Abstract
The most common breast cancer (BC) susceptibility genes beyond BRCA1/2 are ATM and CHEK2. For the purpose of exploring the clinicopathologic characteristics of BC developed by ATM or CHEK2 mutation carriers, we reviewed the archive of our Family Cancer Clinic. Since 2018, 1185 multi-gene panel tests have been performed. Nineteen ATM and 17 CHEK2 mutation carriers affected by 46 different BCs were identified. A high rate of bilateral tumors was observed in ATM (26.3%) and CHEK2 mutation carriers (41.2%). While 64.3% of CHEK2 tumors were luminal A-like, 56.2% of ATM tumors were luminal B-like/HER2-negative. Moreover, 21.4% of CHEK2-related invasive tumors showed a lobular histotype. About a quarter of all ATM-related BCs and a third of CHEK2 BCs were in situ carcinomas and more than half of ATM and CHEK2-related BCs were diagnosed at stage I-II. Finally, 63.2% of ATM mutation carriers and 64.7% of CHEK2 mutation carriers presented a positive BC family history. The biological and clinical characteristics of ATM and CHEK2-related tumors may help improve diagnosis, prognostication and targeted therapeutic approaches. Contralateral mastectomy should be considered and discussed with ATM and CHEK2 mutation carriers at the first diagnosis of BC.
Keywords: breast cancer, ATM, CHEK2, genetic testing, bilateral tumor, mastectomy
1. Introduction
The recent introduction of multigene panel testing for mutations associated with breast and/or ovarian cancer has raised new challenges in the management of both individuals at increased cancer risk and cancer patients. In addition to the known high-penetrance BRCA1/2 mutations, pathogenic variants in other high/moderate-penetrance genes can increase the risk of breast and/or ovarian cancer. Nevertheless, while providing risk assessment, their clinical utility in terms of primary and secondary prevention, prognostication and treatment modalities are still uncertain [1].
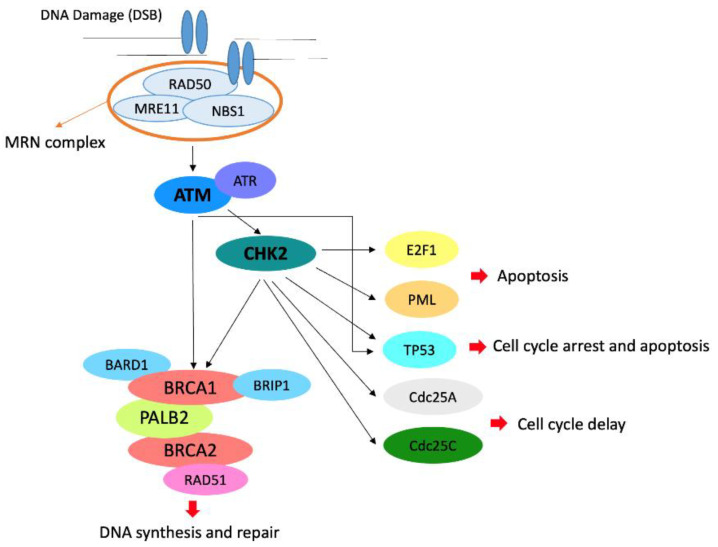
The most common non-BRCA pathogenic or likely pathogenic variants were found in ATM and CHEK2 genes [2]. In particular, the mutation frequency for ATM is 0.78% and for CHEK2 1.08% in unselected breast cancer patients, whereas the prevalence in unaffected women is 0.41% for ATM and 0.42% for CHEK2 [3]. Both ATM and CHEK2 are considered as moderate-penetrance genes and are involved in DNA-double strand break repair mechanisms [4]. In particular, the ATM protein kinase is a critical intermediary in a number of cellular responses to ionizing irradiation and possibly other stresses. In addition, its dysfunction results in abnormal checkpoint responses in multiple phases of the cell cycle [5]. After DNA damage, ATM and DNA-dependent (DNA-PK) protein kinase activate CHK2, which in turn phosphorylates a number of downstream substrates involved in various cellular processes, including cell cycle arrest, apoptosis, DNA repair and mitosis [6] (Figure 1).
Figure 1.
Role of ATM and CHK2 in the pathways of cell cycle arrest, apoptosis, DNA repair and mitosis. In particular, the MRN complex resects DNA at the double-strand break (DSB) and recruits ATM that phosphorylates CHK2 and recruits the BRCA complex.
Individuals carrying heterozygous pathogenic variants in ATM present a 33% cumulative lifetime risk for BC by 80 years of age [7], whereas certain variants in the CHEK2 gene are associated with increased BC risk, with a cumulative lifetime risk ranging from 28% to 37% depending on family history [8,9]. Due to this increased risk, for both ATM and CHEK2 carriers, mammogram with consideration of breast MRI is recommended yearly from 40 years of age according to the current National Comprehensive Cancer Network (NCCN) guidelines [10]. Although only insufficient data are available, furthermore, bilateral risk-reducing mastectomy may be considered, based on family history [10]. Additionally, ATM heterozygous pathogenic variants have also been described in some cases of familial ovarian [11], pancreatic [12], and prostate cancer [13], whereas pathogenic CHEK2 variants were also associated to an increased risk of other malignancies including colon, prostate, kidney, bladder and thyroid cancers, according to specific mutations (frameshift or missense substitutions) [14].
So far, only a few small-sample studies investigated whether BC developed by ATM or CHEK2 mutation carriers includes distinct histopathological features and clinical outcomes from sporadic BC and BRCA1/2 associated tumors. Renault et al. [15] showed that most ATM-associated tumors are luminal B or luminal B/HER2+ tumors. Nizic-Kos et al. [16] reported that the majority of patients with CHEK2 pathogenic or likely pathogenic variants develop luminal A or luminal B BC subtypes. In a recent retrospective case-control study, finally, Bergstrom and colleagues [17] found that BC patients with germline pathogenic variants of ATM, CHEK2, or PALB2 have an increased family history of breast cancer, tumor size >2.0 cm at the time of diagnosis, and potentially an increased risk of recurrence compared to mutation-negative patients. However, lymph nodes, nuclear grade, histology, Ki-67 proliferation and receptor status were not different from sporadic tumors.
The aim of our study was to explore whether the presence of ATM or CHEK2 pathogenic or likely pathogenic germline variants in BC patients is associated with specific clinicopathologic characteristics and prognostic features at our institution.
2. Materials and Methods
2.1. Study Population and Design
The Modena Family Cancer Clinic (MFCC), located in the Emilia Romagna region (Northern Italy), offers genetic counseling to individuals with a personal or family history of BC and/or ovarian cancer (OC) in accordance with regional criteria for BRCA genetic testing [18]. Since the 8 January 2018, counseling has also been given to all patients affected by pancreatic cancer (PC) following Olaparib approval as a first-line maintenance treatment (Table 1). During pre-test counseling, family and personal histories of cancer are collected. At the same time, a family pedigree is drawn including third-degree relatives on both maternal and paternal sides. In particular, healthy women with BC and/or OC family history are referred to the MFCC by general practitioners or radiologists that perform population-based screening mammography. On the other hand, BC, OC and PC patients are referred to the MFCC by oncologists, radiologists, surgeons or gynecologists. Eligible individuals can undergo genetic testing. Then, in case of a positive result, the option of searching for specific pathogenic or likely pathogenic variant can be provided to other family members, in order to access risk-reducing surgery [19], chemoprevention studies [20] or more intensive surveillance programs [21,22]. After post-test counseling, finally, a copy of all patient documents and reports is stored in the MFCC archive.
Table 1.
The MFCC criteria for genetic testing in BC, OC and PC cancer patients.
| BC and OC in the Same Patient or Family. |
| OC, fallopian tube or primary peritoneal cancer (excluding mucinous and borderline) at any age. |
| Male BC |
| Triple negative BC diagnosed ≤60 years. |
| BC diagnosed ≤35 years. |
| PC at any age |
| At least two first-degree blood relatives with BC with at least one diagnosed ≤40 years or bilateral in the same family. |
BC: breast cancer; OC: ovarian cancer; PC: pancreatic cancer.
Between 1998 and 2017, the MFCC offered BRCA1/2 genetic testing to BC and/or OC patients, first according to the Modena Criteria for genetic testing [23,24] and subsequently, according to the criteria recommended by the Emilia Romagna region [18]. On the 8 January 2018, the Clinical Genomics Laboratory of the MFCC started to provide a Next Generation Sequencing (NGS) multigene panel testing to all new patients who met the Regional Criteria for BRCA genetic testing and all PC patients. Furthermore, patients who tested negative for BRCA genes in the previous years were recalled to undergo the new multi-gene panel test. Clinical and pathologic characteristics of BC patients testing positive for variants classified as pathogenic or likely pathogenic in the ATM or CHEK2 genes were then collected. These included age at first diagnosis, histotype, immunohistochemical profile of invasive carcinomas, clinical stage at diagnosis, type of breast and axillary surgery, radiotherapy, chemotherapy and rate of recurrence.
Estrogen Receptor (ER), Progesterone Receptor (PgR) and Human Epidermal growth factor Receptor 2 (HER2) expression were determined according to national pathology guidelines, which closely adhere to international standards [25,26]. According to the ESMO Clinical Practice Guidelines [27], for the purpose of prognostication and treatment decision making, tumors should be grouped into surrogate intrinsic subtypes, defined by routine histology and IHC data. In our study, in line with the 2013 St Gallen Consensus Conference [28] and local laboratory values, luminal A-like tumors have been defined as ER-positive, PgR ≥20%, HER2-negative and Ki67 <20%. On the other hand, luminal B-like tumors are characterized by ER-positive, and either Ki67 high (≥20%) or PgR low (<20%) or HER2-positive.
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the Area Vasta Emilia Nord (Project identification code 125/2021/OSS*/AOUMO, Modena, Italy).
2.2. Procedures for Multi-Gene Panel Testing
Peripheral blood samples (PB) were collected into EDTA tubes, in accordance with the current revision of the Helsinki Declaration, and genomic DNA was extracted with the DNA Midi Kit via QIASymphony platform (Qiagen, Hilden, Germany); nucleic acid quantity/quality were checked by Qubit dsDNA High Sensitivity kit and Nanodrop (Thermo Scientific, Waltham, MA, USA).
Sequencing libraries were prepared using the CE-IVD SOPHiA HCS v1.1 kit, exclusively through the automated procedure implemented on the STARlet platform (Hamilton, https://www.hamiltoncompany.com/press-releases/application-note-automation-of-the-hereditary-cancer-solution-hcs-by-sophia-genetics-on-a-starlet#top, accessed on 20 April 2021). Individual library quantification was performed via fluorometric quantitation by Qubit dsDNA High Sensitivity kit (Thermo Scientific, Waltham, Massachusetts, USA) and quality control analysing the profile of each sample via capillary electrophoresis with Bioanalyzer DNA 1000 (Agilent Technologies, Santa Clara, CA, USA). Samples were run onto a 600-cycle format V3 flow-cell and sequenced via Illumina MiSeq DX platform according to their own and SOPHiA GENETICS’ (Lausanne, Switzerland; Boston, MA, USA) protocols.
The SOPHiA HCS allows for the enrichment of coding and splicing regions of 26 genes (APC, ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, FAM175A, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PIK3CA, PMS2, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53, XRCC2) and the pseudogene PMS2CL. This is known to be associated with increased risk for cancer syndromes. The sequencing data were simultaneously processed for single nucleotide variants (SNVs), indels and copy number variations (CNVs) using the SOPHiA DDM software (DDM) updated to the last available version at the time of sequencing. In accordance with local and international guidelines as well as with the patients’ informed consent, data analysis and variant interpretation were limited to the following actionable gene set: APC, ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53. Genetic variant annotations were also integrated with data from in literature and open source bioinformatics tools customized and validated in the laboratory (Annovar [29] and Variant Effect Predictor (VEP) [30]), and through consultation of specific databases: Leiden Open source Variation Database (https://grenada.lumc.nl/LOVD2/mendelian_genes/home.php? accessed on 20 April 2021), ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/ accessed on 20 April 2021), 1000 Genomes Project (http://www.1000genomes.org/data accessed on 20 April 2021), ExAC (http://exac.broadinstitute.org/ accessed on 20 April 2021), dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/ accessed on 20 April 2021), The Genome Aggregation Database (http://gnomad.broadinstitute.org/ accessed on 20 April 2021), BRCA Share (http://www.umd.be/BRCA1/ http://www.umd.be/BRCA2/ accessed on 20 April 2021). Variants were reported using the international standard HGVS nomenclature and a classification into 5 classes (Pathogenic, Likely Pathogenic, Variant of Uncertain Significance, Likely Benign and Benign), according to the American College of Medical Genetics and Genomics (ACMG, Bethesda, MD, USA) criteria [31].
All gene variants or CNVs interpreted as Pathogenic or Likely Pathogenic were confirmed by Sanger sequencing performed with predesigned primers (the BigDye Direct Cycle Sequencing Kit), analyzed through the Applied Biosystems 3500xL Dx Genetic Analyzer platform and SeqScape3 software (Thermo Scientific), or by MLPA (MRC-Holland, Amsterdam, The Netherlands) and examined through the Coffalyser.Net software (MRC-Holland) updated to the latest available version.
3. Results
3.1. Overall Population
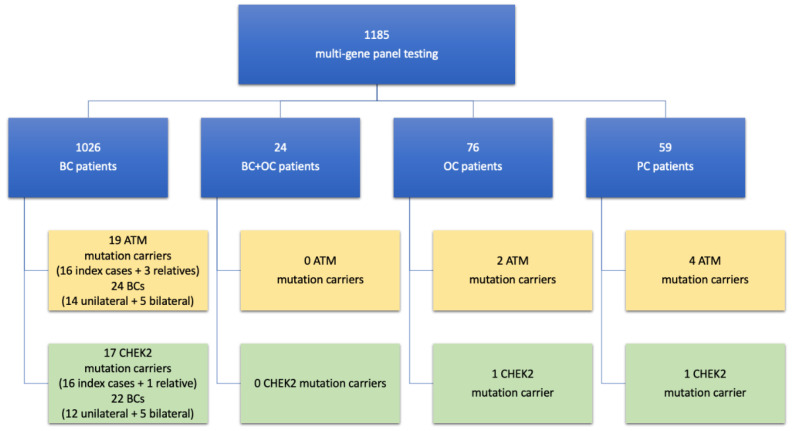
Since the 8 January 2018, 1185 multi-gene panel tests have been performed. Of these tests, 1026 were performed on BC patients (422 of these on those who previously tested negative for BRCA genes). In addition, 24 were performed on BC and OC patients, with 11 of these previously testing negative for BRCA genes. Moreover, 76 tests were conducted on OC patients, 48 of these previously testing negative for BRCA genes, and 59 on PC patients (never tested before). Overall, ATM pathogenic or likely pathogenic germline variants were found in 16 index BC cases (detection rate among BC patients: 1.5%) and in 3 relatives affected by BC (Figure 2). On the other hand, CHEK2 pathogenic or likely pathogenic germline variants were found in 16 index BC cases (detection rate among BC patients: 1.5%) and one relative affected by BC (Figure 2). The likely pathogenic and pathogenic variants of ATM and CHEK2 detected in our population are listed in Table 2.
Figure 2.
Study flow-chart.
Table 2.
Likely pathogenic and pathogenic variants of ATM and CHEK2 in our study population of BC patients.
| Variants of ATM Detected | Variant Classification | Number of BC Patients | |
|---|---|---|---|
| c.6154G>A, p.Glu2052Lys | Likely pathogenic | 5 index cases | 1 relative |
| c.2838+2162_4110-292del | Pathogenic | 1 index case | 1 relative |
| c.5441T>A, p.Leu1814 * | Pathogenic | 1 index case | 1 relative |
| c.(8850+1_8851-1)_(*3591_?)del, p.(?) | Pathogenic | 1 index case | - |
| c.8010+1delG | Likely pathogenic | 1 index case | - |
| c.5697C>A, p.Cys1899 * | Pathogenic | 1 index case | - |
| c.7327C>T, p.Arg2443 * | Pathogenic | 1 index case | - |
| c.2192dupA, p.Tyr731 * | Pathogenic | 1 index case | - |
| c.2135C>G, p.Ser712 * | Likely pathogenic | 1 index case | - |
| c.8395_8404del10, p.Phe2799Lysfs *4 | Pathogenic | 1 index case | - |
| c.8814_8824del, p.Met2938Ilefs * | Pathogenic | 1 index case | - |
| c.5932G>T, p.Glu1978 * | Pathogenic | 1 index case | - |
| Variants of CHEK2 detected | Variant classification | Number of BC patients | |
| c.190G>A, p.Glu64Lys | Likely pathogenic | 5 index cases | - |
| c.470T>C, p.Ile157Thr | Pathogenic | 3 index cases | - |
| c.1169A>C, p.Tyr390Ser | Likely pathogenic | 2 index cases | 1 relative |
| c.1100delC, p.Thr367Metfs *15 | Pathogenic | 2 index cases | - |
| c.1189A>C, p.Tyr390Ser | Likely pathogenic | 1 index case | - |
| c.592+3A>T, p.(?) | Likely pathogenic | 1 index case | - |
| c.549G>C, p.Leu183Phe | Pathogenic | 1 index case | - |
| c.85C>T, p.Gln29 * | Pathogenic | 1 index case | - |
The final analysis included 36 BC patients (19 ATM mutation carriers and 17 CHEK2 mutation carriers) affected by 46 different BCs, with ten patients developing bilateral BC. The characteristics of patients, tumors and treatments are outlined in Table 3.
Table 3.
Patient and tumor characteristics.
| BC in ATM Mutation Carriers (n = 24) | BC in CHEK2 Mutation Carriers (n = 22) | |
|---|---|---|
| Median age at First BC Diagnosis, Years | 46.9 | 46.1 |
| Hystotype (n, %) | (n = 24) | (n = 22) |
| In situ ductal carcinoma | 6 (25) | 6 (30) |
| Invasive ductal carcinoma | 16 (66.7) | 11 (55) |
| Invasive lobular carcinoma | 2 (8.3) | 3 (15) |
| data not available | 0 | 2 |
| Clinical Stage at diagnosis (n, %) | (n = 24) | (n = 22) |
| is | 6 (27.3) | 6 (30) |
| I/II | 13 (59.1) | 11 (55) |
| III | 3 (13.6) | 1 (5) |
| IV | 0 | 2 (10) |
| data not available | 2 | 2 |
| Immunohistochemical profile of invasive carcinomas (n, %) |
(n = 18) | (n = 16) |
| HR+/HER2- | 9 (56.3) | 11 (78.6) |
| Luminal A-like | 4 (25.1) | 9 (64.3) |
| Luminal B-like | 5 (31.2) | 2 (14.3) |
| HR–/HER2+ | 0 | 0 |
| HR+/HER2+ | 4 (25) | 3 (21.4) |
| TNBC | 3 (18.8) | 0 |
| data not available | 2 | 2 |
| Grade of invasive carcinomas (n, %) | (n = 18) | (n = 16) |
| G1-2 | 6 (42.8%) | 8 (57.1%) |
| G3 | 8 (57.1%) | 6 (42.8%) |
| data not available | 4 | 2 |
| Breast Surgery (n, %) | (n = 24) | (n = 22) |
| Mastectomy | 7 (33.3) | 10 (55.6) |
| Conserving surgery | 13 (61.9) | 8 (44.4) |
| No breast surgery | 1 (4.8) | 0 |
| data not available or stage IV | 3 | 4 |
| Axillary Surgery (n, %) | (n = 24) | (n = 22) |
| Sentinel node biopsy | 11 (52.4) | 7 (38.9) |
| Axillary node dissection | 4 (19) | 7 (38.9) |
| No axillary surgery | 6 (28.6) | 4 (22.2) |
| data not available or stage IV | 3 | 4 |
| Radiotherapy (n, %) | (n = 24) | (n = 22) |
| Yes | 16 (84.2) | 7 (41.2) |
| No | 3 (15.8) | 10 (58.8) |
| data not available or stage IV | 5 | 5 |
| Neoadjuvant chemotherapy in invasive carcinomas (n, %) |
(n = 18) | (n = 16) |
| Yes | 6 (42.9) | 1 (9.1) |
| No | 8 (57.1) | 10 (90.9) |
| data not available or stage IV | 4 | 5 |
| Adjuvant chemotherapy in invasive carcinomas (n, %) |
(n = 18) | (n = 16) |
| Yes | 7 (53.8) | 4 (36,4) |
| No | 6 (46.2) | 7 (63,6) |
| data not available or stage IV | 5 | 5 |
| Local or distant recurrence in localized BC at diagnosis (n, %) |
(n = 24) | (n = 20) |
| Yes | 0 (0) | 1 (5) |
| No | 24 (100) | 19 (95) |
| Median follow-up since diagnosis (months) | 106 | 152 |
HR+/HER2-: hormonal-receptor positive and HER2 negative; HR–/HER2+: hormonal-receptor negative and HER2 positive; HR+/HER2+: hormonal-receptor and HER2 positive; TNBC: triple negative breast cancer.
Additionally, in 24 patients affected by both BC and OC, neither ATM nor CHEK2 likely pathogenic or pathogenic variants were detected. In 76 OC patients, two ATM (one index case and one relative) and one CHEK2 (index case) likely pathogenic or pathogenic variants were found. In 59 PC patients, finally, four ATM (all index cases) and one CHEK2 (index case) likely pathogenic or pathogenic variants were detected.
3.2. ATM Mutation Carriers
Median age at first BC diagnosis in ATM mutation carriers was 46.9 years. Five patients (5/19, 26.3%) developed bilateral BC (one of them synchronous). Therefore, 24 tumors were analyzed in ATM carriers. Overall, 6 (25%) tumors were accounted for as ductal carcinoma in situ, while invasive ductal and lobular carcinoma amounted to 16 (66.7%) and 2 (8.3%), respectively. Invasive tumors were hormonal-receptor (HR) positive and HER2 negative (HR+/HER2–) in 9 (56.3%) cases, both HR and HER2 positive (HR+/HER2+) in 4 (25%) cases and triple negative in 3 (18.8%) cases. No HR–/HER2+ tumors were found. Thirteen (59.1%) early-stage BC (I/II stage) and three (13.6%) locally advanced tumors (III stage) were detected. No “de novo” metastatic BC were diagnosed.
Based on available data, seven (33.3%) tumors were treated by mastectomy and 13 (61.9%) through breast conserving surgery. One patient underwent axillary node dissection without breast surgery for CUP syndrome. Eleven (52.4%) sentinel node biopsies and four (19.1%) axillary node dissections were performed (with no axillary surgery in 6 cases). In 16 (84.2%) cases, radiation therapy followed breast surgery. Six out of 14 (42.9%) patients diagnosed with invasive BC underwent neoadjuvant chemotherapy, whereas 7 out of 13 (53.8%) patients underwent adjuvant chemotherapy. After a median follow up of 106 months, no local or distant recurrences were observed.
The most frequent mutation detected in the ATM gene was c.6154G>A, p.Glu2052Lys (5 out of 19 patients, 26.3%). Three of these women developed bilateral BC and five out of eight of these tumors were categorized as DCIS.
Twelve patients (63.2%) had a positive BC family history. In addition, a family history of ovarian, gastric, kidney/bladder and colon cancer were documented for 4 (21%) patients each, while a family history of pancreatic cancer was reported for 3 (15.8%) patients. One patient carrying an ATM pathogenic mutation with BC also developed gastric cancer. Moreover, two cases of epithelial ovarian cancer and four cases of pancreatic cancer were detected in six carriers of ATM pathogenic or likely pathogenic germline variants.
3.3. CHEK2 Mutation Carriers
Median age at first BC diagnosis in CHEK2 mutation carriers was 46.1 years. Five patients (5/17, 29.4%) had bilateral BC (two of them synchronous). Therefore, 22 CHEK2-associated tumors were analyzed. Overall, 6 (30%) tumors were accounted for as ductal carcinoma in situ (Stage 0), while invasive ductal and lobular carcinoma amounted to 11 (55%) and 3 (15%), respectively. Invasive tumors were HR positive and HER2 negative (HR+/HER2–) in 11 (78.6%) cases, both HR and HER2 positive (HR+/HER2+) in 3 (21.4%) cases. No HR–/HER2+ and triple negative BCs were found. Eleven (55%) early-stage BC (I/II stage), one (5%) locally advanced tumor (III stage) and 2 (10%) stage IV cancers were diagnosed.
In patients diagnosed with localized BC, 10 (55.6%) tumors were treated by mastectomy and eight (44.4%) by breast conserving surgery. Seven (38.9%) sentinel node biopsies and seven (38.9%) axillary node dissections were performed (with no axillary surgery in 4 cases). In seven (41.2%) cases, radiation therapy followed breast surgery. One out of 11 (9.1%) patients diagnosed with invasive BC underwent neoadjuvant chemotherapy, whereas 4 out of 11 (36.4%) patients underwent adjuvant chemotherapy. After a median follow up of 152 months, only one local recurrence and no distant recurrences were observed in patients diagnosed with stage I–III BC.
The most frequent mutation detected in the CHEK2 gene was c.190G>A, p.Glu64Lys (5 out of 17 patients, 29.4%). One of these women developed bilateral BC. The second most frequent pathogenic variant was c.470T>C, p.Ile157Thr and one case of bilateral tumor was observed in these women. The founder mutation c.1100delC, p.Thr367Metfs*15 was present in two patients.
Eleven patients (64.7%) had a positive BC family history and 6 (35.3%) patients one of colon cancer, whereas 4 (23.5%) patients had a prostate cancer family history and 4 (23.5%) patients one of kidney/bladder cancer. Six of our BC patients carrying a CHEK2 pathogenic mutation were also diagnosed with thyroid carcinoma, acute myeloid leukemia, colon cancer, malignant melanoma, or uterine endometrial carcinoma. In addition to BC, the following malignancies were detected in five carriers of CHEK2 pathogenic or likely pathogenic germline variants: pancreatic cancer, uterine cancer, prostate cancer, bladder cancer, multiple myeloma, kidney cancer, colon cancer, and thyroid carcinoma.
4. Discussion
The ATM and CHEK2 genes encode proteins that act as tumor suppressors and are involved in the DNA damage response following generation of DNA double-strand breaks (DSBs) [4]. Second to the BRCA1 and BRCA2 genes, the most common germline pathogenic or likely pathogenic variants predisposing to BC were found in the ATM and CHEK2 genes [2,3]. Individuals carrying heterozygous pathogenic variants in ATM or CHEK2 present a 33% and 28–37% cumulative lifetime risk for BC by 80 years of age, respectively [7,8,9]. Nevertheless, while the phenotypes of BRCA-related tumors have been widely characterized, little is known about the clinicopathologic features of ATM and CHEK2-associated tumors, BC in the first place.
Interestingly, CHEK2 has the highest mutation prevalence in individuals of European descent, while the spectrum and frequency of pathogenic variants vary among specific European populations. In particular, the frequency of the founder mutation c.1100delC declines from the north to the south of Europe, whereas the most frequent European CHEK2 variant, p.I157T, has a carrier frequency of around 5% in Poles, Latvians, Hungarians and Russians and around 2–3% in Czechs, Slovaks and Germans [32]. In our study, the most frequent CHEK2 mutation beyond c.470T>C, p.Ile157Thr (p.I157T), was c.190G>A, p.Glu64Lys. This likely pathogenic variant was observed at an allele frequency of 0.03% (38/126,668) in individuals of European ancestry in large population cohorts [33] and has been associated with a personal and/or family history of breast, prostate, ovarian, colorectal, thyroid and pancreatic cancer [34,35,36,37,38,39,40,41]. On the other hand, the most frequent ATM mutation in our population was c.6154G>A, p.Glu2052Lys. This likely pathogenic variant has also previously been reported in individuals with a personal and/or family history of breast and/or ovarian cancer [34,42]. It is noteworthy that one of the ATM mutations described in our population, c.2838+2162_4110-292del, has been recently characterized at a molecular level by our research group [43].
Overall, our study identified 19 ATM mutation carriers with 24 breast tumors and 17 CHEK2 mutation carriers with 22 breast tumors. Median age at first BC onset was 46.9 years for ATM and 46.1 years for CHEK2, in keeping with the literature [44]. Moreover, a high rate of bilateral tumors was observed in ATM (26.3%) and CHEK2 mutation carriers (41.2%). Previous studies differ from one another on the role of ATM mutations in increasing the risk of contralateral BC [45,46,47]. On the other hand, bilateral BC was reported for 3.7–12.1% of the patients harboring a CHEK2 likely pathogenic or pathogenic variant [48,49], whereas a recent analysis has provided evidence of contralateral BC in 19.5% of Slovenian BC patients with CHEK2 mutations [16]. Moreover, a systematic review and meta-analysis by Akdeniz et al. [50] has recently shown a strong association with contralateral BC for carriers of CHEK2 c.1100delC mutation (relative risk, 2.7). In our study, however, only two patients with this variant were included (one of them with synchronous bilateral BC), so that no conclusion can be drawn. Interestingly, the low frequency of the founder mutation c.1100delC variant in our cohort of patients is consistent with a previous analysis reporting this variant in only 1 of 939 (0.11%, 95% CI = 0.00–0.59%) unrelated patients from Italian breast cancer families. These results indicate that the CHEK2 c.1100delC variant has marginal relevance to breast cancer predisposition in the Italian population [51]. In our cohort, interestingly, all patients underwent genetic testing after breast surgery: this could have determined the high rate of contralateral BC since none of these women underwent risk-reducing contralateral mastectomy. Although the evidence on whether contralateral prophylactic mastectomy improves survival for BRCA carriers with BC is conflicting, this procedure reduces the risk of contralateral tumor by 93% [52]. As a result, several international guidelines include this option [10,53]. However, no studies have investigated risk reduction and survival advantage in relation to contralateral prophylactic mastectomy for patients with a diagnosis of BC harboring ATM or CHEK2 likely pathogenic or pathogenic variants. In this regard, therefore, the decision should be shared with patients following a multidisciplinary and personalized approach.
In line with previous research [15,16,54,55,56,57] and unlike BRCA1 and PALB2-associated BCs that commonly present triple-negative subtype [58,59], ATM and CHEK2-related BC in our population mostly resulted in luminal-like subtypes. In particular, 64.3% of the CHEK2 tumors were luminal A-like, whereas most of the ATM tumors were luminal B-like/HER2-negative (56.2%). Interestingly, 81.2% of the ATM tumors and 100% of the CHEK2 tumors were HR positive. In addition, 25% of ATM BCs and 21.4% of CHEK2 BCs were observed to be HER2 positive, while only 18.8% of the ATM BCs and none of the the CHEK2 tumors were accounted for as triple negative BC. Consistent with that, no CHEK2 mutation carriers were observed in a previous analysis of 1824 triple negative breast cancer patients [60]. Contrary to what was described in previous experiences [61,62], it is notable that most of the CHEK2 tumors (57.1%) were associated with lower grades (G1-G2). Finally, 21.4% of the CHEK2-related invasive tumors showed a lobular histotype, a high rate as previously highlighted in the literature [16,63,64]. In our population, however, due to the small sample size, the lobular histotype was not associated with any particular variant. On the other hand, ATM invasive BC in our population showed no particular histological subtype (88.9% were invasive ductal carcinomas and 11.1% were invasive lobular carcinomas), as already observed elsewhere [15].
About a quarter of all ATM-related BCs and a third of CHEK2 BCs were in situ carcinomas and more than half of ATM and CHEK2-related BCs were diagnosed at stage I-II (59.1% and 55%, respectively), whereas 13.6% of the ATM BCs and 15% of the CHEK2 BCs were stage III-IV. Despite the early stages at diagnosis, 55.6% of the CHEK2 BCs were treated by mastectomy and 38.9% by axillary node dissection, since most of these patients underwent surgery in the late 90s when breast surgery was less conservative. A higher rate of ATM-associated BCs was treated with neoadjuvant chemotherapy (42.9%) and/or adjuvant chemotherapy (53.8%) compared to CHEK2 tumors (9.1% and 36.4%, respectively), reflecting the higher percentage of stage III, HER2 positive and triple-negative BC in ATM-related tumors. Confirming good prognosis for luminal-like subtypes and early-stage BCs [65], after more than 8 years of follow up in both groups, only one local recurrence was observed in localized BCs at diagnosis.
As previously described for other cohorts [16,17], 63.2% of ATM mutation carriers and 64.7% of CHEK2 mutation carriers presented a positive BC family history. Nevertheless, both germline ATM and CHEK2 likely pathogenic or pathogenic variants have been linked with susceptibility to several malignancies other than BC. In our population, accordingly, ATM families exhibited ovarian cancer in 21% of cases and pancreatic cancer in 15.8% of cases, besides gastric, kidney/bladder and colon tumors. On the other hand, CHEK2 families presented a recurrence of colon cancer (35.3% of cases), prostate tumors (23.5%) and kidney/bladder cancers (23.5%).
As has already been the case with BRCA-associated tumors, the definition of biological and clinical characteristics of ATM and CHEK2-related tumors may help improve diagnosis, prognostication and targeted therapeutic approaches [66,67,68]. In particular, in light of the high rate of contralateral tumors described in our experience, we believe that contralateral mastectomy should be considered and discussed with ATM and even more with CHEK2 mutation carriers at the first diagnosis of BC. Further studies with larger patient cohorts are needed to confirm our findings and assist both patients and physicians in decision making and management recommendations in this subset of patients.
Acknowledgments
The authors would like to thank the Angela Serra Association for Cancer Research for their support to patients and research activities.
Author Contributions
Conceptualization, A.T. and L.C.; methodology, C.P.; validation, E.B. and E.R.; formal analysis, F.D.; investigation, E.G. and F.C. (Francesca Combi); resources, M.V.; data curation, E.T. (Elena Tenedini) and I.M.; writing—original draft preparation, C.P. and A.T.; writing—review and editing, E.T. (Elena Tenedini) and L.C.; visualization, G.G. and M.V.; supervision, G.T., M.D. and E.T. (Enrico Tagliafico); project administration, F.C. (Federica Caggia). All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the grant Ricerca Finalizzata Giovane Ricercatore 2018 with the project GR-2018-12367239.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the Area Vasta Emilia Nord (Project identification code 125/2021/OSS*/AOUMO).
Informed Consent Statement
Written informed consent was obtained from the patients to publish this paper.
Data Availability Statement
The data presented in this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest associated with this manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Desmond A., Kurian A.W., Gabree M., Mills M.A., Anderson M.J., Kobayashi Y., Horick N., Yang S., Shannon K.M., Tung N., et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 2015;1:943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 2.Kapoor N.S., Curcio L.D., Blakemore C.A., Bremner A.K., McFarland R.E., West J.G., Banks K.C. Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer. Ann. Surg. Oncol. 2015;22:3282–3288. doi: 10.1245/s10434-015-4754-2. [DOI] [PubMed] [Google Scholar]
- 3.Hu C., Hart S.N., Gnanaolivu R., Huang H., Lee K.Y., Na J., Gao C., Lilyquist J., Yadav S., Boddicker N.J., et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y., McCorvie T.J., Yates L.A., Zhang X. Structural basis of homologous recombination. Cell. Mol. Life Sci. 2020;77:3–18. doi: 10.1007/s00018-019-03365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandita T.K., Lieberman H.B., Lim D.S., Dhar S., Zheng W., Taya Y., Kastan M.B. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene. 2000;19:1386–1391. doi: 10.1038/sj.onc.1203444. [DOI] [PubMed] [Google Scholar]
- 6.Pommier Y., Weinstein J.N., Aladjem M.I., Kohn K.W. Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin. Cancer Res. 2006;12:2657–2661. doi: 10.1158/1078-0432.CCR-06-0743. [DOI] [PubMed] [Google Scholar]
- 7.Marabelli M., Cheng S.-C., Parmigiani G. Penetrance of ATM Gene mutations in breast cancer: A meta-analysis of different measures of risk. Genet. Epidemiol. 2016;40:425–431. doi: 10.1002/gepi.21971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cybulski C., Wokołorczyk D., Jakubowska A., Huzarski T., Byrski T., Gronwald J., Masojć B., Deebniak T., Górski B., Blecharz P., et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J. Clin. Oncol. 2011;29:3747–3752. doi: 10.1200/JCO.2010.34.0778. [DOI] [PubMed] [Google Scholar]
- 9.Piombino C., Cortesi L., Lambertini M., Punie K., Grandi G., Toss A. Secondary Prevention in Hereditary Breast and/or Ovarian Cancer Syndromes Other Than BRCA. J. Oncol. 2020;2020:6384190:1–6384190:10. doi: 10.1155/2020/6384190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Comprehensive Cancer Network Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic (Version 2.2021) [(accessed on 17 December 2020)]; Available online: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf.
- 11.Lu H.-M., Li S., Black M.H., Lee S., Hoiness R., Wu S., Mu W., Huether R., Chen J., Sridhar S., et al. Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing. JAMA Oncol. 2019;5:51–57. doi: 10.1001/jamaoncol.2018.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shindo K., Yu J., Suenaga M., Fesharakizadeh S., Cho C., Macgregor-Das A., Siddiqui A., Witmer P.D., Tamura K., Song T.J., et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J. Clin. Oncol. 2017;35:3382–3390. doi: 10.1200/JCO.2017.72.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilié P.G., Johnson A.M., Hanson K.L., Dayno M.E., Kapron A.L., Stoffel E.M., Cooney K.A. Germline genetic variants in men with prostate cancer and one or more additional cancers. Cancer. 2017;123:3925–3932. doi: 10.1002/cncr.30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cybulski C., Górski B., Huzarski T., Masojć B., Mierzejewski M., Debniak T., Teodorczyk U., Byrski T., Gronwald J., Matyjasik J., et al. CHEK2 is a multiorgan cancer susceptibility gene. Am. J. Hum. Genet. 2004;75:1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renault A.-L., Mebirouk N., Fuhrmann L., Bataillon G., Cavaciuti E., Le Gal D., Girard E., Popova T., La Rosa P., Beauvallet J., et al. Morphology and genomic hallmarks of breast tumours developed by ATM deleterious variant carriers. Breast Cancer Res. 2018;20:28. doi: 10.1186/s13058-018-0951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nizic-Kos T., Krajc M., Blatnik A., Stegel V., Skerl P., Novakovic S., Gazic B., Besic N. Bilateral Disease Common Among Slovenian CHEK2-Positive Breast Cancer Patients. Ann. Surg. Oncol. 2020:1–10. doi: 10.1245/s10434-020-09178-y. [DOI] [PubMed] [Google Scholar]
- 17.Bergstrom C., Pence C., Berg J., Partain N., Sadeghi N., Mauer C., Pirzadeh-Miller S., Gao A., Li H., Unni N., et al. Clinicopathological Features and Outcomes in Individuals with Breast Cancer and ATM, CHEK2, or PALB2 Mutations. Ann. Surg. Oncol. 2020 doi: 10.1245/s10434-020-09158-2. [DOI] [PubMed] [Google Scholar]
- 18.Giunta Della Regione Emilia Romagna Rischio Eredo-Familiare Per Il Carcinoma Della Mammella—Approvazione Linee Guida Per Le Aziende Sanitarie Della Regione Emilia Romagna. [(accessed on 17 December 2020)]; Available online: https://salute.regione.emilia-romagna.it/normativa-e-documentazione/leggi/regionali/delibere/dgr.-220-2011.
- 19.Cortesi L., Razzaboni E., Toss A., De Matteis E., Marchi I., Medici V., Tazzioli G., Andreotti A., De Santis G., Pignatti M., et al. A rapid genetic counselling and testing in newly diagnosed breast cancer is associated with high rate of risk-reducing mastectomy in BRCA1/2-positive Italian women. Ann. Oncol. 2014;25:57–63. doi: 10.1093/annonc/mdt422. [DOI] [PubMed] [Google Scholar]
- 20.Razzaboni E., Toss A., Cortesi L., Marchi I., Sebastiani F., De Matteis E., Federico M. Acceptability and adherence in a chemoprevention trial among women at increased risk for breast cancer attending the Modena Familial Breast and Ovarian Cancer Center (Italy) Breast J. 2013;19:10–21. doi: 10.1111/tbj.12045. [DOI] [PubMed] [Google Scholar]
- 21.Cortesi L., Canossi B., Battista R., Pecchi A., Drago A., Dal Molin C., Toss A., De Matteis E., Marchi I., Torricelli P., et al. Breast ultrasonography (BU) in the screening protocol for women at hereditary-familial risk of breast cancer: Has the time come to rethink the role of BU according to different risk categories? Int. J. Cancer. 2019;144:1001–1009. doi: 10.1002/ijc.31794. [DOI] [PubMed] [Google Scholar]
- 22.Cortesi L., De Matteis E., Toss A., Marchi I., Medici V., Contu G., Xholli A., Grandi G., Cagnacci A., Federico M. Evaluation of Transvaginal Ultrasound plus CA-125 Measurement and Prophylactic Salpingo-Oophorectomy in Women at Different Risk Levels of Ovarian Cancer: The Modena Study Group Cohort Study. Oncology. 2017;93:377–386. doi: 10.1159/000479155. [DOI] [PubMed] [Google Scholar]
- 23.Federico M., Maiorana A., Mangone L., Turchetti D., Canossi B., Romagnoli R., Silingardi V. Identification of families with hereditary breast and ovarian cancer for clinical and mammographic surveillance: The Modena Study Group proposal. Breast Cancer Res. Treat. 1999;55:213–221. doi: 10.1023/A:1006192230332. [DOI] [PubMed] [Google Scholar]
- 24.Cortesi L., Turchetti D., Marchi I., Fracca A., Canossi B., Rachele B., Silvia R., Rita P.A., Pietro T., Massimo F. Breast cancer screening in women at increased risk according to different family histories: An update of the Modena Study Group experience. BMC Cancer. 2006;6:210. doi: 10.1186/1471-2407-6-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018;36:2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 26.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch. Pathol. Lab. Med. 2010;134:907–922. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardoso F., Kyriakides S., Ohno S., Penault-Llorca F., Poortmans P., Rubio I.T., Zackrisson S., Senkus E. ESMO Guidelines Committee. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2019;30:1194–1220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 28.Goldhirsch A., Winer E.P., Coates A.S., Gelber R.D., Piccart-Gebhart M., Thürlimann B., Senn H.J., Wood W.C. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K., Li M., Hakonarson H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stolarova L., Kleiblova P., Janatova M., Soukupova J., Zemankova P., Macurek L., Kleibl Z. CHEK2 Germline Variants in Cancer Predisposition: Stalemate Rather than Checkmate. Cells. 2020;9:2675. doi: 10.3390/cells9122675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraus C., Hoyer J., Vasileiou G., Wunderle M., Lux M.P., Fasching P.A., Krumbiegel M., Uebe S., Reuter M., Beckmann M.W., et al. Gene panel sequencing in familial breast/ovarian cancer patients identifies multiple novel mutations also in genes others than BRCA1/2. Int. J. Cancer. 2017;140:95–102. doi: 10.1002/ijc.30428. [DOI] [PubMed] [Google Scholar]
- 35.Desrichard A., Bidet Y., Uhrhammer N., Bignon Y.J. CHEK2 contribution to hereditary breast cancer in non-BRCA families. Breast Cancer Res. 2011;13:R119. doi: 10.1186/bcr3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eliade M., Skrzypski J., Baurand A., Jacquot C., Bertolone G., Loustalot C., Coutant C., Guy F., Fumoleau P., Duffourd Y., et al. The transfer of multigene panel testing for hereditary breast and ovarian cancer to healthcare: What are the implications for the management of patients and families? Oncotarget. 2017;8:1957–1971. doi: 10.18632/oncotarget.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Garay M.L., McGuire A.L., Pereira S., Caskey C.T. Personalized genomic disease risk of volunteers. Proc. Natl. Acad. Sci. USA. 2013;110:16957–16962. doi: 10.1073/pnas.1315934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong X., Wang L., Taniguchi K., Wang X., Cunningham J.M., McDonnell S.K., Qian C., Marks A.F., Slager S.L., Peterson B.J., et al. Mutations in CHEK2 associated with prostate cancer risk. Am. J. Hum. Genet. 2003;72:270–280. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Susswein L.R., Marshall M.L., Nusbaum R., Vogel Postula K.J., Weissman S.M., Yackowski L., Vaccari E.M., Bissonnette J., Booker J.K. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet. Med. 2016;18:823–832. doi: 10.1038/gim.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurgelun M.B., Kulke M.H., Fuchs C.S., Allen B.A., Uno H., Hornick J.L., Ukaegbu C.I., Brais L.K., McNamara P.G., Mayer R.J., et al. Cancer Susceptibility Gene Mutations in Individuals with Colorectal Cancer. J. Clin. Oncol. 2017;35:1086–1095. doi: 10.1200/JCO.2016.71.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirts B.H., Casadei S., Jacobson A.L., Lee M.K., Gulsuner S., Bennett R.L., Miller M., Hall S.A., Hampel H., Hisama F.M., et al. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet. Med. 2016;18:974–981. doi: 10.1038/gim.2015.212. [DOI] [PubMed] [Google Scholar]
- 42.Singh J., Thota N., Singh S., Padhi S., Mohan P., Deshwal S., Sur S., Ghosh M., Agarwal A., Sarin R., et al. Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: Prevalence of BRCA1/2 and non-BRCA mutations. Breast Cancer Res. Treat. 2018;170:189–196. doi: 10.1007/s10549-018-4726-x. [DOI] [PubMed] [Google Scholar]
- 43.Parenti S., Rabacchi C., Marino M., Tenedini E., Artuso L., Castellano S., Carretta C., Mallia S., Cortesi L., Toss A., et al. Characterization of New ATM Deletion Associated with Hereditary Breast Cancer. Genes. 2021;12:136. doi: 10.3390/genes12020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skasko E., Kluska A., Niwińska A., Kwiatkowska E., Bałabas A., Piatkowska M., Dabrowska M., Nowakowska D., Pieńkowski T. Age at onset of bilateral breast cancer, the presence of hereditary BRCA1, BRCA2, CHEK2 gene mutations and positive family history of cancer. Onkologie. 2009;32:182–188. doi: 10.1159/000200930. [DOI] [PubMed] [Google Scholar]
- 45.Tommiska J., Jansen L., Kilpivaara O., Edvardsen H., Kristensen V., Tamminen A., Aittomäki K., Blomqvist C., Børresen-Dale A.-L., Nevanlinna H. ATM variants and cancer risk in breast cancer patients from Southern Finland. BMC Cancer. 2006;6:209. doi: 10.1186/1471-2407-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broeks A., Urbanus J.H., Floore A.N., Dahler E.C., Klijn J.G., Rutgers E.J., Devilee P., Russell N.S., van Leeuwen F.E., van’t Veer L.J. ATM-heterozygous germline mutations contribute to breast cancer-susceptibility. Am. J. Hum. Genet. 2000;66:494–500. doi: 10.1086/302746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein J.L., Haile R.W., Stovall M., Boice J.D., Jr., Shore R.E., Langholz B., Thomas D.C., Bernstein L., Lynch C.F., Olsen J.H., et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J. Natl. Cancer Inst. 2010;102:475–483. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vahteristo P., Bartkova J., Eerola H., Syrjäkoski K., Ojala S., Kilpivaara O., Tamminen A., Kononen J., Aittomäki K., Heikkilä P., et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am. J. Hum. Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuusisto K.M., Bebel A., Vihinen M., Schleutker J., Sallinen S.L. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 2011;13:R20. doi: 10.1186/bcr2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akdeniz D., Schmidt M.K., Seynaeve C.M., McCool D., Giardiello D., van den Broek A.J., Hauptmann M., Steyerberg E.W., Hooning M.J. Risk factors for metachronous contralateral breast cancer: A systematic review and meta-analysis. Breast. 2019;44:1–14. doi: 10.1016/j.breast.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Caligo M.A., Agata S., Aceto G., Crucianelli R., Manoukian S., Peissel B., Scaini M.C., Sensi E., Veschi S., Cama A., et al. The CHEK2 c.1100delC mutation plays an irrelevant role in breast cancer predisposition in Italy. Hum. Mutat. 2004;24:100–101. doi: 10.1002/humu.20051. [DOI] [PubMed] [Google Scholar]
- 52.Teoh V., Tasoulis M.K., Gui G. Contralateral Prophylactic Mastectomy in Women with Unilateral Breast Cancer Who Are Genetic Carriers, Have a Strong Family History or Are just Young at Presentation. Cancers (Basel) 2020;12:140. doi: 10.3390/cancers12010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paluch-Shimon S., Cardoso F., Sessa C., Balmana J., Cardoso M.J., Gilbert F., Senkus E., ESMO Guidelines Committee Prevention and screening in BRCA mutation carriers and other breast/ovarian hereditary cancer syndromes: ESMO Clinical Practice Guidelines for cancer prevention and screening. Ann. Oncol. 2016;27:v103–v110. doi: 10.1093/annonc/mdw327. [DOI] [PubMed] [Google Scholar]
- 54.Cybulski C., Huzarski T., Byrski T., Gronwald J., Debniak T., Jakubowska A., Górski B., Wokołorczyk D., Masojć B., Narod S.A., et al. Estrogen receptor status in CHEK2-positive breast cancers: Implications for chemoprevention. Clin. Genet. 2009;75:72–78. doi: 10.1111/j.1399-0004.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 55.Keeney M.G., Couch F.J., Visscher D.W., Lindor N.M. Non-BRCA familial breast cancer: Review of reported pathology and molecular findings. Pathology. 2017;49:363–370. doi: 10.1016/j.pathol.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Massink M.P., Kooi I.E., Martens J.W., Waisfisz Q., Meijers-Heijboer H. Genomic profiling of CHEK2*1100delC-mutated breast carcinomas. BMC Cancer. 2015;15:877. doi: 10.1186/s12885-015-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domagala P., Wokolorczyk D., Cybulski C., Huzarski T., Lubinski J., Domagala W. Different CHEK2 germline mutations are associated with distinct immunophenotypic molecular subtypes of breast cancer. Breast Cancer Res. Treat. 2012;132:937–945. doi: 10.1007/s10549-011-1635-7. [DOI] [PubMed] [Google Scholar]
- 58.Cybulski C., Kluźniak W., Huzarski T., Wokołorczyk D., Kashyap A., Jakubowska A., Szwiec M., Byrski T., Dębniak T., Górski B., et al. Clinical outcomes in women with breast cancer and a PALB2 mutation: A prospective cohort analysis. Lancet Oncol. 2015;16:638–644. doi: 10.1016/S1470-2045(15)70142-7. [DOI] [PubMed] [Google Scholar]
- 59.Musolino A., Bella M.A., Bortesi B., Michiara M., Naldi N., Zanelli P., Capelletti M., Pezzuolo D., Camisa R., Savi M., et al. BRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: A population-based study. Breast. 2007;16:280–292. doi: 10.1016/j.breast.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 60.Couch F.J., Hart S.N., Sharma P., Toland A.E., Wang X., Miron P., Olson J.E., Godwin A.K., Pankratz V.S., Olswold C., et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kleiblova P., Stolarova L., Krizova K., Lhota F., Hojny J., Zemankova P., Havranek O., Vocka M., Cerna M., Lhotova K., et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int. J. Cancer. 2019;145:1782–1797. doi: 10.1002/ijc.32385. [DOI] [PubMed] [Google Scholar]
- 62.Kilpivaara O., Bartkova J., Eerola H., Syrjakoski K., Vahteristo P., Lukas J., Blomqvist C., Holli K., Heikkila P., Sauter G., et al. Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int. J. Cancer. 2005;113:575–580. doi: 10.1002/ijc.20638. [DOI] [PubMed] [Google Scholar]
- 63.Huzarski T., Cybulski C., Domagała W., Gronwald J., Byrski T., Szwiec M., Woyke S., Narod S.A., Lubiński J. Pathology of breast cancer in women with constitutional CHEK2 mutations. Breast Cancer Res. Treat. 2005;90:187–189. doi: 10.1007/s10549-004-3778-2. [DOI] [PubMed] [Google Scholar]
- 64.Liu C., Wang Y., Wang Q.-S., Wang Y.J. The CHEK2 I157T variant and breast cancer susceptibility: A systematic review and meta-analysis. Asian Pac. J. Cancer Prev. 2012;13:1355–1360. doi: 10.7314/APJCP.2012.13.4.1355. [DOI] [PubMed] [Google Scholar]
- 65.Minicozzi P., Bella F., Toss A., Giacomin A., Fusco M., Zarcone M., Tumino R., Falcini F., Cesaraccio R., Candela G., et al. Relative and disease-free survival for breast cancer in relation to subtype: A population-based study. J. Cancer Res. Clin. Oncol. 2013;139:1569–1577. doi: 10.1007/s00432-013-1478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toss A., Venturelli M., Peterle C., Piacentini F., Cascinu S., Cortesi L. Molecular Biomarkers for Prediction of Targeted Therapy Response in Metastatic Breast Cancer: Trick or Treat? Int. J. Mol. Sci. 2017;18:85. doi: 10.3390/ijms18010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toss A., Cortesi L. Molecular Mechanisms of PARP Inhibitors in BRCA-related Ovarian Cancer. J. Cancer Sci. Ther. 2013;5:11. [Google Scholar]
- 68.Toss A., Piacentini F., Cortesi L., Artuso L., Bernardis I., Parenti S., Tenedini E., Ficarra G., Maiorana A., Iannone A., et al. Genomic alterations at the basis of treatment resistance in metastatic breast cancer: Clinical applications. Oncotarget. 2018;9:31606–31619. doi: 10.18632/oncotarget.25810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available within the article.