ABSTRACT
Selective autophagy receptors have been implicated in the degradation of cellular constituents of various size and rigidity. However, the identity of protein cargo have largely remained elusive. In our recent study, we combined limited proteolysis-enhanced proximity biotinylation and organelle enrichment with quantitative proteomics to map the inventory of autophagosomes in a manner dependent on six different selective autophagy receptors, namely SQSTM1/p62, NBR1, CALCOCO2/NDP52, OPTN, TAX1BP1 and TOLLIP. Conducting this approach under basal and proteostasis-challenged conditions in mammalian cells led to the identification of various new autophagy substrates of which some were degraded through endosomal microautophagy rather than canonical autophagy dependent on the receptors TOLLIP and SQSTM1, respectively.
KEYWORDS: Selective autophagy receptors, APEX2, proximity proteomics, proteostasis challenges, endosomal microautophagy, TOLLIP
Macroautophagy (hereafter autophagy) is the main cellular degradation mechanism for cytosolic cargo, including pathogens, defective mitochondria, organelles or aggregated proteins. Double-membraned phagophores sequester the cargo and expand into autophagosomes involving a machinery of autophagy-related (ATG) proteins before they fuse with lysosomes for degradation of their content through lysosomal hydrolases. The family of human (Hs)Atg8-family proteins plays an important role in this process and comprises the two subfamilies of MAP1LC3 and GABARAP proteins that are both conjugated via a ubiquitin-like conjugation cascade to the concave and convex sides of forming autophagosomes. Selective autophagy receptors harboring an LC3-interacting region (LIR) bind to lipidated HsAtg8-family proteins thereby recruiting the forming autophagosomes to simultaneously bound cargo. The latter is often decorated with polyubiquitin as a degradation signal, which can be recognized by selective autophagy receptors via dedicated ubiquitin-binding domains (UBDs). However, direct interactions are also possible. Due to their structural similarities the six autophagy receptors SQSTM1/p62, NBR1, CALCOCO2/NDP52, OPTN, TAX1BP1 and TOLLIP are referred to after their founding member as SQSTM1-like receptors (SLRs). Many of the SLRs are implicated in selective degradation of protein inclusions following proteostasis-challenging conditions. But the fact that autophagy deficiency leads to the buildup of aggregated or misfolded proteins already under fed conditions raises the possibility that SLRs constitutively mediate phagophore engulfment of natively folded proteins. However, the identity of these autophagic substrates have remained scarce.
In our recent study [1] we combined proximity proteomics and organelle enrichment with quantitative proteomics to study receptor-specific autophagosome content and identify new cargo candidates. Proteinase K treatment is essential to digest all neighborhood proteins outside of closed vesicles in order to exclusively focus the analysis on the autophagosomal content (Figure 1). Briefly, APEX2 was coupled to each of the six SLRs as well as LC3B as a reference HsAtg8-family protein and subjected to autophagosomal content profiling under fed, proteostasis-challenging and autophagy-modulating conditions. Importantly, all APEX2 chimeras localize to mature autophagosomes and yield biotinylation patterns that are partially protected from proteinase K digestion.
Figure 1.
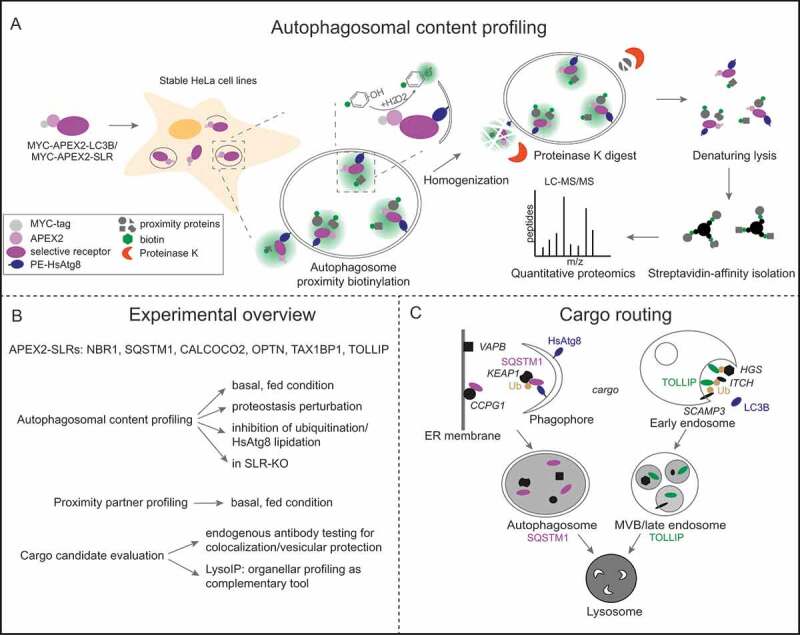
Autophagy cargo mapping. (A) Overview of main steps performed during autophagosomal content profiling in APEX2-LC3B/APEX2-SLRs. (B) Sum-up of conditions and experiments used in the study. (C) TOLLIP and SQSTM1 cargos (marked in italics) are delivered to lysosomes by different routes, namely endosomal microautophagy and canonical autophagy. Ub: ubiquitin
By comparing bafilomycin A1 (BafA1)- and DMSO-treated samples we found that between 500 and 900 proteins are proteinase K protected of which only approximately 14% are BafA1 sensitive. These were defined as cargo candidates and for most of the receptors approximately two thirds are shared with at least one other APEX2 chimera leaving a total of 279 non-redundant cargo candidates. Among those were members of the HsAtg8 family, the receptors themselves, proteins that were previously associated with autophagy (VCP, TBC1D15, UBQLN2, TBK1, TNFAIP3, PRKCI, VAPA and VAPB) and completely new candidates such as PRKAR1A. The receptor whose cargo spectrum stuck out the most was TOLLIP, which only contains SQSTM1 and NBR1 but otherwise lacks known autophagy proteins. Instead among TOLLIP’s cargo candidates are numerous proteins of the endosomal system (e.g. SCAMP3, PDCD6, PDCD6IP, STAM, HGS, ITCH, RAB5C). GO-Term analysis of all cargo candidates revealed intriguing new clusters like ATP/GTP-associated terms and proteins associated in cell adherens, besides the expected autophagy- and unfolded protein-associated categories. Comparison of cargo candidates and neighborhood proteins of SLRs identified by proximity proteomics with and without proteinase K treatment of homogenates surprisingly revealed only a small overlap between these two populations, implying that SLRs might serve functions beyond selective autophagy or only engage additional other substrates in response to specific stimuli. Quantitative proteomics of immunoisolated lysosomes from parental cells or those lacking all 6 SLRs or HsAtg8s independently confirmed numerous proteins as cargo candidates. These included proteins identified across different APEX2 chimeras (LC3B, NOCA4, HLA-A, RAB1A, RAB5C, RAB7A, RAB14 and PLD3) as well as proteins identified by distinct APEX2 baits such as VAPA and VAPB (SQSTM1), SCAMP3 and PDIA3 (TOLLIP), TMEM33 (LC3B), CKAP4 (NBR1), AFG3L2 (OPTN) or ATP6V0D1 (CALCOCO2). Unbiased autophagosome content profiling in SLR knockout cells and biochemical analysis of 25 selected cargo candidates with regard to lysosomal localization, membrane protection and abundance changes provided further proof-of-principle that our experimental strategy facilitated the identification of bona fide SLR-targeted cargo proteins under fed (basal) conditions.
Unexpectedly, autophagosome content profiling in cells treated with inhibitors of the E1 enzymes required for lipidation (Atg8-E1) or ubiquitination (Ub-E1) result only in overall moderate abundance changes of cargo proteins. Whereas 40%–60% of the cargo sampled by APEX2-LC3B or -NBR1 increases upon Ub-E1 treatment, all other APEX2 fusions show decreases for 17%–33% of their substrate proteins. Even smaller changes of cargo candidates are observed for the Atg8-E1 treatment. While these findings certainly call for further in-depth analyses, they nevertheless question the absolute requirement of ubiquitin and LC3/GABARAP conjugation for the phagophore engulfment of a subset of SLR-targeted cargo proteins.
Furthermore, three different settings of proteostasis challenges, namely translation inhibition (by puromycin), proteasome inhibition (by bortezomib) and overexpression of a neurodegenerative disease-relevant aggregation-prone protein (polyGA), only marginally affect basal cargo candidates. However, a considerable but heterogenous population of stimuli-induced new cargo candidates was sampled across the different APEX2 fusions. Examples are the enrichment in ER proteins upon polyGA treatment in APEX2-LC3B cells, whereas mitochondrial proteins are enriched after inhibition of the proteasome in APEX2-NBR1 cells.
Closer inspection of TOLLIP, SQSTM1 and some of their cargo candidates revealed differential subcellular routing of these two receptors in order to deliver their cargo to lysosomes. Briefly, TOLLIP localizes to multivesicular bodies (MVBs) and its cargo is sensitive to inhibition of MVB-related functions but unaffected by blockage of early steps during autophagosome formation. These findings suggest that TOLLIP mediates lysosomal cargo delivery via MVBs rather than canonical autophagosomal structures and therefore serves as a receptor for endosomal microautophagy. In contrast, SQSTM1 cargo candidates neither colocalizes with endosomal markers nor localizes ultrastructurally to MVBs under fed conditions and are insensitive to impairment of MVBs or ESCRTs, indicating that SQSTM1 takes its substrates on a canonical autophagy route to lysosomes (Figure 1).
Our study unveiled an extensive but certainly not complete inventory of SLR-containing autophagic vesicles under basal and proteostasis-challenging conditions and thereby provides new cargo candidates for comprehensively monitoring of autophagy activity in cells and for fueling further mechanistic studies. One very interesting group of cargo candidates contain GTPase proteins (RAB5C, RAB7A, RHOA, SAR1B and RHEB), which are known regulators of membrane dynamics, but their turnover via autophagy is so far unexplored.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft [DFG, German Research Foundation] within the frameworks of the Munich Cluster for Systems Neurology [EXC 2145 SyNergy – ID 390857198] and of the Collaborative Research Center 1177 [ID 259130777].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Reference
- [1].Zellner S, Schifferer M, Behrends C.. Systematically defining selective autophagy receptor–specific cargo using autophagosome content profiling. Mol Cell. 2021. March 18;81(6):1337–1354.e8. [DOI] [PubMed] [Google Scholar]


