ABSTRACT
Structured data formats are gaining momentum in electronic health records and can be leveraged for decision support and research. Nevertheless, such structured data formats have not been explored for clinical coding, which is an essential process requiring significant manual workload in health organisations. This article explores the extent to which fully structured clinical data can support assignment of clinical codes to inpatient episodes, through a methodology that tackles high dimensionality issues, addresses the multi-label nature of coding and optimises model parameters. The methodology encompasses transformation of raw data to define a feature set, build a data matrix representation, and testing combinations of feature selection methods with machine learning models to predict code assignment. The methodology was tested with a real hospital dataset and showed varying predictive power across codes, while demonstrating the potential of leveraging structuring data to reduce workload and increase efficiency in clinical coding.
KEYWORDS: Clinical coding, analytics, data mining, decision support, health systems improvement
1. Introduction
Clinical coding has been conceived with the primary purpose of reporting health statistics in standardised formats, creating a basis for statistical analysis. Using multiple schemes such as the International Classification of Diseases (ICD) (France, 2003), coded data has later been used in many countries as the basis for funding providers through prospective payment systems, such as through diagnosis-related groups (DRGs) (Mateus, 2008). Considering its strong financial implications, ICD coding represents a key process for health organisations. In practice, ICD coding requires manual review of data in clinical records after patient discharge, assigning a set of diagnosis and procedure codes to each episode (Schraffenberger, 2010).
The coding process is typically carried out by certified professionals – who may be physicians (as it happens in Portugal), scribes or other technical staff – making use of dictations, manuals and code look-up tools. Due to its complexity, ICD coding is a resource-intensive and error-prone process. With growing pressures for cost reduction and with the increasing availability of health data in digital formats as a result of the widespread implementation of electronic health record (EHR) systems (AHIMA, 2013; Ford, Menachemi, & Phillips, 2006; Patel, Jamoom, Hsiao, Furukawa, & Buntin, 2013), there has been significant research devoted to develop coding support tools which combined with additional clinical and demographic attributes may be used to determine the patients’ DRG (Gartner, Kolisch, Neill, & Padman, 2015).
EHR systems have changed the paradigm of data collection in health settings and currently produce massive amounts of data (Davidson, Gao, & McCullough, 2015). These large data volumes and their increasing availability for research (Mortenson, Doherty, & Robinson, 2015) enable retrospective analyses (Faber, Konrad, Tang, & Trapp, 2016) and the construction of decision support systems (Capan, Wu, Campbell, Mascioli, & Jackson, 2017; Nadler & Downing, 2010). As such, research has focused on developing coding support methodologies using unstructured EHR data and applying natural language processing (NLP) methods (Stanfill, Williams, Fenton, Jenders, & Hersh, 2010). NLP methods have been used since free-text formats are not machine-readable and therefore not directly usable for decision support (McDonald & Tierney, 1988). Nevertheless, NLP methods cannot be used in many contexts, due to limitations in their generalisability when there is intrinsic variability of medical texts and due to a lack of NLP source tools for non-English languages (Meystre, Savova, Kipper-Schuler, & Hurdle, 2008). In face of these difficulties related to the reuse of unstructured clinical data, EHR systems have evolved towards the use of structured data formats (Hyppönen et al., 2014; Kalra, Fernando, Morrison, & Sheikh, 2013). These structured formats entail potential benefits in data uniformity, easy of reporting and advanced decision support (Bleeker, Derksen-Lubsen, van Ginneken, Van Der Lei, & Moll, 2006). In practice, structured data entry is performed by using controlled formats and terminologies (Fernando, Kalra, Morrison, Byrne, & Sheikh, 2012; Kalra et al., 2013), through pick lists and catalogues, dropdown fields and checkboxes to record clinical data – as opposed to narrative free-text typically used in clinical notes. The majority of studies addressing clinical coding support have been based on NLP applications using traditional free-text EHR data. Structured EHR data has been used in several studies focusing on patient phenotyping and subtyping (i.e., finding patients with certain health characteristics or patterns (Shivade et al., 2013), and in predicting specific diagnoses in ICD formats. While these studies generally indicate that structured data entail potential for predicting clinical codes, many of these studies focus on limited subsets of diagnoses and/or analyse predictions at more aggregate levels (e.g., only at general disease or 3-digit ICD level (Choi, Bahadori, Schuetz, Stewart, & Sun, 2016)). In this article, we sought to develop and test a methodology that would address a wide range of clinical conditions and codes, and to use detailed EHR data, as aligned with the type of information used by coding professionals to assign clinical codes to patient episodes.
This article develops a methodology to assess the extent to which coding can be supported by fully structured EHR data. This methodology is designed to handle dimensionality and multi-label issues, and addresses both data pre-processing tasks (including construction of feature sets), as well as the data mining stage using machine learning models. The applicability of the methodology is illustrated with real EHR data from a public hospital in Portugal. This article contributes to the literature by proposing (and applying to a real case study) a comprehensive coding support methodology using exclusively structured EHR data, as opposed to previous studies which invariably used free-text data. This article sheds light onto the extent to which structured data can assist coding, as a means to reduce manual workload and improve the usage of health care resources. Additionally, it provides useful information for researchers and software developers working in coding support technology on how to handle EHR data to build prediction models. It further raises issues for EHR system designers, implementers and users by identifying potential factors influencing performance of code prediction models.
This article is structured as follows: Section 2 reviews studies proposing methodologies for coding support. Section 3 describes the proposed methodology, and section 4 presents key results from its application in a case study using a real-world dataset. Section 5 discusses key findings, with section 6 presenting main conclusions and lines for future research.
2. Review of studies
Coding support studies are generally based on the active interpretation of clinical record data, proposing a set of codes to be validated by coding professionals (AHIMA, 2013). Extremely varied approaches and contexts of application are found in the literature (Stanfill et al., 2010). Previous studies focusing specifically in the clinical coding process have been largely based on free-text since this format is typically preferred by health professionals in recording clinical information (Stanfill et al., 2010). Amongst these, one study demonstrated the value of incorporating structured EHR data to improve code prediction (Scheurwegs, Luyckx, Luyten, Daelemans, & Van den Bulcke, 2015), indicating the potential value of these formats. Moreover, multiple research studies have leveraged structured EHR data to predict certain patient phenotypes and other characteristics expressed in terms of clinical codes. While not explicitly aiming to support clinical coding, these studies provide insight into the potential of using structured formats for diagnosis prediction.
The utilised coding schemes included several ICD versions and different levels of granularity. Sometimes the 3-digit category (Choi et al., 2016) or the ICD chapter level (Che, Purushotham, Cho, Sontag, & Liu, 2018) are used. Other studies exist that used SNOMED-CT (Cornet & de Keizer, 2008; Lussier, Shagina, & Friedman, 2001), UMLS (Friedman, Shagina, Lussier, & Hripcsak, 2004), ICF (Kukafka, Bales, Burkhardt, & Friedman, 2006) and procedure classification (ICD-10-PCS) (Subotin & Davis, 2014). The corpora of clinical records used in previous studies ranged from admission notes (Gundersen et al., 1996) to radiology or pathology reports (Aronson et al., 2007; Crammer, Dredze, Ganchev, Talukdar, & Carroll, 2007; Farkas & Szarvas, 2008; Goldstein, Arzumtsyan, & Uzuner, 2007; Matykiewicz, Duch, & Pestian, 2006; Oleynik, Patrao, & Finger, 2017; Rizzo, Montesi, Fabbri, & Marchesini, 2015; Suominen et al., 2008; Zhang, 2008), discharge summaries (Delamarre, Burgun, Seka, & Le Beux, 1995; Dinwoodie & Howell, 1973; Franz, Zaiss, Schulz, Hahn, & Klar, 2000; Friedman et al., 2004; Kevers & Medori, 2010; Kukafka et al., 2006; Larkey & Croft, 1995; Li, Chen, & Huang, 2011; Lussier, Shagina, & Friedman, 2000,0; Medori & Fairon, 2010), death certificates (Koopman, Zuccon, Nguyen, Bergheim, & Grayson, 2015,1) and entire medical records (Kavuluru, Rios, & Lu, 2015; Lita, Yu, Niculescu, & Bi, 2008; Morris et al., 2000; Pakhomov, Buntrock, & Chute, 2006; Ruch, Gobeill, Tbahriti, & Geissbühler, 2008), with variable structure and level of curation. Moreover, the majority of studies has been based on English texts, with the exception of particular studies in French (Kevers & Medori, 2010; Medori & Fairon, 2010; Pereira, Névéol, Massari, Joubert, & Darmoni, 2006; Ruch et al., 2008), Spanish (Pérez, Gojenola, Casillas, Oronoz, & de Ilarraza, 2015), Italian (Chiaravalloti, Guarasci, Lagani, Pasceri, & Trunfio, 2014; Rizzo et al., 2015) or German (Franz et al., 2000), while information extraction from Portuguese medical texts is still emmerging (LdS, 2011; Rijo, Silva, Pereira, Gonçalves, & Agostinho, 2014). The scope of clinical conditions comprised in each study also varied greatly, ranging from limited sets of respiratory (Farkas & Szarvas, 2008), cerebrovascular (Li et al., 2011) or coronarography exams (Delamarre et al., 1995) to heterogeneous episodes (Kevers & Medori, 2010). Such variable scope has also been reflected on the range of codes considered – one (principal diagnosis) (Avillach, Joubert, & Fieschi, 2008), five (Lita et al., 2008), six (Li et al., 2011), twenty (Yan, Fung, Dy, & Rosales, 2010) or fifty (Xu et al., 2007) codes, with only one study considering a significantly larger number (more than 1,400) of codes (Medori & Fairon, 2010).
Regarding methodological approaches to assist clinical coding, previous studies typically used NLP in data preparation and transformation stages to extract concepts and achieve a feature-vector representation (e.g. using a bag-of-words model), then applying machine learning models to predict code assignment. These models are frequently coupled with feature selection methods (such as chi-square) to reduce dimensionality by retaining only the most relevant features. Machine learning models used across the literature included support vector machines (SVM) (Aronson et al., 2007; Lita et al., 2008; Perotte et al., 2013; Xu et al., 2007; Yan et al., 2010; Zhang, 2008), Deep Learning (Shi, Xie, Hu, Zhang, & Xing, 2017; Xu et al., 2018; Yao, Mao, & Luo, 2018), naïve Bayes (Medori & Fairon, 2010; Pakhomov et al., 2006), decision trees (Farkas & Szarvas, 2008), (ridge) regression (Lita et al., 2008; Xu et al., 2007) and k-nearest neighbours (Aronson et al., 2007; Larkey & Croft, 1995; Ruch et al., 2008), exhibiting highly variable, yet encouraging, results. Recent studies in related areas of research have started exploring deep learning methods (Oleynik et al., 2017). Frequently used NLP tools include MedLEE (Friedman, Alderson, Austin, Cimino, & Johnson, 1994), MetaMap (Aronson & Lang, 2010), NegEx (Chapman, Bridewell, Hanbury, Cooper, & Buchanan, 2001) and UMLS dictionaries (Lindberg, Humphreys, & McCray, 1993), which have variable availability across languages. In studies leveraging structured EHR data, authors applied – in addition to the methods mentioned above – temporal modelling and deep learning methods such as recurrent and convolutional neural networks based on long short-term memory (LSTM) to predict diagnoses or generically clinical conditions (Choi et al., 2016; Lipton, Kale, Elkan, & Wetzel, 2016).
In spite of the myriad of coding support studies found in the literature, the use of structured EHR data formats for clinical coding support has not been explored in a systematic way, as previous studies often focused on smaller subsets of clinical conditions or predicted ICD codes at a more aggregate level, e.g., 3-code level. Notwithstanding, these studies show promise on the value of using structured data, which can be advantageous in contexts where NLP-based methods are difficult to apply due to language and text quality constraints. Considering the growing interest and availability of structured data formats in EHR systems, the key objective of this article is to investigate the extent to which structured data can be used for coding support. In addition, the use of structured formats in a data mining approach requires a series of data preparation and transformation steps, in line with general knowledge discovery frameworks (Corne, Dhaenens, & Jourdan, 2012). However, literature does not provide specific guidelines on the preparation and transformation of structured EHR data for coding support. As such, we incorporate these data preparation steps into our methodology as described in the next section. The proposed methodology is particularly relevant in contexts where the use of NLP tools is limited – notably when NLP tools are scarce for many languages other than English (this is the case for Portuguese) – and when there is increasing availability of structured EHR data. Moreover, the implementation of structured formats requires system users to adapt to the used of predefined data formats as opposed to using the often preferred free-text. Developing research that directly leverages and realises benefits from these structured formats can also motivate professionals to increase adoption and improve data collection patterns, which in turn can bring benefits care quality and safety, improve clinical documentation and further enable secondary uses of EHR data.
3. Methods
In order to explore whether structured EHR data can assist clinical coding, the proposed methodology follows a general knowledge discovery framework (Corne et al., 2012) which entails the two building blocks represented in Figure 1. The first block represents the EHR data transformation steps through which structured data are extracted, integrated and transformed into a data matrix, providing a format suitable for predictive modelling (Bishop, 2006). These steps construct a set of features (independent variables) and populate the data matrix with corresponding values. This first block is explained in detail in section 3.1. The second block – described in section 3.2– refers to the actual data mining framework, which entails filter feature selection methods to reduce dimensionality, machine learning models to predict code assignment, and the use of cross-validation to evaluate these models.
Figure 1.

Methodology framework: the upper blocks correspond to EHR data transformation steps, and lower blocks represent data mining stages
3.1. EHR data structure
Before delving into the stages of data transformation and data mining, we provide a brief overview of the specificities of the EHR system that inspired the development of this research. This research was conducted on the EHR system Soarian® (Haux et al., 2003), originally developed by SIEMENS and currently owned by Cerner Inc., with numerous implementations in healthcare providers around the world. Despite its own specificities, Soarian® shares the main data elements and formats with most EHR systems from other vendors and also with locally-developed systems. As such, the proposed methodology is generalisable and can be applied to other systems in which essential clinical data is represented in structured formats. Soarian® is a patient-centred system and stores the majority of data in structured formats, ensuring coherence and integration of the different data elements. Table 1 describes the key data elements in which data are conceptually structured, along with the catalogues used for pick list-based entries and the type of features derived from each of these elements. Catalogues consist in system-embedded lists for a specific domain (e.g., diagnoses, medication) from which users select one or multiple relevant entries, either through a search function or scrolling the catalogues. Further details on feature construction (including harmonisation of allergy data) are provided in section 3.2.
Table 1.
Main EHR data elements comprised in the EHR system Soarian®
| Data element | Description | Catalogue | Derived feature type |
|---|---|---|---|
| Demographics | Age and sex information | – | Numerical (age) and binary (sex) |
| Diagnoses | Diagnoses (including principal, co-morbidities and complications) assigned by clinicians, selected from system-embedded catalogues | 3 catalogue options: ICD-9-CM, ICD-10 and a local “working diagnoses” catalogue with preferred terms | Binary |
| Personal history | Personal history conditions selected from a small set configured in the EHR system; these conditions are selected through checkboxes and become assigned to a given patient, being transversal to all episodes from that patient | Local system catalogue | Binary |
| Allergies | Allergy conditions selected from a system-embedded catalogue or written by clinicians as narrative | Local system catalogue + free-text designations | Binary (after free-text harmonisation) |
| Prescriptions | Medical and nursing procedures, diagnostic and imaging exams, laboratory tests | Local system catalogue | Binary |
| Medications | Medications prescribed to the patient | Local system catalogue | Binary |
| Assessments | Clinical forms parametrised for different scopes, such as evaluation of respiration, feeding, fluid balance and elimination, scoring scales, clinical and nursing notes, admission and discharge forms; composed of structured (checkboxes, dropdowns, buttons and pick-lists) and free-text fields for additional information | – | Multiple (numerical, ordinal and categorical); Narrative fields are not considered |
Besides demographic data, EHR data elements can be grouped into two sets. The first group contains diagnoses, personal history, allergies and assessments, and is primarily meant to characterise the patient’s health status. The second group comprises information on medical services (prescriptions and medication) provided during each episode. In our context, an inpatient episode represents a separate contact with the hospital, i.e., a single interval between admission and discharge.
Diagnoses are selected from system-embedded catalogues and provide a visible representation of conditions characterising the patient’s health status, enabling statistical analyses. Nonetheless, diagnosis data produced across EHR systems can vary in structure and content due to the use of either standard vocabularies or local system-specific catalogues. Personal history consists of checkboxes indicating chronic or persistent clinical conditions. After these conditions are assigned to a patient in a given episode, they will remain associated with that patient and will be replicated in all subsequent episodes of the same patient. Allergies are selected similarly to diagnoses, but also allow manual input as short free-text. Lastly, EHR assessments consist of structured forms (with pick-lists, checkboxes, dropdown lists and radio buttons) to record information for a particular scope, such as for capturing respiration function, feeding, fluid balance and elimination, and for scoring scales (e.g., Glasgow), as well as of clinical, nursing, admission and discharge notes. Content-wise, these assessments may be considered equivalent to classic clinical narrative notes and are composed of labelled fields to record data. In certain contexts, the system also allows free-text fields to accommodate additional information needs. Free-text was not considered in our research (with the exception of short allergy designations, as described in section 3.2) due to the assumption that in the EHR system most relevant information is recorded and available in structured formats. The EHR system is, by design, highly focused on structured data entry, in line with industry trends (Kalra et al., 2013).
In terms of care services provided, prescriptions include diagnostic exams such as imaging scans, physiological measurements and laboratory tests, and medical and nursing procedures. Medication refers specifically to drug therapies prescribed to patients. Similarly to diagnoses, prescription and medication entries are made through pick-lists and specifically using locally-defined catalogues. Some EHR systems may use standard catalogues for these components (e.g., LOINC (Huff et al., 1998) for laboratory, RxNorm (Liu, Ma, Moore, Ganesan, & Nelson, 2005) for medication).
Making use of these structured data elements, the first methodological stage addresses the construction of feature sets to be used for developing prediction models, i.e., defining the attributes based on which the EHR dataset is described (Meisel & Mattfeld, 2010). In this context, features are regarded as attributes that characterise each instance (i.e., each episode) in the dataset, such as the allergies, personal history information, clinical observations or prescribed medications. The values of each of these features then allow models to make predictions for a given dependent variable (code assignment in this context). The next section describes the approach to construct the feature set based on the structured EHR data.
3.1.1. Modelling EHR data
The definition of the study objective is the primary step in any data mining framework and guides the subsequent stages of data source identification, data extraction and pre-processing (Olafsson, Li, & Wu, 2008). As such, the data preparation and transformation steps are tailored to support clinical coding. Since the coding process entails a broad review of the medical record, the proposed methodology makes use of all data elements presented in Table 1 (clinician-assigned diagnoses are analysed but are often modified or discarded as the relevance criteria differ between clinicians and coding professionals). This EHR data transformation stage includes both the definition of features and the appropriate pre-processing (transformation) tasks in order to map raw EHR data into the target feature set (Meisel & Mattfeld, 2010). Thereby, we achieved a data matrix format as a basis for subsequent data mining stages. It is important to note that the process of feature set construction requires a combination of domain knowledge (to represent clinical concepts in a meaningful way) and best practices from data analysis, given that literature does not provide guidance on how to effectively use structured EHR data for coding support.
The straightforward approach to construct a feature set from structured EHR data involves exhaustively defining binary features for all catalogue items and categorical/numerical features for all assessment fields. However, this approach is not adequate due to frequent redundancy within EHR data. Redundancy occurs when the same clinical information is recorded in different contexts, for example when catheter information is recorded in different assessments, or when a diagnosis of hyperlipidaemia is recorded in two episodes using different catalogues or different levels of granularity. To mitigate undesired data dispersion and bias, redundant features are collapsed under the same feature.
In addition, assessment fields produce different feature types (nominal, ordinal or numerical) which need to be properly defined and handled according to the underlying clinical concept. These aspects must be taken into account upon constructing a feature set, mapping EHR fields to features and populating a data matrix from raw EHR data. In this article, we address the construction of features from each EHR data element in two groups: catalogue-based data elements (diagnoses, prescriptions, medication, personal history and allergies) and assessment fields.
Firstly, for catalogue-based data elements, a binary feature was defined for each unique catalogue item, assigning value 1 if an entry was present in the episode, or assuming value 0 if the catalogue entry was absent. Subsequently, data were manipulated to mitigate redundancy. Due to the simultaneous use of multiple diagnosis catalogues (see Table 1), equivalence mappings (cross-walks) between catalogues were developed and validated by experts (in our case, ICD-10 and “working diagnoses” catalogues were mapped to the ICD-9-CM catalogue, a similar procedure that has been used by (Gartner et al., 2015) and (Gartner, 2015)). Personal history features were defined directly from system labels provided that these did not exhibit redundancy. For allergy data, free-text labels were harmonised by modifying terms to ensure that all allergies are expressed in terms of allergen (e.g., cat, egg albumin) or active ingredient in the case of drug allergy. For prescriptions, catalogue entries were simplified by taking only the main designation of each diagnostic exam (e.g., removing information on number/axes of X-ray shots), laboratory test or medical/nursing procedure. Lastly, medication entries were simplified by taking only information on active ingredient (removing dosage and administration mode) and decomposing entries with mixtures of active ingredients (e.g., when a solution of potassium and sodium chloride was prescribed, we decomposed this prescription into two entries – one of sodium chloride and a second for potassium chloride).
All the data elements described above – diagnoses, personal history, allergies, prescriptions and medication – were entered through catalogues or pick-lists. The feature construction process generated binary features whose value was based on presence/absence of concepts in each episode and, therefore, did not produce any missing data. This approach is analogous to the presence or absence of concepts/terms in NLP-based methodologies.
For assessment-based data, we performed an exhaustive listing of all field labels from the EHR system, listed the clinical concepts conveyed by these fields and mapped redundant fields to the same concept. While numerical concepts were directly transformed into features (e.g., blood pressure) and assumed the corresponding field value, binary concepts were inferred both from field values (e.g., presence of catheter: yes/no) and from associated fields (e.g., entry with date of catheter insertion indicated the presence of a catheter). Additional specificities of defining features from assessments included:
Creation of dummy variables for all categorical features;
Handling multiple values of the same feature (due to multiple measurements of the same parameter during each episode), by:
(a) Defining value 1 (of dummy variables) for all categories of the same feature occurring in each episode;
(b) Splitting numerical features into two features for maximum and minimum values occurring in each episode;
(2) Handling data missingness according to the feature type and corresponding data entry mechanism:
• For categorical checkbox-based features, absence of record was assumed to represent feature value 0 for each possible feature categories;
• For categorical dropdown/button-based, absence of records was assumed to represent missing data, since these data entry mechanisms imply mandatory data entry by users;
• For numerical features, absence of records was assumed to also represent missing data.
All the methodological stages and decisions outlined above aimed to minimise information loss when consolidating EHR data into a data matrix format. Figure 2 depicts the process of mapping raw EHR database entries to a data matrix. We streamlined this process by designing a mechanism that automatically created and populated a data matrix from raw EHR data. For catalogue-based data, this was performed by listing all unique catalogue entries and removing redundancy. For assessment-based data, we created a feature specification (master) file which defined features, mapped EHR fields (i.e., their system labels) to be inspected in order to determine feature values, and defined the corresponding type, admissible values and missingness pattern. The construction of a data matrix was performed by a custom-made algorithm that read the list of unique catalogue entries and the master file, analysed raw EHR data by looking up mapped EHR fields and populated values in the data matrix. In such configuration, the process of building a data matrix can be automated for new datasets with the same EHR structure. If there are changes in catalogue-based items or in allergies, these would only require analysis of redundancy and free-text harmonisation after listing unique entries. On the other hand, changes in assessment-derived features (e.g., when field labels are changed in the EHR system) would only require these to be added/modified/deleted in the feature specification (master) file, followed by execution of the custom-made algorithm to create a new data matrix.
Figure 2.

EHR data transformation and matrix construction processes to create and populate a data matrix from raw EHR database entries
3.2. Data mining approach for clinical coding
3.2.1. Modelling framework
After performing the data preprocessing steps, our data mining approach was applied to extract patterns from data using inductive learning algorithms (Olafsson et al., 2008). In our context, data mining aimed to model relationships between clinical (EHR) data from each episode and the set of clinical codes assigned to that episode. These relationships may then guide clinical code assignment for future episodes, both for the same and future patients, as the coding patterns are learned. As such, each data point in the dataset contains episode EHR data and the corresponding codes.
In this study, we adopted a supervised learning approach for coding support. Supervised learning can be used to model patterns between features (the independent variables) and labels (dependent variables) in a training set, making use of knowledge from all previous episodes of the overall hospital population. Model predictive power was then evaluated on a test set (Bishop, 2006; Corne et al., 2012). Unsupervised learning, which represents a different machine learning paradigm, would have been unable to perform such pattern modelling. Within supervised learning, classification models were suitable for this study given that we aimed to predict categorical dependent variables (the assignment of each code) and not a numerical variable (in which case regression models would be appropriate). Using the constructed data matrix of inpatient episodes, feature selection was required to reduce dimensionality. Subsequently, it was necessary to choose which machine learning models to apply and optimise corresponding model parameters. Additionally, code prediction required handling the existence of multiple labels for each instance (episode).
Multiple supervised classification models are currently established in the literature (Corne et al., 2012), highly differing in terms of type of features handled, mechanism for modelling feature-label relationships, model training algorithms and interpretability of results (Bishop, 2006). In order to choose which models to use for coding support, one should consider the need to accommodate both numerical and categorical features, be scalable for datasets with high number of features and instances, and preferentially yield interpretable outputs. Since there is no axiomatic guideline as to which model should be applied in each problem, we tested and compared five machine learning methods to predict the assignment of ICD codes: decision trees, naïve Bayes classifiers, logistic regression models, SVM and Deep Neural Networks (DNN, a form of deep learning). These approaches have been widely used, particularly in other ICD coding support studies, and show potential applicability in our context. We adopted a data-driven approach whereby models were selected based on predictive power observed throughout a set of experiments with a case study dataset. We excluded other methods such as -nearest neighbours as these are typically computationally-intensive with a high number of binary features (resulting in artificial distance measures), and neural networks due to the complex process of topology and parameter optimisation that would compromise applicability and scalability.
Since each episode may be assigned one or more codes, clinical coding represents a multi-label classification problem. To tackle this matter, we used a binary relevance method whereby the problem was decomposed into single-label problems (Tsoumakas, Katakis, & Vlahavas, 2009), creating a binary classifier for each code which predicted if a code should be assigned (or not) to each episode.
Moreover, structured EHR data produces a high number of features (i.e., high dimensionality) which are computationally-intensive and prone to overfitting issues. To reduce dimensionality, feature selection methods were applied prior to developing prediction models. We detail below the feature selection methods and supervised learning models tested in this study.
3.2.2. Feature selection
Feature selection methods represent mechanisms to determine a subset of relevant features based on a specific metric (Guyon and Elisseeff 2003). In this article, we adopted filter methods, which analyse dataset characteristics independently from classifiers and are more scalable (Saeys, Inza, & Larran˜aga, 2007). Within the family of filter methods (Lazar et al., 2012), we tested several feature selection methods based on different metrics. To select which methods to test, we performed a literature review using combinations of “feature selection”, “electronic health record” and “filter” search terms. We then selected seven methods that are scalable and able to handle the required feature types: fast correlation-based filter (FCBF) (Yu & Liu, 2004), information gain (IG) and chi-square (Yang & Pedersen, 1997), Relief (Kira & Rendell, 1992), symmetrical uncertainty (SU) (Press et al., 1992), correlation-based feature selection (CFS) (Hall & Holmes, 2003) and minimal-redundancy maximal-relevance (mRMR) (Peng, Long, & Ding, 2005). Some of these methods required setting user-defined parameters upon implementation, see section 4.2.
3.2.3. Overview of selected classification models
The models selected to predict code assignment – decision trees, naïve Bayes classifiers, logistic regression models, SVM and Deep Neural Network models – highly differ in the approach to model patterns in data, using measures based on entropy, likelihood or distances. Similarly to feature selection methods, each prediction model entails specific parameters which are also addressed in section 4.2.
3.2.3.1. Decision trees
Decision trees are suitable for datasets with categorical features and have the key advantage of producing interpretable results (Dreiseitl & Ohno-Machado, 2002). These models recursively partition the dataset based on splitting criteria and are represented in a tree structure (Mitchell et al., 1997). Each instance is classified by evaluating feature values in the specified order and assigning the label of the resulting leaf node. Model building was performed by determining the splitting criterion at each node using the Gini index (Rokach & Maimon, 2005). To mitigate overfitting, two techniques were employed: pre-pruning to avoid excessive tree growth (by imposing a minimum number of instances in leaf nodes), and post-pruning by discarding branches of the final model that resulted in lower performance. In this study, we used the CART (classification and regression trees) variant of decision trees (Breiman et al., 1984).
Classification models can also be built by estimating a posteriori probabilities of an instance belonging to class Ck of k possible classes given its feature values (Mitchell et al., 1997). These probabilities may be estimated using either generative or discriminative approaches. The naïve Bayes classifier is a generative method wherein priors and likelihood values are firstly estimated in order to compute for each class using Bayes rule:
| (1) |
3.2.3.2. Naïve Bayes
In the naïve Bayes model, prior probabilities may be obtained empirically from the training set. While the class-conditional probability estimation is simplified with the assumption of conditional independence, models may still perform well in contexts where this assumption does not hold (Hand & Yu, 2001). In fact, class-conditional joint probabilities can be modelled as the product of the class-conditional probabilities for each feature . In practical terms, the classification decision involved assigning an instance to the positive class if the output was higher than a user-defined threshold, whose manipulation helped compensating for class imbalance (i.e., much lower number of instances of the negative class).
3.2.3.3. Logistic regression
Conversely, logistic regression represents a discriminative approach which models a posteriori probabilities directly from training data to build binary classifiers. The positive class probability () was modelled for a given instance (with N features) using a logistic link function, as represented in eq. (2). Training logistic regression models was performed by estimating the parameters w0 and wj (logistic regression coefficients) which best fitted the training dataset using maximum likelihood estimation (Hosmer and Lemeshow 2000). Fitted models were used to predict class assignment for new instances similarly to the mechanism of naïve Bayes, assigning the positive class if the model output was higher than a specified threshold.
| (2) |
3.2.3.4. Support vector machines
The fourth machine learning model, SVM, defines a hyperplane that separates data points of different classes by maximising the margin of the nearest training instances of different classes. This provides a decision boundary to classify new instances (Cortes & Vapnik, 1995). Since training sets may not be linearly separable, the feature space can be mapped to another space with different dimension, by applying a kernel function . We performed preliminary analyses to compare different kernels and observed that linear kernels yielded consistently better results. In model training, SVM classifiers (represented as vectors w) were obtained by determining the solution to a quadratic optimisation problem for each instance i in a dataset with N instances (Olafsson et al., 2008), as formulated in eq. (3) and subject to the constraints in eq. (4). Parameters , and represent, respectively, the incorrectly classified instances, the bias and the penalty (cost) applied to these misclassifications (reflected in the number of misclassified instances). represents the classifier function (or class assigned) for each instance . Upon training SVM models with linear kernels, it was necessary to manipulate parameter , as described in section 4.2.
| (3) |
| (4) |
3.2.3.5. Deep learning
The fifth method, Deep Neural Networks (DNN), has achieved great success in many application domains including computer vision, natural language processing, and speech recognition (Bhandare, Bhide, Gokhale, & Chandavarkar, 2016). DNN architectures mainly consist of input, multiple hidden and output layers. According to the types of layers and the corresponding learning methods, there are many variants of DNNs, among which typical examples are multi-layer perceptrons (MLP), deep belief networks (DBN) and stacked auto-encoders (SAE), as well as convolutional neural networks (CNN) and recurrent neural networks (RNN). These models are the widely used in biomedical analysis with a certain representative property of model structure and training process. In our classification problem, deep learning was applied using a multi-layer deep neural network MLP and the DL4 J library (Deeplearning4j (2017)) in combination with the WEKA Java API (Witten and Frank (2011)). These models entail multiple hyperparameters, of which the learning rate is the most important.
3.2.4. Parameter selection and optimisation
The selected machine learning models contain one or more specific hyperparameters whose values needed to be manipulated in order to find the combination of values yielding the best results. For this purpose, we made use of a grid search procedure which is suitable for problems with a low number of parameters (Bergstra & Bengio, 2012). Grid search defines a range and interval of variation for each parameter and then systematically testing performance of parameter combinations, selecting the parameter combination which yielded the highest F1-score (see this metric in section 3.2.5). For models where only one hyperparameter was tuned – e.g., the learning rate in deep learning – we used a simple parameter search procedure by testing a given subset of hyperparameter values and observing resulting performance The implementation of the parameter search procedure is described in further detail in section 4.2.
3.2.5. Model evaluation metrics
The selected prediction models produced a set of binary outputs (as many as the number of codes considered) for each episode, indicating the codes to be assigned to that episode. Since the number of assigned codes is unknown for each episode, we predicted code assignment for all codes in the dataset by developing a binary classifier for each code. Model performance was evaluated by comparing model outputs with the known assigned codes (gold standard) in test data. We then counted true positives (TP), false positives (FP) and false negatives (FN). In this case-study, the gold standard consists of the codes assigned to each episode by coding professionals (without any coding assistance), which allowed evaluating models against real-world practice. Human-assigned codes in the gold standard were subject to validation mechanisms (based on inter-code restrictions and admissible primary diagnoses) embedded in the national database of hospital episodes.
Model performance was assessed on each test set using three key metrics found in other coding support studies and based on TP, FP and FN counts. These metrics consist of precision , recall and F1-score , computed for each clinical code . These performance measures were aggregated using macro-averaging (averaging measures obtained for each code) (Tsoumakas et al., 2009).
To produce training and test sets for each experiment, we used 5-fold cross validation whereby the dataset was randomly partitioned into 5 non-overlapping subsets, using 4 of subsets as training sets (to fit prediction models) and then testing models on the remaining (test) set (Kohavi, 1995). For each model, this procedure was performed 5 times, using one of the 5 subsets as test set at a time, and ensuring that each instance was used as test instance exactly once.
3.2.6. Experimental design
The coding scheme used in this study was the 9th Revision, Clinical Modification of ICD (ICD-9-CM) (Bowie & Schaffer, 2012). During the time this study was developed, ICD-9-CM was the national coding standard in the Portuguese National Health Service, firstly to characterise mortality and morbidity statistics, and later as a basis for hospital episode classification. Coding professionals (physicians) were, therefore, required to code all inpatient episodes using ICD-9-CM, as determined by the Ministry of Health. Specific issues related with the use of ICD-9-CM are discussed later in section 5.3.
The first experiment aimed to analyse average and code-by-code performance for the 50 most frequent diagnosis codes (which accounted for approximately 50% of the total code effort), using combinations of the 7 feature selection and the five classification methods, looking into the patterns of variation and comparative model performance across codes. We also computed models using the full feature set so as to analyse the influence of feature selection. These results are presented in section 4.3. For the best performing feature selection method, we further analysed selected feature subsets for high and low performing codes in order to investigate factors influencing performance. These results are presented in section 4.3.
Additionally, we carried out a second experiment encompassing 90% of total code occurrences, aiming to analyse the influence of class imbalance (i.e., the fact that many codes occur in very few episodes, resulting in a much higher proportion of negative examples for each code) and the applicability of the proposed methodology using the same metrics, thereby allowing comparison with previous results. These results are presented in section 4.4.
4. Results
4.1. Dataset
The dataset used in this study contained 5,089 anonymised medical records pertaining to 4,210 different patients (3,595 patients had a single episode) admitted in Internal Medicine, Pneumology, Nephrology, Infectiology and Gastroenterology departments during the first semester of 2013 (note that this particular EHR system had started going live in early 2012, and in 2013 had achieved a considerable maturity of routine use). The dataset was composed exclusively of inpatient episodes. The mean and median patient ages were 67.7 and 72 years, respectively, with 50.5% female and 49.5% male patients. No information about race or ethnicity was collected in the EHR.
Using 5-fold cross-validation, training and test sets are composed of 4,072 and 1,017 instances, respectively. After performing the data pre-processing tasks described in Figure 2, the resulting feature set contained a total of 5,023 features, of which 3,714 were catalogue-based and 1,309 were assessment-based. 203 features exhibited missing values – these refer to assessment-based features (frequently numerical and non-mandatory) that were not filled in for all patients (e.g., patient weight, volumes of drained liquids or level of muscle strength).
Conversely, catalogue-based features did not produce missing values since the absence of a record was defined as feature value 0. Due to the low representativeness of missing features in the dataset (4% of all features), these features were removed, thus resulting in 4820 features. Coding data associated with these episodes contained 39,273 code occurrences in total, corresponding to 2,272 different ICD-9-CM diagnosis codes. The observed occurrence of ICD codes was highly imbalanced (the 50 most frequent codes account for approximately 50% of total code occurrences), as evidenced by the relative frequencies in Figure 3 (in effect, 860 of these 2,272 ICD codes occurred only once in the dataset). The ICD codes were also divided across different groups of clinical conditions, as shown in Table 2.
Figure 3.

Relative frequencies of the 50 most frequent ICD-9-CM diagnosis codes. ICD-9-CM codes described in Table A1 in the appendix. Legend: NOS – Not otherwise specified; NEC – not elsewhere classified
Table 2.
Proportion of code occurrences of Top-50 ICD-9-CM codes, grouped by ICD group (chapter)
| Chapter | Description | % in Top 50 |
|---|---|---|
| 001–139 | Infectious and Parasitic Diseases | 1.16% |
| 240-279 | Endocrine, Nutritional and Metabolic Diseases, And Immunity Disorders | 23.33% |
| 280-289 | Diseases of the Blood and Blood-Forming Organs | 4.30% |
| 290-319 | Mental Disorders | 5.59% |
| 390-459 | Diseases of the Circulatory System | 28.74% |
| 460-519 | Diseases of the Respiratory System | 12.47% |
| 580-629 | Diseases of the Genitourinary System | 7.91% |
| V01-V91 | Supplementary Classification of Factors Influencing Health Status and Contact with Health Services | 15.23% |
| E000-E999 | Supplementary Classification of External Causes of Injury and Poisoning | 1.27% |
4.2. Feature selection and model implementation
This section specifies in further detail the settings and parameters associated with the implementation of the proposed methodology. Firstly, the data pre-processing tasks were carried out using the source files extracted from the EHR database. Since assessment data (the last element in Table 1) contained more than 22 million entries, it required the use of a database management system. We used MySQL to manage raw data and implemented stored procedures to automatically split source data and create a single comma-separated file for each episode. Using the feature specification master file (as mentioned in Figure 2), we implemented a custom-made algorithm in Matlab® which received as input arguments the source data files and the master file, and automatically built and populated the data matrix. The resulting matrix was then coupled with the other matrices derived from demographic, diagnoses, personal history, allergies, prescriptions and medication data. Integration of these matrices was performed using episode identifiers (typically available in a patient-centred system).
For the implementation of feature selection and classification methods, we used available functions or implemented methods as needed, using Matlab® R2013a as well as the DL4 J library (Deeplearning4j (2017)) in combination with the WEKA Java API (Witten and Frank (2011)). Filter feature selection was performed for each code with the parameters described in Table 3 (when no toolbox or source function is specified, the methods were implemented by the authors in Matlab®). Using each filter method, we obtained the 50 most relevant features, ranked by decreasing order of importance according to the relevance criterion underlying each method. The decision of selecting the 50 most relevant features was informed by preliminary analyses in which we did not observe any performance improvement by adding more features. Reducing the feature set helped significantly in keeping the runtimes of computational simulations manageable. These feature subsets were then used to develop prediction models in a stepwise forward selection process, starting by building models using only the most relevant feature and then adding one feature to the model at a time, in decreasing order of relevance.
Table 3.
Implementation parameters for feature selection methods
| Feature selection method | Implementation |
|---|---|
| Fast Correlation Based Filter (FCBF) | • Decreasing order of relevance |
| • SU threshold | |
| • FEAST Toolbox for Matlab® (Brown, Pocock, Zhao, & Luján, 2012) | |
| Information Gain (IG) | • Decreasing order of relevance |
| • Forward selection – first order utility (Brown, 2009) | |
| Relief | • Decreasing feature weight |
| • 10 nearest neighbours | |
| • Matlab® function | |
| Chi-square | • Decreasing order of (chi-square) value |
| Symmetrical uncertainty (SU) | • Decreasing order of SU value |
| Correlation-based Feature Selection (CFS) | • Decreasing order of heuristic merit for each feature subset with features ( and represent the average feature-class and feature-feature correlations, respectively) (Hall & Holmes, 2003): |
| • Correlation based on symmetrical uncertainty (SU) | |
| Minimal Redundancy Maximal Relevance (mRMR) | • Decreasing order of relevance |
| • FEAST Toolbox for Matlab® (Brown et al., 2012) |
Supervised classification models were developed and tested using Matlab® using its Statistics toolbox. In order to select the combination of model-specific parameters yielding best performance, the authors implemented scripts to automatically execute the grid search procedure described in section 3.3.4, using the values and settings shown in Table 4. These values were replicated for each feature subset in the stepwise forward selection approach.
Table 4.
Implementation parameters for the classification methods
| Method | Implementation |
|---|---|
| Decision trees | • Splitting criterion: Gini index |
| • Pre-pruning: minimum of 1, 3 and 5 instances in lead nodes | |
| • Post-pruning: test all admissible prune levels between minimum and maximum values for each tree | |
| Naïve Bayes | • Feature distributions: multivariate multinomial (discrete), kernel estimation (continuous) |
| • Classification threshold: from 0 to 1 in steps of 0.005 | |
| Logistic regression | • Classification threshold: from 0 to 1 in steps of 0.005 |
| Support Vector Machines | • Linear kernels |
| • Penalty parameter (C) to (unitary exponent increments) | |
| Deep Neural Network | • Stochastic Gradient Descent |
| • Number of epochs: 10 | |
| • Softmax activation function | |
| • Learning rate to (unitary exponent increments) |
For each code prediction model developed (i.e., each combination of feature set and model hyperparameter values), we computed precision, recall and F1-score values to measure predictive performance using 5-fold cross validation. The combination of feature set and hyperparameter values yielding the highest F1-score (computed through cross-validation) was selected to report performance results and conduct subsequent analyses presented in sections 4.3, 4.4 and 4.5.
4.3. Average and code-by-code performance
We start by reporting the average performance of the classification methods combined with each feature selection method. This allows us to demonstrate the usefulness of feature selection in combination with classification. Table 5 presents the results obtained for all combinations of feature selection and classification methods, as well as with the full 4,820 feature set (without feature selection). This table shows that logistic regression models achieved the best average results in terms of F1-scores. Decision trees exhibited higher precision (i.e., lower rate of false positives), while SVM models showed higher recall (i.e., lower rate of false negatives). In practice, these results mean that decision trees would be less likely to incorrectly suggest codes, while SVM would be less likely to miss/overlook codes that should be assigned. Note that recall is lowest for the Deep Learning results which can, however be boosted using a filtered classifier. This takes into account the class imbalance in the dataset. The results using this method are shown in Table C1 and demonstrate that deep neural network classification reaches a precision of 0.77. Amongst the tested feature selection methods, mRMR, CFS and FCBF showed consistently better results while for the deep learning results, there was no significant difference in precision and F1-scores with or without attribute selection.
Table 5.
Average precision, recall and F1-score obtained for 50 most frequent ICD codes with combinations of feature selection methods (FSM) and classification methods (CM). Underlined numbers represent the maximum value in each measure across all models. Numbers in boldface represent the highest values within each machine learning model
| Macro |
Micro |
||||||
|---|---|---|---|---|---|---|---|
| CM | FSM | Precision | Recall | F1-score | Precision | Recall | F1-score |
| DT | None | 0.512 | 0.417 | 0.453 | 0.494 | 0.428 | 0.454 |
| FCBF | 0.680 | 0.411 | 0.479 | 0.644 | 0.400 | 0.470 | |
| IG | 0.556 | 0.411 | 0.464 | 0.532 | 0.419 | 0.462 | |
| Relief | 0.574 | 0.424 | 0.476 | 0.539 | 0.425 | 0.467 | |
| Chi-square | 0.680 | 0.411 | 0.479 | 0.589 | 0.440 | 0.494 | |
| SU | 0.632 | 0.443 | 0.506 | 0.603 | 0.437 | 0.495 | |
| CFS | 0.686 | 0.414 | 0.483 | 0.643 | 0.422 | 0.486 | |
| |
mRMR |
0.642 |
0.456 |
0.518 |
0.604 |
0.452 |
0.506 |
| NB | None | 0.201 | 0.400 | 0.254 | 0.238 | 0.416 | 0.293 |
| FCBF | 0.568 | 0.611 | 0.572 | 0.533 | 0.590 | 0.543 | |
| IG | 0.421 | 0.547 | 0.451 | 0.423 | 0.547 | 0.459 | |
| Relief | 0.490 | 0.577 | 0.495 | 0.473 | 0.563 | 0.487 | |
| Chi-square | 0.548 | 0.588 | 0.536 | 0.514 | 0.576 | 0.517 | |
| SU | 0.546 | 0.587 | 0.540 | 0.519 | 0.567 | 0.523 | |
| CFS | 0.573 | 0.603 | 0.572 | 0.540 | 0.579 | 0.546 | |
| |
mRMR |
0.554 |
0.608 |
0.563 |
0.526 |
0.581 |
0.539 |
| LR | None | 0.121 | 0.504 | 0.189 | 0.163 | 0.526 | 0.241 |
| FCBF | 0.578 | 0.618 | 0.577 | 0.542 | 0.600 | 0.551 | |
| IG | 0.543 | 0.561 | 0.533 | 0.423 | 0.552 | 0.523 | |
| Relief | 0.550 | 0.568 | 0.535 | 0.530 | 0.550 | 0.521 | |
| Chi-square | 0.569 | 0.601 | 0.569 | 0.540 | 0.579 | 0.546 | |
| SU | 0.567 | 0.604 | 0.569 | 0.541 | 0.580 | 0.547 | |
| CFS | 0.578 | 0.617 | 0.580 | 0.554 | 0.585 | 0.556 | |
| |
mRMR |
0.588 |
0.612 |
0.585 |
0.558 |
0.586 |
0.559 |
| SVM | None | 0.294 | 0.502 | 0.363 | 0.319 | 0.487 | 0.378 |
| FCBF | 0.499 | 0.659 | 0.520 | 0.480 | 0.632 | 0.511 | |
| IG | 0.543 | 0.522 | 0.138 | 0.524 | 0.472 | 0.187 | |
| Relief | 0.441 | 0.674 | 0.467 | 0.446 | 0.620 | 0.473 | |
| Chi-square | 0.506 | 0.628 | 0.510 | 0.486 | 0.604 | 0.503 | |
| SU | 0.502 | 0.629 | 0.514 | 0.487 | 0.603 | 0.507 | |
| CFS | 0.484 | 0.686 | 0.527 | 0.480 | 0.640 | 0.517 | |
| |
mRMR |
0.479 |
0.684 |
0.516 |
0.475 |
0.642 |
0.512 |
| DL | None | 0.602 | 0.408 | 0.472 | 0.581 | 0.417 | 0.474 |
| FCBF | 0.658 | 0.366 | 0.442 | 0.630 | 0.370 | 0.445 | |
| IG | 0.629 | 0.389 | 0.462 | 0.608 | 0.403 | 0.469 | |
| Relief | 0.598 | 0.341 | 0.411 | 0.577 | 0.346 | 0.412 | |
| Chi-square | 0.622 | 0.396 | 0.467 | 0.603 | 0.408 | 0.473 | |
| SU | 0.626 | 0.400 | 0.471 | 0.607 | 0.411 | 0.477 | |
| CFS | 0.637 | 0.372 | 0.444 | 0.616 | 0.385 | 0.454 | |
| mRMR | 0.640 | 0.369 | 0.442 | 0.618 | 0.384 | 0.453 | |
Figure 4 depicts the performance of the five models broken down by the 50 most frequently occurring ICD codes. This chart shows that model performance has a wide range of variation across codes and does not seem to depend directly on the relative frequency of codes (since it does not decrease steadily as the frequency decreases). For example, code 595.0 (acute cystitis) shows better results than the most frequent code (401.9 – hypertension, not otherwise specified), despite having lower relative frequency. Secondly, Figure 4 also shows that tested models have a similar pattern of variation across codes, as F1-scores do not vary significantly for each code. This finding is interesting considering the fact that the tested prediction models use very different approaches for modelling patterns and predicting code assignment.
Figure 4.
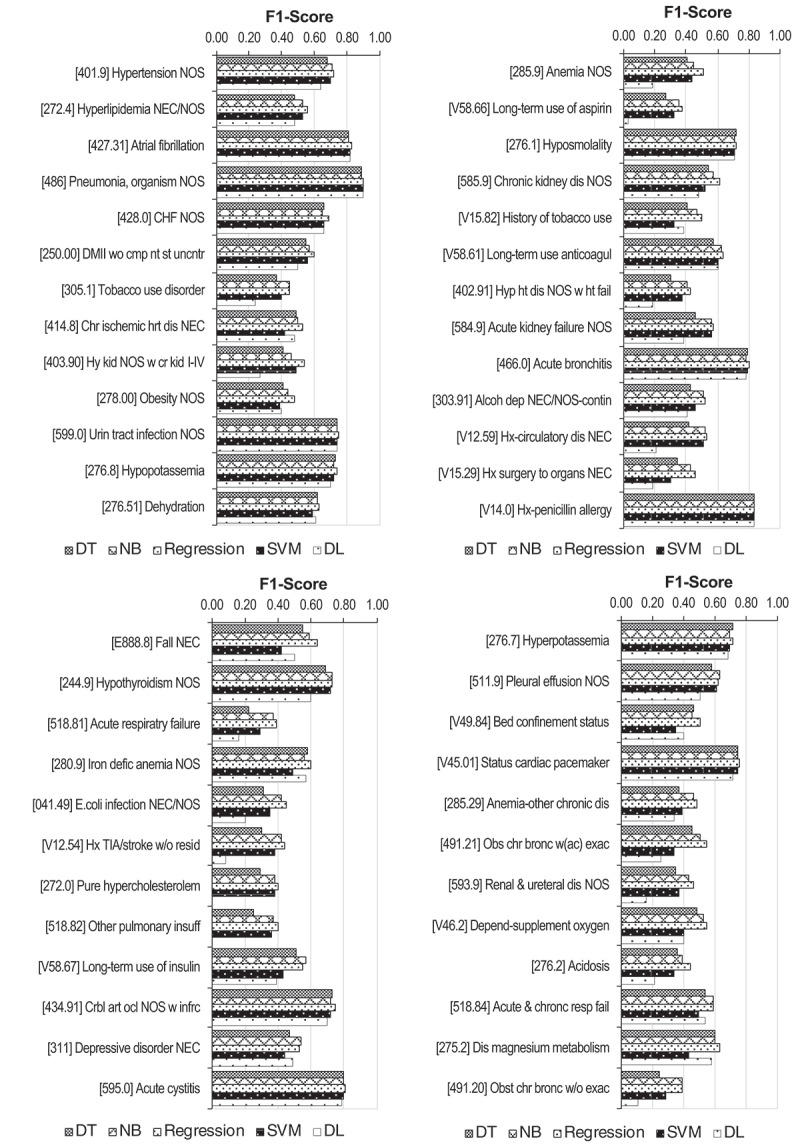
F1-scores obtained with decision trees, naïve Bayes, logistic regression, SVM and deep learning methods, using mRMR feature selection for the 50 most frequent ICD codes (ordered by relative frequency). ICD-9-CM codes described in Table A1 in the appendix
4.3.1. Analysis of selected features
Table 6 presents the six most relevant features selected with the mRMR method, for codes with high performance (466.0 – acute bronchitis; and 486 – pneumonia, organism unspecified) (codes description provided in the Appendix), as well as with low performance (402.91 – hypertensive heart disease, not otherwise specified, with heart failure; and 518.82 – other pulmonary insufficiency, not elsewhere classified). This table shows that selected features are mostly related to diagnoses, medication and prescriptions, showing clinically meaningful correlations with the corresponding code. For each code, this clinical meaningfulness is observed by having features with clinical conditions, medication or tests related to the code being predicted. Having such meaningful correlations appears to be associated with higher model performance, as observed in codes 466.0 and 486. Conversely, unexpected features (such as the malignant neoplasm of ureter used to predict pulmonary insufficiency – 518.82) which are not clinically meaningful (at least directly) are also observed. Furthermore, similar diagnosis-related features with different levels of granularity are seen, showing that information detail is not uniformly recorded within the EHR system. This is the case of hypertensive heart disease (402.91), for which different (yet related) features appear in the feature set, albeit using different modifiers of heart disease or just stating the condition as unspecified.
Table 6.
Six most relevant features selected by the mRMR filter (all exhibited features are binary). Legend: ht – heart; dx – assigned diagnosis; med – prescribed medication; w/- with; w/o – without; unspec – unspecified
| 466.0 (Acute bronchitis) | 486 (Pneumonia, organism unspec) | 402.91 (Hypertensive ht disease NOS with ht failure) | 518.82 (Other pulmonary insufficiency NEC) |
| Acute bronchitis (dx) | Pneumonia, organism NOS (dx) | Hypertensive ht disease NOS w/ht failure (dx) | Acute respiratory failure (dx) |
| Amoxycillin (med) | Clarithromycin (med) | Furosemide (med) | Acute and chronic respiratory failure (dx) |
| Acute laryngotracheitis w/o obstruction (dx) | Other bacterial pneumonia (dx) | Acute lung oedema, unspec (dx) | Ceftriaxone (med) |
| Use of non-invasive mechanical ventilation | Bacterial pneumonia, unspec (dx) | Malignant hypertensive ht disease w/ht failure (dx) | Benign hypertensive ht disease w/ht failure (dx) |
| Ipatropium (med) | Compromised breathing | Benign hypertensive ht disease w/ht failure (dx) | Hypertensive ht disease NOS with ht failure (dx) |
| Acute upper respiratory infection site unspec (dx) | Hemoculture (aerobiosis) | Unspec hypertensive ht disease w/ht failure (dx) | Malignant neoplasm of ureter (dx) |
4.4. Impact of class imbalance on performance
The influence of class imbalance on model performance was also analysed by encompassing a wider range of ICD codes. For this purpose, prediction models for the 544 most frequent codes were developed, covering 90% of code occurrences (as described in Table 7). We tested the prediction models and parameter selection technique earlier described, using all non-redundant features selected with FCBF (due to its faster execution times and elimination of redundant features). The results in Figure 5 show that the average performance decreases more abruptly when more than 60% of occurrences are covered. Comparing to the results in Figure 4, one can argue that this performance decrease was caused by much lower performance for highly imbalanced codes. While more frequent codes did not provide evidence of decreasing performance with relative frequency, performance effectively deteriorates for extremely imbalanced codes.
Table 7.
Occurrences of positive instances of ICD-9-CM codes in relation to the dataset coverage. This table shows the number of top ICD codes (ordered by decreasing frequency) corresponding to various proportions of total code occurrences. # positive examples represents the number of occurrences
| Cumulative occurrences | 5% | 10% | 20% | 30% | 40% | 50% | 60% | 70% | 80% | 90% | 100% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| # Top codes (K) | 1 | 4 | 9 | 19 | 35 | 59 | 98 | 161 | 272 | 544 | 2272 |
| # positive examples ( code) | 2043 | 885 | 492 | 324 | 200 | 133 | 79 | 49 | 25 | 8 | 1 |
Figure 5.

Variation of average F1-scores with the percentage of covered ICD code occurrences
5. Discussion
5.1. Implications for coding support and EHR systems
Despite achieving high performance for several ICD-9-CM codes, average model performance across the spectrum of ICD codes was not fully sufficient to allow direct implementation in real-world settings. As stated by Stanfill et al. (Stanfill et al., 2010), comparing performance across studies is typically challenging due to differences in problem scope and complexity. If we compare our results directly with other studies, we obtained lower performance than Pakhomov et al. (Pakhomov et al., 2006) and Perez et al. (Pérez et al., 2015). Conversely, our results outperform those from Kavuluru et al. (Kavuluru et al., 2015; Lipton et al., 2016). Also, results across codes varied significantly in ranges similar to the ones reported by Xu et al. (Xu et al., 2007). Results are aligned with the studies emphasising that clinical coding is still an extremely challenging research problem and model results are typically lower than results communicated in other machine learning applications – see literature reviewed by (Scheurwegs, Cule, Luyckx, Luyten, & Daelemans, 2017).
Our results exhibited high performance levels for multiple codes with high occurrence rates. The use of these results could potentially entail a considerable relief on coding workload. In comparison, many other studies addressed fewer codes (e.g., ((Avillach et al., 2008; Li et al., 2011; Lita et al., 2008; Yan et al., 2010) and therefore potentially brought about a much lower impact in relieving the coding workload. In spite of the observed performance challenges, the proposed methodology can still produce valuable results with potential in (1) automating coding for high performing codes, (2) suggesting ranked lists of codes to avoid looking up large code lists, and (3) explaining why codes are assigned by analysing associated features. These applications illustrate the potential value of using structured EHR data for coding support. Since there is not a generally acceptable threshold to define high performing codes which would be candidates for automatic coding (e.g., some authors suggest 95% accuracy or higher (Pakhomov et al., 2006)), it would be recommended to implement a manual verification step after automatic code assignment, particularly as this may likely be a quality requirement by healthcare stakeholders.
Since the tested models did not rank consistently in the different metrics, it is not possible to make a straightforward recommendation as to which model is best for coding support based on structured EHR data. The choice of prediction model must account for its impact on clinical coding – as investigated in our study – as well as on episode classification and financing. Our results suggest that in cases where incorrectly suggesting a code may lead to upcoding penalties (i.e., a hospital receiving a fine for classifying and/or billing diagnosis or procedure codes incorrectly, for which there is no clinical evidence), decision trees should be preferred in order to minimise false positives. Conversely, for codes representing losses in clinical data quality and/or funding for care provision, it is important to avoid overlooking codes and in this case SVM models should be chosen to minimise false negatives.
Based on our results, we can argue that the causes explaining variations in performance across codes resided not only in the selection of prediction models and in data imbalance, but also in the clinical concepts underlying each code. The observed results suggest that health professionals do not use the same level of granularity when documenting clinical information, leading to variability in granularity and in use of modifiers. These modifiers represent elements used to add detailed information to the core diagnosis, such as infectious/acute/subacute modifiers used for bronchiolitis. These findings raise questions on how data recording practices of clinicians can influence model performance, notably by introducing dispersion (i.e., identical diagnoses being recorded differently) in the dataset. Given that clinical coding requires using the most granular level, EHR data needs to be recorded with the necessary detail in order to properly support the coding process. The influence of data quality is also evidenced by comparison with the lower results obtained in preliminary works (reference omitted), which were obtained in earlier stages of EHR implementation when system users were expected to have lower levels of proficiency.
5.2. Methodology applicability and scalability
The development of a coding support methodology based on structured EHR data is relevant in light of the evolution of EHR systems towards structured formats. Our study differs from studies found in the literature which are either based on unstructured data, or on leveraged structured data and which are focused on a small subset of diagnoses, on predicting diagnoses at a less granular level and which are less tailored to support the clinical coding process. Our results indicate that the coding process may in fact be supported by using only structured EHR data. This is particularly valuable in contexts where using NLP is deemed impractical, namely NLP resources are lacking for specific languages and many EHR systems are being developed to entail mostly structured data.
In terms of applicability over the ICD spectrum, our methodology reveals pitfalls for heavily imbalanced codes (as seen in Figure 5), exhibiting performance over 0.5 approximately for the most frequent 150 codes. Still, these codes cover around 70% of code occurrences, which is higher than most studies found in the literature. Data imbalance is a pervasive issue in coding support and may give rise to statistical artefacts and lower results (He & Garcia, 2008). It may be worth exploring ensemble learning (Khalilia, Chakraborty, & Popescu, 2011) and bootstrap methods (Dupret & Koda, 2001) to compensate this imbalance.
The applicability of the proposed methodology to datasets from other EHR systems is viable since we have used data elements that are typically found and routinely collected in most EHR systems. Examples of such elements are diagnoses, prescribed exams and therapies, and structured assessments, on which analogous steps of feature construction (Figure 2) can be applied. In such cases, it will be instrumental to adequately transform data to minimise redundancies and define features in terms of their type and missing patterns. The possibility of generalisation to other EHR systems will also be determined by the degree of structuring of EHR data found in such systems. In effect, although structured EHR formats are increasingly more common (Hyppönen et al., 2014; Kalra et al., 2013), free-text is invariably preferred by clinicians in expressing clinical information (Johnson et al., 2008). Some EHR components, such as discharge summaries and descriptive reports from generic medical observations and diagnostic exams, are typically only available in unstructured formats.
The proposed methodology is applicable to multiple coding schemes in use world-wide (Busse, Geissler, & Quentin, 2011). It would be important to tailor the feature set according to the scope and level of granularity of each coding scheme, and thereby minimise eventual information losses when transforming EHR data into a data matrix format.
As the volume of episode data is continuously increasing, it is also important that the proposed methodology is scalable for larger volumes, so that we can take advantage of new data points and incorporate changes in the hospital population over time. Computation times for each code were manageable (see Table B1 in the appendix). Scalability over the range of clinical codes is also ensured via decomposition in binary classification problems, which results in computation times growing linearly with the number of codes. In effect, scalability to contexts with higher volume and complexity may highly benefit from the constant innovation and contributions from the operations research field to the improvement in efficiency of inductive learning algorithms (Corne et al., 2012; Meisel & Mattfeld, 2010; Olafsson et al., 2008).
5.3. Critical assessment of the methodology
In terms of data processing automation, the proposed methodology aimed to balance streamlining tasks while ensuring and preserving clinical meaningfulness. Expert knowledge (both medical and coding-related) and manual review/input are required only in specific tasks, notably mapping diagnosis catalogues and structuring assessment-based features. These efforts are expected to be performed mostly in the first deployment of the proposed methodology to provide the input information necessary for the automatic data processing. Further modifications in the EHR can easily be incorporated in the feature specification (master) file.
Looking into the predictive modelling framework, it is important to address the adequacy of feature selection methods, of machine learning models and of evaluation metrics. Firstly, feature selection was found to be extremely relevant to tackle dimensionality and improve results (as seen in Table 5). The appearance of clinically meaningful features (in Table 6) also corroborates their adequacy. The impact of feature selection was lower for decision trees since these models already entail intrinsic feature selection (upon splitting criteria). Although filter methods may yield sub-optimal performance, we argue that wrapper methods would have been inadequate due to computational limitations. Secondly, the choice of supervised learning models suited the purpose of modelling patterns between EHR data and corresponding codes. Thirdly, we chose not to use other traditional measures, such as the area under the receiving operating characteristic (ROC) curve for being less intuitive, and accuracy due to its propensity to be over-optimistic in highly imbalanced problems.
Notwithstanding limitations of the ICD-9-CM coding scheme – see for example (Bergstra & Bengio, 2012) – in this study we were bound to use it since it was the coding standard dictated by the Portuguese Ministry of Health at the time of this research (episodes in the dataset were all coded in ICD-9-CM). It would be relevant to evaluate the proposed methodology in other coding schemes, e.g., ICD-10-CM, which is increasingly adopted and more widely used worldwide.
As an additional remark, it is important to point out the possibility of leveraging the five-level hierarchical structure of coding schemes. This has been addressed in previous studies and it has produced positive results (Perotte et al., 2013; Zhang, 2008). Although coders must assign codes with the most granular level according to guidelines (Centers for Disease Control, 2011), the proposed methodology could be used, in a first stage of coding support, to predict codes at a less granular level and then let coders decide which codes to assign within that level.
6. Conclusions
In this work, we proposed and applied an end-to-end data mining methodology to predict ICD code assignment using fully structured EHR data. We addressed the stages of transforming EHR data into a data matrix, performing feature selection, building classification models and evaluating performance. We tested the extent to which structured data – which is becoming increasingly common in healthcare settings – can be used for coding support and which challenges arise in the use of these data formats. Our research work differs from other studies predicting diagnoses and/or clinical codes by leveraging a comprehensive scope of structured EHR data across a broad spectrum of diagnoses, specifically aiming to support the clinical coding process,
Our case study experiments revealed higher performance for logistic regression models, while decision trees were able to predict with higher precision and support vector machines with higher recall. Furthermore, combined with a filtered classifier that takes into account the class imbalance, the DNN’s recall can be improved substantially with the drawback of a reduction in precision and F1-score. Results also have shown the positive contribution of feature selection techniques. These promising results and insights provide evidence of the potential of structured EHR data in reducing coding workload, improving human resource utilisation and mitigating coding errors. The proposed methodology can lay a sound groundwork to introduce improvements in the coding process in numerous healthcare settings.
As future research, it would be useful to further develop the building blocks of the proposed methodology, starting with more systematic analysis of data quality and inconsistencies between EHR data and ICD codes, as well as exploring different approaches to handling missing data (Cismondi et al., 2013). It may also be valuable to incorporate additional expert knowledge in validating feature subsets, so as to mitigate artefacts and include additional clinically relevant features. Expert knowledge can also help improving data quality by harmonising feature specificity and including clinical conditions inferred from medication and test results. In order to prioritise research efforts, it may be useful to firstly address codes with higher impact in terms of a) operational workload (with higher frequency), b) health statistics and indicators (Zhan & Miller, 2003), and c) in provider financing (e.g., ICD codes associated with higher reimbursement rates). Moreover, domain knowledge may play a central role in imposing restrictions on code combinations.
In terms of models and algorithms, it may also be pertinent to test the viability of using wrapper feature selection with simpler space search procedures to cope with the high computational demand. It may also be valuable to explore cost-sensitive classification to mitigate the impact of the imbalanced class distribution in the dataset (Dupret & Koda, 2001; He & Garcia, 2008) and include inter-label relationships (Alvares-Cherman, Metz, & Monard, 2012). Another area of future work in terms of methodology is to formulate and solve the classification problem using multi-label classification algorithms (Read, Reutemann, Pfahringer, & Holmes, 2016).
Lastly, it may also be worth exploring more complex evaluation metrics aligned to the actual benefit for coders. This subject has recently been addressed in the literature, namely by Puentes et al. (Puentes, Montagner, Lecornu, & Cauvin, 2013) through usability-related performance measures (from the coder perspective), by Perotte et al. (Perotte et al., 2013) through hierarchy-based distance measures, and by (Chiaravalloti et al., 2014) based on code rankings. Accordingly, future work should consider these advanced performance measures, as well as issues concerning the acceptability and adoption of coding support tools by coding professionals in line with evidence on EHR system adoption (Weeger & Gewald, 2015).
Acknowledgments
The authors wish to thank colleagues from Hospital Professor Doutor Fernando Fonseca for close collaboration and availability throughout this research. The authors also acknowledge the financial support from Fundação para a Ciência e a Tecnologia (grant SFRH/BDE/51605/2011), Siemens Healthcare, the Centre for Management Studies of Instituto Superior Técnico (CEG-IST, University of Lisbon) and the Seedcorn Bursary of the University of Cardiff.The authors sincerely thank the area editor and the anonymous referees for their careful review and excellent suggestions for improvement of this paper. The authors sincerely thank the area editor and the anonymous referees for their careful review and excellent suggestions for improvement of this paper. Also, the Data Innovation Research Institute at Cardiff University provided Seedcorn Funding to support the project and to facilitate new collaborations.
Appendix A. List of ICD-9-CM codes.
Table A1.
List of the 50 most frequent ICD-9-CM codes analysed in this study (NOS – not otherwise specified; NEC – not elsewhere classified; TIA – transient ischaemic attack)
| ICD-9-CM code | Description |
|---|---|
| 401.9 | Hypertension NOS |
| 272.4 | Hyperlipidaemia NEC/NOS |
| 427.31 | Atrial fibrillation |
| 486 | Pneumonia, organism NOS |
| 428.0 | Congestive heart failure NOS |
| 250.00 | DMII without complications, not stated as uncontrolled |
| 305.1 | Tobacco use disorder |
| 414.8 | Chronic ischaemic heart disease NEC |
| 403.90 | Hypertensive chronic kidney disease, unspecified, stage I–IV |
| 278.00 | Obesity NOS |
| 599.0 | Urinary tract infection NOS |
| 276.8 | Hypopotassemia |
| 276.51 | Dehydration |
| 285.9 | Anaemia NOS |
| V58.66 | Long-term use of aspirin |
| 276.1 | Hyposmolality |
| 585.9 | Chronic kidney disease NOS |
| V15.82 | History of tobacco use |
| V58.61 | Long-term use of anticoagulants |
| 402.91 | Unspecified hypertensive heart disease with heart failure |
| 584.9 | Acute kidney failure NOS |
| 466.0 | Acute bronchitis |
| 303.91 | Other and unspecified alcohol dependence, continuous |
| V12.59 | Personal history of other diseases of circulatory system |
| V15.29 | Personal history of surgery to other organs |
| V14.0 | Personal history of allergy to penicillin |
| E888.8 | Fall NEC |
| 244.9 | Hypothyroidism NOS |
| 518.81 | Acute respiratory failure |
| 280.9 | Iron deficiency anaemia NOS |
| 041.49 | E. coli infection NEC/NOS |
| V12.54 | Personal history of TIA and cerebral infarction without residual deficits |
| 272.0 | Pure hypercholesterolaemia |
| 518.82 | Other pulmonary insufficiency |
| V58.67 | Long-term use of insulin |
| 434.91 | Cerebral artery occlusion, unspecified with cerebral infarction |
| 311 | Depressive disorder NEC |
| 595.0 | Acute cystitis |
| 276.7 | Hyperpotassemia |
| 511.9 | Pleural effusion NOS |
| V49.84 | Bed confinement status |
| V45.01 | Status cardiac pacemaker |
| 285.29 | Anaemia of other chronic disease |
| 491.21 | Obstructive chronic bronchitis with (acute) exacerbation |
| 593.9 | Unspecified disorder of kidney and ureter |
| V46.2 | Depend-supplement oxygen |
| 276.2 | Acidosis |
| 518.84 | Acute and chronic respiratory failure |
| 275.2 | Disorders of magnesium metabolism |
| 491.20 | Obstructive chronic bronchitis without exacerbation |
Appendix B. Computational Performance.
Table B1.
Execution times for feature selection and model training algorithms (including parameter optimisation) obtained for a binary classification problem. Training times were obtained with a subset of 50 mRMR features and 4,072 training instances. Simulations were performed using an intel core i5-2520 M CPU 2.50 GHz, 4 GB RAM
| Feature selection | Execution time (ms) |
|---|---|
| FCBF | 18,922 |
| IG | 787,152 |
| Relief | 2,612,942 |
| Chi-square | 4,671 |
| SU | 2,008 |
| CFS | 2,884,391 |
| mRMR |
21,652 |
| Prediction model |
Training time (ms) |
| DT (mRMR) | 30,585 |
| NB (mRMR) | 141,530 |
| Logit (mRMR) | 12,341 |
| SVM (mRMR) | 59,465 |
| DNN (mRMR) | 85,770 |
Appendix C. Deep Learning Classification Results using Class Imbalance Filtering.
Table C1.
Deep learning classification results using class imbalance filtering
| Precision | Recall | F1 | |
|---|---|---|---|
| None | 0.294 | 0.742 | 0.406 |
| FCBF | 0.348 | 0.763 | 0.446 |
| IG | 0.294 | 0.773 | 0.408 |
| Relief | 0.271 | 0.756 | 0.380 |
| Chi2 | 0.298 | 0.774 | 0.413 |
| SU | 0.300 | 0.770 | 0.414 |
| CFS | 0.353 | 0.763 | 0.453 |
| mRMR | 0.353 | 0.761 | 0.453 |
Table C2.
List of abbreviations
| Abbreviation | Description |
|---|---|
| API | Application Programming Interface |
| CART | Classification and regression trees |
| CFS | Correlation-based feature selection |
| CNN | Convolutional neural networks |
| DBN | Deep belief network |
| DRG | Diagnosis-related groups |
| DL | Deep Learning |
| DNN | Deep Neural Network |
| DT | Decision trees |
| EHR | Electronic health record |
| FCBF | Fast correlation-based filter |
| FN | False negative |
| FP | False positive |
| ICD-9-CM | International Classification of Diseases, 9th revision, Clinical Modification |
| ICD-10(-PCS) | International Classification of Diseases, 10th revision (Procedure Classification System) |
| ICF | International Classification of Functioning, Disability and Health |
| IG | Information gain |
| LOINC | Logical Observation Identifiers Names and Codes |
| MLP | Multi-layer perceptron |
| mRMR | Minimal redundancy maximal relevance |
| NEC | Not elsewhere classified |
| NOS | Not otherwise specified |
| NB | Naïve Bayes |
| NLP | Natural language processing |
| RNN | Recurrent neural networks |
| SAE | Stacked auto-encoder |
| SNOMED-CT | Systematised Nomenclature of Medicine – Clinical Terms |
| SU | Symmetrical uncertainty |
| SVM | Support vector machines |
| TIA | Transient ischaemic attack |
| TP | True positive |
| UMLS | Unified Medical Language System |
Funding Statement
This work was supported by the Fundação para a Ciência e a Tecnologia [SFRH/BDE/51605/2011, Instituto Superior Técnico [FCT project UID/GES/00097] and Siemens Healthineers.
Disclosure statement
The authors declare that there are no competing interests regarding this study.
References
- AHIMA . (2013). Delving into computer-assisted coding (AHIMA practice brief). Journal of AHIMA, 75, 48A–48H. Retrieved from http://library.ahima.org/PB/CACGuidance. [PubMed] [Google Scholar]
- Alvares-Cherman, E., Metz, J., & Monard, M. C. (2012). Incorporating label dependency into the binary relevance framework for multi-label classification. Expert Systems with Applications, 39(2), 1647–1655. [Google Scholar]
- Aronson, A. R., Bodenreider, O., Demner-Fushman, D., Fung, K. W., Lee, V. K., Mork, J. G., … Rogers, W. J. (2007). From indexing the biomedical literature to coding clinical text: Experience with MTI and machine learning approaches. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing (pp. 105–112). Prague, Czech Republic: Association for Computational Linguistics. [Google Scholar]
- Aronson, A. R., & Lang, F. M. (2010). An overview of MetaMap: Historical perspective and recent advances. Journal of the American Medical Informatics Association, 17(3), 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avillach, P., Joubert, M., & Fieschi, M. (2008). Improving the quality of the coding of primary diagnosis in standardized discharge summaries. Health Care Management Science, 11(2), 147–151. [DOI] [PubMed] [Google Scholar]
- Bergstra, J., & Bengio, Y. (2012). Random search for hyper-parameter optimization. Journal of Machine Learning Research, 13(Feb), 281–305. [Google Scholar]
- Bhandare, A., Bhide, M., Gokhale, P., & Chandavarkar, R. (2016). Applications of convolutional neural networks. International Journal of Computer Science and Information Technologies, 7(5), 2206–2215. [Google Scholar]
- Bishop, C. (2006). Pattern recognition and machine learning. New York: Springer. [Google Scholar]
- Bleeker, S. E., Derksen-Lubsen, G., van Ginneken, A. M., Van Der Lei, J., & Moll, H. A. (2006). Structured data entry for narrative data in a broad specialty: Patient history and physical examination in pediatrics. BMC Medical Informatics and Decision Making, 6(1), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie, M. J., & Schaffer, R. M. (2012). Understanding ICD-9-CM coding: A worktext. Delmar Cengage Learning. [Google Scholar]
- Breiman, L, Friedman, J., Stone, C. J., & Olshen, R. A. (1984). Classification and regression trees. Chapman and Hall. [Google Scholar]
- Brown, G. (2009). A new perspective for information theoretic feature selection. Artificial intelligence and statistics (pp. 49–56), Clearwater, FL, USA. [Google Scholar]
- Brown, G., Pocock, A., Zhao, M. J., & Luján, M. (2012). Conditional likelihood maximisation: A unifying framework for information theoretic feature selection. Journal of Machine Learning Research, 13(Jan), 27–66. [Google Scholar]
- Busse, R., Geissler, A., & Quentin, W. (2011). Diagnosis-related groups in Europe: Moving towards transparency, efficiency and quality in hospitals. McGraw-Hill Education (UK). [DOI] [PubMed] [Google Scholar]
- Capan, M., Wu, P., Campbell, M., Mascioli, S., & Jackson, E. V. (2017). Using electronic health records and nursing assessment to redesign clinical early recognition systems. Health Systems, 6(2), 112–121. [Google Scholar]
- Centers for Disease Control . (2011). ICD-9-CM official guidelines for coding and reporting. Atlanta, GA: Centers for Medicare & Medicaid Services. [Google Scholar]
- Chapman, W. W., Bridewell, W., Hanbury, P., Cooper, G. F., & Buchanan, B. G. (2001). A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of Biomedical Informatics, 34(5), 301–310. [DOI] [PubMed] [Google Scholar]
- Che, Z., Purushotham, S., Cho, K., Sontag, D., & Liu, Y. (2018). Recurrent neural networks for multivariate time series with missing values. Scientific Reports, 8(1), 6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaravalloti, M. T., Guarasci, R., Lagani, V., Pasceri, E., & Trunfio, R. (2014). A coding support system for the ICD-9-CM standard. 2014 IEEE International Conference on Healthcare Informatics (ICHI), (pp. 71–78). Verona, Italy: IEEE. [Google Scholar]
- Choi, E., Bahadori, M. T., Schuetz, A., Stewart, W. F., & Sun, J. (2016). Doctor AI: Predicting clinical events via recurrent neural networks. In: Doshi-Velez F., Fackler J., Kale D., Wallace B., & Wiens J. (Eds), Proceedings of the 1st Machine Learning for Healthcare Conference, volume 56 of Proceedings of Machine Learning Research (pp. 301–318). LA, Los Angeles, CA, USA: PMLR, Children’s Hospital. [PMC free article] [PubMed] [Google Scholar]
- Cismondi, F., Fialho, A. S., Vieira, S. M., Reti, S. R., Sousa, J. M., & Finkelstein, S. N. (2013). Missing data in medical databases: Impute, delete or classify? Artificial Intelligence in Medicine, 58(1), 63–72. [DOI] [PubMed] [Google Scholar]
- Corne, D., Dhaenens, C., & Jourdan, L. (2012). Synergies between operations research and data mining: The emerging use of multi-objective approaches. European Journal of Operational Research, 221(3), 469–479. [Google Scholar]
- Cornet, R., & de Keizer, N. (2008). Forty years of SNOMED: A literature review. BMC medical informatics and decision making, volume 8. BioMed Central, S2. [DOI] [PMC free article] [PubMed]
- Cortes, C., & Vapnik, V. (1995). Support-vector networks. Machine Learning, 20(3), 273–297. [Google Scholar]
- Crammer, K., Dredze, M., Ganchev, K., Talukdar, P. P., & Carroll, S. (2007). Automatic code assignment to medical text. Proceedings of the workshop on bionlp 2007: Biological, translational, and clinical language processing (pp. 129–136). Prague, Czech Republic: Association for Computational Linguistics. [Google Scholar]
- Davidson, E. J., Gao, G. G., & McCullough, J. S. (2015). Health IT and economics. Health Systems, 4(1), 54. [Google Scholar]
- Deeplearning4j . (2017). Deeplearning4j: open-source distributed deep learning for the JVM.
- Delamarre, D., Burgun, A., Seka, L. P., & Le Beux, P. (1995). Automated coding of patient discharge summaries using conceptual graphs. Methods of Information in Medicine, 34(04), 345–351. [PubMed] [Google Scholar]
- Dinwoodie, H., & Howell, R. (1973). Automatic disease coding: The’fruit-machine’method in general practice. British Journal of Preventive & Social Medicine, 27(1), 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreiseitl, S., & Ohno-Machado, L. (2002). Logistic regression and artificial neural network classification models: A methodology review. Journal of Biomedical Informatics, 35(5–6), 352–359. [DOI] [PubMed] [Google Scholar]
- Dupret, G., & Koda, M. (2001). Bootstrap re-sampling for unbalanced data in supervised learning. European Journal of Operational Research, 134(1), 141–156. [Google Scholar]
- Faber, B., Konrad, R. A., Tang, C., & Trapp, A. C. (2016). Examining the impact of regular physician visits on heart failure patients: A use case with electronic health data. Health Systems, 5(2), 132–139. [Google Scholar]
- Farkas, R., & Szarvas, G. (2008). Automatic construction of rule-based ICD-9-CM coding systems. BMC bioinformatics (Vol. 9, pp. S10). BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando, B., Kalra, D., Morrison, Z., Byrne, E., & Sheikh, A. (2012). Benefits and risks of structuring and/or coding the presenting patient history in the electronic health record: Systematic review. BMJ Quality & Safety, 21(4), 337–346. [DOI] [PubMed] [Google Scholar]
- Ford, E. W., Menachemi, N., & Phillips, M. T. (2006). Predicting the adoption of electronic health records by physicians: When will health care be paperless? Journal of the American Medical Informatics Association, 13(1), 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- France, F. H. R. (2003). Case mix use in 25 countries: A migration success but international comparisons failure. International Journal of Medical Informatics, 70(2–3), 215–219. [DOI] [PubMed] [Google Scholar]
- Franz, P., Zaiss, A., Schulz, S., Hahn, U., & Klar, R. (2000). Automated coding of diagnoses– Three methods compared. Proceedings of the AMIA Symposium. American Medical Informatics Association (p. 250), Los Angeles, CA, USA. [PMC free article] [PubMed] [Google Scholar]
- Friedman, C., Alderson, P. O., Austin, J. H., Cimino, J. J., & Johnson, S. B. (1994). A general natural- language text processor for clinical radiology. Journal of the American Medical Informatics Association, 1(2), 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, C., Shagina, L., Lussier, Y., & Hripcsak, G. (2004). Automated encoding of clinical documents based on natural language processing. Journal of the American Medical Informatics Association, 11(5), 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner, D. (2015). Scheduling the hospital-wide flow of elective patients. Heidelberg: Springer Lecture Notes in Economics and Mathematical Systems. [Google Scholar]
- Gartner, D., Kolisch, R., Neill, D. B., & Padman, R. (2015). Machine learning approaches for early DRG Classification and resource allocation. INFORMS Journal on Computing, 27(4), 718–734. [Google Scholar]
- Goldstein, I., Arzumtsyan, A., & Uzuner, Ö.(2007). Three approaches to automatic assignment of ICD-9-CM codes to radiology reports. AMIA Annual Symposium Proceedings, volume 2007 (p. 279). Chicago, IL: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- Gundersen, M. L., Haug, P. J., Pryor, T. A., van Bree, R., Koehler, S., Bauer, K., & Clemons, B. (1996). Development and evaluation of a computerized admission diagnoses encoding system. Computers and Biomedical Research, 29(5), 351–372. [DOI] [PubMed] [Google Scholar]
- Guyon, I., & Elisseeff, A. (2003). An introduction to variable and feature selection. Journal of Machine Learning Research, 3, 1157–1182. [Google Scholar]
- Hall, M., & Holmes, G. (2003). Benchmarking attribute selection techniques for discrete class data mining. IEEE Transactions on Knowledge and Data Engineering, 15(6), 1437–1447. [Google Scholar]
- Hand, D. J., & Yu, K. (2001). Idiot’s bayes—not so stupid after all? International Statistical Review, 69(3), 385–398. [Google Scholar]
- Haux, R., Seggewies, C., Baldauf-Sobez, W., Kullmann, P., Reichert, H., Luedecke, L., & Seibold, H. (2003). SoarianTM–workflow management applied for health care. Methods of Information in Medicine, 42(01), 25–36. [PubMed] [Google Scholar]
- He, H., & Garcia, E. A. (2008). Learning from imbalanced data. IEEE Transactions on Knowledge & Data Engineering, 9, 1263–1284. [Google Scholar]
- Hosmer, D. W., & Lemeshow, S. (2000). Applied Logistic Regression (2nd ed.). John Wiley & Sons, Inc. [Google Scholar]
- Huff, S. M., Rocha, R. A., McDonald, C. J., De Moor, G. J., Fiers, T., Jr WD, B., … Leavelle, D., et al. (1998). Development of the logical observation identifier names and codes (LOINC) vocabulary. Journal of the American Medical Informatics Association, 5(3), 276–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppönen, H., Saranto, K., Vuokko, R., Mäkelä-Bengs, P., Doupi, P., Lindqvist, M., & Mäkelä, M. (2014). Impacts of structuring the electronic health record: A systematic review protocol and results of previous reviews. International Journal of Medical Informatics, 83(3), 159–169. [DOI] [PubMed] [Google Scholar]
- Johnson, S. B., Bakken, S., Dine, D., Hyun, S., Mendonça, E., Morrison, F., … Stetson, P. (2008). An electronic health record based on structured narrative. Journal of the American Medical Informatics Association, 15(1), 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra, D., Fernando, B., Morrison, Z., & Sheikh, A. (2013). A review of the empirical evidence of the value of structuring and coding of clinical information within electronic health records for direct patient care. Journal of Innovation in Health Informatics, 20(3), 171–180. [DOI] [PubMed] [Google Scholar]
- Kavuluru, R., Rios, A., & Lu, Y. (2015). An empirical evaluation of supervised learning approaches in assigning diagnosis codes to electronic medical records. Artificial Intelligence in Medicine, 65(2), 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevers, L., & Medori, J. (2010). Symbolic classification methods for patient discharge summaries encoding into ICD. International Conference on Natural Language Processing (pp. 197–208). Reykjavik, Iceland: Springer. [Google Scholar]
- Khalilia, M., Chakraborty, S., & Popescu, M. (2011). Predicting disease risks from highly imbalanced data using random forest. BMC Medical Informatics and Decision Making, 11(1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kira, K., & Rendell, L. A. (1992). The feature selection problem: Traditional methods and a new algorithm. Aaai, 2, 129–134. [Google Scholar]
- Kohavi, R. (1995). A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th international joint conference on Artificial intelligence - Volume 2 (IJCAI’95). Morgan Kaufmann Publishers Inc., San Francisco, CA, USA(Vol. 14, pp. 1137–1145). Canada: Montreal. [Google Scholar]
- Koopman, B., Zuccon, G., Nguyen, A., Bergheim, A., & Grayson, N. (2015). Automatic ICD-10 classification of cancers from free-text death certificates. International Journal of Medical Informatics, 84(11), 956–965. [DOI] [PubMed] [Google Scholar]
- Kukafka, R., Bales, M. E., Burkhardt, A., & Friedman, C. (2006). Human and automated coding of rehabilitation discharge summaries according to the international classification of functioning, disability, and health. Journal of the American Medical Informatics Association, 13(5), 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkey, L. S., & Croft, W. B. (1995). Automatic assignment of icd9 codes to discharge summaries. Technical report, Technical report. Amherst, MA: University of Massachusetts at Amherst. [Google Scholar]
- Lazar, C., Taminau, J., Meganck, S., Steenhoff, D., Coletta, A., Molter, C., … Nowe, A. (2012). A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB), 9(4), 1106–1119. [DOI] [PubMed] [Google Scholar]
- LdS, F. (2011). Medical information extraction in European Portuguese. Ph.D. thesis, Universidade de Aveiro. [Google Scholar]
- Li, S. T., Chen, C. C., & Huang, F. (2011). Conceptual-driven classification for coding advise in health insurance reimbursement. Artificial Intelligence in Medicine, 51(1), 27–41. [DOI] [PubMed] [Google Scholar]
- Lindberg, D. A., Humphreys, B. L., & McCray, A. T. (1993). The unified medical language system. Yearbook of Medical Informatics, 2(01), 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton, Z. C., Kale, D. C., Elkan, C., & Wetzel, R. C. (2016). Learning to diagnose with LSTM recurrent neural networks. CoRR abs/1511.03677.
- Lita, L. V., Yu, S., Niculescu, S., & Bi, J. (2008). Large scale diagnostic code classification for medical patient records. Proceedings of the Third International Joint Conference on Natural Language Processing: Volume-II, Hyderabad, India. [Google Scholar]
- Liu, S., Ma, W., Moore, R., Ganesan, V., & Nelson, S. (2005). RxNorm: Prescription for electronic drug information exchange. IT Professional, 7(5), 17–23. [Google Scholar]
- Lussier, Y. A., Shagina, L., & Friedman, C. (2000). Automating icd-9-cm encoding using medical language processing: A feasibility study. Proceedings of the AMIA Symposium (p. 1072). Los Angeles, CA: American Medical Informatics Association. [Google Scholar]
- Lussier, Y. A., Shagina, L., & Friedman, C. (2001). Automating SNOMED coding using medical language understanding: A feasibility study. Proceedings of the AMIA Symposium (p. 418). Washington, DC: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- Mateus, C. (2008). The globalization of managerial innovation in health care, chapter case mix implementation in Portugal. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Matykiewicz, P., Duch, W., & Pestian, J. (2006). Associating Medical Concept Relations with ICD-9-CM Coding Rules.
- McDonald, C. J., & Tierney, W. M. (1988). Computer-stored medical records: Their future role in medical practice. Journal of the American Medical Association, 259(23), 3433–3440. [PubMed] [Google Scholar]
- Medori, J., & Fairon, C. (2010). Machine learning and features selection for semi-automatic ICD-9-CM encoding. Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents (pp. 84–89). Los Angeles, CA: Association for Computational Linguistics. [Google Scholar]
- Meisel, S., & Mattfeld, D. (2010). Synergies of operations research and data mining. European Journal of Operational Research, 206(1), 1–10. [Google Scholar]
- Meystre, S. M., Savova, G. K., Kipper-Schuler, K. C., & Hurdle, J. F. (2008). Extracting information from textual documents in the electronic health record: A review of recent research. Yearbook of Medical Informatics, 17(01), 128–144. [PubMed] [Google Scholar]
- Mitchell, T. M. (1997). Machine learning. New York, NY: McGraw-Hill.
- Morris, W. C., Heinze, D. T., Jr HR, W., Primack, A., Morsch, A., Sheffer, R. E., … Jimmink, M. A. (2000). Assessing the accuracy of an automated coding system in emergency medicine. Proceedings of the AMIA Symposium. (p. 595). Los Angeles, CA: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- Mortenson, M. J., Doherty, N. F., & Robinson, S. (2015). Operational research from taylorism to terabytes: A research agenda for the analytics age. European Journal of Operational Research, 241(3), 583–595. [Google Scholar]
- Nadler, J. J., & Downing, G. J. (2010). Liberating Health Data for Clinical Research Applications. Science Translational Medicine, 2(18), 18cm6–18cm6. [DOI] [PubMed] [Google Scholar]
- Olafsson, S., Li, X., & Wu, S. (2008). Operations research and data mining. European Journal of Operational Research, 187(3), 1429–1448. [Google Scholar]
- Oleynik, M., Patr˜ao, D. F., & Finger, M. (2017). Automated classification of semi-structured pathology reports into ICD-O using SVM in Portuguese. Studies in Health Technology and Informatics, 235, 256–260. [PubMed] [Google Scholar]
- Pakhomov, S. V., Buntrock, J. D., & Chute, C. G. (2006). Automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques. Journal of the American Medical Informatics Association, 13(5), 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, V., Jamoom, E., Hsiao, C. J., Furukawa, M. F., & Buntin, M. (2013). Variation in electronic health record adoption and readiness for meaningful use: 2008–2011. Journal of General Internal Medicine, 28(7), 957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, H., Long, F., & Ding, C. (2005). Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence, 27(8), 1226–1238. [DOI] [PubMed] [Google Scholar]
- Pereira, S., Névéol, A., Massari, P., Joubert, M., & Darmoni, S. (2006). Construction of a semi- automated ICD-10 coding help system to optimize medical and economic coding. Studies in Health Technology and Informatics, 124, 845–850. [PubMed] [Google Scholar]
- Pérez, A., Gojenola, K., Casillas, A., Oronoz, M., & de Ilarraza, A. D. (2015). Computer aided classification of diagnostic terms in spanish. Expert Systems with Applications, 42(6), 2949–2958. [Google Scholar]
- Perotte, A., Pivovarov, R., Natarajan, K., Weiskopf, N., Wood, F., & Elhadad, N. (2013). Diagnosis code assignment: Models and evaluation metrics. Journal of the American Medical Informatics Association, 21(2), 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press W. H., Teukolsky S. A., Vetterling W. T. & Flannery B. P. (1992). Numerical recipes in C. Cambridge, UK: Cambridge University press. [Google Scholar]
- Puentes, J., Montagner, J., Lecornu, L., & Cauvin, J. M. (2013). Information quality measurement of medical encoding support based on usability. Computer Methods and Programs in Biomedicine, 112(3), 329–342. [DOI] [PubMed] [Google Scholar]
- Read, J., Reutemann, P., Pfahringer, B., & Holmes, G. (2016). Meka: A multi-label/multi-target extension to weka. The Journal of Machine Learning Research, 17(1), 667–671. [Google Scholar]
- Rijo, R., Silva, C., Pereira, L., Gonçalves, D., & Agostinho, M. (2014). Decision support system to diagnosis and classification of epilepsy in children. Journal of Universal Computer Science, 20(6), 907–923. [Google Scholar]
- Rizzo, S. G., Montesi, D., Fabbri, A., & Marchesini, G. (2015). ICD code retrieval: Novel approach for assisted disease classification. International Conference on Data Integration in the Life Sciences (pp. 147–161). Marina del Rey, CA: Springer. [Google Scholar]
- Rokach, L., & Maimon, O. (2005). Top-down induction of decision trees classifiers-a survey. IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews), 35(4), 476–487. [Google Scholar]
- Ruch, P., Gobeill, J., Tbahriti, I., & Geissbühler, A. (2008). From episodes of care to diagnosis codes: Automatic text categorization for medico-economic encoding. AMIA Annual Symposium Proceedings, volume 2008 (p. 636). Washington, DC: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- Saeys, Y., Inza, I., & Larran˜aga, P. (2007). A review of feature selection techniques in bioinformatics. bioinformatics, 23(19), 2507–2517. [DOI] [PubMed] [Google Scholar]
- Scheurwegs, E., Cule, B., Luyckx, K., Luyten, L., & Daelemans, W. (2017). Selecting relevant features from the electronic health record for clinical code prediction. Journal of Biomedical Informatics, 74, 92–103. [DOI] [PubMed] [Google Scholar]
- Scheurwegs, E., Luyckx, K., Luyten, L., Daelemans, W., & Van den Bulcke, T. (2015). Data integration of structured and unstructured sources for assigning clinical codes to patient stays. Journal of the American Medical Informatics Association, 23(e1), e11–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraffenberger, L. A. (2010). Basic ICD-9-CM coding. Chicago, IL: American Health Information Management Association. [Google Scholar]
- Shi, H., Xie, P., Hu, Z., Zhang, M., & Xing, E. P. (2017). Towards automated ICD coding using deep learning. arXiv Preprint, arXiv, 1711.04075. [Google Scholar]
- Shivade, C., Raghavan, P., Fosler-Lussier, E., Embi, P. J., Elhadad, N., Johnson, S. B., & Lai, A. M. (2013). A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association, 21(2), 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfill, M. H., Williams, M., Fenton, S. H., Jenders, R. A., & Hersh, W. R. (2010). A systematic literature review of automated clinical coding and classification systems. Journal of the American Medical Informatics Association, 17(6), 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subotin, M., & Davis, A. (2014). A system for predicting ICD-10-PCS codes from electronic health records. Proceedings of BioNLP, 2014, 59–67. [Google Scholar]
- Suominen, H., Ginter, F., Pyysalo, S., Airola, A., Pahikkala, T., Salanterä, S., & Salakoski, T. (2008). Machine learning to automate the assignment of diagnosis codes to free-text radiology reports: A method description. Proceedings of the ICML/UAI/COLT Workshop on Machine Learning for Health-Care ApplicationsHelsinki, Finland. [Google Scholar]
- Tsoumakas, G., Katakis, I., & Vlahavas, I. (2009). Mining multi-label data. In O. Maimon & L. Rokach (Eds.), Data mining and knowledge discovery handbook (pp. 667–685). Boston, MA: Springer. [Google Scholar]
- Weeger, A., & Gewald, H. (2015). Acceptance and use of electronic medical records: An exploratory study of hospital physicians’ salient beliefs about HIT systems. Health Systems, 4(1), 64–81. [Google Scholar]
- Witten, I., & Frank, E. (2011). Data mining: Practical machine learning tools and techniques (3rd ed.). San Francisco: Morgan Kaufmann. [Google Scholar]
- Xu, J., Yu, S., Bi, J., Lita, L. V., Niculescu, R. S., & Rao, R. B. (2007). Automatic medical coding of patient records via weighted ridge regression. In: Sixth International Conference on Machine Learning and Applications (ICMLA 2007) (pp. 260–265), Cincinnati, Ohio, USA. [Google Scholar]
- Xu, K., Lam, M., Pang, J., Gao, X., Band, C., Xie, P., & Xing, E. (2018). Multimodal machine learning for automated ICD coding. arXiv Preprint, arXiv, 1810.13348. [Google Scholar]
- Yan, Y., Fung, G., Dy, J. G., & Rosales, R. (2010). Medical coding classification by leveraging inter-code relationships. In: Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’10 (pp. 193–202). New York, NY, USA: ACM. [Google Scholar]
- Yang, Y., & Pedersen, J. O. (1997). A Comparative Study on Feature Selection in Text Categorization.
- Yao, L., Mao, C., & Luo, Y. (2018). Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. 2018 IEEE International Conference on Healthcare Informatics Workshop (ICHI-W) (pp. 70–71). New York, NY: IEEE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., & Liu, H. (2004). Efficient feature selection via analysis of relevance and redundancy. Journal of Machine Learning Research, 5(1), 1205–1224. [Google Scholar]
- Zhan, C., & Miller, M. (2003). Administrative data based patient safety research: A critical review. BMJ Quality & Safety, 12(suppl 2), ii58–ii63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. (2008). A hierarchical approach to encoding medical concepts for clinical notes. Proceedings of the 46th Annual Meeting of the Association for Computational Linguistics on Human Language Technologies: Student Research Workshop (pp. 67–72). Columbus, OH: Association for Computational Linguistics. [Google Scholar]


