Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic in Brazil was dominated by two lineages designated as B.1.1.28 and B.1.1.33. The two SARS-CoV-2 variants harboring mutations at the receptor-binding domain of the Spike (S) protein, designated as lineages P.1 and P.2, evolved from lineage B.1.1.28 and are rapidly spreading in Brazil. Lineage P.1 is considered a Variant of Concern (VOC) because of the presence of multiple mutations in the S protein (including K417T, E484K, N501Y), while lineage P.2 only harbors mutation S:E484K and is considered a Variant of Interest (VOI). On the other hand, epidemiologically relevant B.1.1.33 deriving lineages have not been described so far. Here we report the identification of a new SARS-CoV-2 VOI within lineage B.1.1.33 that also harbors mutation S:E484K and was detected in Brazil between November 2020 and February 2021. This VOI displayed four non-synonymous lineage-defining mutations (NSP3:A1711V, NSP6:F36L, S:E484K, and NS7b:E33A) and was designated as lineage N.9. The VOI N.9 probably emerged in August 2020 and has spread across different Brazilian states from the Southeast, South, North, and Northeast regions.
Keywords: SARS-CoV-2, E484K, variant of Interest, genomic epidemiology, Brazil
1. Introduction
The SARS-CoV-2 epidemic in Brazil was mainly driven by lineages B.1.1.28 and B.1.1.33 that probably emerged in February 2020 and were the most prevalent variants in most country regions until October 2020 [1,2]. Recent genomic studies, however, bring attention to the emergence of new SARS-CoV-2 variants in Brazil harboring mutations at the receptor-binding site (RBD) of the Spike (S) protein that might impact viral fitness and transmissibility.
So far, one variant of concern (VOC), designated as lineage P.1, and one variant of interest (VOI), designated as lineage P.2, have been identified in Brazil and both evolved from lineage B.1.1.28. The VOC P.1, first described in January 2021 [3], displayed an unusual number of lineage-defining mutations in the S protein (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I) and its emergence was associated with a second COVID-19 epidemic wave in the Amazonas state [4,5]. The VOI P.2, first described in samples from October 2020 in the state of Rio de Janeiro, was distinguished by the presence of the S:E484K mutation in RBD and other four lineage-defining mutations outside the S protein [6]. The P.2 lineage has been detected as the most prevalent variant in several states across the country in late 2020 and early 2021 (https://www.genomahcov.fiocruz.br, accessed on 1 March 2021).
Several B.1.1.33-derived lineages are currently defined by the Pangolin system including: lineage N.1 detected in the US, lineage N.2 detected in Suriname and France, lineage N.3 circulating in Argentina, and lineages N.4 and B.1.1.314 circulating in Chile (https://cov-lineages.org/lineages.html, accessed on 1 March 2021). However, none of these B.1.1.33-derived lineages were characterized by mutations of concern in the S protein. Here, we define the lineage N.9 within B.1.1.33 diversity that harbors mutation E484K in the S protein as was detected in different Brazilian states between November 2020 and February 2021.
2. Materials and Methods
The Fiocruz COVID-19 Genomic Surveillance Network has recovered SARS-CoV-2 lineage B.1.1.33 genomes from 422 positive samples between 12th March 2020 and 27th January 2021 (Supplementary Material). Sequencing protocols were as previously described [7,8]. The FASTQ reads obtained were imported into the CLC Genomics Workbench version 20.0.4 (Qiagen A/S, Denmark), trimmed, and mapped against the reference sequence EPI_ISL_402124 available in EpiCoV database in the GISAID (https://www.gisaid.org/, accessed on 1 March 2021). The alignment was refined using the InDels and Structural Variants module.
Sequences were then combined with 816 B.1.1.33 Brazilian genomes available in the EpiCoV database in GISAID by 1st March 2021 (Supplementary Table S1). Only high quality (<1% of N) complete (>29 kb) SARS-CoV-2 genomes were used. This dataset was then aligned using MAFFT v7.475 [9] and subjected to maximum likelihood (ML) phylogenetic analysis using IQ-TREE v2.1.2 [10] under the GTR + F + G4 nucleotide substitution model, as selected by the ModelFinder application [11]. Branch support was assessed by the approximate likelihood-ratio test based on the Shimodaira–Hasegawa procedure (SH-aLRT) with 1000 replicates. The mutational profile was investigated using the Nextclade tool (https://clades.nextstrain.org, accessed on 1 March 2021) and temporal signal was assessed by the regression analysis of the root-to-tip genetic distance against sampling dates using the program Tempest [12].
A time-scaled phylogenetic tree was estimated using the Bayesian Markov Chain Monte Carlo (MCMC) approach implemented in BEAST 1.10.4 [13]. Bayesian tree was reconstructed using the GTR + F + I + G4 nucleotide substitution model, the non-parametric Bayesian skyline (BSKL) model as the coalescent tree prior and a strict molecular clock model with a uniform substitution rate prior (8 × 10–4–10 × 10–4 substitutions/site/year). Ancestral node states were reconstructed using a reversible discrete phylogeographic model [14] where transitions between sampling locations (Brazilian states) were estimated in a continuous-time Markov chain (CTMC) rate reference prior. Convergence (effective sample size > 200) in parameter estimates was assessed using TRACER v1.7 18. The maximum clade credibility (MCC) tree was summarized with TreeAnnotator v1.10.4. ML and MCC trees were visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 1 March 2021).
3. Results and Discussion
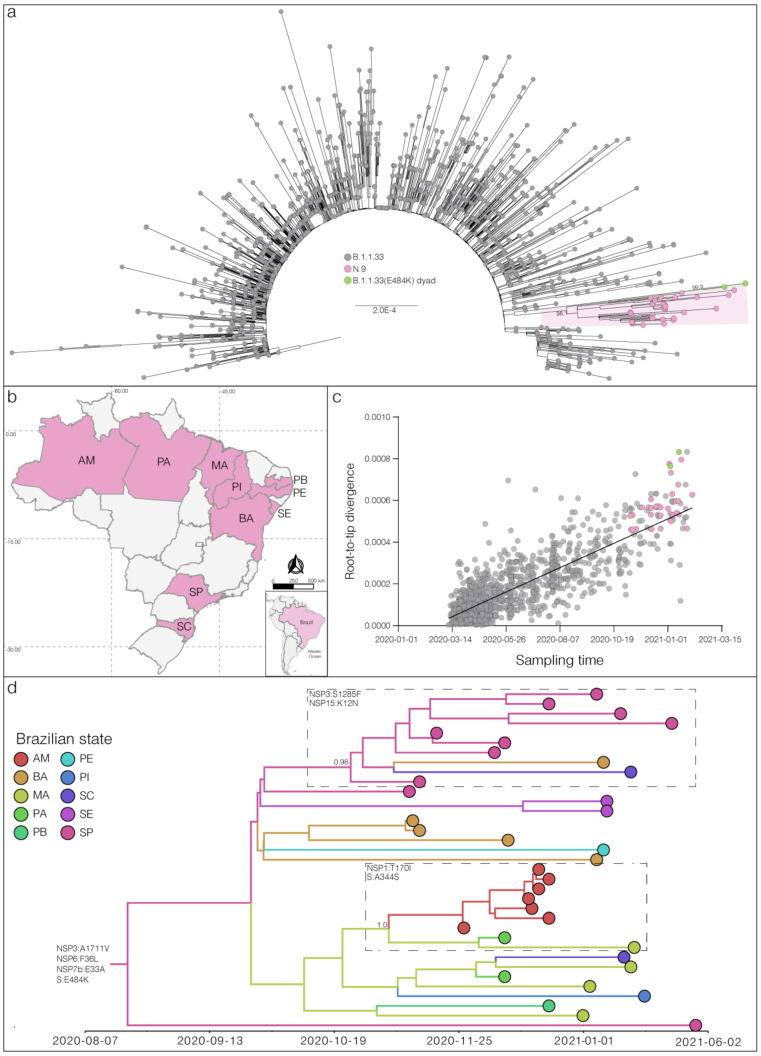
Mutation profile analysis revealed a total of 34 B.1.1.33 sequences harboring the S:E484K mutation. ML phylogenetic analysis revealed that 32 of these sequences branched in a highly supported (SH-aLRT = 98%) monophyletic clade that define a potential new VOI designated as N.9 PANGO lineage [15]. The other two sequences harboring the S:E484K mutation branched separately in a highly supported (SH-aLRT = 100%) dyad (Figure 1a). The VOI N.9 is characterized by four non-synonymous lineage-defining mutations (NSP3:A1711V, NSP6:F36L, S:E484K, and NSP7b:E33A) and also contains a group of three B.1.1.33 sequences from the Amazonas state that has no sequencing coverage in the position 484 of the S protein, but share the remaining N.9 lineage-defining mutations (Table 1), thus forming a cluster of 35 sequences. The B.1.1.33 (S:E484K) dyad comprises two sequences from the Maranhao state and were characterized by a different set of non-synonymous mutations (Supplementary Table S2).
Figure 1.
Lineage N.9 evolutionary origin and spatial-temporal distribution. (a) Maximum likelihood (ML) phylogenetic tree of the B.1.1.33 whole-genome sequences from Brazil. The B.1.1.33 sequences with mutation S:E484K are represented by pink (VOI N.9) and green (B.1.1.33(E484K)) circles. The SH-aLRT support values are indicated in key nodes and branch lengths are drawn to scale with the left bar indicating nucleotide substitutions per site. (b) Geographic distribution of the VOI N.9 identified in Brazil. Brazilian states’ names follow the International Organization for Standardization (ISO) 3166-2 standard. (c) Correlation between the sampling date of B.1.1.33 sequences and their genetic distance from the root of the ML phylogenetic tree. Colors indicate the B.1.1.33 clade as indicated in (a). (d) Bayesian phylogeographic analysis of N.9 lineage. Tips and branches colors indicate the sampling state and the most probable inferred location of their descendent nodes, respectively, as indicated in the legend. Branch posterior probabilities are indicated in key nodes. Boxes highlight two N.9 subclades carrying additional mutations (indicated in each box). The tree was automatically rooted under the assumption of a strict molecular clock, and all horizontal branch lengths are time-scaled.
Table 1.
Synapomorphic mutations of SARS-CoV-2 lineage N.9.
| Genomic Region (Protein) | Nucleotide | Amino Acid |
|---|---|---|
| ORF1a | G1264T | - |
| ORF1a | C7600T | - |
| ORF1a (NSP3) | C7851T | A2529V (A1711V) |
| ORF1a (NSP6) | T11078C | F3605L (F36L) |
| Spike (S) | G23012A | E484K |
| ORF7b (NSP7b) | A27853C | E33A |
Among the 35 genomes identified so far as VOI N.9, 10 Brazilian states were represented, suggesting that this lineage is already highly dispersed in the country. The VOI N.9 was first detected in Sao Paulo state on 11 November 2020, and soon later in other Brazilian states from the South (Santa Catarina), North (Amazonas and Para), and Northeast (Bahia, Maranhao, Paraiba, Pernambuco, Piaui, and Sergipe) regions (Figure 1b). Analysis of the temporal structure revealed that the overall divergence of lineage N.9 is consistent with the substitution pattern of other B.1.1.33 sequences (Figure 1c), thus suggesting no unusual accumulation of mutations in this VOI. Molecular clock analysis estimated the emergence of the VOI N.9 most probably in the states of Sao Paulo (Posterior State Probability (PSP) = 0.42), Bahia (PSP = 0.32) or Maranhao (PSP = 0.18) at 15th August, 2020 (95% High Posterior Density (HPD): 16th June–22th September, 2020) (Figure 1d). This analysis also revealed that some additional mutations were acquired during evolution of VOI N.9 in Brazil, determining two highly supported (PP > 0.95) subclades. One subclade, that mostly contains sequences from Sao Paulo state, probably arose on 16th October (95% HPD: 22th September–5th November) and was defined by additional mutations NSP3:S1285F and NSP15:K12N. The other subclade that mostly comprises sequences from the North region probably arose on 29th October (95% HPD: 5th October–17th November) and was defined by additional mutations NSP1:T170I and S:A344S (Figure 1d).
4. Conclusions
In this study we identified the emergence of a new VOI (S:E484K) within lineage B.1.1.33 circulating in Brazil. The VOI N.9 displayed a low prevalence (~3%) among all Brazilian SARS-CoV-2 samples analyzed between November 2020 and February 2021, but it is already widely dispersed in the country and comprises a high fraction (35%) of the B.1.1.33 sequences detected in that period. Mutation S:E484K has been identified as one of the most important substitutions that could contribute to immune evasion as confers resistance to several monoclonal antibodies and also reduces the neutralization potency of some polyclonal sera from convalescent and vaccinated individuals [16,17,18]. Mutation S:E484K has emerged independently in multiple VOCs (P.1, B.1.351 and B.1.1.7) and VOIs (P.2 and B.1.526) [19] spreading around the world, and it is probably an example of convergent evolution and ongoing adaptation of the virus to the human host.
The onset date of VOI N.9 here estimated around mid-August roughly coincides with the estimated timing of emergence of the VOI P.2 in late-July 6 and shortly precede the detection of a major global shift in the SARS-CoV-2 fitness landscape after October 2020 [20]. These findings indicate that 484K variants probably arose simultaneously in the two most prevalent viral lineages circulating in Brazil around July–August, but may have only acquired some fitness advantages, which accelerated its dissemination, after October 2020. We predict that the Brazilian COVID-19 epidemic during 2021 will be dominated by a complex array of B.1.1.28 (S:E484K), including P.1 and P.2, and B.1.1.33 (S:E484K) variants that will completely replace the parental 484E lineages that drove the epidemic in 2020. Implementation of efficient mitigation measures in Brazil is crucial to reduce community transmission and prevent the recurrent emergence of more transmissible variants that could further exacerbate the epidemic in the country.
Acknowledgments
The authors wish to thank all the health care workers and scientists who have worked hard to deal with this pandemic threat, the GISAID team, and all the EpiCoV database′s submitters, GISAID acknowledgment table containing sequences used in this study is available in Supplementary Table S1. We also appreciate the support of the Fiocruz COVID-19 Genomic Surveillance Network (http://www.genomahcov.fiocruz.br/; accessed on 1 March 2021) members, the Respiratory Viruses Genomic Surveillance. General Coordination of the Laboratory Network (CGLab), Brazilian Ministry of Health (MoH), Brazilian States Central Laboratories (LACEN), Brazilian Ministry of Health (MoH), and the Amazonas surveillance teams for the partnership in the viral surveillance in Brazil.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13050724/s1, Supplementary Material: GISAID accession numbers of genomes lineage B.1.1.33 from this study, Supplementary Table S1: GISAID Acknowledgement Table, Supplementary Table S2: Synapomorphic non-synonymous mutations of the B.1.1.33(S:E484K) dyad isolated in the state of Maranhao.
Author Contributions
Conceptualization, P.C.R., T.G., E.D., G.L.W., F.G.N., G.B.; methodology, P.C.R., T.G., E.D., G.L.W., F.G.N., G.B.; formal analysis, P.C.R., T.G., E.D., G.L.W., F.G.N., G.B.; resources, F.G.N., M.M.S.; data acquisition and curation, A.C.D.P., L.A., R.S.L., A.C.d.F.M., A.S.B.d.R., F.C.M., L.G.L.N., R.K., C.I.d.O., P.S.-M., J.F.B., D.L.F.T., I.R., M.d.C.D., R.R.-R., A.B.L., C.A.d.S., T.S.G., S.B.F., A.F.L.B., A.C.C., F.M., C.S., T.M., C.F.d.C.; writing—original draft preparation, T.G., G.B.; writing—review and editing, P.C.R., T.G., E.D., G.L.W., F.G.N., G.B.; funding acquisition, F.G.N., M.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided by Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM) (PCTI-EmergeSaude/AM call 005/2020 and Rede Genômica de Vigilancia em Saúde-REGESAM); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant 402457/2020–0); CNPq/Ministério da Ciência, Tecnologia, Inovações e Comunicações/Ministério da Saúde (MS/FNDCT/SCTIE/Decit) (grant 403276/2020-9); Inova Fiocruz/Fundação Oswaldo Cruz (Grants VPPCB-007-FIO-18–2–30 and VPPCB-005-FIO-20–2–87), INCT-FCx (465259/2014–6) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (26/210.196/2020). F.G.N, G.L.W, G.B and M.M.S are supported by the CNPq through their productivity research fellowships (306146/2017–7, 303902/2019–1, 302317/2017–1 and 313403/2018-0, respectively). G.B. is also funded by FAPERJ (Grant number E-26/202.896/2018).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the FIOCRUZ-IOC (68118417.6.0000.5248 and CAAE 32333120.4.0000.5190), the Amazonas State University Ethics Committee (CAAE: 25430719.6.0000.5016) and the Brazilian Ministry of the Environment (MMA) A1767C3.
Informed Consent Statement
A waiver of informed consent was obtained from the ethics committee.
Data Availability Statement
All genomes generated in this work were deposited in the EpiCoV database of GISAID (https://www.gisaid.org/, accessed on 1 March 2021). Accession codes are available in Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Disclaimers
The opinions expressed by the authors do not necessarily reflect the opinions of the Ministry of Health of Brazil or the institutions with which the authors are affiliated.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Resende P.C., Delatorre E., Graf T., Mir D., Motta F.C., Appolinario L.R., Dias da Paixao A.C., da Fonseca Mendonca A.C., Ogrzewalska O., Caetano B., et al. Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B.1.1.33 during the early pandemic phase in Brazil. Front. Microbiol. 2021;11:1–14. doi: 10.3389/fmicb.2020.615280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., Fujimoto T., Kuroda M., Wakita T., Ohmagari N. Novel SARS-CoV-2 variant identified in travelers from Brazil to Japan. Emerg. Infect. Dis. 2021;27:1243–1245. doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naveca F., Nascimento V., Souza V., Corado A., Nascimiento F., Silva G., Costa A., Duarte D., Pessoa K., Mejia M., et al. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new Variant of Concern, P.1. [(accessed on 1 March 2021)];Res. Sq. 2021 preprint. Available online: https://www.researchsquare.com/article/rs-275494/v1. [Google Scholar]
- 5.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., da Silva Candido D., Mishra S., Crispim M.A.E., Sales F.C., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1101/2021.02.26.21252554. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voloch C.M., da Silva Francisco R., Jr., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerder A.L., Guimaraes A.P.C., Mariani D., da Costa R.M., Ferreisa O.C., Jr., et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021 doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nascimento V.A., de Lima Guerra Corado A., do Nascimento F.O., da Costa A.K.A., Gomes Duarte D.C., Bessa Luz S.L., Goncalves L.M.F., de Jesus M.S., da Costa C.F., Delatorre E., et al. Genomic and phylogenetic characterization of an imported case of SARS-CoV-2 in Amazonas State, Brazil. Mem. Inst. Oswaldo Cruz. 2020;115:e200310. doi: 10.1590/0074-02760200310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Resende P.C., Motta F.C., Roy S., Applinario L., Fabri A., Xavier J., Harris K., Matos A.R., Caetano B., Orgeswalska M., et al. SARS-CoV-2 genomes recovered by long amplicon tiling multiplex approach using nanopore sequencing and applicable to other sequencing platforms. bioRxiv. 2020 doi: 10.1101/2020.04.30.069039. preprint. [DOI] [Google Scholar]
- 9.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minh B.Q., Schmidt H.A., Chernomor O., Schretmpf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haetseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambaut A., Lam T.T., Carvalho L.M., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baum A., Fulton B.O., Wloga E., Copin R., Pascal L.E., Russo V., Giordano S., Lanza K., Negrom N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone D.K., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:P463–P476.E6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P., Liu L., Iketani S., Nair M.S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Increased resistance of SARS-CoV-2 variants, B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021 doi: 10.1038/s41586-021-03398-2. preprint. [DOI] [PubMed] [Google Scholar]
- 19.Gangavarapu K., Alkuzweny M., Cano M., Andersen K., Haag E., Hughes L., Mullen J., Su A., Latif A.A., Tsueng G., et al. outbreak.info. [(accessed on 1 March 2021)];2020 Available online: https://outbreak.info.
- 20.Martin D.P., Weaver S., Tegally H., San E.L., Shank S.D., Wilkinson E., Giandhari J., Naidoo S., Pullay Y., Singh L., et al. The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. medRxiv. 2021 doi: 10.1101/2021.02.23.21252268. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomes generated in this work were deposited in the EpiCoV database of GISAID (https://www.gisaid.org/, accessed on 1 March 2021). Accession codes are available in Supplementary Material.