Abstract
Purpose:
Biomarkers for disease specific survival (DSS) in early stage melanoma are needed to select patients for adjuvant immunotherapy and accelerate clinical trial design. We present a pathology-based computational method using a deep neural network architecture for DSS prediction.
Experimental design:
The model was trained on 108 patients from four institutions and tested on 104 patients from Yale School of Medicine (YSM). A receiver operating characteristic (ROC) curve was generated based on vote aggregation of individual image sequences, an optimized cutoff was selected, and the computational model was tested on a third independent population of 51 patients from Geisinger Health Systems (GHS).
Results:
Area under the curve (AUC) in the YSM patients was 0.905 (p<0.0001). AUC in the GHS patients was 0.880 (p<0.0001). Using the cutoff selected in the YSM cohort, the computational model predicted DSS in the GHS cohort based on Kaplan-Meier (KM) analysis (p<0.0001).
Conclusions:
The novel method presented is applicable to digital images, obviating the need for sample shipment and manipulation and representing a practical advance over current genetic and IHC-based methods.
Keywords: Tumor microenvironment, Biomarkers/ Imaging biomarkers, Biomarkers/ Prognostic biomarkers, Computational methods
Introduction
There is an urgent need to define prognostic biomarkers in early stage melanoma. This is because, while effective adjuvant therapies to prevent recurrence and death are available, they incur significant toxicity and are very costly.1 Toxicity is tolerable in the advanced disease setting, but it is much less acceptable for otherwise healthy patients who have high probability of living a normal lifespan with good functional status if left untreated. Moreover, treatment lasts one year and costs run over $20,000 per patient per month.2 Given that death rates from melanoma at ten years range from 2%−8% for stage I disease, 12%−25% for stage II disease, and 12%−40% for stage III disease, treating all early stage melanoma patients would result in significant over-treatment and resource expenditure.3,4
The current clinical criterion for evaluating risk of recurrence is the American Joint Committee on Cancer (AJCC) staging system.3,4 The AJCC staging system includes multiple parameters including depth of the primary tumor, ulceration, mitotic rate, and local or nodal metastasis. This system is highly useful but has several limitations. First, it does not account for the relative risk conferred by tumor depth and lymph node spread in that a deeper primary is deadlier than a small nodal metastasis, such that a stage IIIA patient has a better survival rate than does a stage IIC patient. Second, depth can be difficult to estimate accurately in some patients depending on technique, for example if a shave biopsy is performed or the original lesion is incompletely excised.4 Third, complete staging requires examination of lymph nodes, a procedure that is invasive and confers no survival benefit.5 In order to avoid surgery, patients are therefore in some situations incompletely staged. More precise and broadly applicable staging systems are needed to supplement AJCC staging.4
Traditionally, characterization of genomic and proteomic features of primary melanoma tumors has been challenging because the very small size of these tumors necessitates that the entire specimen be formalin fixed and paraffin embedded (FFPE) in almost all circumstances to allow for review by an expert pathologist. Fortunately, newer technologies including the NanoString assay and specialized RNA sequencing methods coupled with quantitative multiplexed immune-fluorescence (QIF) assays have allowed for quantification of RNA transcripts and phenotyping of immune cells within the tumor micro-environment. In melanoma, our group and others have developed and validated genomic signatures, and, most recently, a QIF-based biomarker consisting of the ratio of CD8+ T cells to CD68+ macrophages in tumor stroma.6–10 While these methods show promise, application can be challenging due to complex analysis methods not typically in use in clinical laboratories.
Meanwhile, the application of artificial intelligence (AI) to health care promises to substantially alter how medical care is delivered in the coming decades. While initial applications were primarily outside of medicine, for example in the well-known automated identification of images of cats, machine learning has been successfully applied in multiple health care contexts including interpretation of imaging data for segmentation of anatomical features from MRI data and diagnosis of skin lesions.11 Most recently, machine learning has been applied to pathology imaging, notably to the identification of lymph node metastasis in breast cancer.12 Developing prognostic biomarkers represents a unique challenge because pathologists generally diagnose rather than prognosticate, as prognostication generally includes multiple clinical parameters and is most frequently performed as a collaborative effort between pathologists and clinicians who have interactions with patients in an office setting.
Deep learning, a subset of machine learning, allows the computer to select ways of identifying patterns correlating with a defined outcome. Convolutional neural networks (CNN) are a specific type of deep learning well suited to image analysis tasks that require prediction based on smaller image patches.13,14 Deep learning techniques and CNN in particular have been applied to more complex problems in pathology such as identification of tumor infiltrating lymphocytes (TILs) and, more broadly, characterization of the tumor immune microenvironment.15–18 Further, deep learning promises to offer rapid and efficient methods to identify tumor subsets, correctly “grade” tumors based on cellular atypia, and “predict” gene mutations.19–22
We propose a deep learning method to predict visceral recurrence and DSS in patients with primary melanoma. This method was developed on an image base from 108 patients and applied to two independent validation sets of 104 and 51 patients respectively, yielding AUC values of 0.905 and 0.880. A cutoff selected based on the first validation set was tested in the second validation set and predicted DSS based on Kaplan Meier analysis (p<0.0001). This method is novel and rapidly applicable to standard clinical workflows and could be tested in the prospective setting for application to patient care.
Materials and Methods
Patients, clinical information, and imaging.
This study was approved by Columbia University Irving Medical Center’s (CUIMC) Institutional Review Board (IRB). This study was determined by CUIMC’s IRB to not require written consent from subjects, as it is retrospective and involves minimal risk. This study was conducted in accordance with the ethical guidelines outlined by the Declaration of Helsinki. The training cohort was selected based on availability of H&E slides and clinical information. Patients from databases previously generated for the development and validation of melanoma immune profile (MIP) with at least one available H&E slide and 24 months of clinical follow up for patients who did not die of melanoma during follow up were included. Full patient demographics of the training cohort are provided in Table 1.8,9 Two validation cohorts were tested, the first consisting of 104 patients from Yale School of Medicine (YSM) described in Table 2, and the second including 51 patients from Geisinger Health Systems (GHS) described in Table 3. All slides were reviewed by a pathologist to confirm melanoma content. Slides were scanned using a Leica SCN 400 system with high throughput 384 slide autoloader (SL801) and tiff format files were generated. A separate image was generated for separate pieces of tissue on each slide as is frequently the case for primary melanomas due to tissue sectioning methods. Images were reviewed for quality and excluded due to excessive melanin obscuring cellular features or poor tissue quality (supplementary table S1).
Table 1:
Patient characteristics of the Training cohort.
| (n = 108) | |
|---|---|
| Clinical characteristics | |
| Sex, n (%) | |
| Male | 73 (67.6) |
| Female | 34 (31.5) |
| Unknown | 1 (0.9) |
| Age | |
| Known, n (%) | 103 (95.4) |
| Median, n (range) | 67 (22–96) |
| Unknown, n (%) | 5 (4.6) |
| Location of tumor, n (%) | |
| Trunk | 58 (53.7) |
| Extremity | 48 (44.4) |
| Unknown | 2 (1.9) |
| T-stage, n (%) | |
| T1a or T1b | 18 (16.7) |
| T2a | 11 (10.2) |
| T2b or T3a | 41 (38.0) |
| T3b or T4a | 22 (20.4) |
| T4b | 12 (11.1) |
| Unknown | 4 (3.7) |
| Stage, n (%) | |
| I | 22 (20.4) |
| II | 62 (57.4) |
| III | 24 (22.2) |
| Pathologic characteristics | |
| Depth (mm) | |
| Median, n (range) | 2.30 (0.30–30) |
| Ulceration, n (%) | |
| Absent | 57 (52.8) |
| Present | 47 (43.5) |
| Unknown | 4 (3.7) |
| Microsatellite lesions, n (%) | |
| Absent | 101 (93.5) |
| Present | 6 (5.6) |
| Unknown | 1 (0.9) |
| TILs | |
| Absent | 9 (8.3) |
| Non-brisk | 67 (62.0) |
| Brisk | 23 (21.3) |
| Unknown | 9 (8.3) |
| SLNB status, n (%) | |
| Completed | 66 (61.1) |
| Positive, n (% of completed) | 20 (18.5) |
| Negative, n (% of completed) | 46 (42.6) |
| Not completed | 15 (13.9) |
| SLNB status unknown | 27 (25) |
| Outcome characteristics | |
| Patient follow-up (months) | |
| Median, n (range) | 58 (7–173) |
| DMR, n (%) | |
| Distant recurrence | 34 (31.5) |
| No recurrence or local recurrence only | 74 (68.5) |
| OS, n (%) | |
| Alive (at least 2 years) | 69 (63.9) |
| Dead | 39 (36.1) |
| DSS, n (%) | |
| Alive or NED at death | 78 (72.2) |
| Median follow-up (months) | 65 |
| Dead with melanoma | 30 (27.8) |
| Median follow-up (months) | 34.5 |
| Unknown | 0 (0) |
Abbreviations: DMR, distant metastatic recurrence; DSS, disease-specific survival; NED, no evidence of disease; OS, overall survival
Table 2:
Patients characteristics of the YSM cohort.
| (n = 104) | |
|---|---|
| Clinical characteristics | |
| Sex, n (%) | |
| Male | 51 (49.0) |
| Female | 53 (51.0) |
| Age | |
| Median, n (range) | 61 (25–86) |
| Location of tumor, n (%) | |
| Trunk | N/A |
| Extremity | N/A |
| T-stage, n (%) | |
| T1a or T1b | 23 (22.1) |
| T2a | 13 (12.5) |
| T2b or T3a | 30 (28.9) |
| T3b or T4a | 22 (21.2) |
| T4b | 14 (13.5) |
| Unknown | 2 (1.9) |
| Stage, n(%) | |
| I | N/A |
| II | N/A |
| III | N/A |
| Pathologic characteristics | |
| Depth (mm) | |
| Median, n (range) | 2.35 (0.15–8.30) |
| Ulceration, n (%) | |
| Absent | 63 (60.6) |
| Present | 39 (37.5) |
| Unknown | 2 (1.9) |
| Microsatellite lesions, n (%) | |
| Absent | 76 (73.1) |
| Present | 26 (25.0) |
| Unknown | 2 (1.9) |
| TILs | |
| Absent | 6 (5.8) |
| Non-brisk | 77 (74.0) |
| Brisk | 19 (18.3) |
| Unknown | 2 (1.9) |
| SLNB status, n (%) | |
| Completed | N/A |
| Positive, n (% of completed) | N/A |
| Negative, n (% of completed) | N/A |
| Not completed | N/A |
| Outcome characteristics | |
| Patient follow-up (months) | |
| Median, n (range) | 68.7 (1.4–456.2) |
| DMR, n (%) | |
| Distant recurrence | 46 (44.2) |
| No distant recurrence or local recurrence only | 58 (55.8) |
| OS, n (%) | |
| Alive (at least 2 years) | 26 (25.0) |
| Dead | 78 (75.0) |
| DSS, n (%) | |
| Alive or NED at death | 58 (55.8) |
| Median follow-up (months) | 114.4 |
| Dead with melanoma | 46 (44.2) |
| Median follow-up (months) | 36.7 |
| Unknown | 0 (0) |
Abbreviations: DMR, distant metastatic recurrence; DSS, disease-specific survival; NED, no evidence of disease; OS, overall survival
Table 3:
Patient characteristics of the GHS cohort.
| (n = 51) | |
|---|---|
| Clinical characteristics | |
| Sex, n (%) | |
| Male | 27 (52.9) |
| Female | 24 (47.1) |
| Age | |
| Median, n (range) | 67 (20–90) |
| Location of tumor, n (%) | |
| Trunk | 31 (60.8) |
| Extremity | 20 (39.2) |
| T-stage, n (%) | |
| T1a or T1b | 1 (2.0) |
| T2a | 0 (0) |
| T2b or T3a | 19 (37.3) |
| T3b or T4a | 20 (39.2) |
| T4b | 11 (21.6) |
| Unknown | 0 (0) |
| Stage, n (%) | |
| I | 0 (0) |
| II | 25 (49.0) |
| III | 26 (51.0) |
| Pathologic characteristics | |
| Depth (mm) | |
| Median, n (range) | 3.45 (0.65–13) |
| Ulceration, n (%) | |
| Absent | 23 (45.1) |
| Present | 28 (54.9) |
| Unknown | 0 (0) |
| Microsatellite lesions, n (%) | |
| Absent | 43 (84.3) |
| Present | 7 (13.7) |
| Unknown | 1 (2.0) |
| TILs | |
| Absent | 13 (25.5) |
| Non-brisk | 32 (62.7) |
| Brisk | 5 (9.8) |
| Unknown | 1 (2.0) |
| SLNB status, n (%) | |
| Completed | 47 (92.2) |
| Positive, n (% of completed) | 19 (40.4) |
| Negative, n (% of completed) | 28 (59.6) |
| Not completed | 4 (7.8) |
| Outcome characteristics | |
| Patient follow-up (months) | |
| Median, n (range) | 56 (9–142) |
| DMR, n (%) | |
| Distant recurrence | 29 (56.9) |
| No recurrence or local recurrence only | 22 (43.1) |
| OS, n (%) | |
| Alive (at least 2 years) | 21 (41.2) |
| Dead | 30 (58.8) |
| DSS, n (%) | |
| Alive or NED at death | 25 (49.0) |
| Median follow-up (months) | 93 |
| Dead with melanoma | 19 (37.3) |
| Median follow-up (months) | 28 |
| Unknown | 7 (13.7) |
Abbreviations: DMR, distant metastatic recurrence; DSS, disease-specific survival; NED, no evidence of disease; OS, overall survival
Binary classifier selection.
To generate a binary classifier for training, patients in the training set were characterized based on whether they developed distant metastatic recurrence (DMR). The DMR endpoint was selected because death rates from melanoma have decreased over the past decade due to fundamental advances in immunotherapy such that, fortunately, patients diagnosed today are more likely to survive.23 Thus, over time DMR is a more consistent reflection of biology than is survival. Effective adjuvant therapy, however was not introduced into general practice until 2017 with the FDA approval of nivolumab for resected stage III melanoma.24 Therefore, the time to DMR has remained consistent until very recently. DMR is defined as recurrence beyond the local lymph node basin. Local recurrence, in contrast, comprises growth of residual disease at the resection margin and/or local metastatic recurrence within the anatomic region drained by the local lymph node basin or within the local lymph nodes.25 Patients with isolated local recurrence are at significantly lower risk of dying of melanoma and remain in the stage III category26–28 Patients who only developed local recurrence over the course of follow up were characterized into the favorable group provided they had 24 months of recurrence-free clinical follow up after the local recurrence. Thus, the label of DMR was designed to distinguish patients with aggressive melanoma from those at low risk of death from disease.
Identification of Regions of Interest.
In order to isolate tumor and immune regions for RNN sequence generation, we used QuPath digital pathology software22 to build modules for nuclear segmentation and cell classification. Nuclear segmentation was performed using Watershed cell detection based on segmentation parameters derived from images randomly selected from 9 subjects. Using the cell segmentation, we trained a random forest classifier to differentiate the nuclei into three classes (immune cells, tumor cells, and other, which included non-lymphocyte stromal tissue, areas obscured by melanin, or non-cell objects) based on 33 morphological features (supplementary table S2). The slide was divided into tiles, and thresholds were applied to each tile to determine the presence of relevant cell types. Tile size was empirically fixed to the width of 5 patches. Tiles with more than 65% of raw image pixels as white space background (pixel intensity value above 217) or 80% of segmented objects within the tile area classified as “other” were immediately discarded. Then, points on the slide were randomly sampled from a 2D symmetric Gaussian distribution centered on the tile with a standard deviation equal to 3 times the patch width. A 500×500 patch centered on the randomly sampled point was analyzed, applying thresholds for maximum portion of white space background, minimum number of segmented tumor or immune cell nuclei, and maximum portion of segmented objects classified as “other.” If the patch area passed the empirically determined thresholds, the downsampled area was added to the image sequence. Otherwise, the patch was discarded and a new point was randomly sampled. A maximum of six sequences (of length 20 each) were generated from each tile and if a sequence could not be generated after sampling 10,000 points, then the tile was discarded.
Feature design.
Morphology features measure the ratio of nuclear size in tumor and immune cells within the tile area, and the clustering features measure the ratio of cell density and cluster size based on Delaunay triangulation. The optimal parameters for the feature generation were selected using grid search of Delaunay pixel radius and minimum cluster size (Supplementary figure S1). The optimal features were then computed locally for every sequence based on information computed from all valid tiles immediately adjacent to the tile of the sequence being generated.
Analysis pipeline.
We designed a deep neural network (DNN) architecture consisting of a convolutional neural network (CNN) and a recurrent neural network (RNN). To avoid overfitting, we used the dropout procedure, which randomly sets a specified percentage of input units in every layer to zero and has been shown to outperform other regularization methods.29 In all our experiments, we empirically set the dropout rate to 0.7 and learning rate of 0.005. The CNN input consisted of a 500×500×3 pixel patch from the raw H&E image at 40x magnification, downsampled to 100×100×3 pixels. The CNN output for each patch served as the RNN cell input. We fixed the sequence length to 20 image patches, and every sequence was normalized before input by subtracting the mean pixel intensity values and dividing by the standard deviation. The output of the RNN was appended with the features and processed through a fully connected layer to generate the final result.
Vote aggregation.
We aggregated the classification output from individual sequences across all images for a patient. Every sequence equally contributed to the final decision. The final decision for each patient’s recurrence was made by computing the class (favorable vs. unfavorable) to which the majority of the sequences voted such that the cutoff was 0.5.
Statistics.
Statistical analysis was completed using XLSTAT Version 2019.1.3 on Excel Version 15.0.5127 and GraphPad Prism Version 8.0.1. Statistical significance was defined as P≤0.05. Receiver Operating Characteristic (ROC) curve analyses and standard univariable and multivariable Cox proportional hazards models were generated using the “Survival Analysis” feature on XLSTAT. Kaplan Meier (KM) curves were generated on GraphPad Prism and P values were calculated using Log-rank (Mantel-Cox) test.
Results
Training Population.
Patients from previously generated databases used for prior work in developing Melanoma Immune Profile (MIP) were included in the training set.8–10 In addition, patients with stage I disease for whom clinical data was available were included to broaden applicability. Images from archival slides with available clinical information for 119 patients were screened. 10 patients were excluded because of excessive melanin obscuring cellular features and 1 patient was excluded because the tissue sample was torn. Demographics for the training population are shown in Table 1. All living patients had at least 24 months of follow up. As shown, patients were 22.2% stage III, 57.4% stage II and 20.4% stage I. 80 patients were from Columbia University Irving Medical Center (CUIMC), 14 patients from New York University Medical Center (NYUMC), 6 patients from Geisinger Health Systems (GHS), and 8 patients from the Icahn School of Medicine at Mount Sinai (ISMMS). Patients were 67.6% male and 31.5% female, with one patient of unknown sex. Median age was 67 years. Median follow up was 58 months. Univariable cox analysis shows that depth, ulceration, and stage correlated significantly with DSS showing that the training set was generally representative of melanoma populations in the United States (supplementary table S3).
Method development and training.
Following selection of the labeled dataset for binary classification, we developed an automated data processing pipeline (Figure 1) that takes H&E stained images from a melanoma sample and outputs a DMR status prediction. The pipeline is designed to handle one or more images of varying sizes per patient, which can result from multiple tumor locations, multiple cuts of the same tumor, or multiple 2-D slices. The first issue to resolve is that histopathology images have varying and large sizes (1–15 Gigabytes), and are therefore not suitable for directly processing through a neural network. While standard methods for processing histopathological images with neural networks21,30 involve dividing the image into smaller (ie 512 × 512) patches, this is not a viable strategy for our problem because recurrence risk is mediated by interactions between tumor cells and adjacent host tissues, which in some cases are not present in many quadrants of each slide or in every image patch. Thus, recurrence risk prediction requires incorporation of regional information from the image. Additionally, melanoma histopathology images often contain areas of connective tissue without tumor nuclear information. Tumor information is necessary for an accurate recurrence prediction, and thus image regions lacking cell information must be omitted to reduce noise in the final output.
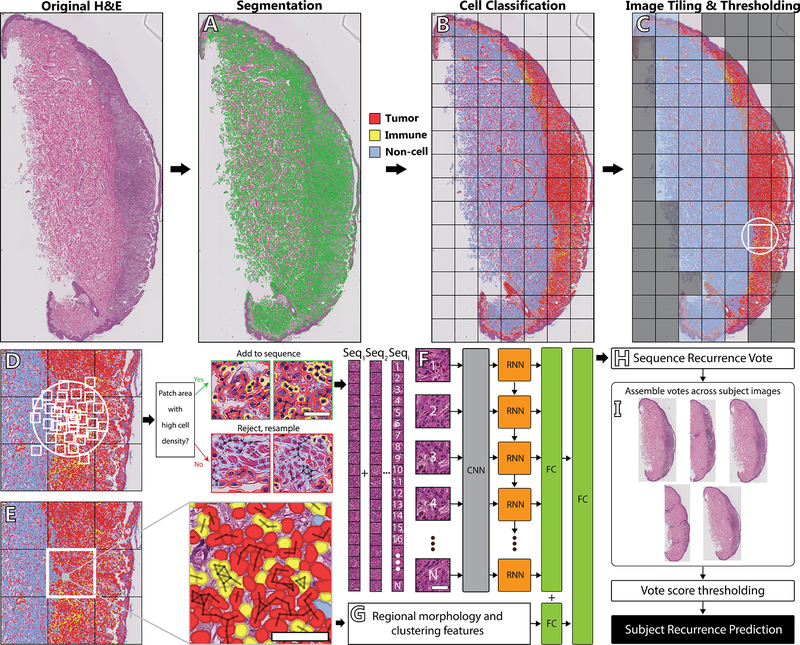
Figure 1: A detailed view of our approach.
(A) A raw H&E scan is first segmented using Watershed Cell Detection. (B) Segmented objects are classified into one of three classes, Tumor, immune, and non-cell object. (C) The total image is split into tiles. Tiles with excess whitespace or non-cell objects are discarded. (D) Of the remaining tiles, points are randomly sampled from a 2D Gaussian distribution centered on the tile. A patch is drawn with the sampled point as the centroid, and the patch area is thresholded for the presence of white space and tumor or immune objects. Patches with high cell density are added to a DNN sequence. (E) All segmented objects within one cell width of another object in the same class (40px at 40x magnification) are clustered. Cell counts, cluster proportions, and nuclear area are calculated within the tile and all adjacent tiles remaining after the initial thresholding. (F) Each patch is processed through a 5-layer CNN followed by RNN with sequence length N (N=20). (G) The features for the tile corresponding to the generated sequence are concatenated with the RNN output and run through a final fully connected layer. (H) The MRP outputs a binary vote for the sequence of “Recurrent” or “Nonrecurrent.” (I) Votes are aggregated equally across all available images for a given subject, and generates the subject’s recurrence prediction based on a majority vote or predefined threshold.
To address the problem of tissue heterogeneity with respect to relevant tumor content, we developed a sampling strategy that is sensitive to cell types. Starting with a raw H&E image, we identified cell objects within the image (Fig. 1B), divided the image into a regular grid, selected grid tiles with minimum tumor and/or lymphocyte density (Fig. 1C), and then randomly sampled spatially localized, fixed-length sequences of patches from each grid tile (Fig 1D). In addition, we augmented regional image information with cell density features that were designed both to characterize the atypia of tumor cells and to summarize a larger immune infiltration context around each tile (Fig. 1E). Sequences consisting of raw image data were then processed by our deep neural network (DNN) (Fig. 1F), first by the CNN, which extracted high-dimensional features from the individual patches, and then by the RNN, which processed the CNN output to identify discriminative spatial patterns. Finally, two fully connected layers combined the output of RNN with the pre-computed regional features (Fig. 1G), resulting in a softmax recurrence probability vote for every sequence (Fig. 1H). To generate the DMR probability for each patient in the test set, votes were aggregated across all available subject images (Fig. 1I), and the percentage of positively classified sequences were counted to generate the final prediction score.
First Test Population.
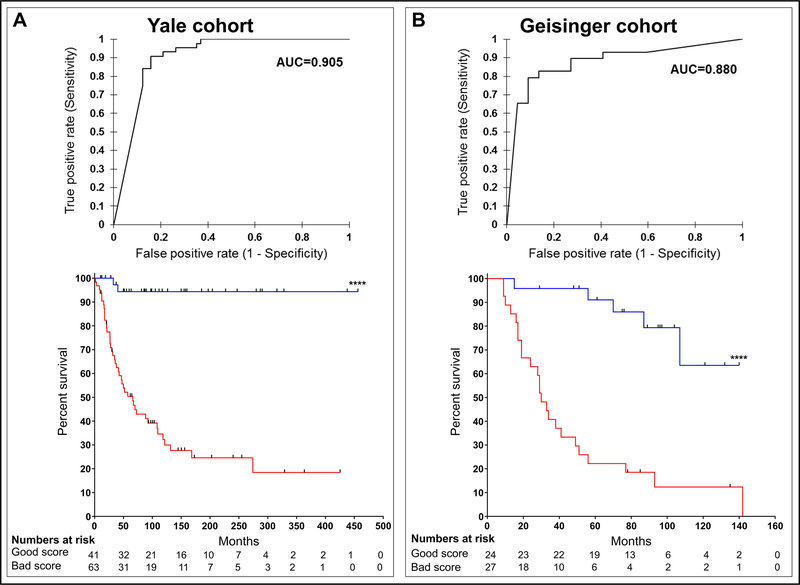
The first test population consisted of 118 samples from Yale School of Medicine. On pre-review, 7 were excluded because of heavy melanin and 7 were excluded because slides were cracked, images were blurred, or tissue was folded. Demographics are shown in Table 2 and Cox survival analysis using standard predictors is shown in Supplementary Table S3. Patients were 49% male and 51% female with a median age of 61 years. Median follow up was 68.7 months. One slide was included for each patient and image sequences were generated followed by a prediction score as described above. A receiver operating characteristic (ROC) analysis was constructed and showed that the predictor strongly correlates with DMR (AUC=0.905). Disease specific survival is a key endpoint for adjuvant clinical trials and is the standard for prognostic biomarkers. We selected a cutoff to maximize sensitivity for recurrence with the goal of identifying a population that could be excluded from clinical trials, thereby increasing efficiency of accrual of patients at risk for death from melanoma, maximizing significance, and minimizing exposure of patients who do not need treatment. When this single cutoff was applied using KM analysis, the DNN classifier correlated significantly with DSS (p<0.0001, Figure 2A). When a multivariable analysis was performed, the DNN predictor correlated with DSS when other clinical predictors were included as co-variables (P < 0.0001, supplementary table S4).
Figure 2: ROC and KM curves for the validation cohort.
(A) ROC curve analysis for YSM cohort (n=104, AUC=0.905, P < 0.0001) and KM curve for recurrence (P < 0.0001) created using AUC cutoff from ROC curve shown at top. (B) ROC curve analysis for GHS cohort (n=51, AUC=0.880, P < 0.0001) and KM curve for recurrence (P < 0.0001) created as in (A).
Second Test population.
The second test population consisted of 56 patients from GHS. On pre-review, 4 patients were excluded because of excessive melanin and 1 patient was excluded due to a lack of tumor in the image. Demographics are shown in Table 3 and univariable Cox survival analysis using standard predictors is shown in supplementary table S3. When the DNN predictor was evaluated in this patient set, the AUC value was 0.880. Using the same cutoff as for the first population, the classifier significantly correlated with DSS using KM analysis (p<0.0001, Figure 2B). 24 patients had a favorable prediction score, of whom 5 had DMR and 27 patients had an unfavorable prediction of whom 24 had DMR. When a multivariable analysis was performed, the DNN predictor correlated with DSS when other clinical predictors were included as co-variables (P < 0.001, table 4). Finally, in order to assess the contribution of regional features to the overall accuracy of the classifier to the second test set, we reran the algorithm excluding each one of the features (supplementary table S5). We found that ratio of lymphocyte area over tumor cell area appeared most important since removing it decreased accuracy to AUC 0.509 (supplemental table S5). In order to exclude the possibility that this feature alone would be sufficient to predict death from melanoma we extracted this feature independently from QuPath. Again, AUC was not predictive (AUC=0.589, data not shown). This shows that, while immune cell content is critical to accuracy it is not sufficient outside of the deep learning algorithm to predict DMR or death from melanoma. This also shows that it is not sufficient to use the histopathological features globally and that our method of localizing the features by computing them within the tiles surrounding the patch and combining them with the DNN sequence leads to discriminatory performance.
Table 4: Cox regression analysis of GHS cohort.
Multivariable Cox regression analysis of disease-specific survival.
| Hazard ratio | 95% CI | P | |
|---|---|---|---|
| Score** | 58.7 | 4.83 to 713 | 0.001 |
| Stage | 1.04 | 0.337 to 3.23 | 0.942 |
| Gender | 2.01 | 0.541 to 7.45 | 0.297 |
| Age | 1.04 | 0.994 to 1.08 | 0.096 |
| TILs | 1.03 | 0.337 to 3.12 | 0.965 |
| Location | 0.607 | 0.178 to 2.07 | 0.425 |
| Depth | 0.966 | 0.729 to 1.28 | 0.812 |
| Ulceration | 1.49 | 0.469 to 4.73 | 0.499 |
Discussion
Here we present a biomarker that stratifies patients with early stage melanoma using only information derived from computational analysis of H&E images using a DNN. This biomarker is easily applicable in a clinical context as it requires no additional tissue processing, such as RNA extraction or immunohistochemical staining. The biomarker was generated based on image analysis of a training set with DMR as the label distinguishing favorable from unfavorable outcomes. This label was selected based on the hypothesis that a subset of melanoma patients who develop local recurrence but not DMR have a more indolent biology, perhaps related to immune surveillance, that is associated with prolonged survival.31 The deep learning-based biomarker was then found to correlate with DSS in two independent validation populations. We believe that, after prospective validation, this tool could be used as a screening tool with value for adjuvant studies and, potentially, included in AJCC staging criteria.
The method applied in this paper is based on a newly designed algorithm and includes adaptations to allow for exclusion of areas with less relevant information, namely both the labeling of irrelevant areas such as those containing high levels of pigment as “other,” and the requirement for a minimal number of tumor and/or lymphocytes in each patch. In addition, the DNN method presented here includes features such as nuclear size and distribution of immune cells within the tumor that have a high probability of being predictive based on previous pathology literature.20,32 One advantage of this method is that it is robust to variable H&E stains from different institutions, demonstrating broad applicability and robustness of the algorithm. Yet another advantage of this method is that it does not depend of pre-identification of tumor location or tumor type in the sample image for predicting DMR and can therefore directly work with image samples of varying tumor types and sizes.
One key aspect of our biomarker is that the DNN incorporates raw imaging features from the RBG matrix as well as pathology features known or hypothesized to impact melanoma prognosis based on the pathology literature, such as density and distributions of lymphocytes as well as morphology of tumor nuclei. These features were calculated based on a matrix surrounding each patch as the patches themselves are too small to allow for accurate calculation of these features. Note that these features are centered on tiles containing a minimal density of cells of interest (either tumor or lymphocyte) and therefore differ from a global estimate of TILs performed visually by the pathologist. Nonetheless, loss of accuracy with removal of the feature describing lymphocyte are over total area in QuPath does diminish accuracy, showing that the classifier includes immune infiltration as an important component of the algorithm. This is consistent with prior work in the field demonstrating that density of lymphocyte infiltration confers prognostic information. 33,34 The fact that the QuPath feature on its own is not an accurate predictor demonstrates that the classifier relies on a combination of multiple parameters for survival prediction.
There are several limitations of this study. First, the two validation sets are retrospective and total under 200 patients. The use of retrospective cohorts is limiting because non-random variables may influence practice patterns and hence the availability of clinical follow up such that prospective validation would be very important to verify clinical applicability of the biomarker. Second, the method requires slides to have tissue quality sufficient to allow for identification of regions of interest containing tumor cells and lymphocytes. Most freshly biopsied samples handled per pathology standards should meet these criteria. In addition, we could not apply this method to samples with high melanin content as cellular features could not be identified. Refinement will need to be made for the small percentage of patients with highly elevated melanin content. One means whereby to achieve this would be to bleach slides from cases with high melanin content, test whether the biomarker accuracy is impacted by bleaching, and make any necessary modifications in QuPath to account for differences created by bleaching. This could be achieved using a large cohort of high melanin content cases. Finally, a high-quality scanner needs to be available to generate images and this may not be available at some centers.
The method proposed in this paper is highly promising with AUC values of 0.905 and 0.880 in two independent validation sets and should be prospectively evaluated in larger studies to develop an accurate AI-based biomarker with clinical application to facilitate stratification for clinical trials and improve the care of patients with early stage melanoma. Such a biomarker would accelerate screening for adjuvant clinical studies for early stage melanoma patients.
Supplementary Material
Supplementary table 1: Subject exclusion criteria.
Supplementary table 2: 33 morphological features used to classify cells.
Supplementary figure 1: Delaunay Grid Search. The overall AUC for each Delaunay pixel radius and small cluster size within the Training cohort for selection of configuration parameters in model training.
Supplementary table 3: Univariable Cox regression analysis in Training, YSM, and GHS cohorts. Univariable Cox regression analysis of disease-specific survival.
Supplementary table 4: Multivariable Cox analysis in YSM cohort. Multivariable Cox regression analysis of disease-specific survival.
Supplementary table 5: Features included in the classifier. List of features included in the classifier and ROC values for feature removal in GHS cohort.
Statement of Translational Relevance.
While effective adjuvant therapies to prevent recurrence and death in early stage melanoma are now available, these therapies often incur significant toxicity and are very costly. As such, avoiding the over-treatment of patients with early stage melanoma is crucial. Precise and more broadly applicable biomarkers would therefore allow clinicians to identify and treat the patients most at risk of death from melanoma, while sparing patients with the lowest risk the cost and toxicities of treatment. Here, we propose a deep learning-based prognostic biomarker to predict visceral recurrence and DSS in patients with primary melanoma. Because our method only requires digital images of hematoxylin and eosin (H&E) slides, this biomarker bypasses the need for sample shipment and is rapidly applicable to standard clinical workflows. Further evaluation of this biomarker in a larger, prospective setting would potentially allow for its application to patient care.
Acknowledgments
This publication was supported by the National Institutes of Health through Grant Numbers R01FD006108 (Y.M. Saenger), UH2CA218149 (Y.M. Saenger), and KL2TR001874 (R.D. Gartrell-Corrado). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This project also received funding from the Melanoma Research Alliance (Y.M. Saenger), Columbia University’s Irving Institute for Clinical and Translational Research (Y.M. Saenger), and Swim Across America (R.D. Gartrell-Corrado). This work was supported by funds from Navigate BioPharma (Novartis subsidiary), Yale SPORE in Lung Cancer (P50CA196530) and Yale Cancer Center (P30CA016359) to D.L. Rimm. The funding sources had no role in study design; collection, analysis, and interpretation of data; preparation of the manuscript; or the decision to submit for publication.
Conflicts of interest
Y.M. Saenger receives research funding from Amgen. D.L. Rimm has served as a consultant, advisor, or served on a Scientific Advisory Board for Amgen, Astra Zeneca, Agendia, Biocept, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, GSK, Merck, NanoString, Perkin Elmer, PAIGE, and Ultivue. He has received research funding from Astra Zeneca, Cepheid, Nanostring, Navigate/Novartis, NextCure, Lilly, Ultivue, and Perkin Elmer.
References
- 1.Palmieri DJ, Carlino MS: Immune Checkpoint Inhibitor Toxicity. Curr Oncol Rep 20:72, 2018 [DOI] [PubMed] [Google Scholar]
- 2.Gordan L, Blazer M, Saundankar V, et al. : Cost differential of immuno-oncology therapy delivered at community versus hospital clinics. Am J Manag Care 25:e66–e70, 2019 [PubMed] [Google Scholar]
- 3.Trinidad CM, Torres-Cabala CA, Curry JL, et al. : Update on eighth edition American Joint Committee on Cancer classification for cutaneous melanoma and overview of potential pitfalls in histological examination of staging parameters. J Clin Pathol 72:265–270, 2019 [DOI] [PubMed] [Google Scholar]
- 4.Taylor L, Hood K, Reisch L, et al. : Influence of variability in assessment of Breslow thickness, mitotic rate and ulceration among US pathologists interpreting invasive melanoma, for the purpose of AJCC staging. J Cutan Pathol 45:588–596, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falk Delgado A, Zommorodi S, Falk Delgado A: Sentinel Lymph Node Biopsy and Complete Lymph Node Dissection for Melanoma. Curr Oncol Rep 21:54, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris LK, Gerami P, Skelsey MK, et al. : Real-world performance and utility of a noninvasive gene expression assay to evaluate melanoma risk in pigmented lesions. Melanoma Res 28:478–482, 2018 [DOI] [PubMed] [Google Scholar]
- 7.Gerami P, Cook RW, Russell MC, et al. : Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J Am Acad Dermatol 72:780–5 e3, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Gartrell RD, Marks DK, Rizk EM, et al. : Validation of Melanoma Immune Profile (MIP), a Prognostic Immune Gene Prediction Score for Stage II-III Melanoma. Clin Cancer Res 25:2494–2502, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivendran S, Chang R, Pham L, et al. : Dissection of immune gene networks in primary melanoma tumors critical for antitumor surveillance of patients with stage II-III resectable disease. J Invest Dermatol 134:2202–2211, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gartrell RD, Marks DK, Hart TD, et al. : Quantitative Analysis of Immune Infiltrates in Primary Melanoma. Cancer Immunol Res 6:481–493, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, et al. : Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. : Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 318:2199–2210, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen D, Wu G, Suk HI: Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 19:221–248, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo SB, Lou SA, Lin JS, et al. : Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans Med Imaging 14:711–8, 1995 [DOI] [PubMed] [Google Scholar]
- 15.Linder N, Taylor JC, Colling R, et al. : Deep learning for detecting tumour-infiltrating lymphocytes in testicular germ cell tumours. J Clin Pathol 72:157–164, 2019 [DOI] [PubMed] [Google Scholar]
- 16.Saltz J, Gupta R, Hou L, et al. : Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 23:181–193 e7, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia D, Casanova R, Machiraju D, et al. : Computationally-Guided Development of a Stromal Inflammation Histologic Biomarker in Lung Squamous Cell Carcinoma. Sci Rep 8:3941, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehteshami Bejnordi B, Mullooly M, Pfeiffer RM, et al. : Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod Pathol 31:1502–1512, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvaniti E, Fricker KS, Moret M, et al. : Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep 8:12054, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casanova R, Xia D, Rulle U, et al. : Morphoproteomic Characterization of Lung Squamous Cell Carcinoma Fragmentation, a Histological Marker of Increased Tumor Invasiveness. Cancer Res 77:2585–2593, 2017 [DOI] [PubMed] [Google Scholar]
- 21.Coudray N, Ocampo PS, Sakellaropoulos T, et al. : Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24:1559–1567, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankhead P, Loughrey MB, Fernandez JA, et al. : QuPath: Open source software for digital pathology image analysis. Sci Rep 7:16878, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henriques V, Martins T, Link W, et al. : The Emerging Therapeutic Landscape of Advanced Melanoma. Curr Pharm Des 24:549–558, 2018 [DOI] [PubMed] [Google Scholar]
- 24.Weber J, Mandala M, Del Vecchio M, et al. : Adjuvant Nivolumab versus Ipilimumab in Resected Stage III or IV Melanoma. N Engl J Med 377:1824–1835, 2017 [DOI] [PubMed] [Google Scholar]
- 25.MacCormack MA, Cohen LM, Rogers GS: Local melanoma recurrence: a clarification of terminology. Dermatol Surg 30:1533–8, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez AB, Baum CL, Brewer JD, et al. : Patterns of failure following the excision of in-transit lesions in melanoma and the influence of excisional margins. J Surg Oncol 118:606–613, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beasley GM, Hu Y, Youngwirth L, et al. : Sentinel Lymph Node Biopsy for Recurrent Melanoma: A Multicenter Study. Ann Surg Oncol 24:2728–2733, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown CD, Zitelli JA: The prognosis and treatment of true local cutaneous recurrent malignant melanoma. Dermatol Surg 21:285–90, 1995 [DOI] [PubMed] [Google Scholar]
- 29.Srivastava N HG, Krizhevsky A, Sutskerver I, Ruslan S: Dropout: A Simple Way to Prevent Nerual Networks from Overfitting. Journal of Machine learning Research 2014:1929–1958, 2014 [Google Scholar]
- 30.Bychkov D, Linder N, Turkki R, et al. : Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep 8:3395, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribatti D: The concept of immune surveillance against tumors. The first theories. Oncotarget 8:7175–7180, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clemente CG, Mihm MC Jr., Bufalino R, et al. : Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 77:1303–10, 1996 [DOI] [PubMed] [Google Scholar]
- 33.Azimi F, Scolyer RA, Rumcheva P, et al. : Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 30:2678–83, 2012 [DOI] [PubMed] [Google Scholar]
- 34.Weiss SA, Hanniford D, Hernando E, et al. : Revisiting determinants of prognosis in cutaneous melanoma. Cancer 121:4108–23, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1: Subject exclusion criteria.
Supplementary table 2: 33 morphological features used to classify cells.
Supplementary figure 1: Delaunay Grid Search. The overall AUC for each Delaunay pixel radius and small cluster size within the Training cohort for selection of configuration parameters in model training.
Supplementary table 3: Univariable Cox regression analysis in Training, YSM, and GHS cohorts. Univariable Cox regression analysis of disease-specific survival.
Supplementary table 4: Multivariable Cox analysis in YSM cohort. Multivariable Cox regression analysis of disease-specific survival.
Supplementary table 5: Features included in the classifier. List of features included in the classifier and ROC values for feature removal in GHS cohort.