Abstract
The efficacy of a novel subunit vaccine candidate, based in the CSFV E2 glycoprotein produced in plants to prevent classical swine fever virus (CSFV) vertical transmission, was evaluated. A Nicotiana benthamiana tissue culture system was used to obtain a stable production of the E2-glycoprotein fused to the porcine Fc region of IgG. Ten pregnant sows were divided into three groups: Groups 1 and 2 (four sows each) were vaccinated with either 100 μg/dose or 300 μg/dose of the subunit vaccine at 64 days of pregnancy. Group 3 (two sows) was injected with PBS. Groups 1 and 2 were boosted with the same vaccine dose. At 10 days post second vaccination, the sows in Groups 2 and 3 were challenged with a highly virulent CSFV strain. The vaccinated sows remained clinically healthy and seroconverted rapidly, showing efficient neutralizing antibodies. The fetuses from vaccinated sows did not show gross lesions, and all analyzed tissue samples tested negative for CSFV replication. However, fetuses of non-vaccinated sows had high CSFV replication in tested tissue samples. The results suggested that in vaccinated sows, the plant produced E2 marker vaccine induced the protective immunogenicity at challenge, leading to protection from vertical transmission to fetuses.
Keywords: classical swine fever virus, E2 glycoprotein, marker vaccine, vertical transmission, plant, protection
1. Introduction
Classical swine fever (CSF) is considered as a highly contagious diseases affecting the Suidae family. The causative agent is a small, enveloped, single-stranded RNA virus known as CSF virus (CSFV), which belongs to the Pestivirus genus of the Flaviviridae family, together with several other viral species, including Bovine viral diarrhea virus and Border disease virus [1,2].
CSF outbreaks have major socio-economic consequences, including serious restrictions on international trade of pigs and pork-derived products [3]. The outbreak of this disease has also been accompanied by a high financial burden due to direct or indirect losses in the pig industry [4]. In addition, the ethical aspects linked to mass stamping out of herds in affected farms have made CSFV a notifiable pathogen by the World Organization for Animal Health (OIE). Although many countries manage this at the national level, the disease is endemic in several regions, such as South and Central America, the Caribbean and Asia [5,6].
Mass vaccination has been implemented in those countries as a mandatory control program for more than 50 years, mainly using live attenuated vaccines. In this regard, the C-strain has been identified as one of the most effective CSF vaccines, which broadly protects pigs from clinical CSF disease against all CSFV genotypes. However, the vaccination using these vaccines are immunologically indistinguishable from CSFV infection. Therefore, it is desirable to develop alternative vaccines such as subunit or modified virus-vectored vaccines, which allow differentiation of infected from vaccinated animals (DIVA) as well as having high efficacy [7].
The CSFV E2 glycoprotein is essential for viral replication and infection, and is the major immunogenic protein for inducing neutralizing antibodies to elicit protective immunity against CSFV [8,9]. Subunit vaccines based on the E2 of CSFV, developed years ago, have been regarded as a very suitable antigenic candidate for the DIVA vaccine.
Though scarcely used in field conditions, there are commercial DIVA vaccines based on the E2 protein, including recombinant E2 proteins expressed in insect cells [10,11,12], BAYOVAC CSF Marker (Bayer, Leverkusen, Germany) and Porcilis Pesti (Intervet, Boxmeer, Netherlands). Although some of those vaccines have been proven to prevent CSFV infection, there are reports to show limitations, especially in regard to early protection and transplacental transmission prevention [13,14]. Currently, a vaccine including the E2 fused with the CD154 (PorVac)(CIGB, Havana, Cuba), expressed in mammalian cells, is being used for disease control in the Caribbean region [15].
Safety and efficacy, as well as supply cost, are very important factors to consider when developing a vaccine for animals, especially livestock. In this respect, a plant-based manufacturing system is an attractive platform. In a previous study, a novel E2 glycoprotein of CSFV fused with the porcine Fc region of IgG was developed for increased stability in the host, and solubility [16]. Considering this, the main objective of the present study was to evaluate the effect of vaccination with this novel subunit marker vaccine produced in plant tissue, to prevent CSFV vertical transmission in pregnant sows.
2. Materials and Methods
2.1. Vaccine
We used a novel plant-produced E2 glycoprotein of CSFV subunit marker vaccine (E2-Bioapp) developed in a previous study [16]. Briefly, the pCAMBIA1300 MELCHE2 construct, containing cellulose binding domain-fused E2 recombinant protein, was prepared through digesting unnecessary domain, followed by ligation with the prepared pFc2 derived from porcine IgG—for better solubility and a longer half-life in the host— resulting in pCAMBIA1300-pmE2:pFc2:HDEL.
The fused construct, pCAMBIA1300-pmE2:pFc2:HDEL, was then introduced into Agrobacterium tumefaciens strain LBA4404, cultured in a Nicotiana benthamiana tissue culture system, harvested, and protein purification conducted. The purified and concentrated pmE2:pFc2 fusion protein was then adjuvanted in Emulsigen®-D adjuvant (MVP Adjuvant®, Omaha, NE, USA).
2.2. Cells and Viruses
Viral stocks were propagated and titrated in the porcine kidney cell line (PK-15, ATCC®:CCL-33, Manassas, VA, USA), cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum pestivirus-free at 37 °C in 5% CO2. The highly virulent CSFV strain Margarita, which belongs to subgroups Genogroup 1.4 was employed for the challenge experiment and neutralization assay [17,18]. Additionally, the Alfort/187 strain of genotype 1.1, was also employed. Virus titration was conducted by end-point dilution using a peroxidase-linked assay (PLA) and the titers calculated according to Reed and Muench [19].
2.3. Experimental Design
At 57 days of gestation, 10 pregnant sows in 2~4 parity (Landrace × Yorkshire) were purchased, which tested negative for Pestivirus. All animals were analyzed again twice at six and two weeks before starting the study. All sows were housed throughout the experiment in an environmentally controlled building with pens over completely slatted floors.
The sows were randomly divided into three groups. The sows in Group 1 (n = 4; Individual identification numbers are set from 1 to 4) were immunized with 100 µg of E2-Bioapp vaccine. The sows in Group 2 (n = 4; individual identification numbers are set from 5 to 8) were immunized with 300 µg of E2-Bioapp vaccine. The sows in Group 3 served as a non-vaccinated control group (n = 2; individual identification numbers are set from 9 to 10).
At 64 days of gestation, Groups 1 and 2 were inoculated intramuscularly on the neck with the E2-Bioapp vaccine, followed by boost immunization at 17 days post vaccination (dpv). At 10 days post second vaccination, the sows in Groups 2 and 3 were challenged intramuscularly on the neck with a 2.5 × 105 tissue culture infective dose (TCID)/mL. On that date, sows in Group 1 were euthanized before viral challenge. Following the methodology used in previous studies, this experimental design did not include an extra group of animals vaccinated with commercial live attenuated CSFV vaccine [10,20,21,22]. Blood, nasal and rectal swab samples were collected at 0, 7, 17, 21 and 28 dpv, and at 4, 8, 14 and 18 days post challenge (dpc) to evaluate the CSFV-specific humoral immune response and virus shedding. At 18 dpc (109 days of gestation and around 2 weeks prepartum), the sows were euthanized, and gross examination was carried out in all fetuses [10]. Representative tissue samples including tonsil, spleen, thymus (only fetuses) and Peyer’s patch (only sows) in particular, were collected during necropsy. Following CSFV inoculation, the clinical signs in the sows were monitored daily by a trained veterinarian in a blinded manner, including rectal temperature. Animals were euthanized before the end of the trial if they presented clinical signs compatible with severe CSF or exhibited prostration behavior.
The experiment was evaluated and approved by the Ethical Committee of the Generalitat of Catalonia, Spain, according to existing European regulations (Project no. 1090).
2.4. Detection of CSFV E2-Specific and Neutralizing Antibodies
The serum samples were tested with a commercially available competitive CSFV Enzyme-Linked Immunosorbent Assay (ELISA) test (IDEXX Laboratories, Bern, Switzerland) for the detection of specific antibodies against the E2. In addition, a neutralization peroxidase-linked assay (NPLA) was also carried out. Neutralizing antibody titers were expressed as the reciprocal dilution of sera that neutralized 100TCID50 of Margarita or Alfort/187 strains in 50% of the culture replicates [23].
2.5. CSFV RNA Detection
Sera and nasal and rectal swabs, as well as tissue samples from sows and fetuses were subjected to RNA extraction for detection of CSFV RNA. The RNA extraction was performed using the IndiMag® Pathogen Kit (Indical bioscience, Leipzig, Germany) according to the manufacturer’s instructions, and stored at −80 °C until use. Quantitative real-time (RT-qPCR) assay for the specific detection of CSFV RNA was performed in an ABI7500 instrument (Applied Biosystems, Foster City, CA, USA) [24]. Threshold cycle values (Ct) equal to, or above 40, were considered to be positive, whereas samples in which fluorescence was undetectable were considered to be negative. As previously described, Ct values above 28 were considered as low, from 23 to 28 as moderate and from 10 to 23, as high RNA viral load [25].
3. Results
3.1. E2-Antibody Response and Clinical Signs Generated in Sows after Vaccination and Challenge
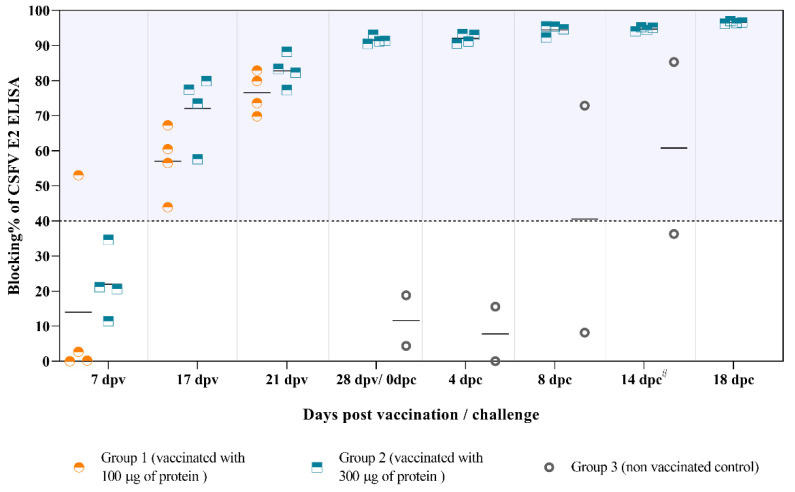
At the time of vaccination (64 days of gestation), sows in all three groups were seronegative against CSFV. E2-specific antibodies started to appear at 7 dpv in one of the sows from Group 1 (Figure 1). All vaccinated sows had already seroconverted before the boost (17 dpv) indicating that single-dose vaccination can effectively induce E2-specific antibodies during 17 days.
Figure 1.
The antibody responses against the E2 glycoprotein after vaccination and challenge. The E2-specific antibody response was analyzed by ELISA and represented as blocking percentage (blocking %). Blocking % values equal to or greater than 40% were considered as positive. #: Sows 9 and 10 were euthanized at 10 dpc due to severe clinical signs and the data are represented at this time-point.
At 21 dpv, the antibody response generated in the animals from Groups 1 and 2 was similar, while all the sows from Group 3 were negative. An increase in antibody response against E2 was observed in all the vaccinated sows after challenge and continued until the end of the experiment. By contrast, both sows in the non-vaccinated group showed fever, mild to moderate apathy, loss of appetite, weight loss, severe constipation, bloody stools and semi-prostration from day 4 after infection. These animals were euthanized at 10 dpc due to severe clinical signs associated with CSF. In addition, low antibody response against E2 was detected in only one of these animals, starting at 8 dpc (Figure 1).
3.2. Protection Levels against CSFV Replication in Vaccinated Sows after Challenge
The vaccinated sows were protected from viremia and viral excretion throughout the trial. However, low viral RNA load was detected in some tissue samples from the vaccinated sows at 18 dpc. All of these animals exhibited viral RNA in tonsil tissue, whereas sows 5, 6, and 8 were also CSFV RNA positive in Peyer’s patch and spleen tissue. By contrast, CSFV RNA was detected in sera and nasal swabs from 4 dpc in non-vaccinated sows (Group 3). In addition, high viral RNA load was detected in all the clinical samples analyzed (sera, nasal and rectal swabs) (Figure 2), as well as in tissues (tonsil, Peyer’s patch and spleen) tested at 10 dpc (Figure 2).
Figure 2.
CSFV RNA detection in tissue samples from either vaccinated or non-vaccinated sows after challenge. CSFV RNA in sera, nasal and rectal swabs and tissue samples including tonsil, Peyer’s patch and spleen from the challenged sows was assessed by qRT-PCR. Ct values less than 40 were considered positive.
3.3. Neutralizing Antibody Response Detected after Vaccination and Challenge
Neutralizing antibody response was present in all vaccinated sows from Group 2 before challenge (28 dpv). Meanwhile, absence of neutralizing activity was found in all sera samples collected from non-vaccinated sows in Group 3. The neutralizing activity against both Margarita (Table 1a) and Alfort/187 strains (Table 1b) detected in all vaccinated sows was progressively increased from 4 dpc until the end of the trial. The NPLA titers were slightly higher against Alfort/187 than those against Margarita strain. Conversely, non-vaccinated sows showed negative or very low NPLA titers against both strains after challenge (Table 1a,b).
Table 1.
CSFV neutralizing peroxidase linked antibody (NPLA) titer: (a) NPLA titer to CSFV strain Margarita, (b) NPLA titer to CSFV strain Alfort/187.
| (a) | |||||||||
| NPLA to Margarita Strain (Challenge Virus) | |||||||||
| Sow ID | Days Post Vaccination (dpv) | Days Post Challenge (dpc) | |||||||
| 0 dpv | 7 dpv | 17 dpv | 21 dpv | 28 dpv/0 dpc | 4 dpc | 8 dpc | 14 dpc * | 18 dpc | |
| Group 1: Sows inoculated with 100 µg of protein | |||||||||
| Sow 1 | Neg ** | Neg | Neg | 1/10 | |||||
| Sow 2 | Neg | Neg | Neg | 1/20 | |||||
| Sow 3 | Neg | Neg | Neg | Neg | |||||
| Sow 4 | Neg | Neg | Neg | Neg | |||||
| Group 2: Sows inoculated with 300 µg of protein | |||||||||
| Sow 5 | Neg | Neg | Neg | 1/10 | 1/40 | 1/80 | 1/1280 | 1/2560 | 1/1280 |
| Sow 6 | Neg | Neg | Neg | Neg | 1/40 | 1/20 | 1/320 | 1/2560 | 1/1280 |
| Sow 7 | Neg | Neg | 1/10 | 1/40 | 1/160 | 1/320 | 1/320 | 1/2560 | 1/2560 |
| Sow 8 | Neg | Neg | Neg | Neg | 1/40 | 1/20 | 1/160 | 1/2560 | 1/640 |
| Group 3: Non vaccinated control animals | |||||||||
| Sow 9 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 1/10 | |
| Sow 10 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | |
| (b) | |||||||||
| NPLA to Alfort 187 CSFV Strain | |||||||||
| Sow ID | Days Post Vaccination (dpv) | Days Post Challenge (dpc) | |||||||
| 0 dpv | 7 dpv | 17 dpv | 21 dpv | 28 dpv/0 dpc | 4 dpc | 8 dpc | 14 dpc * | 18 dpc | |
| Group 1: Sows inoculated with 100 µg of protein | |||||||||
| Sow 1 | Neg ** | Neg | 1/10 | 1/40 | |||||
| Sow 2 | Neg | Neg | 1/10 | 1/320 | |||||
| Sow 3 | Neg | Neg | 1/10 | 1/20 | |||||
| Sow 4 | Neg | Neg | 1/20 | 1/20 | |||||
| Group 2: Sows inoculated with 300 µg of protein | |||||||||
| Sow 5 | Neg | Neg | Neg | 1/10 | 1/1280 | 1/320 | 1/5120 | 1/10240 | 1/10240 |
| Sow 6 | Neg | Neg | Neg | 1/10 | 1/160 | 1/160 | 1/5120 | 1/5120 | 1/5120 |
| Sow 7 | Neg | Neg | 1/20 | 1/160 | 1/2560 | 1/2560 | 1/5120 | 1/10240 | 1/10240 |
| Sow 8 | Neg | Neg | 1/10 | 1/20 | 1/640 | 1/160 | 1/640 | 1/5120 | 1/2560 |
| Group 3: Non vaccinated control animals | |||||||||
| Sow 9 | Neg | Neg | Neg | Neg | Neg | Neg | 1/10 | 1/10 | |
| Sow 10 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | |
* Sow 9 and 10 were measured at 10 dpc, and for ethical reasons, were euthanized due to severe clinical symptoms. ** Neg means negative.
3.4. Protection Levels of Fetuses from Either Vaccinated or Non-Vaccinated Sows after CSFV Challenge
Absence of gross lesions and mummifications were found in the litters from vaccinated sows (Group 2). All fetuses from these animals showed similar size and weight (Figure 3a). On the contrary, litters from unvaccinated sows (Group 3) showed different levels of mummifications (Figure 3b). Sow number 9 carried 18 fetuses, one of which was mummified and the rest were of irregular size, whereas sow 10 carried 20 fetuses, of which 7 were mummified and 13 were of irregular size. All fetuses from the non-vaccinated litters were CSFV RNA positive in one or more of the tissue samples tested (Table 2b). The fetuses of non-vaccinated sows showed obvious CSFV trans-placental transmissions with 84.6% and 92.3% of positivity. In direct contrast, the litters from vaccinated sows tested negative for CSFV replication by RT-qPCR in all samples analyzed, including serum, tonsil, spleen and thymus (Table 2a).
Figure 3.
Representative fetuses of sows challenged with highly virulent CSFV Margarita strain. (a) Litters of sows vaccinated with E2-Bioapp (300 µg/dose) were observed healthy and uniform in size. (b) Fetuses from the unvaccinated sows were mummified and the remaining showed uneven size.
Table 2.
Detection of CSFV RNA in tissue samples of fetuses from either vaccinated or non-vaccinated sows after challenge: (a) Tissue samples of fetuses from vaccinated sows, (b) Tissue samples of fetuses from non-vaccinated sows.
| (a) | |||||||||
| Fetus ID | CSFV RT-qPCR (Ct Value) | Fetus ID | CSFV RT-qPCR (Ct Value) | ||||||
| Serum | Tonsil | Spleen | Thymus | Serum | Tonsil | Spleen | Thymus | ||
| Fetuses from sow 5 | Fetuses from sow 7 | ||||||||
| 1 | Neg * | Neg | Neg | Neg | 1 | Neg | Neg | Neg | Neg |
| 2 | Neg | Neg | Neg | Neg | 2 | Neg | Neg | Neg | Neg |
| 3 | Neg | Neg | Neg | Neg | 3 | Neg | Neg | Neg | Neg |
| 4 | Neg | Neg | Neg | Neg | 4 | Neg | Neg | Neg | Neg |
| 5 | Neg | Neg | Neg | Neg | 5 | Neg | Neg | Neg | Neg |
| 6 | Neg | Neg | Neg | Neg | 6 | Neg | Neg | Neg | Neg |
| 7 | Neg | Neg | Neg | Neg | 7 | Neg | Neg | Neg | Neg |
| 8 | Neg | Neg | Neg | Neg | 8 | Neg | Neg | Neg | Neg |
| 9 | Neg | Neg | Neg | Neg | 9 | Neg | Neg | Neg | Neg |
| 10 | Neg | Neg | Neg | Neg | 10 | Neg | Neg | Neg | Neg |
| 11 | Neg | Neg | Neg | Neg | 11 | Neg | Neg | Neg | Neg |
| 12 | Neg | Neg | Neg | Neg | 12 | Neg | Neg | Neg | Neg |
| 13 | Neg | Neg | Neg | Neg | 13 | Neg | Neg | Neg | Neg |
| Fetuses from sow 6 | Fetuses from sow 8 | ||||||||
| 1 | Neg | Neg | Neg | Neg | 1 | Neg | Neg | Neg | Neg |
| 2 | Neg | Neg | Neg | Neg | 2 | Neg | Neg | Neg | Neg |
| 3 | Neg | Neg | Neg | Neg | 3 | Neg | Neg | Neg | Neg |
| 4 | Neg | Neg | Neg | Neg | 4 | Neg | Neg | Neg | Neg |
| 5 | Neg | Neg | Neg | Neg | 5 | Neg | Neg | Neg | Neg |
| 6 | Neg | Neg | Neg | Neg | 6 | Neg | Neg | Neg | Neg |
| 7 | Neg | Neg | Neg | Neg | 7 | Neg | Neg | Neg | Neg |
| - | - | - | - | - | 8 | Neg | Neg | Neg | Neg |
| - | - | - | - | - | 9 | Neg | Neg | Neg | Neg |
| - | - | - | - | - | 10 | Neg | Neg | Neg | Neg |
| - | - | - | - | - | 11 | Neg | Neg | Neg | Neg |
| - | - | - | - | - | 12 | Neg | Neg | Neg | Neg |
| - | - | - | - | - | 13 | Neg | Neg | Neg | Neg |
| (b) | |||||||||
| Fetus ID | CSFV RT-qPCR (Ct Value) | Fetus ID | CSFV RT-qPCR (Ct Value) | ||||||
| Serum | Tonsil | Spleen | Thymus | Serum | Tonsil | Spleen | Thymus | ||
| Fetuses from sow 9 | Fetuses from sow 10 | ||||||||
| 1 | Neg * | Neg | 36.50 | Neg | 1 | Neg | 35.58 | Neg | 36.56 |
| 2 | Neg | Neg | 36.60 | Neg | 2 | Neg | 31.22 | 26.63 | 23.78 |
| 3 | 33.39 | 30.78 | 24.44 | 23.52 | 3 | Neg | Neg | 30.48 | Neg |
| 4 | Neg | Neg | Neg | 34.51 | 4 | Neg | 33.90 | 30.36 | 27.99 |
| 5 | Neg | 39.83 | Neg | 29.71 | 5 | Neg | 37.23 | 31.91 | 32.03 |
| 6 | Neg | 31.89 | 35.00 | Neg | 6 | Neg | 32.46 | Neg | Neg |
| 7 | Neg | Neg | 35.15 | Neg | 7 | Neg | 34.40 | Neg | 36.50 |
| 8 | Neg | 36.00 | Neg | Neg | 8 | Neg | 34.20 | Neg | 35.65 |
| 9 | Neg | Neg | Neg | Neg | 9 | Neg | Neg | Neg | 30.28 |
| 10 | Neg | 37.85 | Neg | 35.80 | 10 | Neg | 33.85 | 27.17 | 29.16 |
| 11 | Neg | 34.46 | 30.96 | 26.82 | 11 | Neg | Neg | Neg | 30.43 |
| 12 | Neg | Neg | Neg | Neg | 12 | Neg | 29.85 | 36.44 | 33.47 |
| 13 | Neg | 30.64 | 28.92 | 29.56 | 13 | Neg | 32.99 | 27.05 | 26.39 |
* Neg means negative.
4. Discussion
Despite significant efforts to control and eradicate CSF with mandatory vaccination policies, using live attenuated vaccines, the disease is still endemic. Sporadic outbreaks continue to occur in some countries [26,27]. Previous reports have suggested that the virus is conducted by an evolutionary process of circulating strains, due to the implementation of inefficient vaccination programs [27,28].
Low and moderate virulence CSFV strains have been reported to be circulating in pig herds. These strains have the capability to generate weak and persistently infected newborn piglets by trans-placental infection of the fetuses [29,30,31].
A novel E2 glycoprotein of CSFV subunit marker vaccine has been previously developed [16], which was produced in plant tissue and fused with the porcine Fc region of IgG. This production system fits the demand for vaccination in the veterinary field; being safe, effective and cost-affordable. The plant produced subunit marker vaccine candidate against CSFV was administered twice, and induced immunogenicity and protected pregnant sows from clinical signs, as well as reproductive failure. However, animals of Group 1 were not challenged, since before the challenge the antibody titers were similar to those of Group 2.
Neutralizing antibodies (NA) provide the best evidence that protective immunity has been established. The close correlation between NA titer to protection has been widely reported. It is accepted that a reasonable threshold antibody level for protection is 1:32 dilution of serum by NPLA test [32]. The NPLA titers increased rapidly and were high in vaccinated sows, and were shown to protect the animals from the development of CSF disease. Moreover, the vaccination aided in controlling viral replication after challenge, protecting the animals from developing viremia and viral shedding. Interestingly, the NPLA titers before challenge were only present against Alfort/187 strain among vaccinated groups (Group 1 and 2). It might be explained by the fact that the E2 protein of the vaccine formulation has a similar portion with the native fraction of Alfort/187 strain. Likewise, the antibody response generated by the E2-Bioapp vaccine was shown to have neutralizing activity against genotype 1.4 (Margarita) as well as genotype 1.1 (Alfort/187), suggesting the capacity of E2-Bioapp to confer protection against heterologous strains.
However, viral RNA was still detected in lymphoid tissue of vaccinated sows when all other tissues were cleared. The virus was inoculated intramuscularly to the sows that were previously vaccinated, being retained in lymphoid tissue. Since CSFV has a tropism for the lymphoid system, it may explain why the virus is detected in these tissues [33]. Possibly, it would take more than the 18 days to clear viral RNA in lymphoid tissue due to the viral tropism to this tissue.
Notably, fetuses from vaccinated sows were protected from CSFV transplacental transmission, shown by the absence of CSFV RNA measured by RT-qPCR. It is well established that trans-placental transmission of CSFV is generated mainly during mid-gestation [30,34,35]. The outcome of trans-placental infection of fetuses depends largely on the time of gestation and viral virulence [35]. CSFV trans-placental transmission may result in abortion, stillbirth, mummification, malformations or the birth of weak persistently infected piglets [36].
The glycosylation of E2 protein produced in plant tissue was confirmed elsewhere [16] and correct glycosylation of antigenic proteins has been known to affect antigen efficacy [37]. However, the effect of xylose and fucose epitopes on N-glycans produced in plants is controversial and how plant-specific glycans on E2 protein affect antigenicity of the subunit vaccine in pigs remains elusive.
Collectively, the results of the present study support the capacity of E2-Bioapp vaccine to protect against transplacental transmission from the sow to the fetuses after two vaccination doses. Further studies will provide insight to elucidate the efficacy of this vaccine prototype in different age groups. The E2-Bioapp vaccine candidate is an economically feasible strategy, applicable for large scale production in order to guarantee vaccine coverage in large populations. This work opens the window for the development of new strategies for vaccine production in animal health.
5. Conclusions
E2 glycoprotein subunit marker vaccine produced in plant tissue generated a rapid and high antibody response against the E2 glycoprotein. In addition, it is able to safely induce neutralizing antibody in sows, and subsequently protect against vertical transmission of swine fever virus challenge.
Author Contributions
Conceptualization, E.-J.S., S.L., J.K.; methodology, J.A.B., M.W.; validation, J.P., S.G.; resources, L.G., J.A.B., M.W., S.P.; writing-original draft preparation, Y.O.; writing-review and editing, Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
J.A.B has a pre-doctoral fellowship, FPI 2016, Ministerio de Ciencia e Innovación, from the Spanish government. M.W. has a pre-doctoral fellowship, CSC scholarship (2017), from the Chinese government.
Institutional Review Board Statement
For studies with animals, the experiment was conducted in accordance with the existing Spanish and European regulations and was approved by the Ethical Committee of the Generalitat de Catalonia, Spain, under animal experimentation project number 1090.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Smith D.B., Meyers G., Bukh J., Gould E.A., Monath T., Muerhoff A.S., Pletnev A., Rico-Hesse R., Stapleton J.T., Simmonds P., et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017;98:2106–2112. doi: 10.1099/jgv.0.000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindenbach B., Rice C. Flaviviridae: The Viruses and Their Replication. Volume 1 Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2001. [Google Scholar]
- 3.Beltran-Alcrudo D., Falco J.R., Raizman E., Dietze K. Transboundary spread of pig diseases: The role of international trade and travel. BMC Veter Res. 2019;15:1–14. doi: 10.1186/s12917-019-1800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meuwissen M.P., Horst S.H., Huirne R.B., Dijkhuizen A.A. A model to estimate the financial consequences of classical swine fever outbreaks: Principles and outcomes. Prev. Veter Med. 1999;42:249–270. doi: 10.1016/S0167-5877(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 5.Ganges L., Núñez J.I., Sobrino F., Borrego B., Fernández-Borges N., Frías-Lepoureau M.T., Rodríguez F. Recent advances in the development of recombinant vaccines against classical swine fever virus: Cellular responses also play a role in protection. Veter J. 2008;177:169–177. doi: 10.1016/j.tvjl.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Coronado L., Perera C., Rios L., Frías M., Pérez L. A critical review about different vaccines against classical swine fever virus and their repercussions in endemic regions. Vaccines. 2021;9:154. doi: 10.3390/vaccines9020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abid M., Teklue T., Li Y., Wu H., Wang T., Qiu H.-J., Sun Y. Generation and immunogenicity of a recombinant pseudorabies virus co-expressing classical swine fever virus E2 protein and porcine circovirus type 2 capsid protein based on Fosmid library platform. Pathogens. 2019;8:279. doi: 10.3390/pathogens8040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Rijn P.A. A common neutralizing epitope on envelope glycoprotein E2 of different pestiviruses: Implications for improvement of vaccines and diagnostics for classical swine fever (CSF)? Veter Microbiol. 2007;125:150–156. doi: 10.1016/j.vetmic.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Lin H., Ma Z., Chen L., Fan H. Recombinant swinepox virus expressing glycoprotein E2 of classical swine fever virus confers complete protection in pigs upon viral challenge. Front. Veter Sci. 2017;4 doi: 10.3389/fvets.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahrens U. Efficacy of the classical swine fever (CSF) marker vaccine Porcilis® Pesti in pregnant sows. Veter Microbiol. 2000;77:83–97. doi: 10.1016/S0378-1135(00)00265-0. [DOI] [PubMed] [Google Scholar]
- 11.Bouma A., De Smit A., De Kluijver E., Terpstra C., Moormann R. Efficacy and stability of a subunit vaccine based on glycoprotein E2 of classical swine fever virus. Veter Microbiol. 1999;66:101–114. doi: 10.1016/S0378-1135(99)00003-6. [DOI] [PubMed] [Google Scholar]
- 12.Bouma A., De Smit A., De Jong M., De Kluijver E., Moormann R. Determination of the onset of the herd-immunity induced by the E2 sub-unit vaccine against classical swine fever virus. Vaccine. 2000;18:1374–1381. doi: 10.1016/S0264-410X(99)00398-9. [DOI] [PubMed] [Google Scholar]
- 13.Depner K.R., Bouma A., Koenen F., Klinkenberg D., Lange E., De Smit H., Vanderhallen H. Classical swine fever (CSF) marker vaccine. Veter Microbiol. 2001;83:107–120. doi: 10.1016/S0378-1135(01)00410-2. [DOI] [PubMed] [Google Scholar]
- 14.van Oirschot J. Vaccinology of classical swine fever: From lab to field. Veter Microbiol. 2003;96:367–384. doi: 10.1016/j.vetmic.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Sordo-Puga Y., Suárez-Pedroso M., Naranjo-Valdéz P., Pérez-Pérez D., Santana-Rodríguez E., Sardinas-Gonzalez T., Mendez-Orta M., Duarte-Cano C., Estrada-Garcia M., Rodríguez-Moltó M. Porvac® subunit vaccine E2-CD154 induces remarkable rapid protection against classical swine fever virus. Vaccines. 2021;9:167. doi: 10.3390/vaccines9020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park Y., Lee S., Kang H., Park M., Min K., Kim N.H., Gu S., Kim J.K., An D.-J., Choe S., et al. A classical swine fever virus E2 fusion protein produced in plants elicits a neutralizing humoral immune response in mice and pigs. Biotechnol. Lett. 2020;42:1247–1261. doi: 10.1007/s10529-020-02892-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giangaspero M., Zhang S.-Q. Genomic characteristics of classical swine fever virus strains of bovine origin according to primary and secondary sequence-structure analysis. Open Veter J. 2020;10:94–115. doi: 10.4314/ovj.v10i1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postel A., Schmeiser S., Perera C.L., Rodríguez L.J.P., Frias-Lepoureau M.T., Becher P. Classical swine fever virus isolates from Cuba form a new subgenotype 1.4. Veter Microbiol. 2013;161:334–338. doi: 10.1016/j.vetmic.2012.07.045. [DOI] [PubMed] [Google Scholar]
- 19.Reed L., Muench H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 20.Blome S., Gabriel C., Schmeiser S., Meyer D., Meindl-Böhmer A., Koenen F., Beer M. Efficacy of marker vaccine candidate CP7_E2alf against challenge with classical swine fever virus isolates of different genotypes. Veter Microbiol. 2014;169:8–17. doi: 10.1016/j.vetmic.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Dewulf J., Laevens H., Koenen F., Mintiens K., De Kruif A. An experimental infection to investigate the indirect transmission of classical swine fever virus by excretions of infected pigs. J. Veter Med. Ser. B. 2002;49:452–456. doi: 10.1046/j.1439-0450.2002.00593.x. [DOI] [PubMed] [Google Scholar]
- 22.Gabriel C., Blome S., Urniza A., Juanola S., Koenen F., Beer M. Towards licensing of CP7_E2alf as marker vaccine against classical swine fever—Duration of immunity. Vaccine. 2012;30:2928–2936. doi: 10.1016/j.vaccine.2012.02.065. [DOI] [PubMed] [Google Scholar]
- 23.Terpstra C., Bloemraad M., Gielkens A. The neutralizing peroxidase-linked assay for detection of antibody against swine fever virus. Veter Microbiol. 1984;9:113–120. doi: 10.1016/0378-1135(84)90026-9. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann B., Beer M., Schelp C., Schirrmeier H., Depner K. Validation of a real-time RT-PCR assay for sensitive and specific detection of classical swine fever. J. Virol. Methods. 2005;130:36–44. doi: 10.1016/j.jviromet.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 25.Wang M., Liniger M., Muñoz-González S., Bohórquez J.A., Hinojosa Y., Gerber M., López-Soria S., Rosell R., Ruggli N., Ganges L. A polyuridine insertion in the 3’ untranslated region of classical swine fever virus activates immunity and reduces viral virulence in piglets. J. Virol. 2019;94 doi: 10.1128/JVI.01214-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei J.-L., Xia S.-L., Wang Y., Du M., Xiang G.-T., Cong X., Luo Y., Li L.-F., Zhang L., Yu J., et al. Safety and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant expressing the E2 protein of classical swine fever virus in pigs. Immunol. Lett. 2016;174:63–71. doi: 10.1016/j.imlet.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Yoo S.J., Kwon T., Kang K., Kim H., Kang S.C., Richt J.A., Lyoo Y.S. Genetic evolution of classical swine fever virus under immune environments conditioned by genotype 1-based modified live virus vaccine. Transbound. Emerg. Dis. 2018;65:735–745. doi: 10.1111/tbed.12798. [DOI] [PubMed] [Google Scholar]
- 28.Coronado L., Liniger M., Muñoz-González S., Postel A., Pérez L.J., Pérez-Simó M., Perera C.L., Lepoureau M.T.F.-, Rosell R., Grundhoff A., et al. Novel poly-uridine insertion in the 3’UTR and E2 amino acid substitutions in a low virulent classical swine fever virus. Veter Microbiol. 2017;201:103–112. doi: 10.1016/j.vetmic.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Van Oirschot J. Experimental production of congenital persistent swine fever infections: I. Clinical, pathological and virological observations. Vet. Microbiol. 1979;4:117–132. doi: 10.1016/0378-1135(79)90048-8. [DOI] [Google Scholar]
- 30.Van Oirschot J. Experimental production of congenital persistent swine fever infections: II. Effect on functions of the immune system. Vet. Microbiol. 1979;4:133–147. doi: 10.1016/0378-1135(79)90049-X. [DOI] [Google Scholar]
- 31.Muñoz-González S., Ruggli N., Rosell R., Pérez L.J., Frías-Leuporeau M.T., Fraile L., Montoya M., Cordoba L., Domingo M., Ehrensperger F., et al. Postnatal persistent infection with classical swine fever virus and its immunological implications. PLoS ONE. 2015;10:e0125692. doi: 10.1371/journal.pone.0125692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terpstra C., Wensvoort G. The protective value of vaccine-induced neutralising antibody titres in swine fever. Veter Microbiol. 1988;16:123–128. doi: 10.1016/0378-1135(88)90036-3. [DOI] [PubMed] [Google Scholar]
- 33.Gavier-Widén D., Meredith A., Duff J.P. Infectious Diseases of Wild Mammals and Birds in Europe. John Wiley & Sons; Hoboken, NJ, USA: 2012. [Google Scholar]
- 34.Liess B. Persistent infections of hog cholera: A review. Prev. Veter Med. 1984;2:109–113. doi: 10.1016/0167-5877(84)90054-0. [DOI] [Google Scholar]
- 35.Van Oirschot J., Terpstra C. A congenital persistent swine fever infection. I. Clinical and virological observations. Vet. Microbiol. 1977;2:121–132. doi: 10.1016/0378-1135(77)90003-7. [DOI] [Google Scholar]
- 36.Trautwein G. Pathology and Pathogenesis of the Disease. In: Liess B., editor. Classical Swine Fever and Related Viral Infections. Springer; Boston, MA, USA: 1988. pp. 27–54. [Google Scholar]
- 37.Risatti G.R., Holinka L.G., Sainz I.F., Carrillo C., Lu Z., Borca M.V. N-linked glycosylation status of classical swine fever virus strain Brescia E2 glycoprotein influences virulence in swine. J. Virol. 2006;81:924–933. doi: 10.1128/JVI.01824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.