Abstract
Background: The extended-spectrum beta-lactamase (ESBL) Klebsiella pneumoniae is one of the leading causes of health-associated infections (HAIs), whose antibiotic treatments have been severely reduced. Moreover, HAI bacteria may harbor pathogenic factors such as siderophores, enzymes, or capsules, which increase the virulence of these strains. Thus, new therapies, such as antimicrobial photodynamic inactivation (aPDI), are needed. Method: A collection of 118 clinical isolates of K. pneumoniae was characterized by susceptibility and virulence through the determination of the minimum inhibitory concentration (MIC) of amikacin (Amk), cefotaxime (Cfx), ceftazidime (Cfz), imipenem (Imp), meropenem (Mer), and piperacillin–tazobactam (Pip–Taz); and, by PCR, the frequency of the virulence genes K2, magA, rmpA, entB, ybtS, and allS. Susceptibility to innate immunity, such as human serum, macrophages, and polymorphonuclear cells, was tested. All the strains were tested for sensitivity to the photosensitizer PSIR-3 (4 µg/mL) in a 17 µW/cm2 for 30 min aPDI. Results: A significantly higher frequency of virulence genes in ESBL than non-ESBL bacteria was observed. The isolates of the genotype K2+, ybtS+, and allS+ display enhanced virulence, since they showed higher resistance to human serum, as well as to phagocytosis. All strains are susceptible to the aPDI with PSIR-3 decreasing viability in 3log10. The combined treatment with Cfx improved the aPDI to 6log10 for the ESBL strains. The combined treatment is synergistic, as it showed a fractional inhibitory concentration (FIC) index value of 0.15. Conclusions: The aPDI effectively inhibits clinical isolates of K. pneumoniae, including the riskier strains of ESBL-producing bacteria and the K2+, ybtS+, and allS+ genotype. The aPDI with PSIR-3 is synergistic with Cfx.
Keywords: antibiotic resistance, virulence factors, Klebsiella pneumoniae, photodynamic therapy
1. Introduction
Klebsiella pneumoniae is one of the major health-associated infection (HAIs) producers worldwide, including pneumonia, urinary tract, and bloodstream infections [1,2]. Moreover, HAI-producing K. pneumoniae strains progressively accumulate more multiple-drugs resistance (MDR), including extended-spectrum β-lactamases (ESBL) and carbapenemases, such as KPC [3,4,5]. Treatment options for infections caused by MDR strains of K. pneumoniae are severely reduced to colistin and tigecycline [6,7,8]. The high MDR shown by strains of K. pneumoniae does not fully explain its notable success as one of the most important agents of HAIs, suggesting the participation of other factors [9]. An increasing number of authors suggest that increased bacterial survival during infections is related to virulence factors [1,3,9,10,11,12,13,14].
Virulence factors shown by Klebsiella pneumoniae strains are common to enterobacteria, such as siderophores, enzymes, and capsules [9]. These factors may be part of genes conserved amongst all K. pneumoniae, called the core set of genes, but other genes vary in frequency between strains, and they are part of the accessory set of genes [9]. During colonization and infection, K. pneumoniae requires the expression of several core and accessory genes to deal with, for example, the nutritional stress due to nutrient sequestration or the immune response of the host [15]. For instance, during lung infection, the acquisition of iron is facilitated by the siderophores enterobactin (entB) and yersiniabactin (ybtS) genes encoded on the bacterial chromosome [1,16]. Like iron, during infections of the lungs, urinary tract, and bloodstream, access to nitrogen is limited. The allS gene allows K. pneumoniae to use allantoin degradation as an alternative nitrogen source [17,18]. The polysaccharide capsule is involved, among others, in resistance to death by complement-mediated opsonophagocytosis [9], and inhibition of macrophages [19,20]. Some capsular varieties (such as K2) may produce hypervirulent strains with characteristic phenotypes such as the hypermucoviscous [19]. Some hypervirulent phenotypes are associated with the expression of the mucoviscosity-associated gene A (magA) and the regulator of the mucoid phenotype A gene (rmpA) [21]. Moreover, the rmpA gene increases the ability of ESBL-producing strains to resist the bactericidal activity of serum and phagocytosis [22]. Since the virulence factors increase bacterial survival and may influence antibiotic resistance expression, the composition of the pool of the virulence gene of K. pneumoniae strains may be associated with the antibiotic susceptibility pattern [9,13].
Due to the emergence of multi-drug resistance (MDR), the deficit of new antibiotics is one of the most pressing threats to human health in the 21st century [23]. Given the high risk to public health caused by the deficiency of new antibiotics, the use of complementary antimicrobial therapies non-antibiotic-based, such as antibacterial photodynamic inactivation (aPDI), emerges as a promising alternative [24]. The aPDI may support the lack of antibiotic therapies against MDR and KPC strains [24,25]. The aPDI is a procedure based on the use of photosensitizer (PS) compounds to produce light-activated local cytotoxicity (photooxidative stress) [26]. The PSs work by energy absorbs of a specific wavelength in the UV–Vis range, which is then transferred to molecular oxygen in solution to produce reactive oxygen species (ROS) [27]. Molecular O2 can accept this energy together with electrons, or it can only undergo a one-electron reduction to produce superoxide anion radical (O2•−), hydrogen peroxide (H2O2), and hydroxyl radical (HO•) [28,29]. The energy transferred to the O2 without one-electron reduction produces singlet oxygen (1O2) [27]. The ROS production generates photooxidative stress induced by aPDI, which occurs mainly due to the action of 1O2. The oxidative action of 1O2 occurs on organic molecules close to the PS through concerted addition reactions of alkene groups [30]. These organic molecules can be structural proteins or lipids of the bacterial envelope; therefore, the damage occurs to the cell wall plasma membrane or other bacterial structures, leading to nonspecific cell death [27,31]. Previously, a PS compound based on a polypyridine Ir(III) complex (PSIR-3, see Figure 4) demonstrated aPDI activity, inhibiting the bacterial growth of CKP and synergism with imipenem [32,33]. In this work, we determine the frequency of the K2, entB, ybtS, allS, rmpA, and magA virulence genes, in a population of 118 clinical isolates of K. pneumoniae and evaluate their association to the susceptibility pattern to several antibiotics. Our data show that virulence factors K2+, ybtS+, and allS+ were associated with a modification in the bacterial minimum inhibitory concentrations (MICs) and increased ESBL production probability. These virulence genes significantly increased the survival of the bacteria, to innate immunity. These more virulent ESBL bacteria were all susceptible to the aPDI treatment with PSIR-3 and demonstrate synergism with cephotaxime.
2. Materials and Methods
2.1. Bacterial Isolates
The ethics committee of the Faculty of Health Sciences, Central University of Chile, and the Central Metropolitan Health Service of Chile (MHSC), Act number: N 124/07, approved the study protocol and the informed consent form. The clinical isolates of K. pneumoniae were obtained from 122 samples from unrelated patients, received in the bacteriology laboratory of “Hospital el Carmen” (HEC) for a period of six months (2017/2018). The HEC is a complex hospital with 412 beds and serving a population of more than 600,000 inhabitants. All clinical isolates were identified as K. pneumoniae following the protocols of the Institute of Clinical and Laboratory Standards (CLSI) [34]. As controls, the virulent strain (ATCC 43816 KPPR1) of susceptible K. pneumoniae (K2+, ybtS+, and allS+) [35] and the MDR KPC+ ST258 (KP35) (K2−, ybtS− and allS−) [36] strains were also included.
2.2. Antimicrobial Susceptibility Testing
The MIC of the antimicrobial agents were determined in 96-well plates by micro-dilution methodology in cations-adjusted Mueller-Hinton (ca-MHB) broth. Inoculum of 1 × 106 colony forming units (CFUs)/mL of each clinical isolate was mixed with decreasing concentrations of each antibiotic and incubated overnight at 37 °C following the CLSI recommendations. The MIC for each antibiotic was determined as the last dilution in which no bacterial growth occurred, and the susceptibility intervals were assigned based on the cutoff points established by the CLSI (2018) for amikacin (Amk), cefotaxime (Cfx), ceftazidime (Cfz), imipenem (Imp), meropenem (Mer), and piperacillin–tazobactam (Pip–Taz). According to the CLSI protocols [34], clinical isolates were strains considered ESBL-producing when resistant to cefotaxime but susceptible to the combination of Cfx/clavulanic acid. Values are presented as the median in mg/L and interquartile range (IQR).
2.3. DNA Extraction and PCR Amplification
The total DNA was obtained from stationary bacterial cultures in LB broth using the phenol-chloroform methodology. In brief, pelleted bacteria were suspended in 300 µL of PBS and mixed with 300 µL of phenol: chloroform (25:24 vol:vol); the final mix was stirred well in a vortex and centrifuged at 13,000× g at 4 °C for 15 min. The aqueous phase was mixed 1:1 with chloroform and centrifuged as above. Genomic and plasmidial DNA contained in the aqueous phase was precipitated in 0.6 vol of 2-propanol and sedimented at 13,000× g at 4 °C for 20 min. The nucleic acids were washed twice with 70% ethanol and resuspended in Tris-EDTA buffer (10 mM Tris-HCl, 1 mM disodium EDTA, pH 8.0). PCR reactions were performed, using 0.5 µM of each specific primer pair listed in Table 1, in 1× master mix GoTaq (Promega). The amplification was carried out at a final volume of 20 µL in a Veriti (Applied Biosystem) PCR machine with an initial denaturation step of 10 s at 95 °C, followed by 35 cycles of 10 s at 95 °C, 15 s at 58 °C, and 30 s of extension at 72 °C. A final extension step of 7 min at 72 °C was included, and PCR products were visualized on a 1.7% agarose gel.
Table 1.
Primers for genes encoding virulence factors of Klebsiella pneumoniae.
| Gene | Primers | Gene Type | Amplicon Size |
|---|---|---|---|
| ybtS | GACGGAAACAGCACGGTAAA | Siderophores | 242 |
| GAGCATAATAAGGCGAAAGA | |||
| entB | GTCAACTGGGCCTTTGAGCCGTC | Siderophores | 400 |
| TATGGGCGTAAACGCCGGTGAT | |||
| magA | GGTGCTCTTTACATCATTGC | Capsular serotype K1 and hypermucoviscosity phenotype | 128 |
| GCAATGGCCATTTGCGTTAG | |||
| rmpA | CATAAGAGTATTGGTTGACAG | Regulator of mucoid phenotype A | 461 |
| CTTGCATGAGCCATCTTTCA | |||
| K2 | CAACCATGGTGGTCGATTAG | Capsular serotype K2 and hypermucoviscosity phenotype | 531 |
| TGGTAGCCATATCCCTTTGG | |||
| allS | CATTACGCACCTTTGTCAGC | Allantoin metabolism | 764 |
| GAATGTGTCGGCGATCAGCTT | |||
| 16S | ATTTGAAGAGGTTGCAAACGAT | Gene encoding the 16S ribosomal RNA | 133 |
| TTCACTCTGAAGTTTTCTTGTGTTC |
2.4. K. Pneumoniae Survival to Innate Immunity
To evaluate the survival of K. pneumoniae to the innate immunity, the bactericidal activity of normal human serum (NHS), as well as phagocytosis by human macrophages (MΦ), and polymorphonuclear (PMN) cells was determined. Both serum and leukocytes were obtained from blood samples of healthy voluntary donors who had not taken any antibiotic or anti-inflammatory medication for at least ten days before the day of sampling.
2.4.1. Susceptibility to Normal Human Serum
Serum susceptibility was carried out as before [37]; in brief, 75 µL of pooled NHS were mixed with 25 µL suspension containing 2 × 106 CFUs of each isolate in a 96-well plate. As a control, bacterial cultures of each isolate were mixed with PBS. The mixtures were incubated at 37 °C for 3 h, and viable bacteria were enumerated by serial micro-dilution and colony counting on ca-MH agar plates. Serum resistance is expressed as viable bacteria in CFUs/mL compared to untreated isolates controls.
2.4.2. Susceptibility to Phagocytosis by Macrophages and Polymorphonuclear Cells
The separation of mononuclear and polymorphonuclear cells was performed by centrifugation in a gradient density column of Histopaque (Sigma-Aldrich, St. Louis, MO, USA), following the manufacturer instructions. Monocytes were selected from other mononuclear cells by incubation in RPMI-1640 medium (Sigma-Aldrich) without fetal bovine serum (FBS) and differentiated into macrophages incubating during 7–9 days in RPMI-1640 10% FBS at 37 °C with 5% CO2 [38]. The macrophage monolayer was infected with 2.5 × 107 CFUs with a multiplicity of infection (MOI) of 50:1 of each bacterial isolate and centrifuged at 200× g for 5 min, to synchronize phagocytosis. After 2 h of incubation, cells were washed and incubated for an additional 60 min in RPMI-1640 + 100 µg/mL gentamicin. Macrophages were lysed with 0.1% saponin for 10 min at room temperature, and viable bacteria were enumerated as above. For PMN assays [39], 5 × 105 cells were combined with 5 × 106 CFUs of each isolate (MOI 10:1) in serum-free RPMI-1640 and synchronize by centrifugation at 524× g for 8 min at 4 °C. After 3 h, PMNs were lysed with 0.1% saponin, and viable bacteria were enumerated. Control groups of non-phagocyted bacteria were included.
2.5. Synthesis of the PSIR-3 Compound
The structural and photophysical characterization of the PSIR-3 compound was described previously [40]. The complex synthesized can be described by using the following general formula: [Ir(C^N)2(N^N)](PF6), where N^N is the ancillary ligand; and C^N corresponds to a cyclometalating ligand. In this study PSIR-3 is [Ir(ppy)2(ppdh)]PF6 where ppdh is pteridino(7,6-f)(1,10)phenanthroline-1,13(10H,12H)-dihydroxy and ppy is 2-phenylpyridine [41]. The structure and purities of the compound were confirmed by nuclear magnetic resonance (NMR), Fourier-transform infrared spectroscopy (FTIR), and mass spectroscopy (MALDI–MS) measurements. The absorption spectra were measured in acetonitrile (ACN) solutions using a Shimadzu UV–Vis Spectrophotometer UV-1900. The molar extinction coefficients of the characteristic bands were determined from the absorption spectra. Photoluminescence spectra were taken on an Edinburgh Instrument spectrofluorimeter using ACN solutions of the compounds previously degassed with N2 for approximately 20 min. The emission quantum yields (Φem) were calculated according to the description of the literature [42]. Fluorescence lifetimes were measured by using a time-correlated single-photon counting (TC-SPC) apparatus (PicoQuant Picoharp 300) equipped with a sub-nanosecond LED source (excitation at 380 nm) powered by a PicoQuant PDL 800-B variable (2.5−40 MHz) pulsed power supply.
2.6. Antimicrobial Activity of Photosensitizer Compounds
Stock solutions of 2 g/L of the PSIR-3 compound solubilized in ACN were used to prepare working solutions in distilled water. For the antimicrobial assay, the collection of 118 clinical isolates of K. pneumoniae was used, and the control strains of susceptible K. pneumoniae KPPR1 and the MDR strain ST258 were also included. All bacteria were grown as axenic culture in Luria Bertani broth or agar medium as appropriate. PSIR-3 was mixed in 24-well plates at a final concentration of 4 mg/L for photodynamic experiments, with suspensions of 1 × 107 colony forming units (CFUs)/mL of each bacterium, in a final volume of 500 µL of cation-adjusted Mueller-Hinton (ca-MH) broth. Exposure to light was performed for 30 min in a chamber with a white LED lamp at a photon flux of 17 µW/cm2. After exposure to light, the CFUs of the viable bacteria were determined by broth-micro dilution and sub-cultured on ca-MH agar plates. Following the recommendations of the Clinical and Laboratory Standards Institute (CLSI 2017) [34], the agar plates were incubated during 16–20 h at 37 °C, in the dark, and colony count was recorded, using a stereoscopic microscope. Control wells with bacteria culture with no photosensitizer or photosensitizer but not exposed to light were also included.
2.7. Determination of the Synergy between PSIR-3 and Cfx
The fractional inhibitory concentration index (FIC) value was determined using the following formula [43,44]. MICac is the MIC of a compound A, combined with a compound B, and MICbc is the MIC of the compound B combined with the compound A. The MICa and MICb are the MIC of the A and B compounds alone, respectively. Values in the FIC index ≤0.5 are considered synergistic, and values > 4 are considered antagonistic [44].
To determine the MIC-Cfx combined with each PSs, 1 × 107 UFC/mL of ESBL-producing bacteria were aPDI treated for 30 min with 4 mg/L of each PS and mixed with serial dilution (32–0.125 mg/L) of Cfx in ca-MH broth, as above. To determine the MIC-PSs combined with Cfx, 1 × 107 UFC/mL of ESBL-producing bacteria was mixed with serial dilution of each PSs (32–0.125 mg/L) and a fixed concentration of 4 mg/L of Cfx, and then it was subjected to aPDI, as above.
2.8. Statistical Analyses
Statistical analyses were performed by using the Systat 13.2 software (Systat Software, Inc., San Jose, CA, USA) and GraphPad v6.01 (Prism) software. The X2 test or Fisher’s exact test for categorical variables and the Mann–Whitney U test for continuous non-parametric variables were used. The risk of virulence genes to modify the MIC of the antibiotic was established by determining the odds ratio with a CI: 95%.
3. Results
3.1. Demographic Characterization
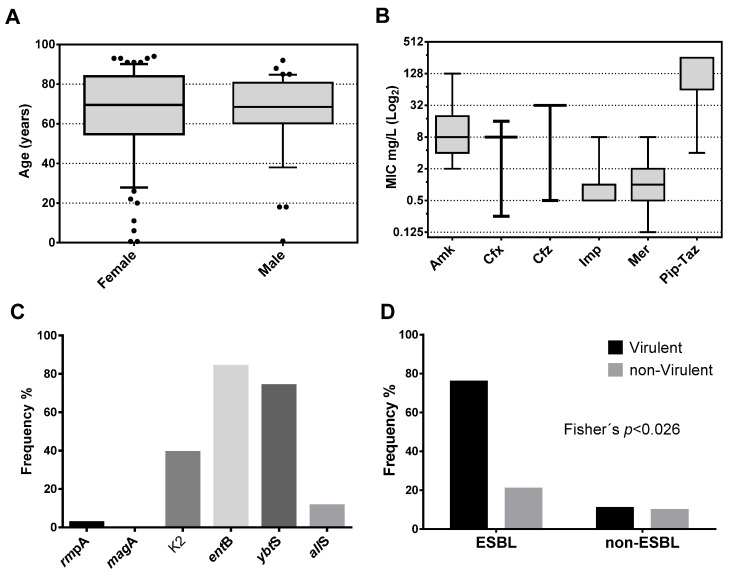
This work seeks to demonstrate the capacity of aPDI to inhibiting the growth of clinical isolates of K. pneumoniae, which are diverse in antimicrobial susceptibility, genotype, and virulence. Phenotypic and genotypic characterization was conducted to determine the frequency of genes encoding virulence factors, the MIC values determined for various antibiotics, and susceptibility to innate immune components. From the clinical isolates of K. pneumoniae received in the laboratory, 118 were selected from different unrelated patients. The isolates were recovered mainly from urine samples, 113 (95.8%), and only five from respiratory samples (three endotracheal aspirates (2.5%) and two expectorations (1.7%)). As shown in Table 2, the samples were obtained from 78 (66%) females and 40 (34%) males, from 12 clinical services, with a higher contribution from the emergency room 41 (34.75%), secondly the unit of medicine 24 (20.34%), third ambulatory 16 (13.56%) and in the fourth place urology 14 (11.86%). Of the 118 clinical isolates, 114 came from adults and 4 from pediatric patients. As shown in Figure 1A, the patient’s age fluctuated between 7 months and 94 years, with a median (IQR: 25–75%) of 69.5 (54.8–84) years for females, and between 10 months and 92 years, with a median of 68.5 (60.3–80.8) years for males. There are no significant differences in age between genders (p = 0.279 Mann–Whitney U test). The results confirm the infections with MDR and not MDR K. pneumoniae mainly affects the elderly population. The elderly are the most susceptible population to present complications derived from infectious diseases [45].
Table 2.
Frequency of K. pneumoniae isolation by hospital unit and genre.
| Service | Female | Male | Total | Percent |
|---|---|---|---|---|
| Outpatient | 12 | 4 | 16 | 13.56% |
| surgery | 5 | 5 | 10 | 8.47% |
| Endocrinology | 1 | 0 | 1 | 0.85% |
| Geriatrics | 1 | 2 | 3 | 2.54% |
| Gynecology | 2 | 0 | 2 | 1.69% |
| Home hospitalization | 1 | 0 | 1 | 0.85% |
| Medicine | 2 | 0 | 24 | 20.34% |
| Medical–surgical service | 13 | 11 | 2 | 1.69% |
| Pediatrics | 1 | 0 | 1 | 0.85% |
| Critical patient unit | 2 | 1 | 3 | 2.54% |
| Emergency room | 31 | 10 | 41 | 34.75% |
| Urology | 7 | 7 | 14 | 11.86% |
| TOTAL | 78 | 40 | 118 | 100% |
Figure 1.
Characterization of patients and clinical isolates of K. pneumoniae. (A) Box plot for the age of patients stratified by gender. Patients show a median of 69.5 years for females and 68.5 years for males. (B) Box plot showing the median in the population of the minimum inhibitory concentration (MIC) of the seven antibiotics commonly used to treat infections by Gram-negative bacteria. (C) Frequency of carrying of the six genes evaluated in the study population. (D) Frequency distribution of carriers of three or more virulence factors stratified by extended-spectrum beta-lactamase (ESBL) production.
3.2. Antibiotic Susceptibility
In the K. pneumoniae population, the median and IQR (25–75%) of the MIC were determined and expressed in mg/L in a log2 box plot. As shown in Figure 1B, the bacterial population was mainly susceptible to Amk with a median (IQR) of 8 (4–20). Similar to Amk, the population was mainly susceptible to carbapenem, Imp, and Mer with a median of 1 (0.5–1) and 1 (0.5–2), respectively. In contrast, most of the population was resistant to the cephalosporins, Cfx, and Cfz, with medians of 8 (8–8) and 32 (32–32). Finally, similar to cephalosporin, almost the entire population was resistant to the Pip–Taz combination with a median of 256 (64–256). From the population, 66 isolates resistant to Cfx were susceptible to the combination Cfx/clavulanic acid. Those isolates were identified as ESBL-producing bacteria.
3.3. Virulence Gene Frequency
In this work, we select certain virulence genes belonging to families with different mechanisms of action. PCR determined the frequency of carrying genes encoding the virulence factors rmpA, magA, K2, entB, allS, and ybtS for each isolate using specific primers (Table 1). These genes were selected for being representatives of different families of virulence factors. As shown in Figure 1C, the harboring frequency was; rmpA [2.54% (3/118)], magA [0% (0/118)], K2 [39% (46/118)], entB [83.9% (99)], allS [11.2% 13/118)] and ybtS [73.8% (87/118)]. The most frequent virulence factor was the entB gene, followed by the ybtS gene. There were no isolates with the magA gene, and only three isolates harbor the rmpA gene. No isolates showed the hypermucoviscosity phenotype (by string test) in agar plates in the population of 118 unrelated isolates.
3.4. Correlation of Virulence Factors with Antibiotic Resistance
As shown in Figure 1D, there is a significantly higher frequency of ESBL strains that harbor three or more virulence factors compared to non-ESBL strains (Fisher’s; p < 0.026). The non-parametric Mann–Whitney U test (Systat 13 software) was used to determine the association that each virulence gene has on the median MIC value assuming a null hypothesis α = 5%. As shown in Table 3, the rmpA gene was not significantly associated with a modification of the median-MIC of any of the antibiotics analyzed in this study (p > 0.05). On the other hand, the allS gene significantly influenced the median-MIC of Cfx, Cfz, and Pip–Taz (p < 0.05). Similarly, the K2 gene significantly influenced the median-MIC of Cfx and Pip–Taz (p < 0.05). The entB gene significantly affected the median-MIC of Amk and Imp (p < 0.05). The ybtS gene significantly influenced the median-MIC of Cfx (p < 0.006). Finally, the allS gene significantly influenced the median-MIC of Cfx and Cfz antibiotics (p < 0.05). The MIC of the antibiotics more sensitive to the presence of virulence factors were the Cfx (sensitive to K2, ybtS, and allS genes) and Pip–Taz (sensitive to the K2 and allS genes). These data show that, regardless of the patient’s conditions, the multi-virulence is effectively an independent risk factor that promotes ESBL-production of clinical populations of K. pneumoniae with a p < 0.026 Fisher’s exact test.
Table 3.
Mann–Whitney U test p-values for modification of antibiotic MIC by virulence genes.
| Amikacin | Cefotaxime | Ceftazidime | Imipenem | Meropenem | Pip–Taz | |
|---|---|---|---|---|---|---|
| rmpA | 0.285 | 0.051 | 0.432 | 0.363 | 0.527 | 0.411 |
| K2 | 0.158 | 0.002 | 0.919 | 0.102 | 0.588 | 0.001 |
| entB | 0.009 | 0.918 | 0.117 | 0.015 | 0.291 | 0.072 |
| ybtS | 0.572 | 0.006 | 0.722 | 0.340 | 0.502 | 0.067 |
| allS | 0.326 | 0.024 | 0.001 | 0.086 | 0.537 | 0.013 |
Values in bold represent a p < 0.05, below the null hypothesis value with α = 5%, which means a significant difference in MIC due to the presence of the virulence factor. Pip–Taz, piperacillin–tazobactam.
As shown in Figure 2, the box plot for each antibiotic stratified by the presence or absence of virulence genes was constructed to verify if the influence is to increase or decrease the median of the MIC. The presence of the entB gene is associated with a decrease in the median-MIC for Amk and Imp. On the other hand, the K2 gene is associated with an increase in the median-MIC for Cfx and Pip–Taz. Similarly, the ybtS gene is associated with an increase in the median-MIC for Cfx. Finally, the allS gene is associated with an increased median-MIC for Cfz and decreased median-MIC for Cfx and Pip–Taz. Some of the virulence factors studied here were associated with the median- MIC modification for several antibiotics. In the K. pneumoniae population tested, the most influencing virulence factors were entB, ybtS, and allS genes. The stratified MICs-box plot made it possible to distinguish how virulence genes modulate antibiotic susceptibility by increasing or decreasing. Remarkably, the sum of the genes of the K2+, ybtS+, and allS+ genes contributes to increasing the median-MICs to values higher than the clinical susceptibility breakpoint established by the CLSI. These increased values occur similarly when efflux pumps are activated in other Gram-negative bacteria [46]. For example, in a population of Pseudomonas aeruginosa, the combined overexpression of the mexA and mexX efflux pump increased the median MIC for ciprofloxacin and cefepime above the cutoff points [46,47].
Figure 2.
Association of virulence genes in the measurement of median MIC. The box diagrams created with the Systat 13 software summarize the MIC measures for six commonly used antibiotics, stratified by the presence or absence of each of the six virulence genes. The data are presented as mg/L in a Log2 scale of MICs for each antibiotic stratified by the presence (+) or absence (−) of each virulence gene. The horizontal lines represent the median and the frequency limits between 25 and 75% of the individuals, the hollow dots represent single individuals, and the asterisks represent small groups of individuals.
3.5. Association of K2+, ybtS+, and allS+ Virulence Genes to Survive the Innate Immunity
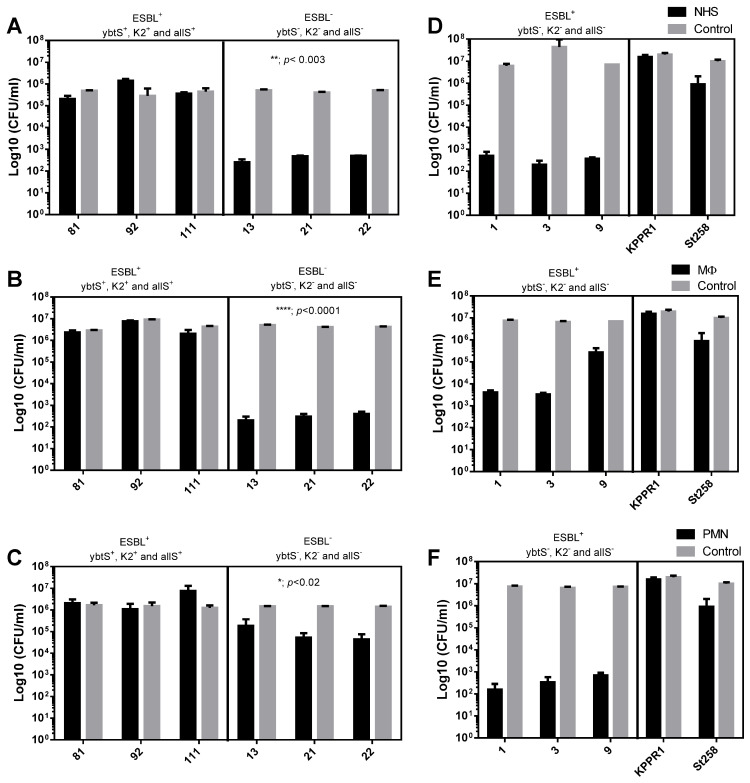
Our results show that the K2 and ybtS virulence genes are risk factors for the production of ESBL and that the allS gene acts as a protective factor. Then, we selected two groups of clinical isolates of K. pneumoniae, which harbor or not these three genes, to assess their susceptibility to innate immunity components. Only three isolates, identified by the numbers 81, 92, and 111, are of the K2+, ybtS+, and allS+ genotype, and unexpectedly all of them are ESBL-producers. From the genotype K2−, ybtS− and allS−, the isolates 13, 21, and 22 were selected as being non-ESBL. As controls, the susceptible but highly virulent K. pneumoniae KPPR1 (K2+, ybtS+, and allS+) and the MDR but less virulent ST258 (K2−, ybtS− and allS−) strains were also included.
To determine the bacterial susceptibility to normal human serum (NHS), they were exposed for 3 h, and then the number of viable bacteria was determined by microdilution and plate counting. As shown in Figure 3A, the presence of all three virulence genes, K2+, ybtS+, and allS+, significantly (** = p < 0.003; two-way ANOVA) increases the survival of ESBL-producing bacteria compared to non-ESBL bacteria lacking all three virulence genes. On average, the serum resistance was improved by four orders of magnitude (4log10). Similar to that observed with serum, ESBL-producing bacteria of the K2+, ybtS+, and allS+ genotype show a significant (**** = p < 0.0001) increase, 4log10, in the survival to the MΦ activity, compared to no- ESBL bacteria that lacks these virulence genes (Figure 3B). Finally, comparable to MΦ, survival to the activity of PMNs of the bacteria of genotype K2+, ybtS+, and allS+ increased significantly (* = p < 0.02), at least one time on average, compared to non-ESBL strains that lack the virulence genes (Figure 3C). As shown in Figure 3, the virulent control KPPR1 strain was more resistant to serum (Figure 3D), macrophages (Figure 3E), and PMN (Figure 3F) compared to the MDR ST258 K. pneumoniae strain. Consistently the clinical control isolates have shown to be more susceptible to the bactericidal activity of the serum (3D) and the phagocytic activity of MΦ (3E) and PMN (3F). The co-occurrence of harboring multiple genes that encode virulence factors and the ESBL-production leads to enhanced virulence. The ESBL-producing strains of K. pneumoniae of the genotype K2+, ybtS+, and allS+ were more resistant to innate immunity, consistently with studies over MDR populations of K. pneumoniae that increased their 30-day mortality over patients undergoing bloodstream infections [1,48]. Moreover, virulence genes, such as siderophores, which have an essential role in bacterial survival and virulence [49,50], have been previously associated with the MDR-K. pneumoniae [16,51]. In this study, ESBL-producer K. pneumoniae of the K2+, ybtS+, and allS+ genotype shown a survival improvement for killing by PMNs. Previously, the PMN has demonstrated a limited binding and uptake capacity for MDR–K. pneumoniae [39]. The activity of the PMN is the most important cellular component of the innate immune response, essential as the first line of defense against bacterial infections [52].
Figure 3.
Effect of virulence genes on the susceptibility of ESBL or non-ESBL strains to components of innate immunity. The susceptibility of ESBL-producing K. pneumoniae clinical isolates bearing the virulence genes ybtS+, K2+, and allS+ was determined and compared with non-ESBL producing bacteria that lack the virulence genes. (A,D) Susceptibility to normal human serum (NHS), (B,E) susceptibility to phagocytosis by macrophages (MΦ), and (C,F) susceptibility to phagocytosis by polymorphonuclear cells (PMN). The results of two independent experiments performed in triplicate are shown (n = 6). Viable bacteria were enumerated by colony count on ca-MH agar after serial micro-dilution. The colony forming units (CFUs)/mL values are presented as means +/− SD, on a log10 scale of treated bacteria (black bars) compared to untreated control bacteria (gray bars). **** = p < 0.0001, ** = p < 0.003 and * = p < 0.02 of two-way ANOVA comparing the proportion of treated/untreated ESBL bacteria with non-ESBL bacteria.
3.6. Susceptibility of Clinical Isolates to aPDI with PSIR-3
3.6.1. Photophysical Properties of the PSIR-3 Compound
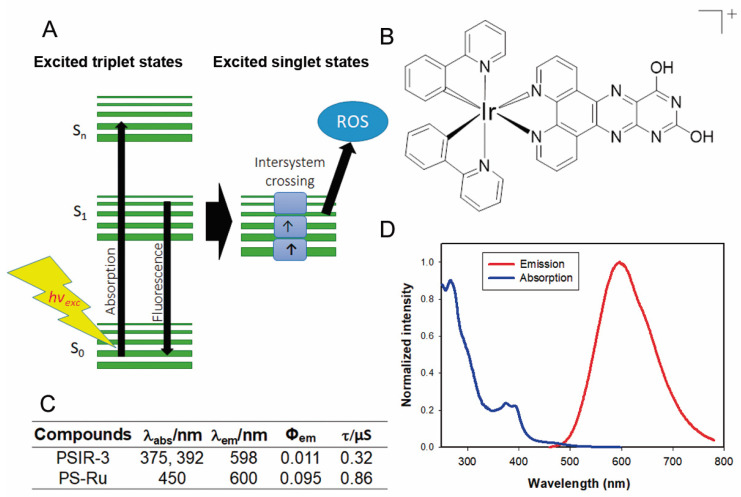
We have previously shown that Ir(III)-based compounds, such as PSIR-3, have photodynamic antimicrobial activity against imipenem-resistant Klebsiella pneumoniae [32,33]. In this work, we tested a coordination compound characterized by a positive charge in the first coordination sphere (Figure 4B). The photophysical evaluation of the PSIR-3 performed in acetonitrile solution [40] revealed absorption processes at 375 and 392 nm attributable at the first instance of charge-transfer transitions (Figure 4C,D) [32]. When the PSIR-3 compound was excited with a wavelength corresponding to the lowest charge-transfer absorption energy, 375 nm, it showed maximum emission at 598 nm (Figure 4C,D). Figure 4C shows the recorded lifetimes of excited states in 0.32 μs and the calculated quantum yield (Φem) in 0.011 [40]. The aPDI activity of the PSIR-3 compound was compared with the positive PS control [Ru(bpy)3](PF6)2 (bpy = 2,2′-bipyridine) called PS-Ru. According to the literature, the PS-Ru shows a charge-transfer absorption process at 450 nm with maximum emission at 600 nm (excited in 450 nm) in acetonitrile [42], with Φem of 0.095 [42], and a lifetime registered of its excited state of 0.855 μs (Figure 4C) [53]. The maximum absorption of PSIR-3 occurs at wavelengths below 400 nm, that although it is more energetic, it penetrates the tissues poorly. Therefore, this compound will activate better if it is directly irradiated, such as in superficial wounds. However, UTI is one of the most common diseases caused by K. pneumoniae, where probes that deliver the light dose within internal surfaces can irradiate the epithelial lining of the bladder [54]. Certain kinds of fiber arrays or inflatable balloons may provide a homogenous light power delivery [55,56]. This catheterization can be applied for inpatients suffering UTI that does not respond to antibiotic treatment.
Figure 4.
The photophysical properties of the PSIR-3 photosensitizer. (A) schematic representation of the absorption–emission process of photosensitizer molecules. Light excites external electrons to accede from a basal state S0 to a higher energetic state S1–Sn. Suppose the electrons return to its ground state; the energy is then released as fluorescence. However, when the excited electron enters an intersystem crossing process, the released energy excites molecular oxygen and converts it to reactive oxygen species (ROS). (B) chemical structure of the Ir(III) compound (PSIR-3, [Ir(ppy)2(ppdh)]PF6). (C) Summary of the photophysical properties of the PSIR-3 and PS-Ru compounds, where λabs = wavelength of absorbance, λem = wavelength of emission, Φem = emission quantum yield, and τ = time in the excited state. Data for PS-Ru were obtained from the literature. Adapted from [42], Elsevier, 2010; Adapted from [53], American Chemical Society, 1983. (D) The absorption and the emission spectra of the PSIR-3 in acetonitrile (ACN).
3.6.2. Antimicrobial Photodynamic Inhibition of the PSIR-3 over Clinical Isolates
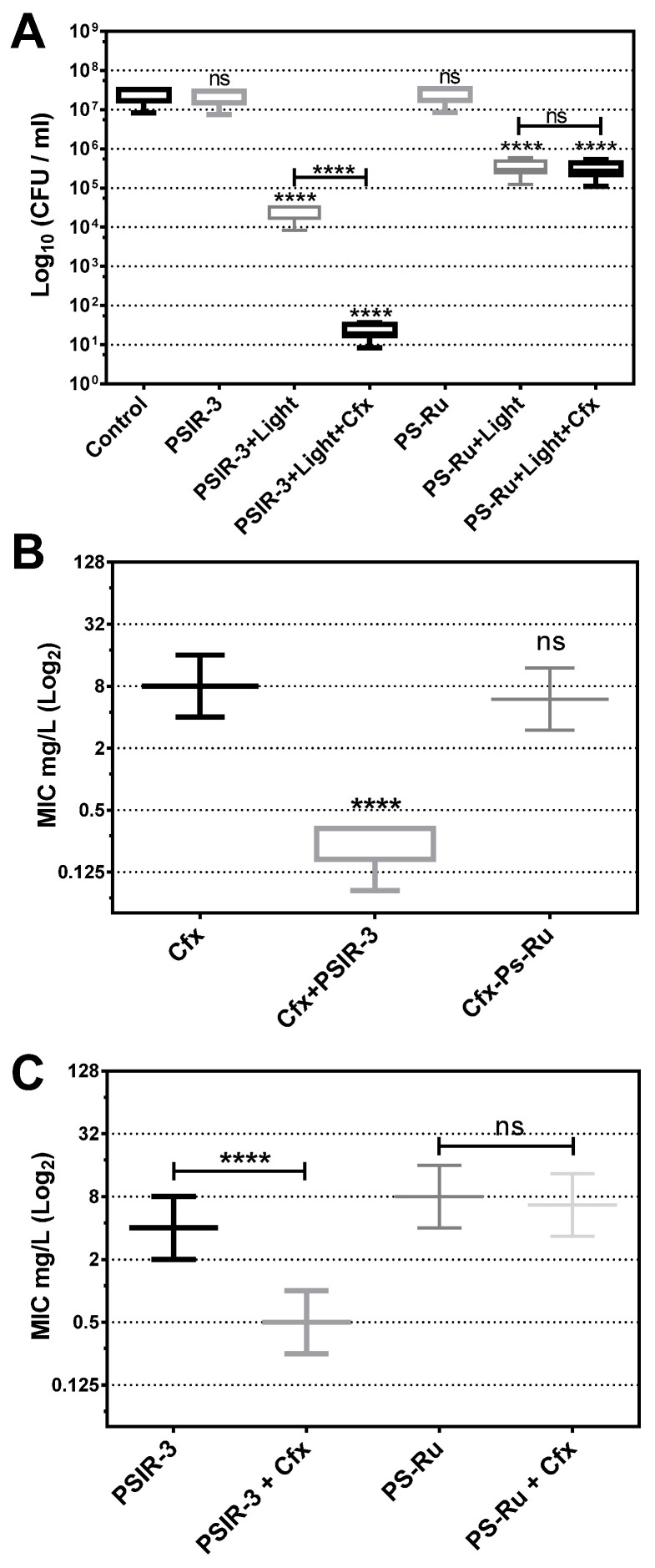
Photodynamic treatment was verified to produce the observed growth inhibition of the 118 clinical isolates of K. pneumoniae compared to the untreated bacteria. The photodynamic activity of the PSIR-3 compound was compared to the activity of the PS-Ru reference compound as a positive control [41,57,58,59]. As seen in Figure 5, compared to the control of untreated bacteria, photodynamic treatment with 4 µg/mL PSIR-3 inhibits bacterial growth > 3 log10 (>99.9%) of clinical isolates of K. pneumoniae (**** p < 0.0001; compared to untreated control). The results show that the bactericidal effect produced by PSIR-3 is light-dependent (ns = p > 0.05; compared to the untreated control). These results are comparable with those obtained with the positive control compound PS-Ru, which has shown that bacterial growth inhibition is light-dependent (**** p < 0.0001; compared to the untreated control).
Figure 5.
Antimicrobial photodynamic inactivation of clinical isolates of K. pneumoniae. (A) Growth inhibition of 118 clinical isolates of K. pneumoniae subjected to antimicrobial photodynamic inactivation (aPDI) with PSIR-3. The bacteria were used at a concentration of 1 × 107 CFUs/mL and mixed in triplicate with 4 mg/L of PSIR-3 or PSIR-4 compounds. For the aPDI, the mixture of bacteria with PS was exposed for 1 h at 17 µW/cm2 of white light. As a control, bacteria combined with the photosensitizers (PSs) not exposed to light (PSIR-3 or PS-Ru) and bacteria not combined with the PSs (control) were included. Colony count enumerated of viable bacteria on ca-MH agar after serial micro-dilution. The CFUs/mL values are presented as means ± SD on a log10 scale. (B) From the clinical isolates, 66 ESBL-producing bacteria were exposed to aPDI, using PSIR-3 or PS-Ru, and MIC for cefotaxime (Cfx) was performed in triplicate, in ca-MH broth, for 16–20 h. (C) For ESBL-producing bacteria, the MIC for PSIR-3 or PS-Ru, were determined in combination with 4 mg/L of Cfx, performed in triplicate in ca-MH agar for 16–20 h. The MIC values are presented as median ± SD of mg/mL on a log2 scale. Not significant (ns) p > 0.05 by Student’s t-test among treated bacteria compared to control; **** p < 0.0001 by Student’s t-test among treated bacteria compared to control.
3.6.3. Synergism between aPDI with PSIR-3 and Cefotaxime
Since PSIR-3 showed synergism combined with imipenem [32], we analyzed whether it shows synergism with cefotaxime in the population of clinical isolates. First, it was determined whether the combined treatment with Cfx increases the inhibition of bacterial growth of aPDI with PSIR-3. The 118 clinical isolates of K. pneumoniae were exposed to the preparation of 4 µg/mL of cefotaxime with 4 µg/mL of PSIR-3 (its MIC). Control bacteria without Cfx and exposure to light were included. As expected, the PSIR-3 compound mixed with cefotaxime significantly (**** p < 0.0001) increased the bactericidal effect on the clinical isolates population from 3 to 6 log10 reduction (Figure 5A). As seen before, a significantly increased inhibitory effect was not observed when combining the cefotaxime with the PS-Ru control compound (ns p > 0.05). Secondly, the set of 66 clinical isolates characterized as ESBL-producing K. pneumoniae were treated with PSIR-3 aPDIfor 1 h, and serial dilutions determined the MIC for Cfx in ca-MH broth. As seen in Figure 5B, a significant reduction (**** p < 0.0001) from 8 µg/mL (8–8) to 0.17 µg/mL (0.17–0.333) on Cfx-MIC was observed compared to the untreated group. The combined treatment also reduced the PSIR-3-MIC, from 4 to 0.5 mg/L (Figure 5C). Similar to previously shown with imipenem [32], compound PSIR-3 produced a significant change in Cfx-susceptibility with a fractional inhibitory concentration (FIC) index of 0.15 (Table 4). Figure 5C shows the control compound, PS-Ru, did not significantly change Cfx-susceptibility with an FIC Index 1.58 (Table 4). Because synergy is defined as an FIC index of ≤0.5 [44], the increased inhibitory effect observed when combining PSIR-3 with Cfx is not summative but synergistic.
Table 4.
Fractional inhibitory concentration (FIC) index calculation.
| Compounds | MIC | MIC Combined | FIC | FIC Index |
|---|---|---|---|---|
| Cfx | 8.00 | |||
| PSIR-3 | 4.00 | 0.17 | 0.02 | 0.15 |
| PSIR-3 * | 0.50 | 0.13 | ||
| PS-Ru | 8.00 | 6.67 | 0.83 | 1.58 |
| PS-Ru * | 6.00 | 0.75 |
MIC values are de median for the ESBL-producing K. pneumoniae, n = 66. * The MIC value modified by Cfx.
The behavior exhibited by PSIR-3 must be related, as mentioned in previous reports [32,33], to the external substituents bonded to polypyridine ligand structures and affinity to bacterial envelope [41]. This synergism is also comparable to other photosensitizers; for example, rose bengal showed an increase in the susceptibility of Acinetobacter baumannii for a range of antibiotics used along with aPDI [60]. In another example, conventional antibiotics and alternative compounds reported synergism in a murine model for pathogens of the ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) [61], using anti-biofilm peptides [62]. For now, it is difficult to accurately calculate the dose of light necessary to activate PSIR-3 effectively; however, we observed in vitro that with low doses of energy (17 µW/cm2 of white light), it is bactericidal. Compounds with optimum absorbance at higher wavelengths, bordering the 630–750 nm, would improve the exposure of PS to light into the tissues, but being less energetic, the PSs must have a triplet excited state of easy access to promote energy transfer [63]. We, therefore, need to perform a better characterization of its antimicrobial activity when activated with defined wavelengths, before starting in vivo studies, in urinary infection models and to evaluate the need to deliver light through intraurethral catheters [64].
4. Conclusions
In this work, we saw that multi-drug resistance and virulence are significant factors in clinical isolates of K. pneumoniae. However, the increase in MICs can be neutralized by aPDI, turning resistant strains susceptible. APDI is effective in treating multidrug-resistant bacteria and more virulent strains, as well as strains that combine both characteristics. The aPDI then becomes a great support to antimicrobial therapy in a shortage of new effective antibiotics. The photophysical characterization of PS indicates that its maximum absorption occurs at wavelengths lower than 400 nm, which could constitute a problem for its use in infections of internal organs due to low penetration. In UTI, optical fibers can be used through a catheter to deliver the dose of light [54,55]. Moreover, compounds with optimum absorbance at higher wavelengths, bordering the 630–750 nm, would improve the exposure of PS to light in the tissues.
Acknowledgments
We would like to thank Myriam Pizarro, head of the “Hospital El Carmen de Maipú” laboratory, for her collaboration and provision of the clinical isolates of Klebsiella pneumoniae. We also would like to thank Susan Bueno from Pontificia Universidad Católica de Chile for providing the control strains of K. pneumoniae, KPPR1, and the typo strain ST258.
Author Contributions
Conceptualization, C.E.P. and I.A.G.; methodology, C.E.P. and I.A.G.; software, C.E.P. and I.A.G.; validation, C.E.P., P.D. and I.A.G.; formal analysis, C.E.P., P.D. and I.A.G.; investigation, C.N. and A.P.; resources, C.E.P., P.D. and I.A.G.; data curation, C.E.P. and I.A.G.; writing—original draft preparation, C.E.P.; writing—review and editing, C.E.P.; visualization, C.E.P. and I.A.G.; supervision, C.E.P. and I.A.G.; project administration, C.E.P.; funding acquisition, C.E.P., P.D. and I.A.G. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are supported by UCEN grants CIP2019007 (awarded to C.E.P.), Fondecyt grants 1180673 and 1201173 (awarded to I.A.G. and P.D., respectively), Office of Naval Research (ONR) grant N62909-18-1-2180 (awarded to I.A.G.) and USM PM_I_2020_31 project of P.D.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Ethics Committee of the Faculty of Health Sciences, of the Universidad Central de Chile N: 01/2017 approved on 01-04-2017.
Informed Consent Statement
The informed consent form was approved by the ethics committee of the Faculty of Health Sciences of the Central University of Chile and by the Bioethics Committee of the Central Metropolitan Health Service of Chile (MHSC), Act number: N 124/07.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available because they are confidential data of patients protected by the informed consent protocol.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim D., Park B.Y., Choi M.H., Yoon E.J., Lee H., Lee K.J., Park Y.S., Shin J.H., Uh Y., Shin K.S., et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J. Antimicrob. Chemother. 2019;74:190–199. doi: 10.1093/jac/dky397. [DOI] [PubMed] [Google Scholar]
- 2.Zhen X., Lundborg C.S., Sun X., Hu X., Dong H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resist. Infect. Control. 2019;8:137. doi: 10.1186/s13756-019-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lev A.I., Astashkin E.I., Kislichkina A.A., Solovieva E.V., Kombarova T.I., Korobova O.V., Ershova O.N., Alexandrova I.A., Malikov V.E., Bogun A.G., et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012–2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog. Glob. Health. 2018;112:142–151. doi: 10.1080/20477724.2018.1460949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tofas P., Skiada A., Angelopoulou M., Sipsas N., Pavlopoulou I., Tsaousi S., Pagoni M., Kotsopoulou M., Perlorentzou S., Antoniadou A., et al. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections in neutropenic patients with haematological malignancies or aplastic anaemia: Analysis of 50 cases. Int. J. Antimicrob. Agents. 2016;47:335–339. doi: 10.1016/j.ijantimicag.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Li Y., Sun Q.L., Shen Y., Zhang Y., Yang J.W., Shu L.B., Zhou H.W., Wang Y., Wang B., Zhang R., et al. Rapid Increase in Prevalence of Carbapenem-Resistant Enterobacteriaceae (CRE) and Emergence of Colistin Resistance Gene mcr-1 in CRE in a Hospital in Henan, China. J. Clin. Microbiol. 2018:56. doi: 10.1128/JCM.01932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO . WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. WHO; Geneva, Switzerland: 2017. [Google Scholar]
- 7.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 8.Pragasam A.K., Shankar C., Veeraraghavan B., Biswas I., Nabarro L.E., Inbanathan F.Y., George B., Verghese S. Molecular Mechanisms of Colistin Resistance in Klebsiella pneumoniae Causing Bacteremia from India-A First Report. Front. Microbiol. 2016;7:2135. doi: 10.3389/fmicb.2016.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin R.M., Bachman M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonda T., Kumburu H., van Zwetselaar M., Alifrangis M., Mmbaga B.T., Lund O., Kibiki G.S., Aarestrup F.M. Molecular epidemiology of virulence and antimicrobial resistance determinants in Klebsiella pneumoniae from hospitalised patients in Kilimanjaro, Tanzania. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018;37:1901–1914. doi: 10.1007/s10096-018-3324-5. [DOI] [PubMed] [Google Scholar]
- 11.Khaertynov K.S., Anokhin V.A., Rizvanov A.A., Davidyuk Y.N., Semyenova D.R., Lubin S.A., Skvortsova N.N. Virulence Factors and Antibiotic Resistance of Klebsiella pneumoniae Strains Isolated from Neonates with Sepsis. Front. Med. 2018;5:225. doi: 10.3389/fmed.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rastegar S., Moradi M., Kalantar-Neyestanaki D., Ali Golabi D., Hosseini-Nave H. Virulence Factors, Capsular Serotypes and Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae and Classical Klebsiella pneumoniae in Southeast Iran. Infect. Chemother. 2019 doi: 10.3947/ic.2019.0027. [DOI] [PubMed] [Google Scholar]
- 13.Derakhshan S., Hosseinzadeh A. Resistant Pseudomonas aeruginosa carrying virulence genes in hospitalized patients with urinary tract infection from Sanandaj, west of Iran. Gene Rep. 2020:100675. doi: 10.1016/j.genrep.2020.100675. [DOI] [Google Scholar]
- 14.Imtiaz W., Syed Z., Rafaque Z., Andrews S.C., Dasti J.I. Analysis of Antibiotic Resistance and Virulence Traits (Genetic and Phenotypic) in Klebsiella pneumoniae Clinical Isolates from Pakistan: Identification of Significant Levels of Carbapenem and Colistin Resistance. Infect. Drug Resist. 2021;14:227–236. doi: 10.2147/IDR.S293290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. MMBR. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachman M.A., Oyler J.E., Burns S.H., Caza M., Lepine F., Dozois C.M., Weiser J.N. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.J., Kim S.I., Kim Y.R., Wie S.H., Lee H.K., Kim S.Y., Park Y.J. Virulence factors and clinical patterns of hypermucoviscous Klebsiella pneumoniae isolated from urine. Infect. Dis. 2017;49:178–184. doi: 10.1080/23744235.2016.1244611. [DOI] [PubMed] [Google Scholar]
- 18.French J.B., Neau D.B., Ealick S.E. Characterization of the structure and function of Klebsiella pneumoniae allantoin racemase. J. Mol. Biol. 2011;410:447–460. doi: 10.1016/j.jmb.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadasy K.A., Domiati-Saad R., Tribble M.A. Invasive Klebsiella pneumoniae syndrome in North America. Clin. Infect. Dis. 2007;45:e25–e28. doi: 10.1086/519424. [DOI] [PubMed] [Google Scholar]
- 20.Chuang Y.P., Fang C.T., Lai S.Y., Chang S.C., Wang J.T. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2006;193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 21.Cubero M., Grau I., Tubau F., Pallares R., Dominguez M.A., Linares J., Ardanuy C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013) Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016;22:154–160. doi: 10.1016/j.cmi.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Lin H.A., Huang Y.L., Yeh K.M., Siu L.K., Lin J.C., Chang F.Y. Regulator of the mucoid phenotype A gene increases the virulent ability of extended-spectrum beta-lactamase-producing serotype non-K1/K2 Klebsiella pneumonia. J. Microbiol. Immunol. Infect. Wei Mian Yu Gan Ran Za Zhi. 2016;49:494–501. doi: 10.1016/j.jmii.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 24.Valenzuela-Valderrama M., Gonzalez I.A., Palavecino C.E. Photodynamic treatment for multidrug-resistant Gram-negative bacteria: Perspectives for the treatment of Klebsiella pneumoniae infections. Photodiagn. Photodyn. Ther. 2019;28:256–264. doi: 10.1016/j.pdpdt.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Tosato M.G., Schilardi P., Lorenzo de Mele M.F., Thomas A.H., Lorente C., Minan A. Synergistic effect of carboxypterin and methylene blue applied to antimicrobial photodynamic therapy against mature biofilm of Klebsiella pneumoniae. Heliyon. 2020;6:e03522. doi: 10.1016/j.heliyon.2020.e03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agostinis P., Berg K., Cengel K.A., Foster T.H., Girotti A.W., Gollnick S.O., Hahn S.M., Hamblin M.R., Juzeniene A., Kessel D., et al. Photodynamic therapy of cancer: An update. CA Cancer J. Clin. 2011;61:250–281. doi: 10.3322/caac.20114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briviba K., Klotz L.O., Sies H. Toxic and signaling effects of photochemically or chemically generated singlet oxygen in biological systems. Biol. Chem. 1997;378:1259–1265. [PubMed] [Google Scholar]
- 28.Winterbourn C.C., Kettle A.J. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 29.Alvarez L.A., Kovacic L., Rodriguez J., Gosemann J.H., Kubica M., Pircalabioru G.G., Friedmacher F., Cean A., Ghise A., Sarandan M.B., et al. NADPH oxidase-derived H2O2 subverts pathogen signaling by oxidative phosphotyrosine conversion to PB-DOPA. Proc. Natl. Acad. Sci. USA. 2016;113:10406–10411. doi: 10.1073/pnas.1605443113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziegelhoffer E.C., Donohue T.J. Bacterial responses to photo-oxidative stress. Nat. Rev. Microbiol. 2009;7:856–863. doi: 10.1038/nrmicro2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trindade A.C., de Figueiredo J.A., de Oliveira S.D., Barth Junior V.C., Gallo S.W., Follmann C., Wolle C.F., Steier L., Morgental R.D., Weber J.B. Histopathological, Microbiological, and Radiographic Analysis of Antimicrobial Photodynamic Therapy for the Treatment of Teeth with Apical Periodontitis: A Study in Rats’ Molars. Photomed. Laser Surg. 2017;35:364–371. doi: 10.1089/pho.2016.4102. [DOI] [PubMed] [Google Scholar]
- 32.Valenzuela-Valderrama M., Carrasco-Véliz N., González I.A., Dreyse P., Palavecino C.E. Synergistic effect of combined imipenem and photodynamic treatment with the cationic Ir(III) complexes to polypyridine ligand on carbapenem-resistant Klebsiella pneumoniae. Photodiagn. Photodyn. Ther. 2020;31:101882. doi: 10.1016/j.pdpdt.2020.101882. [DOI] [PubMed] [Google Scholar]
- 33.Bustamante V., Gonzalez I.A., Dreyse P., Palavecino C.E. The mode of action of the PSIR-3 photosensitizer in the photodynamic inactivation of Klebsiella pneumoniae is by the production of type II ROS which activate RpoE-regulated extracytoplasmic factors. Photodiagn. Photodyn. Ther. 2020;32:102020. doi: 10.1016/j.pdpdt.2020.102020. [DOI] [PubMed] [Google Scholar]
- 34.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. 2017. [(accessed on 2 January 2021)]. Available online: https://clsi.org/media/3481/m100ed30_sample.pdf. [Google Scholar]
- 35.Broberg C.A., Wu W., Cavalcoli J.D., Miller V.L., Bachman M.A. Complete Genome Sequence of Klebsiella pneumoniae Strain ATCC 43816 KPPR1, a Rifampin-Resistant Mutant Commonly Used in Animal, Genetic, and Molecular Biology Studies. Genome Announc. 2014;2 doi: 10.1128/genomeA.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn D., Penaloza H., Wang Z., Wickersham M., Parker D., Patel P., Koller A., Chen E.I., Bueno S.M., Uhlemann A.C., et al. Acquired resistance to innate immune clearance promotes Klebsiella pneumoniae ST258 pulmonary infection. JCI Insight. 2016;1:e89704. doi: 10.1172/jci.insight.89704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahly H., Aucken H., Benedi V.J., Forestier C., Fussing V., Hansen D.S., Ofek I., Podschun R., Sirot D., Tomas J.M., et al. Increased serum resistance in Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 2004;48:3477–3482. doi: 10.1128/AAC.48.9.3477-3482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ares M.A., Fernandez-Vazquez J.L., Rosales-Reyes R., Jarillo-Quijada M.D., von Bargen K., Torres J., Gonzalez-y-Merchand J.A., Alcantar-Curiel M.D., De la Cruz M.A. H-NS Nucleoid Protein Controls Virulence Features of Klebsiella pneumoniae by Regulating the Expression of Type 3 Pili and the Capsule Polysaccharide. Front. Cell. Infect. Microbiol. 2016;6:13. doi: 10.3389/fcimb.2016.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi S.D., Porter A.R., Dorward D.W., Brinkworth A.J., Chen L., Kreiswirth B.N., DeLeo F.R. Phagocytosis and Killing of Carbapenem-Resistant ST258 Klebsiella pneumoniae by Human Neutrophils. J. Infect. Dis. 2016;213:1615–1622. doi: 10.1093/infdis/jiw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González I., Gómez J., Santander-Nelli M., Natali M., Cortés-Arriagada D., Dreyse P. Synthesis and photophysical characterization of novel Ir(III) complexes with a dipyridophenazine analogue (ppdh) as ancillary ligand. Polyhedron. 2020;186:114621. doi: 10.1016/j.poly.2020.114621. [DOI] [Google Scholar]
- 41.González I., Natali M., Cabrera A.R., Loeb B., Maze J., Dreyse P. Substituent influence in phenanthroline-derived ancillary ligands on the excited state nature of novel cationic Ir(iii) complexes. New J. Chem. 2018;42:6644–6654. doi: 10.1039/C8NJ00334C. [DOI] [Google Scholar]
- 42.Ishida H., Tobita S., Hasegawa Y., Katoh R., Nozaki K. Recent advances in instrumentation for absolute emission quantum yield measurements. Coord. Chem. Rev. 2010;254:2449–2458. doi: 10.1016/j.ccr.2010.04.006. [DOI] [Google Scholar]
- 43.MacNair C.R., Stokes J.M., Carfrae L.A., Fiebig-Comyn A.A., Coombes B.K., Mulvey M.R., Brown E.D. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 2018;9:458. doi: 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stokes J.M., MacNair C.R., Ilyas B., French S., Cote J.P., Bouwman C., Farha M.A., Sieron A.O., Whitfield C., Coombes B.K., et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017;2:17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng N.P. Aging of the immune system: How much can the adaptive immune system adapt? Immunity. 2006;24:495–499. doi: 10.1016/j.immuni.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riou M., Avrain L., Carbonnelle S., El Garch F., Pirnay J.P., De Vos D., Plesiat P., Tulkens P.M., Van Bambeke F. Increase of efflux-mediated resistance in Pseudomonas aeruginosa during antibiotic treatment in patients suffering from nosocomial pneumonia. Int. J. Antimicrob. Agents. 2016;47:77–83. doi: 10.1016/j.ijantimicag.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 47.EUCAST EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. [(accessed on 25 December 2020)];2016 Available online: https://www.eucast.org/resistance_mechanisms/
- 48.Brady M., Cunney R., Murchan S., Oza A., Burns K. Klebsiella pneumoniae bloodstream infection, antimicrobial resistance and consumption trends in Ireland: 2008 to 2013. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2016;35:1777–1785. doi: 10.1007/s10096-016-2727-4. [DOI] [PubMed] [Google Scholar]
- 49.Marques C., Menezes J., Belas A., Aboim C., Cavaco-Silva P., Trigueiro G., Telo Gama L., Pomba C. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: Population structure, antimicrobial resistance and virulence genes. J. Antimicrob. Chemother. 2019;74:594–602. doi: 10.1093/jac/dky499. [DOI] [PubMed] [Google Scholar]
- 50.Russo T.A., Olson R., MacDonald U., Beanan J., Davidson B.A. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 2015;83:3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vargas J.M., Moreno Mochi M.P., Nunez J.M., Caceres M., Mochi S., Del Campo Moreno R., Jure M.A. Virulence factors and clinical patterns of multiple-clone hypermucoviscous KPC-2 producing K. pneumoniae. Heliyon. 2019;5:e01829. doi: 10.1016/j.heliyon.2019.e01829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLeo F.R., Nauseef W.M. Granulocytic Phagocytes. In: Bennett J.E., Dolin R., Blaser M.J., editors. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Volume 1. Elsevier; Philedelphia, PA, USA: 2014. pp. 78–92. [Google Scholar]
- 53.Rodgers M.A.J. Solvent-induced deactivation of singlet oxygen: Additivity relationships in nonaromatic solvents. J. Am. Chem. Soc. 1983;105:6201–6205. doi: 10.1021/ja00358a001. [DOI] [Google Scholar]
- 54.Svanberg K., Bendsoe N., Axelsson J., Andersson-Engels S., Svanberg S. Photodynamic therapy: Superficial and interstitial illumination. J. Biomed. Opt. 2010;15:041502. doi: 10.1117/1.3466579. [DOI] [PubMed] [Google Scholar]
- 55.Baran T.M., Foster T.H. Comparison of flat cleaved and cylindrical diffusing fibers as treatment sources for interstitial photodynamic therapy. Med. Phys. 2014;41:022701. doi: 10.1118/1.4862078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beyer W. Systems for light application and dosimetry in photodynamic therapy. J. Photochem. Photobiol. B Biol. 1996;36:153–156. doi: 10.1016/S1011-1344(96)07363-0. [DOI] [PubMed] [Google Scholar]
- 57.Wachter E., Heidary D.K., Howerton B.S., Parkin S., Glazer E.C. Light-activated ruthenium complexes photobind DNA and are cytotoxic in the photodynamic therapy window. Chem. Commun. 2012;48:9649–9651. doi: 10.1039/c2cc33359g. [DOI] [PubMed] [Google Scholar]
- 58.Edwards L., Turner D., Champion C., Khandelwal M., Zingler K., Stone C., Rajapaksha R.D., Yang J., Ranasinghe M.I., Kornienko A., et al. Photoactivated 2,3-distyrylindoles kill multi-drug resistant bacteria. Bioorganic Med. Chem. Lett. 2018;28:1879–1886. doi: 10.1016/j.bmcl.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li F., Collins J.G., Keene F.R. Ruthenium complexes as antimicrobial agents. Chem. Soc. Rev. 2015;44:2529–2542. doi: 10.1039/C4CS00343H. [DOI] [PubMed] [Google Scholar]
- 60.Hachey A.C., Havrylyuk D., Glazer E.C. Biological activities of polypyridyl-type ligands: Implications for bioinorganic chemistry and light-activated metal complexes. Curr. Opin. Chem. Biol. 2021;61:191–202. doi: 10.1016/j.cbpa.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wozniak A., Rapacka-Zdonczyk A., Mutters N.T., Grinholc M. Antimicrobials Are a Photodynamic Inactivation Adjuvant for the Eradication of Extensively Drug-Resistant Acinetobacter baumannii. Front. Microbiol. 2019;10:229. doi: 10.3389/fmicb.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pletzer D., Mansour S.C., Hancock R.E.W. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 2018;14:e1007084. doi: 10.1371/journal.ppat.1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kachynski A.V., Pliss A., Kuzmin A.N., Ohulchanskyy T.Y., Baev A., Qu J., Prasad P.N. Photodynamic therapy by in situ nonlinear photon conversion. Nat. Photonics. 2014;8:455–461. doi: 10.1038/nphoton.2014.90. [DOI] [Google Scholar]
- 64.Paez P.L., Bazan C.M., Bongiovanni M.E., Toneatto J., Albesa I., Becerra M.C., Arguello G.A. Oxidative stress and antimicrobial activity of chromium(III) and ruthenium(II) complexes on Staphylococcus aureus and Escherichia coli. BioMed Res. Int. 2013;2013:906912. doi: 10.1155/2013/906912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available because they are confidential data of patients protected by the informed consent protocol.