Abstract
Mounting evidence has demonstrated that microRNAs (miRNAs or miRs) play significant roles in various types of human tumors, including retinoblastoma (RB). However, the biological role and regulatory mechanisms of miRNAs in RB remain to be fully elucidated. The present study was designed to identify the regulatory effects of miRNAs in RB and the underlying mechanisms. Differentially expressed miRNAs in RB tissue were screened out based on the Gene Expression Omnibus (GEO) dataset, GSE7072, which revealed that miR-153 in particular, displayed the highest fold change in expression. It was identified that miR-153 was significantly downregulated in RB tissues, and its downregulation was closely associated with a larger tumor base and differentiation. Functional analysis revealed that the overexpression of miR-153 inhibited RB cell proliferation, migration and invasion, and promoted the apoptosis of WERI-RB-1 and Y79 cells. In addition, insulin-like growth factor 1 receptor (IGF1R) was identified as a target of miR-153 in RB cells. More importantly, it was demonstrated that miR-153 upregulation inhibited the expression of its target gene, IGF1R, which inhibited the activation of the Raf/MEK and PI3K/AKT signaling pathways. Collectively, the present study demonstrates for the first time, to the best of our knowledge, that miR-153 functions as a tumor suppressor in RB by targeting the IGF1R/Raf/MEK and IGF1R/PI3K/AKT signaling pathways. Collectively, the findings presented herein demonstrate that miR-153 targets IGF1R and blocks the activation of the Raf/MEK and PI3K/AKT signaling pathway, thus preventing the progression of RB. Thus, this miRNA may serve as a novel prognostic biomarker and therapeutic target for RB.
Keywords: miR-153, retinoblastoma, apoptosis, migration and invasion, insulin-like growth factor 1 receptor, IGF1R/Raf/MEK and IGF1R/PI3K/AKT signaling pathways
Introduction
Retinoblastoma (RB) is the most common type of primary intraocular malignant tumor that occurs either genetically in children or sporadically in adults (1). Apart from its occult onset, RB can easily metastasize, and can rapidly fill much of the eyeball and can subsequently spread to the other parts of body, such as the brain and lymph nodes (2). Although marked improvements have been made in the treatment of RB, including enucleation, radiation, focal techniques and systemic chemotherapy, RB is associated with a high metastatic rate and poor prognosis (3,4). Therefore, there is an urgent need to explore the underlying mechanisms involved in RB tumorigenesis, development and prognosis.
MicroRNAs (miRNAs or miRs), normally containing ~21-25 nucleotides, are a class of small non-coding RNA molecules involved in the regulation of gene expression by targeting mRNAs for translational repression or cleavage (5,6). There is increasing evidence to indicate that miRNAs play critical roles in regulating cell apoptosis, proliferation, invasion and migration (7). A number of studies have indicated the role of miRNAs in various types of tumors (8). MicroRNA-153-3p (miR-153), an extensively studied miRNA, has been reported to serve as a tumor suppressor in a variety of cancer types, such as breast cancer (9), gastric cancer (10), osteosarcoma (11) and lung cancer (12). For example, miR-153 has been shown to target the inhibitor of growth protein 2 to inhibit acute lymphoblastic leukemia cell proliferation, migration and invasion (13). It has also been reported that miR-153 can inhibit the MCL1 gene, thus suppressing the progression of ovarian carcinoma in vitro and in vivo (14). In addition, miR-153 has been reported to inhibit the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells (15). However, the function of miR-153 in RB remains unknown.
In the present study, miR-153 expression was analyzed in RB tissues and cell lines by reverse transcription-quantitative PCR (RT-qPCR) assays. In vitro experiments were performed to investigate the functional role of miR-153 in RB cells and the underlying mechanisms. The findings of these experiments suggested that miR-15 may be a potential diagnostic and therapeutic target for RB treatment.
Materials and methods
Tissue samples
A total of 50 RB tissues samples were obtained from patients who received enucleation or enucleation + chemotherapy ± radiation therapy at the Department of Ophthalmology, the First People's Hospital of Shangqiu, Shangqiu, China between February, 2017 and November, 2018. A total of 10 normal retinas were obtained from patients who had passed away due to conditions other than ophthalmologic diseases at the First People's Hospital of Shangqiu. Written consent for tissue donation for research purposes was obtained from the donor or family members prior to tissue collection. The present study was approved by the Research Ethics Committee of the First People's Hospital of Shangqiu. The samples were snap-frozen in liquid nitrogen and stored at −80°C.
RT-qPCR
Total RNA was extracted from tissues and cells by TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the PrimeScript RT reagent kit (Promega Corporation). For the detection of miR-153, qPCR was conducted using the MicroRNAs Quantitation PCR kit [Sangon Biotech (Shanghai) Co., Ltd.]. U6 was used as an internal control for the detection of miR-153 expression. The sequences of the primers used for miR-153 and U6, as well as those for insulin-like growth factor 1 receptor (IGF1R) and GAPDH are listed in Table I. The thermocycling conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 10 min. Fold changes in the expression of each gene were calculated using the 2−∆∆Cq method (16).
Table I.
Sequences of the primers used in the present study.
| Gene | Primer sequence |
|---|---|
| miR-153-3p | F: 5′-ACACTCCAGCTGGGTTGCATAGTCACAAA-3′ |
| R: 5′-CAGTGCGTGTCGTGGAGT-3′ | |
| U6 | F: 5′-TCGCACAGACTTGTGGGAGAA-3′ |
| R: 5′-CGCACATTAAGCCTCTATAGTTACTAGG-3′ | |
| IGF1R | F: 5′-GGGGCTCCTGTTTCTCTCC-3′ |
| R: 5′-GCCTTGGAGATGAGCAGGAT-3′ | |
| GAPDH | F: 5′-AGCTTGTCATCAACGGGAAG-3′ |
| R: 5′-TTTGATGTTAGTGGGGTCTCG-3′ |
F, forward; R, reverse.
Cell lines and cell culture
WERI-RB-1 (ATCC® HTB-169) and Y79 (ATCC® HTB-18), a normal retinal epithelial cell line (ARPE-19) (ATCC® CRL-2302) cells were obtained from American Type Culture Collection (ATCC), while the RB cell line SO-RB50 was obtained from the Zhongshan Ophthalmic Center, Sun Yat-sen University (Zhongshan, China). All the cells were cultured in Dulbecco's modified Eagle's medium (DMEM, HyClone; Cytiva) supplemented with 10% fetal bovine serum (HyClone; Cytiva), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a 5% CO2 atmosphere.
Cell transfection
miR-153 mimics (5′-UUGCAUAGUCACAAAAGUGAUC-3′), mimics negative control (NC) (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-153 inhibitor (5′-GAUCACUUUUGUGACUAUGCAA-3′) and inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′) were obtained from Shanghai GenePharma Co., Ltd. pcDNA3.1-IGF1R, pcDNA3.1 vector, as well as small interfering RNA (si-RNA) specific for IGF1R (si-IGF1R) (5′-AGGCGGGCTTCCGGGAGGTCTCCTT-3′) or si-Scramble (5′-GGAAGAGGTGGAGGCTCGCTCGCGG-3′) were obtained from Invitrogen; Thermo Fisher Scientific, Inc.. The WERI-RB-1 and Y79 cells (5×105/well) were seeded in a six-well plate overnight, and the cells were then transfected with miR-153 mimics (20 nM), mimics NC (20 nM), miR-153 inhibitor (20 nM), inhibitor NC (20 nM), pcDNA-IGF1R (2 μg) or si-IGF1R (100 nM) (Shanghai GenePharma Co., Ltd.) using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h following transfection, cells were harvested, and protein and RNA were then extracted for analyses.
Microarray datasets
The miRNA dataset (GSE7072) was searched and downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). GSE7072 was analyzed through the GPL4879 platform (Agilent-019118 Human miRNA Microarray 2.0) and consisted of three samples. Differentially expressed miRNAs (DEmiRNAs) between normal and RB samples were screened based on GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r/), an interactive web tool. DE-miRNAs were then identified based on the fold change in expression. A heatmap of DE-miRNAs was created using a method of hierarchical clustering by GeneSpring GX, version 7.3 (Agilent Technologies, Inc.).
Cell viability assay
At 48 h following transfection, cell viability in 96-well plates was evaluated using the Cell Counting Kit-8 (CCK8; Dojindo Laboratories, Inc.) assay. Briefly, 10 μl CCK-8 solution (Dojindo Laboratories, Inc). were added to each well and incubated at 37°C for a further 2 h. The absorbance at 560 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc.). The experiments were repeated three times.
Detection of apoptosis by flow cytometry
Cell apoptosis was evaluated using an Annexin V/propidium iodide (PI) apoptosis-detection kit (KeyGen BioTech, Co., Ltd.) according to the manufacturer's protocols. Briefly, at 48 h following trans- fection, cells were centrifuged at 4°C and washed with PBS, and stained with Annexin V and PI for 15 min at room temperature in the dark. The stained cells were then analyzed with EPICS XL-MCL FACScan (Becton-Dickinson and Company).
Bioinformatics analysis
miRNA target prediction tools, including PicTar version 2007 (https://pictar.mdc-berlin.de/) and TargetScan Release 7.0 (http://targetscan.org/) were used to search for the putative targets of miR-153.
Vector construction and luciferase assays
Luciferase reporters were generated based on the Firefly luciferase expressing vector pGL3-control (Promega Corporation). The 3′-UTR fragment of the IGF1R gene and its mutant of the theoretical miR-153 binding site were cloned into the pGL3 control vector (Promega Corporation) to form the reporter vector, wild-type (wt) and mutant-type (mut) of IGF1R, respectively. To construct pGL3-IGF1R-3′UTR, a partial 3′UTR of the IGF1R segment of human IGF1R mRNA containing the putative miR-153 binding sites was amplified and cloned into the vector pGL3-control. Mutations within potential miR-153 binding sites were introduced using the QuikChange Site-Directed Mutagenesis kit (Life Technologies; Thermo Fisher Scientific, Inc.). When the Y79 cells grew to 60-70% confluency, the cells were co-transfected with 100 ng luciferase plasmid and 50 ng Renilla luciferase plasmid along with 100 ng miR-153 mimics/inhibitor or miR-NC using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation for 48 h at 37°C, the luciferase activity was assessed using the dual luciferase reporter kit (Beyotime Institute of Biotechnology). Renilla activity was used to normalize Firefly luciferase activity.
Western blot analysis
At 48 h following transfection, the total protein from cells was obtained using RIPA lysis buffer (Santa Cruz Biotechnology, Inc.) and quantified using a BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). The proteins in the lysates were separated by 10% SDS-PAGE gels and transferred to PVDF membranes (GE Healthcare; Cytiva). After being blocked with a 5% skim milk solution for 1 h at room temperature, the membranes were incubated with specific primary antibodies at 4°C overnight, including E-cadherin (cat. no. ab40772, 1:1,000 dilution), N-cadherin (cat. no. ab202030, 1:1,000 dilution), Vimentin (cat. no. ab45939, 1:1,000 dilution), fibronectin (cat. no. ab2413, 1:1,000 dilution), Snail (cat. no. ab216347, 1:1,000 dilution), Twist (cat. no. ab50581, 1:1,000 dilution), zinc finger E-box binding homeobox 1 (ZEB1; cat. no. ab203829, 1:1,000 dilution) (all from Abcam), IGF1R (cat. no. sc81464, 1:1,000 dilution), PI3K (cat. no. S365290, 1:1,000 dilution), phosphorylated (p)-AKT (cat. no. sc-514032, 1:1,000 dilution), p-MEK1/2 (cat. no. sc-81503, 1:1,000 dilution) (both from Santa Cruz Biotechnology, Inc.), p-Raf/1 (Ser338, 1:1,000; cat. no. 9427; Cell Signaling Technology, Inc.) and β-actin (cat no. sc-84322, Santa Cruz Biotechnology, Inc., 1:1,000 dilution). Subsequently, the mouse anti-rabbit IgG-HRP secondary antibody (cat no. sc2537, Santa Cruz Biotechnology, Inc., 1:1,000 dilution) was added to the membranes followed by incubation for 2 h at room temperature. The protein bands were visualized using an ECL detection system (Thermo Fisher Scientific, Inc.). Semi-quantification was performed using ImageJ version 1.46 (NIH).
Immunofluorescence analysis and immunohistochemistry (IHC)
For immunofluorescence, the cells on the coverslips were fixed with 4% paraformaldehyde for 10 min at 48 h following transfection, and subsequently, the cells were incubated at 4°C overnight in a solution containing primary antibodies specific for caspase-3 (cat. no. sc-7272, Santa Cruz Biotechnology, Inc., 1:200). The cells were then stained with mouse anti-goat IgG-FITC (cat. no. sc2356, Santa Cruz Biotechnology, Inc., 1:200). The cells were counterstained with DAPI for 15 min at room temperature to identify the nuclei and imaged with a confocal laser-scanning microscope (Axiovert 200 M, Zeiss GmbH).
For IHC, the expression of caspase-3 was evaluated by IHC staining as previously described (17) with the following primary antibodies: Caspase-3 (cat. no. sc-7272, Santa Cruz Biotechnology, Inc., 1:100). Samples were photographed under a light Leica DMD 108 microscope (Leica Microsystems GmbH).
Transwell assay
Briefly, a total of 3×104 transfected cells in DMEM without serum were added to the top Matrigel-coated chambers (pore size, 8 μm; Corning, Inc.), while DMEM with 20% FBS (600 μl) was then added to the lower chamber. Following 24 h of incubation at 37°C, cells on the upper side of each membrane were cleaned with a cotton swab. The membranes were then fixed in 20% methanol for 15 min and stained with 0.2% crystal violet (Sigma-Aldrich; Merck KGaA, Germany) at room temperature for 30 min. A total of five visual fields of each insert were randomly selected and photographed under a light microscope (Olympus Corporation) at ×200 magnification.
Wound healing assay
At 48 h following transfection, the cell layer at 80-90% confluency was gently and slowly scratched with a new 200 μl pipette tip across the center of the well. The cells were then washed twice with phosphate-buffered saline (PBS) and incubated in free-serum DMEM. The wound healing images (magnification, ×200) were obtained at the time of wounding (time 0), and at 24 and 48 h after scratching. The wound closure process was observed and photographed under a microscope (Olympus Corporation).
Statistical analysis
The SPSS 23.0 software package (SPSS Inc.) was applied to analyze the data. All data are presented as the mean ± SD. An unpaired Student's t-test was performed for two-group comparisons. Comparisons between multiple groups were analyzed by one-way ANOVA followed by Tukey's post hoc test. The correlation between IGF1R and miR-153 expression was analyzed using Pearson's correlation coefficient. The Chi-squared test was used for association analysis between clinicopathological features of patients with retinoblastoma and miR-153-3p expression profiles. The Kaplan-Meier method was applied to calculate the 5-year overall survival (OS), and the log-rank test was used to evaluate whether there were statistically significant differences in OS. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-153 expression is downregulated in RB tissues and cell lines
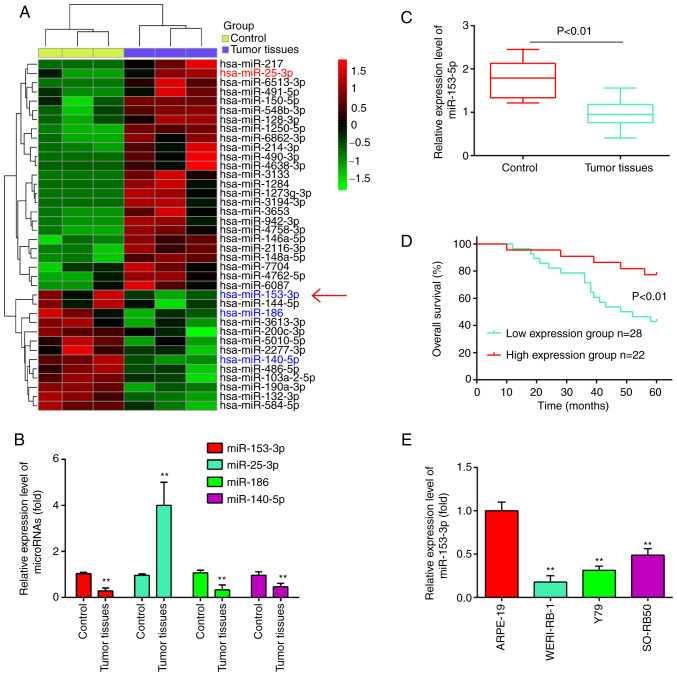
To identify RB-associated miRNAs, the differentially expressed miRNAs (DEmiRNAs) in the GSE dataset (GSE7072) were analyzed using bioinformatics analysis. As shown in Fig. 1A, a number of miRNAs were found to be differentially expressed between these RB tissues and normal retinal tissues. Out of these miRNAs, it was found that miR-25-3p was upregulated, while miR-186 and miR-140-5p were downregulated in the RB tissues (Fig. 1B), which was consistent with the findings of previous studies (18-20). Of particular interest, miR-153 was one of the most significantly downregulated miRNA in the RB tissues (Fig. 1B). Previous studies have indicated that miR-153 functions as a tumor suppressor in several malignancies, such as non-small cell lung cancer (NSCLC) (21) and ovarian carcinoma (OC) (14). Therefore, miR-153 was selected for further analyses.
Figure 1.
miR-153-3p is downregulated in human RB tissues and cell lines. (A) Heatmap of miRNA expression was created using a method of hierarchical clustering by GeneSpring GX, version 7.3. Rows represent groups and columns represent miRNAs. The color key indicates the miRNA expression value; green indicates low expression levels, while red indicates high expression levels. (B) Relative expression of miR-25-3p, miR-186 miR-140-5p and miR-153-3p in the RB tissues. The results (mean ± SD) are from three independent experiments (**P<0.01 vs. control group). (C) Relative expression of miR-153-3p in 50 human RB tissues and 10 normal retinal tissues. (D) Overall survival was compared between patients with RB with low expression level of miR-153-3p and those with a high level of miR-153-3p. (E) Relative expression of miR-153-3p in RB cells and ARPE-19 cells. **P<0.01 vs. control or ARPE-19 cells. RB, retinoblastoma.
Using RT-qPCR assay, miR-153 was detected in 50 RB tissues and 10 normal retinal tissues. As shown in Fig. 1C, the results revealed that the expression levels of miR-153 were significantly lower in the tumor tissues than those in the normal tissues (P<0.01). Moreover, the patients with RB were divided into a high miR-153 expression group and a low expression group based on the median value of miR-153 expression as a cut-off point. As shown in Fig. 1D, patients with a low expression of miR-153 had an evidently shorter overall survival time than those patients with a high expression of miR-153 (P<0.01). In addition, the expression levels of miR-153 in RB cancer cell lines were measured by RT-qPCR. Compared with that in ARPE-19 cells, miR-153 expression was significantly downregulated in the three RB cancer cell lines (Fig. 1E). Based on these findings, miR-153 may be involved in the development of RB.
Subsequently, the association between miR-153 expression and the clinicopathological characteristics of patients with RB was analyzed. As shown in Table II, a low expression of miR-153 was significantly associated with the largest tumor base and differentiation; however, no significant association was observed between the miR-153 expression levels and the patient age, sex, tumor enucleation location, T classification and clinical stage. On the whole, these data indicated that miR-153 expression in RB tumor tissues may serve as a diagnostic and prognostic marker for RB.
Table II.
Association between miR-153-3p and clinicopathological features of patients with retinoblastoma.
| Feature | Total n=50 | miR-153-3p expression
|
P-value | |
|---|---|---|---|---|
| High | Low | |||
| Sex | 0.4741 | |||
| Male | 21 | 8 | 13 | |
| Female | 29 | 14 | 15 | |
| Age at presentation (years) | 0.1212 | |||
| ≤5 | 31 | 11 | 20 | |
| >5 | 19 | 11 | 8 | |
| Tumor enucleation location | 0.1443 | |||
| Right | 24 | 8 | 16 | |
| Left | 26 | 14 | 12 | |
| Largest tumor base (mm) | 0.0199a | |||
| ≤15 | 32 | 18 | 14 | |
| >15 | 18 | 4 | 14 | |
| T classification | 0.1830 | |||
| T1-2 | 28 | 10 | 18 | |
| T3-4 | 22 | 12 | 10 | |
| Clinical stage | 0.2008 | |||
| I-II | 20 | 11 | 9 | |
| III-IV | 30 | 11 | 19 | |
| Differentiation | 0.0025b | |||
| Well and moderate | 16 | 12 | 4 | |
| Poor | 34 | 10 | 24 | |
P<0.05;
P<0.01. The Chi-squared test was used for association analysis between clinicopathological features of patients with retinoblastoma and miR-153-3p expression profiles.
miR-153 overexpression inhibits RB cell proliferation and promotes apoptosis
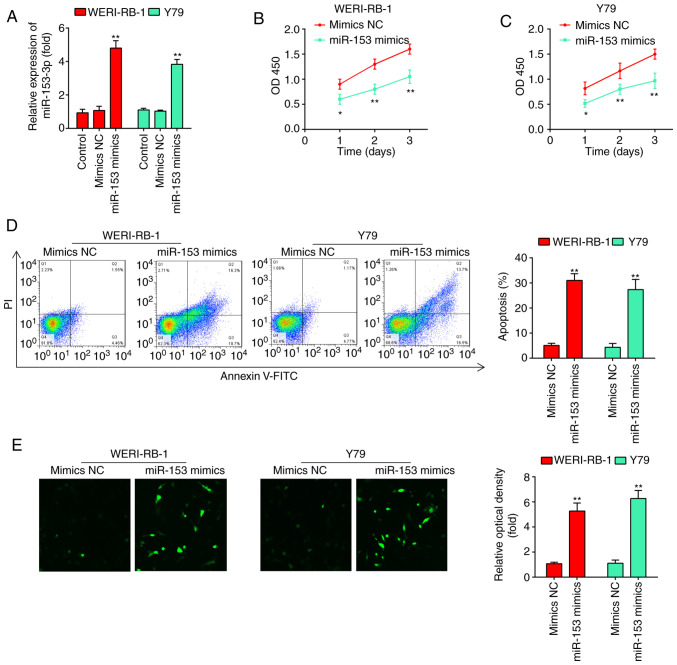
To examine the effects of miR-153 on RB cell proliferation and apoptosis, miR-153 mimics and mimics NC were transfected into the WERI-RB-1 and Y79 cell lines, which exhibited the lowest levels of miR-153 expression among the four cell lines examined. As shown in Fig. 2A, miR-153 expression was significantly increased compared with that in the mimics NC-transfected cells (P<0.01). According to the results of the CCK-8 assay, miR-153 overexpression significantly inhibited the proliferation of the WERI-RB-1 and Y79 cells compared with that in the mimics NC group (Fig. 2B and C). Flow cytometric analyses were subsequently performed to examine the effects of miR-153 on cell apoptosis. It was found that miR-153 overexpression significantly increased the percentage apoptosis in comparison with the mimics NC group (WERI-RB-1 cells: 30.8±1.49 vs. 4.76±0.191%, P=0.01; Y79 cells: 26.8±1.13 vs. 3.36±0.179%, P=0.01; Fig. 2D). In addition, whether miR-153 modulates the expression of apoptosis-associated proteins, such as caspase-3 was determined by immunofluorescence. The expression level of caspase-3 in the cells transfected with miR-153 mimics was evidently increased compared with that in the mimics NC-transfected cells (Fig. 2E). Taken together, these results suggest that miR-153 inhibits the proliferation and induces the apoptosis of RB cell lines.
Figure 2.
miR-153-3p inhibits the proliferation and promotes the apoptosis of RB cells. (A) RT-qPCR was performed to determine the expression levels of miR-153 following miR-153 mimic and miR-153 inhibitor transfection into WERI-RB-1 and Y79 cells. (B and C) Cell viability was measured using a CCK-8 assay following miR-153 mimic and miR-153 inhibitor transfection into WERI-RB-1 and Y79 cells. The results (mean ± SD) are from three independent experiments (*P<0.05, **P<0.01 vs. mimics NC). (D) Apoptosis of WERI-RB-1 and Y79 cells following transfection with miR-153 mimic or mimics NC was determined by flow cytometric analysis (**P<0.01 vs. mimics NC). (E) The expression of caspase-3 in WERB-RI-1 and Y79 cells following transfection with miR-153 mimic or mimics NC was detected by immunofluorescence (**P<0.01 vs. mimics NC). RB, retinoblastoma.
Overexpression of miR-153 inhibits invasion, migration and the epithelial-mesenchymal transformation (EMT) process in RB cells
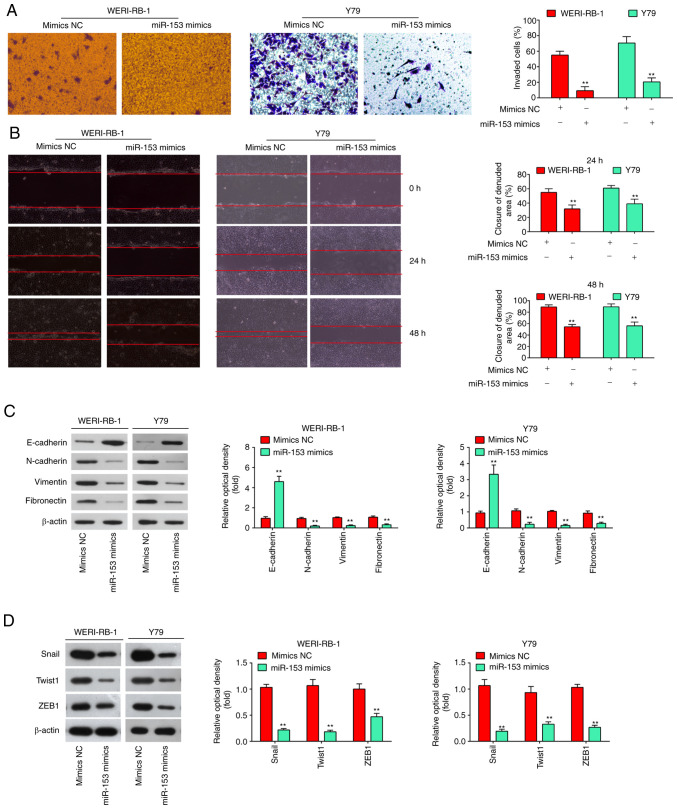
It is well known that cell invasion, migration and the EMT process are crucial for tumor development. Thus, in the present study, Transwell and wound healing assays were performed to examine the effects of miR-153 on cell invasion and migration. It was found that the overexpression of miR-153 in WERI-RB-1 and Y79 cells markedly suppressed cell invasion, compared with the cells transfected with mimics NC (P<0.01, Fig. 3A). As was expected, the WERI-RB-1 and Y79 cells transfected with miR-153 mimics also exhibited a decreased migration rate, compared with the mimics NC group (P<0.01, Fig. 3B). E-cadherin and vimentin are the major markers of EMT, and EMT plays an important role in tumor metastases (22). Thus, the present study first detected whether miR-153 modulates the expression of EMT markers. It was observed that the level of E-cadherin (epithelial marker) was significantly increased, while the levels of N-cadherin, Vimentin, fibronectin (mesenchymal markers) were markedly decreased in the miR-153 mimics group compared with the mimics NC group (Fig. 3C). It is well known that EMT-promoting transcription factors, such as Snail, Twist and ZEB1 play a key role in the regulation of the EMT process (23). Thus, the present study also examined the effects of miR-153 on these EMT-promoting transcription factors. As shown in Fig. 3D, the expression levels of Snail, Twist and ZEB1 were significantly decreased in the miR-153 mimics group compared with the mimics NC group. Taken together, these findings demonstrated that miR-153 suppresses the invasion and migration of RB cells in vitro probably by inhibiting the EMT process.
Figure 3.
miR-153-3p inhibits the invasion, migration and EMT process in RB cells in vitro. (A) Transwell assay was used to determine the invasion of RB cells following transfection of miR-153 mimics into WERI-RB-1 and Y79 cells. The results (mean ± SD) are from three independent experiments (**P<0.01 vs. mimics NC). (B) Wound healing assay was used to determine the migration of RB cells following transfection of miR-153 mimics into WERI-RB-1 and Y79 cells. The results (mean ± SD) are from three independent experiments (**P<0.01 vs. mimics NC). (C) Western blot analysis was used to determine the expression level of EMT-associated proteins following transfection of miR-153 mimics into WERI-RB-1 and Y79 cells. (D) The expression levels of Snail, Twist and ZEB1 were measured by western blot analysis following transfection of miR-153 mimics into WERI-RB-1 and Y79 cells. The results (mean ± SD) are from three independent experiments (**P<0.01 vs. mimics NC). RB, retinoblastoma; EMT, epithelial-mesenchymal transition; ZEB1, zinc finger E-box binding homeobox 1.
IGF1R is a direct target of miR-153 in RB cells
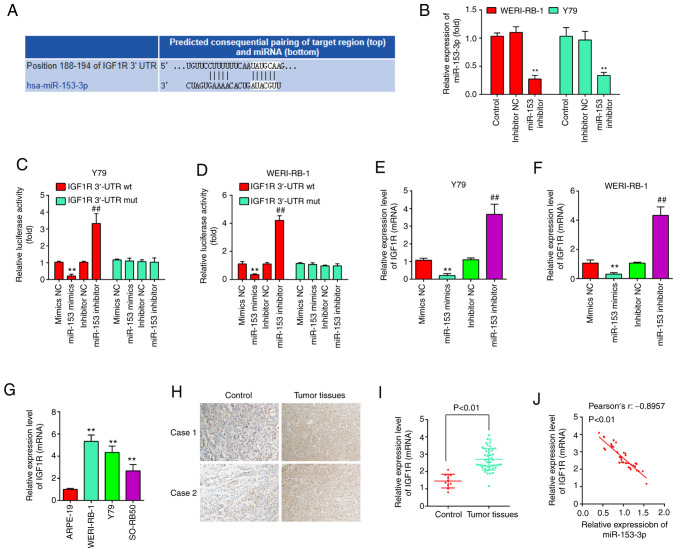
To explore the potential mechanisms through which miR-153 functions as a tumor suppressor in RB, potential target genes of miR-153 were screened using TargetScan 7.0 and PicTar. Bioinformatics analysis indicated that one potential binding site for miR-153 was found in the 3′-UTR region of IGF1R mRNA (Fig. 4A). To verify whether miR-153 directly binds to IGF1R, a dual luciferase reporter assay was performed using the Y79 and WERI-RB-1 cells. First, the WERI-RB-1 and Y79 cells were transfected with miR-153 inhibitor and inhibitor-NC as a control, and RT-qPCR was then performed to determine the transfection efficiency. The results revealed that miR-153 expression was significantly decreased in the WERI-RB-1 and Y79 cells following transfection with miR-153 inhibitor (Fig. 4B). The results of luciferase reporter assay then revealed that transfection with miR-153 mimics significantly inhibited the luciferase activity combined with the IGF1R-3′-UTR wt reporter, while transfection with miR-153 inhibitor led to an increase in luciferase activity; however, no significant changes were observed using the IGF1R 3′-UTR mut reporter with miR-153 mimics or inhibitor (Fig. 4C and D). In addition, to confirm whether miR-153 regulate IGF1R expression, qRT-PCR was performed. It was observed that IGF1R was significantly reduced following miR-153 mimics transfection, whereas increased by miR-153 inhibitor in Y79 cells and WERI-RB-1 cells (Fig. 4E and F). In addition, the expression levels of IGF1R were detected in RB tissues from two patients with RB by IHC assay. Compared with the control group, IGF1R expression was markedly increased in RB tissues (Fig. 4G). Moreover, the IGF1R levels were also measured in RB cell lines, as well as in 50 pairs of RB tissues and normal tissue by RT-qPCR. The results revealed that the IGF1R expression level was higher in both RB cells lines and tumor tissues compared with the APRE-19 cells and normal tissues (Fig. 4H and I). Additionally, an obvious inverse correlation was observed between the expression of IGF1R and miR-153 in the tissue samples (r=-0.8957, P<0.01, Fig. 4J). On the whole, these data suggest that IGF1R may be a functional target of miR-153.
Figure 4.
IGF1R is a target of miR-153 and is upregulated in RB tissues and cells. (A) Prediction of IGF1R as a target of miR-153-3p. (B) The transfection efficiency of miR-153 inhibitor was assessed by RT-qPCR. **P<0.01 vs. control. (C and D) Firefly luciferase reporter activity in Y79 cells and WERI-RB-1 cells transfected with mimics NC or miR-153 mimics or inhibitor NC or miR-153 inhibitor following transfection with wt-IGF1R-3′-UTR reporter or mut-IGF1R-3′-UTR reporter plasmid, respectively (**P<0.01 vs. mimics NC, ##P<0.01 vs. inhibitor NC). (E and F) mRNA level of IGF1R in Y79 cells and WERI-RB-1 cells transfected with miR-153 inhibitor and miR-153 mimics measured by RT-qPCR (**P<0.01 vs. mimics NC, ##P<0.01 vs. inhibitor NC). (G) The relative expression levels of IGF1R in two paired RB tissues and normal retinal tissues were measured by immunohistochemistry (**P<0.01 vs. ARPE-19). (H) Relative expression of IGF1R in RB cells compared to ARPE-19 cells. (I) Relative expression of IGF1R in RB tissues compared to normal retinal tissues. (J) Correlation between IGF1R and miR-153 expression analyzed using Pearson's correlation coefficient in RB tissues. RB, retinoblastoma; IGF1R, insulin-like growth factor 1 receptor.
Knockdown of IGF1R reverses the phenotypes induced by miR-153
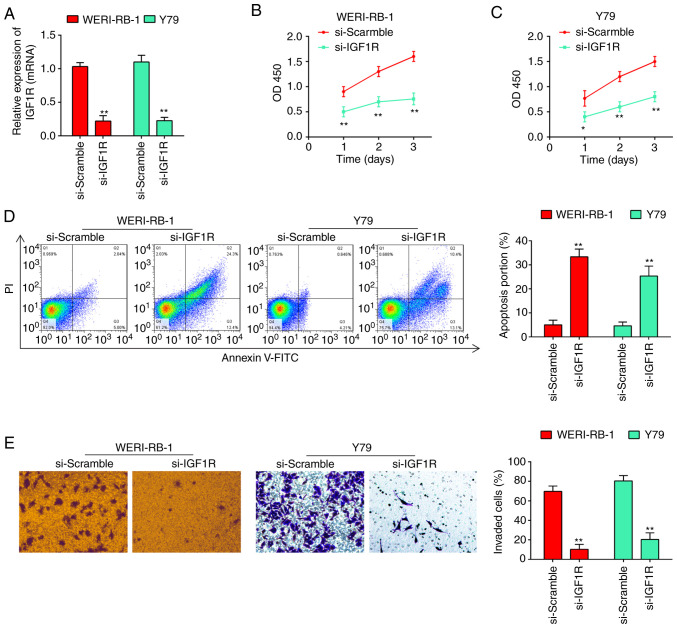
As is known, the IGF1R oncogene is involved in the carcinogenesis of several human cancers (24-26). In the present study, given that IGF1R was upregulated in RB tissues, the possible effects of IGF1R on RB cell biological behaviors were then investigated. The results of RT-qPCR revealed that the expression of IGF1R was knocked down by transfection of the WERI-RB-1 and Y79 cells with siRNA (Fig. 5A). CCK-8 assay revealed that the knockdown of IGF1R led to a marked reduction in the viability of WERI-RB-1 (Fig. 5B) and Y79 cells (Fig. 5C). Additionally, as shown by flow cytometric analysis, the cell apoptotic rate was significantly increased following transfection with si-IGF1R (from 4.24±0.332% to 32.7±1.27% for WERI-RB-1 cells, and from 3.83±0.311% to 25.1±1.31% for Y79 cells, P<0.01; Fig. 5D). As was expected, the invasive abilities of the WERI-RB-1 and Y79 cells were also inhibited following transfection with si-IGF1R (Fig. 5E). Collectively, these findings suggested that miR-153 probably inhibited RB cell proliferation and invasion, and promoted apoptosis by downregulating IGF1R.
Figure 5.
Silencing of IGF1R reverses the phenotypes caused by miR-153. (A) siRNA efficiency of si-Scramble, si-IGF1R, respectively, in WERI-RB-1 and Y79 cells. (B) Viability of WERI-RB-1 cells was measured following transfection with si-Scramble or si-IGF1R by CCK-8 assay. The results (mean ± SD) are from three independent experiments (**P<0.01. vs. si-Scramble). (C) Viability of Y79 cells was measured following transfection with si-Scramble or si-IGF1R by CCK-8 assay. The results (mean ± SD) are from three independent experiments (*P<0.05, **P<0.01. vs. si-Scramble). (D) Apoptosis of WERI-RB-1 and Y79 cells following transfection with si-Scramble or si-IGF1R was determined by flow cytometric analysis. Shown in the right panel are the percentages of apoptotic cells (**P<0.01. vs. si-Scramble). Prior to flow cytometry, the tested cells were firstly stained with Annexin V-fluorescein isothiocyanate. (E) Silencing of IGF1R attenuated the invasion of RB cells. Transwell assay was used to determine the invasion of RB cells following transfection of si-Scramble, and si-IGF1R, respectively, in WERI-RB-1 and Y79 cells. The results (mean ± SD) are from three independent experiments (**P<0.01. vs. si-Scramble). RB, retinoblastoma; IGF1R, insulin-like growth factor 1 receptor.
miR-153 inhibits retinoblastoma proliferation, invasion and migration by targeting IGF1R
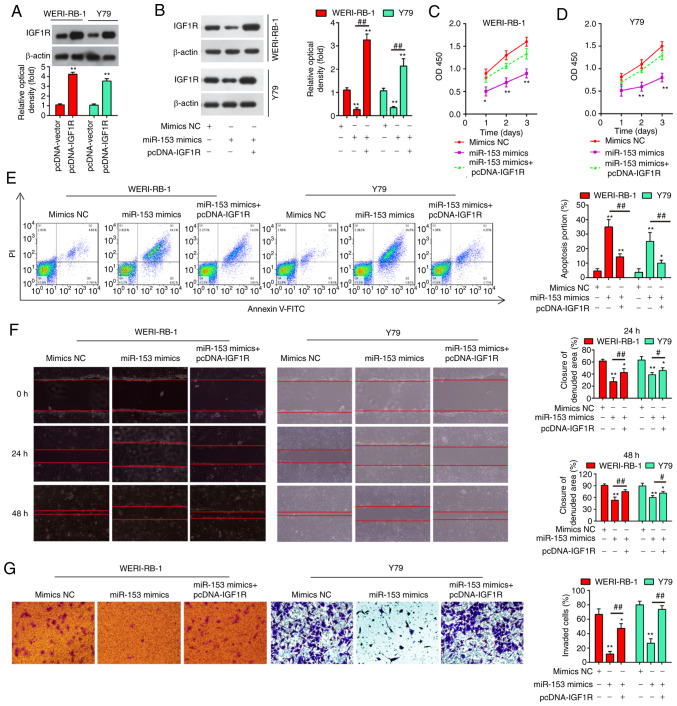
To explore whether IGF1R mediates the antitumor effects of miR-153, the WERI-RB-1 and Y79 cells were co-transfected with miR-153 mimics and pcDNA-IGF1R. Firstly, the protein expression of IGF1R was examined by western blot analysis in the WERI-RB-1 and Y79 cells following pcDNA-IGF1R transfection. As shown in Fig. 6A, in both the cell lines tested, the expression levels of IGF1R were significantly increased compared with the pcDNA-vector-transfected cells. Moreover, IGF1R expression was significantly suppressed in the cells transfected with miR-153 mimics compared with the mimics NC-transfected cells (P<0.01), while the suppression of IGF1R was fully reversed in the cells transfected with pcDNA-IGF1R, with a significantly upregulated expression level of IGF1R (Fig. 6B). Consistently, the suppressive effects of miR-153 on cell proliferation were evidently impaired by transfection with pcDNA-IGF1R (Fig. 6C and D), while the apoptotic rate of the RB cells was markedly decreased (Fig. 6E). Additionally, the results of Transwell and wound healing assay demonstrated that the suppression of RB cell invasion and migration by miR-153 was also significantly attenuated by the overexpression of IGF1R (Fig. 6F and G). These findings thus demonstrate that IGF1R is involved in the tumor-suppressive roles of miR-153 in RB cells.
Figure 6.
Overexpression of IGF1R impairs the effects of miR-153 on RB cells. (A) Protein expression of IGF1R was examined by western blot analysis in WERI-RB-1 and Y79 cells following pcDNA-IGF1R transfection. The results (mean ± SD) are from three independent experiments (**P<0.01 vs. pcDNA vector group). (B) Expression level of IGF1R in RB cells determined by western blot analysis. WERI-RB-1 and Y79 cells were co-transfected with miR-153 mimics, mimics NC, or pcDNA-IGF1R. The results (mean ± SD) are from three independent experiments (**P<0.01 vs. mimics NC group; ##P<0.01 vs. miR-153 mimics group). (C) Cell viability was measured in WERI-RB-1 cells transfected with mimics NC or miR-153 mimics or miR-153 mimics plus pcDNA-IGF1R. The results (mean ± SD) are from three independent experiments (*P<0.05 and **P<0.01 vs. miR-153 mimics group). (D) Cell viability was measured in Y79 cells transfected with mimics NC or miR-153 mimics or miR-153 mimics plus pCDNA-IGF1R. The results (mean ± SD) are from three independent experiments (*P<0.05 and **P<0.01 vs. miR-153 mimics group). (E) Apoptosis of WERI-RB-1 and Y79 cells transfected with mimics NC or miR-153 mimics or miR-153 mimics plus pCDNA-IGF1R, was measured by flow cytometric analysis. The results (mean ± SD) are from three independent experiments (*P<0.05 and **P<0.01 vs. mimics NC group; ##P<0.01 vs. miR-153 mimics group). (F) Invasion of WERI-RB-1 and Y79 cells transfected with mimics NC or miR-153 mimics or miR-153 mimics plus pCDNA-IGF1R, was measured by Transwell assay. The results (mean ± SD) are from three independent experiments (*P<0.05 and **P<0.01 vs. mimics NC group; #P<0.05 and ##P<0.01 vs. miR-153 mimics group). (G) Migration of WERI-RB-1 and Y79 cells transfected with mimics NC or miR-153 mimics or miR-153 mimics plus pCDNA-IGF1R, was measured by wound healing assay. The results (mean ± SD) are from three independent experiments (*P<0.05 and **P<0.01 vs. mimics NC group; ##P<0.01 vs. miR-153 mimics group). RB, retinoblastoma; IGF1R, insulin-like growth factor 1 receptor.
miR-153-3p deactivates the PI3K/Akt and MEK pathways in RB via IGF1R
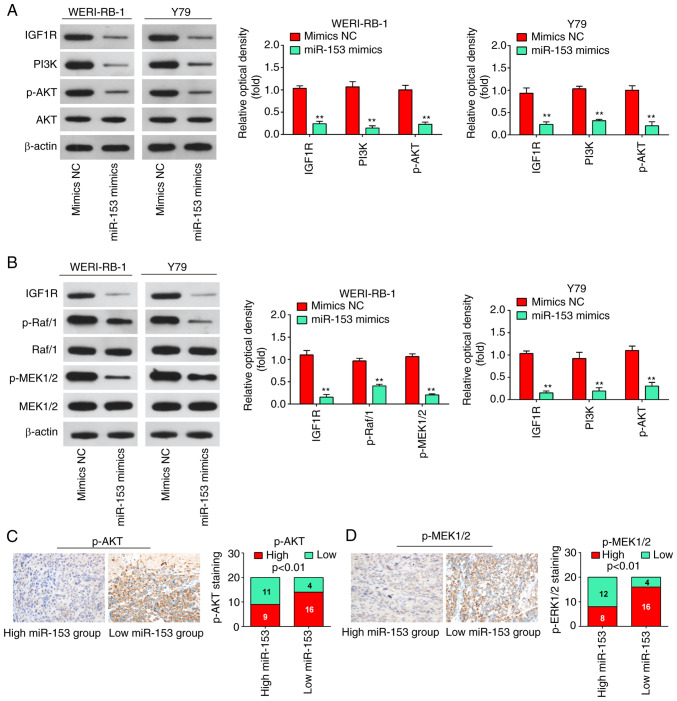
IGF1R has recently been implicated in the activation of the PI3K/Akt pathway and Raf1/MEK pathway, which affects the carcinogenesis and development of numerous types of cancer (27,28). Therefore, the present study attempted to identify whether miR-153-3p is able to deactivate the PI3K/AKT and Raf1/MEK pathways in RB cells. The results of western blot analysis revealed that the expression levels of IGF1R, PI3K, p-AKT, p-Raf/1 and p-MEK1/2 in the WERI-RB-1 and Y79 cells were significantly decreased following miR-153 mimics transfection (Fig. 7A and B), suggesting that miR-153 may have inactivated the PI3K/Akt and MEK pathway in RB cells by targeting IGF1-R. It was also found that a lower miR-153 expression in RB tissues was associated with an increased p-AKT and p-MEK1/2 expression (P<0.01, Fig. 7C and D). Collectively, these data suggest that miR-153 probably suppresses RB cell growth and metastasis via the suppression of the IGF1R/Raf/MEK and IGF1R/PI3K/AKT signaling pathways.
Figure 7.
miR-153 deactivates the PI3K/AKT and Raf/MEK pathway in WERI-RB-1 and Y79 cells by decreasing IGF1R expression. (A) Following transfection, western blot analysis was performed to measure IGF1R, PI3K, p-Akt and Akt expression. The bands were semi-quantitatively analyzed by using ImageJ software, normalized to β-actin density. **P<0.01 vs. mimics NC group. (B) Following transfection, western blot analysis was performed to measure IGF1R, Raf/1, p-Raf/1, p-MEK1/2, and MEK1/2 expression. The bands were semi-quantitatively analyzed by using ImageJ software, normalized to β-actin density. **P<0.01 vs. mimics NC group. (C) Immunohistochemistry was conducted to detect p-AKT in RB tissues with high or low miR-153 expression. Bar graphs demonstrated a significant inverse association between miR-153 and p-AKT expression in RB tissues (n=50). Data are presented as the mean ± standard deviation of three individual experiments. (D) Immunohistochemistry was conducted to detect p-MEK1/2 in RB tissues with high or low miR-153 expression. Bar graphs demonstrated a significant inverse association between miR-153 and p-MEK1/2 expression in RB tissues (n=50). Data are presented as the mean ± standard deviation of three individual experiments. RB, retinoblastoma; IGF1R, insulin-like growth factor-1 receptor; MEK, mitogen activated protein kinase kinase; miR, microRNA; p, phosphorylated.
Discussion
In the present study, it was found that miR-153 was significantly downregulated in RB tissues and cell lines, and a low miR-153 expression was closely associated with a larger tumor base and differentiation. The overexpression of miR-153 inhibited RB cell proliferation and promoted apoptosis, and suppressed the migration and invasion of the WERI-RB-1 and Y79 cell lines. In addition, IGF1R was identified as a direct target of miR-153 in RB cells. Notably, the results of the present study demonstrate that miR-153 may exert its antitumor effects on RB cells by blocking the activation of the PI3K/AKT and MEK pathway. The findings presented herein provide valuable clues towards the understanding of the specific tumor suppressive function and regulatory mechanisms of miR-153 in RB.
Accumulating evidence has indicated that miRNAs, such as miR-124 (29) and miR-25-3p (9), play important roles in the development of human RB. However, the functional significance of the unique role of miRNAs has yet to be elucidated in RB. In the present study, miRNA microarray profiling was performed using a GSE dataset (GSE7072), which was searched and downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo). miR-153 was found to be one of the most significantly downregulated miRNAs in RB tissues. Furthermore, it was confirmed that miR-153 was frequently downregulated in RB tissues and cell lines. In addition, the association between miR-153 expression and the clinicopathological parameters of patients with RB was investigated, and the data indicated that miR-153 in RB was significantly associated with a larger tumor base and differentiation, while no significant association was observed between miR-153 expression levels and patient age, sex, tumor enucleation location, T classification and clinical stage. Moreover, a low tumorous miR-153 expression was found to be associated with a poor overall survival of patients with RB. Taken together, these data suggest that miR-153 may be an effective biomarker for RB prognosis.
Several studies have focused on comprehensive investigations into the biological functions of miR-153 in various types of cancer. For example, Li et al found that miR-153 inhibited EMT by targeting metadherin in breast cancer (30). Zuo et al demonstrated that miR-153 may play a suppressive role in breast tumor growth and metastasis via direct targeting of RUNX2 (9). Another study demonstrated that the upregulation of miR-153 inhibited the proliferation, invasion and migration of triple-negative breast cancer (TNBC) cells (31). However, the specific function of miR-153 in RB progression has not been fully elucidated. In the present study, the results revealed that the overexpression of miR-153 inhibited the proliferation and invasion, and promoted the apoptosis of RB cells. The present study, for the first time, to the best of our knowledge, confirmed that miR-153 functions as a tumor suppressor in RB. These findings are consistent with those of previous studies demonstrating the tumor suppressive role of miR-153 in other types of tumors (32,33).
It has been well documented that IGF1R is involved in the carcinogenesis of a number of human cancers by activating the PI3K/AKT and MEK pathways (27,34). Recent studies have demonstrated that it acts as an important target of several miRNAs in RB, including miR-98 (35) and miR-145 (36). Recently, using a microarray assay, Song et al found that miR-153 was significantly downregulated in venous smooth muscle cells (VSMCs) under conditions of stretch stress, and an enhanced miR-153 expression reduced IGF-1R expression, contributing to VSMC proliferation (37). However, the association between miR-153 and IGF1R in RB has not yet been clarified. In the present study, miR-153 exerted a potent inhibitory effect on the luciferase activity of the pGL3-report vector linked to IGF1R 3′UTR. In addition, it was found that the levels of IGF1R were significantly upregulated in RB tissues and cell lines, and its expression was inversely correlated with miR-153 expression in RB tissues. Furthermore, the overexpression of IGF1R reversed the suppressive effects induced by miR-153 upregulation in RB cells. Taken together, these data indicate that miR-153 exerts its antitumor effects by targeting IGF1R.
As a vital oncogene in the development and maintenance of cancers, IGF1R triggers numerous downstream signaling cascades, including the PI3K/AKT and Raf1/MEK1/2/ERK1/2 signaling pathways, which are involved in the tumorigenesis, apoptosis and metastasis of a variety of cancer types (38-40). In the present study, the results revealed that the upregulation of miR-153 reduced the levels of key PI3K/AKT and Raf1/MEK1/2/ERK1/2 pathway proteins by suppressing IGF1R. The data suggested that miR-153 suppressed IGF1R to inhibit PI3K/AKT and Raf1/MEK1/2/ERK1/2 pathway activity, thus suppressing the malignancy of RB cells.
Retinoblastoma gene (RB1), located at chromosome 13q14, was the first described tumor suppressor gene and its critical role in cancer has attracted increasing attention (41). It has been reported that the loss of the RB1 gene in RB confers limitless replicative potential to retinoblasts and is a rate-limiting step for RB tumorigenesis (42). When both alleles of the RB1 gene in RB are lost, the function of RB protein (pRB) is curtailed, which results in abnormal cell proliferation and tumor formation (43,44). Previous studies have reported that the loss of function of pRB is not only associated with RB, but also with multiple other non-ocular malignancies, such as prostate cancer and hepatocellular carcinoma (45,46). In view of the association between RB1 genes and RB, the authors aim to further study whether miR-153 exert antitumor effects through the regulation of the RB1 gene in RB in the future.
There are some limitations to the present study. For example, IGF1R is not unique as a target gene of miR-153; there are other genes as well. In the future, other target genes of miR-153 or other differentially expressed miRNAs found in the present study also need to be carefully examined for their role in RB development. In addition, the expression level of miR-153 was detected in the RB cell lines, Y79 and WERI-RB-1, which have somewhat different characteristics, with the Y79 cells exhibiting inherent metastatic properties and the WERI-RB-1 cells exhibiting non-metastatic properties (47). The results of the present study revealed that the expression levels of miR-153 in the Y79 cells were higher than those in the Weri-RB1 cells (Fig. 1E), suggesting that the miR-153 expression level may not be associated with the aggressiveness of those cell lines. In the future, the authors aim to examine the expression of miR-153 in more metastatic and non-metastatic RB tissues samples in order to assess whether miR-153 is directly associated with the aggressiveness of RB.
In conclusion, the present study demonstrated that miR-153 inhibited the proliferation, migration and invasion, and promoted the apoptosis of WERI-RB-1 and Y79-45 cell lines by inhibiting the IGF1R/PI3K/AKT and IGF1R/Raf/MEK signaling pathways. These findings suggest that miR-153 may serve as a potential biomarker for the prognosis and a therapeutic target for patients with RB.
Acknowledgments
Not applicable.
Funding Statement
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LG was responsible for main conception of the study and the draft of the manuscript. YB, TN, YL, RC, SJ and SL assisted in the design of the study and performed the statistical analysis. YB and TN assisted with the revision of the manuscript and participated in its design. LG and YB confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All individuals provided written informed consent for the use of human specimens for clinical research. The present study was approved by The First People's Hospital of Shangqiu Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dimaras H, Dimba EA, Gallie BL. Challenging the global retinoblastoma survival disparity through a collaborative research effort. Br J Ophthalmol. 2010;94:1415–1416. doi: 10.1136/bjo.2009.174136. [DOI] [PubMed] [Google Scholar]
- 2.Narang S, Mashayekhi A, Rudich D, Shields CL. Predictors of long-term visual outcome after chemoreduction for management of intraocular retinoblastoma. Clin Exp Ophthalmol. 2012;40:736–742. doi: 10.1111/j.1442-9071.2012.02757.x. [DOI] [PubMed] [Google Scholar]
- 3.Shields CL, Shields JA. Basic understanding of current classification and management of retinoblastoma. Curr Opin Ophthalmol. 2006;17:228–234. doi: 10.1097/01.icu.0000193079.55240.18. [DOI] [PubMed] [Google Scholar]
- 4.Chen HM, Ong SJ, Chao AN, Liou KL, Jung SM, Kao LY. Histopathologic findings after selective ophthalmic arterial injection of melphalan for retinoblastoma. Taiwan J Ophthalmol. 2019;9:262–266. doi: 10.4103/tjo.tjo_119_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 8.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuo Z, Ye F, Liu Z, Huang J, Gong Y. MicroRNA-153 inhibits cell proliferation, migration, invasion and epithelial-mesenchymal transition in breast cancer via direct targeting of RUNX2. Exp Ther Med. 2019;17:4693–4702. doi: 10.3892/etm.2019.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Sun J, Bai Z, Li H, He S, Chen R, Che X. MicroRNA-153 acts as a prognostic marker in gastric cancer and its role in cell migration and invasion. Onco Targets Ther. 2015;8:357–364. doi: 10.2147/OTT.S78236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu G, Li B, Sun L, An C. MicroRNA-153 inhibits osteosarcoma cells proliferation and invasion by targeting TGF-β2. PLoS One. 2015;10:e0119225. doi: 10.1371/journal.pone.0119225. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Chen WJ, Zhang EN, Zhong ZK, Jiang MZ, Yang XF, Zhou DM, Wang XW. MicroRNA-153 expression and prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:8671–8675. [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J, Liu Y, Zhao Y, Tian F, Wang G. miR-153-3p suppresses inhibitor of growth protein 2 expression to function as tumor suppressor in acute lymphoblastic leukemia. Technol Cancer Res Treat. 2019;18:1533033819852990. doi: 10.1177/1533033819852990. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Li C, Zhang Y, Zhao W, Cui S, Song Y. miR-153-3p regulates progression of ovarian carcinoma in vitro and in vivo by targeting MCL1 gene. J Cell Biochem. 2019;120:19147–19158. doi: 10.1002/jcb.29244. [DOI] [PubMed] [Google Scholar]
- 15.Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, Chen C. miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis. 2018;21:849–860. doi: 10.1007/s10456-018-9630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Villar J, Cabrera-Benitez NE, Valladares F, García-Hernández S, Ramos-Nuez Á, Martín-Barrasa JL, Muros M, Kacmarek RM, Slutsky AS. Tryptase is involved in the development of early ventilator-induced pulmonary fibrosis in sepsis-induced lung injury. Crit Care. 2015;19:138. doi: 10.1186/s13054-015-0878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan W, Wan W, Long Y, Li Q, Jin X, Wan G, Zhang F, Lv Y, Zheng G, Li Z, Zhu Y. MiR-25-3p promotes malignant phenotypes of retinoblastoma by regulating PTEN/Akt pathway. Biomed Pharmacother. 2019;118:109111. doi: 10.1016/j.biopha.2019.109111. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Han M, Zhang C. Overexpression of microRNA-186 inhibits angiogenesis in retinoblastoma via the hedgehog signaling pathway by targeting ATAD2. J Cell Physiol. 2019;234:19059–19072. doi: 10.1002/jcp.28545. [DOI] [PubMed] [Google Scholar]
- 20.Liao Y, Yin X, Deng Y, Peng X. MiR-140-5p suppresses retinoblastoma cell growth via inhibiting c-Met/AKT/mTOR pathway. Biosci Rep. 2018;38:BSR20180776. doi: 10.1042/BSR20180776. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Shan N, Shen L, Wang J, He D, Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem Biophys Res Commun. 2015;456:385–391. doi: 10.1016/j.bbrc.2014.11.093. [DOI] [PubMed] [Google Scholar]
- 22.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 23.Guarino M. Epithelial-mesenchymal transition and tumour invasion. Int J Biochem Cell Biol. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Hakuno F, Takahashi SI. IGF1 receptor signaling pathways. J Mol Endocrinol. 2018;61:T69–T86. doi: 10.1530/JME-17-0311. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S, Soutto M, Chen Z, Blanca Piazuelo M, Kay Washington M, Belkhiri A, Zaika A, Peng D, El-Rifai W. Activation of IGF1R by DARPP-32 promotes STAT3 signaling in gastric cancer cells. Oncogene. 2019;38:5805–5816. doi: 10.1038/s41388-019-0843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Pan W, Shen Y, Chen Z, Zhang L, Zhang Y, Luo Q, Ying X. IGF1/IGF1R and microRNA let-7e down-regulate each other and modulate proliferation and migration of colorectal cancer cells. Cell Cycle. 2018;17:1212–1219. doi: 10.1080/15384101.2018.1469873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kooijman R, Lauf JJ, Kappers AC, Rijkers GT. Insulin-like growth factor induces phosphorylation of immunoreactive insulin receptor substrate and its association with phosphatidylinositol-3 kinase in human thymocytes. J Exp Med. 1995;182:593–597. doi: 10.1084/jem.182.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu HW, Lin X, Zhu YX, Lan X, Kuang Y, Wang YZ, Ke ZG, Yuan T, Chen P. Circ-IGF1R has pro-proliferative and anti-apoptotic effects in HCC by activating the PI3K/AKT pathway. Gene. 2019;716:144031. doi: 10.1016/j.gene.2019.144031. [DOI] [PubMed] [Google Scholar]
- 29.Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem. 2018;119:3853–3863. doi: 10.1002/jcb.26464. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Zhai L, Zhao C, Lv S. MiR-153 inhibits epithelial-mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat. 2015;150:501–509. doi: 10.1007/s10549-015-3346-y. [DOI] [PubMed] [Google Scholar]
- 31.Shi D, Li Y, Fan L, Zhao Q, Tan B, Cui G. Upregulation of miR-153 inhibits triple-negative breast cancer progression by targeting ZEB2-mediated EMT and contributes to better prognosis. Onco Targets Ther. 2019;12:9611–9625. doi: 10.2147/OTT.S223598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo J, Zhao M, Fan Z, Liu B, Wang Y, Li Y, Lv P, Xing L, Zhang X, Shen H. MicroRNA-153-3p regulates cell proliferation and cisplatin resistance via Nrf-2 in esophageal squamous cell carcinoma. Thorac Cancer. 2020;11:738–747. doi: 10.1111/1759-7714.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng HF, Yan S, Wu SF. MicroRNA-153-3p suppress cell proliferation and invasion by targeting SNAI1 in melanoma. Biochem Biophys Res Commun. 2017;487:140–145. doi: 10.1016/j.bbrc.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 34.Scheidegger KJ, Du J, Delafontaine P. Distinct and common pathways in the regulation of insulin-like growth factor-1 receptor gene expression by angiotensin II and basic fibroblast growth factor. J Biol Chem. 1999;274:3522–3530. doi: 10.1074/jbc.274.6.3522. [DOI] [PubMed] [Google Scholar]
- 35.Guo L, Bai Y, Ji S, Ma H. MicroRNA-98 suppresses cell growth and invasion of retinoblastoma via targeting the IGF1R/k-Ras/Raf/MEK/ERK signaling pathway. Int J Oncol. 2019;54:807–820. doi: 10.3892/ijo.2019.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Yang H, Nie Y, Xing Y. miR-145 regulates the proliferation and apoptosis of Y79 human retinoblastoma cells by targeting IGF-1R. Int J Clin Exp Pathol. 2018;11:4331–4338. [PMC free article] [PubMed] [Google Scholar]
- 37.Song L, Duan P, Guo P, Li D, Li S, Xu Y, Zhou Q. Downregulation of miR-223 and miR-153 mediates mechanical stretch-stimulated proliferation of venous smooth muscle cells via activation of the insulin-like growth factor-1 receptor. Arch Biochem Biophys. 2012;528:204–211. doi: 10.1016/j.abb.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Knowlden JM, Hutcheson IR, Barrow D, Gee JM, Nicholson RI. Insulin-like growth factor-I receptor signaling in tamoxifen-resistant breast cancer: A supporting role to the epidermal growth factor receptor. Endocrinology. 2005;146:4609–4618. doi: 10.1210/en.2005-0247. [DOI] [PubMed] [Google Scholar]
- 39.Rozen F, Pollak M. Inhibition of insulin-like growth factor I receptor signaling by the vitamin D analogue EB1089 in MCF-7 breast cancer cells: A role for insulin-like growth factor binding proteins. Int J Oncol. 1999;15:589–594. doi: 10.3892/ijo.15.3.589. [DOI] [PubMed] [Google Scholar]
- 40.LeRoith D, Werner H, Neuenschwander S, Kalebic T, Helman LJ. The role of the insulin-like growth factor-I receptor in cancer. Ann N Y Acad Sci. 1995;766:402–408. doi: 10.1111/j.1749-6632.1995.tb26689.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhu L. Tumour suppressor retinoblastoma protein Rb: A transcriptional regulator. Eur J Cancer. 2005;41:2415–2427. doi: 10.1016/j.ejca.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Mu G, Liu H, Zhou F, Xu X, Jiang H, Wang Y, Qu Y. Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 down-regulation in retinoblastomas. Hum Pathol. 2010;41:493–502. doi: 10.1016/j.humpath.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 43.Dalgard CL, Gonzalez M, de Niro JE, O'Brien JM. Differential microRNA-34a expression and tumor suppressor function in retinoblastoma cells. Invest Ophthalmol Vis Sci. 2009;50:4542–4551. doi: 10.1167/iovs.09-3520. [DOI] [PubMed] [Google Scholar]
- 44.Ganguly A, Shields CL. Differential gene expression profile of retinoblastoma compared to normal retina. Mol Vis. 2010;16:1292–1303. [PMC free article] [PubMed] [Google Scholar]
- 45.Maddison LA, Sutherland BW, Barrios RJ, Greenberg NM. Conditional deletion of Rb causes early stage prostate cancer. Cancer Res. 2004;64:6018–6025. doi: 10.1158/0008-5472.CAN-03-2509. [DOI] [PubMed] [Google Scholar]
- 46.Hui AM, Li X, Makuuchi M, Takayama T, Kubota K. Over-expression and lack of retinoblastoma protein are associated with tumor progression and metastasis in hepatocellular carcinoma. Int J Cancer. 1999;84:604–608. doi: 10.1002/(SICI)1097-0215(19991222)84:6<604::AID-IJC11>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Guihurt Santiago J, Burgos-Tirado N, Lafontaine DD, Mendoza Sierra JC, Camacho RH, Vecchini Rodríguez CM, Morales-Tirado V, Flores-Otero J. Adhesion G protein-coupled receptor, ELTD1, is a potential therapeutic target for retinoblastoma migration and invasion. BMC Cancer. 2021;21:53. doi: 10.1186/s12885-020-07768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.