Abstract
Objectives
Detection and surveillance of SARS-CoV-2 is of eminent importance, particularly due to the rapid emergence of variants of concern (VOCs). In this study we evaluated if a commercially available quantitative real-time PCR (qRT-PCR) assay can identify SARS-CoV-2 B.1.1.7 lineage samples by a specific N gene dropout or Ct value shift compared with the S or RdRp gene.
Methods
VOC B.1.1.7 and non-B.1.1.7 SARS-CoV-2-positive patient samples were identified via whole-genome sequencing and variant-specific PCR. Confirmed B.1.1.7 (n = 48) and non-B.1.1.7 samples (n = 58) were analysed using the Allplex™ SARS-CoV-2/FluA/FluB/RSV™ PCR assay for presence of SARS-CoV-2 S, RdRp and N genes. The N gene coding sequence of SARS-CoV-2 with and without the D3L mutation (specific for B.1.1.7) was cloned into pCR™II-TOPO™ vectors to validate polymorphism-dependent N gene dropout with the Allplex™ SARS-CoV-2/FluA/FluB/RSV™ PCR assay.
Results
All studied B.1.1.7-positive patient samples showed significantly higher Ct values in qRT-PCR (Δ6–10, N gene dropout on Ct values > 29) of N gene than the corresponding values of S (p ≤ 0.0001) and RdRp (p ≤ 0.0001) genes. The assay reliably discriminated B.1.1.7 and non-B.1.1.7 positive samples (area under the curve = 1) in a receiver operating characteristic curve analysis. Identical Ct value shifts (Δ7–10) were detected in reverse genetic experiments, using isolated plasmids containing N gene coding sequences corresponding to D3 or 3L variants.
Discussion
An N gene dropout or Ct value shift is shown for B.1.1.7-positive samples in the Allplex™ SARS-CoV-2/FluA/FluB/RSV™ PCR assay. This approach can be used as a rapid tool for B.1.1.7 detection in single assay high throughput diagnostics.
Keywords: B.1.1.7, Diagnostic quantitative RT-PCR, Mutation, SARS-CoV-2, VOC
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the cause of the life-threatening COVID-19 disease, has rapidly spread worldwide since December 2019. Since the virus undergoes adaptive evolution, resulting genetic variations pose a challenge particularly in molecular diagnostics, because detection is based on small regions in the viral genome. To this date, several major variants of concern (VOCs) of SARS-CoV-2 have been reported. Currently, B.1.351 (first detected in South Africa) [1], P.1, (Brazil) [2] and B.1.1.7 (UK) [3] are among the most prevalent ones. The B.1.1.7 variant has been associated with increased mortality [4] and risk of transmission [5] and is rapidly expanding its geographic range worldwide. Therefore, early identification of this variant in patients may help reduce further spread of infection. Each lineage is characterized by a combination of single mutations across the SARS-CoV-2 genome, and while some are shared between aforementioned lineages, others are specific for one individual lineage. For example, amino acid exchange N501Y in the SARS-CoV-2 spike protein (S) is present in all three mentioned lineages, whereas the additional deletion H69/V70 in the same protein is present in B.1.1.7, but not in the other two VOCs B.1.351 and P.1 [3]. B.1.1.7 also has a specific single nucleotide polymorphism in the N gene (D3L) which can be targeted by PCR assays.
Multiplex PCR is considered as the reference standard for SARS-CoV-2 detection, and dual or triple target approaches are recommended [6,7]. However, identification of viral variants is not intended with regular PCR kits. The presence of SARS-CoV-2 variants in a patient sample can potentially change the performance of a SARS-CoV-2 assay. Some studies have reported dropouts of various genes targeted by SARS-CoV-2 multiplex PCR approaches affecting the S gene [8,9], N gene [10,11] or E gene [12], which may allow presumptive identification of specific lineages. While spread of VOCs is an imminent danger worldwide, rapid detection of VOCs is limited by the turnaround time and costs, or availability of methods such as next-generation sequencing or variant-specific PCR, respectively. Here, we report an approach which allows for presumptive identification of the B.1.1.7 lineage by calculating a score from the Ct values of the target genes included in the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay (Seegene, South Korea), thus greatly speeding up the time to result of laboratory diagnosis of this lineage.
Materials and methods
Nucleic acid extraction from patient swabs was performed on Seegene Nimbus IVD using the STARMag 96 X 4 Viral DNA/RNA 200 C Kit (Seegene). Amplification and detection of SARS-CoV-2 via qRT-PCR was performed with Allplex™ SARS-CoV-2/FluA/FluB/RSV or Allplex™ 2019-nCoV assays, respectively (Seegene).
Isolated RNA (one confirmed UK variant B.1.1.7 and one non-B.1.1.7) was reversed transcribed using SuperScript™ IV First-Strand Synthesis System (ThermoFisher Scientific) with specific primer AS-CoV2_N_3' (5′-TTAGGCCTGAGTTGAGTCAGC-3′). Subsequent amplification of the entire N gene region was achieved using Platinum™ Taq DNA Polymerase High Fidelity (ThermoFisher Scientific) and forward primer S–CoV2_N_D3_5' (5′-CAAACTAAAATGTCTGATAATGGACCCCAAAATC-3′) or S–CoV2_N_3L_5' (5′-CAAACTAAAATGTCTCTAAATGGACCCCAAAATC-3′) each combined with reverse primer AS-CoV2_N_3'. PCR products were cloned into the pCR™II-TOPO™ vector backbone using TOPO™ TA Cloning™ Kit (ThermoFisher Scientific) and propagated by transformation of TOP10 competent Escherichia coli. The presence of D3 or 3L variant was confirmed via Sanger sequencing (Fig. S1).
For validation of D3L polymorphism specific Ct value shift in the N gene, each plasmid was diluted to an initial copy number of 1.25 × 108 and, subsequently diluted to 1:10, 1:100 and 1:1000 dilutions. To imitate extraction and test conditions, 0.5 μl of each dilution was then mixed with 500 μL of viral transport medium containing a negative tested nasopharyngeal swab and treated as described above.
Patient samples were identified by variant-specific PCR (Novaplex™ SARS-CoV-2 Variants I assay (Seegene)) or by whole-genome sequencing using Illumina MiSeq and Novaseq 6000 (LGL, Oberschleißheim, Germany; Synlab MVZ, Weiden, Germany).
The study was approved by the institutional review board of the Paracelsus Medical University, Nuremberg, Germany (reference number IRB 2021-008).
Statistical analysis was performed using GraphPad Prism version 9.1.0 for Windows (GraphPad Software, San Diego, CA, USA).
Results
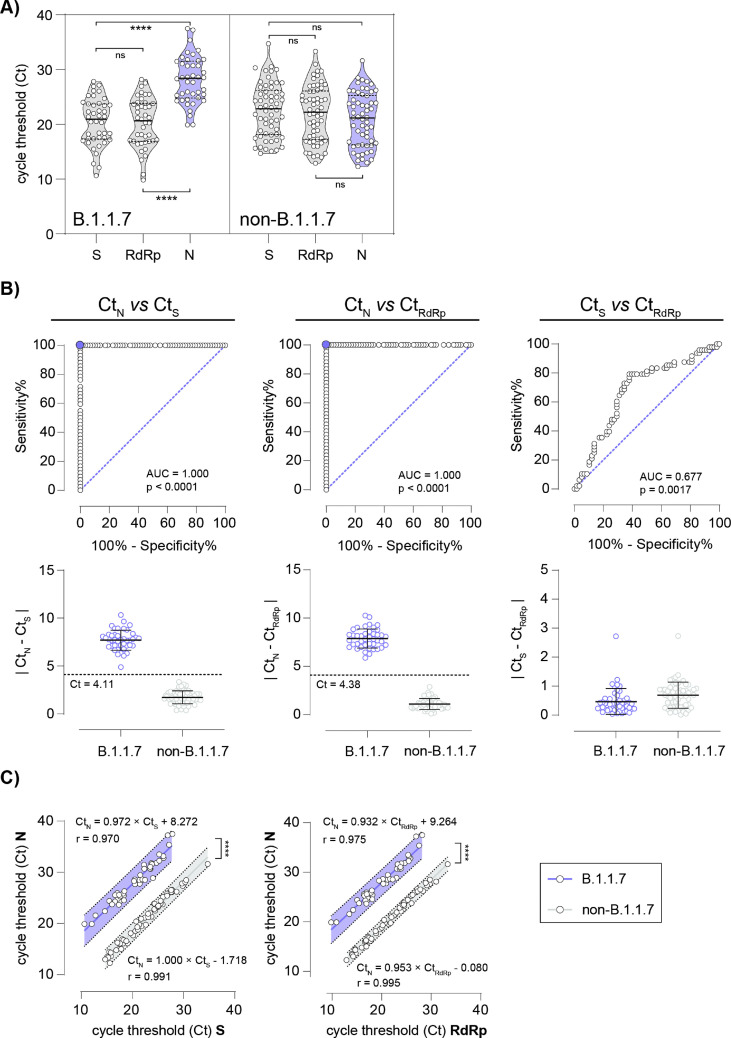
A qRT-PCR assay, based on a triple target approach is routinely used for SARS-CoV-2 detection (S gene, RdRp gene and N gene). Using this method, we noticed significant N gene-specific changes in Ct values for the N target compared with results for S gene and RdRp gene (p ≤ 0.0001), initially occurring in early 2021 (Fig. 1 A). A total of 48 consecutive samples, all showing N gene dropouts (n = 6) or N gene cycle threshold (Ct) value shifts (n = 42) (ΔCt N/RdRp or ΔCt N/S > 6) were collected for further analysis aiming at lineage-specific assignment of the respective SARS-CoV-2 strains. In addition, a selection of 58 samples with unsuspicious ΔCt N/RdRp or ΔCt N/S (<6) were included as controls. To this end, either whole-genome sequencing (n = 23) or variant-specific PCR (n = 25) were performed. These analyses revealed that all of the samples which had previously yielded N gene-specific Ct value shifts or dropouts (ΔCt N/RdRp or ΔCt N/S score > 6) belonged to the B.1.1.7 lineage, whereas the remaining 58 SARS-CoV-2 positive samples with ΔCt N/RdRp or ΔCt N/S < 6 were all classified as non-B.1.1.7 variants (B.1.1.317 (n = 2), B.1.351 (n = 1), N501Y+, E484K+ (n = 1), N501Y-, E484K+ (n = 1) and N501Y-, Del. H69/V70- (n = 53), reflecting the local epidemiology in early 2021 (Tables S1 and S2).
Fig. 1.
The Allplex™ SARS-CoV-2/FluA/FluB/RSV assay reliably discriminates SARS-CoV-2 B.1.1.7 from non-B.1.1.7 positive patient specimens. (A) Violin plots with median (solid black lines), quartiles (dashed black lines) and individual data points (open circles) of determined cycle threshold (Ct) values for B.1.1.7 and non-B.1.1.7 positive patient specimens in qRT-PCR for three genes, S, RdRp and N. ∗∗∗∗p ≤ 0.0001 in one-way ANOVA followed by Tukey's multiple comparison test, ns = not significant. (B) Receiver operating characteristic (ROC) curves discriminating absolute differences in Ct values of B.1.1.7 and non-B.1.1.7 positive patient specimens between N gene and S gene (left panel), N gene and RdRp gene (middle panel) and S gene and RdRp gene (right panel). Lower respective panels display differences in Ct values; dashed lines indicate Ct with 100% sensitivity and specificity (95% CI sensitivity 91.62–100% and CI specificity 93.79–100%) as determined in ROC curves. (C) Pearson correlation (r) and comparative linear regression of Ct values of N gene and S gene (left panel) and N gene and RdRp gene of B.1.1.7 (blue line and 99% prediction band) and non-B.1.1.7 (grey line and 99% prediction band) positive patient specimens, respectively. No significant difference in slope is determined (CtN/CtS p 0.474, CtN/CtRdRp p 0.446), y-intercepts (elevations) differ significantly (∗∗∗∗p ≤ 0.0001).
The observed N gene Ct value shift or N gene dropout was detectable regardless of whether samples were strongly or weakly positive for SARS-CoV-2, as evident from the broad range of Ct values for B.1.1.7 and non-B.1.1.7 (Fig. 1A). For S or RdRp targets we did not observe any shifts in Ct values (Fig. S2). Delta Ct values of 4.11 and 4.38 were identified in a receiver operating characteristic (ROC) curve analysis to discriminate B.1.1.7 and non-B.1.1.7 variants with 100% sensitivity and specificity using ΔCt N/RdRp (95% CI sensitivity 91.62–100% and 95% CI specificity 93.79–100%), and ΔCt N/S (95% CI sensitivity 91.62–100% and 95% CI specificity 93.79–100%), respectively (Fig. 1B). Similar shifts in ΔCt for S/RdRp were not observed (Fig. 1B). In addition, comparative linear regression analysis of ΔCt N/S or ΔCt N/RdRp values revealed no significant differences in slopes (ΔCt N/S p 0.474, ΔCt N/RdRp p 0.446), while elevations of lines (y-intercepts) differed significantly (ΔCt N/S p ≤ 0.0001, ΔCt N/RdRp p ≤ 0.0001) (Fig. 1C). Concluding, ΔCt value shifts observed for both N/RdRp and N/S allow for meaningful classification of B.1.1.7 versus non-B.1.1.7 (no overlap/outliers).
A subset of the samples with N gene Ct value shift or N gene dropout were tested by two other commercially available SARS-CoV-2 PCR assays. Neither the Allplex™ 2019-nCoV (Seegene) nor the VIASURE SARS-CoV-2 (CerTest, for BD MAX™ System) showed N gene Ct value shift or N gene dropout.
To elucidate the underlying molecular mechanism resulting in the observed N gene dropout or Ct value shift, we cloned the N gene coding sequence with and without D3L mutation into pCR™II-TOPO™ vectors which were then used for PCR analysis.
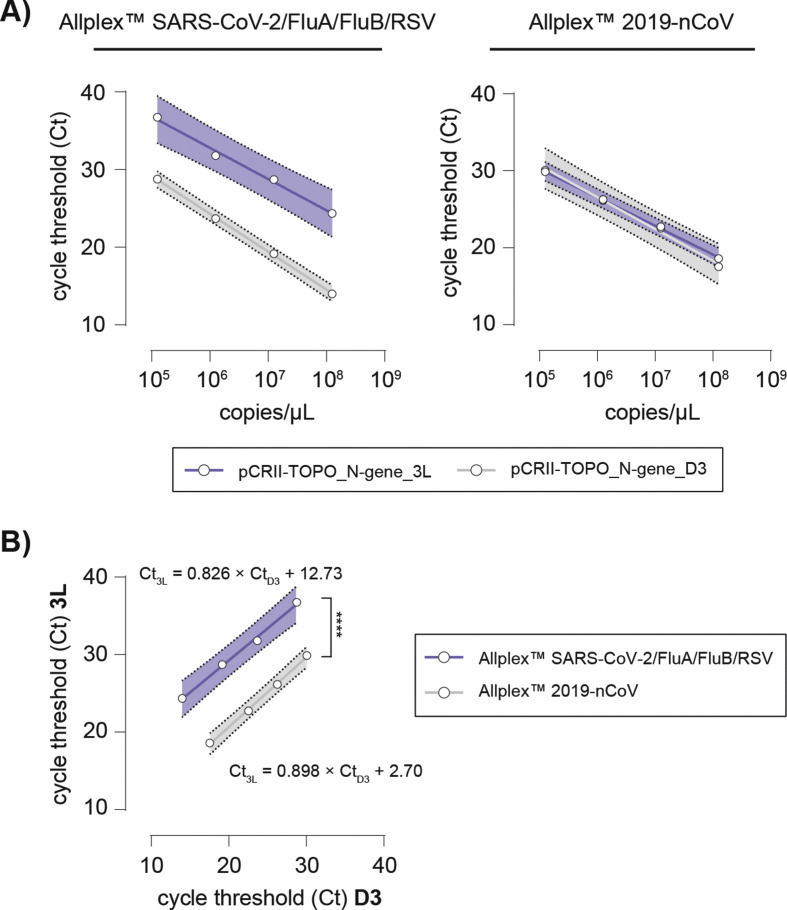
Again, we found a shift in Ct values ΔCt N D3/N 3L 7.1-10.1 for the 3L but not for D3 variant N gene using the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay, whereas Allplex™ 2019-nCoV did not show any shift in Ct (ΔCt N D3/N 3L 0.1–1.07) values for both variants (Fig. 2 A, Table S3). Here, comparative linear regression analysis identified no significant discrepancy in slopes of the curves (Ct N D3/N 3L p 0.212), while intercepts differed significantly (Ct N D3/N 3L p ≤ 0.0001) in the two assays (Fig. 2B). This confirmed that the D3L mutation can be identified by the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay, but not by the Allplex™ 2019-nCoV test (Fig. 2).
Fig. 2.
The Allplex™ SARS-CoV-2/FluA/FluB/RSV assay discriminates SARS-CoV-2 N gene 3L from D3 plasmids, while the Allplex™ 2019-nCoV Assay does not. (A) Standard curves of pCRII-TOPO_N-gene_3L (blue line and 99% prediction band) and _D3 (grey line and 99% prediction band) plasmid titrations using the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay (left panel) and the Allplex™ 2019-nCoV assay (right panel), respectively. (B) Pearson correlation (r) and comparative linear regression of Ct values of pCRII-TOPO_N-gene_3L and _D3 assessed using the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay (blue line and 99% prediction band) and the Allplex™ 2019-nCoV assay (grey line and 99% prediction band). No significant difference in slope is determined (Ct3L/CtD3 p 0.212), y-intercepts (elevations) differ significantly (∗∗∗∗p ≤ 0.0001).
Discussion
N gene dropout or Ct value shift as indicator for B.1.1.7 positive samples
We were able to demonstrate that all confirmed B.1.1.7 samples have an N gene dropout or Ct value shift compared with the S or RdRp gene for SARS-CoV-2 by the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay. These N gene dropouts or Ct value shifts were caused by a D3L mutation in the N gene of SARS-CoV-2 which was confirmed by reverse genetics. In contrast, strains of other lineages were not affected by N gene dropouts or Ct value shifts. Mutations in the N gene are known to occur less often than in other regions of the SARS-CoV-2 genome [[13], [14], [15], [16], [17]]. Nevertheless, mutations of N gene D3 also occur in other lineages, where D3 is replaced by an amino acid other than leucine. For example, D3Y is present in the variant of interest B.1.525, which currently represents less than 1% of the SARS-CoV-2 population [18,19]. While S gene dropouts have been reported for the TaqPath COVID-19 assay (ThermoFisher, USA), to the best of our knowledge, this is the first report of N gene dropout or Ct value shift with diagnostic relevance as a marker for VOC differentiation (Allplex™ SARS-CoV-2/FluA/FluB/RSV assay). However, the N gene dropout has a higher specificity for the B.1.1.7 lineage, as it does not occur in other VOCs and most other lineages.
Allplex™ SARS-CoV-2/FluA/FluB/RSV assay can be used as fast detection tool for presumptive identification of the B.1.1.7 lineage
Detection of SARS-CoV-2 variants has become an additional task for many laboratories, which can be a time critical challenge. As whole-genome sequencing is the reference standard to detect, track and monitor virus variants (surveillance), this method is neither accessible everywhere nor easy to perform. Especially in clinical contexts, fast identification of VOC lineages is crucial for patient management in both inpatient and outpatient settings (infection control and prevention measures including contact tracing). While by now most laboratories have implemented workflows to deliver SARS-CoV-2 qRT-PCR results in a timely manner, variant-specific discrimination of positive samples requires additional resources and thus delays reporting of VOCs. This counteracts public health strategies aiming at slowing down the fast spread of VOCs which due to their increased transmissibility, tends to occur at higher rates.
This study has some limitations. Although N gene shift or dropout was detectable in all 48 B.1.1.7-positive specimens, sample size was rather small. Second, the assay is probably not 100% specific for B.1.1.7 because mutations of the N gene can also occur in other variants like B.1.525. In addition to the detection of four viral respiratory pathogens (SARS-CoV-2, Influenza A and B plus RSV), the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay allows for presumptive assignment of SARS-CoV-2 positive samples to the B.1.1.7 lineage in a one-step approach by using the proposed ΔCt N/RdRp or N/S score. However, confirmation of this presumptive result is warranted as the Allplex™ SARS-CoV-2/FluA/FluB/RSV assay is not intended for detection of VOCs by the manufacturer.
In conclusion, this approach can be used as a rapid tool for B.1.1.7 indication and surveillance in high throughput diagnostics.
Transparency declaration
J.S. received lecture honoraria from Pfizer and Gilead, unrelated to the current study. All other authors declare no conflict of interest. Part of the work was funded by the Bavarian State Ministry of Health and Care, Germany (Verbundprojekt “BAY-VOC”). The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board, Paracelsus Medical University, Nuremberg, Germany (reference number IRB-2021-008; 22 March 2021).
Author contributions
Designed experiments: W.P., G.N., T.D., P.S., S.E., S.J. Performed experiments: W.P., G.N., T.D. Provided important samples and data: W.P., G.N., T.D., P.S., B.T., S.A., D.A., A.N., K.K., E.A., S.E., B.M., S.J. Wrote the manuscript: W.P., G.N., T.D., B.M., S.J.
Acknowledgement
We would like to thank all members of the Institute of Clinical Hygiene, Medical Microbiology and Infectiology, Paracelsus Medical University for support and fruitful discussion.
Editor: F. Allerberger
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.05.025.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. Preprints. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 2.Faria N.R., Claro I.M., Candido D., Franco L.A.M., Andrade P.S., Thais M., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. VirologicalOrg. 2021:1–9. [Google Scholar]
- 3.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A. 2020. Preliminary genomic characterisation of an emergent SARS-CoV- 2 lineage in the UK defined by a novel set of spike mutations.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- 4.Davies N.G., Jarvis C.I., Group C.C.-W., Edmunds W.J., Jewell N.P., Diaz-ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;3055:1–16. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus ( 2019-nCoV ) by. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., et al. Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021;20 doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kidd M., Richter A., Best A., Mirza J., Percival B., Mayhew M., et al. S-variant SARS-CoV-2 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-QPCR. MedRxiv. 2020 doi: 10.1101/2020.12.24.20248834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washington N.L., White S., Barrett K.M.S., Cirulli E.T., Bolze A., Lu J.T.S. Gene dropout patterns in SARS-CoV-2 tests suggest spread of the H69del/V70del mutation in the US. MedRxiv. 2020 doi: 10.1101/2020.12.24.20248814. [DOI] [Google Scholar]
- 10.Ziegler K., Steininger P., Ziegler R., Steinmann J., Korn K., Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Eurosurveillance. 2020;25:1–4. doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasan M.R., Sundararaju S., Manickam C., Mirza F., Al-Hail H., Lorenz S., et al. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by RT-qPCR. J Clin Microbiol. 2021 doi: 10.1128/jcm.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., Boreux R., et al. A recurrent mutation at position 26,340 of SARS-CoV-2 is associated with failure of the E-gene qRT-PCR utilised in a commercial dual-target diagnostic assay. MedRxiv. 2020:1–15. doi: 10.1101/2020.04.28.20083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1405. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 14.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 15.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27 doi: 10.1016/j.chom.2020.03.002. 671–80.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y., Liu M., Zhao W., Zhang J., Zhang X., Wang K., et al. Isolation of virus from a SARS patient and genome-wide analysis of genetic mutations related to pathogenesis and epidemiology from 47 SARS-CoV isolates. Virus Genes. 2005;30:93–102. doi: 10.1007/s11262-004-4586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ascoli C.A. Could mutations of SARS-CoV-2 suppress diagnostic detection? Nat Biotechnol. 2021;422555:14–15. doi: 10.1038/s41587-021-00845-3. [DOI] [PubMed] [Google Scholar]
- 18.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Toole A.H. PANGO lineages; 2021. International lineage report.https://cov-lineages.org/lineages.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.