Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been spreading worldwide, causing a global pandemic. Bat-origin RaTG13 is currently the most phylogenetically related virus. Here we obtained the complex structure of the RaTG13 receptor binding domain (RBD) with human ACE2 (hACE2) and evaluated binding of RaTG13 RBD to 24 additional ACE2 orthologs. By substituting residues in the RaTG13 RBD with their counterparts in the SARS-CoV-2 RBD, we found that residue 501, the major position found in variants of concern (VOCs) 501Y.V1/V2/V3, plays a key role in determining the potential host range of RaTG13. We also found that SARS-CoV-2 could induce strong cross-reactive antibodies to RaTG13 and identified a SARS-CoV-2 monoclonal antibody (mAb), CB6, that could cross-neutralize RaTG13 pseudovirus. These results elucidate the receptor binding and host adaption mechanisms of RaTG13 and emphasize the importance of continuous surveillance of coronaviruses (CoVs) carried by animal reservoirs to prevent another spillover of CoVs.
Keywords: COVID-19, SARS-CoV-2, RaTG13, intermediate horseshoe bat, ACE2, RBD
Graphical abstract
Structural and molecular analysis of the receptor binding domain of RaTG13, a coronavirus phylogenetically closely related to SARS-CoV-2, bound to the human receptor ACE2 as well as ACE2 orthologs in 24 other species provides a framework to understand its host range as well as the basis of antibody cross-reactivity between the two viruses.
Introduction
Emerging and re-emerging pathogens threaten global public health and cause tremendous economic losses (Gao, 2018). The new coronavirus (CoV) severe acute respiratory syndrome CoV 2 (SARS-CoV-2) was detected and then isolated in early 2020 (Tan et al., 2020; The 2019-nCoV Outbreak Joint Field Epidemiology Investigation Team and Li, 2020; Wang et al., 2020a; Zhu et al., 2020). SARS-CoV-2 later spread worldwide, causing a global pandemic. As of March 5, 2021, the World Health Organization (WHO; https://covid19.who.int/) has recorded more than 114 million confirmed cases of CoV disease 2019 (COVID-19) globally and more than 2.5 million related deaths.
SARS-CoV-2 belongs to the family Coronaviridae, enveloped RNA viruses in the order Nidovirales (https://talk.ictvonline.org/taxonomy/?Redirected=true). CoVs are classified into four genera: alpha, beta, gamma, and delta CoVs. Bats are recognized as natural reservoirs of alpha and beta CoVs (Latinne et al., 2020). Increasing evidence indicates that bat CoVs (bCoVs) are the evolutionary sources of SARS-CoV (beta CoV), Middle East respiratory syndrome CoV (MERS-CoV; beta CoV), swine acute diarrhea syndrome CoV (SADS-CoV; alpha CoV), human CoV (HCoV)-OC43 (beta CoV), HCoV-NL63 (alpha CoV), and HCoV-229E (alpha CoV) (Corman et al., 2015; Gao, 2018; Huynh et al., 2012; Latinne et al., 2020; Lau et al., 2005; Li et al., 2005; Smith and Wang, 2013; Wang et al., 2018), most of which can infect humans and cause disease (Su et al., 2016). SARS-CoV and MERS-CoV have caused global outbreaks in the past two decades and confirmed cases of MERS-CoV infection are still growing in the Middle East region.
SARS-CoV-2 is closely related to several CoVs from bats (Chan et al., 2020; Liu et al., 2021; Malik et al., 2020). RaTG13, sequenced from Rhinolophus affinis (intermediate horseshoe bat) in Yunnan province is the closest relative to SARS-CoV-2, with 96.2% identity of the overall genome sequence and 89.3% amino acid identity in the receptor binding domain (RBD) (Boni et al., 2020; Wu et al., 2020a; Zhou et al., 2020b). One variance of RaTG13 compared with SARS-CoV-2 is the absence of the four-residue (PRRA) insertions at the S1/S2 cleavage site on the spike (S) protein. However, such residue insertions can occur naturally in CoVs, which may contribute to RaTG13 evolving into the causative pathogen of the next potential pandemic. For example, RmYN02, another bCoV, detected in Rhinolophus malayanus, shares 93.3% nucleotide identity with the SARS-CoV-2 genome and contains three-amino-acid-residue (PAA) insertions at the S1/S2 cleavage site of its S protein, which is similar to that in SARS-CoV-2 (Zhou et al., 2020a). Considering the similarity between RaTG13 and SARS-CoV-2 and the devastation caused by SARS-CoV-2, RaTG13 may pose a significant threat to humans and other animals. To access this risk, the host range of RaTG13 needs to be investigated.
Receptor binding is the key step for a virus to achieve cross-species infection and transmission (Li et al., 2020b). The gain of function to interact with the receptor orthologs of another species is also a prerequisite for inter-species transmission (Su et al., 2016). Thus, evaluating the interactions between viral ligands and receptor orthologs of various animals may lead to identification of susceptible hosts. For SARS-CoV-2, angiotensin-converting enzyme 2 (ACE2) is required for human cell entry by binding to the SARS-CoV-2 RBD on the S protein (Lan et al., 2020; Shang et al., 2020; Wang et al., 2020b). We and other groups independently discovered ACE2 as the receptor and investigated the interactions between the SARS-CoV-2 RBD and ACE2 orthologs of other species. Our previous work revealed that SARS-CoV-2 might have a broad host range, including cats and dogs, which is consistent with the findings of epidemiological studies. Thus, quantifying the interactions between RaTG13 RBD and ACE2 orthologs of domestic and wild animals that are in close proximity to humans could provide clues for determining the host range of RaTG13.
Currently, the structures of SARS-CoV-2 RBD in complex with human ACE2 (hACE2), cat ACE2 (cACE2), bat ACE2 (bACE2), and pangolin ACE2 (pACE2) have been determined (Lan et al., 2020; Liu et al., 2021; Shang et al., 2020; Wang et al., 2020b; Wu et al., 2020b, 2021). These structures revealed that the SARS-CoV-2 RBD utilizes a similar binding mechanism with all four ACE2s, but it forms fewer H-bonds in the interactions with the latter three ACE2s than with hACE2, providing the molecular basis for the decreased binding affinity between the SARS-CoV-2 RBD and the three animal ACE2s. Previous work has reported that RaTG13 RBD binds to hACE2 with a much lower affinity than the SARS-CoV-2 RBD (Wrobel et al., 2020). However, the mechanism of interaction between the RaTG13 RBD and hACE2 remains unclear.
Here the complex structure of RaTG13 RBD with hACE2 was determined, and the underlying mechanism for the lower binding affinity of the RaTG13 RBD compared with that of the SARS-CoV-2 RBD to hACE2 was resolved. Moreover, the binding ability of 24 ACE2 orthologs, together with hACE2, to the RaTG13 RBD was quantified. We also evaluated the effects of six substitutions between the RaTG13 RBD and SARS-CoV-2 RBD on the interaction with hACE2 and orthologs. Specifically, 501 is most likely the key residue that determines the potential host range of RaTG13. Coincidentally, residue 501 is also the key mutation of the SARS-CoV-2 strains that have been spreading rapidly in many countries, including variant of concern (VOC) lineages 20I/501Y.V1 in the United Kingdom, 20H/501Y.V2 in South Africa, and 20J/501Y.V3 in Brazil and Japan, and also the key mutation of a mouse adaptive SARS-CoV-2 virus strain, indicating that mutation at this residue should be watched closely (Gu et al., 2020). Moreover, we also found that SARS-CoV-2 could induce a strong cross-reactive immune response to RaTG13 and identified a SARS-CoV-2 monoclonal antibody (mAb), CB6, that could cross-neutralize the RaTG13 pseudovirus. These results broaden our knowledge of the molecular mechanisms of RaTG13 RBD binding to hACE2 and the potential host adaption of RaTG13, providing evidence emphasizing the importance of continuous surveillance of CoVs carried by other potential animal reservoirs, monitoring RaTG13 and its related CoVs especially closely.
Results
Complex structure of RaTG13 RBD with hACE2
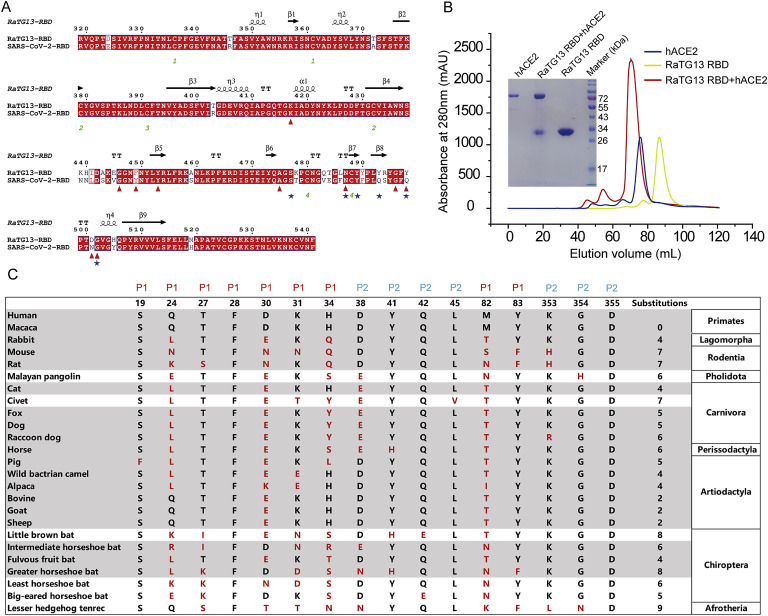
Sequence alignments of the RaTG13 RBD and SARS-CoV-2 RBD were performed. Overall, the RBDs of RaTG13 and SARS-CoV-2 showed an 89.3% amino acid identity (Figure S1 A). The complex structure indicates that the SARS-CoV-2 RBD forms 14 hydrogen bonds through 10 amino acids in its interaction with hACE2. Only 3 of 10 amino acids differed between the RaTG13 RBD and SARS-CoV-2 RBD (Figure S1A). However, the binding affinity of the RaTG13 RBD to hACE2 (K D = 3.86 μM) was approximately 2-log lower than that of SARS-CoV-2 RBD to hACE2 (K D = 21.1 nM) (Figures 1A and 1B). To investigate whether RaTG13 infects host cells by binding to hACE2, a vesicular stomatitis virus (VSV)-based pseudotyped RaTG13, which incorporates the S protein of RaTG13, was used to attempt to transduce HeLa cells expressing hACE2 (HeLa-hACE2s). A similar amount of SARS-CoV-2 pseudovirus (as determined by quantitative real-time PCR) was used as a positive control. The RaTG13 pseudovirus and SARS-CoV-2 pseudovirus were unable to transduce HeLa cells; however, both pseudoviruses were able to transduce HeLa-hACE2s, but the RaTG13 pseudovirus displayed a substantially lower transduction efficiency than the SARS-CoV-2 pseudovirus (Figure 1C).
Figure S1.
Structure-based sequence alignment and protein expression, related to Figure 1
(A) Structure-based sequence alignment of SARS-CoV-2 RBD and RaTG13 RBD. Coils indicate α helices, and black arrows indicate β strands. Conserved residues are highlighted in red. Residues of SARS-CoV-2 RBD or RaTG13 RBD contact with hACE2 through hydrogen bonds are marked with red triangle or blue star, respectively. Sequence alignment is generated with ClustalX and ESPript. (B) Gel filtration profiles of hACE2 (blue), RaTG13 RBD (yellow) and the RaTG13 RBD/hACE2 complex (red) were analyzed and displayed. The separation profiles of each pooled samples on SDS-PAGE are shown in reducing conditions (+DTT). (C) Characteristics of the RaTG13 RBD-binding residues of ACE2s. The conserved residues among 25 ACE2 orthologs are shown as black letters and the residue substitutions in the ACE2 of 24 species comparing with human ACE2 are shown as red letters.
Figure 1.
Structural basis of binding between the RaTG13 RBD and hACE2
(A and B) SPR characterization of the SARS-CoV-2 RBD (A) and RaTG13 RBD (B) interacting with hACE2.
(C) Entry of the SARS-CoV-2 and RaTG13 pseudovirus into HeLa cells expressing the hACE2 (HeLa-hACE2s). Data represent the mean ± SD of six replicates. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (two-tailed unpaired t test).
(D) Overall structure of the RaTG13 RBD in complex with hACE2. Boxes indicate the patches of the RaTG13 RBD/hACE2 complex.
(E and F) The hydrogen bond networks of patch 1 (E) and patch 2 (F). The complex structure is shown as a cartoon, and residues taking part in hydrogen bond formation are shown as sticks.
(G and H) Residues involved in the interaction of hACE2 with the RaTG13 RBD or SARS-CoV-2 RBD are listed and connected by solid lines. Black lines indicate vdw contacts, and red lines represent an H-bond or salt bridge.
(I) The binding surface of hACE2 with the RaTG13 RBD (left) or SARS-CoV-2 RBD (right).
(J) Six different residues between RaTG13 RBD and SARS-CoV-2 RBD among RaTG13 RBD binding sites are labeled.
See also Figure S1.
To analyze the molecular mechanism behind this difference, the complex protein of RaTG13 RBD with hACE2 was prepared by co-infection of insect cells with baculoviruses expressing the RaTG13 RBD and hACE2 (Figure S1B). The stable complex was further purified and crystallized. Finally, we obtained diffraction data and determined the complex structure at a resolution of 3.1 Å (Table 1 ). There are two RBD and hACE2 complex molecules in one asymmetric unit. The electron densities of residues S19–A614 of hACE2 and residues T333–P527 of RaTG13 RBD were observed clearly. Five N-glycans linked to N53, N90, N103, N322, and N546 on hACE2 and one N-glycan linked to N343 of RaTG13 RBD were identified (Figure 1D).
Table 1.
Crystallographic data collection and refinement statistics
| SARS2-CoV-2 RBD/dACE2 | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 81.28, 122.88, 110.74 |
| α, β, γ (o) | 90.00, 95.07, 90.00 |
| Resolution (Å) | 50.00–3.10 (3.21–3.10) |
| Unique reflections | 39622 (3906) |
| Completeness (%) | 99.7 (99.5) |
| Rmerge | 0.106 (1.310) |
| I /σI | 13.9 (1.1) |
| CC1/2 | 0.985(0.685) |
| Redundancy | 6.8 (6.9) |
| Refinement | |
| Resolution (Å) | 48.94–3.10 |
| No. of reflections | 39,578 |
| Rwork/Rfree | 0.2260/0.2500 |
| No. of atoms | |
| Protein | 12,914 |
| Ligand/ion | 2 |
| Water | 0 |
| B-factors | |
| Protein | 118.8 |
| Ligand/ion | 219.8 |
| Water | |
| RMSD | |
| Bond lengths (Å) | 0.002 |
| Bond angles (o) | 0.478 |
| Ramachandran statistics (%) | |
| Favored | 96.44 |
| Allowed | 3.56 |
| Disallowed | 0.00 |
Values in parentheses are for highest-resolution shell.
Overall, the complex structure of the RaTG13 RBD with hACE2 is extremely similar to that of the SARS-CoV-2 RBD with hACE2 (Lan et al., 2020; Shang et al., 2020; Wang et al., 2020b; Yan et al., 2020), with a root-mean-square deviation (RMSD) of 1.012 Å. Similar to other beta CoV RBDs, RaTG13 RBD consists of a core subdomain and an external subdomain (Han et al., 2017). The external subdomain is dominated by a disulfide bond-stabilized flexible loop that connects two small β strands (Figure 1D). The binding interface of hACE2 with RaTG13 RBD is distributed into two patches. Patch 1 is located on the N-terminal α1 and α2 helixes, and Patch 2 is on the conformational surface consisting of the residues from α1 together with a β-hairpin constituted by β3 and β4. In patch 1, residues S19 and K31 from α1 and Y83 from α2 form a hydrogen bond network with residues S477, Y493, N487, and Y489 of the RaTG13 RBD (Figures 1E and 1G; Table 2 ). In patch 2, residues D38, Q42, and Y41 from α2 and K353 from the loop linking β3 and β4 form a hydrogen bond network with residues Y498, T500, and G502 (Figures 1F and 1H; Table 2).
Table 2.
Comparison of RaTG13 RBD and SARS-CoV-2 RBD binding to hACE2
| RaTG13 RBD/SARS-CoV-2 RBD | hACE2 (RaTG13 RBD) | hACE2 (SARS-CoV-2 RBD) |
|---|---|---|
| K417 | D30 (1) | D30 (3,1) |
| G446 | Q42 (2,1) | |
| F449/Y449 | D38 (4) | D38 (5,1), Q42 (2,1) |
| Y453 | H34 (3) | H34 (3,1) |
| L455 | H34 (5), D30 (1) | H34 (4) |
| F456 | T27 (5), D30 (2), K31 (1) | T27 (5), D30 (1), K31 (1) |
| A475 | Q24 (1) | S19 (3,1), Q24 (1), T27 (1) |
| G476 | S19 (2) | |
| S477 | S19 (1,1) | |
| L486/F486 | M82 (1) | M82 (4), Y83 (7) |
| N487 | Q24 (8), Y83 (5,2) | Q24 (7,1), Y83 (4,1) |
| Y489 | T27 (2), F28 (3), K31(2), Y83 (1,1) | T27 (2), F28 (4), Y83 (1) |
| F490 | K31 (1) | |
| Y493/Q493 | K31 (6,1) | H34 (3), E35 (3) |
| G496 | K353 (2) | D38 (1), K353 (5,1) |
| Y498/Q498 | Y41 (6), Q42 (3,1), D38 (5,1), K353 (2), L45 (1) | Y41(5), Q42(5,3), L45(1) |
| T500 | Y41 (7,1), L45 (1), D355 (5) | Y41 (6), N330 (3), D355 (6), R357 (3) |
| D501/N501 | Y41 (5), K353 (5) | Y41 (5,1), K353 (5) |
| G502 | K353 (3,1), 354 (3) | K353 (3,1), G354 (5) |
| H505/Y505 | K353 (11), G354 (2) | E37 (3), K353 (14), G354 (2) |
| Total | 113, 9 | 141, 14 |
The numbers without underline in parentheses of hACE2 (RaTG13 RBD) and hACE2 (SARS-CoV-2 RBD) residues represent the number of vdw contacts between the indicated hACE2 residues with the RaTG13 RBD or SARS-CoV-2 RBD. Underlined numbers with bold format suggest numbers of potential H-bonds between the pairs of residues. vdw contact was analyzed at a cutoff of 4 Å and H-bonds at a cutoff of 3.3 Å.
Comparison of the complex structures of the SARS-CoV-2 RBD with hACE2 and the RaTG13 RBD with hACE2
The total contacts of the RaTG13 RBD with hACE2 (113 contacts) are significantly less than those of SARS-CoV-2 with the same receptor (141 contacts). Specifically, there are 48 contacts between the RaTG13 RBD and hACE2 in patch 1, which is fewer than those in the SARS-CoV-2 RBD with the hACE2 complex (60 contacts) (Figure 1G). Similarly, in patch 2, fewer contacts are formed between the RaTG13 RBD and hACE2 than between the SARS-CoV-2 RBD and hACE2 (65 versus 81) (Figure 1H). Moreover, there are substantially fewer hydrogen bonds in patch 2 of the RaTG13 RBD with hACE2 than of the SARS-CoV-2 RBD with the hACE2 complex (4 versus 9) (Figure 1H).
The buried surface area of the RaTG13 RBD bound to hACE2 is 1604 Å2, which is smaller than that of the SARS-CoV-2 RBD with the hACE2 complex (1,773 Å2). Specifically, residues G446 and F490 of the SARS-CoV-2 RBD contributed to contact with hACE2, whereas the corresponding residues of the RaTG13 RBD (G446 and Y490) did not (Figure 1I). On the binding surface with hACE2, the RaTG13 RBD displays six differences compared with the SARS-CoV-2 RBD: Y449F, F486L, Q493Y, Q498Y, N501D, and Y505H (Figure 1J).
The binding capacity of RaTG13 RBD to ACE2 orthologs from 24 different species
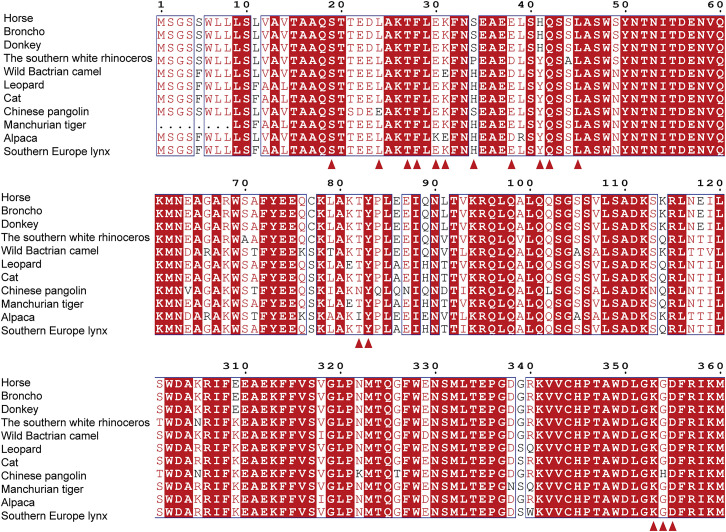
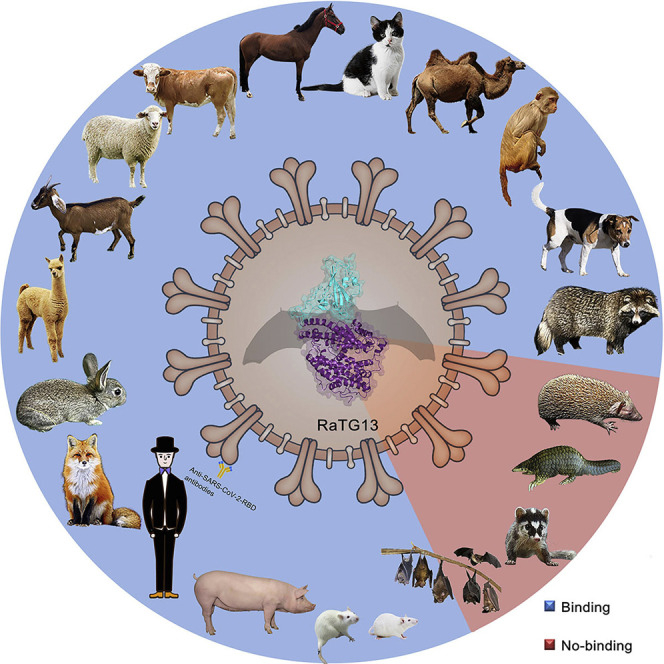
To evaluate the potential host range of RaTG13, we analyzed the sequence of hACE2 and of 24 other animals belonging to nine orders, including primates (human and monkey), Lagomorpha (rabbit), Rodentia (mouse and rat), Pholidota (Malayan pangolin), Carnivora (cat, civet, fox, dog, and raccoon dog), Perissodactyla (horse), Artiodactyla (pig, wild Bactrian camel, alpaca, bovine, goat, and sheep), Chiroptera (intermediate horseshoe bat, least horseshoe bat, little brown bat, fulvous fruit bat, greater horseshoe bat, and big-eared horseshoe bat), and Afrotheria (lesser hedgehog tenrec) (Figure S1C). The 16 key residues in hACE2 responsible for the interaction with the RaTG13 RBD were highlighted and compared with the 24 ACE2 orthologs. Sequence alignment showed a range of 0–9 residue substitutions in the ACE2 orthologs. Overall, residues distributed in patch 1 are more diverse than residues in patch 2 (Figure S1C). Specifically, residues F28 and D355 are fully conserved among all ACE2 orthologs compared in this study. Residues S19, Y41, Q42, L45, and G354 of hACE2 were substituted in no more than three ACE2 orthologs among the 24 species (Figure S1C).
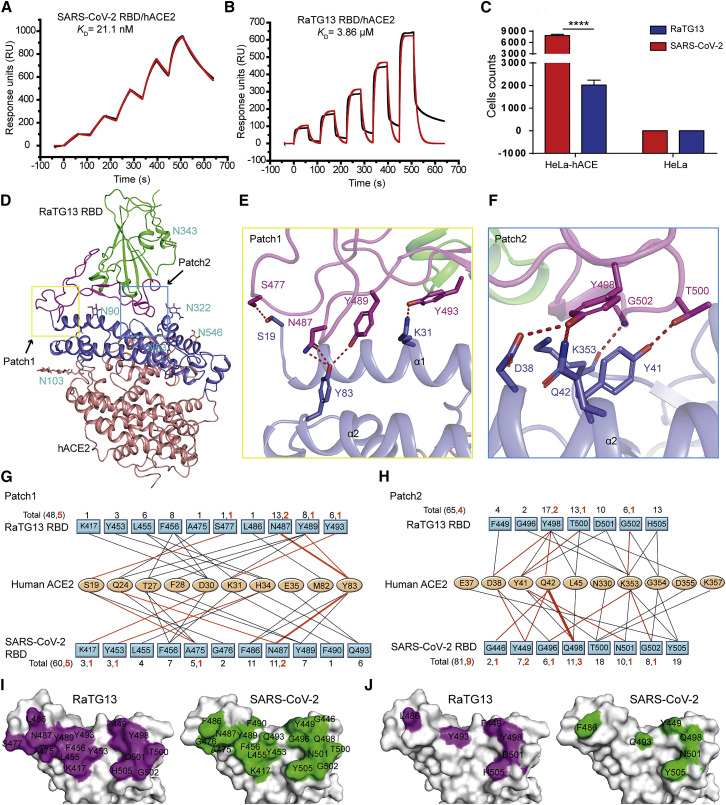
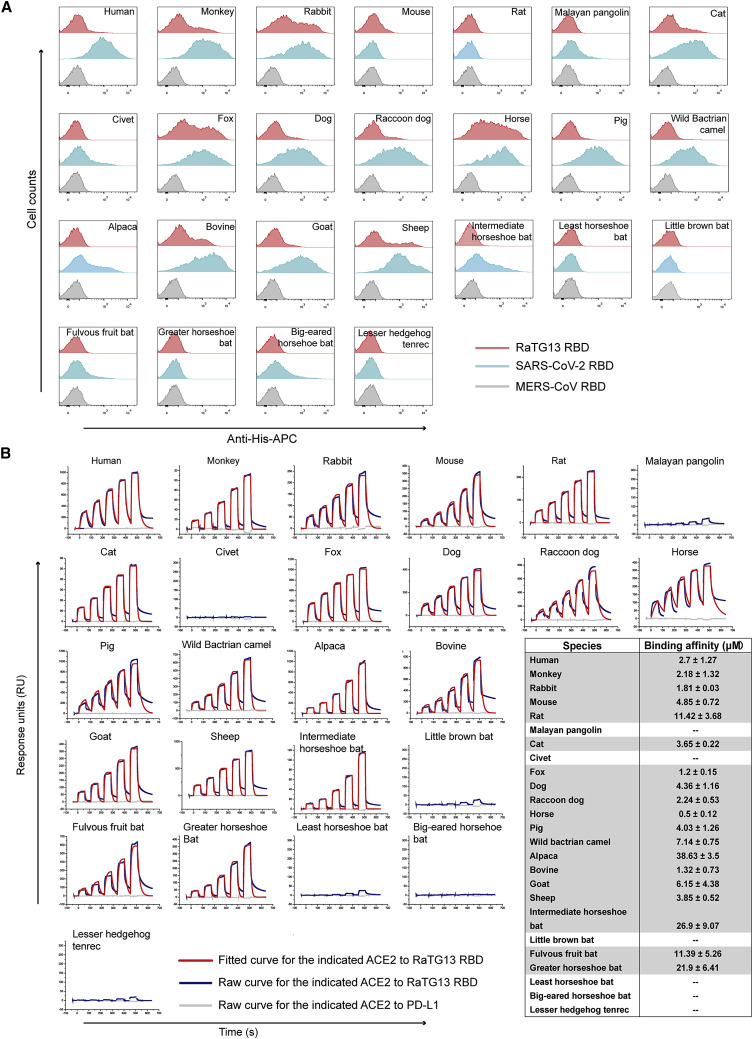
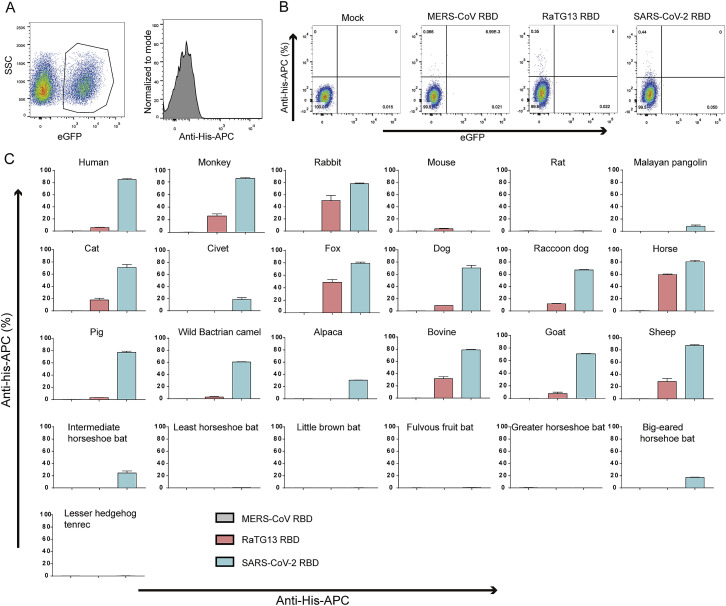
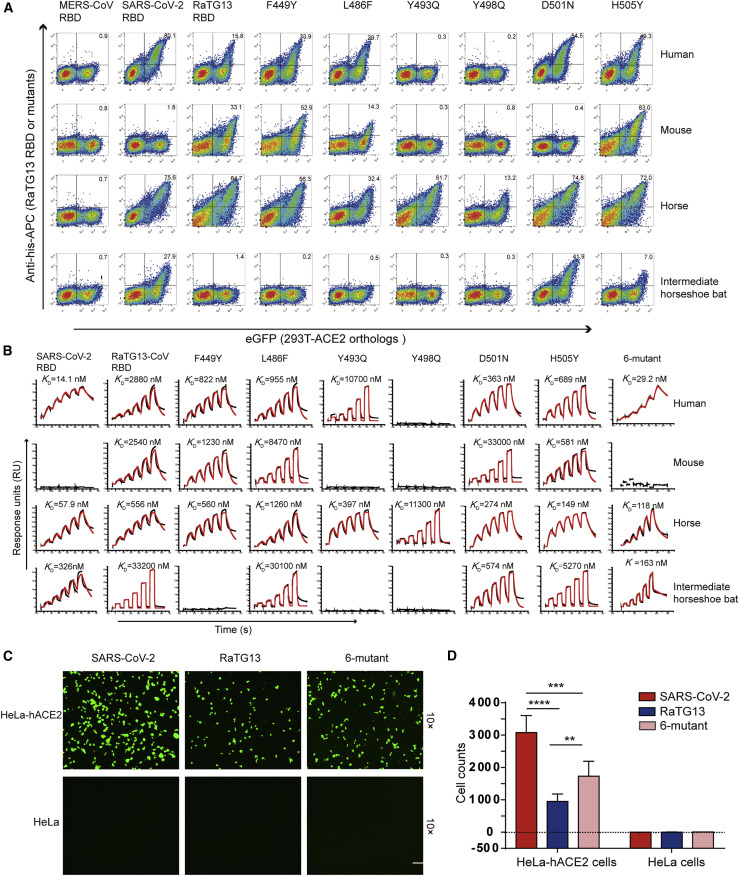
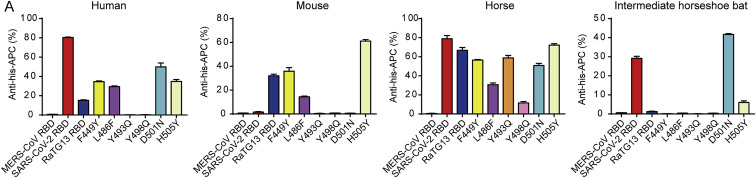
We subsequently tested the binding of ACE2 orthologs with the RaTG13 RBD or SARS-CoV-2 RBD using eGFP-fused ACE2 orthologs expressed on the cell surface via flow cytometry. The MERS-CoV RBD was the negative control. The RaTG13 RBD interacted with cells expressing primate (human and monkey), Lagomorpha (rabbit), Carnivora (cat, fox, dog, and raccoon dog), Perissodactyla (horse), and Artiodactyla (pig, wild Bactrian camel, bovine, goat, and sheep) ACE2s (Figures 2 A and S2 ). The SARS-CoV-2 RBD also interacted with the abovementioned ACE2s, albeit with a higher fluorescence shift. Additionally, the SARS-CoV-2 RBD also interacted with Malayan pangolin, alpaca, intermediate horseshoe bat, and big-eared horseshoe bat ACE2s, as reported previously, whereas the RaTG13 RBD does not (Figures 2A and S2). Another differing binding feature was that the RaTG13 RBD, but not the SARS-CoV-2 RBD, showed positive binding to mouse ACE2 (Figures 2A and S2). No detectable interactions were observed between either RBDs with rat, least horseshoe bat, little brown bat, fulvous fruit bat, greater horseshoe bat, or lesser hedgehog tenrec ACE2 orthologs (Figures 2A and S2).
Figure 2.
Binding between ACE2 orthologs and the SARS-CoV-2 RBD or RaTG13 RBD
(A) Flow cytometry analysis of binding between 25 ACE2 orthologs and the SARS-CoV-2 RBD or RaTG13 RBD. HEK293T cells expressing EGFP-fused ACE2s are stained with the indicated His-tagged proteins (RaTG13 RBD, SARS-CoV-2 RBD, and MERS-CoV RBD). An anti-His/Allophycocyanin (APC) antibody is used to detect His-tagged proteins. The MERS-CoV RBD is used as a negative control.
(B) SPR characterizations of the binding between 25 ACE2 orthologs and the RaTG13 RBD. ACE2s with a mouse Fc (mFc) tag are immobilized on a CM5 chip. SPR characterizations of the binding affinity between the RaTG13 RBD and each ACE2 ortholog are shown. PD-L1 is used as a negative control. Raw and fitted curves are displayed as blue and red lines, respectively. Data represent the mean ± SD of three independent experiments.
See also Figures S2 and S3 and Tables S1 and S2.
Figure S2.
Gating strategy and statistics for flow cytometric analysis of the binding between ACE2s and the SARS-CoV-2 RBD or RaTG13 RBD, related to Figure 2
(A) Gating strategy for flow cytometric analysis of the binding between ACE2s and SARS-CoV-2 RBD or SARS-CoV RBD. eGFP-positive HEK293T cells are gated first, followed by analysis of anti-his-APC positive cells. (B) Dot plot of untransfected HEK293T cells stained by MERS-CoV RBD, RaTG13 RBD and SARS-CoV-2 RBD proteins. (C) Frequency of SARS-CoV-2 RBD or RaTG13 RBD positive HEK293T cells expressing hACE2. MERS-CoV RBD is used as negative control. Data represent the results of three replicates, and error bars show the SD of each measurement.
To clarify the interactions between the RaTG13 RBD and ACE2 orthologs from different animals, we quantified their binding affinities via surface plasmon resonance (SPR) assay. The RaTG13 RBD was capable of binding to human, monkey, rabbit, cat, fox, dog, raccoon dog, horse, pig, bovine, wild Bactrian camel, goat, sheep, and mouse ACE2, with equilibrium dissociation constant (K D) values of 0.5–7.14 μM, but were unable to interact with Malayan pangolin, civet, little brown bat, least horseshoe bat, big-eared horseshoe bat, or lesser hedgehog tenrec ACE2s (Figure 2B). Notably, the RaTG13 RBD displayed an ~5-fold stronger affinity to horse ACE2 than to hACE2 (0.5 ± 0.12 μM versus 2.7 ± 1.27 μM) (Figure 2B). We performed a BLAST search to identify the 10 ACE2 orthologs that are most phylogenetically related to horse ACE2 and observed that most residues interacting with the RaTG13 RBD are conserved (Figure S3 ). Three of these ten ACE2 orthologs, wild Bactrian camel, cat, and alpaca ACE2, were included here, all of which showed binding to the RaTG13 RBD (Figure 2B). Although flow cytometry results showed no interactions between RaTG13 RBD and ACE2 orthologs of rat, alpaca, intermediate horseshoe bat, fulvous fruit bat, or greater horseshoe bat (Figures 2A and S2), SPR results showed RaTG13 can bind to these ACE2s, with K D values of 11.42–38.63 μM) (Figures 2A and 2B).
Figure S3.
Structure-based sequence alignment of horse ACE2 and 10 ACE2 orthologs most closely related to horse ACE2, related to Figure 2
Coils indicate α helices, and black arrows indicate β strands. Conserved residues are highlighted in red. Residues highlighted in blue boxes are highly (80%) conserved, with consensus amino acids in red. Residues of hACE2 marked with blue star indicate key binding residues with RaTG13 RBD. Sequence alignment is generated with ClustalX, and ESPript.
Distinctive binding sites between the RaTG13 RBD and SARS-CoV-2 RBD
To evaluate the roles of six substitutions between the SARS-CoV-2 RBD and RaTG13 RBD in binding to the human receptor, we mutated the residues in the RaTG13 RBD to those of their SARS-CoV-2 RBD counterparts. Thus, we prepared six RaTG13 RBD mutants containing F449Y, L486F, Y493Q, Y498Q, D501N, and H505Y, respectively. Another mutant, called the 6 mutant, where all six RaTG13 residues were exchanged for their SARS-CoV-2 RBD counterparts, was also prepared. We evaluated their ability to interact with hACE2. Compared with the wild-type RaTG13 RBD, the F449Y, L486F, D501N, and H505Y mutants showed stronger interactions with hACE2, with binding affinity increased by about ~3.5-, ~3.1-, ~8.0-, and ~4.0-fold respectively, whereas the Y493Q mutation substantially decreased the binding affinity to hACE2, and the Y498Q mutation abolished the interaction with hACE2 (Figures 3A, 3B, and S4 ). More importantly, the binding affinity of the 6 mutant to human, mouse, horse, and intermediate horseshoe bat ACE2 orthologs showed similar binding characteristics to those of the SARS-CoV-2 RBD (Figure 3B). The pseudovirus infection assay also indicated that the RaTG13 pseudovirus integrating the S protein with the 6 mutant increased transduction to HeLa-hACE2 cells, although this was still significantly weaker than the SARS-CoV-2 pseudotyped virus (Figures 3C and 3D).
Figure 3.
Mutational analysis of the key residues in RaTG13 involved in interaction with hACE2
(A) Flow cytometry analysis of binding between the six RaTG13 RBD mutants and ACE2s from human, mouse, horse, or intermediate horseshoe bat. SARS-CoV-2 RBD and MERS-CoV RBD are used as control.
(B) SPR analysis of the binding affinity between wild-type/mutated RaTG13 RBD with ACE2s from human, mouse, horse, or intermediate horseshoe bat, respectively. Raw and fitted curves are displayed as blue and red lines, respectively.
(C) Entry of the pseudovirus of SARS-CoV-2, RaTG13, and 6-mutant RaTG13 into HeLa-hACE2s. Green fluorescent HeLa-hACE2s indicate pseudovirus-transducing-cells. Untransfected HeLa cells were used as negative controls. The scale bar indicates 150 μm.
(D) Statistics for transduction of the SARS-CoV-2, RaTG13, and 6-mutant RaTG13 pseudoviruses. Data represent the results of six replicates. All data are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (two-tailed unpaired t test).
Figure S4.
Percentage of ACE2-positive cells among total EGFP-positive cells, related to Figure 3
The percentage of SARS-CoV-2 RBD and RaTG13 RBD positive cells in HEK293T cells with different ACE2 othologs. Data represent the results of three replicates, and error bars show the SD of each measurement.
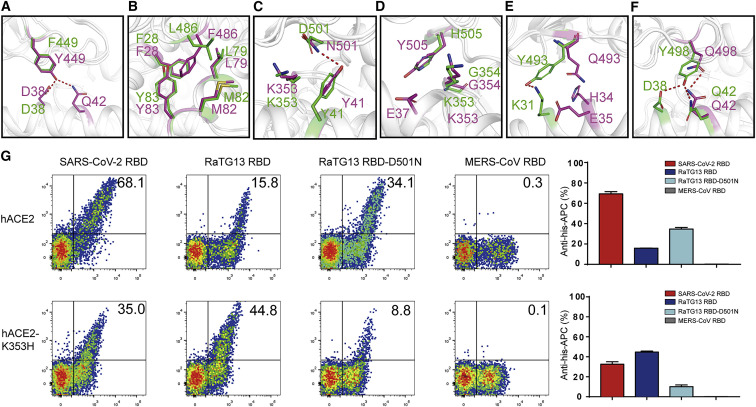
Structural analysis revealed the basis of why the RaTG13 RBD has a lower binding affinity with hACE2 compared with that of the SARS-CoV-2 RBD. Some differing residues on the RaTG13 RBD form fewer contacts than their SARS-CoV-2 RBD counterparts, including F449, L486, D501, and H505. Specifically, RaTG13 RBD F449 contacts the D38 of hACE2 through van der Waals (vdw) force (4 contacts), whereas Y449 of the SARS-CoV-2 RBD forms stronger interactions with two potential hydrogen bonds (with D38 and Q42 of hACE2) and seven vdw contacts (Figure 4 A). Moreover, SARS-CoV-2 RBD F486 closely packs against the F28, L79, M82, and Y83 residues of hACE2 and forms a small patch of hydrophobic interactions at the interface, and N501 forms a hydrogen bond with Y41 of hACE2. In contrast, RaTG13 RBD L486 interacts with M82 through only one vdw contact (Figure 4B), and D501 contacts Y41 through vdw contacts (Figure 4C). H505 of the RaTG13 RBD and Y505 of SARS-CoV-2 are in contact with K353 and G354, but SARS-CoV-2 Y505 also binds to an extra E37 on hACE2 (Figure 4D).
Figure 4.
Structural and functional analysis of the role of each key residue between ACE2 orthologs and the RaTG13 RBD or SARS-CoV-2 RBD
(A–F) Structural comparison of the binding between hACE2 and the RaTG13 RBD or SARS-CoV-2 RBD. Substituted residues on the RaG13 RBD and SARS-CoV-2 RBD are shown in green and purple, respectively. Key residues are shown as sticks with corresponding colors.
(G) Flow cytometric analysis of the effect of D501 and N501 of the RaTG13 RBD and K353 and H353 of hACE2. Bar graphs indicate the ratio of anti-His-positive cells in HEK293T cells with hACE2 expression.
Some other substituted residues on RaTG13 RBD form weaker interactions compared with their SARS-CoV-2 RBD counterparts. RaTG13 RBD Y493 interacts with hACE2 K31 through a hydrogen bond, whereas SARS-CoV-2 RBD Q493 only interacts with hACE2 H34 and E35 through vdw interactions because of different conformations (Figure 4E). Additionally, although RaTG13 RBD Y498 and SARS-CoV-2 RBD Q498 are able to form two potential hydrogen bonds with hACE2 D38 and Q42, RaTG13 RBD Y498 is 0.1 Å closer to hACE2 Q42 compared with SARS-CoV-2 RBD Q498. Thus, substitution from Y498 to Q498 potentially weakened the interactions between RaTG13 RBD Y498 and hACE2, explaining the inability of the RaTG13 RBD Y498Q mutant to interact with the human receptor (Figure 4F).
Key amino acids of RaTG13 and SARS-CoV-2 RBDs for binding to different ACE2s and the role of residue 501
To further evaluate changes in binding affinity because of the six RaTG13 mutants, we chose the ACE2 orthologs from horse, mouse, and intermediate horseshoe bat for in-depth analysis. Horse ACE2 was chosen because it binds to the RaTG13 RBD with a higher affinity than hACE2, whereas mouse ACE2 was chosen for its distinct binding feature to the RaTG13 RBD and SARS-CoV-2 RBD. As the origin of RaTG13, intermediate horseshoe bat ACE2 was also included in this analysis.
Our results showed a broad spectrum of binding abilities to these ACE2 orthologs among the RaTG13 RBD mutants. Specifically, the H505Y mutant increased the binding affinity with the three chosen ACE2s by ~4.3-fold (mouse ACE2), ~4.6-fold (horse ACE2), and ~7.3-fold (intermediate horseshoe bat), respectively (Table S3). The D501N mutant displayed increased binding affinities to the ACE2s of human and intermediate horseshoe bat by 1 and 2 orders of magnitude, respectively, and decreased binding affinity with mouse ACE2 by ~14-fold (Table S3). The L486F mutant also displayed decreased interaction with mouse ACE2 (~1.8-fold) but favored the interaction with hACE2 (Table S3). Unlike D501N and L486F, which increased adaptation to the human ACE2 and decreased affinity for the mouse ACE2, F449Y showed benefit to the association to hACE2 (~2.7-fold) and mouse ACE2 (~2.5-fold) but abolished binding to its natural host ACE2 (Table S3; Figures 3A, 3B, and S4). The Y493Q mutant could not interact with ACE2s from mouse or intermediate horseshoe bat and displayed substantial decreased interaction with human ACE2 (~3.7-fold) but favored association with horse ACE2 (2.1-fold increase) (Figures 3A, 3B, and S4). The Y498Q mutant maintained binding to horse ACE2 with substantially decreased binding affinity (~13.6-fold), whereas the other four mutants (F449Y, L486F, Y493Q, and D501N) had little effect on the interaction with the horse receptor, with the K D fluctuating within a ~3-fold range (Figures 3A, 3B, and S4).
Differential amino acids on ACE2 orthologs may also have a significant effect on the interaction with viral RBDs. The complex structure revealed that the Y41 and K353 residues of hACE2 mediate the contacts with RaTG13 RBD D501. However, Y41 is relatively conserved among the 25 ACE2s analyzed, suggesting that it may not play an important role in determining binding affinity. In contrast, K353 is present in the majority of the ACE2 orthologs, but not in mouse and rat ACE2, where it has been substituted with H353. Thus, considering that K353 of hACE2 is also a key residue for binding with the SARS-CoV-2 RBD and that SARS-CoV-2 is unable to infect mice and rats, we introduced K353H into hACE2 to investigate the relevance for the ability of the SARS-CoV-2 RBD to bind to mouse and rat ACE2s. Through flow cytometry analysis, we found that the SARS-CoV-2 RBD (containing N501) is more likely to bind to cells expressing hACE2 (containing K353) compared with cells expressing the hACE2 K353H mutant. In contrast, RaTG13 RBD (containing D501) binds preferentially to the K353H mutant of hACE2 (Figure 4G). Introducing D501N to RaTG13 RBD shifted the binding preference of the parental RBD from H353 to K353 (Figure 4G). These results indicate that RaTG13 RBD D501 and SARS-CoV-2 RBD N501 favor binding with H353 and K353, respectively, suggesting that RaTG13 RBD D501 plays a key role in the ability to bind to mouse and rat ACE2s and providing an explanation for the inability of SARS-CoV-2 to interact with mouse and rat ACE2s.
The cross-reactive immune response of SARS-CoV-2 to RaTG13
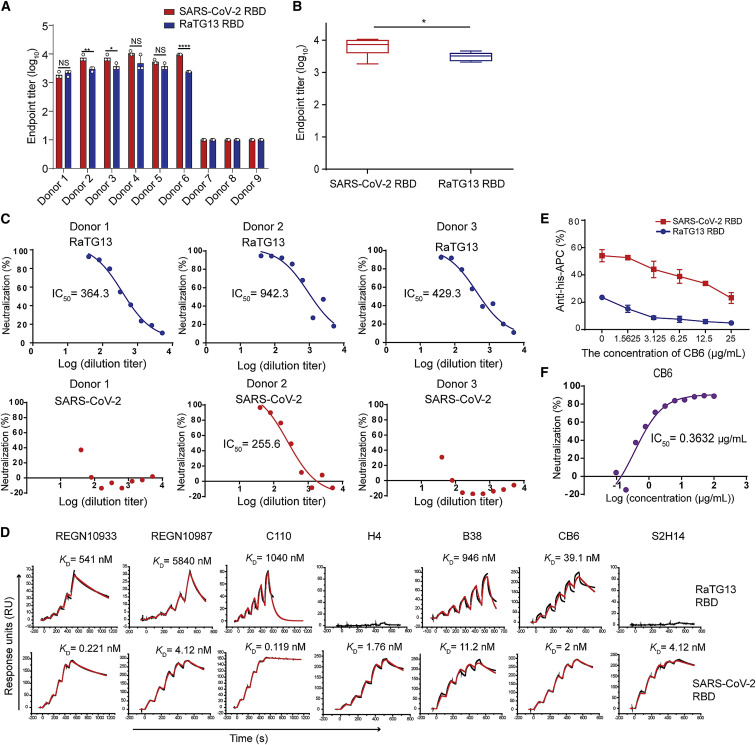
To assess the cross-reactive immune response of SARS-CoV-2 to RaTG13, we tested binding of the RaTG13 RBD with six COVID-19 convalescent sera by enzyme-linked immunosorbent assay (ELISA). The SARS-CoV-2 antibodies in all six convalescent donors’ sera could cross-recognize the RaTG13 RBD with an endpoint titer of 103–104, which was slightly lower than that of the SARS-CoV-2 RBD (Figures 5A and 5B). In contrast, sera from three healthy donors showed no binding to the RaTG13 RBD or SARS-CoV-2 RBD (Figures 5A and 5B). We then evaluated the neutralization activity of three COVID-19 convalescent sera against SARS-CoV-2 and RaTG13 pseudoviruses. Interestingly, although all three sera showed effective neutralization against the RaTG13 pseudovirus, only one completely neutralized the SARS-CoV-2 pseudovirus, whereas the other two only partially neutralized the SARS-CoV-2 pseudovirus at the highest concentration (Figure 5C).
Figure 5.
The cross-reactive immune response of SARS-CoV-2 to RaTG13
(A) Enzyme-linked immunosorbent assay (ELISA) measurement of the titers of SARS-CoV-2 RBD- and RaTG13 RBD-specific immunoglobulin G (IgG) in serum samples.
(B) Statistic of the titers of SARS-CoV-2 RBD- and RaTG13 RBD-specific IgG in six convalescent individuals’ serum samples. Data are presented as mean ± SD. ∗p < 0.05 (two-tailed unpaired t test).
(C) Neutralization of the RaTG13 or SARS-CoV-2 pseudovirus by antisera from three convalescent individuals.
(D) SPR characterization of the binding affinities of the SARS-CoV-2 RBD or RaTG13 RBD with the indicated antibodies.
(E) The ratio of SARS-CoV-2 RBD- and RaTG13 RBD-binding cells among Baby Hamster Syrian Kidney cells with hACE2 expression (BHK-hACE2 cells) in the presence of CB6. Data represent the results of three replicates. Data are presented as mean ± SD.
(F) Neutralization of the RaTG13 or SARS-CoV-2 pseudovirus by the CB6 antibody.
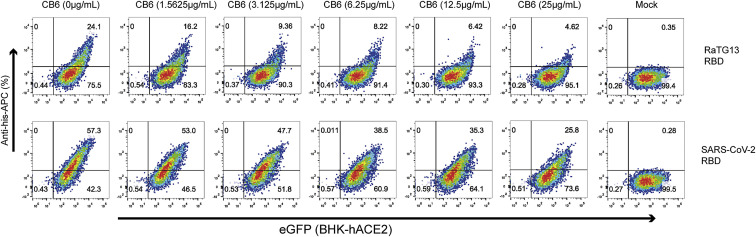
To assess the ability of SARS-CoV-2 RBD mAbs to cross-react with the RaTG13 RBD, we tested the binding affinity of six SARS-CoV-2 RBD mAbs (REGN10933, REGN10987, C110, H4, B38, CB6, and S2H14) with the RaTG13 RBD (Barnes et al., 2020; Hansen et al., 2020; Piccoli et al., 2020; Shi et al., 2020; Wu et al., 2020c) to the RaTG13 RBD. Our data showed that CB6 binds to the RaTG13 RBD with the highest binding affinity (39.1 nM) among these six mAbs (Figure 5D). The binding affinities of REGN10933, REGN10987, C110, and B38 with the RaTG13 RBD were decreased 100- to 10,000-fold compared with their corresponding affinities for the SARS-CoV-2 RBD (Figure 5D). H4 and S2H14, which bind to the SARS-CoV-2 RBD with high affinities (1.76 nM and 4.12 nM, respectively), lost the ability to interact with the RaTG13 RBD (Figure 5D). Flow cytometry analysis indicates that, in the presence of the CB6 antibody, the numbers of events in which hACE2-expressing cells bind to SARS-CoV-2 or RaTG13 were decreased substantially in a CB6 concentration-dependent manner (Figures 5E and S5 ). Finally, by applying CB6 to the RaTG13 pseudovirus, we found that CB6 could effectively neutralize the RaTG13 pseudovirus, with a half maximal inhibitory concentration (IC50) of 0.3632 μg/mL (Figure 5F).
Figure S5.
Blocking of the binding between HeLa-hACE2 cells and the SARS-CoV-2 RBD or RaTG13 RBD by CB6, related to Figure 5
CB6 was serially diluted by 2 folds, followed by incubation with SARS-CoV-2 RBD or RaTG13 RBD. Blocking efficacies were analyzed through staining BHK-hACE2 cells. Cells stained with SARS-CoV-2 RBD or RaTG13 RBD proteins was used as a positive control. Unstained HeLa-hACE2 cells were used as negative controls.
Discussion
Increasing studies indicate that SARS-CoV-2 shares a common ancestor with bCoVs, and bats are suspected to be the reservoir host. So far, RaTG13, sequenced from bats, is the virus most closely related to SARS-CoV-2 (Zhou et al., 2020b). Quantification of the association between RaTG13 and its human receptor and those of wild and domestic animals is critical for evaluating the potential inter-species transmission of RaTG13 to control and prevent related infectious diseases, especially in humans (Su et al., 2016).
The binding affinity between the RaTG13 RBD and hACE2 is approximately 70-fold lower than that between the SARS-CoV-2 RBD and hACE2 because of the substantially reduced area of the buried surface and decreased network of hydrogen bonds in the RaTG13 RBD-hACE2 complex structure. In addition, substituting six residues on the RaTG13 RBD (F449Y, L486F, Y493Q, Y498Q, D501N, and H505Y) with their counterparts on the SARS-CoV-2 RBD rescued the binding affinity of RaTG13 with hACE2. Such observation is consistent with a recent report that identified the importance of these RaTG13 mutations for binding to hACE2 (Conceicao et al., 2020). Here we provide substantially more data indicating the effect of these mutations on binding with ACE2s from mouse, horse, and intermediate horseshoe bat.
The cryoelectron microscopy (cryo-EM) structure of the RaTG13 S protein was reported recently, and through structural alignments of the RaTG13 S protein and SARS-CoV-2 S protein, RaTG13 RBD Y493 was speculated to confer a potential steric clash to hACE2 (Wrobel et al., 2020). However, the structure of the RaTG13 RBD in complex with hACE2 reported here indicates that RaTG13 RBD Y493 forms a hydrogen bond with hACE2 K31.
Most of the 24 ACE2 orthologs have weaker binding affinity for the RaTG13 RBD than the SARS-CoV-2 RBD, with ACE2s from mouse and rat as two exceptions. Through analysis of mutations, we identified D501 on RaTG13 as the key residue to bind to ACE2s of mouse and rat, which also explains why SARS-CoV-2 (N501) is unable to interact with mouse and rat ACE2s. N501Y was identified in a mouse adaptive SARS-CoV-2 virus strain that enhances the interaction between SARS-CoV-2 RBD and mouse ACE2 (Gu et al., 2020). SARS-CoV-2 carrying the N501Y mutation has been spreading widely in many countries with different mutations, including VOCs 20I/501Y.V1 (United Kingdom), 20H/501Y.V2 (South Africa), and 20H/501Y.V3 (Brazil/Japan), indicating that this mutation is the “hotspot” for host adaptation. It is believed that human infection with novel CoVs is related to a “hot jump” of the viruses from reservoir (bat)-intermediate-host. Our work here with bat-origin RaTG13 further confirms that residue 501 is the key amino acid position in the S protein for bat-intermediate-host adaptation, which implicates that RaTG13 has the potential for interspecies transmission.
Among the ACE2 orthologs from humans and 24 animals, horse ACE2 has the highest binding affinity with the RaTG13 RBD even after introduction of the Y498Q substitution on the RaTG13 RBD, which abolishes binding with ACE2s from human, mouse, and intermediate horseshoe bat. Our previous study indicates that horse ACE2 also possesses high binding affinity for the SARS-CoV-2 RBD (Wu et al., 2020b). However, so far, no evidence indicating susceptibility of horse ACE2 to SARS-CoV-2 has been reported. Other host factors may evolve, and further studies are needed. Because non-SARS-CoV-2 beta CoVs are widespread in horses (Haake et al., 2020), which could potentially result in recombination between CoVs, horses and other species with similar ACE2s, especially ones in close proximity to bats, should be monitored closely to prevent inter-species CoV transmission, which could ultimately result in human infection.
Interestingly, our data, together with previous reports, indicate that RaTG13 possesses different binding spectra to ACE2 orthologs compared with those of SARS-CoV-2, suggesting that RaTG13 has undergone diverse evolutionary routes compared with SARS-CoV-2. For example, although civets and pangolins have been proposed to play roles in transmission of SARS-CoV and SARS-CoV-2, respectively (Guan et al., 2003; Lam et al., 2020; Xiao et al., 2020), our data showed that the ACE2 orthologs of these species do not interact with the RaTG13 RBD. Our previous work showed that Rodentia (mouse and rat) ACE2s were unable to bind to the SARS-CoV-2 RBD (Wu et al., 2020b), whereas the data presented here indicate that they can bind to the RaTG13 RBD.
The intermediate horseshoe bat is the host from which RaTG13 was originally sequenced, but its ACE2 only binds weakly to the RaTG13 RBD. Interestingly, a similar phenomenon has been observed in other CoVs, including SARS-CoV (Wu et al., 2020b), SARS-related CoVs (Guo et al., 2020), and MERS-CoV (Yuan et al., 2020), possibly because of the unique immune system of bats. Specifically, some pivotal receptors for natural killer (NK) cell activation are not expressed in bat NK cells, and the signaling pathway of type I interferons (IFNs) is suppressed by reduction of the stimulator of IFN genes (STING) pathway in bats (Pavlovich et al., 2018; Xie et al., 2018; Zhang et al., 2013). Besides the suppressed innate immune response, bats also have a special adaptive immune system, exemplified by the unusual peptide presentation features brought by specific 3-amino-acid insertion in major histocompatibility complex (MHC) class I (Lu et al., 2019). In addition, polymorphisms of Rhinolophus sinicus ACE2 confer varied susceptibility to SARS-related CoV infection (Guo et al., 2020). The polymorphism of ACE2 may also exist in intermediate horseshoe bats, which show different binding capacities to the RaTG13 RBD.
RaTG13 has a broad host range, which suggests that acquisition of other adaptive mutations can further broaden its host range. Thus, RaTG13 and related CoVs can possibly jump the species barrier to humans and have pandemic potential when they acquire adaptive mutations required for human infection. Evaluating the cross-reactive immune response of SARS-CoV-2 to RaTG13 revealed that serum samples from SARS-CoV-2 convalescent donors could more efficiently cross-neutralize the RaTG13 pseudovirus than the SARS-CoV-2 pseudovirus, probably because of the lower binding affinity between hACE2 and the RaTG13 RBD than the SARS-CoV-2 RBD. Thus, lower concentrations of neutralizing antibodies are required to block binding of the receptor to the RaTG13 RBD than to the SARS-CoV-2 RBD; therefore, fewer neutralizing antibodies are needed to neutralize the RaTG13 pseudovirus than the SARS-CoV-2 pseudovirus. Moreover, our studies using SARS-CoV-2 RBD monoclonal antibodies show that many of these antibodies interact with RaTG13 with a 103-fold reduced binding affinity. One exception with high binding affinity with RaTG13 is CB6, a SARS-CoV-2 mAb identified previously by our group that could bind to RaTG13 with high affinity and neutralize RaTG13 pseudovirus infection of HeLa-hACE2s.
Limitations of the study
Our approach for analyzing the host range of RaTG13 by studying the interaction between the RaTG13 RBD and ACE2 is limited because we only examine the binding affinity for the ACE2 receptor. RaTG13 may potentially infect cells through other receptors besides ACE2. Further studies are required to identify whether RaTG13 utilizes other receptors for cell entry. Cell entry of SARS-CoV-2 also requires S protein priming by cellular proteases, such as transmembrane protease serine 2 (TMPRSS2) and furin (Hoffmann et al., 2020), which should be addressed in the future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli (E. coli) strain DH5α | TIANGEN | Cat# CB101-02 |
| Escherichia coli (E. coli) strain BL21 (DE3) | Novagen | Cat# 69450 |
| MAX Efficiency DH10Bac Competent E. coli | Invitrogen | Cat# 10361-012 |
| Chemicals, antibodies, and recombinant proteins | ||
| PEI | Alfa | A04043896-1g |
| Anti-His/APC | Miltenyi Biotec | Cat# 130-119-820; RRID: AB_2751870 |
| SARS-CoV-2 RBD protein with his-tag, spike residues 319-541, accession number: EPI_ISL_402119 | This paper | N/A |
| Goat anti-human IgG-HRP | Thermo Fisher | Cat# 31410 |
| SARS-CoV-2 RBD-F449Y | This paper | N/A |
| SARS-CoV-2 RBD-L486F | This paper | N/A |
| SARS-CoV-2 RBD-Q493Y | This paper | N/A |
| SARS-CoV-2 RBD-Q498Y | This paper | N/A |
| SARS-CoV-2 RBD-D501N | This paper | N/A |
| SARS-CoV-2 RBD-H505Y | This paper | N/A |
| SARS-CoV RBD protein with his-tag, spike residues 306-527, accession number: NC_004718 | This paper | N/A |
| cACE2 protein, residues 18-740, accession number: NP_001034545.1 | This paper | N/A |
| Critical commercial assays | ||
| HisTrap HP 5 mL column | GE Healthcare | Cat# 17524802 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat# 28989335 |
| Series S Sensor Chip CM5 | GE Healthcare | Cat# 29149603 |
| Sensor Chip Protein A | GE Healthcare | Cat# 29127556 |
| Membrane concentrator | Millipore | UFC901096 |
| Deposited data | ||
| RaTG13 RBD/hACE2 complex | This paper | Protein Data Bank: 7DRV |
| Experimental models: Cell lines | ||
| Sf9 Cells, SFM Adapted | Invitrogen | Cat# 11496015 |
| High Five cells | Invitrogen | Cat# B85502 |
| HEK293T cells | ATCC | ATCC CRL-3216 |
| Recombinant DNA | ||
| pEGFP-N1 | MiaoLingPlasmid | Cat# P0133 |
| pEGFP-N1-hACE2, accession number: BAJ21180 | This paper | N/A |
| pEGFP-N1-monkey ACE2, accession number: A0A2K5X283 | This paper | N/A |
| pEGFP-N1-rabbit ACE2, accession number: G1TEF4 | This paper | N/A |
| pEGFP-N1-mouse ACE2, accession number: Q8R0I0 | This paper | N/A |
| pEGFP-N1-rat ACE2, accession number: Q5EGZ1 | This paper | N/A |
| pEGFP-N1-malayan pangolin ACE2, accession number: XP_017505746 | This paper | N/A |
| pEGFP-N1-catACE2, accession number: Q56H28 | This paper | N/A |
| pEGFP-N1-civet ACE2, accession number: Q56NL1.1 | This paper | N/A |
| pEGFP-N1-fox ACE2, accession number: XP_025842512.1 | This paper | N/A |
| pEGFP-N1-dog ACE2, accession number: J9P7Y2 | This paper | N/A |
| pEGFP-N1-raccoon dog ACE2, accession number: ABW16956.1 | This paper | N/A |
| pEGFP-N1-horse ACE2, accession number: F6V9L3 | This paper | N/A |
| pEGFP-N1-pig ACE2, accession number: A0A220QT48 | This paper | N/A |
| pEGFP-N1-wild Bactrian camel ACE2, accession number: XP_006194263.1 | This paper | N/A |
| pEGFP-N1-alpaca ACE2, accession number: XP_006212709.1 | This paper | N/A |
| pEGFP-N1-bovine ACE2, accession number: Q58DD0 | This paper | N/A |
| pEGFP-N1-goat ACE2, accession number: XP_005701129.2 | This paper | N/A |
| pEGFP-N1-sheep ACE2, accession number: W5PSB6 | This paper | N/A |
| pEGFP-N1-intermediate horseshoe bat ACE2, accession number: W5PSB6 | This paper | N/A |
| pEGFP-N1-Least horseshoe bat ACE2, accession number: W5PSB6 | This paper | N/A |
| pEGFP-N1-little brown bat ACE2, accession number: G1PXH7 | This paper | N/A |
| pEGFP-N1-fulvous fruit bat ACE2, accession number: D8WU01 | This paper | N/A |
| pEGFP-N1-greater horseshoe bat ACE2, accession number: B6ZGN7 | This paper | N/A |
| pEGFP-N1-big-eared horseshoe bat ACE2, accession number: XP_007538670.1 | This paper | N/A |
| pEGFP-N1-lesser hedgehog tenrec ACE2, accession number: XP_004710002.1 | This paper | N/A |
| pFastbac1 | Invitrogen | 10360014 |
| pFastbac-SARS-CoV-2 RBD -His, residues 319-541, accession number: EPI_ISL_402119 | This paper | N/A |
| pFastbac- SARS-CoV RBD -His, residues 306-527, accession number: NC_004718 | This paper | N/A |
| pCAGGS | MiaoLingPlasmid | Cat# P0165 |
| pET21a | Novagen | Cat# 69740 |
| pET21a-hACE2, residues 18-740, accession number: NP_001034545.1 | This paper | N/A |
| Software and algorithms | ||
| PyMOL software | Molecular Graphics System, Version 1.8 Schrödinger | https://pymol.org/2/ |
| BIAcore 8K Evaluation software | GE Healthcare | N/A |
| FlowJo V10 | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads |
| Motioncor2 | Zheng et al., 2017 | N/A |
| COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/peemsley/coot/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| MolProbity | N/A | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Qihui Wang (wangqihui@im.ac.cn).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
The number of replicates carried out for each experiment is described in the figure/table legends.
Data and code availability
The atomic coordinates for the crystal structure of the RaTG13 RBD and hACE2 complex have been deposited in the Protein Data Bank (https://www.rcsb.org/) (PDB: 7DRV).
Experimental model and subject details
Cells
HEK293T cells (ATCC CRL-3216), HeLa cells (ATCC CCL-2), BHK-21 cells (ATCC CCL-10), HeLa-hACE2 cells (prepared in this study), and BHK-hACE2 (prepared in this study) were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Sera samples
The information of convalescent and healthy donors is included in the Table S5.
Method details
Gene cloning
The full-length coding sequence of the 25 ACE2 orthologs was synthesized and cloned into the pEGFP-N1 vector for flow cytometry (Table S1). The extracellular domain of these 25 ACE2 orthologs fused with the Fc domain of mouse IgG (mFc) were cloned into the pCAGGS vector for protein expression. The coding sequences of RaTG13 RBD (residues 319-541, GenBank: QHR63300.2), SARS-CoV-2 RBD (residues 319-541, GISAID: EPI_ISL_402119), MERS-CoV RBD (residues 367-606, GenBank: JX869059) and hACE2 (residues 19-615, NCBI Reference Sequence: NP_001358344.1) were cloned into pFastBac vectors (Dai et al., 2020; Lu et al., 2013). The wild-type RaTG13 RBD (residues 319-541, GenBank: QHR63300.2) and mutated RaTG13 RBDs (F449Y, L486F, Q493Y, Q498Y, D501N and H505Y) were cloned in pCAGGS vectors. The coding sequence of RaTG13 S (residues 1-1233) and SARS-CoV-2 S (residues 1-1255) were cloned into pCAGGS vectors. The variable region of MAbs, REGN10933 (PDB: 6XDG), REGN10987 (PDB: 6XDG), C110 (PDB: 7K8P), and S2H14 (PDB: 4JX3) fused with the constant region of IgG1 were cloned into pCAGGS vectors. The extracellular domain of PD-L1 was constructed as previously described.
Protein expression and purification
The 25 ACE2 orthologs fused with mFc and 7 MAbs were expressed and purified from the culture supernatants of HEK293F cells using a Protein A affinity column (GE Healthcare) and further purified by gel filtration using a Superdex™ 200 10/300 GL (GE Healthcare). Purified proteins were stored in a buffer containing 20 mM Tris-HCl and 150 mM NaCl (pH 8.0). Proteins for SPR assay were transferred to PBST (1.8 mM KH2PO4, 10 mM Na2HPO4 (pH 7.4), 137 mM NaCl, 2.7 mM KCl, and 0.005% (v/v) Tween 20) buffer.
The recombinant baculovirus-expressed proteins were expressed and purified as previously described (Tan et al., 2017). Briefly, the RaTG13 RBD, SARS-CoV-2 RBD, MERS-CoV RBD, and hACE2 proteins were expressed by infecting Hi5 cells with recombinant baculovirus containing the corresponding coding sequences. The RaTG13 RBD/hACE2 complex protein was expressed by co-infecting Hi5 cells with recombinant baculovirus containing the coding sequence of RaTG13 RBD and hACE2. The wild-type RaTG13 RBD (residues 319-541, GenBank: QHR63300.2) and mutated RaTG13 RBDs (F449Y, L486F, Q493Y, Q498Y, D501N and H505Y) cloned in pCAGGS were expressed in HEK293F cells. Cell culture supernatants were collected, filtered with a 0.22 μm filter, purified by His-Trap HP column (GE Healthcare), and further purified by Superdex™ 200 Increase 10/300 GL column (GE Healthcare). Purified proteins were stored in protein buffer (20 mM Tris-HCl, pH 8.0 and 150 mM NaCl).
Flow cytometry assay
Plasmids containing the 25 ACE2 orthologs fused with eGFP were transfected into HEK293T cells. Cells were harvested after 24 h post-transfection, washed thrice, and incubated with 100 μL 30 μg/mL test proteins (SARS-CoV-2 RBD, RaTG13 RBD and MERS RBD with histidine tag) at 37°C for 30 min. Subsequently, the cells were washed thrice with PBS to remove redundant protein and incubated with anti-His/APC antibodies (1:500, Miltenyi Biotec) at 37°C for 30 minutes. Finally, cells were washed for thrice before being analyzed using a BD FACS Calibur Flow Cytometer (BD Biosciences).
The plasmid containing recombinant hACE2 or mutated ACE2s was stably transfected into BHK-21 cells (BHK-hACE2). A mixture containing SARS-CoV-2 RBD (10 μg/mL), RaTG13 RBD (10 μg/mL) and CB6 antibody were pre-incubated at 4°C for 30 min, and then incubated with the BHK-hACE2 cells at 4°C for 30 min. Subsequently, cells were washed with PBS thrice and stained with APC mouse anti-his secondary antibody for 30 min before being analyzed using BD FACS Canto Flow Cytometer (BD Biosciences). The data of all samples were analyzed using FlowJo 7.6 (TreeStar Inc., Ashland, OR, USA).
SPR analysis
The 25 ACE2-mFC fusion proteins were transferred into HBST buffer (20 mM HEPES (pH 7.4), 150 mM NaCl, and 0.005% (v/v) Tween 20) and immobilized on the CM 5 chip (Table S2). Then, serially diluted wild-type or mutated RaTG13 RBD proteins flowed over the chip in HBST buffer. PD-L1 protein was used as negative control. Binding affinities were measured using a BIAcore 8K (GE Healthcare) at 25°C in the single-cycle mode. Binding kinetics were analyzed with Biacore™ Insight software (GE healthcare) using a 1:1 Langmuir binding model.
The seven recombinant antibodies (2 μg/mL) were first captured on flow cell 2 of the protein A sensor chip (GE Healthcare) at more than 500 response units (Table S4). Flow cell 1 was used as the negative control. Then, serially diluted SARS-CoV-2 RBD or RaTG13 RBD proteins flowed over the chip in PBST buffer. Response Units (RU) were measured with a BIAcore 8K (GE Healthcare) at 25°C in single-cycle mode. The antibodies were regenerated with 10 mM glycine-HCl (pH 1.5). The equilibrium dissociation constants (K D) of each pair of interactions were calculated using BIAcore® 8K Evaluation Software (GE Healthcare) by fitting to a 1:1 Langmuir binding model.
Enzyme-linked immunosorbent assay
The SARS-CoV-2 and RaTG13 RBD proteins were first diluted to 2 μg/mL with 0.05 M carbonate-bicarbonate buffer (pH 9.6) and then coated onto 96-well ELISA plates (Corning, USA) by overnight incubation at 4°C. Plates were blocked in 5% skim milk in PBST and incubated with serially diluted serum samples. Each serum sample was prepared in triplicate. Subsequently, plates were incubated with goat anti-human IgG-HRP antibody (Thermo Fisher, USA) and developed with 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate. The reactions were stopped with 2 M sulfuric acid. Absorbance at 450 nm was measured using a microplate reader (PerkinElmer, USA). The endpoint titer was defined as the highest reciprocal dilution of serum to give an absorbance greater than 2.1-fold that of the background values. Antibody titers below the limit of detection were determined to be half the limit of detection.
Production and quantification of pseudoviruses
The RaTG13 and SARS-CoV-2 pseudoviruses were constructed with a GFP-encoding replication-deficient vesicular stomatitis virus (VSV) vector backbone (VSV-ΔG-GFP) and the coding sequence of corresponding spike proteins, as previously described (Li et al., 2020a; Muik et al., 2021). HEK293T cells were transfected with 30 μg of the spike protein expression plasmids. The VSV-ΔG-GFP pseudovirus was added 24h post-transfection. The inoculum was removed after incubation for 1 h at 37°C. The culture medium were then changed into DMEM supplemented with 10% FBS and 10 μg/mL of anti-VSV-G antibody (I1Hybridoma ATCC® CRL2700) after washing the cells with PBS. The pseudoviruses were harvested 20 h post-inoculation, passed through a 0.45 μm filter (Millipore, Cat#SLHP033RB) before aliquoted and stored at −80°C.
All pseudoviruses were treated with 0.5U/μL BaseMuncher endonuclease (Abcam, ab270049) for 1.5 h at 37°C to remove unpackaged RNA before quantification. Viral RNA was extracted (Bioer Technology, Cat# BYQ6.6.101711-213) and quantified by quantitative RT-PCR (qPCR) using 7500 fast Real-Time PCR System (Applied Biosystems) with the primers and probe for detecting the P protein coding sequence of VSV.
Pseudovirus infection assay
The pseudovirus particles of SARS-CoV-2, wild-type RaTG13 and mutated RaTG13 were normalized to the same amount for quantitation by qRT-PCR. Then, 100 μL of each pseudovirus was added to each well of the 96 well plate containing HeLa-hACE2 cells. Untransfected HeLa cells were used as controls. The plates were imaged 15 h post-transfection. The imaging and analysis of fluorescent cells was determined using a CQ1 confocal image cytometer (Yokogawa, Japan). Each group contains 6 replicates.
Neutralization assay
Neutralization assay was performed as previously described (Dai et al., 2020). Briefly, 50 μL of serially diluted human sera or monoclonal antibody were incubated with 50 μL of pseudoviruses at 37°C for 1 h before being added onto pre-plated HeLa-hACE2 cells. The transducing units (TU) numbers were calculated on a CQ1 confocal image cytometer (Yokogawa).
Crystallization
The sitting-drop method was used to obtain high-resolution RaTG13 RBD/hACE2 complex crystals. In detail, the purified complex proteins were concentrated to 10 mg/mL. Then, 0.8 μL protein was mixed with 0.8 μL reservoir solution. The resulting solution was sealed and equilibrated against 100 μL of the reservoir solution at 18°C. High-resolution RaTG13 RBD/hACE2 complex crystals were grown in 0.1 M succinic acid (pH 7.0), 0.1 M bicine pH 8.5, and 30% v/v polyethylene glycol monomethyl ether 550.
Data collection and structure determination
Reservoir solution supplemented with 20% (v/v) glycerol was prepared as anti-freezing buffer for freezing crystals. Crystals were picked up from the groove by using a mini loop and soaked in anti-freezing buffer for a few seconds. Then, crystals were picked up and frozen by soaking in liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) BL19U. The dataset was processed with HKL2000 software as previously described (Otwinowski and Minor, 1997). The structure of the RaTG13 RBD/hACE2 complex was determined by the molecular replacement method using Phaser (Adams et al., 2010) with a previously reported complex structure of the SARS-CoV-2-RBD complex with human ACE2 (PDB: 6LZG). The atomic models were completed using Coot (Emsley and Cowtan, 2004) and refined with phenix.refine in Phenix (Adams et al., 2010), and the stereochemical qualities of the final models were assessed using MolProbity . Data collection, processing, and refinement statistics are summarized in Table 1. All structural figures were generated using Pymol software (https://pymol.org/2/).
Quantification and statistical analysis
Binding studies
KD values for SPR experiments were obtained with BIAcore 8K Evaluation Software (GE Healthcare), using a 1:1 binding model. The values shown are the mean ± SD of three replicates.
Flow cytometry analysis
All experiments were performed in triplicate; one representative result was shown in Figures 2, 3, and 4.
Acknowledgments
We are grateful to William Ningda Gao for revising the manuscript and Hillary Yida Zhang for constructing the RaTG13 6-mutant plasmid. We are grateful to the Pathogenic Microbiology and Immunology Public Technology Service Center for its support regarding the flow cytometry assay. We acknowledge the staff of beamline BL19U1 at the Shanghai Synchrotron Radiation Facility for assistance during data collection. We also thank Y. Chen, B. Zhou, and Z. Yang from the Institute of Biophysics, Chinese Academy of Sciences, for technical support with the SPR assay. We acknowledge Z. Shi from Wuhan Institute of Virology, Chinese Academy of Sciences, for providing the ACE2 sequence of Rhinolophus affinis. This work was supported by the Ministry of Science and Technology of the People's Republic of China; the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010202, XDB29040203, and XDB37030204); an intramural special grant for SARS-CoV-2 research from the Chinese Academy of Sciences, National Natural Science Foundation of China (81922044), and the National Science and Technology Major Project (2018ZX10101004-001 and 2018ZX09711003-002-001). Q.W. is supported by the Youth Innovation Promotion Association of the CAS (2018119). G.F.G is supported by the Yanqi Lake Meeting organized by the academic divisions of the CAS.
Author contributions
J.Q., G.F.G., and Q.W. initiated and designed the project. K.L. and X.P. purified the proteins and grew the crystals with help from Y.M., Y.Z., L.W., and C.S. X.P. and L.L. performed the SPR analysis with help from Y.J., S.N., and C.Q. K.L., X.P., and L.L. conducted the flow cytometry assay with help from A.Z., Q.C., and L.W. F.Y. and J.Q. collected the structural data and solved the structures. K.L., F.Y., S.T., and J.Q. analyzed the data and prepared the figures with help from D.M. and X.M. A.Z., P.H., and X.Z. performed pseudovirus-related assays. D.L. and Z.C. collected the serum samples. K.L., P.D., Q.W., and G.F.G. wrote and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.05.031.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao C., Thakur N., Human S., Kelly J.T., Logan L., Bialy D., Bhat S., Stevenson-Leggett P., Zagrajek A.K., Hollinghurst P., et al. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 2020;18:e3001016. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733.e11. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gao G.F. From “A”IV to “Z”IKV: attacks from emerging and re-emerging pathogens. Cell. 2018;172:1157–1159. doi: 10.1016/j.cell.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guo H., Hu B.J., Yang X.L., Zeng L.P., Li B., Ouyang S., Shi Z.L. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes. J. Virol. 2020;94:e00902–e00920. doi: 10.1128/JVI.00902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake C., Cook S., Pusterla N., Murphy B. Coronavirus infections in companion animals: virology, epidemiology, clinical and pathologic features. Viruses. 2020;12:1023. doi: 10.3390/v12091023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Qi J., Song H., Wang Q., Zhang Y., Wu Y., Lu G., Yuen K.Y., Shi Y., Gao G.F. Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein. Virology. 2017;507:101–109. doi: 10.1016/j.virol.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Chmura A.A., Field H.E., Zambrana-Torrelio C., Epstein J.H., et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11:4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang H., Tang X., Fang S., Ma D., Du C., Wang Y., Pan H., Yao W., Zhang R., et al. SARS-CoV-2 and three related coronaviruses utilize multiple ACE2 orthologs and are potently blocked by an improved ACE2-Ig. J. Virol. 2020;94:e01283. doi: 10.1128/JVI.01283-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Tan S., Niu S., Wang J., Wu L., Sun H., Zhang Y., Pan X., Qu X., Du P., et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020216118. e2020216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Liu K., Zhang D., Yue C., Lu Q., Cheng H., Wang L., Chai Y., Qi J., Wang L.F., et al. Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats. PLoS Biol. 2019;17:e3000436. doi: 10.1371/journal.pbio.3000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M., Tiwari R., Chaicumpa W. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pavlovich S.S., Lovett S.P., Koroleva G., Guito J.C., Arnold C.E., Nagle E.R., Kulcsar K., Lee A., Thibaud-Nissen F., Hume A.J., et al. The egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell. 2018;173:1098–1110.e18. doi: 10.1016/j.cell.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Smith I., Wang L.F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Zhang H., Chai Y., Song H., Tong Z., Wang Q., Qi J., Wong G., Zhu X., Liu W.J., et al. An unexpected N-terminal loop in PD-1 dominates binding by nivolumab. Nat. Commun. 2017;8:14369. doi: 10.1038/ncomms14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Zhao X., Ma X., Wang W., Niu P., Xu W., Gao G., Wu G. A novel coronavirus genome identified in a cluster of pneumonia cases-Wuhan, China 2019-2020. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- The 2019-nCoV Outbreak Joint Field Epidemiology Investigation Team. Li Q. Notes from the Field: An Outbreak of NCIP (2019-nCoV) Infection in China — Wuhan, Hubei Province, 2019−2020. China CDC Weekly. 2020;2:79–80. [PMC free article] [PubMed] [Google Scholar]
- Wang L., Su S., Bi Y., Wong G., Gao G.F. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol. 2018;26:466–470. doi: 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Chen Q., Liu K., Wang J., Han P., Zhang Y., Hu Y., Meng Y., Pan X., Qiao C., et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Su J., Niu S., Chen Q., Zhang Y., Yan J., Shi Y., Qi J., Gao G., Wang Q. Molecular basis of pangolin ACE2 engaged by COVID-19 virus. Chin. Sci. Bull. 2021;66:73–84. [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Xie J., Li Y., Shen X., Goh G., Zhu Y., Cui J., Wang L.F., Shi Z.L., Zhou P. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23:297–301.e4. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Qi J., Peng R., Li C., Lu G., Yan J., Wang Q., Gao G.F. Molecular basis of binding between Middle East respiratory syndrome coronavirus and CD26 from seven bat species. J. Virol. 2020;94:e01387. doi: 10.1128/JVI.01387-19. e01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W., et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates for the crystal structure of the RaTG13 RBD and hACE2 complex have been deposited in the Protein Data Bank (https://www.rcsb.org/) (PDB: 7DRV).