Abstract
Objectives
To compare the clinical and epidemiological aspects associated with different predominant lineages circulating in Marseille from March 2020 to January 2021.
Methods
In this single-centre retrospective cohort study, characteristics of patients infected with four different severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants were documented from medical files. The outcome was the occurrence of clinical failure, defined as hospitalization (for outpatients), transfer to the intensive care unit (inpatients) and death (all).
Results
A total of 254 patients were infected with clade 20A (20AS), 85 with Marseille-1 (M1V), 190 with Marseille-4 (M4V) and 211 with N501Y (N501YV) variants. 20AS presented a bell-shaped epidemiological curve and nearly disappeared around May 2020. M1V reached a very weak peak, then disappeared after six weeks. M4V appeared in July presented an atypical wave form for 7 months. N501YV has only recently appeared. Compared with 20AS, patients infected with M1V were less likely to report dyspnoea (adjusted odds ratio (OR) 0.50, p 0.04), rhinitis (aOR 0.57, p 0.04) and to be hospitalized (aOR 0.22, p 0.002). Patients infected with M4V were more likely to report fever than those with 20AS and M1V (aOR 2.49, p < 0.0001 and aOR 2.30, p 0.007, respectively) and to be hospitalized than those with M1V (aOR 4.81, p 0.003). Patients infected with N501YV reported lower rate of rhinitis (aOR 0.50, p 0.001) and anosmia (aOR 0.57, p 0.02), compared with those infected with 20AS. A lower rate of hospitalization was associated with N501YV infection compared with 20AS and M4V (aOR 0.33, p < 0.0001 and aOR 0.27, p < 0.0001, respectively).
Conclusions
The four lineages have presentations that differ from one another, epidemiologically and clinically. This supports SARS-CoV-2 genomic surveillance through next-generation sequencing.
Keywords: Coronavirus disease 2019, Marseille, Mutation, N501Y, Severe acute respiratory syndrome coronavirus 2, Variant
Introduction
Very little is known about viral factors underlying the clinical presentation and severity of coronavirus disease 2019 (COVID-19). To date, most studies showed that the viral mutations, especially the D614G mutation in the spike protein, correlated with a higher infectivity than the wild-type virus. However, the evidence of an association between viral mutations and severity of the disease is scant [1].
In Marseille, France, the first episode was predominantly due to clade 20A (20AS) strains originating from Wuhan, China. The second episode was linked to Marseille-1 variant (M1V), which had an African origin and specific clinical and epidemiological profiles [2]. Marseille-4 variant (M4V), probably originating from a mink farm in the north of France, was observed soon after [3], and became prevalent across most of Europe [[4], [5], [6], [7]]. Finally, the third episode due to N501Y variants (N501YV) previously identified in the UK, South Africa and Brazil [[4], [5], [6], [7]] started in early 2021.
We compared the clinical and epidemiological aspects of the diseases associated with four different predominant lineages circulating in Marseille.
Materials and methods
Data source and study design
A single retrospective cohort study was conducted at the Méditerranée Infection Institute (Institut Hospitalo-Universitaire) in Marseille, Southern France. All available severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome sequences obtained by our institute between March 2020 and January 2021 were reviewed. Those selected were sequences of 20AS obtained from respiratory samples collected before June 2020 (the predominant variant of the first epidemic in Marseille), sequences of M1V or M4V (predominant variants during the second phase of the epidemic), and sequences harbouring the N501Y substitution within the spike, which is the main variant circulating during the third episode of the current epidemic in Marseille. A second filter was applied to include only patients with available clinical status and follow up.
Patients
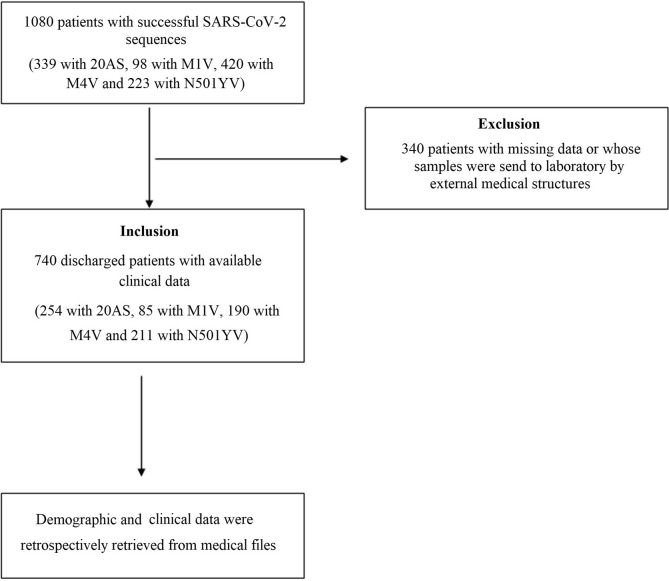
Individuals seen by our institute's medical team at the outpatient clinic or hospitalized in the conventional infectious disease units were included, regardless of age and disease severity. Demographic and clinical data including co-morbidities were retrospectively retrieved from medical files, including the main symptoms, inpatient/outpatient status, transfer to an intensive care unit and death. At the time of study, all patients were discharged (either recovered or died). In addition, possible death occurring post-discharge (90 days for the last included patient) was checked using the national data on mortality related to COVID-19 [8]. All patients with available clinical data were included. Patients with missing clinical data were patients whose samples were sent to our laboratory by external medical organizations and were excluded in further analysis. Fig. 1 shows the flowchart of the study.
Fig. 1.
Flowchart of the study; 20AS, clade 20A strain; M1V, Marseille-1 variant; M4V, Marseille-4 variant; N501YV, N501Y variant.
Genome sequencing and assembling
SARS-CoV-2-positive patients identified by real-time PCR with a Ct value < 30 [9] were selected for next-generation sequencing. Whole-genome sequencing was performed for 20S, M1V and M4V, as previously described [2], from 200 μL of nasopharyngeal swab fluid after viral RNA extraction with the EZ1 Virus Mini Kit v2.0, followed by reverse transcription by SuperScript IV (ThermoFisher Scientific, Waltham, MA, USA), cDNA second-strand synthesis using Klenow Fragment DNA polymerase (New England Biolabs, Beverly, MA, USA) and the generation of purified DNA with Agencourt AMPure XP beads (Beckman Coulter, Villepinte, France). Genome next-generation sequencing used Illumina technology on a MiSeq instrument and the Illumina Nextera XT Paired end strategy (Illumina Inc., San Diego, CA, USA). Genome assemblies were performed with the CLC Genomics workbench v.7 software by mapping on the SARS-CoV-2 genome GenBank Accession no. NC_045512.2 (Wuhan-Hu-1 isolate). Recovered genomes were compared with sequences from the GISAID database (https://www.gisaid.org/). Phylogeny reconstructions were performed with GISAID TreeTool v2.0 (https://www.gisaid.org/epiflu-applications/upcoming-features-in-v20/treetool-app/). N501YV was identified by specific quantitative PCR [10].
Statistics
Statistical analyses were carried out using R (R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2020. URL: https://www.Rproject.org/) and Stata version 15.1 (StataCorps, College Station, TX, USA). Qualitative variables were presented by percentage. Univariate and multivariate analyses were conducted to evaluate the different characteristics and clinical profiles of patients infected with different lineages. We aim to evaluate the predictive factors associated with infection with different SARS-CoV-2 variants. The dependent variables were the genetic characteristics of SARS-CoV-2 lineages (20AS, M1V, M4V and N501YV). The primary outcome was the occurrence of clinical failure, defined as hospitalization (for outpatients), transfer to the intensive care unit (inpatients) and death (all). Unadjusted associations between multiple covariate factors and infection with different lineages were investigated using χ2 test. Crude odds ratio (OR) and 95% CI were calculated using a two-by-two frequency table to evaluate the association of each predictive factor and infection with different SARS-CoV-2 variants. Multivariate analysis was performed using logistical regression. Covariate variables with general proportion <5% were excluded from statistical analysis, because of low effectiveness. Variables with a p value < 0.2 in the univariate analysis were included in the multivariate analysis [11]. The ϕ coefficient was used to test for multicollinearity among the independent variables. For pairs of variables that were highly correlated (absolute value of correlation coefficient >0.7), only one variable was entered into the multivariate model. Multivariable logistic regression (created by stepwise regression) was used to predictive factors associated with infection with different SARS-CoV-2 variants. The results were presented by adjusted odds ratio (aOR) with a 95% CI. A p value < 0.05 was considered as statistically significant.
Ethics statement
Whole-genome sequencing was performed on nasopharyngeal samples that were collected in the context of routine diagnosis. No additional samples were collected for this study. Clinical data were retrospectively retrieved from medical files and anonymized before analysis. Ethical approval was obtained from the Marseille Institutional Review Board and Ethics Committee (No. 2020-016-03).
Results
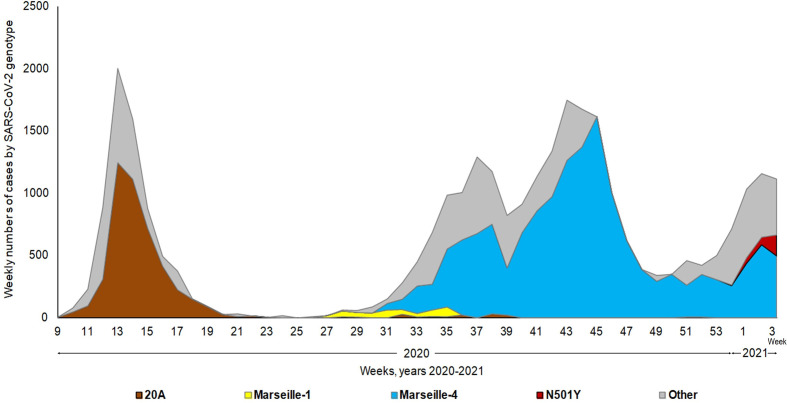
Between 29 February 2020 and 31 January 2021, 15 variants circulated in Marseille. We identified 1080 patients infected with four principal lineages: 20AS (n = 339), M1V (n = 98), M4V (n = 420) and N501YV (n = 223). Fig. 2 shows the extrapolated timeline of SARS-CoV-2 variants identified in our institute. 20AS presented a bell-shaped epidemiological curve characteristic of seasonal respiratory infections and nearly disappeared around May 2020. The M1V reached a very weak peak but represented up to 100% of infections during part of the month of July, then disappeared after six weeks [2]. The M4V, which appeared in July, presented an atypical wave form and continued to represent a significant proportion of the cases in February, indicating a duration of 7 months, which is not comparable with that of the other two epidemics. The N501YV variant has only recently appeared, and conclusions cannot yet be reached as to the epidemic form it will take. Clinical data were available from 740 patients whose virus genome was documented and who were treated at our institute (Table 1 ). Twenty per cent (148/740) of patients were aged ≥60 years and 49.5% (366/740) were male. A total of 20.5% (152/740) of patients were hospitalized, 3.2% (24/740) were transferred to an intensive care unit and 4.3% (32/704) died. It should be noted that no patient infected with the M1V variant was transferred to an intensive care unit or died.
Fig. 2.
Weekly numbers of cases of infections with the different genotypes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The weekly numbers of cases of infections with the different genotypes of SARS-CoV-2 were extrapolated from the actual numbers of detection of each of the genotypes by relating them to the weekly numbers of SARS-CoV-2 diagnoses. The grey zone corresponds to the extrapolated number of cases with other genotypes than clade 20A strain, Marseille-1, Marseille-4 and N501Y variants. These genotypes included clades 19, 20B, 20C, and Marseille-2, -3, -5, -6, -7, -8, -9 and -10 variants. SARS-CoV-2 genotypes were determined by genomic sequencing and specific quantitative PCR. The grey zone corresponds to the extrapolated number.
Table 1.
Sociodemographic characteristics, co-morbidities, clinical signs and disease severity of patients infected with clade 20A strains and with the Marseille-1, Marseille-4 and N501Y SARS-CoV-2 variants
| Variables | 20A (n = 254) |
Marseille-1 (n = 85) |
Marseille-4 (n = 190) |
N501Y (n = 211) |
p value |
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | ||
| Agea | |||||
| Median | 45.5 | 34.0 | 54.0 | 49.0 | <0.0001 |
| Interquartile | 32–59 | 24–48 | 36–70 | 35–61 | |
| Range | 0–95 | 13–90 | 11–97 | 13–100 | |
| <45 years | 125 (49.2) | 59 (69.4) | 63 (33.2) | 91 (43.1) | <0.0001 |
| 45–64 years | 81 (31.9) | 20 (23.5) | 69 (36.3) | 84 (39.8) | |
| ≥65 years | 48 (18.9) | 6 (7.1) | 58 (30.5) | 36 (17.1) | |
| Sex (male) | 113 (44.5) | 48 (56.5) | 102 (53.7) | 103 (48.8) | 0.13 |
| Hypertension | 51 (20.1) | 8 (9.4) | 47 (24.7) | 36 (17.1) | 0.02 |
| Diabetes | 18 (7.1) | 4 (4.7) | 21 (11.1) | 12 (5.7) | 0.14 |
| Cancer | 10 (3.9) | 2 (2.4) | 15 (7.9) | 15 (7.1) | 0.13 |
| Chronic respiratory diseases | 23 (9.1) | 7 (8.2) | 20 (10.5) | 27 (12.8) | 0.53 |
| Chronic heart diseases | 27 (10.6) | 4 (4.7) | 29 (15.3) | 20 (9.5) | 0.06 |
| Obesity | 22 (8.7) | 8 (9.4) | 10 (5.3) | 18 (8.5) | 0.49 |
| Fever | 67 (26.4) | 22 (25.9) | 87 (45.8) | 97 (46.0) | <0.0001 |
| Cough | 123 (48.4) | 39 (45.9) | 73 (38.4) | 97 (46.0) | 0.20 |
| Dyspnoea | 72 (28.3) | 13 (15.3) | 42 (22.1) | 48 (22.8) | 0.08 |
| Rhinitis | 106 (41.7) | 26 (30.6) | 37 (19.5) | 56 (26.5) | <0.0001 |
| Anosmiab | 76 (29.9) | 26 (30.6) | 35 (18.5) | 43 (20.4) | 0.01 |
| Ageusiab | 71 (28.0) | 23 (27.1) | 34 (18.0) | 44 (20.9) | 0.06 |
| Hospitalization | 53 (20.9) | 5 (5.9) | 67 (35.3) | 27 (12.8) | <0.0001 |
| Transfer to ICUbc | 5 (2.0) | 0 (0) | 10 (5.3) | 9 (4.3) | 0.06 |
| Deathc | 10 (3.9) | 0 (0) | 15 (7.9) | 7 (3.3) | 0.02 |
Fifteen patients were less than 18 years old.
Data were not available for two children aged <11 years.
Transfer to intensive care unit and death were not considered in later analyses because of their low proportion (<5%).
In univariate analysis, significant differences were observed between patients infected with different lineages of SARS-CoV-2 with regards to age, certain co-morbidities and symptoms and disease severity (Table 2 ). In multivariate analyses, some characteristics appeared to be independent factors associated with infection with different virus lineages (Table 3 ). Notably, lower rates of dyspnoea, rhinitis and hospitalization were seen in patients infected with the M1V compared with those infected with 20AS. Compared with those infected with 20AS or the M1V, patients infected with the M4V were older and more likely to present with fever. In addition, a lower rate of rhinitis was observed in M4V infection compared with 20AS and a higher hospitalization rate was associated with M4V infection compared with M1V. Higher rates of cancer and fever, and lower rates of rhinitis and anosmia were seen N501YV-infected individuals compared with those infected with the 20AS variant. Older age and fever were more associated with N501YV infections compared with M1V. Finally, a lower rate of hospitalization was associated with N501YV infection compared with 20AS and M4V.
Table 2.
Sociodemographic characteristics, co-morbidities, clinical signs and disease severity of patients infected with different severe acute respiratory syndrome coronavirus 2 lineages, univariate analysis
| Variables | Marseille-1 vs 20A (reference = 20A) |
Marseille-4 vs 20A (reference = 20A) |
Marseille-4 vs Marseille-1 (reference = Marseille-1) |
N501Y vs 20A (reference = 20A) |
N501Y vs Marseille-1 (reference = Marseille-1) |
N501Y vs Marseille-4 (reference = Marseille-4) |
|---|---|---|---|---|---|---|
| OR (95% CI) p value |
OR (95% CI) p value |
OR (95% CI) p value |
OR (95% CI) p value |
OR (95% CI) p value |
OR (95% CI) p value |
|
| Age | ||||||
| <45 years | reference | reference | reference | reference | reference | reference |
| 45–64 years | 0.52 (0.29–0.93) 0.03 |
1.69 (1.09–2.63) 0.02 |
3.23 (1.75–5.95) <0.000 |
1.42 (0.95–2.14) 0.09 |
2.72 (1.51–4.90) 0.001 |
0.84 (0.54–1.32) 0.46 |
| ≥65 years | 0.26 (0.11–0.65) 0.004 |
2.40 (1.47–3.90) <0.000 |
9.05 (3.64–22.55) <0.000 |
1.03 (0.62–1.71) 0.91 |
3.89 (1.54–9.80) 0.004 |
0.43 (0.25–0.73)< 0.002 |
| Sex (male) | 1.62 (0.99–2.66) 0.06 |
1.45 (0.99–2.11) 0.06 |
0.89 (0.53–1.50) 0.67 |
1.19 (0.83–1.72) 0.35 |
0.74 (0.44–1.22) 0.23 |
0.82 (0.55–1.22) 0.33 |
| Hypertension | 0.41 (0.19–0.91) 0.03 |
1.31 (0.83–2.05) 0.24 |
3.16 (1.42–7.03) 0.005 |
0.82 (0.51–1.31) 0.41 |
1.98 (0.88–4.46) 0.10 |
0.62 (0.38–1.02) 0.06 |
| Diabetes | 0.65 (0.21–1.97) 0.44 |
1.63 (0.84–3.15) 0.15 |
2.52 (0.84–7.57) 0.10 |
0.79 (0.37–1.68) 0.54 |
1.22 (0.38–3.90) 0.74 |
0.49 (0.23–1.02) 0.06 |
| Cancer | 0.59 (0.13–2.74) 0.50 |
2.09 (0.92–4.76) 0.08 |
3.56 (0.80–15.92) 0.10 |
1.87 (0.82–4.25) 0.14 |
3.18 (0.71–14.20) 0.13 |
0.89 (0.42–1.88) 0.77 |
| Chronic respiratory diseases | 0.90 (0.37–2.18) 0.82 |
1.18 (0.63–2.22) 0.60 |
1.31 (0.53–3.23) 0.56 |
1.47 (0.82–2.66) 0.20 |
1.64 (0.68–3.91) 0.27 |
1.25 (0.67–2.31) 0.48 |
| Chronic heart diseases | 0.42 (0.14–1.22) 0.11 |
1.51 (0.86–2.66) 0.15 |
3.65 (1.24–10.73) 0.02 |
0.88 (0.48–1.62) 0.68 |
2.12 (0.70–6.40) 0.18 |
0.58 (0.32–1.07) 0.08 |
| Obesity | 1.10 (0.47–2.56) 0.83 |
0.59 (0.27–1.27) 0.18 |
0.53 (0.20–1.41) 0.21 |
0.98 (0.51–1.89) 0.96 |
0.90 (0.37–2.15) 0.81 |
1.68 (0.75–3.73) 0.20 |
| Fever | 0.97 (0.56–1.71) 0.93 |
2.36 (1.58–3.51) <0.000 |
2.42 (1.38–4.25) 0.002 |
2.37 (1.61–3.50) <0.000 |
2.44 (1.40–4.25) 0.002 |
1.01 (0.68–1.49) 0.97 |
| Cough | 0.90 (0.55–1.48) 0.69 |
0.66 (0.45–0.97) 0.04 |
0.74 (0.44–1.23) 0.25 |
0.91 (0.63–1.31) 0.60 |
1.01 (0.61–1.66) 0.99 |
1.36 (0.92–2.03) 0.13 |
| Dyspnoea | 0.46 (0.24–0.87) 0.02 |
0.72 (0.46–1.11) 0.14 |
1.57 (0.79–3.11) 0.19 |
0.74 (0.49–1.14) 0.17 |
1.63 (0.83–3.20) 0.15 |
1.04 (0.65–1.66) 0.88 |
| Rhinitis | 0.62 (0.36–1.04) 0.07 |
0.34 (0.22–0.52) <0.000 |
0.55 (0.31–0.98) 0.04 |
0.50 (0.34–0.75) 0.001 |
0.82 (0.47–1.43) 0.48 |
1.49 (0.93–2.39) 0.10 |
| Anosmia | 1.03 (0.60–1.75) 0.92 |
0.53 (0.34–0.83) 0.006 |
0.52 (0.28–0.93) 0.03 |
0.60 (0.39–0.92) 0.02 |
0.58 (0.33–1.03) 0.06 |
1.13 (0.69–1.85) 0.64 |
| Ageusia | 0.95 (0.55–1.65) 0.86 |
0.56 (0.35–0.89) 0.01 |
0.59 (0.32–1.08) 0.09 |
0.68 (0.44–1.04) 0.07 |
0.71 (0.40–1.27) 0.25 |
1.20 (0.73–1.98) 0.47 |
| Hospitalization | 0.24 (0.09–0.61) 0.003 |
2.07 (1.35–3.16) 0.001 |
8.72 (3.37–22.56) <0.000 |
0.56 (0.34–0.92) 0.02 |
2.35 (0.87–6.32) 0.09 |
0.27 (0.16–0.44) <0.000 |
Table 3.
Sociodemographic characteristics, co-morbidities, clinical signs and disease severity of patients infected with different severe acute respiratory syndrome coronavirus 2 lineages, multivariate analysisa
| Variables | Marseille-1 vs 20A (reference = 20A) |
Marseille-4 vs 20A (reference = 20A) |
Marseille-4 vs Marseille-1 (reference = Marseille-1) |
N501Y vs 20A (reference = 20A) |
N501Y vs Marseille-1 (reference = Marseille-1) |
N501Y vs Marseille-4 (reference = Marseille-4) |
|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) p value |
Adjusted OR (95% CI) p value |
Adjusted OR (95% CI) p value |
Adjusted OR (95% CI) p value |
Adjusted OR (95% CI) p value |
Adjusted OR (95% CI) p value |
|
| Age | ||||||
| <45 years | reference | reference | ||||
| 45–64 years | — | 1.61 (1.01–2.56) 0.04 |
2.61 (1.38–4.92) 0.003 |
— | 2.68 (1.48–4.87) 0.001 |
— |
| ≥65 years | — | 1.93 (1.15–3.23) 0.01 |
4.12 (1.49–11.38) 0.006 |
— | 3.70 (1.45–9.42) 0.006 |
— |
| Cancer | — | — | — | 2.73 (1.09–6.84) 0.03 |
— | — |
| Fever | — | 2.49 (1.64–3.80) <0.000 |
2.30 (1.26–4.23) 0.007 |
2.58 (1.72–3.86) <0.000 |
2.35 (1.33–4.15) 0.003 |
— |
| Dyspnoea | 0.50 (0.26–0.98) 0.04 |
— | — | — | — | — |
| Rhinitis | 0.57 (0.33–0.97) 0.04 |
0.36 (0.23–0.57) <0.000 |
— | 0.50 (0.33–0.76) 0.001 |
— | — |
| Anosmia | — | — | — | 0.57 (0.36–0.90) 0.02 |
— | — |
| Hospitalization | 0.22 (0.09–0.58) 0.002 |
— | 4.81 (1.69–13.69) 0.003 |
0.33 (0.18–0.60) <0.000 |
— | 0.27 (0.17–0.45) <0.000 |
All variables with a p value < 0.20 in the univariate analysis were included in the logistical regression, but only significant variables (in multivariate analysis) were presented.
Discussion
These four lineages have presentations that are quite different from each other on certain points. These variants look completely different in the same location at different times.
Clinically, the age of the patients is different. Patients infected with the M1V are clearly younger than those with the initial virus or those with the M4V and the N501YV. This is also associated with lower disease severity. Differences in the frequency of fever, dyspnoea, rhinitis and anosmia were observed according to lineages. Hospitalization rates also varied with lineages. The N501YV is known to associate with an increased infectivity, but the correlation with the severity of the disease is unclear. Initially, it was described as associating with increased hospitalization and mortality rates [[12], [13], [14]]. An analysis of syndromic community testing and death records showed that this variant was associated with an increase in deaths from 2.5 to 4.1 per 1000 detected cases. By contrast, in a recent clinical study, no association of this variant with disease severity was observed [15]. The increase in severity of N501YV was only found in studies based on a community-based testing data set, whereas our study and that of Frampton et al. [15] were conducted among patients seen at hospital. Although these large community studies found a significant difference in mortality at a population level, the absolute risk increase affecting individual patients is probably minimal.
This work has some limitations. Several biomarkers known to be associated with severity, including lymphopenia, thrombocytopenia, D-dimer counts, troponin level and lactate dehydrogenase, were not considered in this analysis. In addition, we cannot provide information on duration of symptoms, which could be different by variant. Furthermore, we analysed only patients seen at our hospital, requiring medical care and that could introduce a major selection bias. A large proportion of patients (not seen at our hospital) were excluded from our analysis because of missing data. The patients not captured here could be young and asymptomatic or pauci-symptomatic and the real severity of each variant may differ from our results. Finally, the variants changed over time, as did our clinical understanding, management and treatment of COVID-19. There is a secular trend toward decreasing hospitalization and mortality, such that the rates reported here by variant may be either overestimated or underestimated. Nevertheless, these clinical and epidemiological nuances represent a definite interest when considering the clinical and epidemiological specificity of the variants, which appeared extremely clear to us in the field. We must remember that all of these patients were admitted to the same institute and data were collected by doctors who observed the patients directly. This underlines the fact that COVID-19 has presented variability since July 2020 that is associated with epidemiological modifications such as the source (the M1V undoubtedly came from Africa, the 20AS from China, the N501YV from England and the source of M4V is unknown but is believed to be of European origin) [2,3].
Hence, it is difficult to use the same name to qualify a disease, the variants of which present epidemiological origins, epidemic curves and clinical manifestations that are distinct from one other.
In practice, as for influenza virus, SARS-CoV-2 seems to present mutants with a severity and an epidemiological course that is specifically linked to these mutations. The severity is also different as M1V infections were less severe than those caused by other lineages, the N501YV infection appeared to be less severe than M4V infection, which seemed to have had the greatest severity compared with the others. This is despite the fact that patient care from diagnosis to discharge took place under the same circumstances, in the same institute and under the same conditions, providing perfectly comparable data, in contrast to most other studies, in particular multicentric studies.
Author contributions
TLD, VTH and PG contributed to experimental design, data analysis, statistics, interpretation and writing. TLD, VTH, NNN and JCL contributed to collecting the data of patients. JD analysed the sequencing results. PC and AL contributed to interpretation of sequencing results. HC contributed to statistical analysis. DR, HC, PC, AL and JCL contributed to critically reviewing the manuscript. PG coordinated the work.
Transparency declaration
All authors have declared that they have no conflicts of interest.
Funding
This study was supported by the Institut Hospitalo-Universitaire Méditerranée Infection, the French National Research Agency under the Investissements d'avenir programme, reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European FEDER PRIMI funding.
Acknowledgements
Our thanks go to Ludivine Brechard, Vera Esteves-Vieira, Elsa Prudent, Marielle Bedotto, Linda Houhamdi and all staff at the Institut Hospitalo-Universitaire Méditerranée Infection for their support with molecular techniques. Thanks are also due to Yolande Obadia, Laurent Mayer and Loutfia Assoumani for their help in recovering patient data.
Editor: A. Huttner
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.05.029.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dao T.L., Hoang V.T., Colson P., Lagier J.C., Raoult D., Levasseur A., et al. 2020. Severity of COVID-19 infection and different SARS-CoV-2 variants: current evidence. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colson P., Levasseur A., Gautret P., Fenollar F., Thuan Hoang V., Delerce J., et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: an investigational study. Travel Med Infect Dis. 2021;40:101980. doi: 10.1016/j.tmaid.2021.101980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier P.E., Colson P., Levasseur A., Gautret P., Bedotto M., Filosa V., et al. 2021. Genome sequence analysis enabled deciphering the atypical evolution of COVID-19 epidemics in Marseille, France. Preprints. [DOI] [Google Scholar]
- 4.European Centre for Disease Prevention and Control . 2021. Risk Assessment: risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA—first update.https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update Available at: [Google Scholar]
- 5.Centers for Disease Control and Prevention . 2020. COVID-19 and your health.https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html Available at: [PubMed] [Google Scholar]
- 6.PAHO/WHO Pan American Health Organization Epidemiological update: occurrence of variants of SARS-CoV-2 in the Americas—26 january 2021. https://www.paho.org/en/documents/epidemiological-update-occurrence-variants-sars-cov-2-americas-26-january-2021 Available at:
- 7.Faria N.R., Carlo I.M., Candido D., Moyses Franco L.A., Andrade P.S., Coletti T.M., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virol Org. 2021 https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 Available at: [Google Scholar]
- 8.Institut National de la Statistique et des Etudes Economiques (INSEE) Fichier des personnes décédées. https://www.data.gouv.fr/fr/datasets/fichier-des-personnes-decedees/ Available at:
- 9.Amrane S., Tissot-Dupont H., Doudier B., Eldin C., Hocquart M., Mailhe M., et al. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France, January 31st to March 1st, 2020: a respiratory virus snapshot. Travel Med Infect Dis. 2020;36:101632. doi: 10.1016/j.tmaid.2020.101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedotto M., Fournier P.E., Houhamdi L., Colson P., Raoult D. Implementation of an in-house real-time reverse transcription-PCR assay to detect the emerging SARS-CoV-2 N501Y variants. J Clin Virol. 2021;140:104868. doi: 10.1016/j.jcv.2021.104868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bursac Z., Gauss C.H., Williams D.K., Hosmer D.W. Purposeful selection of variables in logistic regression. Source Code Biol Med. 2008;3:17. doi: 10.1186/1751-0473-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group, Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D., et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. 2021;26:2100256. doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021;S1473–3099:170–175. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.