Abstract
Objectives
To evaluate four sample treatments in a safe and straightforward procedure to detect SARS-CoV-2 in saliva.
Methods
Four sample treatments were evaluated in a 3-step procedure to detect SARS-CoV-2 in saliva: 1) heating at 95 °C for 5 min for sample inactivation; 2) sample treatment; 3) analysis by reverse-transcription loop-mediated isothermal amplification (LAMP). Saliva samples used were from infected individuals or were spiked with known quantities of viral particles.
Results
Three treatments had a limit of detection (LOD) of 500.000 viral particles per ml of saliva and could be used to detect individuals with potential to transmit the disease. The treatment of phosphate buffer, dithiothreitol, ethylenediaminetetraacetic acid and proteinase K, with an additional 95 °C heating step, yielded a lower LOD of 95; its sensitivity ranged from 100% in patients with nasopharyngeal swab reverse-transcriptase polymerase chain reaction cycle threshold values <20 to 47.8% for values >30.
Conclusions
This report highlights the importance of an adequate sample treatment for saliva to detect SARS-CoV-2 and describes a flexible procedure that can be adapted to point-of-care. Although its sensitivity when LAMP is used is lower than reverse-transcriptase polymerase chain reaction, this procedure can contribute to COVID-19 control by detecting individuals able to transmit the disease.
Abbreviations: PCR, polymerase chain reaction; LAMP, loop-mediated isothermal amplification; PK, proteinase K; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid
Keywords: SARS-CoV-2, COVID-19, Saliva, RT-LAMP, Assay
Introduction
Control strategies for the COVID-19 worldwide pandemic rely on early detection and identification of infected individuals. Polymerase chain reaction (PCR) is the gold standard for SARS-CoV-2 identification. However, PCR techniques require complex equipment and experienced laboratory technicians to perform the assays, preventing its use as a point-of-care technique and limiting its applications in low-income areas (Chow et al., 2020).
Loop-mediated isothermal amplification (LAMP) has been developed as a novel molecular method that amplifies genetic material with high specificity using the strand-displacement activity of DNA polymerase and an unique primer design to enable amplification at a single temperature (Notomi et al., 2000). Its isothermal nature has various advantages including compatibility with simple instruments such as a heat block or water bath, and the ability to read results directly with minimum equipment (Notomi et al., 2000). In particular, LAMP with simultaneous reverse-transcription (RT-LAMP) allows a fast and straightforward detection of nucleic acids (Tanner et al., 2015).
Naso- and oropharyngeal swabs are the primary specimens for SARS-CoV-2 diagnosis. However, obtaining these swabs requires a trained healthcare worker, which has a potential risk for nosocomial transmission, and causes discomfort to the person sampled. Therefore, their use is not always practical, particularly in serial monitoring or mass screening programs. These drawbacks may be reduced by using saliva samples since these are not painful or stressful to obtain and can be self-collected by the patient, even at home. The use of saliva for the diagnosis of SARS-CoV-2 was suggested early in the pandemic, and it is now being considered a suitable alternative with various tests approved by the US Food and Drug Administration (Ceron et al., 2020).
Several procedures, with different sample processing and treatment conditions, have been developed for the diagnosis of COVID-19 in saliva without the need for an extraction step (Lalli et al., 2020, Rabe and Cepko, 2020, Vogels et al., 2020). Most procedures require a direct manipulation of saliva that has to be diluted and/or treated with chemicals before its inactivation. The requirement for direct manipulation may limit the widespread use of these methods for routine surveillance or testing outside of the clinical laboratory, due to concerns about the risk of infection during the analytical process. Methods in which the manipulation of saliva occurs after inactivation would be safer and more practical; therefore, information about their efficacy and possible use might be beneficial to efforts aimed at expanding testing.
In this report, we evaluated 4 different sample treatments in a procedure developed in our laboratory to detect SARS-CoV-2 in saliva. This procedure “SAFE-SAL” consists of 3 steps: (1) heat inactivation, (2) sample treatment involving the addition of a chemical solution, and in some cases heating, (3) virus detection, that in this report was performed by a RT-LAMP. The 4 sample treatments ranged from a simple phosphate buffer solution to a more complex treatment involving a mixture of chemicals including dithiothreitol (DTT), Proteinase K (PK) and ethylenediaminetetraacetic acid (EDTA), and additional heating.
This SAFE-SAL procedure has 3 main characteristics: (1) low risk of disease transmission because the first step is sample inactivation, (2) a simple and flexible sample processing, requiring only a heat source and a treatment solution, (3) flexibility at the detection, since different methods can be used at this step.
Materials and methods
Sample collection and spiked sample preparation
In all cases, saliva samples were self-collected by individuals in a sterile sputum container. In samples from clinical cases, saliva was frozen at −80 °C until analysis. Informed consent was obtained for all samples. The study was approved by the Ethical Committees of the University of Murcia and IMIB-Arrixaca.
For initial testing to set up the sample treatments and calculate the limit of detection (LOD), saliva confirmed to be SARS-CoV-2 negative by reverse-transcriptase polymerase chain reaction (RT-PCR) was spiked with known amounts of heat-inactivated SARS-CoV-2 virions (VR-1986HK, ATCC, Barcelona, Spain) at different concentrations: 1 × 106, 0.5 × 106, 0.2 × 106, 0.1 × 106 and 0.05 × 106 per mL of saliva.
Procedure for analysis
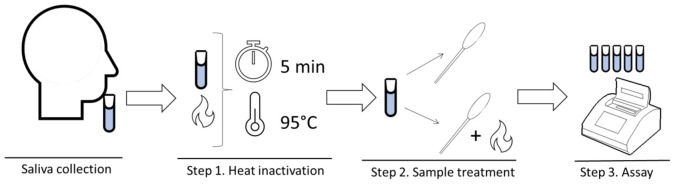
The procedure for analysis has 3 steps (Figure 1 ).
Figure 1.
Overview of the SAFE-SAL procedure.
Step 1. Heat inactivation Saliva samples were inactivated at 95 °C for 5 min in a heat block. After heating, saliva samples were kept at ambient temperature for 2 min to avoid droplet production, then kept on ice during the analytical process.
Step 2. Sample treatment Addition of the treatment solution to the sample in 1:1 volume and, in some cases, additional heating.
The treatments tested for the calculation of the LODs were (final concentration in the sample):
* 5 mM phosphate buffer at pH 7.7
* 5 mM phosphate buffer at pH 7.7, 2.5 mM DTT (Sigma–Aldrich, St. Louis, MO, USA) and 1 mM EDTA (Sigma–Aldrich). The mix is vortexed for 20 s, incubated at 37 °C for 30 min, then heated at 95 °C for 5 min.
* 5 mM phosphate buffer at pH 7.7, 2.5 mM DTT and 1 mM EDTA. The mix is vortexed for 20 s, then heated at 95 °C for 5 min.
* 5 mM phosphate buffer at pH 7.7, 2.5 mM DTT, PK (Thermo Fisher Scientific, Waltham, MA, USA) at 25 μg/mL and 1 mM EDTA. The mix is vortexed for 20 s, then heated at 95 °C for 5 min.
A final volume of 3 μL of treated sample was used for the RT-LAMP.
Step 3. Assay
An RT-LAMP assay was used in this step. The total volume of the reaction was 20 μL, including 3 μL of treated sample, 2 μL of DEPC-treated water, 2.5 μL of 10X primer mix N2 (Taki et al., 2020), and 12.5 μL of WarmStart Colorimetric LAMP 2X Master Mix (New England Biolabs, M1800 L). Table 1 describes the primer mix.
Table 1.
N2 primer.
| F3 | ACCAGGAACTAATCAGACAAG |
| B3 | GACTTGATCTTTGAAATTTGGATCT |
| FIP | TTCCGAAGAACGCTGAAGCGGAACTGATTACAAACATTGGCC |
| BIP | CGCATTGGCATGGAAGTCACAATTTGATGGCACCTGTGTA |
| LF | GGGGGCAAATTGTGCAATTTG |
| LB | CTTCGGGAACGTGGTTGACC |
Reactions were kept on ice, and the sample was added as the last component, then mixed by inversion and spin down. Mixtures were incubated at 65 °C for 30 min in a thermal cycler according to manufacturer’s instructions (Applied Biosystems VeritiTM Thermal Cycler, Applied Biosystems, Foster City, CA, USA). The mix was left for 5 min at room temperature to improve color contrast. A smartphone was used to obtain pictures of the reaction.
A negative control (buffer) and a positive control (saliva with 1 × 106 per mL viral particles added) were used in all experiments.
Limit of detection of the procedure with the different treatments at step 2
To determine the LOD of the procedure under different sample treatments (step 2), saliva spiked with different concentrations of virus particles was analyzed in 10 replicates. The dilutions were made by spiking known amounts of heat-inactivated SARS-CoV-2 virions into fresh human saliva confirmed to be SARS-CoV-2 negative by RT-PCR.
Initially, we tested 1 × 106 particles per mL, and then dilutions of this limit representing different numbers of particles: 0.5 × 106, 0.2 × 106, 0.1 × 106, and 0.05 × 106 per mL were analyzed.
Pilot study
The sample treatment which gave the best LOD (DTT, EDTA, PK and heating at 95 °C for 5 min) was applied in the pilot study of the procedure. The study aimed to evaluate if this procedure could detect individuals previously identified as positive or negative by RT-PCR from a nasopharyngeal swab (NPS) and compare the results of the SAFE-SAL using a LAMP assay and a commercially available RT-PCR in testing saliva samples.
A total of 57 positive saliva samples were collected from individuals confirmed positive for SARS-CoV-2 from NPS by an assay involving RNA extraction and quantification by RT-PCR with a commercial kit (FTD SARS-CoV-2, Siemens, Madrid, Spain). These individuals had different degrees of disease severity (ranging from severe to asymptomatic). The NPS samples had different cycle thresholds (Ct) ranging from <20 to >30. Saliva samples were obtained after confirmation of the diagnosis and within 24 h of NPS collection. Samples were stored at the Biobank of the Imib Center at Murcia Region. A total of 39 saliva samples collected in routine check-ups from individuals negative for SARS-CoV-2 by quantitative RT-PCR in NPS were used as the control group. All these samples were processed and measured by the SAFE-SAL procedure using LAMP as detection method. Results of the procedure were interpreted by the visual inspection by 2 researchers who were unaware of the NPS results. There were no discrepances between the readers.
Of these saliva samples, those with enough volume (48 positive for SARS-CoV-2, 33 negative) were also processed with the SAFE-SAL procedure with the treatment of DTT, EDTA, PK and heating at 95 °C for 5 minutes and in the third step of the procedure were analyzed with a commercially available RT-PCR assay (AllPlex 2019-noCoV assay, Seegene Inc). In a previous study, this assay had the best sensitivity for saliva specimens compared with other commercially available assays (Delaney et al., 2021).
Statistical analysis
The LOD was defined as the last sample target concentration at which all 10 replicates tested positive for the respective target. The number of viral particles per mL of saliva that could be detected by each treatment with a probability of 0.95 (LOD95) was calculated by using a probit regression model. In the pilot study, sensitivity was calculated with Excel and SPSS 24.0.
Results
Limit of detection of the different sample treatments
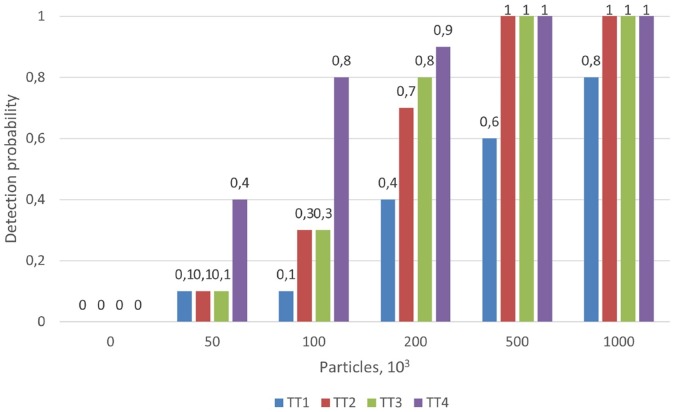
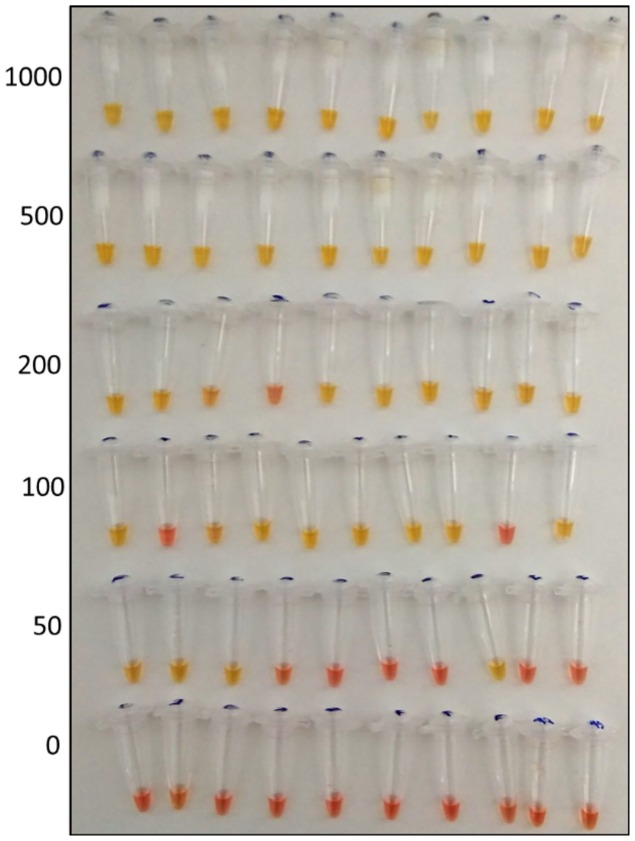
The results obtained for LOD are shown as the probability of detection for each sample treatment, for different viral particle concentrations in saliva samples, in Figure 2. The results of the treatment consisting of DTT, EDTA and PK and heating at 95 °C for 5 min appear in Figure 3 as an example outcome.
Figure 2.
Probability of detection with each sample treatment across different viral particle concentrations in saliva (from 0 to 1000 × 103 per mL). TT1: Phosphate buffer. TT2: DTT and EDTA, incubation at 37 °C for 30 min and heated at 95 °C for 5 min TT3: DTT and EDTA, heated at 95 °C for 5 min. TT4: DTT, EDTA and PK, heated at 95 °C for 5 min.
Figure 3.
Results of the assays to evaluate the limit of detection of treatment DTT + PK + EDTA (left: number of viral particles × 103 per mL; yellow = positive, red = negative).
No LOD was calculated for the treatment with phosphate buffer since the highest concentration tested, 1 × 106 per mL, produced 2 negative samples. The other 3 treatments gave a lower LOD of 0.5 × 106 per mL. The lowest concentration of target viral particles that could be detected with a probability of 0.95 (LOD95), calculated by using a probit regression model, were: 386.39 × 103 per mL for the treatment of DTT and EDTA with incubation at 37 °C for 30 min and heated at 95 °C for 5 min; 326.76 × 103 per mL for the treatment of DTT and EDTA, heated at 95 °C for 5 min; and 224.21 × 103 per mL for treatment of DTT, EDTA and PK, heated at 95 °C for 5 min.
Pilot study
Table 2 shows the results of the pilot study with saliva samples obtained from patients with a positive SARS-CoV-2 result by RT-PCR of NPS. The treatment of DTT, EDTA and PK and heating at 95 °C for 5 min and the LAMP assay was used to analyze these samples. When samples were divided according to the Ct values of the RT-PCR of NPS, the sensitivity was 100% in samples with Ct <20, 71.4% in samples with Ct 20–30, and 47.8% in samples with Ct >30.
Table 2.
SAFE-SAL results using detection by LAMP in saliva samples obtained from individuals with a positive SARS-Cov-2 result by RT-PCR of NPS.
| Ct value RT-PCR |
SAFE-SAL |
|||
|---|---|---|---|---|
| Positive | Negative | Sensitivity | Number of samples | |
| Positive | 57 | |||
| <20 | 13 | 0 | 100% | 13 |
| 20–30 | 15 | 6 | 71.4% | 21 |
| >30 | 11 | 12 | 47.8 % | 23 |
In all 39 RT-PCR negative samples the results of our procedure were also negative.
A total of 81 saliva samples (NPS: 48 positive, 33 negative) were also analyzed by RT-PCR after the treatment. Results of RT-LAMP and RT-PCR were consistent with the exception of 5 samples that tested negative by RT-LAMP and positive with RT-PCR; all had a Ct value >30.
Discussion
In this report, different sample treatments are evaluated for application in a procedure for detecting SARS-CoV-2 in saliva developed by our laboratory. This procedure consists of 3 main steps: (1) initial heating for sample inactivation, (2) sample treatment based on the addition of different solutions and, in some cases, additional heating, (3) an assay for the detection that in this case was a RT-LAMP. One of the main advantages of this procedure is that saliva inactivation is performed as the first step; therefore, the other steps can be taken with a significantly reduced risk of transmission. This reduction of risk is significant since biosafety is of paramount importance in dealing with the analysis of COVID-19 in biological samples, particularly with saliva that is highly contagious (Kutti-Sridharan et al., 2020). As well as inactivating the virus, heating the sample at 95 °C allows for the detection of RNA in the sample without the need for extraction procedures, and it potentially inactivates components that can inhibit PCR or LAMP reactions (Smyrlaki et al., 2020, Chin et al., 2020).
The first treatment evaluated was a dilution with phosphate buffer. The result of the RT-LAMP used in the procedure is based on the decrease in pH caused by the amplification of a target. When the pH is lower than 7, the color of the mix turns from pink to yellow. The mix of the sample with a buffer phosphate with pH 7.7 allowed us to (1) increase the saliva pH in cases of individuals with acidic pH because saliva pH can range from 6.2–7.6 (Baliga et al., 2013) and therefore avoid false positives, (2) ensure that pH did not exceed 8, as this could inhibit the reaction. This could explain why in setting up the procedure, when we tested phosphate buffer at pH 8.4 and borate buffer at pH 8.2, we found false negatives. We also found that the addition of Triton X-100 and Np-40 interfered with the colorimetric lamp even at low concentrations (0.5%). Phosphate buffer as a sample treatment can be used with PCR assays for COVID-19 diagnosis, with the caution that high concentrations can inhibit the assay (Smyrlaki et al., 2020). Despite being a very low-cost treatment, in our conditions, phosphate buffer gave false negatives with all the viral concentrations analyzed; therefore, the risk of false negatives should be considered when used. Other conditions or treatments, such as additional heating at step 2 with phosphate buffer, could be tested to determine if they could increase its sensitivity.
The other 3 treatments tested used DTT and EDTA. DTT is a sulphydryl reagent that specifically reduces mucoprotein disulfide bonds, being a mucolytic agent widely used for sample homogenization in sputum or other viscous fluids (Rabe and Cepko, 2020, Yu et al., 2018). The mucus or viscosity in saliva can cause virus-containing cell components to be trapped in mucus, leading to low yield RNA. It can also cause pipetting errors, clot formation, and failed amplification; therefore, it should be avoided for the analysis of SARS-CoV-2 (Peng et al., 2020). The chelating agent EDTA was added to increase DTT stability since DTT oxidation is catalyzed by free metals (Stevens et al., 1983), and to sequester cations necessary for RNAse activity, preserving the viral RNA (Rabe and Cepko, 2020).
At pH 7.5 and 20 °C, the half-life of DTT is 10 h, and its stability can be increased with the addition of chelating agents. For example, DTT had less than 15% oxidation in 1 week at 4% in the presence of ethylene glycol tetraacetic acid (Getz et al., 1999). Therefore, in the conditions of our procedure, we postulate that DTT could be stable for at least 1 day. Ideally, storage at −20 °C in aliquots and use of fresh solution daily would be recommended until specific data on the stability of the solution used in this procedure is generated. Tris(2-carboxyethyl)phosphine (TCEP) has been proposed as an alternative to DTT as a reducing agent in saliva since it is described as a more stable solution (Rabe and Cepko, 2020). However, in our study, we used DTT because its lower cost and because TCEP stability can decrease with phosphate buffers and chelating agents (Getz et al., 1999).
The 3 different treatments tested in step 2 using DTT combined with EDTA and heating for 5 min at 95 °C gave a detection limit of 500.000 viral particles/mL and a LOD95 lower than this value. No evident improvements were observed in the DTT treatment with a previous incubation for 30 min at 37 °C, so just one heating step at 95 °C can be performed when DTT is used at step 2. Further studies should be done to elucidate the mechanisms involved in the positive effect of the additional heating at 95 °C on step 2 of this procedure. The addition of PK to the treatment solution produced the lowest LOD95. PK is a serine protease that degrades RNAases in samples preventing RNA degradation and homogenizing sputum samples (Sung et al., 2016). Possibly the combination of DTT and PK allows for a larger decrease in viscosity of the sample and the removal of substances that inhibit amplification, thereby improving the detection of virus RNA.
It has been reported that samples collected from patients with <1.000.000 viral copies per mL contain minimal or non-measurable infectious virus that cannot be cultured and, consequently, have a potentially lower risk of infection (Larremore et al., 2020, Wölfel et al., 2020). On this basis, our procedure, using any of the 3 treatments tested at step 2 that had a LOD of 500.000 viral particles, could have a practical use for detecting individuals that could transmit the disease (Mina et al., 2020). However, the procedure using LAMP would be less sensitive than RT-PCR (for example, the RT-PCR used in our study for comparative purposes has a LOD of 250 viral copies per mL in saliva (Delaney et al., 2021)) and, therefore, would have limited sensitivity at the very early stages of infection.
The sensitivity of our procedure using the RT-LAMP assay with the better sample treatment option ranged from 100% in saliva samples from patients with results of RT-PCR in NPS of Ct <20, who could be expected to have high concentrations of the virus in saliva, to 47.8% in samples from patients with Ct >30. Our results of 100% sensitivity in samples with high viral load agreed with previous authors who used similar RT-LAMP assays in pharyngeal swab samples; however, we obtained a higher sensitivity than previous reports in samples with Ct >30 (Mautner et al., 2020), probably due to our sample treatment. In any case, as indicated in previous reports that tested saliva with RT-LAMP (Yokota et al., 2020), and as we observed in our study, some samples with low viral loads detected by RT-PCR cannot be detected with LAMP. This major limitation should be carefully considered when the assay is applied. The lower sensitivity could be due to the inability of RT-LAMP reactions to detect short RNA fragments of historical and no longer contagious infection, unlike RT-PCR (Blackmore, 2020). Therefore, further studies should be made to elucidate if RT-LAMP could have the advantage of potentially identifying most of the individuals with a high enough viral load to transmit the disease.
This study has various limitations. One is the lack of a full clinical validation with a larger number of samples. However, the number of clinical samples tested in our pilot study is in line with previous preliminary studies. For example, Taki et al. (2020) used 17 positive and 14 negative samples, and Lalli et al. (2020) used 20 positive and 10 negative samples. Further studies should include a larger number of patients and evaluate this procedure for detecting asymptomatic patients who could spread the virus. In these studies, comparison between the use of LAMP and RT-PCR would be of high interest. In addition, NSP and saliva samples were obtained within 1 day of difference; therefore, a direct comparison of the clinical value between both samples could not be undertaken and should be done in the future. Regarding sampling, it would be of interest in the future to establish recommendations for saliva collection to minimize the risk of disease transmission. The use of a straw or any collection device that avoids the creation and expansion of drops should be encouraged (Ceron et al., 2020). Finally, it is important to point out that the data presented in this paper is specific to the procedure and assay used. The treatments evaluated could have different effects if tested in other assay procedures. Results could also vary if other sample types are used, such as sputum, which can have a higher viral load than saliva.
This procedure can be subject to improvements. Further trials could test possible new sample treatments at step 2 that could increase the sensitivity of the procedure. Step 3 if using LAMP could be adapted to microtiter plates, increasing the throughput of the assays. Although the set of primers used in our study was one of the most sensitive according to a previous report, the use of other primers or primer combinations could enhance the sensitivity of the assay (Zhang et al., 2020), and other types of LAMP or different types of assays for SARS-CoV-2 detection could be used in step 3. An important consideration for molecular diagnostic assays, particularly LAMP, is the sensitivity to contamination (Tomita et al., 2008). The use of separate spaces and equipment for the different phases of LAMP preparation is recommended where possible, and the inclusion of 2′-Deoxyuridine 5′-Triphosphate (dUTP) and uracil-DNA glycosylase in the reaction to help prevent contamination.
Conclusions
In this report, we described an optimized sample treatment for a procedure for detecting SARS-CoV-2 in saliva. This procedure is safe and flexible since it can work with modifications in various steps. The main limitation is its lower sensitivity when LAMP is used compared with RT-PCR; however, it can contribute to COVID-19 control by detecting individuals able to transmit the disease. Also, it is low-cost, and it could be applied in contexts of reduced economic resources and limited access to high-cost equipment, personnel, or reagents. In addition, it has the advantage of using saliva as a sample and the potential of being adapted to point-of-care use. It is expected that the analytical procedure presented in this report could be a useful resource that can help in COVID-19 management.
Author contributions
Conceptualization, JJC, CPR, EB and ATC; Methodology, CPR, LFM, CSR, SMS, AT, IM, MJA, MRV; writing—original draft preparation, JJC, CP, AT, ATC; writing—review and editing, CPR, LFM, SMS, AT, ATC, IM, EB, MJA, MRV, JJC; All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Program for Research Groups of Excellence of the Seneca Foundation, Murcia, Spain (grant 19894/GERM/15) and from the University of Murcia to support special needs of research groups due to the COVID-19 pandemic. LFM was granted with predoctoral contract ‘FPU’ of the University of Murcia, Spain. AT has a post-doctoral fellowship “Ramón y Cajal” (RYC-2017-22992) supported by the Ministerio de Economía y Competitividad, Agencia Estatal de Investigación (AEI), Spain, and The European Social Fund (ESF).
Institutional review board statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Murcia University (protocol code: CBE 337/220) and the Ethics Committee of the IMIB-Arrixaca: (protocol code 2020-9-2 HCUVA).
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Data availability statement
Data is available under reasonable request.
Conflicts of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgements
The authors would like to thank Dr. Nathan Tanner for his technical support. The support of the Garcia Cugat Foundation to research activities in the field of saliva analysis of Interlab-UMU is acknowledged. We want to particularly acknowledge the patients and the Biobank of Region of Murcia Network, IMIB Biobank (PT17/0015/0038) integrated into the Spanish National Biobanks Network for their collaboration. This article is based upon work from COST Action CA16215, supported by COST (European Cooperation in Science and Technology).
Footnotes
Part of the results from this study was presented to PortASAP 2021 Meeting (10-11 February 2021, web conference) as a poster presentation: An Open and Portable Procedure for the Detection of SARS-CoV-2 in Saliva.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.05.053.
Appendix A. Supplementary data
References
- Baliga S., Muglikar S., Kale R., Salivary P.H. A diagnostic biomarker. J Indian Soc Periodontol. 2013;17:461–465. doi: 10.4103/0972-124X.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore C. The role of population-wide saliva sampling for COVID-19. BMJ. 2020:371. [Google Scholar]
- Ceron J., Lamy E., Martinez-Subiela S., Lopez-Jornet P., Capela-Silva F., Eckersall P. Use of saliva for diagnosis and monitoring the SARS-CoV-2: a general perspective. J Clin Med. 2020;9:1491. doi: 10.3390/jcm9051491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow F.W.-N., Chan T.T.-Y., Tam A.R., Zhao S., Yao W., Fung J. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int J Mol Sci. 2020;21:5380. doi: 10.3390/ijms21155380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney M., Simpson J., Thomas B., Ralph C., Evangalista M., Moshgriz M. The use of saliva as a diagnostic specimen for SARS CoV-2 molecular diagnostic testing for pediatric patients. Acta Sci Microbiol. 2021;4:2581–3226. [Google Scholar]
- Getz E.B., Xiao M., Chakrabarty T., Cooke R., Selvin P.R. A comparison between the sulfhydryl reductants tris(2- carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- Kutti-Sridharan G., Vegunta R., Vegunta R., Mohan B.P., Rokkam V.R.P. SARS-CoV2 in different body fluids, risks of transmission, and preventing COVID-19: a comprehensive evidence-based review. Int J Prev Med. 2020;11:97. doi: 10.4103/ijpvm.IJPVM_255_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli M.A., Langmade S.J., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem. 2020;67(2):415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2020;7(1):eabd5393. doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner L., Baillie C.K., Herold H.M., Volkwein W., Guertler P., Eberle U. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J. 2020;17 doi: 10.1186/s12985-020-01435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina M.J., Parker R., Larremore D.B. Rethinking Covid-19 test sensitivity — a strategy for containment. N Engl J Med. 2020;383:e120. doi: 10.1056/nejmp2025631. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Lu Y., Song J., Vallance B.A., Jacobson K., Yu H.B. Direct clinical evidence recommending the use of proteinase K or dithiothreitol to pretreat sputum for detection of SARS-CoV-2. Front Med. 2020;7 doi: 10.3389/fmed.2020.549860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe B.A., Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci U S A. 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., Vondracek M. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R., Stevens L., Price N. The stabilities of various thiol compounds used in protein purifications. Biochem Educ. 1983;11:70. doi: 10.1016/0307-4412(83)90048-1. [DOI] [Google Scholar]
- Sung H., Yong D., Ki C.S., Kim J.S., Seong M.W., Lee H. Comparative evaluation of three homogenization methods for isolating Middle East Respiratory Syndrome Coronavirus nucleic acids from sputum samples for real-time reverse transcription PCR. Ann Lab Med. 2016;36:457–462. doi: 10.3343/alm.2016.36.5.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki K., Yokota I., Fukumoto T., Iwasaki S., Fujisawa S., Takahashi M. SARS-CoV-2 detection by fluorescence loop-mediated isothermal amplification with and without RNA extraction. J Infect Chemother. 2020;27(2):410–412. doi: 10.1016/j.jiac.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using PH-sensitive dyes. BioTechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Vogels C.B.F., Watkins A.E., Harden C.A., Brackney D.E., Shafer J., Wang J. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. 2020;12:263–280. doi: 10.1016/j.medj.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yokota I., Shane P.Y., Okada K., Unoki Y., Yang Y., Inao T. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020;25:ciaa1388. doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Qiu T., Zeng Y., Wang Y., Zheng S., Chen X. Comparative evaluation of three preprocessing methods for extraction and detection of influenza A virus nucleic acids from sputum. Front Med. 2018;5 doi: 10.3389/fmed.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ren G., Buss J., Barry A.J., Patton G.C., Tanner N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques. 2020;69:179–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available under reasonable request.