., 2016; Barreaux et al., 2018; Gunathilaka et al., 2019).
Larval mosquito SG polytene chromosomes have been used extensively for locus mapping for individual traits, validation of genome sequencing, species identification and discrimination, chromosome conformation analysis and DNA damage studies (Canalis et al., 1956; Service, 1970; Green, 1972; Rabbani and Kitzmiller, 1975; Sharma et al., 1978; 1987; Redfern, 1981; Xu et al., 1983; Sun and Coluzzi, 1989; Sharakhova et al., 2006; 2010; McAbee et al., 2007; Kamali et al., 2011; George et al., 2014; Bondarenko et al., 2017; Artemov et al., 2018; George et al., 2020). Accounts of Anopheles SG development consist of embryonic and larval sectioning and drawing (Clements, 1963; Imms, 1908; Ivanova-Kazas, 1949), larval microscopy (Favia et al., 2007; Neira Oviedo et al., 2009) and gene expression analysis (Linser et al., 2009; 2012; Neira Oviedo et al., 2009). In Aedes mosquitoes, one account suggested similarities and differences in the action of a transcription factor (TF), CrebA, known to regulate secretory capacity in Drosophila melanogaster (Andrew et al., 1997; Abrams and Andrew, 2005; Fox et al., 2010; Barbosa et al., 2013; Nguyen et al., 2013; Fox and Andrew, 2015). Flybase gene entries indicate that orthologues of all major Drosophila SG regulatory TFs are present in Anopheles mosquitoes (Thurmond et al., 2018). Disrupting SG development, or maturation, could block the route pathogens take to exit the mosquito during transmission.
Larval Anopheles stephensi SG features were investigated previously by multiple groups (Jensen and Jones, 1957; Rishikesh, 1959). These studies described the organization of this organ, including the position and structure of the duct, adult precursor cells, proximal and distal sacs, and the constriction separating the secretory sacs (Jensen and Jones, 1957; Rishikesh, 1959). Rishikesh documented when and where secretion began and was highest, as well as when and where histolysis of the glands began, in preparation for metamorphosis (Rishikesh, 1959). Rishikesh showed that larval SG chromosomes are polytenized, including a correlation between band intensity and DNA concentration (Rishikesh, 1959). These studies also determined the number of cells in proximal and distal sacs and documented cell and nuclear size at the different stages (Jensen and Jones, 1957; Rishikesh, 1959). These important foundational overviews described broad biological aspects of larval Anopheles SGs.
Gene expression in mixed sex larval Anopheles SGs was investigated by Neira Oviedo and others (Neira Oviedo et al., 2009). They conducted complementary whole-mount imaging of larval organs and microarray gene expression analysis (Neira Oviedo et al., 2009). Ultimately, they suggest that genes with a role in digestion are enriched in larval SGs (Neira Oviedo et al., 2009). Although this work supports a function for larval SGs in digestion, questions concerning its relatedness to other tissues and to SGs at other life stages remained unaddressed. Mosquito feeding differs greatly between life stages. Larvae are filter feeders and develop in water, whereas adults are aerial and terrestrial, feeding on plant nectar and blood. Substantial differences in cell morphology, cellular processes and gene expression are expected to accommodate these feeding differences.
Previous studies (Orr et al., 1961; Wright, 1969; Janzen and Wright, 1971; Barrow et al., 1975; Beckett, 1977; 1988; 1990; Wells and Andrew, 2015; Wells et al., 2017) have led to appreciable knowledge of adult mosquito SG structure and function across genera. In Anopheles and Culex mosquitoes, male SGs are composed of a single lobe and female SGs contain two relatively slender lateral lobes (subdivided into proximal and distal regions) surrounding a rounder medial lobe. In both sexes of Aedes SGs, each lobe described previously trifurcates. Within each basement membrane-bound lobe resides a monolayer of cup-shaped epithelial cells, a central lumen and in most areas, a chitinous salivary duct.
SG tissue and cellular morphometry are well-understood (Moreira-Ferro et al., 1999; Wells and Andrew, 2015), as are the components of adult saliva (Arcà et al., 2017). Each region of the SG is thought to contribute different molecules to the pooled saliva, and the male saliva is most similar to the proteins produced in the female proximal lateral lobe (Poehling, 1979). Very early after eclosion, SG cells are nearly cuboidal in shape. About 36 h post eclosion, a subcellular, perinuclear compartment comprised of many vesicles containing secretory material and lipid staining [reminiscent of a multivesicular body (Denzer et al., 2000; Hanson and Cashikar, 2012)] is observed, which resolves within the next 12 h, by apical membrane fusion of either budded vesicles or the entire secretory compartment, expanding the newly defined lumen-coincident secretory cavity that the cup-shaped cell surrounds (Wells et al., 2017). Over days three and four post-eclosion, the secretory cavities continue to grow, accentuating the cupshape of the secretory cells (Wells et al., 2017). Multiple accounts have described the relationship between mosquito SG architecture and pathogen transmission (Pimenta et al., 1994; Ando et al., 1999; Mueller et al., 2010; Wells and Andrew, 2019).
In this study, we sought to better understand larval Anopheles gambiae SGs with respect to their adult counterparts using a novel, optimized method for dissection, staining and confocal imaging of larval SGs and analysis of available gene expression data sets. Our work offers several key findings relevant to mosquito-borne disease: (1) We reveal new candidate targets for SG disruption; (2) We describe a new approach (larval SG staining followed by confocal imaging) for determining the efficacy of treatments and interventions on larval and adult stage SGs and (3) We showcase the remarkable cell biology of this system and the biological changes that occur across developmental stages in mosquito SGs.
Results
Larval SG morphologies during lumen formation and expansion
To better describe larval stage 4 SG cells in Anopheles gambiae, nearly 3000 glands were visualized (500 imaged) using our optimized dissection, staining, mounting and confocal imaging protocol (see Experimental Procedures). Each of the two SGs per individual is composed of a proximal, cellular duct region ending in a broader, triangular bud region (duct bud; DB), a secretory cell-containing proximal sac and a larger secretory cell-containing distal sac (Fig. 1A).
Figure 1. Architecture of stage 4 larval salivary glands.
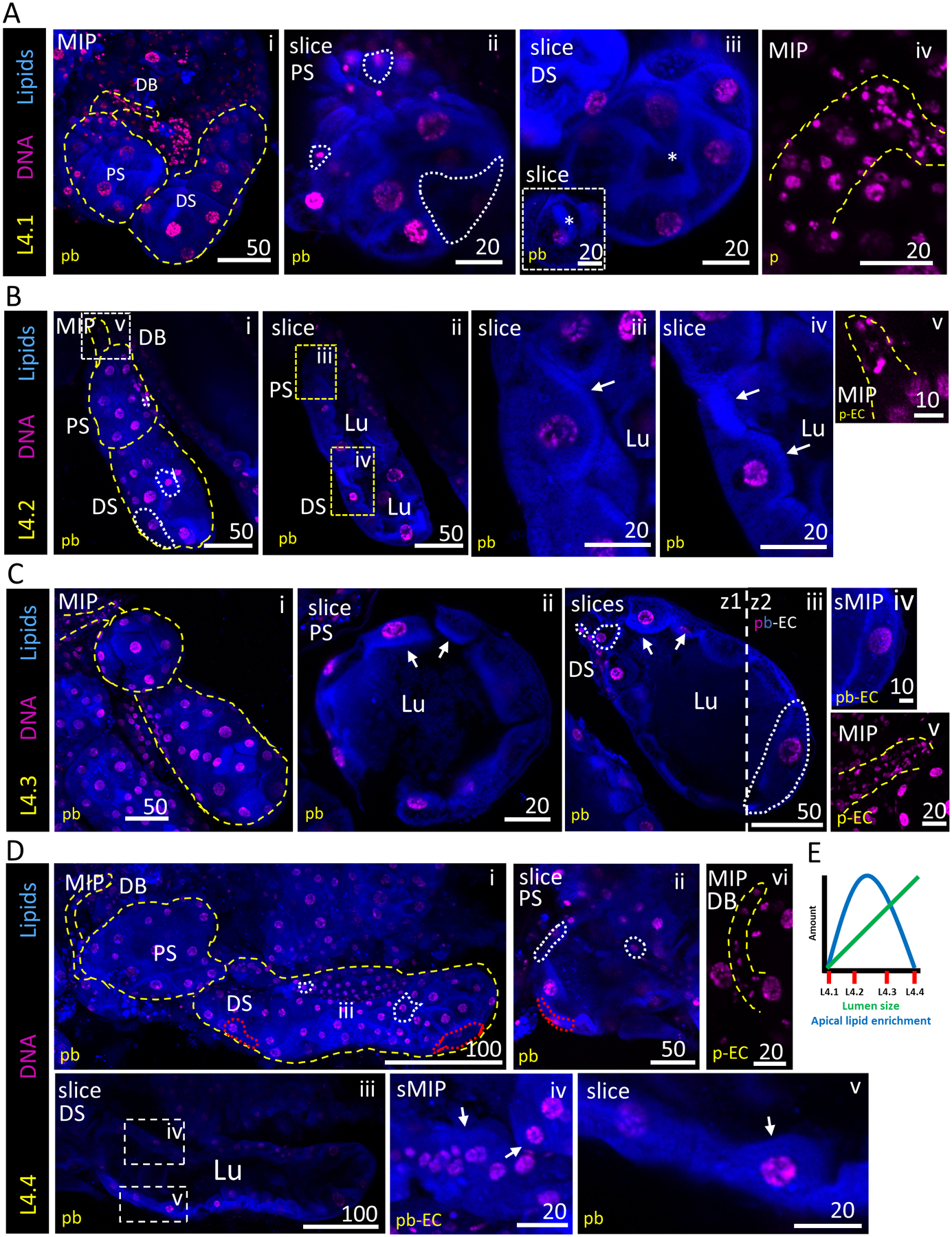
Representative images from dissected larval stage 4 (L4) salivary glands stained with Hoechst (DNA, purple) and Nile Red (lipids, blue). Cell number and morphology were variable across all sub-stages. (A) Characteristic morphology of an L4.1 SG. A slice through the centre of the PS reveals no lumen and a missing cell (white outline) (ii). The DS also lacks a lumen (iii); the inset shows a more basal focal plane of the asterisk-containing region (iii, inset). (B) Characteristic morphology of an L4.2 SG. A slice through the centre of the PS and DS demonstrates the presence of a narrow lumen (ii). A magnified image of L4.2 SG cells shows an apical lipid enrichment (white arrows, iii, iv). (C) Characteristic morphology of an L4.3 SG. A slice through the centre of the PS reveals a broad lumen (ii). A slice through the centre of the DS reveals a fuller lumen, as well as heterogeneous cell shapes and sizes (white dashed line, iii, iv). Apical lipid enrichment persists (white arrows, ii, iii). (D) Characteristic morphology of an L4.4 SG. Cells are noticeably more laterally elongated (red dashed line, i, ii). A slice through the PS and DS reveal a very full lumen (ii, iii). Apical lipid enrichment has largely, or completely, disappeared, and the lipid signal is now scattered more broadly throughout the cytoplasm (white arrows, iv, v). (E) Schematic graph showing the trends observed in apical lipid enrichment (blue) and lumen size (green) across SGs during larval stage 4. Substages 4.1–4.4 are noted on the x-axis.
We binned L4 SGs into four sub-stages (L4.1–L4.4; see Experimental Procedures) based on two prominent morphological features: (1) lumen presence and size, and (2) presence of a lipid-enriched apical cell boundary, reminiscent of a brush border-like structure (Crawley et al., 2014). In L4.1 SGs, SG structure was condensed and compact, and no lumen was visible (Fig. 1Ai–iii). Images from a central plane within L4.1 SGs showed adjacent cells filling up the entire sac volume and no lipid enrichment at the cell membrane (Fig. 1Aii, iii). Instead, lipid signal was broadly distributed throughout the cytoplasm. In L4.2 SGs, a small, partial lumen was present in both distal and proximal sacs (Fig. 1Bi–ii, iv), with strong Nile Red staining enriched at the apical plasma membrane (Fig. 1Aiii, iv). In L4.3 SGs, the lumen was fuller and lipid enrichment was seen at the apical boundary of the majority of cells (Fig. 1Ci–iv). Regions of the gland appeared to have grown in an unequal, non-uniform manner, with high variability in cell sizes and shapes at this stage. Many small cells were often positionally mixed with larger, laterally elongated cells, especially in the distal sac (Fig. 1Ciii). Secretory cell cytoplasms ranged from minimal (barely extending beyond the nucleus) to taking up an entire sector of the SGs as large as 6–8 typical cells (Fig. 1Ciii). Although lipid enrichment at the brush border was still present, the Nile Red signal was weaker at this stage. In L4.4 SGs, lipid enrichment is no longer observed, and the Nile Red signal is broadly distributed throughout the cytoplasm (Fig. 1Di, ii, iv, v). Cell shapes remained variable and irregular (Fig. 1Dii, white dashed outlines). The typical lumen was uniformly full, to the point of cell distension (Fig. 1Diii). Apical lipid accumulation/loss and lumen expansion trends are plotted across the L4 stage (Fig. 1E). In all L4 substages, duct bud morphology was variable and duct cell numbers varied widely (Fig. 1Aiv, Bv, Cv, Dvi). In summary, L4 stage Anopheles gambiae SGs exhibit extensive variability in DNA, cell size and tissue organization (lumen size and apical lipid enrichment) across individuals during lumen expansion.
Larval SG morphometry and cell quantification
To measure a representative L4.2 SG, we counted individual cells and nuclei in the proximal sac, distal sac and duct bud (Fig. S1i). This SG contained 300 duct bud cells, 20 proximal sac cells and 80 distal sac cells. A range of cell and DNA shapes and sizes were observed (Fig. S1Ai, ii, B) and measured (Fig. S1C–E). In many instances, DNA was likely in condensed, prophase stage organization (Fig. S1B). In a late stage SG, we were able to clearly visualize the interface between proximal and distal secretory sacs (Fig. S1F). Notably, lamin C staining was more broadly distributed than expected (Fig. S1Fiii); our control larval and adult staining data (Figs S3, S4), our previous use of this antibody in adult Anopheles stephensi SGs showing perinuclear localization (Wells and Andrew, 2015) and previous validation (Riemer et al., 1995) suggest this reagent is functional and the result is likely valid (see Discussion for further information). A row of cells (Fig. S1Fii, iii) forms the boundary between regions (white arrows, ii, iii) and the lumen narrows slightly between the sacs (white arrows, iv, v).
Continuity between duct bud and proximal sac regions
We next focused on the interface between the duct bud and proximal sac (Fig. 2). Unlike in the adult SGs (Wells et al., 2017), no salivary duct was visible in the secretory portion of the L4 SG (Fig. 2A, B, C, F). Differential interference contrast (DIC) imaging showed us the basement membrane surrounding of the salivary duct bud region (Fig. 2B). Duct bud cells (the precursors that form the adult SG) were clustered together at the interface between the salivary duct and the proximal sac. They were uniform in size with little cytoplasm (Fig. 2C, E–G). Slice images revealed an open lumen extending from the duct, through the duct bud and into the proximal-most portion of the proximal sac (Fig. 1C, D). Cell size was notably variable at the interface between the duct bud and the proximal sac (Fig. 2E–G). There were no identifiable boundaries to indicate a true separation between the duct bud and proximal sac (Fig. 2). Slight lipid enrichment beginning from the distal ends of the duct bud into the proximal part of the proximal sac (Fig. 2F, arrows) highlighted duct bud extension into the proximal sac, a potential blueprint structure for formation of the adult salivary duct (Wells et al., 2017). Mucin (saliva protein) staining was evident in both duct bud cells and cells of the proximal sac (Fig. 2G, H), suggesting that duct bud cells could already have a secretory cell identity during L4 stage. In support of this cell identity conclusion, we investigated Notch signalling in larval SGs to gauge multipotency, using antisera against two components (Notch and Delta; Fig. S2). Minimal localization was found in about three cells of a representative SG (Fig. S2, arrows). We conclude that Notch signalling is not strongly present in SG cells, and thus duct bud cells are likely committed to a secretory cell fate. In summary, these data affirm the connectedness of the proximal sac and duct bud regions, and they suggest that duct bud cells have secretory cell properties.
Figure 2. Duct Bud Morphology and Connection to the Proximal Sac.
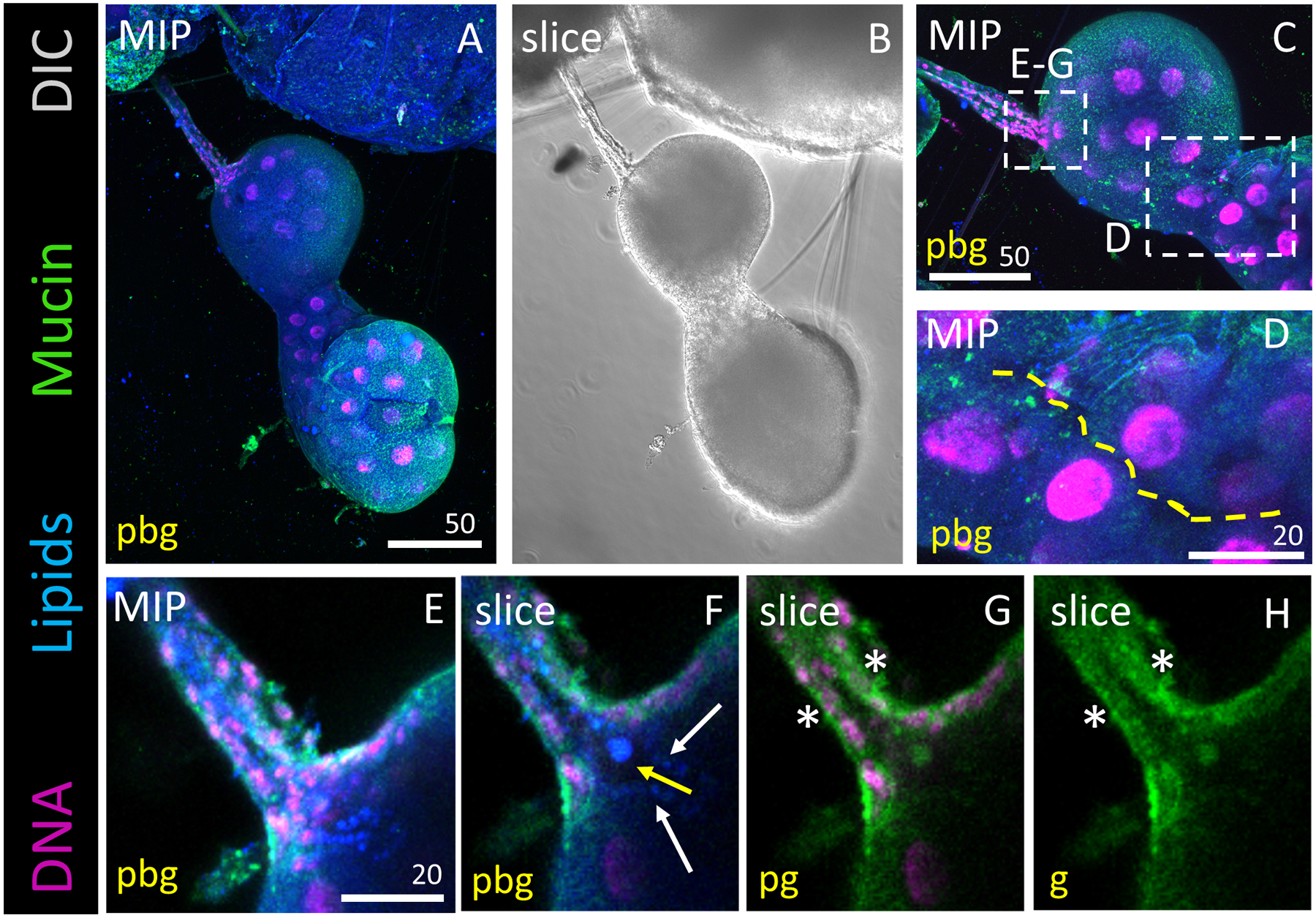
Representative images of an L4 salivary gland stained with Hoechst (DNA, purple), Nile Red (lipids, blue) and antisera against Mucin (Muc2; green). DIC is shown in grayscale (ii). (A, B) Overview of entire salivary gland. (C) Zoom of the duct bud and proximal sac regions. (D) Zoom of the narrow passageway (yellow dashed line) between proximal and distal sacs at this point in the L4 stage. (E) Zoomed projection image of the salivary duct bud. (F) Slice image showing an open connection extending from the lumen into the salivary duct bud region, suggesting they are connected. Note the lipid-rich protrusions extending into the lumen. (G, H) Mucin, a saliva protein, staining is observed in the cytoplasm of duct bud cells (asterisks).
Broad cytoplasmic localization of larval SG secretory pathway organelles
To examine L4 SG secretory function, we visualized the distribution of secretory cell organelle markers. We used a variety of antisera to stain for the endopoasmic reticulum (ER; KDEL), Golgi (GM130), Nucleolus (Fibrillarin), mitochondria (mtTFA) and cytoskeletal elements (lamin C, alpha-tubulin). Fibrillarin staining was tightly localized in a subset of the nucleus in only some cells (Fig. 3Diii–vi), while in other cells the staining was more broadly diffuse (Fig. 3Di, ii). Alphatubulin staining was enriched sub-apically, just basal to the brush border-like apical lipid enrichments (Fig. 3Eii–v). Secretory organelle marker (mtTFA, KDEL, GM130; Fig. 3A–C, arrows) staining was a haze with puncta broadly distributed across secretory sac cell cytoplasms and duct bud cells. Control staining for several of these markers supports the validity of these broad localization patterns (Figs S3, S4). These data suggest that larval SG organelles, and therefore, secretory activities, are broadly distributed across secretory cell cytoplasms.
Figure 3. Subcellular organization of salivary gland cell organelles.
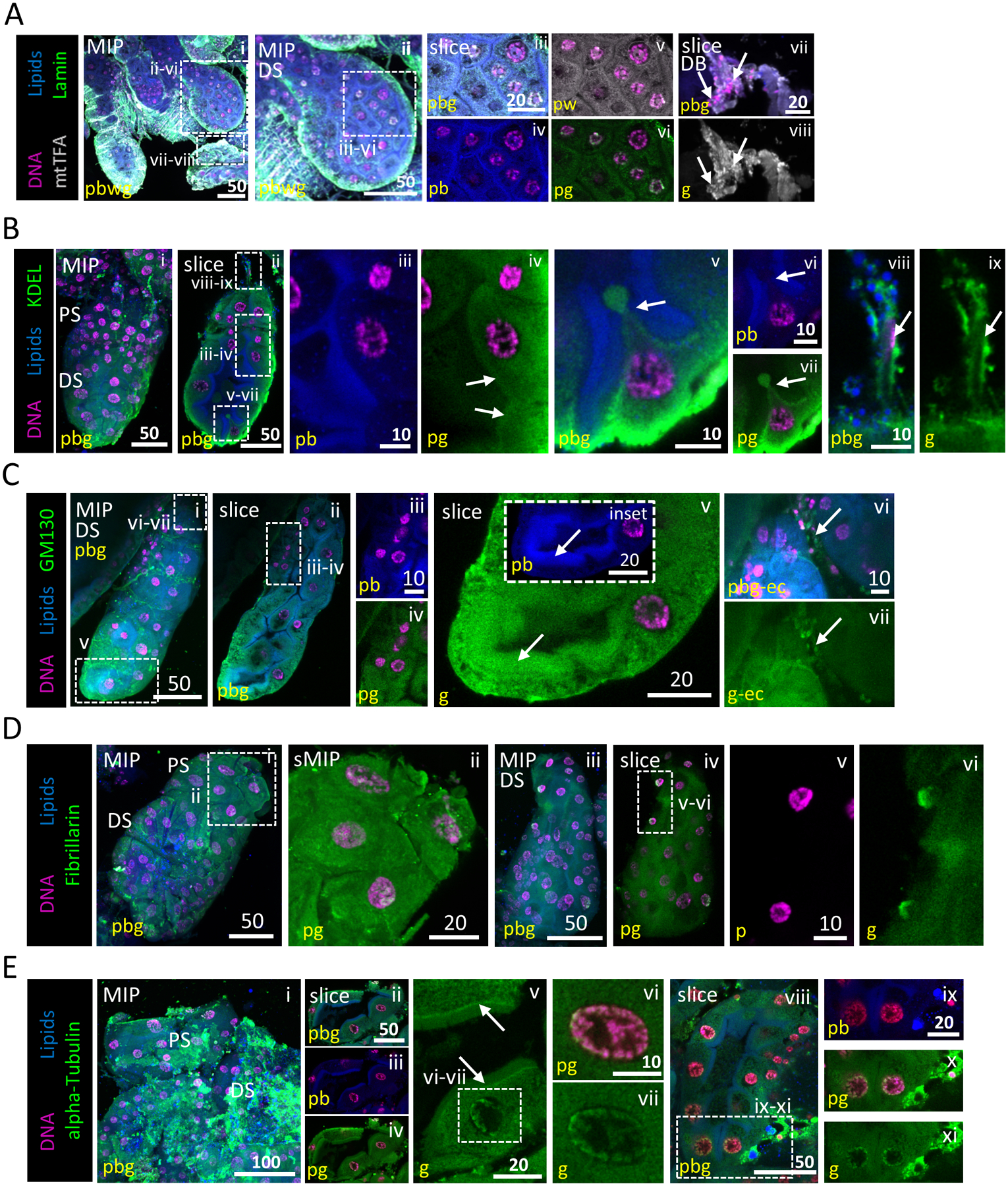
Representative images from dissected L4 larval salivary glands stained with Hoechst (DNA, purple), Nile Red (lipids, blue) and antisera against mtTFA (mitochondria, grey; A), lamin (nuclear membrane, green; A), KDEL (ER, green; B), GM130 (Golgi, grey; C), Fibrillarin (nucleolus, green; D) or alpha-tubulin (cytoskeleton, green; E). (A) mtTFA and lamin are broadly distributed within the cytoplasm. No intact nuclear envelope was observed in secretory sac nuclei (v). Some perinuclear lamin and mtTFA enrichment were observed, likely representing ER localization (iv, v). (B) KDEL is broadly distributed within the cytoplasm (ii, iv). A cell pinched between two adjacent apical brush border-like structures (v–vii) with a small cytoplasmic bud. Images from this L4 SG were also used in Fig. 5C. (C) GM130 is broadly distributed within cytoplasm (iv) and enriched (arrow, v) at the apical brush border-like structure (arrow, v inset). mtTFA (Avii, viii, white), KDEL (Bviii, ix, green) and GM130 (Cvi, vii, green) staining (arrows) are present in duct bud cells. (D) Fibrillarin staining is both diffuse cytoplasmic and in close association with multiple perinuclear sites (likely either fragmented nucleoli or ER association; ii, vi). (E) alpha-Tubulin staining is diffuse and striated through the secretory sac cell cytoplasm (i–iv). Signal enrichment was observed at the junction between the apical cytoplasmic face and the brush border-like structure (arrows, v). A halo of alphatubulin was seen in secretory sac nuclei, surrounding the chromosomes (v–vii, viii, x, xi).
Presence of SG, muscle, and gut transcription factors in larval SG cells
To better understand the regulators that may govern to gene expression in this tissue, we localized a variety of TFs to the nuclei of larval SG cells (Fig. 4). Staining for CrebA (using rabbit antisera), a master regulator of secretory capacity (Abrams and Andrew, 2005; Fox and Andrew, 2015; Johnson et al., 2020), was observed to variable degrees in all larval SG nuclei, along with localization of Sage (Fig. 4A; using rat antisera), an SG-specific TF (Fox et al., 2013). Strong lipid enrichment was present at the apical boundary of cells in this gland, suggesting that lumen expansion (Fig. 1) was ongoing. Satisfyingly, RNA Pol II control staining was localized to the nucleus of all SG cells (Fig. 4B). Two additional TFs (Cut, dMEF2) with no known SG role were investigated due to the similarity in morphology between the larval SG and larval gut compartments (which is not similarly observed in the respective adult counterparts). Cut, a hindgut/Malpighian tubule specification homeobox-containing TF (Liu et al., 1991; Hatton-Ellis et al., 2007), localized to the nuclei of secretory sac and duct bud cells (Fig. 4C). CrebA colocalized with Cut in these cells at the boundary of each nucleus. In SGs stained with antisera against a muscle TF, dMEF2 (Nguyen et al., 1994), localization was observed in secretory sac nuclei (Fig. 4Di–vi) and the centre of duct bud cells (Fig. 4Dvii–x). In totality, we conclude that a variety of TFs and RNA polymerase are localized to SG cell DNA.
Figure 4. Diverse Transcription Factor and RNA Pol II localization to SG cell nuclei.
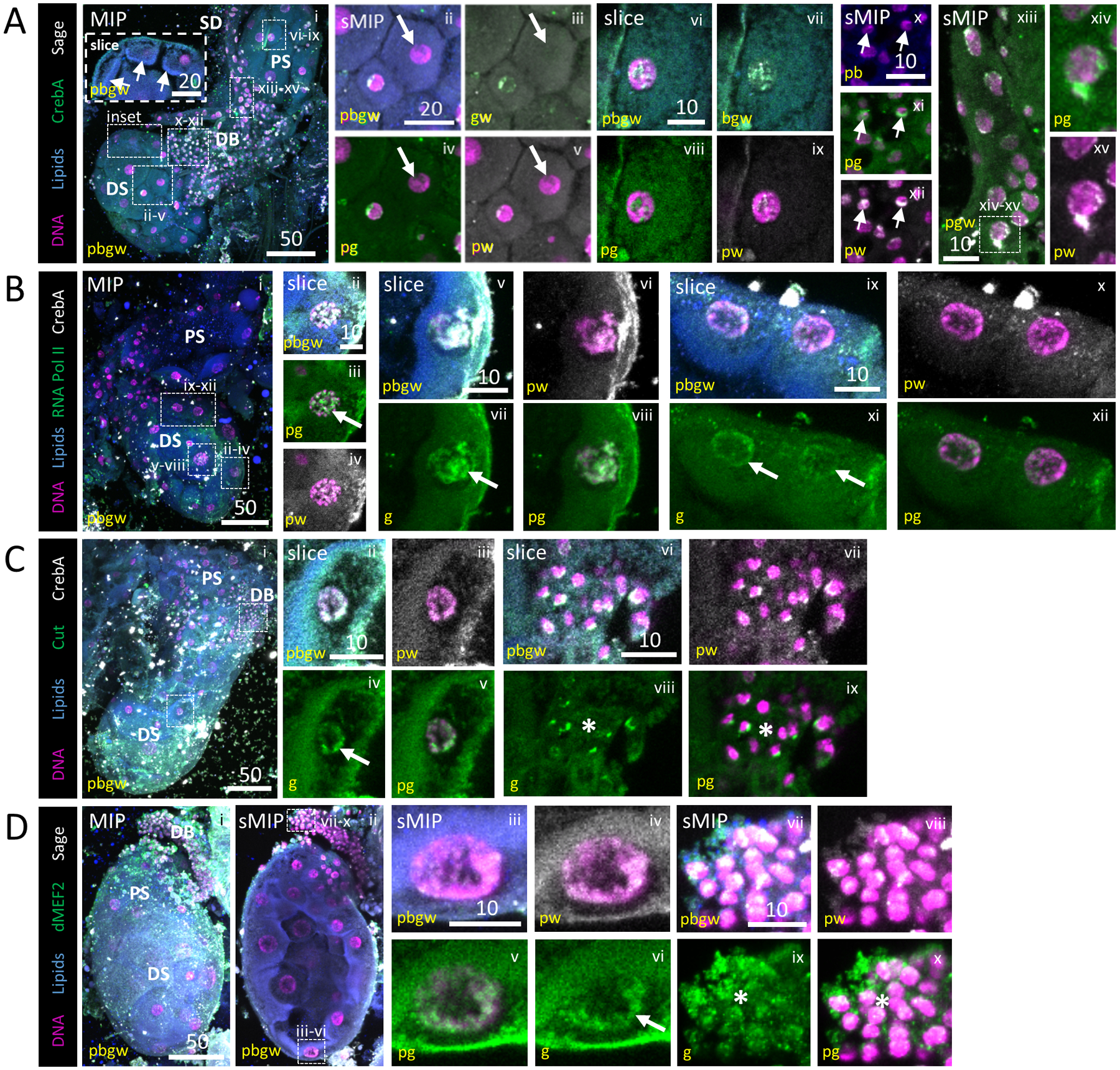
Representative images from dissected L4 larval salivary glands stained with Hoechst (DNA, purple), Nile Red (lipids, blue), antisera against the SG TFs CrebA (A, green; B, C, grey) or Sage (A, D, grey) and antisera against either RNA polymerase II (B, green), the gut TF Cut (C, green) or the muscle TF dMEF2 (D, green). (A) Two SGs (i). Both Sage and CrebA enrichment are observed in nuclei of the distal sac (ii–v), proximal sac (vi–ix), duct bud (arrow, x–xii) and salivary duct (xiii, xv) cells. Some cells have no enrichment (arrow, ii–v). Low level cytoplasmic signal is also observed. (B) RNA Pol II staining was variably enriched [high (arrow), ii–viii; low (arrows), ix–xii] across sac (distal, ii–viii) and duct bud (ix–xii) nuclei. (C) Cut enrichment was observed in sac (arrow, ii–v) and duct bud (asterisk, vi–ix) nuclei. (D) dMEF2 was also enriched on the DNA of sac (arrow, iii–vi) and duct bud (asterisk, vii–x) nuclei.
Validations of larval SG immunostaining results
In support of our antibody staining results, we performed a series of control analyses. We first stained SGs with secondary dyes (Mouse 647, Rabbit 488, Rat 647, Goat 488) in the absence of primary antisera (Fig. S3A). The absence of signal confirmed that secondary dye signals in SGs stained with primary antisera localized to primary antibodies, not non-specific locations. Our initial GM130 (Golgi) and fibrillarin (nucleolus) staining showed broad overlap between the DNA, GM130 and fibrillarin markers (Fig. 3C, D) with enrichment at the nuclear periphery. We pre-incubated these broadly localizing antisera with 0–2 h Drosophila embryos, which display very little protein expression, to remove general primary antisera stickiness. Doing so did not alter the broad localization of these markers (Fig. S3B). We also used markers of apical polarity in Drosophila and mosquito epithelia (Cad99c, Na+/K+ ATPase and H+ V-ATPase; Fig. S3C–E) to demonstrate that staining in larval gastric cecae (Fig. S3Cii, Dvii, Dix, Ev–vii) was consistent with prior reports (Dow et al., 1997; Filippova et al., 1998; Lebovitz et al., 1989; Okech et al., 2008; Hua et al., 2013; Wend et al., 2013; Chung and Andrew, 2014; Chen et al., 2020). We showed that larval SG secretory cells appeared to be only weakly apically polarized (Fig. S3C–E). Staining with two different polyclonal antisera targeting CrebA (rabbit, rat) raised against full-length protein completely overlapped (Fig. S4A). The presence of colocalized signals suggests that both CrebA antisera recognize the same feature in these cells, and our staining result is likely not an artefact linked to a single reagent. A lamin C and Sage co-stain showed overlap in the nuclei of individual SGs as well as at the basement membrane of the gland (Figs 3 and 4). We found similarly broad localization of signal in SGs for lamin, and Sage following pre-incubation (Fig. S4B), suggesting that this broad localization is likely real staining. We also stained with pre-immune serum from animals in which we made Drosophila CrebA or Sage antibodies (Fig. S4C, D). When we stained with pre-immune serum alone, there was little to no signal, which meant that this animal was not already exposed in some way to CrebA or Sage, giving us greater confidence in our staining results. Finally, we confirmed that Sage and CrebA did not localize to the nuclei of GC, but they did localize to the SG nuclei of the same animals (Fig. S4E, F).
Larval SG secretion may involve extracellular vesicles
We used Rab11 (vesicles related to recycling endosomes) to visualize secretory vesicles in mid-L4 SGs (Fig. 5). The lumen shape was irregular throughout both sacs and cell sizes were variable (Fig. 5Aiii–v). Strong apical lipid enrichments were observed (Fig. 5Aiv–vii). However, lipid enrichment at the apical membrane was not continuous across all cells; fading was observed primarily at the connections between adjacent cells (Fig. 5Av, vi). Vesicle structures accumulated just proximal to the lipid enrichments, outlining the border of SG cells completely (Fig. 5Axii). Intraand extracellular vesicles were seen in the open region connecting the proximal and distal sacs (Fig. 5Av–vii). Rab11 signal was found intracellularly and in the lumen (Fig. 5Av–xii), suggesting vesicle distribution throughout the tissue. Nuclear territory size was variable throughout the gland, sometimes filling up much of the cell volume (Fig. 5Aiii–vii, xiv–xvi), particularly in cell clumps found at the external boundary of glands (Fig. 5Axiv–xvi). High magnification imaging of these cells showed strong localization of fibrillarin (nucleolus) signal (Fig. 5Aix–xvi). Nuclear enrichment and broad cytoplasmic localization of Mucin staining were observed (Fig. 5B).
Figure 5. An apical brush border is visible during lumen formation and SG TFs localize to SG secretory and duct cell DNA.
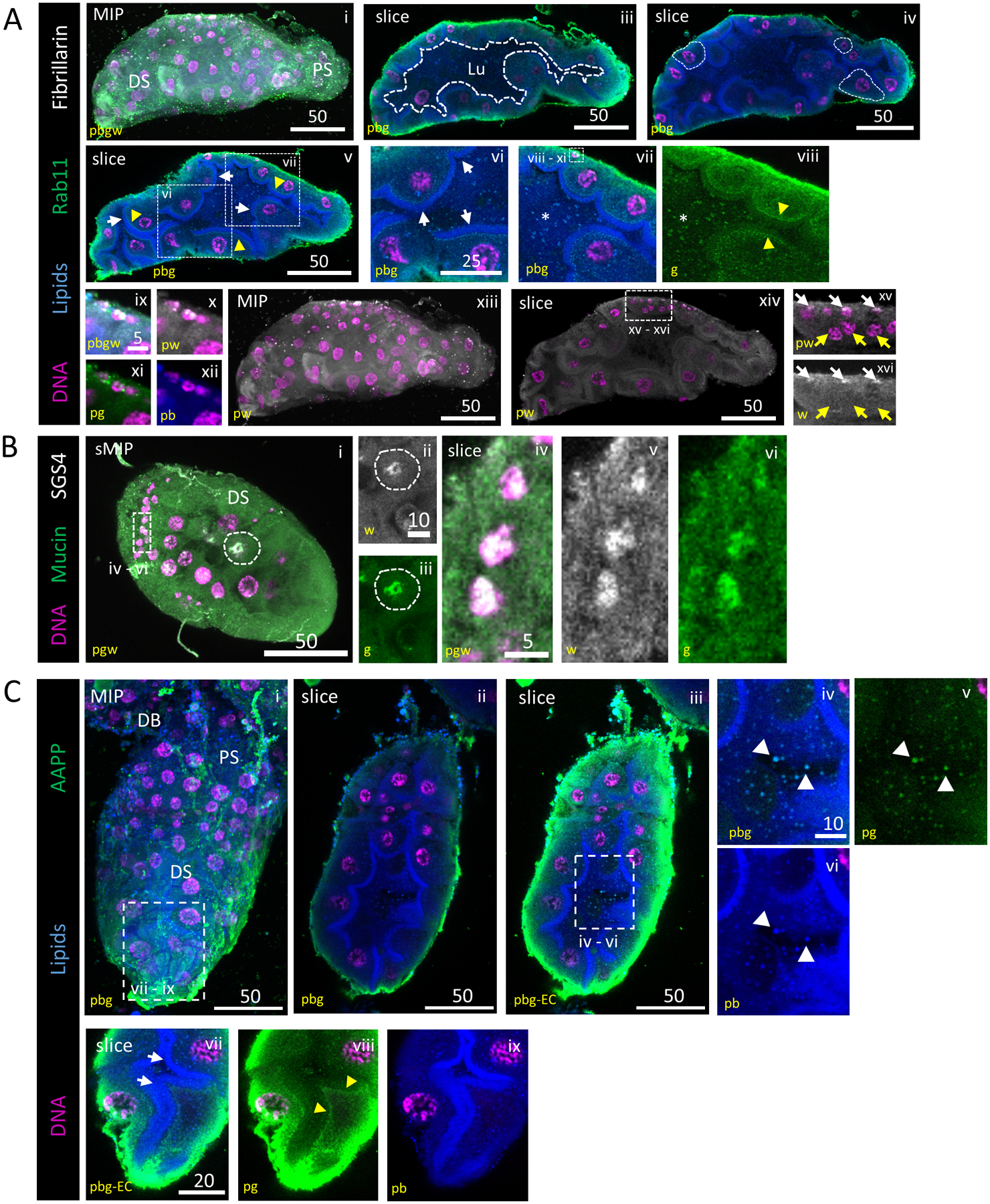
Representative images from dissected L4 larval salivary glands stained with Hoechst (DNA, purple), Nile Red (lipids, blue) and antisera against Rab11 (vesicles, green) and either Fibrillarin (nucleoli, grey; A) or Sage (transcription factor, grey; B). (A) Secretory architecture of larval SGs. 3D and slice overviews are shown (i, ii). A growing, irregularly-shaped lumen (iii; white dashed outline) was seen. Larval SG cell size (iv; white dashed outlines) was variable. A brush border (vi; arrows), comprised of an apical lipid enrichment in many, but not all, cells, was present, confirming that these cells are polarized and specialized for secretion. Vesicle-like puncta were observed in the cytoplasm of secretory cells and in the lumen (vii; asterisk). Rab11-positive vesicles were enriched sub-apically, basal to the brush border (viii; arrowhead). Fibrillarin localization was only apparent in the smallest nuclei (ix–xvi) at the basal surface (xv–xvi; white basal arrows, yellow central arrows). (B) Mucin and SGS4(saliva proteins) staining were localized to a perinuclear enrichment, the broader secretory cell cytoplasm and the lumen. (C) AAPP, a saliva protein, is seen enriched at the basal surface of secretory cells, in intracellular vesicles and in extracellular vesicles (white arrows, iv–vi). AAPP-containing vesicles were seen enriched just basal to the apical lipid enrichment (yellow arrows, vii–ix). Different mages from this L4 SG were also included in Fig. S3B.
We also visualized mucin (an SG-secreted glycoprotein component of mucus) and additional saliva proteins with functional roles identified only in adult blood feeding (Yoshida et al., 2008; King et al., 2011). SGS4 (saliva) enrichment was found in the same regions of the nuclei that Mucin was localized (Fig. 5B), suggesting proteins with known roles in blood feeding are present in the developmental stages of the SG. AAPP (saliva) signal was enhanced at the apical boundaries of cells (Fig. 5Cviii) and present in the lumen within vesicles (Fig. 5Civ–ix), similar to Rab11 localization in relation to lipids (Fig. 5Av–viii). From these images, we describe properties of larval mosquito SG secretion, and we show that saliva proteins functioning in blood feeding are present in the larval SG. These secretion data (Fig. 5) and the broad organelle marker localization observed (Fig. 3), support the conclusion that we have indeed identified extracellular vesicles as a secretion mechanism contributing to larval SG saliva.
Larval SG gene expression compared to other stages and tissues
In order to objectively compare larval SG gene expression patterns to that of adult SGs or larval gut compartments, we conducted a meta-analysis of diverse existing microarray datasets (Neira Oviedo et al., 2008; 2009; Baker et al., 2011) using Chipster (Kallio et al., 2011). Following normalization of the raw data, gene-level results for each sample were binned as either “expressed” or “not expressed” prior to comparisons to other stages and/or tissues. Gene expression status was the total scope of our analysis; gene expression levels were not considered. Importantly, about half of all genes (6706 out of 13 039) were not expressed in the SG (Fig. 6A). Many genes (3332 out of 13 039) were expressed in all three SG tissues (Fig. 6A). Gene ontology (GO) term functional analysis of all 3332 co-expressed SG genes showed enrichment for ribosomes, consistent with a role for this organ in high level protein production and secretion (Fig. 6B). Interestingly, each SG type had a large category of specifically expressed genes (female-894, male-749, larval-812; Fig. 6A).
Figure 6. Comparisons of salivary gland gene expression profiles and functional category analysis.
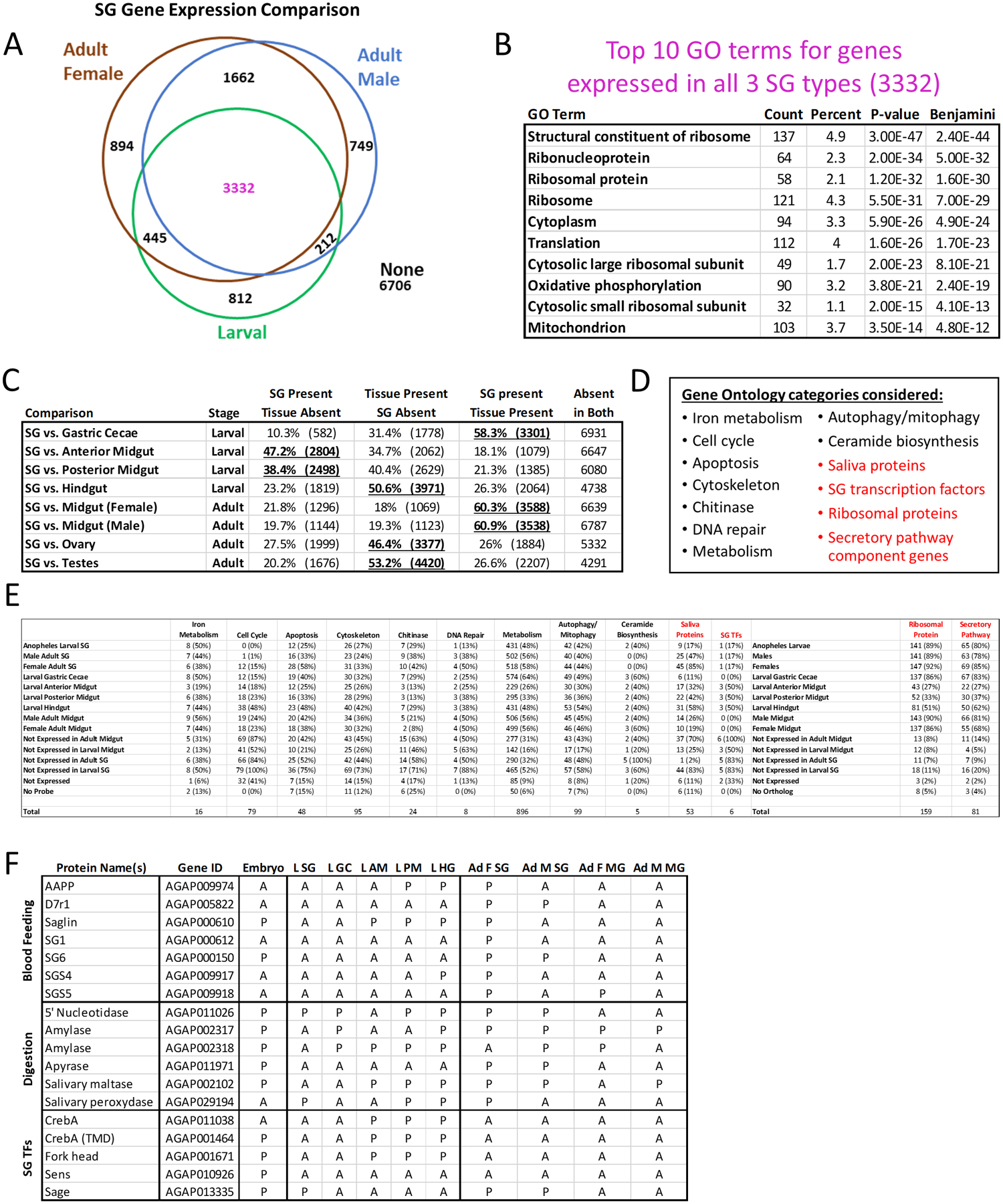
Microarray data from adult female, adult male and larval Anopheles gambiae SGs were downloaded, normalized using Chipster and compared (A–F). (A) Venn diagram depicting numbers of overlapping genes flagged as expressed or not expressed. (B) Top 10 gene ontology (GO) terms from analysis of the 3 332 genes expressed in all three SG tissues (magenta font). (C) Whole genome expression overlaps between larval or adult SGs and other tissues. Bolded, underlined formatting emphasizes striking results. (D) List of GO terms identified from analysis of the other classes of genes from (A; black font) or of known interest in SG biology (red font). From these categories, a list of 587 (474 unique) genes was created (Table S2). (E) Breakdown of genes expressed within GO term categories by tissue. The “Ribosomal Protein” and “Secretory Pathway” genes were originally referenced to Drosophila, while the other categories were referenced to Anopheles, via KEGG pathway diagrams. (F) Gene level expression data (A, absent; P, present) in three categories (blood feeding, digestion or SG TFs) across development and multiple tissues.
To assess the degree of similarity between SG gene expression and that of other tissues, we compared expressed gene sets in larval or adult tissues (Fig. 6C). Among all organs of the larval alimentary canal, the larval SGs shared the greatest number of expressed genes with the gastric cecae (Fig. 6C). We also noticed a loose correlation among the alimentary organ data suggesting that neighbouring organs had more similar expression patterns than organs further from each other (Fig. 6C). By comparison, the adult SGs shared a similar degree of gene expression overlap with the adult midgut, far greater than with sexmatched reproductive tissues (Fig. 6C).
We combined GO term results obtained from the gene categories (Fig. 6A) expressed in a subset of SG tissue types (Fig. 6D, E, black text), as well as SG genes of interest (Fig. 6D, E, red text). A complete list of these genes is located in Table S2. From these functional categories, we assembled lists of all related genes using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway diagrams and compared the fractions of expressed genes within categories across tissue types (Fig. 6E). These data suggest an overall dearth of cell cycle and apoptosis related gene expression, consistent with the developmental timing of these studies occurring prior to, or after, pupal SG histolysis and adult SG formation (Fig. 6E). We also find that mosquito orthologues of known Drosophila SG TFs are not expressed in larval gastric cecae and that many saliva protein genes with known roles only in this context are also expressed in non-SG tissues (Fig. 6E).
We examined expression of three classes of genes in more detail (blood feeding, digestive and SG TFs), in order to discern broader differences in tissue level gene regulation. During the larval stage, blood feeding genes were primarily detected in posterior sections of the gut. Digestive and SG TF gene expression was detected throughout the larval SG and gut compartments. While this analysis allowed us to better understand gene expression, we do not have necessary information related to protein turnover rates (eg SG TFs produced in the embryo or earlier larvae) in order to better understand ongoing gene regulation. In adults, we see SG-biased expression for blood feeding genes, digestive gene expression in both SGs and midgut, and expression of sage (TF) in the SGs.
Gene expression meta-analysis led to identification of 496 unique genes expressed in all included data (Fig. S6A). Of the 57 genes that are well annotated (included common names), all were housekeeping genes. We also uncovered 86 genes expressed only in larval SGs among all analysed tissue data sets (Fig. S6B). These genes represent a novel list of candidate targets for future strategies like mutation and RNA interference that could reduce larval SG viability or function, likely impacting adult SGs and transmission ability.
To support our gene expression meta-analysis approach, which involved binary (on, off) analysis of datasets collected at different times in different labs, we conducted reverse transcriptase PCR (Fig. S6C) and quantitative reverse transcriptase PCR abundance analysis (Fig. S6D) of: two control genes (actin and elongation factor), as well as three SG TFs (CrebA, fkh and sage), two saliva proteins important for adult blood feeding (SG6 and saglin) and three digestive enzymes (50 nucleosidase, maltase and peroxidase) in pooled Anopheles gambiae (G3 strain) L4 SG, GC and midgut replicates. The primers used for PCR are described in Table S3. Results demonstrated that for all targets except sage, which was not present in midguts, every gene was expressed in every tissue sample in one or more of the five biological replicates, to widely varying levels. These data support staining results for SG TFs (Fig. 4) and saliva proteins (Fig. 5) and suggest continuity across labs and strains. In summary, we suggest that it is valid to compare these PCR data with staining data and microarray data from multiple sources solely on the basis of genes expressed versus not expressed.
Discussion
Summary of L4 SG morphological and secretory findings
A depiction of what larval SG morphologies and localizations were hypothesized, and what was observed, is presented in our summary diagram (Fig. 7). Our idealized projection of a 3D SG (Fig. 7A) included uniform layers of SG cells throughout the proximal and distal sacs. Nuclear size was the same across the entire hypothetical gland. In cross sectional views of the ideal gland, we expected uniform lumen expansion and a singular layer of cells at the border. Observed SGs (Fig. 7B) allowed for visualization of actual morphologies of cells. Cell size, cell shape, nuclear size and nuclear shape varied: cytoplasm took up a larger portion of single cells in some cells and not others. Cross section views of the proximal and distal sacs showed nonuniform lumenal cavities. A monolayer of cells bordering the gland was not actualized, nor were cells only attached from one, basal side: cells were also sometimes found in the central gland lumen. After observation of many signals marking organelles (cytoskeleton, nucleus, protein synthesis), we produced a diagram for better visualization of their localizations (Fig. 7C). L4.2 and L4.3 SGs showed apical lipid enrichment (Fig. 7D), whereas L4.1 and L4.4 SGs showed no such enrichment (Fig. 7E). Duct bud-localized TFs (Fig. 7F) included CrebA [a TF that upregulates secretory capacity (Abrams and Andrew, 2005; Fox et al., 2010)], Cut [a TF involved in hindgut specification (Hatton-Ellis et al., 2007)], dMEF2 [a TF acting in muscle (Nguyen et al., 1994)], RNA Pol II and Sage [an SG-enriched TF that promotes SG viability and expression of saliva proteins (Fox et al., 2013)].
Figure 7. Summary of Larval Salivary Gland Morphology.
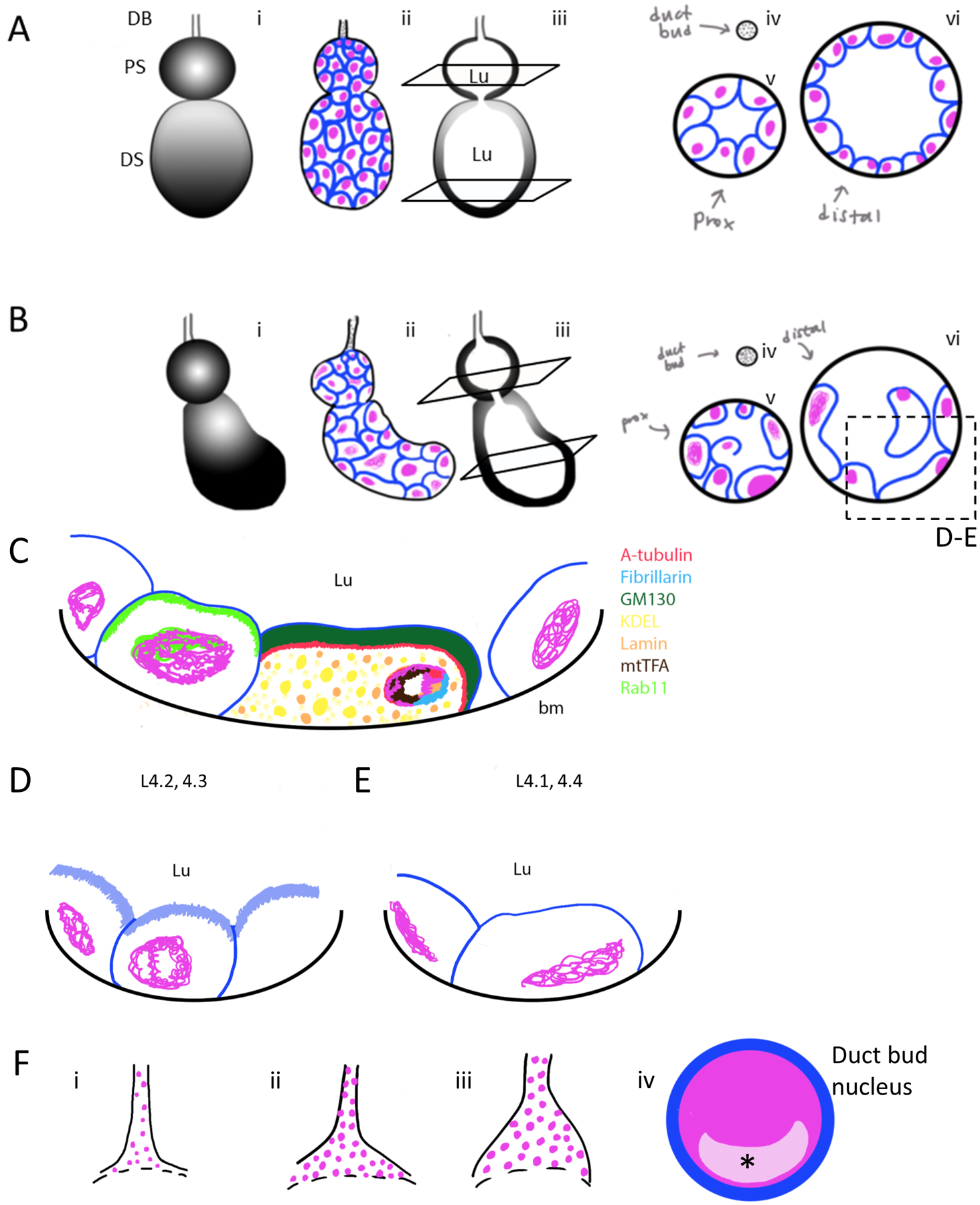
DNA is coloured magenta and individual cells (lipids) are coloured blue. (A) A 3D cell architecture model of an hypothetical, idealized Anopheles gambiae L4 larval salivary gland (SG) cell architecture (i). Schematic diagram for basal view of squamous secretory cell organization; proximal sac is smaller and rounder than the distal sac, nuclear and cell size are uniform in both (ii). A sagittal cross section depicts a connection between the proximal and distal sac; the lumenal compartment is full of saliva (iii). Coronal cross-section views of the duct bud, proximal sac and distal sac show a uniform arrangement of a monolayer of cells; nuclear and cellular size are uniform (iv–vi). (B) A realistic 3D model of observed Anopheles gambiae L4 larval SG cell architecture post dissection (i). Basal view of secretory cell organization shows a proximal sac smaller and rounder than the distal sac; the distal sac juts out in a curve away from the middle. Cell shape is varied, DNA morphology suggests these cells are probably in interphase or prophase cell cycle stages. Large and small cells containing large or small amounts of nuclear DNA are observed (ii). A sagittal cross-section depicts a connection between the proximal and distal sacs, and the lumen extends throughout the SG (iii). Coronal cross-section views of the duct bud, proximal sac and distal sac show various cell shapes and sizes (iv–vi). An apical lipid enrichment (brush border-like structure) is present on cells attached to the outer border of the SG but disappears on those attached only to other cells (v). DNA levels vary and are observed in interphase and prophase. Cells of the distal sac are frequently larger than those in the proximal sac. (C) Localization of different antibodies marking organelles on slice image of SG include alpha-tubulin (cytoskeleton, red), fibrillarin (nucleolus, blue), GM130 (golgi, dark green), KDEL (ER, yellow), lamin (nuclear envelope, orange), mtTFA (mitochondria, brown), rab11 (vesicles, bright green). bm-basal membrane. (D–F) Magnification of L4 SG cells at L4.2–L4.3 stages have increased lipid enrichment (light blue, D), while L4.1 and L4.4 stages have greatly decreased, or no, lipid enrichment. (G) Duct bud variability was observed, ranging from smaller ducts containing few cells (i) to elongated, larger stems (ii) to wider connection at the opening of the proximal sac (iii). Transcription factors found to be localized in the nuclei, enriched in a single subnuclear territory (asterisk/light pink area) of duct bud cells included CrebA, Cut, dMEF2, RNA PolII and Sage.
Confidence in staining data
Our novel larval SG dye and antibody staining method performed robustly. We were surprised when our adult SG staining method (Wells and Andrew, 2015; 2019; Wells et al., 2017) did not allow the markers to penetrate the basement membrane (data not shown). It is possible that there could be differences in lipid composition across life stages that could affect basement membrane permeability. We supported our staining methodology with multiple, stringent control tests (Figs S3, S4). Many of the antibodies we utilized were purchased from the Developmental Studies Hybridoma Bank (DSHB; https://dshb.biology.uiowa. edu/), which contains extensive antibody validation information for these reagents.
Stage comparisons in SG cell architecture
The cellular and tissue differences that we identified between larval and adult stage An. gambiae SGs were striking. Moreover, differences we observed within L4 sub-stages and within, and among, individual SGs at a given substage were also noteworthy. We previously described how adult SG cells change shape from cuboidal to cup-shaped cells through creation and apical fusion of a presumptive secretory compartment during the first 4 days of adulthood (Wells et al., 2017). In larval SGs, we find secretory cells that are highly variable in size and shape (Figs 1, S1). This intraregional variation existed in the secretory sacs of the larval SG, but not the duct bud (Fig. S1); cell size appears much more regular within each region of adult SGs (Wells and Andrew, 2015; 2019; Wells et al., 2017). In adult SGs, every nucleus displayed diffuse DNA-staining consistent with interphase (Wells and Andrew, 2015; 2019; Wells et al., 2017); however, in larval SGs, we often saw discrete condensed (Hoechst-enriched) DNA regions consistent with prophase chromosome condensation (eg Fig. S1B). RNA Pol II localized to all regions of the nucleus (Fig. 4B), but it appeared enriched at sites where Hoechst was also high (Fig. 4BBiii, vii, xi). This enrichment suggests it may be unlikely that Hoechst staining here simply correlates with the degree of heterochromatinization (Zielke and Saumweber, 2014).
In larval SGs, we find that the size of a passageway that connects the proximal and distal sac lumens correlates with lumen size (Figs 1, 2 and S1). In addition, we saw that the salivary duct (only barely visible interior to the duct cells apical to the duct bud region) does not extend into the proximal sac (Fig. 2). In contrast, within adult SGs, a continuous lumen is formed very early after eclosion (Wells et al., 2017), and the salivary duct extends a short distance (medial lobe) or over halfway (lateral lobes) into the lumens (Wells et al., 2017). Salivary duct extension may impact function of the SGs in adults, but it is worth noting that the biological importance of larval SGs is only partially elucidated (Neira Oviedo et al., 2009).
Larval SG cells possess an apical, lipid-rich brush border during the time of L4 stage when the SG lumen is expanding (Fig. 1), a feature either not elaborated/visible or not present at adult stage (Wells and Andrew, 2015; Wells et al., 2017). An elaborate apical brush border, associated with both secretion and absorption, has been described previously in Anopheles midgut cells (Lehane and Billingsley, 1996). Lipid signalling related to this apical enrichment and Nile Red-positive vesicular secretion could be an important indicator of mosquito health, nutrient levels, lipid homeostasis, metabolism, immunity, cellular signal transduction (including hormone signalling), secretion itself and other biological processes. Elongated cell shapes, especially in L4.4 SGs (Figs 1D, S1F, S2, S4Bv–ix, S4E), could be a result of cell movement and/or stretching.
Structure and fate insights related to the SG duct bud
The mosquito SG DB region is retained during pupal histolysis of the secretory sacs, and it goes on to form the adult SG. This appears largely analogous to Drosophila, in which imaginal ring cells construct the long, narrow, tubular adult SG (Chung et al., 2014; Wells and Andrew, 2015; Wells et al., 2017). DB cells are homogenous in size and shape, have a small cytoplasm and likely possess a low DNA copy number. DB cell morphology differs greatly from the extensively variable size and shape of secretory sac cells. Moreover, DB DNA was only observed in a decondensed, interphase-like organization, while secretory sac DNA was found in both interphase-like (Figs 1, 2 and S1), potentially prophase-like conformation. DB cells, unlike secretory sac cells, never showed an apical enrichment of lipids consistent with an apical brush border-like structure. Interestingly, we observed no chitin [wheat germ agglutinin (WGA) staining] enrichment in the DB region closest to the proximal sac, where the salivary duct of the adult SG resides; there is, however, a shadow visible by lipid staining and light microscopy (Fig. 2) that could indicate a scaffold for building the future adult duct.
It was previously unclear to what extent DB cells and secretory cells vary, and whether DB cells have stem cell identity. We found that duct bud nuclei express SG TFs, Mucin and other saliva proteins (Figs 2 and 5), and not the multipotency factors Notch and Delta (Fig. S2) indicating that these cells are likely committed to a secretory cell (perhaps even an SG cell) lineage during larval stage 4. Notably, SG TF and saliva protein signal were consistently enriched in a discrete, nuclear subdomain (Figs 4 and 5). Proximal and distal sac cells could send signalling cues to duct bud differentiation prior to histolysis. Interestingly, we found no evidence of a boundary between the duct bud and the proximal sac (Fig. 2). How the DB region is selectively preserved in pupae is of great interest moving forward. Very little is known regarding the pupal SGs in Anopheles. By comparison, Drosophila pupal SGs are known to secrete subcuticular fluid (Sarmiento and Mitchell, 1982). Pupal stage mosquito SG studies are warranted in order to discern the molecular signalling controlling adult SG development and maturation.
Gene expression meta-analysis uncovered described relationships and new targets
Our detailed gene expression meta-analysis (Fig. 6) identified extensive similarities and differences between larval and adult SGs. While each tissue is secretory in nature (satisfyingly, the GO terms associated with secretion and translation were identified in the category of genes expressed in all three SG types), the saliva produced indeed varies by not only sex, but also by life stage. This variation is partially attributable to variation between cells, variation among individuals, temporal differences and differences in physiological events or experiences (eg feeding behaviour and outcomes). Larval SG gene expression aligned substantially with the gastric cecae, but not other gut compartments, while adult SG gene expression was much more similar to the adult midgut than sex-matched reproductive tissue. Our imaging included a large number of larval gastric cecae, allowing us to discern their comparatively low cell size variability, but similar subcellular organization, to larval SG cells (Fig. S1). Staining for two TFs acting in gut cells [Cut and dMEF2 (Liu et al., 1991; Nguyen et al., 1994)] was observed in both SGs and gastric cecae (Fig. 4C, D).
Beyond gene expression, larval and adult stage SG and gastric cecae/midgut cells have several notable similarities (including apical membrane elaborations, intra- and extracellular secretory vesicles, and secretory organizing compartments) within their cytoplasms and precursor cells populations. Chief among their differences are the TFs acting in each tissue type and the secretions produced by each tissue. Consistent with this, no staining for the orthologues of the Drosophila SG TFs CrebA and Sage was observed in gastric cecae (Fig. 4E, F). Perhaps this distinction drives some portion of the expression differences between these two tissues. Across all larval tissues included, gene expression was most similar among neighbouring tissues along the anterior–posterior axis [Fig. 6, (Neira Oviedo et al., 2008)]. Although not well studied in Anopheles, this correlation may indicate that Hox and other developmental patterning genes (Powers et al., 2000; Devenport et al., 2000; Goltsev et al., 2004; 2007; Ahanger et al., 2013; Heffer and Pick, 2013) drive segmental gene expression differences in larval mosquitoes. We were also able to derive a list of genes expressed across all samples studied (Fig. S6A) or only in larval SGs (Fig. S6B); the latter list constitutes a novel group of candidate targets for SG genetic intervention in future studies.
SG TF localization
Binding of the conserved SG TFs Sage and CrebA in larval SG nuclei varied between cells, from faintly diffuse (Fig. 4Aiii, v) to strong foci (Fig. 4Aii–xv). This variation might indicate different states of secretory activity and suggest that larval mosquito SG cells differentiate and/or function individually, not as a single collective tissue, as is the case in Drosophila (Myat et al., 2019). We also observed cytoplasmic SG TF signal in larval SG cells (Fig. 4ii–ix). One possible explanation could relate to our observations of Rab11-positive vesicles located both intra- and extracellularly (Fig. 5A).
Mechanism of larval SG secretion
It was recently hypothesized the mosquito saliva may contain extracellular vesicles (Arcà et al., 2019). Mosquito midgut cells are known to secrete via multiple exocrine mechanisms (Lehane and Billingsley, 1996), and localization of multiple proteins that are tightly enriched in Drosophila tissues (Filippova et al., 1998; Lebovitz et al., 1989; Chung and Andrew, 2014) is more broadly localized in mosquito gut cells (Filippova et al., 1998; Okech et al., 2008; Hua et al., 2013; Chen et al., 2020). As an example from the current study, the broad lamin C localization observed (Figs S1F, 3A) could indicate either a mutation present in Anopheles gambiae that is partially disrupting function, or lack of expression of a necessary partner protein (Vaughan et al., 2001). In the case of tardigrades, disruption of lamin localization by loss of a partner protein has led to a broader, compensatory cytoplasmic localization (Hering et al., 2016).
Canonical exocrine secretion has been the major mechanism described to date in Drosophila SGs (Massarwa et al., 2009), but apocrine secretion is thought to occur in late larval/early Drosophila pupal glands undergoing histolysis (Farkaš et al., 2014). If apocrine secretion indeed operates in homeostatic Anopheles SGs, protein localization would likely be broader in general than expected from Drosophila (organelle markers, SG TFs, secreted proteins; Figs 3–5) since these vesicles can contain any cellular contents (Gurunathan et al., 2019). CREB3L2, a CrebA orthologue, was identified in exosomes derived from human endothelial cells (van Balkom et al., 2015). Extracellular shuttling of cellular material between SG cells could contribute to functional coordination between cells. Apical lipid enrichments (Fig. 1) and a broad cellular distribution of organelles (Fig. 3) may be indicative of vesicle transport functionality both intra- and extra-cellularly in L4 stage SGs. Secretion in adult An. gambiae SGs are remarkably different, instead relying on fusion of a subapical compartment to the apical surface of cells that places the secretion machinery at the cellular interface with the secretory cavities/lumen (Wells et al., 2017). These findings highlight the importance of further study of mosquito saliva as a delivery mechanism for anti-malarial interventions to parasites in mosquitoes or to mammals. Approaching the larval SGs with a single-cell multi-omics approach could allow us to directly address these questions in future studies.
Broader impacts
It is of keen interest to study the secretory function of these SGs and the roles of SG TFs further in the context of Anopheles to devise mechanistically diverse, novel strategies to disrupt the SGs and/or block pathogen transmission in adults. In support of that goal, this study: (1) uncovered new candidate targets for SG disruption (Fig. S6B) by employing an under-utilized approach (meta-analysis of gene expression data); (2) described a new staining approach applicable to efficacy testing of larval and adult genetic, molecular and chemical treatments and interventions and (3) highlighted the remarkable cell biology of this system and the changes that happen across developmental stages in this medically relevant context.
Experimental procedures
Mosquito maintenance
Anopheles gambiae (Keele) mosquitoes were obtained from the Johns Hopkins University Bloomberg School of Public Health Malaria Research Institute Insectary as L4 larvae maintained by Insectary staff at 80–100% humidity and 28 degrees Celsius as described previously (Das et al., 2007).
Dissection, fixation and staining
L4 larvae of both sexes were collected together at various times during both days of this stage, indiscriminately, in bulk from 1600 × 2400 plastic rearing trays using a plastic transfer pipette. Larvae were transferred into room temperature water in a small, plastic box during dissections. Individual larvae were transferred from the plastic box to a dissection plate (petri dish filled with Xiameter silicone curing agent) spotted with either water or 25% EtOH. The heads and attached SGs were removed from the body using fine-tipped tweezers by gripping the head (non-dominant hand) and just below the neck (dominant hand) and gently pulling in opposite directions with minimal constant force. Larval heads with SGs were then gently transferred on top of 1 ml of 1X phosphate buffered saline (PBS) in microcentrifuge tubes sitting partially in ice. Each aliquot of 40–50 “heads with SGs” was subsequently processed for fixation and staining. The PBS solution was drained from the microcentrifuge tube and replaced with a 3:1 ratio of methanol to glacial acetic acid. The tube was placed at 4 degrees Celsius for a 19-h fixation. (Day 2) Next, the fixative was removed, and the SGs were washed with cold acetone for 90 s. The SGs were then washed briefly and gently with 1X PBS. Primary antibodies in fresh 1X PBS were added to the SGs, and samples were incubated overnight at 4 degrees Celsius. (Day 3) The antibody solution was removed and secondary antibodies in fresh 1X PBS were added to the tube (without any intermediate washes) before incubating at room temperature (shielded from light) for 150 min. After 105 min of this incubation, 3 μl of Nile Red (lipids) and 2 μl of Hoechst (DNA) were added and incubation continued for 45 min. The glands were briefly washed in 1X PBS three times, and a final 200 μl 1X PBS was added.
Mounting
Stained glands were transferred from the microcentrifuge tube to a VWR Superfrost Plus microscope slide with a soft brush. Two hundred microliters of 100% glycerol were added to the sample. Glands were then positioned perpendicular to the long axis of the slide under glycerol, using ultra-fine-tipped forceps. With one tweezer holding the head of the larvae, the larval glands were separated from the head (double dissection method). The discarded heads were thrown away and this process was repeated for all larval glands. A coverslip was added. Slides were sealed with nail polish and stored at 4 degree Celsius.
Adult SG staining
Day seven adult Anopheles gambiae female SG labelling (Fig. S3F) with 40,6-diamidino-2-phenylindole (DAPI; DNA) and anti-Lamin C antisera (Fig. S3F) was conducted as described previously (Wells and Andrew, 2015) using the Anopheles gambiae protocol variation (Wells et al., 2017).
Confocal microscopy
Imaging was conducted using a Zeiss LSM700 laser scanning confocal microscope housed in the Johns Hopkins University School of Medicine Microscope Facility. The step size used in 3D image stack captures was either 0.7 or 1 micron. Each figure is composed of representative images from between six and 20 imaged glands, selected from over 400 dissected and stained glands. Each gland dissected, stained and fixed was processed on one to four different days, depending on the markers used. Image processing was completed in Zeiss Zen 2010 and Adobe Photoshop CS4. Scale bar lengths are given in microns. SG lobes were outlined with a dashed yellow line in overview images, and lobe morphology labels were abbreviated as: DB-duct bud, DS-distal sac, Lu-lumen, PS-proximal sac. Gastric cecae (Figs S1, S2) were labelled: GC. The fluorescent channels displayed are denoted in each image by yellow letters referring to the marker labels at the left of each figure part (p-purple, b-blue, g-green, w-white). “-ec” was added when the contrast of an image was uniformly enhanced to improve clarity. Image view depth and projections were noted in each figure [MIP-maximum intensity projection over half the SG z-axis depth, sMIP-maximum intensity projection over an SG depth shorter than half, slice(s)-single z plane image(s) spliced with a dashed white line when necessary (eg Fig. 1Ciii)].
Antibodies and dyes
The antibodies and dyes employed in this study were: Hoechst dye (bisBenzimide H 33342 trihydrochloride; 1:200, Millipore Sigma, B2261), Nile Red dye (1:50; Sigma 19 123), WGA dye (1:40, Vector Laboratories RL-1022), Mucin (Muc2; 1:100, Proteintech 27 675–1-AP), Lamin C (mouse, 1:100, LC28.26, DSHB), mtTFA (rabbit, 1:100, H-203, Santa Cruz Biotechnology), KDEL (mouse, 1:100, ab69659, Abcam), GM130 (rabbit, 1:100, ab30637, Abcam), Fibrillarin (38F3; mouse, 1:100, ab4566, Abcam), alpha-tubulin (mouse, 1:100, 12G10, DSHB), Sage [rat, 1:100, Andrew lab (Fox et al., 2013)], Sage [guinea pig, 1:100, Andrew lab (unpublished)], CrebA [rabbit, 1:100, Andrew lab (Fox et al., 2010)], CrebA [rat, 1:100, Andrew lab (Andrew et al., 1997)], RNA polymerase II (POLI; rabbit, 1:100, Proteintech 13 635–1-AP), Cut (mouse, 1:100, 2A10, DSHB), dMEF2 [rabbit, 1:100, gift from Elizabeth Chen (Nguyen et al., 1994)], Rab11 [rabbit, 1:100, Andrew Lab (unpublished)], SGS4 [rabbit, 1:100, gift from Julian Hillyer (King et al., 2011)], Anopheles antiplatelet protein (AAPP) [rabbit, 1:100, gift from Hiroyuki Matsuoka (Yoshida et al., 2008)], Cad99c antibody [mouse, 1:100, gift from Christian Dahmann (Schlichting et al., 2005)], H+/V-ATPase (mouse, 1:100, 224–256–2, DSHB), Na+/K+ ATPase (mouse, 1:100, a5, DSHB), Notch (mouse, 1:100, C17.9C6, DSHB), Delta (mouse, 1:100, C594.9B, DSHB). LC28.26 was deposited to the DSHB by Fisher, P.A. (DSHB Hybridoma Product LC28.26). 12G10 anti-alpha-tubulin was deposited to the DSHB by Frankel, J. / Nelsen, E.M. (DSHB Hybridoma Product 12G10 antialpha-tubulin). 2B10 was deposited to the DSHB by Rubin, G.M. (DSHB Hybridoma Product 2B10). 224-256-2 was deposited to the DSHB by Gerisch, G. (DSHB Hybridoma Product 224-256-2). a5 was deposited to the DSHB by Fambrough, D.M. (DSHB Hybridoma Product a5). Secondary antibodies (anti-mouse 647, anti-rabbit 488, anti-rat 647 and antigoat 488 secondary antibodies (1:200) were purchased from ThermoFisher Scientific.
Developmental staging
Imaged SGs were ascribed a stage (L4.1-L4.4) based on two prominent cellular features: (1) apical lipid accumulation, and (2) lumen presence/size (see Fig. 1E and Results section). Of over 500 total SGs imaged (representative of the entire range of SGs available at the time of collection, which was purposely varied, by experimental replicate, across both days of L4 to sample all of this stage), 16% were L4.1, 39% were L4.2, 39% were L4.3 and 6% were L4.4. The following is a list of the stage of each SG shown in this study: L4.1: 1A, 3Di, ii. L4.2: 1B, 2, 3A, 3Diii–vi, 4A, 4D, 5A, 5B, S1B, S3A, S3C, S3Di–v, S4Bi–iv, S4G. L4.3: 1C, 3B, 3C, 3E, 4B, 4C, 5C, S1A, S3B, S3Dvi–ix, S3E, S4A, S4F, S4H. L4.4: 1D, S1F, S2, S4Bv–ix, S4E.
Quantitative image analysis
Nuclei were counted manually by eye using microscope software using every z slice from full SG depth z stacks. Nuclei volume and cell volume were calculated using the Zeiss Zen 2.0 software.
Microarray data analysis
Data from prior studies (Neira Oviedo et al., 2008; 2009; Baker et al., 2011) were downloaded from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) using Accession numbers (Table S1) obtained from the Vectorbase website (https://www.vectorbase. org/). Raw replicate.CEL files were imported into Chipster (Kallio et al., 2011), accessed through the European Bioinformatics Institute (EBI) marketplace, then combined and normalized using the gcRMA method and Bonferroni-corrected P values. Expression flags denoting certain expression, possible expression and not expressed were added to the data by Chipster during normalization. Results were exported and analysed further using Microsoft Excel 365, which included current gene name standardization using a Refseq gene ID table obtained from the UCSC Genome Table Browser (Kuhn et al., 2013). Gene lists were used as input for Gene Ontology analysis using The Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009a; 2009b). The proportional Venn diagram (Fig. 6A) was created using Venn Diagram Plotter (https://omics.pnl.gov/software/venn-diagram-plotter), then traced in Microsoft PowerPoint 365 to allow for editing of colours, line thickness, and annotations.
Reverse-transcriptase polymerase chain reaction
As independent validation in support of the gene expression metaanalysis approach, SG, GC and midguts from Groups of 30 L4 larvae were dissected and processed as a pool (two technical replicates across five biological replicates). RNA extractions were carried out using an RNeasy Micro kit (Qiagen, Germantown, MD). For gene expression analysis, complementary DNA was produced using 0.5 μg of RNA, using a High-Capacity cDNA Reverse transcription kit (Applied Biosystems, Foster City, CA) for a 20 μl reaction. A semi-quantitative PCR amplification was performed using the AccuStart II PCR Supermix (Quantabio, MA), 10 μM primers (Table S3) and 1 μl of cDNA, with an annealing temperature of 55 degrees C and 35 cycles. PCR products were visualized in 1.5% agarose gels stained with ethidium bromide.
RT-qPCR was performed using the Power SYBR-green PCR master MIX (Applied Biosystems) and the same sets of primers as for the semi-quantitative PCR, in the QuantStudio6 Real Time PCR System (Applied Biosystems). Gene expression was calculated relative to the Elongation Factor housekeeping gene (VectorBase: AGAP005128) and expressed as Abundance [2−ΔCT]. Primers used are listed in Table S3.
Supplementary Material
Acknowledgements
We extend our deepest gratitude to Deborah Andrew for funding (NIH grant numbers 5RO1DE013899 and R21AI153588 to Deborah J. Andrew), including salary support (MC, MW), reagent and equipment costs and use of lab-generated antibodies), intellectual input, project planning and comments on the manuscript. We humbly thank the Johns Hopkins Malaria Research Institute for funding through a JHMRI Postdoctoral Fellowship (to MW) and bridge funding (to Deborah J. Andrew, for MC) to support completion of this work. We are grateful to the other 16 M. Chiu et al. © 2020 The Royal Entomological Society, 1–21members of the Andrew lab for their analysis and discussion of this work, as well as many JHMRI member faculty and trainees for fruitful conversations and advice. We thank the JHMRI Insectary and manager Chris Kizito for access to, and rearing of, Anopheles gambiae larvae. We deeply appreciate assistance from the JHU Microscope Facility staff and applicable NIH grant support for the microscope used (NIH Grant #: S10OD016374). We sincerely appreciate kind gifts of antisera aliquots from Elizabeth Chen, Julian Hillyer, Hiroyuki Matsuoka, and Christian Dahmann. Multiple aliquots of various antisera were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. We thank Giuseppe La Rocca (EGI) for kind assistance with the EGI Marketplace access of Chipster (Kallio et al., 2011). We sincerely acknowledge the work of multiple labs in creation of the microarray data (Baker et al., 2011; Neira Oviedo et al., 2008; 2009) meta-analysed in this study.
Footnotes
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Appendix S1: Supplementary Information.
References
- Abrams EW and Andrew DJ (2005) CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development (Cambridge, England), 132(12), 2743–2758. 10.1242/dev.01863. [DOI] [PubMed] [Google Scholar]
- Ahanger SH, Srinivasan A, Vasanthi D, Shouche YS and Mishra RK (2013) Conserved boundary elements from the Hox complex of mosquito, Anopheles gambiae. Nucleic Acids Research, 41(2), 804–816. 10.1093/nar/gks1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alout H, Roche B, Dabiré RK and Cohuet A (2017) Consequences of insecticide resistance on malaria transmission. PLoS Pathogens, 13(9), e1006499. 10.1371/journal.ppat.1006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K et al. (1999) Sporozoite invasion of Plasmodium berghei, rodent malaria parasite, to the salivary glands of the vector mosquito, Anopheles stephensi: an electron microscopic study. Japanese Journal of Tropical Medicine and Hygiene, 27(1), 7–12. 10.2149/tmh1973.27.7. [DOI] [Google Scholar]
- Andrew DJ, Baig A, Bhanot P, Smolik SM and Henderson KD (1997) The Drosophila dCREB-A gene is required for dorsal/ventral patterning of the larval cuticle. Development, 124(1), 181–193. [DOI] [PubMed] [Google Scholar]
- Arcà B et al. (2017) Anopheline salivary protein genes and gene families: an evolutionary overview after the whole genome sequence of sixteen Anopheles species. BMC Genomics, 18 (1), 1–27. 10.1186/s12864-017-3579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcà B et al. (2019) MicroRNAs from saliva of anopheline mosquitoes mimic human endogenous miRNAs and may contribute to vector-host-pathogen interactions. Scientific Reports, 9(1), 2955. 10.1038/s41598-019-39880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemov GN et al. (2018) The development of cytogenetic maps for malaria mosquitoes. Insects, 9(3), 121. 10.3390/insects9030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA et al. (2011) A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics, 12(1), 296. 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Balkom BWM, Eisele AS, Pegtel DM, Bervoets S and Verhaar MC (2015) Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. Journal of Extracellular Vesicles, 4, 26760. 10.3402/jev.v4.26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa S, Fasanella G, Carreira S, Llarena M, Fox R, Barreca C et al. (2013) An orchestrated program regulating secretory pathway genes and Cargos by the transmembrane transcription factor CREB-H. Traffic, 14(4), 382–398. 10.1111/tra.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreaux AMG et al. (2018) The relationship between size and longevity of the malaria vector Anopheles gambiae (s.s.) depends on the larval environment. Parasites & Vectors, 11 (1), 485. 10.1186/s13071-018-3058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow PM, McIver SB and Wright KA (1975) Salivary glands of female Culex pipiens: morphological changes associated with maturation and bloodfeeding. The Canadian Entomologist, 107 (11), 1153–1160. 10.4039/Ent1071153-11. [DOI] [Google Scholar]
- Beckett EB (1977) Peripheral cells in the salivary glands of female Aedes aegypti and A. togoi mosquitoes. Journal of Insect Physiology, 23, 805–807. [DOI] [PubMed] [Google Scholar]
- Beckett EB (1988) Development and ageing of the salivary glands of adult male Aedes aegypti (L.) and Aedes togoi (Theobald) mosquitoes (Diptera: Culicidae). Journal of Insect Morphology and Embryology, 17(4), 327–333. [Google Scholar]
- Beckett EB (1990) Development and ageing of the salivary glands of adult female Aedes aegypti (L.) and Aedes togoi (Theobald) (Diptera: Culicidae). International Journal of Insect Morphology and Embryology, 19(5), 277–290. [Google Scholar]
- Bondarenko SM, Artemov GN, Sharakhov IV and Stegniy VN (2017) Tissue-specific features of the X chromosome and nucleolus spatial dynamics in a malaria mosquito, Anopheles atroparvus. PLoS One, 12(2), 1–12. 10.1371/journal.pone.0171290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis A et al. (1956) New instructions for the study of Anopheles species of the maculipennis group in a DDT-sprayed malaria zone; importance of salivary gland chromosomes of the larvae and their preparation and interpretation technic. Rivista di malariologia, 35(1–3), 21–37. [PubMed] [Google Scholar]
- Capone A et al. (2013) Interactions between Asaia, Plasmodium and Anopheles: new insights into mosquito symbiosis and implications in malaria symbiotic control. Parasites & Vectors, 6(1), 182. 10.1186/1756-3305-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aimanova KG and Gill SS (2020) Aedes cadherin receptor that mediates Bacillus thuringiensis Cry11A toxicity is essential for mosquito development. PLOS Neglected Tropical Diseases, 14(2), e0007948. 10.1371/journal.pntd.0007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S and Andrew DJ (2014) Cadherin 99C regulates apical expansion and cell rearrangement during epithelial tube elongation. Development (Cambridge), 141(9), 1950–1960. 10.1242/dev.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Hanlon CD and Andrew DJ (2014) Building and specializing epithelial tubular organs: the Drosophila salivary gland as a model system for revealing how epithelial organs are specified, form and specialize. Wiley Interdisciplinary Reviews: Developmental Biology, 3(4), 281–300. 10.1002/wdev.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements AN (1963) In: The Physiology of Mosquitoes. Oxford: New York, Paris: Pergamon Press. [Google Scholar]
- Crawley SW, Mooseker MS and Tyska MJ (2014) Shaping the intestinal brush border. The Journal of Cell Biology, 207 (4), 441–451. 10.1083/jcb.201407015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Garver L and Dimopoulos G (2007) Protocol for mosquito rearing (A. gambiae). Journal of Visualized Experiments, 5, 5–6. 10.3791/221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer K et al. (2000) Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. Journal of Cell Science, 113(19), 3365–3374. http://jcs.biologists.org/content/113/19/3365.abstract. [DOI] [PubMed] [Google Scholar]
- Devenport MP, Blass C and Eggleston P (2000) Characterization of the Hox gene cluster in the malaria vector mosquito, Anopheles gambiae. Evolution and Development, 2(6), 326–339. 10.1046/j.1525-142X.2000.00074.x. [DOI] [PubMed] [Google Scholar]
- Dow JA et al. (1997) Molecular genetic analysis of V-ATPase function in Drosophila melanogaster. The Journal of Experimental Biology, 200(Pt 2), 237–245. [DOI] [PubMed] [Google Scholar]
- Farkaš R, Ďatková Z, Mentelová L, Löw P, Beňová- Liszeková D, Beňo M et al. (2014) Apocrine secretion in drosophila salivary glands: subcellular origin, dynamics, and identification of secretory proteins. PLoS One, 9(4), e94383. 10.1371/journal.pone.0094383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favia G et al. (2007) Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proceedings of the National Academy of Sciences of the United States of America, 104(21), 9047–9051. 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova M, Ross LS and Gill SS (1998) Cloning of the VATPase B subunit cDNA from Culex quinquefasciatus and expression of the B and C subunits in mosquitoes. Insect Molecular Biology, 7(3), 223–232. 10.1111/j.1365-2583.1998.00069.x. [DOI] [PubMed] [Google Scholar]
- Fox RM and Andrew DJ (2015) Changes in organelle position and epithelial architecture associated with loss of CrebA. Biology Open, 4(3), 317–330. 10.1242/bio.201411205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Hanlon CD and Andrew DJ (2010) The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. The Journal of Cell Biology, 191 (3), 479–492. 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Vaishnavi A, Maruyama R and Andrew DJ (2013) Organ-specific gene expression: the bHLH protein Sage provides tissue specificity to Drosophila FoxA. Development (Cambridge), 140(10), 2160–2171. 10.1242/dev.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P, Sharma A and Sharakhov IV (2014) 2D and 3D chromosome painting in malaria mosquitoes. Journal of Visualized Experiments, 83, 1–11. 10.3791/51173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P et al. (2020) Three-dimensional organization of polytene chromosomes in somatic and germline tissues of malaria mosquitoes. Cells, 9(2), 339. 10.3390/cells9020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G et al. (2014) Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infection, Genetics and Evolution, 28, 715–724. 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Hsiong W, Lanzaro G and Levine M (2004) Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Developmental Biology, 275(2), 435–446. 10.1016/j.ydbio.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Fuse N, Frasch M, Zinzen RP, Lanzaro G and Levine M (2007) Evolution of the dorsal-ventral patterning network in the mosquito, Anopheles gambiae. Development, 134 (13), 2415–2424. 10.1242/dev.02863. [DOI] [PubMed] [Google Scholar]
- Green CA (1972) Cytological maps for the practical identification of females of the three freshwater species of the Anopheles gambiae complex. Annals of Tropical Medicine and Parasitology, 66(1), 143–147. 10.1080/00034983.1972. 11686806. [DOI] [PubMed] [Google Scholar]
- Gunathilaka N, Upulika H, Udayanga L and Amarasinghe D (2019) Effect of larval nutritional regimes on morphometry and vectorial capacity of Aedes aegypti for dengue transmission. BioMed Research International, 2019, 3607342. 10.1155/2019/3607342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S et al. (2019) Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells, 8(4), 307. 10.3390/cells8040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI and Cashikar A (2012) Multivesicular body morphogenesis. Annual Review of Cell and Developmental Biology, 28(1), 337–362. 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- Hatton-Ellis E, Ainsworth C, Sushama Y, Wan S, VijayRaghavan K and Skaer H (2007) Genetic regulation of patterned tubular branching in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 104(1), 169–174. 10.1073/pnas.0606933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffer A and Pick L (2013) Conservation and variation in hox genes: how insect models pioneered the Evo-Devo field. Annual Review of Entomology, 58(1), 161–179. 10.1146/annurev-ento-120811-153601. [DOI] [PubMed] [Google Scholar]
- Hering L et al. (2016) Novel origin of lamin-derived cytoplasmic intermediate filaments in tardigrades. eLife, 5, e11117. 10.7554/eLife.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua G, Zhang Q, Zhang R, Abdullah AM, Linser PJ and Adang MJ (2013) AgCad2 cadherin in Anopheles gambiae larvae is a putative receptor of Cry11Ba toxin of Bacillus thuringiensis subsp. jegathesan. Insect Biochemistry and Molecular Biology, 43(2), 153–161. 10.1016/j.ibmb.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT and Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13. 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT and Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imms AD (1908) On the larval and pupal stages of anopheles maculipennis, meigen. Parasitology, 1(2), 103–133. 10.1017/S0031182000003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova-Kazas O (1949) Embryonic development of Anopheles maculipennis MG. Izvestiia Akademii Nauk Sepiia Biologicheskaia, 2, 140–170. [Google Scholar]
- James AA (2003) Blocking malaria parasite invasion of mosquito salivary glands. Journal of Experimental Biology, 206(21), 3817–3821. 10.1242/jeb.00616. [DOI] [PubMed] [Google Scholar]
- Janzen HG and Wright KA (1971) The salivary glands of Aedes aegypti (L.): an electron microscope study. Canadian Journal of Zoology, 49(10), 1343–1345. 10.1139/z71-200. [DOI] [PubMed] [Google Scholar]
- Jensen DV and Jones JC (1957) The development of the salivary glands in Anopheles albimanus Wiedemann (Diptera, Culicidae). Annals of the Entomological Society of America, 50(5), 464–469. 10.1093/aesa/50.5.464. [DOI] [Google Scholar]
- Johnson DM, Wells MB, Fox R, Lee JS, Loganathan R, Levings D et al. (2020) CrebA increases secretory capacity through direct transcriptional regulation of the secretory machinery, a subset of secretory cargo, and other key regulators. Traffic, 21(9), 560–577. 10.1111/tra.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MA et al. (2011) Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics, 12, 507. 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamali M, Sharakhova MV, Baricheva E, Karagodin D, Tu Z and Sharakhov IV (2011) An integrated chromosome map of microsatellite markers and inversion breakpoints for an Asian malaria mosquito, Anopheles stephensi. Journal of Heredity, 102(6), 719–726. 10.1093/jhered/esr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JG, Vernicks KD and Hillyer JF (2011) Members of the salivary gland surface protein (SGS) family are major immunogenic components of mosquito saliva. Journal of Biological Chemistry, 286(47), 40824–40834. 10.1074/jbc.M111.280552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D and Kent WJ (2013) The UCSC genome browser and associated tools. Briefings in Bioinformatics, 14(2), 144–161. 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz RM, Takeyasu K and Fambrough DM (1989) Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. The EMBO Journal, 8(1), 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane MJ and Billingsley PF (1996) In: Biology of the Insect Midgut. London: Chapman & Hall. [Google Scholar]
- Lestinova T et al. (2017) Insights into the sand fly saliva: bloodfeeding and immune interactions between sand flies, hosts, and Leishmania. PLoS Neglected Tropical Diseases, 11(7), e0005600. 10.1371/journal.pntd.0005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linser PJ, Smith KE, Seron TJ and Neira Oviedo M (2009) Carbonic anhydrases and anion transport in mosquito midgut pH regulation. Journal of Experimental Biology, 212(11), 1662–1671. 10.1242/jeb.028084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linser PJ, Neira Oviedo M, Hirata T, Seron TJ, Smith KE, Piermarini PM et al. (2012) Slc4-like anion transporters of the larval mosquito alimentary canal. Journal of Insect Physiology, 58(4), 551–562. 10.1016/j.jinsphys.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, McLeod E and Jack J (1991) Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics, 127(1), 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R, Schejter ED and Shilo B-Z (2009) Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Developmental Cell, 16(6), 877–888. 10.1016/j.devcel.2009.04.010. [DOI] [PubMed] [Google Scholar]
- McAbee RD, Christiansen JA and Cornel AJ (2007) A detailed larval salivary gland polytene chromosome photomap for Culex quinquefasciatus (Diptera: Culicidae) from Johannesburg, South Africa. Journal of Medical Entomology, 44(2), 229–237. 10.1603/0022-2585(2007)44[229:adlsgp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Moreira-Ferro CK, Marinotti O and Bijovsky AT (1999) Morphological and biochemical analyses of the salivary glands of the malaria vector, Anopheles darlingi. Tissue and Cell, 31 (3), 264–273. 10.1054/tice.1999.0057. [DOI] [PubMed] [Google Scholar]
- Mueller AK, Kohlhepp F, Hammerschmidt C and Michel K (2010) Invasion of mosquito salivary glands by malaria parasites: prerequisites and defense strategies. International Journal for Parasitology, 40(11), 1229–1235. 10.1016/j.ijpara.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R et al. (2019) In: Marselle MR, et al. (Eds.) Vector-Borne Diseases BT—Biodiversity and Health in the Face of Climate Change. Cham: Springer International Publishing, pp. 67–90. 10.1007/978-3-030-02318-8_4. [DOI] [Google Scholar]
- Myat MM et al. (2019) Regulators of cell movement during development and regeneration in Drosophila. Open Biology, 9(5), 180245. 10.1098/rsob.180245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira Oviedo M, VanEkeris L, Corena-Mcleod MDP and Linser PJ (2008) A microarray-based analysis of transcriptional compartmentalization in the alimentary canal of Anopheles gambiae (Diptera: Culicidae) larvae. Insect Molecular Biology, 17(1), 61–72. 10.1111/j.1365-2583.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- Neira Oviedo M, Ribeiro JMC, Heyland A, VanEkeris L, Moroz T and Linser PJ (2009) The salivary transcriptome of Anopheles gambiae (Diptera: Culicidae) larvae: a microarray-based analysis. Insect Biochemistry and Molecular Biology, 39(5–6), 382–394. 10.1016/j.ibmb.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT et al. (1994) D-mef2: a Drosophila mesodermspecific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proceedings of the National Academy of Sciences of The United States of America, 91 (16), 7520–7524. 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C et al. (2013) Functional genetic characterization of salivary gland development in Aedes aegypti. EvoDevo, 4(1), 1. 10.1186/2041-9139-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norsworthy NB, Sun J, Elnaiem D, Lanzaro G and Soong L (2004) Sand Fly saliva enhances Leishmania amazonensis infection by modulating interleukin-10 production. Infection and Immunity, 72(3), 1240–1247. 10.1128/IAI.72.3.1240-1247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okech BA, Boudko DY, Linser PJ and Harvey WR (2008) Cationic pathway of pH regulation in larvae of Anopheles gambiae. Journal of Experimental Biology, 211(6), 957–968. 10.1242/jeb.012021. [DOI] [PubMed] [Google Scholar]
- Oliveira F, de Carvalho AM and de Oliveira CI (2013) Sand-fly saliva-leishmania-man: the trigger trio. Frontiers in immunology, 4, 375. 10.3389/fimmu.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr CWM, Hudson A and West AS (1961) The salivary glands of Aedes aegypti: histological-histochemical studies’. Canadian Journal of Zoology, 39(3), 265–272. 10.1139/z61-029. [DOI] [Google Scholar]
- Papadopoulos NT, Carey JR, Ioannou CS, Ji H, Müller HG, Wang JL et al. (2016) Seasonality of Post-capture Longevity in a Medically-Important Mosquito (Culex pipiens). Frontiers in Ecology and Evolution, 4, 63. https://www.frontiersin.org/article/10.3389/fevo.2016.00063. [Google Scholar]
- Pimenta PF, Touray M and Miller L (1994) The journey of malaria Sporozoites in the mosquito salivary gland. Journal of Eukaryotic Microbiology, 41(6), 608–624. 10.1111/j.1550-7408.1994.tb01523.x. [DOI] [PubMed] [Google Scholar]
- Poehling HM (1979) Distribution of specific proteins in the salivary gland lobes of Culicidae and their relation to age and blood sucking. Journal of Insect Physiology, 25(1), 3–8. 10.1016/0022-1910(79)90029-5. [DOI] [Google Scholar]
- Powers TP, Hogan J, Ke Z, Dymbrowski K, Wang X, Collins FH et al. (2000) Characterization of the Hox cluster from the mosquito Anopheles gambiae (Diptera: Culicidae). Evolution & Development, 2(6), 311–325. 10.1046/j.1525-142x.2000.00072.x. [DOI] [PubMed] [Google Scholar]
- Price DP, Schilkey FD, Ulanov A and Hansen IA (2015) Small mosquitoes, large implications: crowding and starvation affects gene expression and nutrient accumulation in Aedes aegypti. Parasites & vectors, 8, 252. 10.1186/s13071-015-0863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MG and Kitzmiller JB (1975) Studies on x-ray induced chromosomal translocations in Anopheles albimanus. I. Chromosomal translocations and genetic control. The American Journal of Tropical Medicine and Hygiene, 24(6 Pt 1), 1019–1026. 10.4269/ajtmh.1975.24.1019. [DOI] [PubMed] [Google Scholar]
- Redfern CP (1981) Homologous banding patterns in the polytene chromosomes from the larval salivary glands and ovarian nurse cells of Anopheles stephensi Liston (Culicidae). Chromosoma, 83(2), 221–240. 10.1007/BF00286791. [DOI] [PubMed] [Google Scholar]
- Riemer D et al. (1995) Expression of drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. Journal of Cell Science, 108(10), 3189–3198. [DOI] [PubMed] [Google Scholar]
- Rishikesh N (1959) Morphology and development of the salivary glands and their chromosomes in the larvae of Anopheles stephensi sensu stricto. Bulletin of the World Health Organization, 20(1), 47–61. [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Tchouakui M, Mugenzi L, Menze BD and MuChun Chiang CSW (2018) Insecticide resistance in malaria vectors: an update at a global scale. In: Manguin S, (Ed.) Towards Malaria Elimination—A Leap Forward, Vol. 7. London, UK: Intechopen, pp. 1218. 10.5772/intechopen.78375. [DOI] [Google Scholar]
- Rohousova I and Volf P (2006) Sand fly saliva: effects on host immune response and Leishmania transmission. Folia Parasitologica, 53(3), 161–171. https://folia.paru.cas.cz/artkey/fol-200603-0001.php. [PubMed] [Google Scholar]
- Sarmiento LA and Mitchell HK (1982) Drosophila salivary gland proteins and pupation. Developmental Genetics, 3(3), 255–272. 10.1002/dvg.1020030308. [DOI] [Google Scholar]
- Schlichting K, Demontis F and Dahmann C (2005) Cadherin Cad99C is regulated by Hedgehog signaling in Drosophila. Developmental Biology, 279(1), 142–154. 10.1016/j.ydbio.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Service MW (1970) Identification of the Anopheles gambiae complex in Nigeria by larval and adult chromosomes. Annals of Tropical Medicine and Parasitology, 64(2), 131–136. 10.1080/00034983.1970.11686674. [DOI] [PubMed] [Google Scholar]
- Shapiro LLM, Murdock CC, Jacobs GR, Thomas RJ and Thomas MB (2016) Larval food quantity affects the capacity of adult mosquitoes to transmit human malaria. Proceedings of the Royal Society B: Biological Sciences, 283(1834), 20160298. 10.1098/rspb.2016.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Xia A, Mcalister SI and Sharakhov IV (2006) A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. Journal of Medical Entomology, 43(5), 861–866. 10.1603/0022-2585. [DOI] [PubMed] [Google Scholar]
- Sharakhova MV, George P, Brusentsova IV, Leman SC, Bailey JA, Smith CD et al. (2010) Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics, 11(1), 459. 10.1186/1471-2164-11-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GP et al. (1978) A preliminary map of the salivary gland chromosomes of Aedes (stegomyia) aegypti (Culicadae, Diptera). Cytobios, 22(87–88), 169–178. [PubMed] [Google Scholar]
- Sharma GP et al. (1987) Effect of mitomycin-C and cyclophosphamide on the polytene chromosomes of Anopheles stephensi (Culicidae: Diptera). Cytobios, 52(210–211), 153–160. [PubMed] [Google Scholar]
- Styer LM et al. (2007) Mosquitoes do senesce: departure from the paradigm of constant mortality. The American Journal of Tropical Medicine and Hygiene, 76(1), 111–117. [PMC free article] [PubMed] [Google Scholar]
- Sun YC and Coluzzi M (1989) A paracentric inversion in the salivary gland chromosome 3R of Anopheles sinensis. Chinese Journal of Parasitology & Parasitic Diseases, 7(4), 288. [PubMed] [Google Scholar]
- Thornton J (2020) Covid-19: Keep essential malaria services going during pandemic, urges WHO. BMJ, 369, m1637. 10.1136/bmj.m1637. [DOI] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ et al. (2018) FlyBase 2.0: the next generation. Nucleic Acids Research, 47(D1), D759–D765. 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A et al. (2001) Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. Journal of Cell Science, 114(Pt 14), 2577–2590. [DOI] [PubMed] [Google Scholar]
- Wells MB and Andrew DJ (2015) Salivary gland cellular architecture in the Asian malaria vector mosquito Anopheles stephensi. Parasites and Vectors, 8(1), 1–19. 10.1186/s13071-015-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MB and Andrew DJ (2019) Anopheles salivary gland architecture shapes plasmodium sporozoite availability for transmission. mBio, 10(4), 1–17. 10.1128/mBio.01238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MB, Villamor J and Andrew DJ (2017) Salivary gland maturation and duct formation in the African malaria mosquito Anopheles gambiae. Scientific Reports, 7(1), 1–12. 10.1038/s41598-017-00672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend P, Fang L, Zhu Q, Schipper JH, Loddenkemper C, Kosel F et al. (2013) Wnt/β-catenin signalling induces MLL to create epigenetic changes in salivary gland tumours. EMBO Journal, 32(14), 1977–1989. 10.1038/emboj.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2019) WHO malaria report 2019. https://www.who.int/publications-detail/world-malaria-report-2019.
- Wright KA (1969) The anatomy of salivary glands of Anopheles stephensi Liston. Canadian Journal of Zoology, 47(4), 579–587. 10.1139/z69-101. [DOI] [Google Scholar]
- Xu XF, Wang ZW and Su SZ (1983) Comparative observations on the salivary gland chromosomes of the fourth instar larva of Anopheles sinensis in various districts. Chinese Journal of Parasitology & Parasitic Diseases, 1(2), 91–94. [PubMed] [Google Scholar]
- Yoshida S, Sudo T, Niimi M, Tao L, Sun B, Kambayashi J et al. (2008) Inhibition of collagen-induced platelet aggregation by anopheline antiplatelet protein, a saliva protein from a malaria vector mosquito. Blood, 111(4), 2007–2014. 10.1182/blood-2007-06-097824. [DOI] [PubMed] [Google Scholar]
- Zielke T and Saumweber H (2014) Dissection of open chromatin domain formation by site-specific recombination in Drosophila. Journal of Cell Science, 127(10), 2365–2375. 10.1242/jcs.147546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


