Here, we report near-complete genome sequences of Sicinivirus from U.S. poultry flocks in 2003 to 2005 and Mexico in 2019. They show highest nucleotide identity (84.5 to 85.5%) with other members of the Sicinivirus genus. These sequences update knowledge on diversity and contribute to a better understanding of the molecular epidemiology of Sicinivirus.
ABSTRACT
Here, we report near-complete genome sequences of sicinivirus from U.S. poultry flocks in 2003 to 2005 and Mexico in 2019. They show highest nucleotide identity (84.5 to 85.5%) with other members of the Sicinivirus genus. These sequences update knowledge on diversity and contribute to a better understanding of the molecular epidemiology of sicinivirus.
ANNOUNCEMENT
The Picornaviridae are a family of small, icosahedral viruses with single-stranded, positive-sense RNA genomes (1). The family comprises 47 genera, with 13 of these identified from avian species (1–3). Avian picornaviruses of the Sicinivirus genus have been identified in both healthy and diseased chickens, and the viruses’ role in disease remains unclear (4–9). Here, we report the sequences of five near-complete genomes of sicinivirus from North America.
Fecal samples from broiler chickens with severe hypoglycemia were collected at commercial farms in the state of Georgia in 2003 to 2005, and bursal swab samples (clinical signs were not provided) were collected from a commercial farm in Mexico in 2019 and preserved on Flinders Technology Associates (FTA) cards (Whatman, USA) (Table 1). Feces were first diluted 3:7 in sterile phosphate-buffered saline and then centrifuged for 10 min at 3,200 rpm. The supernatants were passed sequentially through a 1.2-μm- and a 0.45-μm-pore-size filter (Merck Millipore, USA) to remove bacteria and large-cell-size particles and were DNase treated using the TURBO DNA-free kit (Ambion, USA) to remove host DNA. Total nucleic acids were isolated using the DNeasy blood and tissue kit (Qiagen, Germany). For the FTA card bursal swabs, first, nucleic acids were eluted from 24 3-mm discs punched out from the card by incubation in Tris-EDTA (TE) buffer, and then total RNA was extracted from the TE eluate using the MagMAX total RNA isolation kit (Thermo Fisher, USA). Sequence-independent single-primer amplification (10) was used to produce random amplicons. Briefly, first-strand cDNA was synthesized using the random octamer primer tagged with a fixed-sequence K-8N and SuperScript IV reverse transcriptase (Invitrogen, USA), followed by second-strand synthesis using Klenow polymerase (New England Biolabs, USA). Finally, random primer amplification was conducted using Phusion DNA polymerase (New England Biolabs) and the primer consisting of the fixed portion of the random K-8N primer. DNA libraries were subsequently prepared with the Nextera XT DNA library preparation kit (Illumina, USA). Paired-end (2 × 150-bp) sequencing was performed on an Illumina MiSeq instrument. A customized workflow on the Galaxy platform (11) was used to perform preprocessing and assembly of the raw sequencing reads, as described previously (12, 13). Briefly, raw read quality was assessed using FastQC v0.63 (14), and residual adapter sequences were trimmed using Cutadapt v1.6 (15). Sequence data were assembled de novo utilizing MIRA3 v0.0.1 (16). Default parameters were used for all software. The MiSeq runs generated 1,096,430 to 2,446,408 total paired-end reads per sample (Table 1). All final consensuses were called from the raw reads aligned to the de novo-generated contigs using the Burrows-Wheeler Aligner MEM algorithm (BWA-MEM) (17). Open reading frames were identified using Geneious v11.1.5 and confirmed by alignment with published sicinivirus genomes using MEGA v7.0.26. The genomes of all clinical samples had 100% complete coding sequences and showed the typical genetic structure of siciniviruses, with a single polyprotein cleaved into smaller nonstructural and structural capsid proteins, flanked by untranslated regions at both termini (1). Phylogeny based on 3Dpol amino acid sequences confirmed that all viruses clustered together with other members of the Sicinivirus genus (Fig. 1). The highest BLASTp homology score for the 3Dpol protein of all viruses was 96.4% to 97.7% amino acid sequence identity to the Ireland/UCC001 strain (NCBI RefSeq accession number NC_023861.1) (5). All viruses also possessed conserved amino acid motifs in 2Chel (GPPGCGKS, DDVGQ) and 3Dpol (KDELR, GGNPSG, YGDD, and FLKR) proteins. The Georgian viruses additionally demonstrated the conserved motifs in 3Cpro (QFKDL, GLCG) (5, 18, 19). Across the entire polyprotein gene, the strains designated GA/1472/2004, GA/1477/2005, GA/1478/2003, GA/1479/2004, and MEX/B1203/2019 showed the highest (85.5%, 85.3%, 84.9%, 84.5%, and 84.6%, respectively) nucleotide identity to isolate UCC001, from broiler chickens in Ireland (GenBank accession number KF741227.1) (5). These near-complete genome sequences of sicinivirus collected from North America provide molecular epidemiological data needed to explore the evolution, epidemiology, and detailed pathogenesis of chicken picornaviruses globally.
TABLE 1.
Samples, sampling locations, dates, sequencing metrics, and accession numbers of the sicinivirus genomes and contigs in this report
| Isolate name | Collection date (mo/day/yr) | Host | Total no. of raw read pairs | No. of mapped reads | Median coverage depth (reads) | Mean read length (nt)a | Consensus length (bp) | GC content (%) | GenBank accession no. | SRA accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| GA/1472/2004 | 10/5/2004 | 22-day-old broiler | 2,088,901 | 45,973 | 434 | 138 | 9,775 | 53.9 | MN873045 | SRR10500281 |
| GA/1477/2005 | 9/29/2005 | 16-day-old broiler | 2,446,408 | 45,379 | 399 | 121 | 9,687 | 54.5 | MN873046 | SRR10566436 |
| GA/1478/2003 | 4/28/2003 | 18-day-old broiler | 1,303,453 | 132,008 | 2,040 | 127 | 9,670 | 54.1 | MN873047 | SRR10566435 |
| GA/1479/2004 | 6/8/2004 | 15-day-old broiler | 2,212,775 | 8,649 | 76 | 157 | 9,806 | 54.1 | MN873048 | SRR10586503 |
| MEX/B1203/2019 | 6/11/2019 | Chicken bursa | 1,096,430 | 22,464 | 366 | 131 | 9,706 | 54.9 | MT345550 | SRR11542284 |
nt, nucleotides.
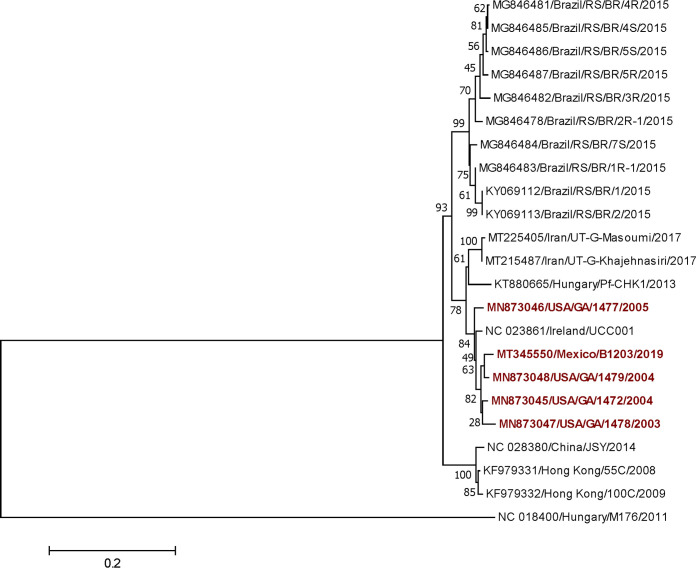
FIG 1.
Phylogenetic analysis of sicinivirus sequences based on the complete amino acid sequence of the 3Dpol protein. The evolutionary history was inferred using the maximum likelihood method based on the Jones-Taylor-Thornton matrix-based model in MEGA v7.0.26. The tree with the highest log likelihood (–3,357.96) is shown. The percentage of trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying the neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with the superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 23 amino acid sequences (the sequence of Gallivirus isolate NC_018400 is included as an outgroup). All positions containing gaps and missing data were eliminated. There was a total of 472 positions in the final data set. The samples used in this study are shown in red.
Data availability.
The complete coding sequences of all four Georgian sicinivirus samples have been deposited in GenBank under the accession numbers MN873045 to MN873048. The raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA590745. The Mexican sequence was deposited in GenBank under accession number MT345550. The raw sequence data were deposited in the SRA under BioProject number PRJNA625289.
ACKNOWLEDGMENT
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. This study was supported by USDA CRIS project 6040-32000-072 and USDA-AFRI grant 2015-68004-23131.
REFERENCES
- 1.Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, Lindberg AM, Pallansch MA, Palmenberg AC, Reuter G, Simmonds P, Skern T, Stanway G, Yamashita T, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Picornaviridae. J Gen Virol 98:2421–2422. doi: 10.1099/jgv.0.000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boros Á, Pankovics P, Reuter G. 2014. Avian picornaviruses: molecular evolution, genome diversity and unusual genome features of a rapidly expanding group of viruses in birds. Infect Genet Evol 28:151–166. doi: 10.1016/j.meegid.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Gerber PF, Shen H, Zheng Y, Li G, Lobato ZIP, Opriessnig T. 2019. Genomic sequence of a Megrivirus strain identified in laying hens in Brazil. Microbiol Resour Announc 8:e01438-18. doi: 10.1128/MRA.01438-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lima D, Cibulski S, Finkler F, Teixeira T, Varela A, Cerva C, Loiko M, Scheffer C, dos Santos H, Mayer F, Roehe P. 2017. Faecal virome of healthy chickens reveals a large diversity of the eukaryote viral community, including novel circular ssDNA viruses. J Gen Virol 98:690–703. doi: 10.1099/jgv.0.000711. [DOI] [PubMed] [Google Scholar]
- 5.Bullman S, Kearney K, O’Mahony M, Kelly L, Whyte P, Fanning S, Morgan JG. 2014. Identification and genetic characterization of a novel picornavirus from chickens. J Gen Virol 95:1094–1103. doi: 10.1099/vir.0.061085-0. [DOI] [PubMed] [Google Scholar]
- 6.Boros Á, Pankovics P, Adonyi Á, Fenyvesi H, Day JM, Phan TG, Delwart E, Reuter G. 2016. A diarrheic chicken simultaneously co-infected with multiple picornaviruses: complete genome analysis of avian picornaviruses representing up to six genera. Virology 489:63–74. doi: 10.1016/j.virol.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Lima D, Cibulski S, Tochetto C, Varela A, Finkler F, Teixeira T, Loiko M, Cerva C, Junqueira D, Mayer F, Roehe P. 2019. The intestinal virome of malabsorption syndrome-affected and unaffected broilers through shotgun metagenomics. Virus Res 261:9–20. doi: 10.1016/j.virusres.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Devaney R, Trudgett J, Trudgett A, Meharg C, Smyth V. 2016. A metagenomic comparison of endemic viruses from broiler chickens with runting-stunting syndrome and from normal birds. Avian Pathol 45:616–629. doi: 10.1080/03079457.2016.1193123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Zhu S, Quan R, Wang J, Wei L, Yang B, Xu F, Wang J, Chen F, Liu J. 2015. Identification and genome characterization of the first Sicinivirus isolate from chickens in mainland China by using viral metagenomics. PLoS One 10:e0139668. doi: 10.1371/journal.pone.0139668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chrzastek K, Lee DH, Smith D, Sharma P, Suarez DL, Pantin-Jackwood M, Kapczynski DR. 2017. Use of sequence-independent, single-primer-amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509:159–166. doi: 10.1016/j.virol.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrov KM, Sharma P, Volkening JD, Goraichuk IV, Wajid A, Rehmani SF, Basharat A, Shittu I, Joannis TM, Miller PJ, Afonso CL. 2017. A robust and cost-effective approach to sequence and analyze complete genomes of small RNA viruses. Virol J 14:72. doi: 10.1186/s12985-017-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goraichuk IV, Kapczynski DR, Seal BS, Suarez DL. 2020. Complete genome sequence of an Avian Metapneumovirus subtype B strain from Hungary. Microbiol Resour Announc 9:e00177-20. doi: 10.1128/MRA.00177-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 3 February 2020.
- 15.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 16.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56. In Computer science and biology. The German Conference on Bioinformatics (GCB ’99), Hanover, Germany. [Google Scholar]
- 17.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau SK, Woo PC, Yip CC, Li KS, Fan RY, Bai R, Huang Y, Chan KH, Yuen KY. 2014. Chickens host diverse picornaviruses originated from potential interspecies transmission with recombination. J Gen Virol 95:1929–1944. doi: 10.1099/vir.0.066597-0. [DOI] [PubMed] [Google Scholar]
- 19.Gorbalenya AE, Koonin EV, Wolf YI. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett 262:145–148. doi: 10.1016/0014-5793(90)80175-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete coding sequences of all four Georgian sicinivirus samples have been deposited in GenBank under the accession numbers MN873045 to MN873048. The raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA590745. The Mexican sequence was deposited in GenBank under accession number MT345550. The raw sequence data were deposited in the SRA under BioProject number PRJNA625289.