ABSTRACT
Immunotherapy that block PD-1–PD-L1 pathway can induce durable tumor control and result in the long-term survival of patients with advanced bladder cancers. However, these responses only occur in a subset of patients. We study gene expression profiles in 1763 muscle-invasive bladder cancers (MIBCs) and 11,835 solid tumors from TCGA. We establish an immune-based classification on the basis of the composition of the tumor microenvironment and identify six distinct phenotypes. The class F was characterized by a strong tertiary lymphoid structures (TLSs) related gene expression signature. Pan-cancer gene expression analysis of tertiary lymphoid structure markers in 11,835 solid tumors from TCGA unveiled the heterogeneity of TLSs abundance both within and between human cancer types. The class F group demonstrated improved survival and a high response rate to PD1 blockade. This work confirms the immune subtypes in patients with MIBC, and unravels the potential of TLS signatures to guide clinical decision-making and treatments.
KEYWORDS: Tertiary lymphoid structures, bladder cancer, immunotherapy, tumor microenvironment, microarray gene expression
1. Introduction
Immunotherapy that block PD-1–PD-L1 pathway can induce durable tumor control and result in the long-term survival of patients with metastatic urothelial cancer .1–4 However, these responses only occur in a subset of patients. Clarifying the determinants of response and resistance is key to improving survival and exploring new therapeutic target. Thorsson et al.5 performed immunogenomics analysis on more than 10,000 tumors, identified six immune subtypes covering multiple cancer types, and based on this, determined the immune response patterns that affected the prognosis. Recently published works showed the potential role of B cells and tertiary lymphoid structures (TLSs) in the response to immunotherapy response in melanoma6,7 and sarcoma.8 A prior study with comprehensive analysis of tumor immune microenvironment of muscle-invasive bladder cancers (MIBCs) patients9 showed that high TLSs amounts were associated with an inflamed phenotype and improved patient survival. These important studies add to the immunotherapy toolbox by providing new methods for predicting prognosis.
Here, we developed a new classification of muscle-invasive bladder cancers (MIBCs), based on the composition of the tumor microenvironment (TME) in large cohorts of MIBC, using the microenvironment cell populations (MCP)-counter method. We found that the TLSs signature – a hallmark of an immune-high class we called F, correlated with an improved survival of patients with MIBC. Finally, we showed that class F exhibited the highest response rate to PD1 blockade therapy and improved overall survival.
2. Results
2.1. Immune classification of MIBC
The composition of the tumor microenvironment (TME) from 14 independent discovery MIBC datasets (n = 1763, Table S1-2, Fig. S1) with publicly available gene expression profiles were analyzed by MCP-counter package .10 In order to select the best number of clusters, we used the ConsensusClusterPlus software package to evaluate the stability of the cluster, which supported the existence of 6 robust subtypes of MIBC in a meta-cohort (Figure 1a–c). Through this analysis, an immune-based MIBC classification was established, and the tumor was classified into one of six MIBC immune classes (MIC), labeled A, B, C, D, E and F, with highly distinct profiles (Figure 1d). The TME composition differs significantly between MICs (Figure 1d, Fig. S2). MIC F, exhibited a strong TLS related gene expression signature with an inflamed phenotype, was characterized by the highest expression of genes specific to immune populations such as B lineage (Figure S2A), CD8 + T cells (Figure S2B), cytotoxic lymphocytes (Figure S2C), myeloid dendritic cells (Figure S2F), natural killer cells (Figure S2I), and T cells (Figure S2J). MIC E, moderate immune and TLS related gene expression signature, was characterized by the moderate expression of genes specific to immune populations such as B lineage and myeloid dendritic cells, CD8 + T cells, natural killer cells and cytotoxic lymphocytes, high expression of genes specific to fibroblasts (Figure S2G). MIC C, immune desert, was characterized by the lowest expression of gene signatures related to immune cells, as well as low fibroblasts. MICs A was characterized by high expression of genes specific to fibroblasts and immune-low profile. MICs B and D were characterized by heterogeneous but generally immune-low and immune-high profiles, respectively.
Figure 1.
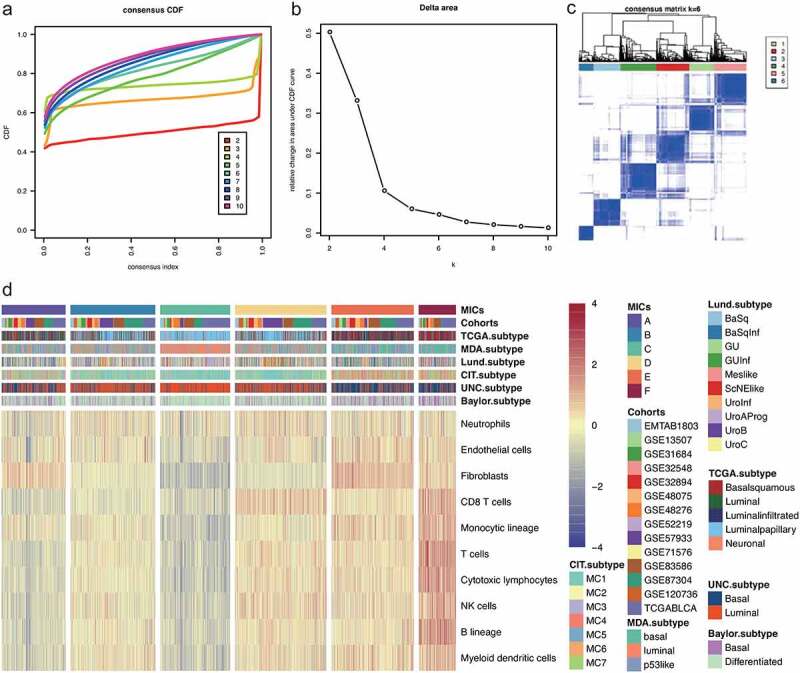
Consensus clustering based on 10 TME cell types of 1763 MIBCs in 14 cohorts. (a) The cumulative distribution function (CDF) shows the cumulative portion of all sample co-clustering at the given consensus index (1.0 = co-clustered 100% of the time) for each tested k. Generally, a more step-like curve is associated with more stable clustering. The relative increase in total area under the CDF is shown, where generally a greater area may suggest more stable clustering. (b) Delta area curve of consensus clustering, indicating the relative change in area under the cumulative distribution function (CDF) curve for each category number k compared with k − 1. The horizontal axis represents the category number k, and the vertical axis represents the relative change in area under the CDF curve. (c) Color-coded heat map corresponding to the consensus matrix for k = 6 obtained by applying consensus clustering. The color gradients were from 0 to 1, representing the degree of consensus, with white corresponding to 0 and dark blue to 1. (d) Composition of the TME by MIC as defined by the MCP-counter Z scores. The patient annotations include MIBC cohorts, MICs, Baylor subtype, UNC subtype, CIT subtype, Lund subtype, MDA subtype, and TCGA subtype. Rows represent TME cells, and columns represent samples
The 8 cohorts with available survival data (n = 843) were pooled to study the clinical outcome of the six MICs (Figure 2a; Table S1). Patients with MIC F exhibited the longest overall survival compared with group A or B patients (P = .006 and P = .019, respectively, Figure 2a). The TLSs were defined as CD20 + B cell follicles juxtaposed with CD3 + T cell aggregates containing at least one LAMP3+ mature dendritic cell .11–14 Representative images of TLSs in bladder cancers detected in formalin-fixed paraffin-embedded tumor sections by immunohistochemistry staining showing CD20 + B cell zones (Figure 2b–c) and LAMP+ (brown) DC cell zones (Figure 2d–e), which were downloaded from Human Pathology Atlas .15
Figure 2.
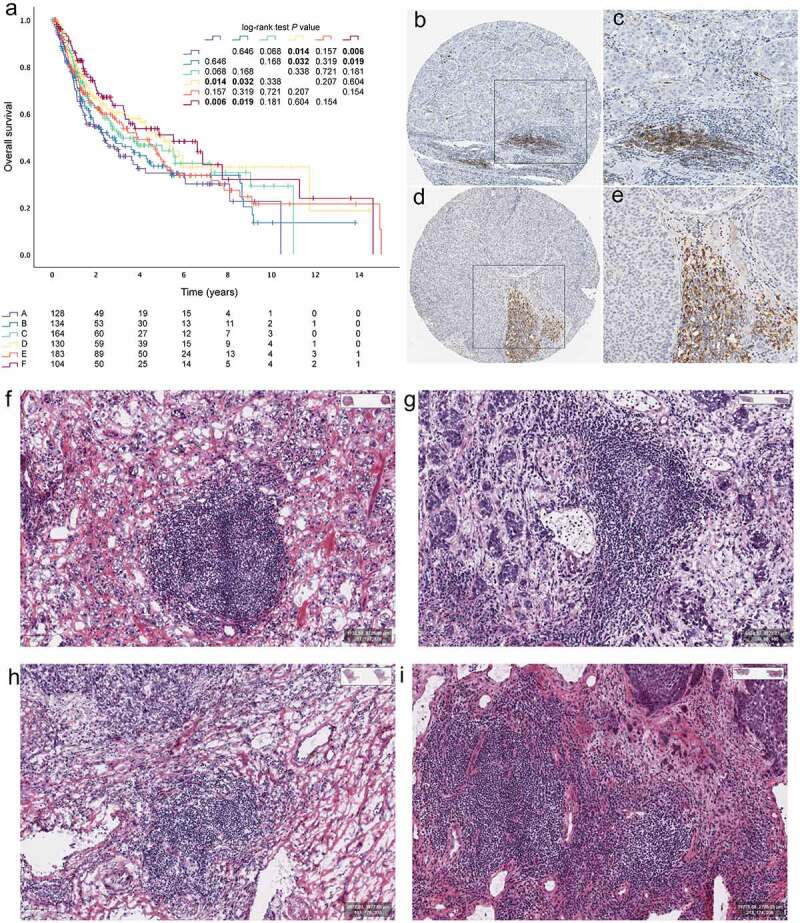
Tertiary lymphoid structures in bladder cancers. (a) Kaplan–Meier curves for OS of 14 cohorts showing the association between MICs and OS. Representative images of tertiary lymphoid structures (TLSs) detected in formalin- fixed paraffin-embedded bladder cancers sections by immunohistochemistry staining showing CD20+ (brown) B cell zones (b and c) and LAMP+ (brown) DC cell zones (d-e). TLSs could be recognized as “small” lymph node like structures in HE slides of MIBC from TCGA (f-i)
In order to verify whether the algorithm could really identify cases with high TLSs infiltration, it was verified by analyzing MIBC HE slides in TCGA (n = 403, Figure 2f–i). We found that 4.7% (n = 19) of MIBC patients with TLSs in class F, 4.0% (n = 16) in class E, 2.0% (n = 8) in class D, 0.7% (n = 3) in class C, 1.0% (n = 4) in class B, and 0.0% (n = 0) in class A, which reached statistical significance (table S3, P < .001). The 9-gene TLS signature (CD79B, CD1D, CCR6, LAT, SKAP1, CETP, EIF1AY, RBP5, and PTGDS) was derived from genes specifically upregulated in CD8+ CD20+ metastasized melanoma tumors .6 As for TCGA HE scans, the expression of 9-gene TLS signature in the TLS+ group was significantly higher than that in the TLS- group. (P < .0001, Fig. S2K)
2.2. Pan-cancer gene expression analysis of TLSs markers in 11,835 solid tumors from TCGA
The t- distributed stochastic neighbor embedding (tSNE) map of 11,835 solid tumors (excluding hematological tumors) from TCGA based on the MCP-counter (Figure 3a). Expression levels of three MCP- counter cell signatures (B lineage, CD8 + T cells and myeloid dendritic cells) and the 9-gene TLS signature had a certain similarity in distribution on the tSNE map (Figure 3b–e).
Figure 3.
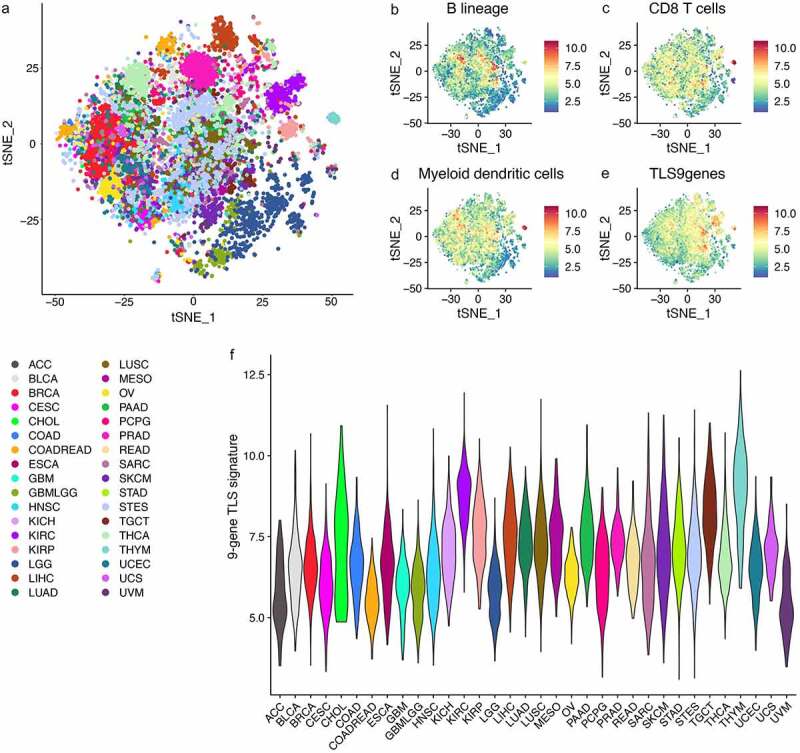
Pan-cancer gene expression analysis of tertiary lymphoid structure markers in 11,835 solid tumors from TCGA
(A) The t- distributed stochastic neighbor embedding (tSNE) map of 11,835 solid tumors (excluding hematological tumors) from TCGA based on the MCP-counter. (B-D) The three graphs on the right display the expression level of three MCP-counter cell signatures related to tertiary lymphoid structures. (E) Expression levels of the TLS-9-genes signature on the tSNE map presented. (F) The violin plot showing the expression of the 9-gene TLS signature in the various cancer types as probability densities. ACC, adrenocortical carcinoma; BLCA, bladder carcinoma; BRCA, breast carcinoma; CESC, cervical squamous carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; COADREAD, colorectal adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma; GBMLGG, lower grade glioma and glioblastoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear- cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, lower- grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; STES, stomach and esophageal carcinoma; TGCT, testicular germ cell tumor; THYM, thymoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
It could be seen that the expression of 9-gene TLSs signatures related to the presence of TLS reveals similar characteristics, where TLSs signatures are strongly expressed in tumors with higher levels of myeloid dendritic cells in B lineage cells and CD8 + T cells. The cancer type-specific expression of the 9-gene TLSs signature (Figure 3f) showed that in most types of cancer, the distribution is extremely uneven, and some tumors express the signature at a high level. For example, thymoma (THYM) showed strong expression of TLSs signatures. However, in some malignant tumors, most tumors did show high expression of TLSs signature. This is especially true for tumors that occur in the sites with immunology privilege, such as the eye (UVM, uveal melanoma) and the brain (GBM, glioblastoma; GBMLGG, lower grade glioma and glioblastoma;). Adrenocortical carcinoma (ACC) also show very low expression of TLSs signature, since corticosteroids secreted by ACC have recently been shown to inhibit TLS regeneration .14 Similarly, as observed in advanced urothelial carcinoma16 and lung cancer, 14 the development of TLSs was suppressed in patients receiving corticosteroids for immunotherapeutic toxicity. All in all, this transcriptomic analysis revealed the heterogeneity of TLSs abundance between different human cancer types.
2.3. MIC and immune-checkpoint-related genes
Among the TCGA datasets, the expression of immune-checkpoint-related genes (Figure 4a) followed that of immune infiltrates, with high expression of the genes (CD274, PDCD1LG2, CTLA4, PDCD1, LAG3, HAVCR2, and TIGIT, respectively) in MIC F followed by MIC D tumors, and low-to very- low expression in MICs E, A, B and C tumors. The expression of genes associated with antigen processing machinery (B2M, HLA-A, HLA-B, HLA-C, TAP1, and TAP2, respectively) was high in MICs F and D, intermediate in MIC E, and very low in MICs A, B and C (Figure 4a). In addition, patients with MIC F exhibited the better overall survival compared with group D patients (P = .022, Figure 4b).
Figure 4.
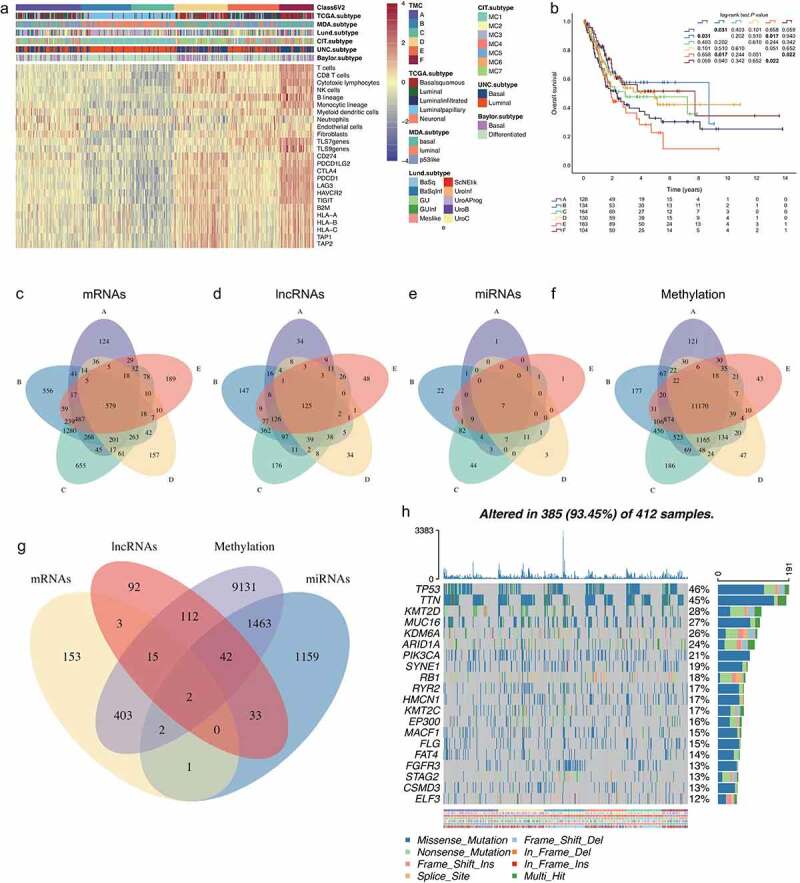
Mutational and epigenetic landscape of the BLCA immune phenotype. (a) Unsupervised clustering of TME cells in the TCGA cohort. MICs, Baylor subtype, UNC subtype, CIT subtype, Lund subtype, MDA subtype, and TCGA subtype are shown as patient annotations. (b) Kaplan–Meier curves for OS of TCGA cohort showing the association between MICs and OS. (c) The Venn diagram of mRNA DEGs, the circle represents mRNA DEGs between MICs F and other MICs; (d) the Venn diagram of lncRNA DEGs; (e) the Venn diagram of miRNA DEGs; (f) the Venn diagram of methylation DEGs. (g) Venn diagrams show the amount of DEGs with MICs F affected by at least one of the indicated genetic (mRNA) or epigenetic (lncRNA, miRNA, methylation) events. (h) The oncoPrint of MICs. Individual patients represented in each column. The top bar plot indicates TMB, whereas the right bar plot shows the mutation frequency of each gene. MICs, Baylor subtype, UNC subtype, CIT subtype, Lund subtype, MDA subtype, and TCGA subtype are shown as patient annotations
2.4. Differentially expressed RNAs in MIBC related to the tertiary lymphoid structures
Among the TCGA datasets, expression data for 20,475 mRNAs, 7656 lncRNAs, 1,881 miRNAs and 396,605 methylated probes were extracted from TCGA. We compared Class F with other classes (A-E), respectively. When we combined these five groups and analyzed for differentially expressed RNAs, 579 mRNAs (−1> log2 FC >1, FDR < 0.01) (Figure 4c; Figure S3; Table S4-S8), 125 lncRNAs (−1> log2 FC >1, FDR < 0.01) (Figure 4d; Figure S4; Table S9-S13), 7 miRNAs (−1> log2 FC >1, FDR < 0.01) (Figure 4e; Figure S5; Table S14-S18), and 11,170 methylated probes (−0.5> log2 FC >0.5, FDR < 0.05) (Figure 4f; Table S19-S23) showed consistently differential expression. Based on these data, these differentially expressed mRNAs (n = 579), lncRNAs (n = 125), miRNAs (n = 7), and methylated probes (n = 11,170) were selected for further analysis.
We hypothesized that differentially expressed genes affected by different patterns of genetic and epigenetic regulation might represent a key driving force for the establishment of tumor immune phenotypes. GO enrichment analysis of these 579 mRNAs (Figure 4g) showed that overexpression of genes involved in tumor immunity pathways, for example, cytokine-cytokine receptor interaction, Th1 and Th2 cell differentiation, Th17 cell differentiation, B cell receptor signaling pathway, T cell receptor signaling pathway, PD-L1 expression and PD-1 checkpoint pathway in cancer, and Toll-like receptor signaling pathway (Fig. S6A–B). As for differentially expressed lncRNAs (n = 125), target mRNA prediction revealed 299 lncRNA-mRNA links (Figure 4g), according to the ENCORI database (Interaction Number ≥ 1, Experiment Num ≥ 1). GO enrichment analysis of these 299 mRNAs showed that overexpression of genes involved in tumor immunity pathways, for example, antigen processing and presentation, Th1 and Th2 cell differentiation, natural killer cell mediated cytotoxicity, Th17 cell differentiation, and PD-L1 expression and PD-1 checkpoint pathway in cancer (Fig. S7A–B). As for differentially expressed miRNAs (n = 7), target mRNA prediction revealed 2702 miRNA-mRNA links (Figure 4g), according to the ENCORI database (strict stringency ≥ 5). GO enrichment analysis of these 2702 mRNAs showed that overexpression of genes involved in cellular senescence, and proteoglycans in cancer (Fig. S8A–B), which are involved in fundamental molecular and cell biology processes occurring in cancer, such as cell immune modulation and metastasis formation .17 As for differentially expressed methylated probes (n = 11,170) (Figure 4g), GO enrichment analysis of these 11,170 mRNAs showed that overexpression of genes involved in PI3K-Akt signaling pathway, Ras signaling pathway, and MAPK signaling pathway (Fig. S9A–B).
2.5. Differences in somatic mutations related to the immune phenotype
To unravel relevant genetic alterations, we next investigated the distributions of somatic alterations and observed different patterns among bladder cancer clusters in terms of gene mutations (Figure 4h). Since the survival differences were related with the expressional alterations in the identified clusters, drugs targeting on the genes with high mutation rates for classifying the subtypes may generate distinctive effects on the subtypes. To identify potential drug targets, potential druggable genes were mapped based on known inhibitors cataloged in the Drug Gene Interaction Database .18 These plot (Fig. S10) shows potential druggable gene categories along with upto top 5 genes involved in them. As results, we found different druggable genomes (MIC A: EP300, ERBB2, FAT4, MUC16 and PIK3CA; MIC B: EP300, FGFR3, HMCN1, MUC16 and MUC17; MIC C: ATM, BIRC6, FAT3, FGFR3 and HMCN1; MIC D: ATM, EP300, FAT1, HMCN1 and KMT2A; MIC E: HMCN1, KMT2A, MUC16, MUC17 and PIK3CA; MIC F: ATM, EP300, FAT4, HMCN1 and MUC16.) may be highly contributable to the survival differences between the identified subtypes. For MIBC patients, more attention should be paid on these drugs since patients from different subtypes may have distinctive responses to these drugs.
2.6. Tertiary lymphoid structures are associated with immunotherapy response in metastatic urothelial cancer
Here we examined whether MICs can predict the patient response to checkpoint blockade therapy from a large cohort of patients with metastatic urothelial cancer (IMvigor21019), from which pre-treatment tumor samples to study the clinical activity of PD-L1 blockers were used for comprehensive evaluation. Different kinds of tissue samples (n = 298) including bladder, kidney, liver, lung, lymph node ureter and others, were performed transcriptome RNA sequencing (RNA-seq) in the IMvigor210 cohort. Here only bladder tissue samples (n = 168) were involved in the following analysis. The patients who had a complete or partial response to treatment were classified as responders, and compared with non-responders who showed stable or progressive disease. The complete response rate (CR) was 8.9% (15 out of 168) in the overall cohort. MICs showed substantial variation in complete response rate (CRR), with MIC F patients exhibiting the highest CRR (21.4%, 3 out of 14), followed by MIC D (16.1%, 5 out of 31) and MIC A (11.1%, 2 out of 18) (Figure 5a). Patients with MIC F tumors also exhibited improved overall survival compared with patients with MIC A (P = .015), MIC C or MIC E (P = .118 and P = .157, respectively, these P values were borderline but trending toward significance). (Figure 5b).
Figure 5.
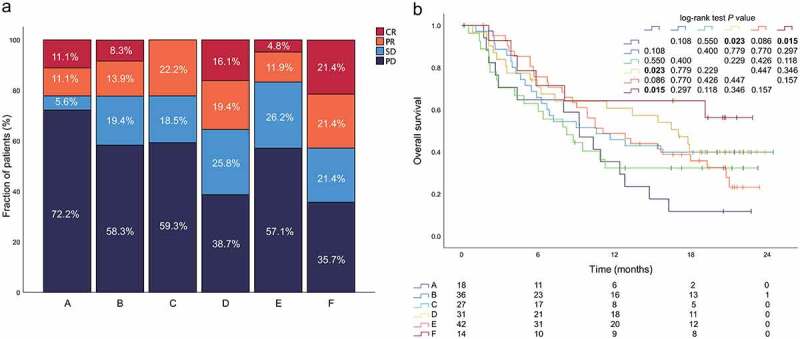
MICs predict patient response to PD1 blockade. This Fig. refers to the IMvigor210 cohort (n = 168). (a) Relationship between MIC and response to PD1 blockade in the IMvigor210 cohort. (b) Ovreall survival of patients by tumor MIC (n = 168). CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease
3. Discussion
Recent study showed that activation of oncogenic pathways in tumor cells could impair induction or execution of a local antitumor immune response .20
A study found that PI3K inhibitors could improve anti‑PD1 efficacy in a clinically relevant breast cancer mouse model .21 In addition, tumors of the non‑T cell-inflamed subset contained FGFR3-activating mutations .22 Interestingly, tumors of the claudin-low subset expressed immune gene signatures at high levels and had decreased frequencies of FGFR3 mutations .23 In our study, we found PIK3CA had high mutation in the subtypes of MIC A and MIC E, and FGFR3 had high mutation in the subtypes of MIC B and MIC C. Therefore, more attention should be paid on the combination of immunotherapy and these druggable genomes, which may help change the tumor immune microenvironment.
Previous study3,24 reported that PD-L1 expression on immune cells was significantly associated with response. By contrast, PD-L1 expression on tumor cells was not associated with response .19 We found that many differentially expressed RNAs (mRNA, lncRNA and miRNA) in MIBC were related to tertiary lymphatic structure, which were mainly involved in tumor immune pathways, such as PD-L1 expression and PD-1 checkpoint pathway in cancer. In addition, we found the CD8 + T cell signature and PD1 were expressed in class F MIBC, which were associated with improved survival, providing high infiltration of TLSs. The integrative analysis demonstrated that TLSs were the key discriminative feature of a group of patients with improved survival. We found that this TLSs-high subgroup was found to respond better to PD1 blockade therapy. In addition, van Dijk16 et al. showed that there was no correlation between baseline TLS numbers and the response of combined CTLA-4 plus PD-1 blockade, although immature TLSs were higher in non-complete-response advanced urothelial cancer .16 Gao et al.25 observed a higher density of TLSs in pre-treatment (neoadjuvant combination anti-PD-L1 plus anti-CTLA-4) tumor tissues of responder patients as compared to non-responder patients of high-risk urothelial carcinoma. However, the underlying mechanism needs further research, but the possible explanation is that TLS is the site of anti-tumor immunity, where B cells indicate T cells (especially CD8 + T cells) to recognize tumor-associated antigens .26 Overall, our findings lay the foundation for risk stratification of MIBC patients and identifying patients who are more likely to benefit from immunotherapy.
The main limitations of our study are that the analyses were based on transcriptome subset due to comparisons across gene expression quantification technologies. Further studies are needed large cohorts across tumor types and stage of disease, as well as with therapeutic regimens. Pre-clinical models will help lend statistical power to the notion that TLSs independently contribute to antitumor immune function in the context of immunotherapies.
4. Conclusions
Our data present multiomic data that support a role for TLS signatures in the response to immunotherapies in patients with bladder cancer. Although the distinct mechanisms through which TLSs contribute are incompletely understood, this finding opens avenues for therapeutic strategies that aim at enhancing TLSs formation and function, which could result in improved clinical outcomes and responses to cancer immunotherapy.
5. Patients and methods
5.1. Inclusion criteria
We downloaded the publicly available bladder cancer gene expression datasets. A total of 14 cohorts were collected (Table S1). Cohorts with <20 tumors, without MIBC, post-treatment (chemotherapy) tumor samples, or <10000 genes, or cohorts hybridized on older or two-color microarray platforms, were removed from meta-cohort compilation (Table S2).
5.2. Data sources and preprocessing
Raw data from the microarray datasets generated using Affymetrix® and Illumina® were downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The raw data for the dataset from Affymetrix® were processed using the RMA algorithm for background adjustment using the “Affy” package .27 The raw data for the dataset from Illumina® were processed using the “lumi” package. The “ComBat” algorithm28 was applied to reduce the likelihood of batch effects from non-biological technical biases. We performed a quality check, platform-specific normalization, and combined them by ComBat (Fig. S1A-C).
5.3. Deconvolution of the cellular composition with MCP-counter
We used the microenvironment- cell-populations (MCP)-counter10 method on the basis of specific molecular markers for eight major immune cell types (CD3 + T cells, CD8 + T cells, cytotoxic lymphocytes, natural killer cells, B lymphocytes, monocytic lineage cells, myeloid dendritic cells and neutrophils), endothelial cells and fibroblasts. The MCP counter method measures the absolute abundance of immune cell subtypes that can be compared between samples.
5.4. Evaluation TLSs by hematoxylin and eosin staining in TCGA cohort
The sections were screened for the presence of TLSs based on morphologic features. Follicular aggregates of lymphatic cells were defined as TLS.
5.5. Consensus clustering for TME-infiltrating cells
Tumors with qualitatively different TME cell infiltration patterns were grouped using hierarchical agglomerative clustering (based on Euclidean distance and Ward’s linkage). Unsupervised clustering methods (K-means) for dataset analysis were used to identify TME patterns and classify patients for further analysis. A consensus clustering algorithm was applied to determine the number of clusters in the meta-dataset to assess the stability of the discovered clusters. This procedure was performed using the ConsensuClusterPlus R package and was repeated 1000 times to ensure the stability of classification. The criteria to determine the number of clusters were as follows: relatively high consistency within clusters, relatively low variation coefficient, and no appreciable rise in the area under the cumulative distribution function (CDF) curve. The category number was selected as the area under the CDF curve and showed no significant change.
5.6. Six published MIBC molecular classifications by BLCAsubtyping
We used BLCAsubtyping29 (R package) six to assign each sample to a subtype in each of the six published MIBC molecular classifications (Baylor, 30 University of North Carolina (UNC), 31 MD Anderson Cancer Center (MDA), 32 the Cancer Genome Atlas (TCGA), 33 Cartes d’Identité des Tumeurs (CIT)-Curie, 34 and Lund .35)
5.7. Pan-cancer gene expression analysis of tertiary lymphoid structure markers in 11,835 solid tumors from TCGA
The t- distributed stochastic neighbor embedding (tSNE) map of 11,835 solid tumors from TCGA based on the MCP-counter software scores estimating the composition of the tumor microenvironment (TME). MCP-counter software was applied to all samples, and the tSNE map was constructed on the estimates with the R.
5.8. Differentially expressed genes (DEG) associated with the MICs
To identify genes associated with MICs patterns, we grouped patients into six subtypes based on immune-cell infiltration. DEGs among these groups were determined using the R package (DESeq2: mRNA, miRNA and lncRNA; ChAMP :36 methylated probes).
5.9. Functional and pathway enrichment analysis
Gene annotation enrichment analysis using the clusterProfiler R package37 was performed on differentially expressed genes. Gene Ontology (GO) terms were identified with a strict cutoff of P < .01 and false discovery rate (FDR) of less than 0.05.37
5.10. Drug and gene interactions
The mutation data for 405 TCGA samples were obtained from UCSC Xena and analyzed by using maftools package. All drug and gene interaction information were obtained from the DGIdb18 which included both the known and reported drug-gene interactions.
5.11. Genomic and clinical data sets with immune-checkpoint blockade
Here we examined whether MICs can predict the patient response to checkpoint blockade therapy from a large cohort of patients with metastatic urothelial cancer (IMvigor21019), from which pre-treatment tumor samples investigating the clinical activity of PD-L1 blockade with atezolizumab in metastatic urothelial cancer (mUC) were used for an integrated evaluation.
5.12. Statistical analysis
The chi-square test or Fisher exact was used for categorical variables, and the t-test or Wilcoxon rank-sum test for continuous variables. Kaplan-Meier analysis was used to determine OS. Log-rank test was used to compare survival between subgroups. Statistical analyses were performed with SPSS, version 26.0 (IBM, Armonk, NY), and R software packages, version 4.0.1 (The R Foundation for Statistical Computing, http://www.r-project.org/). A two-sided P value of less than 0.05 was considered to be statistically significant for all reports.
Supplementary Material
Funding Statement
This study was sponsored by Shanghai Sailing Program [19YF1440300], Shanghai Jiaotong University School of Medicine Multi-center Clinical Research Project [DLY201809], and Innovative research team of high-level local universities in Shanghai [19DZ2204000]. All these study sponsors have no roles in the study design, in the collection, analysis, and interpretation of data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital.
Data availability statement
Data are available upon reasonable request.
Authors’ contributions
LZ, BX, and YL for acquisition of data, analysis and interpretation of data, statistical analysis and drafting of the manuscript; BX and YL for technical and material support; LZ, BX, YL and ZW for study concept and design, analysis and interpretation of data, drafting of the manuscript, obtained funding and study supervision. All authors read and approved the final manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellmunt J, De Wit R, Vaughn DJ, Fradet Y, Lee J-L, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376(11):1015–10. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg JE, Hoffman-Censits J, Powles T, Van Der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830d.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, Johansson I, Phung B, Harbst K, Vallon-Christersson J, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 7.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petitprez F, De Reyniès A, Keung EZ, Chen TWW, Sun C-M, Calderaro J, Jeng Y-M, Hsiao L-P, Lacroix L, Bougoüin A, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 9.Pfannstiel C, Strissel PL, Chiappinelli KB, Sikic D, Wach S, Wirtz RM, Wullweber A, Taubert H, Breyer J, Otto W, et al. The tumor immune microenvironment drives a prognostic relevance that correlates with bladder cancer subtypes. Cancer Immunol Res. 2019;7(6):923–938. doi: 10.1158/2326-6066.CIR-18-0758. [DOI] [PubMed] [Google Scholar]
- 10.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman C, Fridman WH, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218–237. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sautes-Fridman C, Petitprez F, Calderaro J, Fridman WH.. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19(6):307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- 12.Dieu-Nosjean M-C, Goc J, Giraldo NA, Sautès-Fridman C, Fridman WH. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 2014;35(11):571–580. doi: 10.1016/j.it.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Posch F, Silina K, Leibl S, Mündlein A, Moch H, Siebenhüner A, Samaras P, Riedl J, Stotz M, Szkandera J, et al. Maturation of tertiary lymphoid structures and recurrence of stage II and III colorectal cancer. Oncoimmunology. 2018;7(2):e1378844. doi: 10.1080/2162402X.2017.1378844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silina K, Soltermann A, Attar FM, Casanova R, Uckeley ZM, Thut H, Wandres M, Isajevs S, Cheng P, Curioni-Fontecedro A, et al. Germinal centers determine the prognostic relevance of tertiary lymphoid structures and are impaired by corticosteroids in lung squamous cell carcinoma. Cancer Res. 2018;78(5):1308–1320. doi: 10.1158/0008-5472.CAN-17-1987. [DOI] [PubMed] [Google Scholar]
- 15.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 16.Van Dijk N, Gil-Jimenez A, Silina K, Hendricksen K, Smit LA, De Feijter JM, Van Montfoort ML, Van Rooijen C, Peters D, Broeks A, et al. Preoperative ipilimumab plus nivolumab in locoregionally advanced urothelial cancer: the NABUCCO trial. Nat Med. 2020;26(12):1839–1844. doi: 10.1038/s41591-020-1085-z. [DOI] [PubMed] [Google Scholar]
- 17.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 18.Cotto KC, Wagner AH, Feng -Y-Y, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–d1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel III EE, Koeppen H, Astarita JL, Cubas R, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18(3):139–147. doi: 10.1038/nrc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sai J, Owens P, Novitskiy SV, Hawkins OE, Vilgelm AE, Yang J, Sobolik T, Lavender N, Johnson AC, McClain C, et al. PI3K inhibition reduces mammary tumor growth and facilitates antitumor immunity and Anti-PD1 responses. Clin Cancer Res. 2017;23(13):3371–3384. doi: 10.1158/1078-0432.CCR-16-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sweis RF, Spranger S, Bao R, Paner GP, Stadler WM, Steinberg G, Gajewski TF. Molecular drivers of the non-T-cell-inflamed tumor microenvironment in urothelial bladder cancer. Cancer Immunol Res. 2016;4(7):563–568. doi: 10.1158/2326-6066.CIR-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kardos J, Chai S, Mose LE, Selitsky SR, Krishnan B, Saito R, Iglesia MD, Milowsky MI, Parker JS, Kim WY, et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. Jci Insight. 2016;1(3). doi: 10.1172/jci.insight.85902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng S-L, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Navai N, Alhalabi O, Siefker-Radtke A, Campbell MT, Tidwell RS, Guo CC, Kamat AM, Matin SF, Araujo JC, et al. Neoadjuvant PD-L1 plus CTLA-4 blockade in patients with cisplatin-ineligible operable high-risk urothelial carcinoma. Nat Med. 2020;26(12):1845–1851. doi: 10.1038/s41591-020-1086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen JS, Sahota RA, Milne K, Kost SE, Nesslinger NJ, Watson PH, Nelson BH. CD20+tumor-infiltrating lymphocytes have an atypical CD27−memory phenotype and together with CD8+T cells promote favorable prognosis in ovarian cancer. Clin Cancer Res. 2012;18(12):3281–3292. doi: 10.1158/1078-0432.CCR-12-0234. [DOI] [PubMed] [Google Scholar]
- 27.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 28.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 29.Kamoun A, De Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, Hoadley KA, Groeneveld CS, Al-Ahmadie H, Choi W, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol. 2020;77(4):420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo Q, Nikolos F, Chen F, Tramel Z, Lee Y-C, Hayashi K, Xiao J, Shen J, Chan KS. Prognostic power of a tumor differentiation gene signature for bladder urothelial carcinomas. J Natl Cancer Inst. 2018;110(5):448–459. doi: 10.1093/jnci/djx243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111(8):3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi W, Porten S, Kim S, Willis D, Plimack E, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee I-L, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25(2):152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebouissou S, Bernard-Pierrot I, De Reynies A, Lepage M-L, Krucker C, Chapeaublanc E, Herault A, Kamoun A, Caillault A, Letouze E, et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci Transl Med. 2014;6(244):244ra91. doi: 10.1126/scitranslmed.3008970. [DOI] [PubMed] [Google Scholar]
- 35.Marzouka NA, Eriksson P, Rovira C, Liedberg F, Sjödahl G, Höglund M. A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Sci Rep. 2018;8(1):3737. doi: 10.1038/s41598-018-22126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE. ChAMP: updated methylation analysis pipeline for illumina beadchips. Bioinformatics. 2017;33(24):3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.


