Abstract
Periprosthetic joint infections (PJI) represent one of the most catastrophic complications following total joint arthroplasty (TJA). The lack of standardized diagnostic tests and protocols for PJI is a challenge for arthroplasty surgeons.
Next generation sequencing (NGS) is an innovative diagnostic tool that can sequence microbial deoxyribonucleic acids (DNA) from a synovial fluid sample: all DNA present in a specimen is sequenced in parallel, generating millions of reads. It has been shown to be extremely useful in a culture-negative PJI setting.
Metagenomic NGS (mNGS) allows for universal pathogen detection, regardless of microbe type, in a 24–48-hour timeframe: in its nanopore-base variation, mNGS also allows for antimicrobial resistance characterization.
Cell-free DNA (cfDNA) NGS, characterized by lack of the cell lysis step, has a fast run-time (hours) and, together with a high sensitivity and specificity in microorganism isolation, may provide information on the presence of antimicrobial resistance genes.
Metagenomics and cfDNA testing have reduced the time needed to detect infecting bacteria and represent very promising technologies for fast PJI diagnosis.
NGS technologies are revolutionary methods that could disrupt the diagnostic paradigm of PJI, but a comprehensive collection of clinical evidence is still needed before they become widely used diagnostic tools.
Cite this article: EFORT Open Rev 2021;6:236-244. DOI: 10.1302/2058-5241.6.200099
Keywords: next generation sequencing, periprosthetic joint infections, PJI
Introduction
Musculoskeletal infection is a leading cause of chronic pain and major disability. The incidence of musculoskeletal infection, including periprosthetic joint infection (PJI), is increasing in association with an ageing population having serious comorbidities such as diabetes, obesity, chronic heart disease and immunodepression.1,2 Recent reports place the overall infection rate following orthopaedic surgery at approximately 5%, including a PJI incidence of up to 2%.3 Periprosthetic joint infection is one of the most catastrophic and difficult to manage complications following total joint arthroplasty (TJA), as it can result in knee arthrodesis or above-the-knee amputation. In the last ten years, the number of PJIs has dramatically increased, having a major impact on sustainability. The five-year mortality rate following PJI has increased to values similar to oncology patients4 and hospital readmission rates following total hip arthroplasty explants are already double those for many cardiac and oncologic procedures.4 Periprosthetic joint infections present two major challenges to the scientific community: (1) identification of the infecting microorganisms due to the increasing number of culture-negative PJIs, and (2) treatment of the infection due to increasing antibiotic resistance. Early identification and treatment are key to avoid an epidemic escalation of PJI, and musculoskeletal infection in general.
Costs associated with the management of musculoskeletal infection vary widely but are considerably higher than those associated with preceding interventions, such as elective TJA. Infections requiring prompt surgical intervention, such as septic arthritis, have higher costs than those without intervention.5 The average hospital cost for a patient suffering from a musculoskeletal infection may be up to $185,000, leading to an estimated cost of $32 billion dollars to treat hospital-acquired infections worldwide.6 Moreover, managing TJA infections caused by antibiotic-resistant bacteria is more costly than for antibiotic-sensitive strains.7–9 Staphylococcus aureus is the most common culturable pathogen present in culture-positive musculoskeletal infection, and methicillin resistance in this organism (MRSA) is increasing in frequency.10–17 Anderson et al10 showed that MRSA infections were associated with increased hospital charges of $20,000 or more and a 2.6-times higher mortality rate within three months of surgery. Parvizi et al7 showed a significantly higher cost of care for treatment of MRSA PJI compared with antibiotic-sensitive strains ($107,000 vs. $68,000 per case). This places a substantial burden on the United States healthcare system.14,18
The clinical appearance of PJI depends on the virulence of the pathogenic agent, the nature of the infected tissue, the infection route, and the length of disease evolution. Periprosthetic joint infection might present acutely with severe pain, high fever, local warmness, and sometimes surgical wound secretions, while the presenting signs of chronic infections are progressive pain, the formation of cutaneous fistulae and/or drainage of purulent secretions. Currently, surgeons and infectious disease and internal medicine physicians seeking to diagnose PJI use a multidisciplinary test battery that includes: tests to detect local inflammation, such as synovial fluid white blood cell (WBC) count and synovial tissue histology;19 measurement of the levels of systemic markers of inflammation, such as serum C-reactive protein (CRP), erythrocyte sedimentation rate (ESR) and interleukin-6 (IL-6);20 imaging studies such as standard radiographs, technetium-labelled bone scans, magnetic resonance imaging, computed tomography (CT), and positron-emission tomography;21 and several bacterial identification and isolation techniques (e.g., Gram stain, culture). Because early detection and treatment of PJI are major goals, several scientific societies in the field of orthopaedics and many authors, including the current authors, recently published algorithms on how to approach a painful TJA in the presence of a suspected PJI.22–24
The diagnosis of PJIs continues to be a moving target and is subject to extensive debate. This is in part due to the lack of a single ‘gold standard’ test, and the marked heterogeneity in the design and conduct of studies evaluating the accuracy of different diagnostic modalities. The Musculoskeletal Infection Society (MSIS) convened a workgroup in 2011 in an effort to create a standardized definition for PJI.22 This definition was later modified in 2013 and 2019 as a result of an International Consensus Meeting on PJI:25–28 the final document of this joint effort proposed a new stepwise approach to PJI diagnosis which included serological, microbiological and histological tests without demonstrating the superiority of any one test. A very recent literature review29 tried to identify gaps and limitations in the current literature and set forth recommendations for the design of future PJI diagnostic algorithms. This literature review was prompted by many findings. First, many patients with normal levels of systemic markers of inflammation, such as serum CRP and ESR, are infected. Furthermore, due to the lack of homogeneity across studies, index test and reference standard domains showed high risk of bias for WBC and the utility histological analysis, respectively.30–32 Additionally, leukocyte esterase testing lacked standardization with regard to the strip reagent used, and the exclusion of bloody samples limited sample sizes.33
The consensus in the current literature25–28 is that identification of the causative microorganism is the main determinant for success in diagnosing and treating PJIs. Standard culture of periprosthetic tissue specimens on agars and in broths, traditionally used for the detection of causative microorganisms in patients suspected as having PJI, has shown low sensitivity34,35 ranging from 39% to 70% in several reports.36,37 Several factors are associated with decreased yield with culture and failure to isolate a microorganism. First, premature administration of antibiotics may compromise culture yield; thus, ideally, antibiotic treatment should be withheld until organisms are grown in culture.38,39 Second, culturing techniques may also influence culture yield, particularly for less-virulent organisms such as Cutibacterium acnes or coagulase-negative Staphylococcus.40 Extending the incubation period and obtaining a sufficient number of samples may increase the sensitivity of culture.40,41 Culture-negative PJIs represent a ‘clinical nightmare’ for the treatment team. A preferred method of treating culture-negative PJI has not been determined and several studies have shown outcomes for culture-negative PJIs were worse than for culture-positive PJIs,42,43 with a treatment failure rate of up to 73%.44 Thus, culture-negative PJIs are relatively frequent and have an unacceptable rate of treatment failure. A recent review by Rothenberg et al45 showed that implant sonicate culture enhances PJI diagnostic accuracy by identifying pathogens that are inaccessible to traditional intraoperative tissue and synovial fluid cultures.
Molecular techniques that can identify DNA in a sample have been recently proposed as a solution to overcome the challenge of diagnosing and treating PJI.46 Next generation sequencing (NGS), which has the ability to quickly sequence DNA, represents a modern and innovative diagnostic methodology because of the potential to promptly and efficiently identify the infecting organism and its antibiotic resistance capabilities in order to tailor medical and surgical treatments. The purpose of this article is to review the current, sparse literature on the use of the next generation sequencing technology in a PJI setting and to evaluate its sensitivity and specificity in comparison with standard diagnostic protocols. MEDLINE, Scopus and Google Scholar were searched (keywords: NGS, mNGS, next generation sequencing, PJI, metagenomics, nanopore) for randomized clinical trials, quasi-randomized clinical trials, controlled clinical trials and observational studies that assessed sensibility and specificity of next generation sequencing as a diagnostic tool for PJI.
Next generation sequencing (NGS)
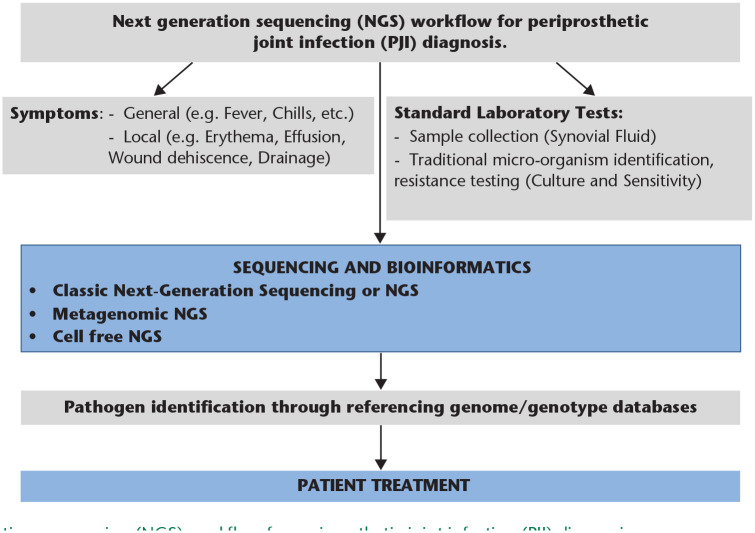
Next generation sequencing (NGS) is a technique where all or a subset of the deoxyribonucleic acids (DNA) present in a specimen, from the host or from microorganism(s), are sequenced in parallel, generating millions of reads per instrument run. Reads are the basic element produced by DNA sequencing and are composed of a series of sequential bases (A-adenine; G-guanine; T-thymine; C-cytosine) reflecting the sequence of the input DNA fragment. The length of each read may vary from 75 to ~10,000 bp, and technologies that produce the longest reads are ideal for assembling complete genome sequences. DNA sequencing allows for culture-free detection and identification of single or multiple microorganisms without the need for a priori knowledge of a PJI. The transformation of raw sequence data into clinically applicable information for PJI treatment requires rigorous analyses (Fig. 1) and early limitations in sequence accuracy have been mitigated by recent improvements in hardware and reagents.
Fig. 1.
Next generation sequencing (NGS) workflow for periprosthetic joint infection (PJI) diagnosis.
NGS shows promise in the context of PJI for organism detection since Tarabichi et al47 first demonstrated the utility of 16S-amplicon targeted NGS for detecting Streptococcus canis in a culture-negative PJI patient. In fact, recent evidence suggests that NGS may be more sensitive at identifying organisms (at a rate of up to 89% for culture-negative PJI) than conventional culture (Table 1).48–51 The 16S rRNA gene has become the most used region for bacterial identification because it is present in all bacteria; because smaller portions of the gene, called variable regions, can be sequenced instead of the entire genome; and, finally, because tools that have been developed for the analysis of 16S rRNA sequences are relatively straightforward even for novice bioinformaticians.52,53
Table 1.
Sensitivity and specificity comparison between next generation sequencing (NGS), standard culture and culture from sonication fluid from all studies included in the systematic review
| NGS (sensitivity and specificity) | Standard culture (sensitivity and specificity) | Sonication fluid (culture) (sensitivity and specificity) |
|
|---|---|---|---|
| Wang et al, 201959 | sens = 94% spec = 95% |
||
| Ivy et al, 201854 | sens = 84% spec = 100% |
sens = 92% spec = 100% |
|
| Tarabichi et al, 201848 | sens (any) = 89.3% spec (any) = 73.0% sens > 59.5% single organism = 71.4% spec > 59.5% single organism = 94.6% |
Deep-tissue specimens sens = 60.7% spec = 97.3% |
|
| Street et al, 201755 | sens = 88% spec = 88% |
sens = 68% spec = 82% |
Note. sens, sensitivity; spec, specificity.
While previously cost prohibitive, the price of this diagnostic technique has dramatically decreased in recent years, making it accessible for clinical use. The technique may be particularly useful when there is strong clinical suspicion of PJI and cultures or other diagnostic tests are negative,41,48 which is an extremely frequent scenario in PJIs.
Metagenomic next generation sequencing (mNGS)
Unbiased metagenomic next generation sequencing (mNGS), which has the ability to broadly detect all classes of organisms directly from patient samples, represents a method that allows for universal pathogen detection regardless of microbe type (bacteria, fungi, parasites and viruses). All the DNA of a specimen is sequenced in parallel resulting in isolation and amplification of both host and pathogen nucleic acid within 24–48 hours of specimen collection. This timeframe differs significantly from the 3–7 days usually necessary to run standard serologic assays for synovial fluid bacterial detection.
The generalized workflow (Fig. 2) for mNGS in PJI diagnosis includes two components: (a) a wet lab protocol in which samples are collected and processed, then DNA is extracted, prepared into a sequencing library, and sequenced; (b) a dry lab computational pipeline that includes microbial identification, statistical analysis, and interpretation.50
Fig. 2.
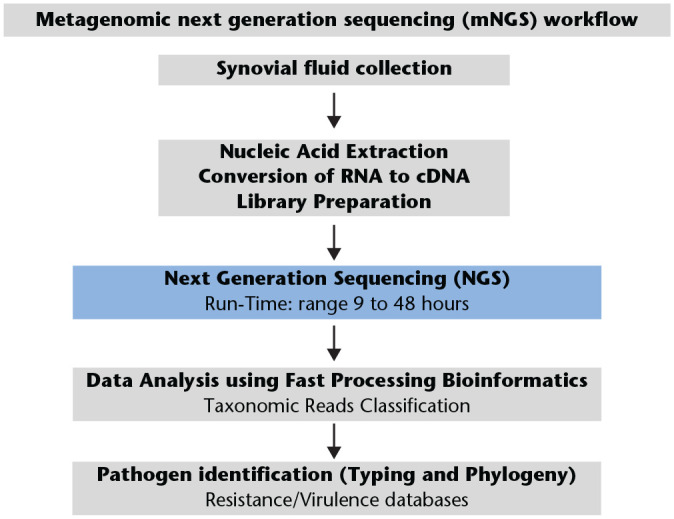
Metagenomic next generation sequencing (mNGS) workflow.
Interestingly, the length of the sequencing process depends on the platform used, the length of the reads and the amount of data generated (Table 2); the turnaround time relative to conventional methods plays a major role in determining the clinical relevance of NGS results at the time of decision making.
Table 2.
The five major sequencing platforms: advantages and disadvantages in terms of accuracy, efficiency and cost
| Sequencing platform |
Chemistry | Avg read length (bp) | Advantages | Disadvantages |
|---|---|---|---|---|
| Illumina | Sequencing by synthesis | ≤300 | High A accuracy |
Short reads, Long turnaround High cost |
| Thermo Fisher | Sequencing by synthesis | ≤400 | High accuracy | Short reads, Long turnaround High cost |
| Pacific Biosciences | Sequencing by synthesis | ≥500 | Long reads | Variable accuracy High cost Long turnaround |
| Oxford Nanopore | Measures the changes in current as molecules pass through the nanopore | ≥500 | Long reads Short turnaround Low cost |
Low accuracy |
| 454 GS Junior (Roche) | Pyro-sequencing | ≥500 | Long reads Short turnaround |
High error rate in homopolymer |
Metagenomic NGS (mNGS) is even more promising than standard NGS as a screening tool for PJI. Ivy et al54 showed, in a series of 168 synovial fluid samples collected from subjects with culture-positive or culture-negative PJIs, that mNGS yielded additional pathogens not detected by culture. Street et al sequenced sonication fluid from infected orthopaedic implants, including prosthetic devices, and showed that mNGS had 88% species-level sensitivity versus sonication fluid culture.55 Huang et al, in a consecutive series of 49 PJI, showed that the sensitivity of mNGS for diagnosing PJI was 95.9%, which was significantly higher than that of comprehensive culture (79.6%) while the specificity between mNGS and comprehensive culture was similar (95.2% and 95.2%, respectively).56 In the same study,56 mNGS was found to be most useful in identifying organisms that usually require special culture conditions (i.e. Mycoplasma and Mycobacterium) and in a scenario where patients had received antibiotics within two weeks prior to resection arthroplasty.
Weaver et al57 demonstrated, in the synovial fluid from seven PJIs, that whole-genome shotgun sequencing (WGSS) is an ideal tool to detect strains when culture did not, notably for dormant, culture-resistant and rare microbes. Interestingly, in all their samples, multiple microorganisms with multiple virulent factors were present. This and other reports58,59 suggest that PJIs are polymicrobial at the microbial DNA level in a significant proportion of sequenced PJI cases, increasing the fear in the adult reconstruction orthopaedic community. This fear was confirmed by Namdari et al58 who showed, in a series of 44 revision shoulder arthroplasties, that NGS data demonstrated that bacterial loads in revision arthroplasty are most commonly polymicrobial and a definition of infection that uses cultures is more prone to ‘probable contaminants’ than NGS. Because of this, further data regarding ‘normal’ microbiota in the shoulder, knee or hip joints are necessary to determine which organisms are truly pathogenic and which are ‘regular’ commensals.
Wang et al59 utilized mNGS to evaluate the efficacy of targeted antibiotics for the treatment of culture-negative PJI in comparison to an empirical antibiotic therapy, showing a better infection control rate, lower antibiotic-related complications and a shorter duration of systemic antibiotic therapy when targeted antibiotics were used according to mNGS results.
Modern NGS platforms (Table 2) have revolutionized biomedical research, and the technology is continually improving, exemplified by a novel approach to NGS using nanopore technologies (Oxford Nanopore Technologies – ONT) that was introduced to the research market. Instead of using a sequencing-by-synthesis approach, an ionic current is passed across the flow cell during sequencing, allowing for the different bacterial nucleotide bases (A-adenine; G-guanine; T-thymine; C-cytosine) to be determined by the changes in current as they pass through the nanopores.60 This technology allows for real-time analysis while sequencing is ongoing, significantly reducing the turnaround time (Table 2). Wang et al61 compared the classic metagenomic NGS, nanopore-based metagenomic sequencing and classical culture for the diagnosis of prosthetic joint infections from collected joint fluid, periprosthetic tissue and sonication fluid. Interestingly, the two metagenomic methods showed a similar sensitivity rate but the nanopore-based metagenomic sequencing method showed a better specificity and a quicker turnaround time (14–22 hours). The use of nanopore sequencing for PJI diagnosis has also allowed for the characterization of antimicrobial resistance (AMR), fundamental information in order to improve patient outcomes. Both Petersen et al52 and Wang et al59 showed that long-read nanopore sequencing was able to identify and map the presence or absence of resistance genes, from which the phenotype of resistance can be inferred. Interestingly, in the Wang et al study,59 nanopore sequencing detected more AMR reads with broader coverage than NGS sequencing, downplaying the role of the classical antibiogram.
Cell-free DNA next generation sequencing (cfNGS)
A very recent innovation in the field of NGS is represented by cell-free DNA next generation sequencing (cfNGS). This method differs from mNGS62 by lacking a cell lysis step, which serves to break open microbial cells in order to isolate DNA. In the case of cfNGS, DNA is isolated directly from the sample without first lysing cells. Sequencing of cell-free DNA (cfDNA) has recently been shown to enable non-invasive diagnosis of several indications that previously required invasive procedures, including the diagnosis of foetal abnormalities, detection of transplanted organ rejection, and characterization or monitoring of cancer.63–68
CfNGS has been shown to be sensitive enough to identify pathogens in patients pre-treated with antibiotics up to 30 days prior to initial sample collection.64 The major advantage and unique characteristic of cfNGS is the fast run-time, which typically can be completed within hours. Direct real-time sequencing of samples provides accurate information compared to laboratory culture, can detect additional unculturable organisms, and provides information regarding the presence of antimicrobial resistance genes.63,64 Interestingly, cfNGS has not been tested on synovial fluid to diagnose PJI but represents a very appealing new frontier.67 Following a previously published workflow for microbial cfDNA sequencing in plasma,64 a proposed workflow for cfDNA sequencing of PJI synovial fluid is shown in Fig. 3.
Fig. 3.
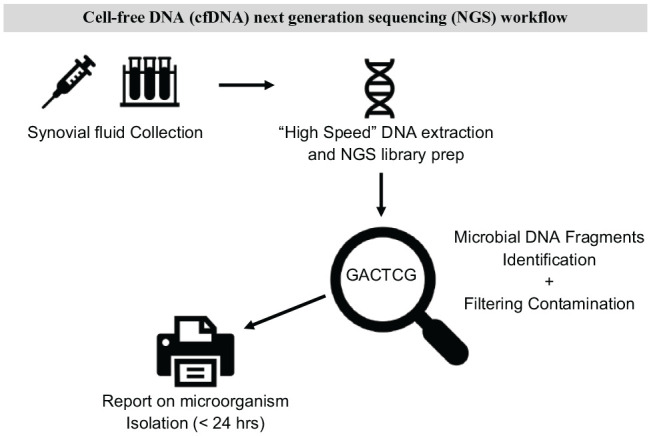
Cell-free DNA (cfDNA) next generation sequencing (NGS) workflow.
Limitations
The ability to quickly and broadly detect all classes of organism from patient synovial fluid is the major advantage of NGS and mNGS as PJI diagnostic tools. However, NGS, even if promising, has several limitations. Significant concerns exist regarding the performance, validity and clinical significance of the detected organisms. Despite NGS-based methods having increased sensitivity for identifying organisms compared to culture,48,62,63 trade-offs exist in their clinical application.
A major limitation of the use of NGS technologies for infectious disease and PJI diagnostics is represented by the possibility of false-positives due to contamination. Unfortunately, detection of normal human body flora or contaminating organisms in laboratory reagents can lead to organism misidentification. Moreover, the yield of DNA was shown to be lower in gram-positive cocci and fungi compared to bacilli,56 indicating that mNGS requires strict storage and transport conditions to decrease the risk of nucleic acid degradation. More studies are needed to determine optimal storage and transport conditions and to identify contaminants.
While this review has focused on NGS sensitivity, a low specificity, resulting in a high rate of false-positives,46,48 has been demonstrated. However, negative mNGS results may reassure the treating orthopaedic surgeon that a PJI is unlikely to be present: this ‘rule-out’ test characteristic may be helpful in a total joint arthroplasty revision scenario (single-stage vs. double-stage determination). Unfortunately, because of their turnaround times, mNGS and nanopore-based metagenomic sequencing are not yet available for intraoperative use, and this represents a major limitation of these technologies.
Another limitation is the fact that there are not yet FDA-cleared or approved tests using NGS technologies for infectious diseases.69 The current guidelines for NGS/mNGS testing have been developed in clinical fields (e.g. oncology) that differ from infectious diseases and have only recently been adapted to infectious disease diagnostics.69
NGS is also more expensive than culturing and other traditional microbiological tests,70 even when taking into account the multiple cultures recommended for diagnosis of PJI.23 Regardless, the current authors suggest that the cost-effectiveness of NGS/mNGS assays for PJI pathogen detection should be compared to the costs of intensive care unit stays and more invasive diagnostic procedures.
Another limitation of NGS/mNGS technologies is represented by the run-time. Classic NGS requires three to five days41,46–48,68,70–73 to identify microorganisms, sometimes reducing the clinical applicability of NGS. However, ‘in-house’ mNGS can have a much shorter turnaround time of less than two days. Unfortunately, to meet this timeframe while also minimizing the risk for contamination,68 standard laboratories will need space dedicated to and designed for NGS, highly specialized equipment, trained lab technicians and bioinformatics expertise.
Perspective
Preoperative, perioperative and intraoperative diagnosis and identification of pathogens involved in PJI represent a challenge, as up to 70% of cases that meet the criteria for PJI are synovial fluid culture-negative.63 PJI treatment is accomplished through targeted surgical and microbiological management, which is contingent on an accurate and strictly perioperative diagnosis and pathogen identification. Standard clinical management in the absence of identified aetiology drives up the use of empiric, broad-spectrum antibiotics in an attempt to encompass all possible pathogens and risks not treating an organism and even missing actual PJI cases. Different NGS technologies have shown the capability of identifying pathogen DNA in synovial fluid: mNGS could become, in the near future, a cost-effective and time-efficient method to diagnose and treat culture-negative PJI in particular and musculoskeletal infections in general.
The current authors recognize the major technical challenge of NGS testing: the vast majority of sequenced DNA is human, from host contamination,68 despite efforts to reduce this in the laboratory preparation. Depletion of host DNA contamination will eventually facilitate greater pathogen genome sequencing coverage. An optimized DNA extraction protocol still needs to be developed in order to sequence longer fragments and to implement a fast-read method,55 which could allow real-time selective sequencing of pathogen DNA over human DNA normally present in the synovial fluid, increasing the proportion of bacterial reads and improving diagnostic power. In fact, multiple studies62,73 have shown that the mere detection of bacterial DNA does not mean infection, and thus, it is clinically important to identify the microorganisms that are normal flora in a total joint arthroplasty setting (‘joint microbiota’).58 The fact that these microorganisms may not necessarily be involved and active in the disease process, and the ultimate ability of NGS technologies to identify true infecting microorganisms, all need to be determined by future studies.
Conclusion
NGS is a revolutionary technology that could disrupt the diagnostic paradigm of PJI. New types of NGS are exciting, their rapid evolution often outpaces a mandatory and comprehensive collection of clinical evidence. Multicentre trials evaluating patient outcomes will be necessary to translate NGS into a useful clinical test.
Footnotes
ICMJE Conflict of interest statement: PFI reports consultancy for Exactech and grants/grants pending from Zimmer Biomet, all outside the submitted work.
SG reports employment as an attending surgeon by Istituto Clinico Sant’ambrogio, Milan, Italy, outside the submitted work.
BV reports consultancy for Smith & Nephew and Exactech, employment by Istituto Clinico Sant’Ambrogio, IRCCS Galeazzi, Milan, Italy, payment for lectures including service on speakers bureaus by Smith & Nephew and Lima Orthopaedics, royalties from Lima Orthopaedics, and stock/stock options in Stryker Corporation and Zimmer Biomet, all outside the submitted work.
DFA reports grants from NIH-NCATS and OREF, related to PJI, not NGS, related to the submitted work, and consultancy for Stryker, Exactech, Depuy, Zimmer- Biomet, Haraeus and Medcura, expert testimony for the Expert Institute, grants from NIH-NCATS and OREF, payment for manuscript preparation from Medscape, patents owned by Arthology Consulting, Arthrology Designs and Stanford, royalties from Exactech, and stock/stock options in nSight Surgical, QT Ultrasound and Recoup Fitness, all unrelated to the submitted work.
OA licence text: This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (CC BY-NC 4.0) licence (https://creativecommons.org/licenses/by-nc/4.0/) which permits non-commercial use, reproduction and distribution of the work without further permission provided the original work is attributed.
Funding statement
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1. Patel A, Calfee RP, Plante M, Fischer SA, Arcand N, Born C. Methicillin-resistant Staphylococcus aureus in orthopaedic surgery. J Bone Joint Surg Br 2008;90:1401–1406. [DOI] [PubMed] [Google Scholar]
- 2. Trampuz A, Widmer AF. Infections associated with orthopedic implants. Curr Opin Infect Dis 2006;19:349–356. [DOI] [PubMed] [Google Scholar]
- 3. Senthi S, Munro JT, Pitto RP. Infection in total hip replacement: meta-analysis. Int Orthop 2011;35:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shahi A, Tan TL, Chen AF, Maltenfort MG, Parvizi J. In-hospital mortality in patients with periprosthetic joint infection. J Arthroplasty 2017;32:948–952.e1. [DOI] [PubMed] [Google Scholar]
- 5. Rubin RJ, Harrington CA, Poon A, Dietrich K, Greene JA, Moiduddin A. The economic impact of Staphylococcus aureus infection in New York City hospitals. Emerg Infect Dis 1999;5:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osler T, Glance LG, Hosmer DW. Complication-associated mortality following trauma: a population-based observational study. Arch Surg 2012;147:152–158. [DOI] [PubMed] [Google Scholar]
- 7. Parvizi J, Pawasarat IM, Azzam KA, Joshi A, Hansen EN, Bozic KJ. Periprosthetic joint infection: the economic impact of methicillin-resistant infections. J Arthroplasty 2010;25:103–107. [DOI] [PubMed] [Google Scholar]
- 8. Resch A, Wilke M, Fink C. The cost of resistance: incremental cost of methicillin-resistant Staphylococcus aureus (MRSA) in German hospitals. Eur J Health Econ 2009;10:287–297. [DOI] [PubMed] [Google Scholar]
- 9. Rosner AJ, Becker DL, Wong AH, Miller E, Conly JM. The costs and consequences of methicillin-resistant Staphylococcus aureus infection treatments in Canada. Can J Infect Dis Med Microbiol 2004;15:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson DJ, Sexton DJ, Kanafani ZA, Auten G, Kaye KS. Severe surgical site infection in community hospitals: epidemiology, key procedures, and the changing prevalence of methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 2007;28:1047–1053. [DOI] [PubMed] [Google Scholar]
- 11. Cayce KO, IV, Galloway MT. Infection. In: Fischgrund JS. editor. Orthopaedic knowledge update 9. Rosemont; American Academy of Orthopaedic Surgeons; 2008;241-257. [Google Scholar]
- 12. Riesgo AM, Liporace FA. Strategies for management of periprosthetic joint infection. Bull Hosp Jt Dis (2013) 2018;76:55–61. [PubMed] [Google Scholar]
- 13. Parvizi J, Shohat N, Gehrke T. Prevention of periprosthetic joint infection: new guidelines. Bone Joint J 2017;99-B:3–10. [DOI] [PubMed] [Google Scholar]
- 14. Kurtz SM, Lau E, Watson H, Schmier JK, Parvizi J. Economic burden of periprosthetic joint infection in the United States. J Arthroplasty 2012;27:61–65.e1. [DOI] [PubMed] [Google Scholar]
- 15. Bozic KJ, Kurtz SM, Lau E, et al. The epidemiology of revision total knee arthroplasty in the United States. Clin Orthop Relat Res 2010;468:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bozic KJ, Kurtz SM, Lau E, Ong K, Vail TP, Berry DJ. The epidemiology of revision total hip arthroplasty in the United States. J Bone Joint Surg Am 2009;91:128–133. [DOI] [PubMed] [Google Scholar]
- 17. Jaekel DJ, Ong KL, Lau EC, Watson HN, Kurtz SM. The epidemiology of total hip and knee arthroplasty infection. In: Springer BD, Parvizi J, eds. Periprosthetic joint infection of the hip and knee. Vol 1. New York: Springer, 2014:1–14. [Google Scholar]
- 18. Hackett DJ, Rothenberg AC, Chen AF, et al. The economic significance of orthopaedic infections. J Am Acad Orthop Surg 2015;23:S1–S7. [DOI] [PubMed] [Google Scholar]
- 19. Bedair H, Ting N, Jacovides C, et al. The Mark Coventry Award: diagnosis of early postoperative TKA infection using synovial fluid analysis. Clin Orthop Relat Res 2011;469:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Della Valle CJ, Sporer SM, Jacobs JJ, Berger RA, Rosenberg AG, Paprosky WG. Preoperative testing for sepsis before revision total knee arthroplasty. J Arthroplasty 2007;22:90–93. [DOI] [PubMed] [Google Scholar]
- 21. Sofka CM, Potter HG, Adler RS, Pavlov H. Musculoskeletal imaging update: current applications of advanced imaging techniques to evaluate the early and long-term complications of patients with orthopedic implants. HSS J 2006;2:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Workgroup Convened by the Musculoskeletal Infection Society. New definition for periprosthetic joint infection. J Arthroplasty 2011;26:1136–1138. [DOI] [PubMed] [Google Scholar]
- 23. Parvizi J, Zmistowski B, Berbari EF, et al. New definition for periprosthetic joint infection: from the Workgroup of the Musculoskeletal Infection Society. Clin Orthop Relat Res 2011;469:2992–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volpe L, Indelli PF, Latella L, Poli P, Yakupoglu J, Marcucci M. Periprosthetic joint infections: a clinical practice algorithm. Joints 2015;2:169–174. [PMC free article] [PubMed] [Google Scholar]
- 25. Parvizi J, Adeli B, Zmistowski B, Restrepo C, Greenwald AS. Management of periprosthetic joint infection: the current knowledge: AAOS exhibit selection. J Bone Joint Surg Am 2012;94:e104. [DOI] [PubMed] [Google Scholar]
- 26. Parvizi J, Gehrke T, Chen AF. Proceedings of the International Consensus on Periprosthetic Joint Infection. Bone Joint J 2013;95-B:1450e2. [DOI] [PubMed] [Google Scholar]
- 27. Parvizi J, Gehrke T. Proceedings of the Second International Consensus Meeting on Musculoskeletal Infection. J Arthroplasty 2019;34:S1–S496. [DOI] [PubMed] [Google Scholar]
- 28. Youssef B, Pavlou G, Tsiridis E. Philadelphia 2013: international consensus meeting on periprosthetic joint infection. Hip Int 2014;24:3–4. [DOI] [PubMed] [Google Scholar]
- 29. Saleh A, George J, Sultan AA, Samuel LT, Mont MA, Higuera-Rueda CA. The quality of diagnostic studies in periprosthetic joint infections: can we do better? J Arthroplasty 2019;34:2737–2743. [DOI] [PubMed] [Google Scholar]
- 30. Dinneen A, Guyot A, Clements J, Bradley N. Synovial fluid white cell and differential count in the diagnosis or exclusion of prosthetic joint infection. Bone Joint J 2013;95-B:554–557. [DOI] [PubMed] [Google Scholar]
- 31. Mason JB, Fehring TK, Odum SM, Griffin WL, Nussman DS. The value of white blood cell counts before revision total knee arthroplasty. J Arthroplasty 2003;18:1038–1043. [DOI] [PubMed] [Google Scholar]
- 32. Stroh DA, Johnson AJ, Naziri Q, Mont MA. How do frozen and permanent histopathologic diagnoses compare for staged revision after periprosthetic hip infections? J Arthroplasty 2012;27:1663–1668.e1. [DOI] [PubMed] [Google Scholar]
- 33. Wetters NG, Berend KR, Lombardi AV, Morris MJ, Tucker TL, Della Valle CJ. Leukocyte esterase reagent strips for the rapid diagnosis of periprosthetic joint infection. J Arthroplasty 2012;27:8–11. [DOI] [PubMed] [Google Scholar]
- 34. Kheir MM, Tan TL, Ackerman CT, Modi R, Foltz C, Parvizi J. Culturing periprosthetic joint infection: number of samples, growth duration, and organisms. J Arthroplasty 2018;33:3531–3536.e1. [DOI] [PubMed] [Google Scholar]
- 35. van den Bijllaardt W, van der Jagt OP, Peijs M, Janssens M, Buiting AG, Reuwer AQ. Culturing periprosthetic tissue in blood culture bottles results in isolation of additional microorganisms. Eur J Clin Microbiol Infect Dis 2019;38:245–252. [DOI] [PubMed] [Google Scholar]
- 36. Larsen LH, Lange J, Xu Y, Schønheyder HC. Optimizing culture methods for diagnosis of prosthetic joint infections: a summary of modifications and improvements reported since 1995. J Med Microbiol 2012;61:309–316. [DOI] [PubMed] [Google Scholar]
- 37. Peel TN, Spelman T, Dylla BL, et al. Optimal periprosthetic tissue specimen number for diagnosis of prosthetic joint infection. J Clin Microbiol 2016;55:234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berbari EF, Marculescu C, Sia I, et al. Culture-negative prosthetic joint infection. Clin Infect Dis 2007;45:1113–1119. [DOI] [PubMed] [Google Scholar]
- 39. Malekzadeh D, Osmon DR, Lahr BD, Hanssen AD, Berbari EF. Prior use of antimicrobial therapy is a risk factor for culture-negative prosthetic joint infection. Clin Orthop Relat Res 2010;468:2039–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parvizi J, Erkocak OF, Della Valle CJ. Culture-negative periprosthetic joint infection. J Bone Joint Surg Am 2014;96:430–436. [DOI] [PubMed] [Google Scholar]
- 41. Zmistowski B, Della Valle C, Bauer TW, et al. Diagnosis of periprosthetic joint infection. J Arthroplasty 2014;29:77–83. [DOI] [PubMed] [Google Scholar]
- 42. Yoon H-K, Cho S-H, Lee DY, et al. A review of the literature on culture-negative periprosthetic joint infection: epidemiology, diagnosis and treatment. Knee Surg Relat Res 2017;29:155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choi HR, Kwon YM, Freiberg AA, Nelson SB, Malchau H. Periprosthetic joint infection with negative culture results: clinical characteristics and treatment outcome. J Arthroplasty 2013;28:899–903. [DOI] [PubMed] [Google Scholar]
- 44. Huang R, Hu CC, Adeli B, Mortazavi J, Parvizi J. Culture-negative periprosthetic joint infection does not preclude infection control. Clin Orthop Relat Res 2012;470:2717–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rothenberg AC, Wilson AE, Hayes JP, O’Malley MJ, Klatt BA. Sonication of arthroplasty implants improves accuracy of periprosthetic joint infection cultures. Clin Orthop Relat Res 2017;475:1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shohat N, Tarabichi M, Goswami K, Parvizi J. Can next generation sequencing play a role in detecting pathogens in synovial fluid? Bone Joint J 2018;100-B:127–133. [DOI] [PubMed] [Google Scholar]
- 47. Tarabichi M, Alvand A, Shohat N, Goswami K, Parvizi J. Diagnosis of Streptococcus canis periprosthetic joint infection: the utility of next-generation sequencing. Arthroplast Today 2017;4:20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tarabichi M, Shohat N, Goswami K, et al. Diagnosis of periprosthetic joint infection: the potential of next generation sequencing. J Bone Joint Surg Am 2018;100:147–154. [DOI] [PubMed] [Google Scholar]
- 49. Camargo JF, Ahmed AA, Lindner MS, et al. Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000Res 2019;8:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 2019;14:319–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019;4:663–674. [DOI] [PubMed] [Google Scholar]
- 52. Petersen LM, Martin IW, Moschetti WE, Kershaw CM, Tsongalis GJ. Third-generation sequencing in the clinical laboratory: exploring the advantages and challenges of nanopore sequencing. J Clin Microbiol 2019;58:e01315–e01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ivy MI, Thoendel MJ, Jeraldo PR, et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J Clin Microbiol 2018;56:e00402–e00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Street TL, Sanderson ND, Atkins BL, et al. Molecular diagnosis of orthopedic-device-related infection directly from sonication fluid by metagenomic sequencing. J Clin Microbiol 2017;55:2334–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang Z, Li W, Lee GC, et al. Metagenomic next-generation sequencing of synovial fluid demonstrates high accuracy in prosthetic joint infection diagnostics: mNGS for diagnosing PJI. Bone Joint Res 2020;9:440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weaver AA, Hasan NA, Klaassen M, Karathia H, Colwell RR, Shrout JD. Prosthetic joint infections present diverse and unique microbial communities using combined whole-genome shotgun sequencing and culturing methods. J Med Microbiol 2019;68:1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Namdari S, Nicholson T, Abboud J, Lazarus M, Ramsey ML, Williams G, Parvizi J. Comparative study of cultures and next-generation sequencing in the diagnosis of shoulder prosthetic joint infections. J Shoulder Elbow Surg 2019;28:1–8. [DOI] [PubMed] [Google Scholar]
- 59. Wang C, Huang Z, Li W, Fang X, Zhang W. Can metagenomic next-generation sequencing identify the pathogens responsible for culture-negative prosthetic joint infection? BMC Infect Dis 2020;20:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Laver T, Harrison J, O’Neill PA, Moore K, Farbos A, Paszkiewicz K, Studholme DJ. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif 2015;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang CX, Huang Z, Fang W, et al. Preliminary assessment of nanopore-based metagenomic sequencing for the diagnosis of prosthetic joint infection. Int J Infect Dis 2020;97:54–59. [DOI] [PubMed] [Google Scholar]
- 62. Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, et al. Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin Infect Dis 2018;67:1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan TL, Kheir MM, Shohat N, et al. Culture-negative periprosthetic joint infection: an update on what to expect. JB JS Open Access 2018;3:e0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shah P, Ruffin F, Seng H, et al. Direct detection and quantification of bacterial cell-free DNA in patients with infective endocarditis (IE) using the Karius Plasma Next Generation Sequencing (NGS) Test. Open Forum Infect Dis 2018;5:S12. [Google Scholar]
- 65. Judge K, Harris SR, Reuter S, Parkhill J, Peacock SJ. Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes. J Antimicrob Chemother 2015;70:2775–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Acharya K, Khanal S, Pantha K, Amatya N, Davenport RJ, Werner D. A comparative assessment of conventional and molecular methods, including MinION nanopore sequencing, for surveying water quality. Sci Rep 2019;9:15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol 2017;28:2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Miller S, Chiu C, Rodino KG, Miller MB. Point-counterpoint: should we be performing metagenomic next-generation sequencing for infectious disease diagnosis in the clinical laboratory? J Clin Microbiol 2020;58:e01739-e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Food and Drug Administration. FDA draft guidance: infectious disease next generation sequencing based diagnostic devices: microbial identification and detection of antimicrobial resistance and virulence markers: draft guidance for industry and Food and Drug Administration Staff. Rockville, MD: Food and Drug Administration, 2016. [Google Scholar]
- 70. Torchia MT, Austin DC, Kunkel ET, et al. Next-generation sequencing vs culture-based methods for diagnosing periprosthetic joint infection after total knee arthroplasty: a cost-effectiveness analysis. J Arthroplasty 2019;34:1333–1341. [DOI] [PubMed] [Google Scholar]
- 71. Jacovides CL, Kreft R, Adeli B, Hozack B, Ehrlich GD, Parvizi J. Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J Bone Joint Surg Am 2012;94:2247–2254. [DOI] [PubMed] [Google Scholar]
- 72. Lee MS, Chang WH, Chen SC, et al. Molecular diagnosis of periprosthetic joint infection by quantitative RT-PCR of bacterial 16S ribosomal RNA. ScientificWorldJournal 2013;2013:950548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Goswami K, Parvizi J. Culture-negative periprosthetic joint infection: is there a diagnostic role for next-generation sequencing? Expert Rev Mol Diagn 2020;20:269–272. [DOI] [PubMed] [Google Scholar]