Abstract
The COVID-19 pandemic caused by SARS-CoV-2 has affected millions of people since its beginning in 2019. The propagation of new lineages and the discovery of key mechanisms adopted by the virus to overlap the immune system are central topics for the entire public health policies, research and disease management. Since the second semester of 2020, the mutation E484K has been progressively found in the Brazilian territory, composing different lineages over time. It brought multiple concerns related to the risk of reinfection and the effectiveness of new preventive and treatment strategies due to the possibility of escaping from neutralizing antibodies. To better characterize the current scenario we performed genomic and phylogenetic analyses of the E484K mutated genomes sequenced from Brazilian samples in 2020. From October 2020, more than 40% of the sequenced genomes present the E484K mutation, which was identified in three different lineages (P.1, P.2 and B.1.1.33 - posteriorly renamed as N.9) in four Brazilian regions. We also evaluated the presence of E484K associated mutations and identified selective pressures acting on the spike protein, leading us to some insights about adaptive and purifying selection driving the virus evolution.
Keywords: COVID-19, E484K, Severe acute respiratory syndrome coronavirus 2, Infectious diseases, Viral evolution
Graphical abstract
1. Introduction
The etiological agent of COVID-19, named SARS-CoV-2, belongs to the Coronaviridae family composed of enveloped positively oriented single-stranded RNA viruses (Zhu et al., 2020). The first cases of the disease were reported in Wuhan province, China, in December 2019. Since then the number of infected people has increased exponentially worldwide and the World Health Organization (WHO) declared it a pandemic on 11 March 2020. As of 24 January 2021, the number of confirmed cases globally has scaled up to 99 million, with over 2 million deaths. Brazil is the third country in total number of confirmed cases, already exceeding 8.8 million infected people and 216,445 deaths (Johns Hopkins Coronavirus Resource Center, 2021). Compared to Latin America, which represents ~18% of the world cases, Brazil accounts for almost half of the COVID-19 notifications. However, the number of cases per million people(~41,600) is comparable to other countries such as Argentina (~41,300), Colombia (~39,600), Costa Rica (~37,160), Chile (~36,570), and Peru (~33,170) (Ritchie et al., 2021).
Nearly 410,000 genomes (up to January 2021) sequenced by researchers from several countries are available on the Global Initiative on Sharing All Influenza Data (GISAID) (Shu and McCauley, 2017), which is crucial to investigate the SARS-CoV-2 genomic epidemiology. A standardized nomenclature was proposed to reflect genetic characteristics and the viral geographical spread patterns. Two ancestral lineages, A and B, have been described containing sublineages that are distinguished by different recurrent mutations and phylogenetic profiles (Rambaut et al., 2020a). More recently, special attention has been directed to the understanding of aspects that might impact the virulence and transmissibility of SARS-CoV-2 (Korber et al., 2020; Li, 2020; Toyoshima et al., 2020; Volz et al., 2020) and the influence of different mutations in the effectiveness of COVID-19 vaccines and immunotherapies (Andreano et al., 2020; Pinto et al., 2020; Yu et al., 2020).
Some viral lineages have been studied in more detail, mainly those carrying mutations in the spike (S) glycoprotein, since it is the binding site to the human ACE2 (hACE2) receptor, an essential step to invade the host cell. Currently, there are three lineages of major worldwide concern: B.1.1.7, B.1.351, and P.1. The former emerged in England in mid-September 2020 and it is characterized by 14 lineage-specific amino acid substitutions, 7 deletions (Rambaut et al., 2020b) and has been rapidly spread across the UK and Europe ever since its first appearance (Rambaut et al., 2020c). Two substitutions present in this lineage deserve special attention: N501Y in the Receptor Binding Domain (RBD) of S1 and P681H near the polybasic RRAR sequence in the furin-like cleavage region. N501Y is one of the key contact residues interacting with hACE2 and P681H is one of four residues comprising the insertion that creates a furin-like cleavage site between S1 and S2, which is not found in closely-related coronaviruses (Xia et al., 2020). The second lineage probably emerged in South Africa in August 2020 and harbors three mutations in RBD: K417N, E484K, and N501Y (Tegally et al., 2020). The most recent lineage is P.1, derived from B.1.1.28, which was recently identified in returning travelers from Manaus (Amazonas state, Brazil) who arrived in Japan. It has almost the same three mutations present in RBD as the South African lineage, except for the substitution in amino acid site 417, where the original lysine (K) is substituted for a threonine (T), instead of an asparagine (N). Its appearance is likely to have arisen independently (Faria et al., 2021). Without previous introductions in other Latin American countries, which have been affected by their own specific mutations, such as the convergent evolution of the S:T1117I in Costa Rica (Molina-Mora et al., 2021) the E484K mutation arose independently in Brazil and was identified in the Rio de Janeiro state (Southeast Brazil) in early-October carried by the P.2 lineage (Voloch et al., 2021).
In January 2021, the E484K mutation was identified in several viral genomes from Brazil. Due to its rapid spread, the independent origins and the potential implications in vaccination and passive immune therapies, E484K has received particular attention ever since. In Qatar, 14 days after the second vaccine dose (Pfizer–BioNTech), the effectiveness against the B.1.351 variant was 75.0% (Abu-Raddad et al., 2021); in South Africa, the AstraZeneca ChAdOx1 vaccine presented only 10% protection against mild-to-moderate disease associated with the B.1.351, while 75% of protection was obtained against B.1.1.7 in the U.K. (Gupta, 2021). However, in Brazil, the CoronaVac/Sinovac effectiveness was estimated at around 50% against symptomatic infection P.1 variant (Gupta, 2021). Importantly, B.1.351, P.1, and P.2 carry this substitution associated with escape from neutralizing antibodies (Baum et al., 2020; Greaney et al., 2021; Weisblum et al., 2020). Recently, this mutation was also identified in a sample from a reinfected patient (Nonaka et al., 2021).
Moreover, B.1.1.7, B.1.351, and P.1 also harbor N501Y mutation, which is associated with enhanced receptor affinity (Starr et al., 2020) and increased infectivity and virulence in a mouse model (Gu et al., 2020). The presence of E484K and N501Y substitutions in the same SARS-CoV-2 genome may be particularly relevant for viral evolution. This combination was shown to induce more conformational changes than the N501Y mutant alone, potentially altering antibodies' complementarity to this region resulting in the above mentioned immune escape phenomena (Nelson et al., 2021).
Structural analysis points to E484K as potentially the most crucial mutation so far in brazilian SARS-CoV-2 genomes. It creates a new site for the amino acid 75 hACE-2 binding. This interaction seems even stronger than the binding between hACE-2 and the original main site located at position 501 (at RBD and hACE-2 interface) (Nelson et al., 2021). We speculate that the consequent neutralization escape due to E484K alone or as a part of a larger array of distinct mutations might act as a common evolutionary solution for several different viral lineages. Due to the independent origin of two current known lineages harboring E484K in Brazil, we aimed to describe the mutation patterns of these lineages, investigate phylogenetic relationships and perform positive selection tests to identify if adaptive evolution has acted as a major evolutionary force leading to the increase of amino acid variability in the RBD sites.
2. Results
2.1. Comparative genomic analyses
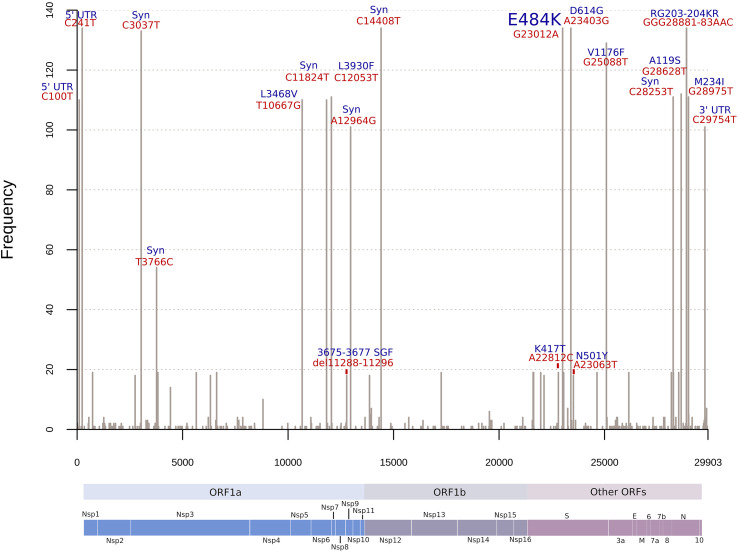
Firstly detected in the South African B.1.351 lineage, the S1 protein mutation E484K is now present in new emerging variants from Brazil. The analysis of genomes containing E484K, downloaded from the GISAID database, showed a distribution of 169 amino acid residues corresponding to nonsynonymous mutations and four amino acid residue deletions in 134 Brazilian samples (Table S1) collected between October and December 2020 (Fig. S1). Regarding synonymous mutations, 217 genome positions had transitions (Fig. 1 and Table S2) and 113 genome positions had associated transversions (Fig. 1 and Table S2). The arising of E484K mutated genomes with at least four different associated mutation patterns for lineages P.1, P.2, and N.9 (B.1.1.33.9) can be seen over time in the aligned genomes (Fig. S1 and Table S1).
Fig. 1.
Histogram of frequent mutations observed in the Brazilian SARS-CoV-2 genomes harboring E484K mutation. Red labels above the bars indicate absolute nucleotide position and the blue labels indicate effects of these mutations in the corresponding proteins. As P.1 has only 19 genomes represented and multiple mutations, only main mutations of concern were highlighted.
UTR: Untranslated region; Syn: Synonymous substitution; del: deletion; ORF: Open Reading Frame; Nsp: Non-structural protein; S: Spike; E: Envelope; M: Membrane; N: Nucleocapsid.
B.1.1 defining mutations were widespread in almost all sequences as expected (e. g. S:D614G, N:R203K, N:G204R, 5’UTR:C241T, and synonymous substitutions in nucleotide positions C3037T and C14408T) (Fig. 1). As B.1.1.28 is the ancestral lineage of P.1 and P.2, sequences from these three lineages harbor the G25088T (S:V1176F) mutation. Four sequences classified as N.9 carry B.1.1.33 lineage-defining mutations (T27299C (NS6:I33T) and T29148C (N:I292T)), but have also five missense mutations in ORF1ab and one in ORF7b (E33A). This pattern indicates a new lineage derived from B.1.1.33 (posteriorly defined as B.1.1.33.9 or N.9 lineage), which also possesses the E484K replacement, as P.1 and P.2 sequences. A hallmark of these Brazilian lineages is the presence of multiple lineage-defining mutations per lineage (Table 1 ).
Table 1.
Lineage-defining mutations of each of the three Brazilian lineages carrying the E484K mutation. These mutations do not necessarily reflect pangolin lineage assignment defining-mutations, but were extracted based on their representativeness in the majority of sequences of each lineage (https://github.com/cov-lineages/pangolin).
| Lineage/Mutation | N.9 (B.1.1.33 with E484K) | P.1 | P.2 |
|---|---|---|---|
| 5′ UTR | – | – | C100T |
| ORF1ab | G1264T (Nsp2) C6573T (Nsp3: S2103F) C7600T (Nsp3) C7851T (Nsp3: A2529V) T11078C (Nsp6: F3605L) C19602T (Nsp14: T6446I) G19656T (Nsp15: R6464M) |
T733C (Nsp1) C2749T (Nsp3) C3828T (Nsp3: S1188L) A5648C (Nsp3: K1795Q) A6319G (Nsp3) A6613G (Nsp3) del11288–11,296 (Nsp6: 3675–3677 SGF) C12778T (Nsp9) C13860T (Nsp12: T4532I) G17259T (Nsp13: S5665I) |
T10667G (Nsp5: L3468V) C11824T (Nsp6) A12964G (Nsp9) |
| S | G23012A (E484K) | C21614T (L18F) C21621A (T20N) C21638T (P26S) G21974T (D138Y) G22132T (R190S) A22812C (K417T) G23012A (E484K) A23063T (N501Y) C23525T (H655Y) C24642T (T1027I) |
G23012A (E484K) |
| ORF3a | – | T26149C (S253P) | – |
| E | – | – | – |
| M | – | – | – |
| ORF6 | – | – | – |
| ORF7a | – | – | – |
| ORF7b | A27853C (E33A) | – | – |
| ORF8 | – | G28167A (E92K) insG28262GAACA (ORF8) |
C28253T (F120F) |
| N | – | C28512G (P80R) AGTAGGG28877-83TCTAAAC (RG203KR) |
G28628T (A119S) G28975T (M234I) |
| ORF10 | – | – | – |
| 3′ UTR | C29722T | – | C29754T |
ORF1ab mutations are represented by its amino acid positions relative to ORF1a (Nsp1-Nsp11) and ORF1b (Nsp12-Nsp16). ins: insertion. The E484K mutation in the lineages is indicated in bold.
The number of genetic changes associated with each E484K Brazilian lineage is highly diverse. N.9 carries a mean of 19.2 (range: 17–22) mutations (considering single nucleotide polymorphisms, insertion and deletion as a single event), while P.1 possesses on average 30.1 changes (range: 24–33). Despite harboring a lower number of mutations (mean: 18.5), P.2 genomes have the highest standard deviation (SD = 2.2) and range (15–25) among these lineages. These data suggest that both P.1 and P.2 have been circulating in Brazil for a longer period and might be fastly evolving.
The analysis of the E484K mutated sequence EPI_ISL_832010, early detected in the municipality of Esteio, Rio Grande do Sul, shows a simpler set of nonsynonymous mutations (n = 10) when compared to others. This genome combines all the prevalent substitutions D614G from spike protein, N:R203K, N:G204R, and Nsp12:P323L, allied to other very frequent mutations (S:V1176F, N:A119S, N:M234I, Nsp5:L205V, and Nsp7:L71F). The presence of spike S:D614G, N:R203K, N:G204R, and Nsp12:P323L in all sequenced E484-containing genomes was observed.
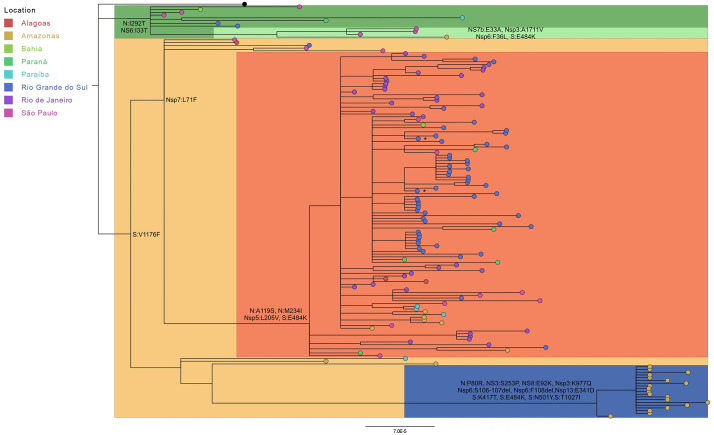
The last samples associated with the recent public health crisis in the state of Amazonas accumulates a higher number of divergences than other lineages (26 nonsynonymous substitutions and 3 amino acid deletions - Table S1) when compared with the SARS-CoV-2 reference genome (NC_045512.2). Regarding the lineage-defining mutations from P.1 lineages, 14 of a total of 19 genomes from the Amazonas monophyletic group present the spike mutations L18F, T20N, P26S, D138Y, and R190S. In this way, the spike protein of P.1 lineages in these Brazilian E484K mutated genomes is characterized by the presence of a variable number of modified sites, without the fixation of all known mutations. In fact, the P.1 and P2. clades are part of a larger monophyletic group with B.1.1.28, which is separated from the B.1.1.33 and N.9 (B.1.1.33 with E484K) clades and the reference genome. Interestingly, the B.1.1.28 genomes grouped in the clades of P.1 and P.2 lineages according to the presence of the Nsp7:L71F mutation. The monophyletic group formed by the B.1.1.28 and P.2 genomes presents these mutations, while the B.1.1.28 genomes grouped with P.1 sequences do not. These results corroborate the clusterization of these genomes in different lineages (Fig. 2 ).
Fig. 2.
Bayesian phylogenetic inference of the 134 Brazilian E484K mutated genomes. Tips were colored by Brazilian state and the reference genome NC_045512.2 is represented in black. The branches were highlighted by lineage: green (B.1.1.33), light green (N.9), beige (B.1.1.28), light red (P.2) and blue (P.1). Mutations occurring in all analyzed genomes for each lineage were described next to the respective nodes. The asterisks indicate the SARS-CoV-2 genomes sequenced by our research group.
Lineage-defining mutations as S:K471T, S:N501Y, S:T1027I, N:P80R, Nsp6:S106-107del, Nsp6:F108del, and NS8:E92K were found only in the P.1 group and reported for all 19 P.1 sequences. Other mutations such as S:V1176F were not found only in N.9 (B.1.1.33.9) lineage. There are also those mutations that are not known as lineage markers but were found in all lineages (n = 19) as Nsp3:K977Q, Nsp13:E341D, and NS3:S253P. New specific single substitutions (S:A27V, N:T16M, N:P151L, N:A267V, Nsp13:T216N, and Nsp14:P443S) were also evaluated. Specifically, S:A27V is located in the N-terminal domain.
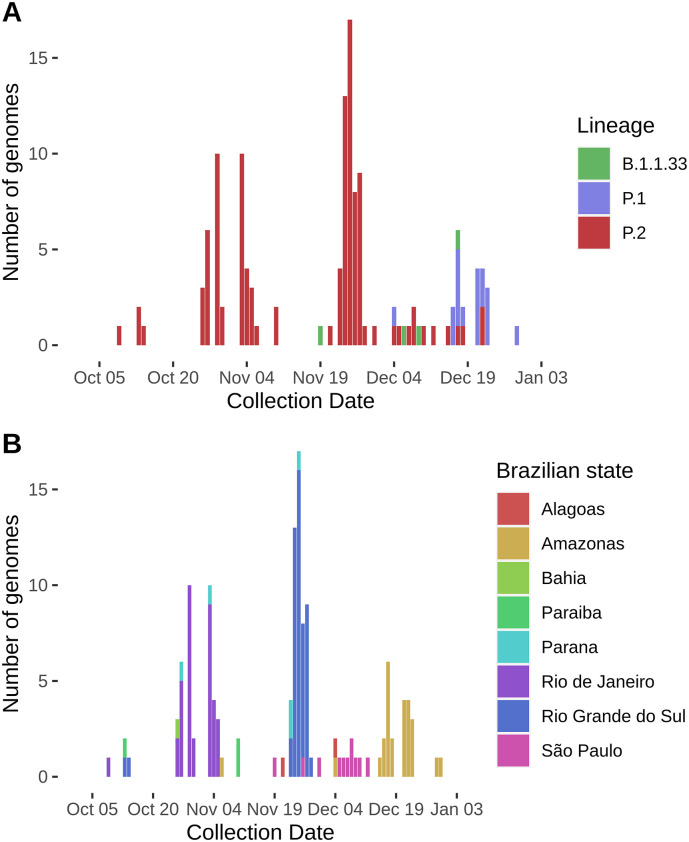
The N.9 (B.1.1.33.9) lineage carrying the E484K mutation was found in São Paulo (n = 3) and Amazonas (n = 1), while P.1 sequences were found only in the Amazonas state (n = 19). All other sequences (n = 111) belong to the P.2 lineage, the most widespread lineage considering sequenced data available so far. The Northeast region was represented by sequences from Alagoas (n = 2), Paraiba (n = 3), and Bahia (n = 1). The North region is represented by Amazonas sequences (n = 6). Southeast sequenced 44 P.2 genomes, 36 from Rio de Janeiro and 8 from São Paulo. Southern Brazil classified 55 genomes as P.2, 5 from Parana and 50 from Rio Grande do Sul, reinforcing its probable dissemination to all regions of Brazil (Fig. 3 ).
Fig. 3.
Distribution of genomes harboring E484K mutation across different lineages (A) and Brazilian states (B) from October to December 2020.
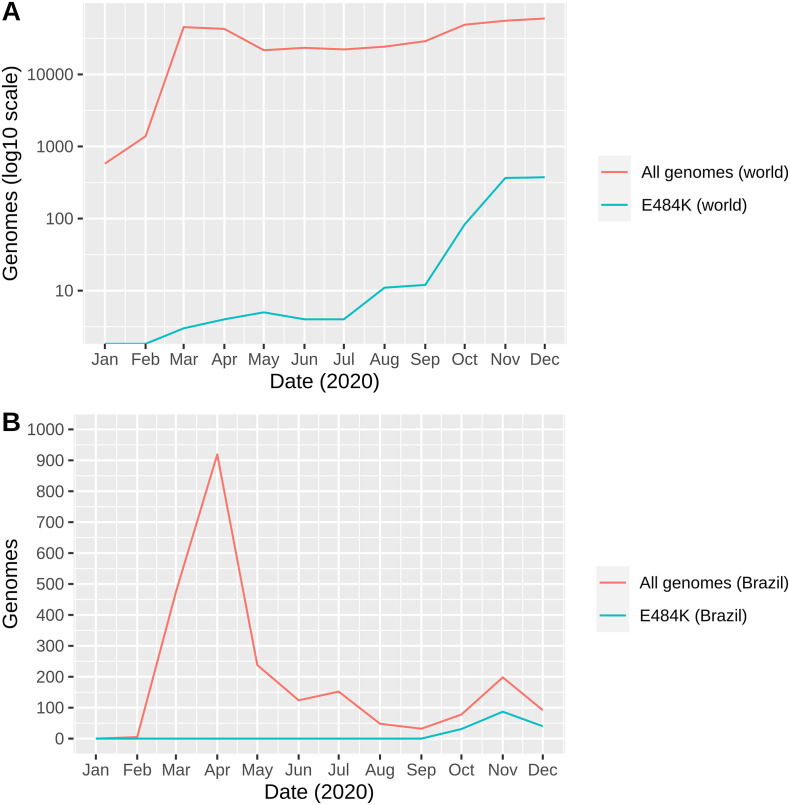
The worldwide emergence of E484K began in March 2020, with three sequences firstly represented. A significant increase was observed in October (n = 86), followed by successive increases in November (n = 366) and December (n = 374) (Fig. 4A). A similar trend was observed in Brazil, as E484K was observed in sequences obtained from October (n = 31), November (n = 87), and December (n = 40). Most importantly, the proportion of genomes carrying this replacement was 39.7%, 43.9%, and 43.5% from October to December in comparison to all genomes sequenced from Brazil (Fig. 4B). We believe this apparent slow-growing pattern in Brazil is due to heterogeneous and limited initiatives of sequencing in the country, and probably this substitution is already widespread through Brazilian states, as its harbored by three distinct and apparently independent evolving lineages.
Fig. 4.
Monthly presence of the E484K mutation considering worldwide available data (A) and Brazilian genomes (B). For clarity, the number of genomes in (A) are represented in log10 scale.
2.2. Selection analysis
In order to obtain a reliable detection of sites submitted to positive/adaptive or negative/purifying selection, a random set of Brazilian genomes was tested with different approaches. Regarding individual site models, the Bayesian inference FUBAR (Fast, Unconstrained Bayesian AppRoximation) identified eight sites evolving under adaptive selection (positive selection) in the spike sequence (Table 2 ), with calculated synonymous and nonsynonymous average rates of 1.227 and 0.857, respectively. For these residues under adaptive pressure, six are included in known mutation sites of spike protein, including E484K (L5F, S12F, P26S, D138Y, A688V).
Table 2.
FUBAR site table for positively selected sites.
| Site | ɑ | β | Prob[ɑ < β] |
|---|---|---|---|
| 5 | 1.880 | 20.275 | 0.9559 |
| 12 | 1.896 | 21.520 | 0.9584 |
| 26 | 1.879 | 20.481 | 0.9564 |
| 138 | 2.300 | 19.109 | 0.9214 |
| 155 | 2.266 | 17.643 | 0.9158 |
| 484 | 3.850 | 25.932 | 0.9109 |
| 677 | 2.931 | 19.933 | 0.9083 |
| 688 | 1.917 | 23.771 | 0.9628 |
FUBAR inferred the sites submitted to diversifying positive selection with a posterior probability ≥0.9.
The analysis with the codeml program from the PAML package confirmed all the positively selected sites predicted by FUBAR. The M3 vs. M0 (p < 0.001) comparison indicated a highly variable selective pressure among sites, while the M2 vs. M1 and M8 vs. M8a models comparison (p < 0.05) indicated some sites submitted to positive selection (Table 3, Table 4, Table 5 ). The results for the M3 model indicated a kappa value (ts/tv) of 2.82630 and a site proportion of 93.156% with an estimated omega (ω) value of 0.25424 (negative selection), while 3.536% of the sites were estimated to have ω = 4.90450 (positive selection) (Table 3). Differently from M2 that is compared with M1 to detect sites under positive selection, the M3 model for discrete evolution also indicates sites under a neutral selection (p1, ω1), being more recommended to evaluate variable selective pressures among sites. The proportion of sites and the ω values obtained for positive selection by M2 and M3 models were almost equal (M2: ω = 4.90459). In the case of M7 vs. M8 and M8a vs. M8 comparisons, the rejection of the alternative model (M8) by the absence of a significant likelihood ratio suggests a beta distribution of the ω. When applied to the analysis of the spike protein, the results of M8a, which is a more refined and conservative model, allows us to confirm that some residues of this protein are actually under adaptive selection.
Table 3.
PAML (codeml): Parameters estimates and log-likelihood values under models of variable ω ratios among sites.
| Model | Parametersa | lnL | Sites showing indications of positive selection |
|---|---|---|---|
| M0 | ω = 0.40548 | −6617.469118 | None |
| M1 | p0 = 0.64290, p1 = 0.35710 ω0 = 0, ω1 = 1 |
−6607.96566 | Not allowed |
| M2 | p0 = 0.96461, p1 = 0.00003, p2 = 0.03536 ω0 = 0.25444, ω1 = 1, ω2 = 4.90459 |
−6604.579083 | 5, 12, 26, 138, 155, 222, 484, 626, 677, 688, 1263 |
| M3 | p0 = 0.93156, p1 = 0.03308, p2 = 0.03536 ω0 = 0.25424, ω1 = 0.25458, ω2 = 4.90450 |
−6604.578953 | 5, 12, 26, 138, 155, 222, 484, 626, 677, 688, 1263 |
| M7 | p = 0.00986, q = 0.01846 | −6608.318528 | Not allowed |
| M8 | p0 = 0.91883, p = 0.00675, q = 0.04374, p1 = 0.08117, ω = 2.79061 | −6605.852845 | 5, 12, 14, 20, 26, 27, 52, 54, 68, 138, 145, 153, 155, 190, 218, 221, 222, 231, 235, 263, 344, 417, 439, 484, 561, 583, 626, 658, 670, 677, 688, 776, 791, 879, 936, 1005, 1065, 1071, 1072, 1076, 1099, 1104, 1118, 1152, 1162, 1238, 1259, 1263, 1264, 1272 |
ω = dN/dS = average over sites; p0,p1 and p2 indicate the proportions of groups 0, 1 and 2 in each model, respectively; ω0, ω1 and ω2 indicate the ω values of groups 0, 1 and 2 in each model, respectively. p and q are beta parameters.
Table 4.
PAML (codeml): Likelihood ratio statistics (2Δ/) for some comparisons between selection models.
| Comparison | 2Δ/ | Probability values (p) |
|---|---|---|
| M3 vs. M0 | 25.78033 | <0.001 |
| M2 vs. M1 | 6.773154 | <0.05 |
| M8 vs. M7 | 4.931366 | <0.1 |
| M8 vs. M8a | 4.225314 | <0.05 |
The degrees of freedom used comparing models M3 vs. M0, M2 vs. M1, M8 vs. M7 and M8a vs. M8 are 4, 2, 2 and 1, respectively. Probability values ≤ 0.05 are considered as statistically significant.
Table 5.
PAML (codeml) site table for positively selected sites.
| Site | NEB probabilities |
BEB probabilities |
||
|---|---|---|---|---|
| Prob (ω > 1) | post mean for ω | Prob (ω > 1) | post mean ± SE for ω | |
| 5 L | 0.988* | 4.850 | 0.892 | 2.153 ± 0.516 |
| 12 S | 0.988* | 4.849 | 0.892 | 2.153 ± 0.517 |
| 26 P | 0.989* | 4.854 | 0.894 | 2.156 ± 0.514 |
| 138 D | 0.758 | 3.779 | 0.741 | 1.922 ± 0.719 |
| 155 S | 0.782 | 3.893 | 0.749 | 1.935 ± 0.712 |
| 222 A | 0.775 | 3.857 | 0.746 | 1.930 ± 0.714 |
| 484 E | 0.841 | 4.163 | 0.770 | 1.968 ± 0.692 |
| 626 A | 0.860 | 4.252 | 0.778 | 1.980 ± 0.685 |
| 677 Q | 0.909 | 4.483 | 0.802 | 2.017 ± 0.659 |
| 688 A | 0.985* | 4.835 | 0.886 | 2.145 ± 0.525 |
| 1263 P | 0.880 | 4.348 | 0.787 | 1.994 ± 0.675 |
M2 selection model: Naive Empirical Bayes (NEB) analysis. Positively selected sites (*: P > 95%; **: P > 99%). Bayes Empirical Bayes (BEB).
The ω estimative (ω = d N /d S) is a common method to detect positive selection, since it assumes that synonymous substitutions are neutral and the nonsynonymous are subject to selection. Consequently, a ω statistically higher than 1 would indicate the action of positive selection or a relaxed selective constraint, whereas low d N /d S values would mean conservation of the gene product due to purifying selection (Tennessen, 2008). Additionally to the FUBAR results, three sites were also considered with ω > 1 (Table 3, Table 5): 222, 626, and 1263. Of these, only site 222 is not related to the E484K-presenting lineages (A626S and P1263L/S are known). The list of positively selected sites identified by the M8 model is available on Table S4.
To avoid the potential missense effects of the function lost by the sequence mutation, the purifying selection acts as a protective model. The FEL (Fixed Effects Likelihood) approach detected 19 sites under negative selection (Table 6 ). Three of them (189, 191, and 564) are near residues presenting known nonsynonymous mutations in the E484K mutated genomes, as S:R190S and S:F565L. The SLAC (Single-Likelihood Ancestor Counting) method identified four sites (55, 856, 943, and 1215), two of which were already predicted by FEL. The prevalence of negative selection across the spike protein is consistent with the low genome-wide mutation rate inferred for SARS-CoV-2 (van Dorp et al., 2020).
Table 6.
FEL site table for negatively selected sites.
| Site | ɑ | β | LRT | Prob[ɑ > β] |
|---|---|---|---|---|
| 55 | 18.379 | 0.000 | 4.058 | 0.0440 |
| 91 | 27.071 | 0.000 | 2.792 | 0.0947 |
| 132 | 121.449 | 0.000 | 4.395 | 0.0360 |
| 180 | 121.798 | 0.000 | 4.401 | 0.0359 |
| 189 | 33.258 | 0.000 | 5.432 | 0.0198 |
| 191 | 120.557 | 0.000 | 4.393 | 0.0361 |
| 266 | 27.216 | 0.000 | 2.794 | 0.0946 |
| 324 | 118.682 | 0.000 | 4.370 | 0.0366 |
| 428 | 26.763 | 0.000 | 2.852 | 0.0913 |
| 475 | 16.429 | 0.000 | 3.268 | 0.0707 |
| 564 | 121.798 | 0.000 | 4.159 | 0.0414 |
| 821 | 21.710 | 0.000 | 3.065 | 0.0800 |
| 897 | 42.768 | 0.000 | 4.241 | 0.0395 |
| 910 | 42.680 | 0.000 | 3.305 | 0.0691 |
| 1120 | 43.397 | 0.000 | 3.598 | 0.0578 |
| 1126 | 26.763 | 0.000 | 3.495 | 0.0616 |
| 1215 | 18.327 | 0.000 | 2.861 | 0.0907 |
| 1228 | 42.759 | 0.000 | 4.136 | 0.0420 |
| 1251 | 42.680 | 0.000 | 3.305 | 0.0691 |
Sites identified under negative selection at p ≤ 0.1. Grey rows: significant sites also identified by SLAC.
3. Discussion
The permanence of a pathogen inside a population host depends on its efficiency in key processes such as the replication, the escaping of the immune system and the binding to the cell receptor. Mutations that create an advantageous scenario towards the host response to infection enhance the pathogen fitness under the natural selection pressure. Consequently, the host-pathogen coevolution represents an important mechanism to understand the establishment and the prognostics of pathogenesis, since the infections are possibly the major selective pressure acting on humans (Sironi et al., 2015). Therefore, the circulation of low and moderate pathogens provides time for the pathogen adaptations, in such a way that the modulation of the immunity may possibly promote molecular convergence in different lineages over the time (Longdon et al., 2014).
The presence of spike S:D614G, N:R203K, N:G204R, and Nsp12:P323L in all sequenced E484 mutated samples, reaching three different lineages, might suggest that these lineages show increased viral replication. Considering that E484K enhances escape from immune system antibodies, these may potentially lead to a viral advantage. The occurrence of simultaneous mutations as N:R203K and N:G204R is already known in the SARS-CoV-2 literature. However, the fixation of these mutations, as well as Nsp12:P323L and D614G in all the E484K evaluated genomes may indicate a novel adaptive relationship among these modifications resulting in viral evolutionary success.
Even as an independent evolutionary event, the potential fixation of mutations such as E484K across lineages may indicate active mechanisms of adaptive selection and are very relevant in planning future therapeutic strategies (for example, newer vaccines and immunotherapy platforms). The deletion of three amino acids in the second helix of the transmembrane protein Nsp6 in P.1 genomes may affect the virus-induced cellular autophagy and the formation of double-membrane vesicles for the viral RNA synthesis (Benvenuto et al., 2020).
The important role of subsequent stabilization of the flexible NTD by mutations has been speculated (Laha et al., 2020). Typically, NTD can harbor a larger number of evolutionary events than RBD, including mutations, insertions and deletions that could act allosterically altering the binding affinities between RBD and hACE-2 and inducing immune evasion. It is known that distinct positions may have a linked relationship in the final protein structure and may display some advantage by acting together to achieve increased stability, adaptability, viability and/or transmission efficiency (Laha et al., 2020). The potential causality or influence of the E484K and other substitutions for the effectiveness of neutralizing antibodies that bind the N-terminal domain of the spike protein (Chi et al., 2020) remains uncertain.
Concerning the selection analysis, the FUBAR method was the first one to be tested. According to Murrell et al. (2013), the FUBAR method may have more detection power than methods like FEL, in particular when positive selection is present but relatively weak. The analysis of the South African clade V501·V2 (Pond et al., 2020) found some similar results for FUBAR evaluation, with the detection of adaptive selection at the sites 5 (L5F), 12 (S12F), and 484 (E484K). The residues 155 and 677 are not associated with the E484K-presenting lineages, however, recent data found evidence of evolutionary convergence of this mutation in at least six distinct sub-lineages, which could improve proteolytic processing, cell tropism, and transmissibility (Hodcroft et al., 2021). The amino acid residue 677 is next to the furin-like cleavage site (678 to 688). Interestingly, some studies suggest that the emergence of the SARS-CoV-2 in human species resulted from a five amino acid change in the critical S glycoprotein binding site (Fam et al., 2020).
To the best of our knowledge, the impact of E484K in different lineages has not been deeply explored. It was structurally demonstrated that, at least in combination with K417N and N501Y, the substitution has profound impact in shifting the main site of contact between viral RBD and hACE-2 residues (Nelson et al., 2021). However, we still do not know if this holds true for other sets of mutations associated with E484K.
In summary, we have demonstrated widespread dissemination of mutants harboring E484K replacement in geographically diverse regions in Brazil, as well as the potential fixation of the E484K mutation despite a short time (three months) after its first arising. This substitution was disseminated in our country as early as October 2020, although phylodynamics inferences placed its emergence in July (Voloch et al., 2021). The fact that E484K was found in the context of different mutations and lineages is suggestive that this particular substitution may act as a common solution for viral evolution in different genotypes. This hypothesis may be related to the profound impact of the mutation, which changes a negatively charged amino acid (glutamic acid) for a positively charged amino acid (lysine). Since this position is present in a highly flexible loop, it has been proposed that the presence of such mutation could create a strong ionic interaction between lysine (K) in RBD and amino acid 75 of the hACE-2 receptor shifting the major sites of binding in S1 from positions 497–502 to 484 (Nelson et al., 2021).
Mutations in RBD could theoretically decrease the neutralizing serum activity of patients receiving vaccines against SARS-CoV-2 with unknown clinical consequences. Arguably the effect of E484K could be particularly relevant. In a previous publication, this mutation was associated with complete abolishment of all neutralizing activity in a high proportion of convalescent serum tested (Wibmer et al., 2021). Jangra et al. (2021) showed that the E484K affects the binding of serum polyclonal neutralizing antibodies and decreases the neutralization efficiency for serum with low or moderate IgG for the SARS-CoV-2 spike protein. Moreover, in the study of Xie et al. (2021), the combination of the mutations E484K + N501Y + D614G generated lower neutralization titers than the N501Y virus or the virus with three mutations from the UK variant (Δ69/70 + N501Y + D614G). When taken together, this growing body of evidence suggests that E484K should be the target of intense virologic surveillance. Studies testing the activity of serum from vaccinated patients against viruses or pseudoviruses with the aforementioned substitution should be considered a high public health priority. Second generation immune therapies and vaccines focusing on more conserved domains (for instance, in S2 fusion domain) may deserve special attention to assure continuous therapeutic efficacy.
4. Methods
4.1. Sequencing data retrieval
For the graphic evaluation of Brazilian lineages, all genomes available on the GISAID database until January 18th, 2021, and presenting the E484K mutation were included in the analysis. Genomes from Brazil and other countries submitted until January 24th, 2021, and with collected dates between January 1st and December 31st, 2020, were selected. The counting of genomes containing E484K was performed by the specification of the mutation in the search fields. The monthly count was defined by the values between the first and the last day of each month.
4.2. Comparative genomic analyses
The comparative genomic analysis of the E484K mutated genomes was performed with all the 134 genomes (Table S1) presenting the E484K mutation for Brazil in GISAID until January 18th, 2021. For these, a genomic multiple sequence alignment was performed on the MAFFT web server (Katoh and Standley, 2013) using default options and 1PAM = k/2 scoring matrix. Single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs) were assessed by using snippy variant calling pipeline v4.6.0 (https://github.com/tseemann/snippy). The mapping of the aligned sequences to the reference genome (NC_045512.2) features was created by the software GENEIOUS 2021.0.3 (https://www.geneious.com). Histogram of mutations was generated using a modified code from Lu et al., 2020 (https://github.com/laduplessis/SARS-CoV-2_Guangdong_genomic_epidemiology/).
4.3. Phylogenetic analysis
The previously aligned genomic sequences were used as input for the phylogenetic analysis. The reference genome NC_045512.2 was added as an outgroup and 16 other sequences from B.1.1.28 and B.1.1.33 lineages were added to analyze the phylogeny patterns with the E484K-containing genomes. The inference of the best evolutionary model was performed by ModelTest-NG (Darriba et al., 2020), which identified GTR + I (Generalised time-reversible model + proportion of invariant sites). The tree was built by Bayesian inference in MrBayes v3.2.7 (Huelsenbeck and Ronquist, 2001) with the GTR + I model and 5 million generations. The data convergence was evaluated with Tracer v1.7.1 (Rambaut et al., 2018) and tree visualization was created by FigTree software (http://tree.bio.ed.ac.uk/software/figtree/).
4.4. Selection analysis
In total, 780 SARS-CoV-2 genomes, collected from Brazil samples between February 1st and December 31st, 2020, available on GISAID, were used for this analysis. After an initial filtering step of undefined nucleotides in the region of spike protein, with deletion of genomes with a N ratio > 0.01, 589 genomes were selected. The second step of subsampling, with Augur toolkit (Huddleston et al., 2021) kept 498 genomes (approximately 119 representative genomes per lineage) for the multiple sequence alignment (Table S3). The genomic multiple sequence alignment was performed by MAFFT (Katoh and Standley, 2013) with default options and 1PAM = k/2 scoring matrix. The selection of the genome location for the spike protein was performed with the software UGENE (Okonechnikov et al., 2012), using the ORF coordinates of the NC_045512 reference genome as parameter. GTR + I was inferred by ModelTest-NG (Darriba et al., 2020) as the best evolutionary model for the spike sequences. ModelTest-NG also indicated identical sequences, which were removed resulting in 161 sequences. The Bayesian phylogenetic inference was performed by MrBayes v3.2.7 using those 161 unique spike selected sequences and 5 million generations. The data convergence was evaluated with Tracer v1.7.1.
The analysis of sites under positive and negative selection was performed by HyPhy v2.5.23 (Pond et al., 2005) according to different approaches: (i) FUBAR (Unconstrained Bayesian AppRoximation) (Murrell et al., 2013), (ii) FEL (Fixed Effects Likelihood) (Kosakovsky Pond and Frost, 2005), and (iii) SLAC (Single-Likelihood Ancestor Counting) (Kosakovsky Pond and Frost, 2005). Finally, the PAML (Phylogenetic Analysis by Maximum Likelihood) (Yang, 2007) program was used for confirmation of possibly selected sites with the codeml package. All models were run using the F3x4 option in the PAML program, where expected codon frequencies were based upon nucleotide frequencies occurring at the three codon positions. The one-ratio model (M0) assumes one ω ratio for all sites. The neutral model (M1) presupposes a proportion p0 of conserved sites with ω0 = 0 and p1 = 1 - p0 of neutral sites with ω1 = 1, as would occur if almost all non-synonymous substitutions were either deleterious or neutral. The positive selection model (M2) adds an additional class of sites with frequency p2 = 1 - p0 - p1 and ω2 is estimated from the data. In the discrete model (M3), the probabilities (p0, p1 and p2) of each site which was submitted to purifying selection, neutral selection and positive selection, respectively, and their corresponding ω ratios (ω0, ω1, ω2) are inferred from the data. The beta model (M7) is a null test for positive selection, assuming a beta distribution with ω between 0 and 1. By last, the beta & ω (M8) model adds one extra class with the same ratio ω1. The LRTs (Likelihood Ratio Tests) were tested to investigate whether ω was significantly different from 1 for each pairwise comparison: M1a vs. M2a, M0 vs. M3, and M7 vs. M8. LRT performs the comparison both with the constraint of ω = 1 and without such constraint: LRT = 2 (ln1−ln2). It follows a chi-square distribution, where the number of degrees of freedom is equal to the number of additional parameters in the more complex model. For rejection of the null hypothesis of neutrality, we considered p ≤ 0.05. Finally, we applied the Naive Empirical Bayes (NEB) and Bayes Empirical Bayes (BEB) approaches available on the PAML package to calculate the posterior probability that each site belongs to the positively selected class.
The following are the supplementary data related to this article.
Brazilian sequenced genomes containing the E484K mutation and GISAID list of acknowledgements.
Single Nucleotide Polymorphisms identified in the multiple genomes comparison.
Brazilian sequenced genomes used for selection analysis (499 sequences) - GISAID list of acknowledgements.
Positively selected sites identified by PAML with the M8 selection model.
Supplementary material
Funding
Scholarships and Fellowships were supplied by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001 and Universidade Federal de Ciências da Saúde de Porto Alegre. The funders had no role in the study design, data generation and analysis, decision to publish or the preparation of the manuscript.
Author contributions
Patrícia A. G. Ferrareze: Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Visualization, Writing - Original Draft, Writing - Review & Editing. Vinicius B. Franceschi: Formal analysis, Investigation, Visualization, Writing - Original Draft, Writing - Review & Editing. Amanda M. Mayer: Writing - Original Draft, Writing - Review & Editing. Gabriel D. Caldana: Writing - Original Draft, Writing - Review & Editing. Ricardo A. Zimerman: Conceptualization, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing. Claudia E. Thompson: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Writing - Original Draft, Writing - Review & Editing, Supervision, Project administration. All authors have read and approved the manuscript.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
We thank the administrators and curators of the GISAID database and research groups across the globe for supporting the rapid and transparent sharing of genomic data during the COVID-19 pandemic. We also thank the Mayor's Office, Health Department and São Camilo Hospital (Esteio, RS, Brazil), Leonardo Duarte Pascoal and Ana Regina Boll for their work in combating Covid-19 and for supporting the work developed by our research group. A full table acknowledging the authors and corresponding labs submitting sequencing data used in this study can be found in Supplementary Files 1 and 3.
References
- Abu-Raddad L.J., Chemaitelly H., Butt A.A. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. New Engl. J. Med. 2021 doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., Manna R., Casa E., Hyseni I., Benincasa L., Montomoli E., Amaro R.E., McLellan J.S., Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1073/pnas.2103154118. 2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., Ciccozzi M., Cassone A. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J. Inf. Secur. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang Jun, Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang Jinlong, Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C., Boshier F., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infec. Genet. Evol.: J. Mol. Epidemiol. Evolut. Genet. Infect. Dis. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam Bibiana S.O., Vargas-Pinilla Pedro, Amorim Carlos Eduardo G., Sortica Vinicius A., Bortolini Maria Cátira. ACE2 diversity in placental mammals reveals the evolutionary strategy of SARS-CoV-2. Genet. Mol. Biol. 2020;43(2) doi: 10.1590/1678-4685-GMB-2020-0104. (Epub June 08, 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N., Claro I.M., Candido D., Franco L.A.M., Andrade P.S., Coletti T.M., Silva C.A.M., Fraiji N.A., Esashika Crispim M.A., Carvalho M. do P.S.S., Rambaut A., Loman N., Pybus O.G., Sabino E.C. Virological; 2021. Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings [WWW Document]https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 URL. (accessed 1.14.21) [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1) doi: 10.1016/j.chom.2020.11.007. 44-57.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Yuchang, Li X.-F., Li J., Zhang N.-N., Yang Xiaolan, Chen S., Guo Y., Zhao G., Wang X., Luo D.-Y., Wang H., Yang Xiao, Li Yan, Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.-F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021 doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Domman D.B., Oguntuyo K., Snyder D.J., Van Diest M., Densmore K.H., Schwalm K.C., Femling J., Carroll J.L., Scott R.S., Whyte M.M., Edwards M.D., Hull N.C., Kevil C.G., Vanchiere J.A., Lee B., Dinwiddie D.L., Cooper V.S., Kamil J.P. Emergence in late 2020 of multiple lineages of SARS-CoV-2 spike protein variants affecting amino acid position 677. medRxiv. 2021 2021.02.12.21251658. [Google Scholar]
- Huddleston J., Hadfield J., Sibley T.R., Lee J., Fay K., Ilcisin M., Harkins E., Bedford T., Neher R.A., Hodcroft E.B. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. J. Open Source Softw. 2021;6:2906. doi: 10.21105/joss.02906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Chernet R.L., Eaker L.Q., Ferreri E.D., Floda D.L., Gleason C.R., Kleiner G., Jurczyszak D., Matthews J.C., Mendez W.A., Mulder L.C.F., Russo K.T., Salimbangon A.-B.T., Saksena M., Shin A.S., Sominsky L.A., Srivastava K., Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins Coronavirus Resource Center . Johns Hopkins Coronavirus Resour. Cent; 2021. COVID-19 Map [WWW Document]https://coronavirus.jhu.edu/map.html URL. (accessed 01.24.21) [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.043. 812-827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S.L., Frost S.D.W. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Hua X. Modifiable lifestyle factors and severe COVID-19 risk: evidence from Mendelian randomization analysis. BMC Med. Genet. 2021;14(38) doi: 10.1186/s12920-021-00887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B., Brockhurst M.A., Russell C.A., Welch J.J., Jiggins F.M. The evolution and genetics of virus host shifts. PLoS Pathog. 2014;10(11) doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., François S., Kraemer M.U.G., Faria N.R., McCrone J.T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Hu J., Liu T., Ma W., Li W., Su J., Zheng H., Peng B., Fang S., Su W., Li K., Sun R., Bai R., Tang X., Liang M., Quick J., Song T., Rambaut A., Loman N., Raghwani J., Pybus O.G., Ke C. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.023. 997-1003.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Mora J.A., Cordero-Laurent E., Godínez A., Calderón-Osorno M., Brenes H., Soto-Garita C., Pérez-Corrales C., Drexler J.F., Moreira-Soto A., Corrales-Aguilar E., Duarte-Martínez F. SARS-CoV-2 genomic surveillance in Costa Rica: evidence of a spanergent population and an increased detection of a spike T1117I mutation. Infect. Genet. Evol. 2021;92:104872. doi: 10.1016/j.meegid.2021.104872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B., Moola S., Mabona A., Weighill T., Sheward D., Kosakovsky Pond S.L., Scheffler K. FUBAR: a fast, unconstrained Bayesian AppRoximation for inferring selection. Mol. Biol. Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G., Buzko O., Spilman P., Niazi K., Rabizadeh S., Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 2021.01.13.426558. [Google Scholar]
- Nonaka C.K.V., Franco M.M., Gräf T., Mendes A.V.A., de Aguiar R.S., Giovanetti M., Souza B.S. de F. Genomic evidence of a Sars-Cov-2 reinfection case with E484K spike mutation in Brazil. Preprints. 2021;2021010132 [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M., UGENE team Unipro UGENE: a unified bioinformatics toolkit. Bioinforma. Oxf. Engl. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Pond S., Wilkison E., Weaver S., James S.E., Tegally H., Oliveira T., Martin D. Visualizing Preliminary Selection Analysis Results for Evolution of the South African South Africa V501.V2 clade. [WWW Document] Observable. 2020. https://observablehq.com/@spond/zav501v2#table1 URL. (accessed 01.27.21)
- Pond S.L.K., Frost S.D.W., Muse S.V. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D., Volz E. Virological; 2020. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations [WWW Document]https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 URL. (accessed 1.4.21) [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E. Virological; 2020. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations [WWW Document]https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 URL. (accessed 5.17.21) [Google Scholar]
- Ritchie H., Ortiz-Ospina E., Beltekian D., Mathieu E., Hasell J., Macdonald B., Giattino C., Appel C., Rodés-Guirao L., Roser M. Our World in Data; 2021. Brazil: Coronavirus Pandemic Country Profile [WWW Document]https://ourworldindata.org/coronavirus/country/brazil URL. (accessed 5.17.21) [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M., Cagliani R., Forni D., et al. Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 2015;16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182 doi: 10.1016/j.cell.2020.08.012. 1295-1310.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally, H., Wilkinson, E., Giovanetti, M., Iranzadeh, A., Fonseca, V., Giandhari, J., Doolabh, D., Pillay, S., San, E.J., Msomi, N., Mlisana, K., von Gottberg, A., Walaza, S., Allam, M., Ismail, A., Mohale, T., Glass, A.J., Engelbrecht, S., Zyl, G.V., Preiser, W., Petruccione, F., Sigal, A., Hardie, D., Marais, G., Hsiao, M., Korsman, S., Davies, M.-A., Tyers, L., Mudau, I., York, D., Maslo, C., Goedhals, D., Abrahams, S., Laguda-Akingba, O., Alisoltani-Dehkordi, A., Godzik, A., Wibmer, C.K., Sewell, B.T., Lourenço, J., Alcantara, L.C.J., Pond, S.L.K., Weaver, S., Martin, D., Lessells, R.J., Bhiman, J.N., Williamson, C., de Oliveira, T., (2020). Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640.
- Tennessen J.A. Positive selection drives a correlation between non-synonymous/synonymous divergence and functional divergence. Bioinformatics. 2008;24(12):1421–1425. doi: 10.1093/bioinformatics/btn205. [DOI] [PubMed] [Google Scholar]
- Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C.M., Francisco R.da S., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P.de C., Mariani D., Costa R.M.da, Ferreira O.C., Covid19-UFRJ Workgroup, LNCC Workgroup, Cavalcanti A.C., Frauches T.S., de Mello C.M.B., Leitão I.de C., Galliez R.M., Faffe D.S., Castiñeiras T.M.P.P., de Vasconcelos A.T.R. Genomic characterization of a novel sars-cov-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95(10) doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2020;184(1) doi: 10.1016/j.cell.2020.11.020. 64-75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., Da Silva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5:1–3. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., Menachery V.D., Weaver S.C., Dormitzer P.R., Shi P.-Y. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nature Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007 Aug;24(8):1586–1591. doi: 10.1093/molbev/msm088. PMID: 17483113. [DOI] [PubMed] [Google Scholar]
- Yu F., Xiang R., Deng X., Wang L., Yu Z., Tian S., Liang R., Li Y., Ying T., Jiang S. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:1–12. doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Brazilian sequenced genomes containing the E484K mutation and GISAID list of acknowledgements.
Single Nucleotide Polymorphisms identified in the multiple genomes comparison.
Brazilian sequenced genomes used for selection analysis (499 sequences) - GISAID list of acknowledgements.
Positively selected sites identified by PAML with the M8 selection model.
Supplementary material