ABSTRACT
Mitophagy is an essential mechanism in maintaining cellular homeostasis, in which damaged and superfluous mitochondria are selectively degraded by the autophagy-lysosome pathway. Our recent study revealed that SPATA33 functions as a novel receptor for mitophagy in the priming of mitochondria for degradation in male germline cells. SPATA33 directly mediates the interaction of the outer mitochondrial membrane protein VDAC2 with the autophagy machinery component ATG16L1 during mitophagy. Upon starvation induction, SPATA33 can promote mitophagy as an autophagy receptor. Thus, SPATA33 confers cargo selectivity during mitophagy in germline cells. These findings provide new insights into selective autophagy and mitochondrial homeostasis.
KEYWORDS: Autophagy, mammals, mitochondria, SPATA33, spermatogenesis
Spermatogenesis is a highly regulated developmental process to produce sperm cells by both mitosis and meiosis. This process is essential to maintain cellular homeostasis by autophagic degradation of undesired organelles and macromolecules during development. Loss of autophagy related genes, for example Atg5 and Atg7, impair spermatogenesis and eventually led to male infertility.
Mitochondrion-specific autophagy (mitophagy) maintains the quantity and stability of mitochondria by the selective removal of damaged and superfluous mitochondria. Thus, mitophagy is an important mechanism for cellular differentiation in a variety of developmental processes. Accumulating evidence shows that dysregulation of mitophagy contributes to various types of human diseases, including Parkinson's disease and metabolic disorders.
Several proteins and related pathways that participate in the regulation of mitophagy have been identified. In particular, a number of receptor proteins mediate autophagic degradation of mitochondria by mitophagy pathways in cellular differentiation and the development of disease. For example, CALCOCO2/NDP52 and OPTN are mitophagy receptors involved in neurodegenerative diseases, and BNIP3L is essential for erythroid differentiation and plays a role in priming for the clearance of redundant mitochondria.
Despite the characterization of these mitophagy receptors, tissue-specific receptors for mitophagy under various physiological and pathological conditions and related regulatory mechanisms are not well known. We have previously identified a novel gene, SPATA33/4732415M23Rik/C16orf55, which is specifically expressed in testis and is conserved in human and mouse. SPATA33 is associated with male infertility in humans. To explore a possible link between this novel gene and autophagy, we recently investigated SPATA33 partners and found that SPATA33 functions as an autophagy receptor in priming mitophagy in the male germline [1]. Immunofluorescence revealed SPATA33 co-expression with the key autophagy protein ATG16L1 in supporting cells (Sertoli and Leydig) and spermatogenic cells in mouse testis. In addition, it colocalizes with both LC3B and ATG16L1, and the mitochondrial outer membrane protein VDAC2 in the mid-piece region (mitochondria region) of spermatozoa from the epididymis of adult mice. The mitochondrial colocalization was further confirmed by MitoTracker costaining in the germline cells. Co-immunoprecipitation and deletion analysis showed that SPATA33 interacts with the C terminus of ATG16L1. Moreover, starvation induction increases the number of colocalized puncta of SPATA33 with ATG16L1 and LC3B. These data suggest that SPATA33 is potentially associated with mitophagy in male germline cells.
Spata33 knockout in both Sertoli cells (TM4) and spermatogenic cells (GC-1) delays the time of LC3B puncta formation in comparison with wild-type cells under starvation conditions, whereas Spata33 re-expression rescues the delayed time phenotype in the knockout cells. Consistent with this, upon starvation induction, the LC3B-II level is significantly decreased in the Spata33 knockout cells, and protein levels of both the mitochondrial outer membrane VDAC2 and inner membrane COX4I1/COX-IV are also decreased in the knockout cells, compared to wild-type cells. In addition, Atg16l1 knockout reveals a similar effect on autophagy inhibition as Spata33 knockout. Transmission electron microscopy showed that damaged mitochondria are engulfed by autophagic membranes under combined treatment of starvation and CCCP (an inducer of mitophagy). Autophagy flux analysis indicated that SPATA33 plays a role in autophagosome formation, but not in fusion of autophagosomes with lysosomes. Further fluorescence microscopy using a pmCherry-GFP-FIS1[101-152] tandem reporter (FIS1 is a mitochondrial outer membrane protein) showed that the mitophagy level is decreased in Spata33 knockout cells, whereas Spata33 overexpression enhances mitophagy. These data suggested that mitochondria are selective cargos whose sequestration is mediated by SPATA33 for autophagic degradation.
To further investigate the mechanism of SPATA33-mediated mitophagy, co-immunoprecipitation and deletion mapping were used to analyze the interaction among SPATA33, ATG16L1 and VDAC2. The C terminus of SPATA33, but not the N terminus, can interact with VDAC2, whereas ATG16L1 interacts with the SPATA33 N terminus. In addition, obvious colocalized puncta among mCherry-SPATA33, GFP-VDAC2 and GFP-ATG16L1 are also detected under starvation and CCCP combined treatment. These data suggested that SPATA33 is a novel autophagy receptor in the cytosol for mitophagy through directly mediating the interaction of the outer mitochondrial membrane protein VDAC2 with ATG16L1 (Figure 1A). Thus, SPATA33 confers the cargo selectivity during mitophagy in germline cells.
Figure 1.
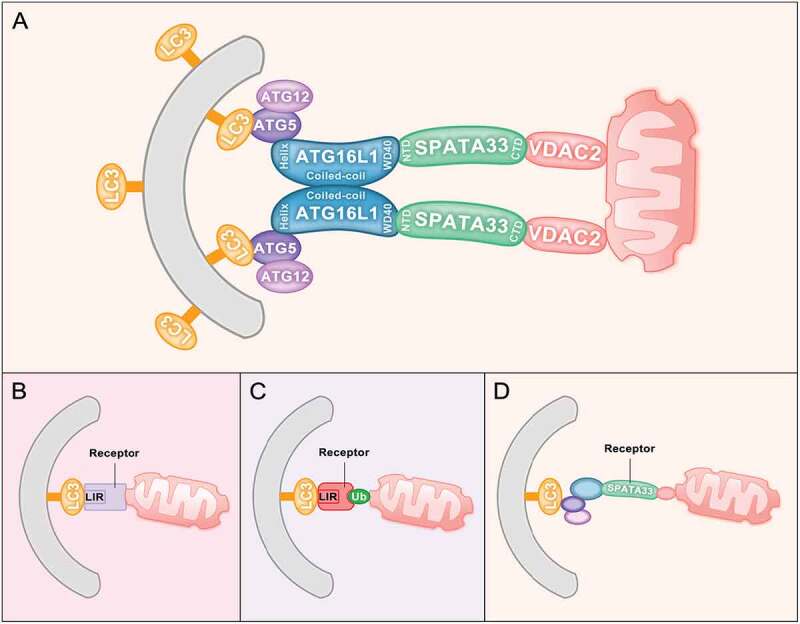
Mitophagy receptors. (A) SPATA33 functions as a mitophagy receptor. The C terminus of SPATA33 can interact with the mitochondrial outer membrane protein VDAC2. The N terminus of SPATA33 can bind to the WD40 region of ATG16L1. Upon starvation stress, damaged mitochondria are engulfed by autophagic membranes through SPATA33 linking VDAC2 and ATG16L1 to initiate mitophagy. (B) Ubiquitin-independent mitophagy. The autophagy receptors are outer mitochondrial membrane proteins, which have a common LIR/AIM motif that can bind to LC3B/Atg8 on the phagophore. (C) PINK-PRKN- or ubiquitin-dependent mitophagy. Mitophagy receptors are cytosolic proteins (CALCOCO2/NDP52 and OPTN), which bind ubiquitin. Accordingly, cytosolic receptors CALCOCO2/NDP52 and OPTN have both an AIM/LIR motif and a ubiquitin-binding domain. (D) The receptor SPATA33 mediates mitophagy via bridging between ATG16L1 and VDAC2, which is ATG16L1 dependent
Most mitophagy receptors are the outer mitochondrial membrane proteins, including BCL2L13, BNIP3, BNIP3L/NIX, FKBP8, and FUNDC1 in mammals, and Atg32 and Atg43 in yeast. These proteins have a common LIR/AIM motif, which can bind to LC3B/Atg8 on the phagophore and do not depend on ubiquitination of cargo proteins during mitophagy (Figure 1B). Another type of mitophagy receptor is seen with cytosolic proteins, including CALCOCO2/NDP52 and OPTN, which are PINK-PRKN-dependent and ubiquitin-mediated. They work as mitophagy receptors in the PINK1-PRKN-mediated pathway. PRKN can ubiquitinate many mitochondrial proteins upon mitochondrial damage or depolarization. Cytosolic receptors CALCOCO2/NDP52 and OPTN have both AIM/LIR motifs and ubiquitin-binding domains, which can recognize ubiquitinated mitochondrial proteins and bind to LC3/Atg8 on the phagophore. Thus, these cytosolic proteins function as mitophagy receptors when mitochondria are ubiquitinated (Figure 1C). SPATA33 is another type of novel receptor for mitophagy in the male germline (Figure 1A,D). SPATA33 directly interacts with the autophagic machinery ATG16L1, instead of LC3B; thus, SPATA33-mediated mitophagy is ATG16L1 dependent. Identification of SPATA33 as a mitophagy receptor will help our understanding of selective autophagy in maintaining mitochondrial homeostasis in the male germline.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31771487, 31771370, and 31970539].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics statement
All animal experiments and methods were performed in accordance with the relevant approved guidelines and regulations, as well as under the approval of the Ethics Committee of Wuhan University.
Reference
- [1].Zhang Y, Xu X, Hu M, et al. SPATA33 is an autophagy mediator for cargo selectivity in germline mitophagy. Cell Death Differ. 2021. March;28(3):1076–1090. PubMed PMID: 33087875. [DOI] [PMC free article] [PubMed] [Google Scholar]


