Abstract
Arenaviral infections often result lethal hemorrhagic fevers, affecting primarily in African and South American regions. To date, there is no FDA-approved licensed vaccine against arenaviruses and treatments have been limited to supportive therapy. Hence, the study was employed to design a highly immunogenic cross-reactive vaccine against Arenaviridae family using reverse vaccinology approach. The whole proteome of Lassa virus (LASV), Lymphocytic Choriomeningitis virus (LCMV), Lujo virus and Guanarito virus were retrieved and assessed to determine the most antigenic viral proteins. Both T-cell and B-cell epitopes were predicted and screened based on transmembrane topology, antigenicity, allergenicity, toxicity and molecular docking analysis. The final constructs were designed using different adjuvants, top epitopes, PADRE sequence and respective linkers and were assessed for the efficacy, safety, stability and molecular cloning purposes. The proposed epitopes were highly conserved (84%–100%) and showed greater cumulative population coverage. Moreover, T cell epitope GWPYIGSRS was conserved in Junin virus (Argentine mammarenavirus) and Sabia virus (Brazilian mammarenavirus), while B cell epitope NLLYKICLSG was conserved in Machupo virus (Bolivian mammarenavirus) and Sabia virus, indicating the possibility of final vaccine construct to confer a broad range immunity in the host. Docking analysis of the refined vaccine with different MHC molecules and human immune receptors were biologically significant. The vaccine-receptor (V1-TLR3) complex showed minimal deformability at molecular level and was compatible for cloning into pET28a(+) vector of E. coli strain K12. The study could be helpful in developing vaccine to combat arenaviral infections in the future. However, further in vitro and in vivo trials using model animals are highly recommended for the experimental validation of our findings.
Keywords: Arenavirus, Hemorrhagic fevers, Nucleoprotein, Recombinant vaccine, Molecular docking
Arenavirus; Hemorrhagic fevers; Nucleoprotein; Recombinant vaccine; Molecular docking
1. Introduction
Arenaviral infections, primarily affecting South American and African regions, have traditionally been neglected as tropical diseases [1]. Although most infections are mild, transmission occurs through rodents to humans, sometimes resulting in severe Hemorrhagic Fevers (HF) with high death rates [2]. Arenaviruses are divided into two categories, Old World (eastern hemisphere) and New World (western hemisphere) arenavirus. Old World arenaviruses and the diseases they cause include Lassa virus (Lassa fever), Lymphocytic Choriomeningitis virus (meningitis, encephalitis, congenital fetal infection, multiple organ failure) and Lujo virus (Lujo-hemorrhagic fever). The latter class includes Junin virus (Argentine-hemorrhagic fever), Machupo virus (Bolivian-hemorrhagic fever), Guanarito virus (Venezuelan-hemorrhagic fever), Sabia virus (Brazilian-hemorrhagic fever) and Chapare virus [3].
Lassa fever is a zoonotic, acute viral illness which is endemic in parts of West Africa including Sierra Leone, Liberia, Guinea and Nigeria. Neighboring countries are also at risk, as the animal vector for LASV, multimammate rat is distributed throughout the region. It was estimated that the virus infects roughly 300,000–500,000 individuals per year yielding approximately 5,000 deaths [4]. In some areas of Sierra Leone and Liberia, 10–16% of people admitted to hospitals annually have Lassa fever, demonstrating the serious impact the disease has on those regions. About 15–20% of patients hospitalized for Lassa fever die from the illness and the case-fatality rate may reach 50% if associated with occasional epidemics [4]. The only available drug, ribavirin is effective if administered within the first 6 days after disease onset [5]. Lymphocytic Choriomeningitis virus (LCMV) is another rodent-borne prototypic virus of Arenaviridae family that can cause substantial neurological problems, including meningitis, encephalitis, and neurologic birth defects, particularly among prenatal and immune compromised humans. LCMV infections have been reported in Europe, America, Australia, Japan, and may occur wherever infected rodent hosts of the virus are found. Several serologic studies performed in urban areas revealed that the prevalence of LCMV antibodies in human populations range from 2% to 5% [6]. A meta-analysis of congenital LCMV infections demonstrated a mortality rate of 35% by 21 months of age [6, 7]. An effective antiviral therapy for LCMV has not yet been developed [8, 9]. Recently, Lujo virus was isolated as a newly discovered novel arenavirus associated with a viral hemorrhagic fever outbreak in southern Africa in 2008. It was found to cause a fulminant viral hemorrhagic fever (LUHF) syndrome characterized by nonspecific symptoms such as fever, malaise, myalgias, sore throat, nausea, vomiting and non-bloody diarrhea followed with variable retrosternal or epigastric pain, usually progressing to bleeding, shock and multiorgan failure [10]. This virus has been associated with an outbreak in Zambia and South Africa with 80% fatality rate [11, 12]. Guanarito virus, on the contrary, is the etiological agent of Venezuelan haemorrhagic fever, another rodent borne zoonosis which is endemic in the Northern America [13]. Infections are characterized by having the onset of pulmonary congestion and edema, renal and cortical necrosis, and hemorrhage in different sites like mucous membranes, major internal organs, digestive and urinary tracts [14]. It was first recognized as a distinct clinical entity in 1989 during an outbreak of hemorrhagic fever that began in the Municipality of Guanarito in Southern Portuguesa with 33.3% fatality rate [13, 15].
Although arenaviral infections are restricted to certain geographic areas, known cases of exportation of arenaviruses from endemic regions and socioeconomic challenges to locally control the rodent reservoirs raised serious concerns about the potential for larger outbreaks in future. Currently, there are no FDA-approved vaccines for arenaviruses and treatments have been limited to supportive therapy or use of non-specific nucleoside analogs like Ribavirin [1, 16]. The live attenuated Candid#1 strain of Junin virus has been shown to be an effective vaccine against Argentine Hemorrhagic Fever [17]. Researchers found that Junin virus vaccine antibodies are also effective against Machupo virus [18]. However, the preventive measures to combat infections caused by Lassa virus (LASV), Lymphocytic Choriomeningitis virus (LCMV), Lujo virus and Guanarito virus has not yet gained considerable success [5, 8]. Although, ML29, an investigational live attenuated vaccine has been shown to provide effective protection in vivo against LASV [19], the mechanism for ML29 attenuation remains unknown [20]. Therefore, the incorporation of a limited number of additional mutations into the ML29 genome may result in viruses with enhanced virulence [21]. Though successful in many cases, traditional vaccines such as live-attenuated or inactivated vaccines are associated with several demerits [22, 23]. There are evidence of non-virulent organisms getting converted into virulent strains [24, 25]. In contrast, vaccines produced by reverse vaccinology approach offer some benefits, as well as reduce the time and cost of production [26]. Such genome based prescreening strategies are independent of growing organisms in vitro and allow researchers to narrow down possible protein targets for further analysis by traditional tools [26]. Hence, the study was designed to develop a highly antigenic polyvalent vaccine against the deadly viruses of Arenaviridae family causing severe HF in human.
2. Material and methods
2.1. Retrieval of viral proteomes and antigenic protein selection
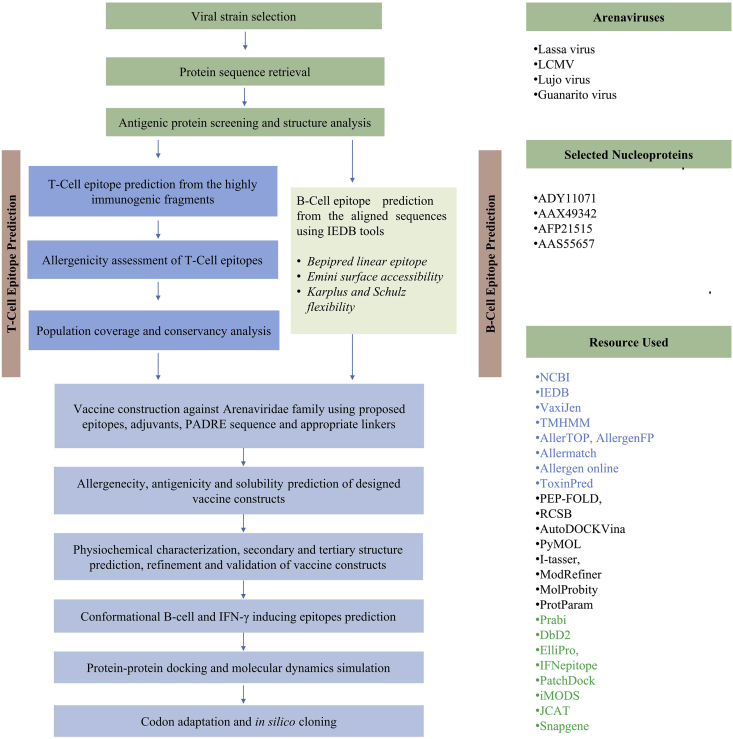
The entire protocol for designing a cross-reactive next generation vaccine against Arenaviridae family was illustrated in Figure 1. The whole genomes of LASV, LCMV, Lujo virus and Guanarito virus were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/). Study of viral genus, family, host, disease, transmission, and genome were performed by using ViralZone (https://viralzone. ExPASy.org/). The most potent immunogenic proteins were identified for all the viruses after determining the antigenicity score via VaxiJen v2.0 server [27]. Different physiochemical parameters of the proteins were analyzed through ProtParam tool [23].
Figure 1.
Schematic presentation of the procedures used for the development of cross-reactive vaccine candidate against Arenaviridae family.
2.2. Retrieval of homologous protein sets and identification of conserved regions
The selected proteins from each virus were used as query and homologous sequences were retrieved using BLASTp tool from NCBI. Multiple sequence alignment (MSA) was further performed to find out the common fragments for each set of protein using CLUSTAL Omega with 1000 bootstrap value and other default parameters to fabricate the alignment [28]. Conserved fragments were screened via VaxiJen v2.0 to find the immunogenic capacity [27]. The regions were subjected TMHMM v0.2 server for transmembrane topology prediction [29].
2.3. Prediction of T-cell epitopes, transmembrane topology screening and antigenicity analysis
Only the common fragments were used to predict the highly immunogenic T-cell epitopes through utilizing MHC-I (http://tools.iedb.org/mhci/) and MHC-II prediction tools (http://tools.iedb.org/mhcii/) from IEDB database [30]. The TMHMM server predicted the transmembrane helices in proteins [29]. Again, VaxiJen v2.0 server [27] was used to determine the antigenicity of predicted fragments to MHC-I (for CTL epitopes) and MHC-II peptides (for HTL epitopes).
2.4. Population coverage, allergenicity, toxicity and conservancy analysis of T-cell epitopes
Population coverage for each individual epitope was analyzed by IEDB population coverage calculation tool and resource (http://tools.iedb.org/population/). The allergenicity patterns of the predicted epitopes were determined through four distinct servers i.e., AllerTOP [31], AllergenFP [32], Allergen Online [33] and Allermatch [34], while the toxicity level was demonstrated using ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/). Conservancy analysis is an important tool to find the degree of distributions of epitopes in the homologous proteins. IEDB's epitope conservancy analysis tool (http://tools.iedb.org/conservancy/) was selected to find out the conservancy level of top epitopes among different viral strains.
2.5. Designing three-dimensional (3D) epitope structure and molecular docking analysis
The superior epitopes were modelled and retrieved using PEP-FOLD server [35]. Docking was conducted using AutoDOCKVina program at 1.00˚A spacing, with exhaustiveness parameters at 8.00 [36]. OpenBabel v2.3.1 was used to convert the output PDBQT files in PDB format. The docking interaction was visualized with the PyMOL molecular graphics system [23].
2.6. B-cell epitope prediction and screening
Three different algorithms i.e., Bepipred linear epitope prediction [37], Emini surface accessibility [38] and Kolaskar & Tongaonkar antigenicity scale analysis [39] from IEDB were used to identify the most potent B-cell epitopes of the selected antigenic proteins. The top epitopes were ranked based on their allergenicity pattern and VaxiJen score.
2.7. Epitope cluster analysis and vaccine construction
Epitope cluster analysis tool from IEDB was used to identify the overlapping peptides among the top CTL, HTL and B-cell epitopes at minimum sequence identity threshold of 100%. Both the clusters and unique epitopes were used and recombined to construct the vaccine molecules. Each construct started with an adjuvant followed by the top T-cell and B-cell epitopes. Interactions of adjuvants with toll like receptors (TLRs) induce robust immune reactions in the host [40]. 3 distinct adjuvants (beta defensin, L7/L12 ribosomal protein and HABA protein from Mycobacterium tuberculosis- AGV15514) were utilized in the present study. PADRE sequence was incorporated along with the adjuvant and peptides to reduce the problem caused by highly polymorphic HLA alleles. EAAAK, GGGS, GPGPG and KK linkers were used to conjugate the adjuvant, CTL, HTL and B-cell epitopes respectively.
2.8. Allergenicity, antigenicity and solubility prediction of different vaccine constructs
Allergenicity pattern of the designed vaccines were determined by AlgPred v.2.0 (http://crdd.osdd.net/raghava/algpred/). The superior candidate was identified by investigating the immunogenic potential of the peptide molecules using VaxiJen v2.0 [27]. Protein-sol software analyzed the solubility score of the proposed vaccine by calculating the surface charge, hydrophobicity and the stability at 91 different combinations of pH and ionic strength [41].
2.9. Physicochemical characterization and secondary structure analysis
ProtParam, a tool provided by ExPASy server was used to functionally characterize the vaccine proteins [23]. Instability and Aliphatic index, isoelectric pH, Molecular weight, GRAVY values, hydropathicity, and estimated half-lives of the vaccine molecules were investigated. The Prabi server (https://npsa-prabi.ibcp.fr/) predicted the alpha helix, beta sheet and coil structure of the vaccine constructs through GOR4 secondary structure prediction method.
2.10. Vaccine tertiary structure prediction, refinement, validation and disulfide engineering
I-TASSER server performed 3D modeling of the designed vaccines depending on the level of similarity between target protein and available template structure in PDB [42]. Refinement was conducted using ModRefiner to improve the accuracy of the predicted 3D modeled structure [43]. The refined protein structure was further validated by Ramachandran plot assessment through MolProbity software [44]. DbD2, an online tool was used to design disulfide bonds for the designed construct [45]. The value of chi3 considered for the residue screening was between –87 to +97 while the energy value was set <2.5.
2.11. Conformational B-cell and IFN-γ inducing epitopes prediction
The conformational B-cell epitopes in the vaccine were predicted via ElliPro server with minimum score 0.5 and maximum distance 7 Å [46]. Moreover, IFN-γ inducing epitopes within the vaccine were predicted by IFNepitope with motif and SVM hybrid prediction approach [47].
2.12. Protein-protein docking
Different Pattern Recognition Receptors (PRRs) including membrane associated Toll-like receptors (TLR3, TLR7) and cytoplasmic RIG-I-like receptors (RIG-I, MDA5) can recognize infections caused by the members of Arenaviridae [48]. Subsequent studies also revealed that α-dystroglycan (αDG) expressed at high levels in skeletal muscle acts as a primary receptor for Old World arenaviruses [49]. The 3D structure of different MHC molecules and human receptors (TLR3, RIG-I, MDA5, αDG) were retrieved from RCSB protein data bank. Protein-protein docking was conducted to determine the binding affinity of designed vaccines with different HLA alleles and human immune receptors via PatchDock [50]. Docked complexes from PatchDock were further subjected to the FireDock server to refine the complexes.
2.13. Molecular dynamics and normal mode analysis (NMA)
Normal mode analysis (NMA) was performed to strengthen the prediction via iMODS server [51]. The structural dynamics of protein complex (V1-TLR3) was investigated due to much faster and effective assessments than other molecular dynamics (MD) simulations tools [52]. The server explained the collective motion of proteins by analyzing the normal modes (NMA) in internal coordinates. Stability was determined by comparing the essential dynamics of proteins to their normal modes [23]. The eigenvalue, motion stiffness, elastic network model and covariance matrix was also investigated.
2.14. Codon adaptation and in silico cloning
Codon adaptation was performed to accelerate the expression of construct V1 in E. coli strain K12 by JCAT server. The cleavage sites of chosen restriction enzymes (BglI and BglII) and prokaryote ribosome-binding site were avoided during the process. The optimized codons for construct V1 were reversed and inserted within specific restriction site at N-terminal and C-terminal ends. To conjugate the adapted sequence into pET28a(+) vector between the BglII (401) and BglI (2187), SnapGene restriction cloning module was utilized [23].
3. Results
3.1. Retrieval of viral proteomes and antigenic protein selection
The proteomes of LASV, LCMV, Lujo and Guanarito virus were extracted in FASTA format. Arenaviruses encode four proteins which are components of virion particles. Among the rescued proteins, Nucleoproteins from Lassa virus (ADY11071), LCMV (AAX49342), Guanarito virus (AAS55657) and Lujo virus (AFP21515) were selected for vaccine candidacy on the basis of highest antigenicity score and different physiochemical parameters were analyzed (Supplementary file 1).
3.2. Retrieval of homologous protein sets and identification of conserved regions
In the present study, the conserved regions of nucleoproteins among the isolates of individual arenaviruses were identified. Different homologous protein sets for LASV (97 sequences with >95% identity), LCMV (39 sequences) and Guanarito virus (22 sequences) were generated after BLASTp search using NCBI BLAST tools (Supplementary file 2). Multiple sequence alignment reveled 5, 12 and 9 conserved fragments among the LASV, LCMV and Guanarito virus nucleoproteins respectively (Table 1). Finally, 4, 6, and 6 fragments from the respective nucleoproteins passed the recommended threshold value through VaxiJen v2.0 server (Table 1). Moreover, transmembrane topology revealed, among the immunogenic conserved regions 3, 5 and 5 sequences met the criteria of exomembrane characteristics (Table 1). There is only one NP sequence available at NCBI database for Lujo virus at this moment. That is why we could not perform organism specific BLASTp in this case, rather used the whole sequence to predict antigenic epitopes.
Table 1.
Identified conserved regions among different homologous protein sets of LASV, LCMV and Guanarito virus.
| Virus | Protein | Conserved region | Vaxijen | Topology |
|---|---|---|---|---|
| Lassa virus | Nucleoprotein | YCSNIKLQVVKDAQALLHGLDFSEV | 0.5300 | inside |
| NNQFGTMPSLTLACLTKQGQVDLND | 1.0428 | outside | ||
| SGYNFSLGAAVKAGACMLDGGNMLET | 0.6470 | outside | ||
| PGERNPYENILYKICLSGDGWPYIASRTSI | 0.0358 | outside | ||
| WMDIEGRPEDPVE | 1.0407 | outside | ||
| LCMV | Nucleoprotein | KSFQWTQALRRELQ | 0.5501 | inside |
| LLNGLDFSEVSNVQRIMRKE | 0.5746 | outside | ||
| DKDLQRLRSLNQTV | 0.3230 | inside | ||
| DSSLLNNQFGTMPSLTM | 0.4840 | outside | ||
| LNDVVQALTDLGLLYTVKYPNL | 0.6929 | outside | ||
| KLNMFVSDQVGDRNPYE | 0.3663 | outside | ||
| NFSLGAAVKAGAALL | 1.2041 | outside | ||
| GGNMLESILIKPSN | 0.2419 | outside | ||
| NILYKVCLSGEGWPYIACRT | 0.2895 | outside | ||
| MLLKDLMGGIDPN | 0.5306 | outside | ||
| GALPQGMVLSCQGS | 0.1074 | outside | ||
| TPHCALMDCIIFESA | 0.0003 | outside | ||
| Guanarito virus | Nucleoprotein | DAKLIADSLDFTQVS | 0.9498 | outside |
| PRMYMGNLTQSQLEKRAGILR | 0.1864 | inside | ||
| AGGVVRLWDVSDPSKLNNQFGSMPALTIA | 0.5374 | outside | ||
| CMTVQGGETMNNVVQALTSLGLLYTVKYPNLDDLEKLTLEHDCLQIITKDESALNISGYN | 0.5873 | outside | ||
| FSLSAAVKAGASLIDGGNMLETI | 0.5427 | outside | ||
| LLYKLCLSGEGWPYIGSRSQI | 0.3678 | outside | ||
| TIWMDIEGPPTDPVELAVFQPSSG | -0.0436 | outside | ||
| TVQGADDIKKLFDIHGRKDLKLVDVRLTGE | 0.9209 | inside | ||
| VWEKFGHLCRAHNGVIVPKKK | 0.9542 | outside |
3.3. Prediction of T-cell epitopes, transmembrane topology screening and antigenicity analysis
A plethora of immunogenic epitopes from the conserved sequences were generated that could bind maximum number of HLA cells with high binding affinity (Supplementary file 3 and Supplementary file 4). Top epitopes from each protein were selected as putative T cell epitope candidates based on their transmembrane topology screening and antigenicity score (Table 2).
Table 2.
Predicted T-cell (CTL and HTL) epitopes of LASV, LCMV, Lujo and Guanarito virus Nucleoproteins.
| Types | Virus | Epitope | Start | End | VaxiJen Score |
No. of interacting HLAs | Conservancy (identity ≤100%) |
|---|---|---|---|---|---|---|---|
| MHC-I petides (CTL epitopes) | Lassa Virus (LASV) | IEGRPEDPV | 4 | 12 | 1.1593 | 81 | 100.00% (97/97) |
| NFSLGAAVK | 4 | 12 | 1.8014 | 27 | 100.00% (97/97) | ||
| PSLTLACLT | 8 | 16 | 1.5130 | 27 | 100.00% (97/97) | ||
| YNFSLGAAV | 3 | 11 | 1.5124 | 54 | 100.00% (97/97) | ||
| SLGAAVKAG | 6 | 14 | 1.4140 | 27 | 100.00% (97/97) | ||
| MPSLTLACL | 7 | 15 | 1.3792 | 81 | 100.00% (97/97) | ||
| FSLGAAVKA | 5 | 13 | 1.3262 | 81 | 100.00% (97/97) | ||
| GYNFSLGAA | 2 | 10 | 1.2890 | 54 | 100.00% (97/97) | ||
| DIEGRPEDP | 3 | 11 | 1.2557 | 27 | 100.00% (97/97) | ||
| SLGAAVKAGA | 6 | 15 | 1.1754 | 27 | 100.00% (97/97) | ||
| LCMV | NFSLGAAVK | 1 | 9 | 1.8014 | 54 | 84.62% (33/39) | |
| LDFSEVSNV | 5 | 13 | 1.7221 | 81 | 97.44% (38/39) | ||
| NGLDFSEVS | 3 | 11 | 1.6145 | 81 | 97.44% (38/39) | ||
| GLDFSEVSN | 4 | 13 | 1.5849 | 27 | 97.44% (38/39) | ||
| DLGLLYTVK | 10 | 18 | 1.4806 | 81 | 94.87% (37/39) | ||
| SLGAAVKAG | 3 | 12 | 1.1754 | 27 | 89.74% (35/39) | ||
| LNGLDFSEV | 2 | 10 | 1.3833 | 81 | 97.44% (38/39) | ||
| DFSEVSNVQ | 6 | 14 | 1.3277 | 81 | 97.44% (38/39) | ||
| FSLGAAVKA | 2 | 10 | 1.3262 | 81 | 89.74% (35/39) | ||
| TDLGLLYTV | 9 | 17 | 1.2339 | 81 | 94.87% (37/39) | ||
| Lujo Virus | WDVKDSSLL | 5 | 13 | 1.7861 | 54 | 100.00% (1/1) | |
| VWDVKDSSL | 4 | 12 | 1.7406 | 27 | 100.00% (1/1) | ||
| VWDVKDSSLL | 4 | 13 | 1.4658 | 27 | 100.00% (1/1) | ||
| DVKDSSLLN | 6 | 14 | 1.2269 | 54 | 100.00% (1/1) | ||
| RVWDVKDSSL | 3 | 12 | 0.9679 | 27 | 100.00% (1/1) | ||
| KDSSLLNNQ | 8 | 16 | 0.8111 | 54 | 100.00% (1/1) | ||
| RVWDVKDSS | 3 | 11 | 0.7777 | 54 | 100.00% (1/1) | ||
| QFGTMPSLT | 16 | 24 | 0.7616 | 27 | 100.00% (1/1) | ||
| LNNQFGTMP | 13 | 21 | 0.7367 | 27 | 100.00% (1/1) | ||
| KDSSLLNNQF | 8 | 17 | 0.6510 | 27 | 100.00% (1/1) | ||
| Guanarito Virus | DSLDFTQVS | 7 | 15 | 1.8576 | 27 | 100.00% (22/22) | |
| ADSLDFTQVS | 6 | 15 | 1.5975 | 27 | 100.00% (22/22) | ||
| GWPYIGSRS | 11 | 19 | 1.5322 | 27 | 100.00% (22/22) | ||
| LKLVDVRLTG | 20 | 29 | 1.4348 | 27 | 100.00% (22/22) | ||
| ADSLDFTQV | 6 | 14 | 1.4384 | 27 | 100.00% (22/22) | ||
| KYPNLDDLE | 27 | 35 | 1.4366 | 27 | 100.00% (22/22) | ||
| VWEKFGHLC | 1 | 9 | 1.3953 | 27 | 100.00% (22/22) | ||
| LVDVRLTGE | 22 | 30 | 1.3388 | 27 | 100.00% (22/22) | ||
| TSLGLLYTVK | 18 | 27 | 1.3006 | 27 | 100.00% (22/22) | ||
| SLGLLYTVK | 19 | 27 | 1.2711 | 27 | 100.00% (22/22) | ||
| MHC-II petides (HTL epitopes) | Lassa Virus (LASV) | PSLTLACLTKQGQVD | 8 | 22 | 1.1813 | 27 | 100.00% (97/97) |
| NFSLGAAVKAGACML | 4 | 18 | 1.1070 | 27 | 100.00% (97/97) | ||
| SGYNFSLGAAVKAGA | 1 | 15 | 1.0922 | 27 | 100.00% (97/97) | ||
| NNQFGTMPSLTLACL | 1 | 15 | 1.0193 | 27 | 100.00% (97/97) | ||
| FSLGAAVKAGACMLD | 5 | 19 | 0.9323 | 27 | 100.00% (97/97) | ||
| LTLACLTKQGQVDLN | 10 | 24 | 1.4060 | 27 | 100.00% (97/97) | ||
| SLTLACLTKQGQVDL | 9 | 23 | 1.3579 | 27 | 100.00% (97/97) | ||
| YNFSLGAAVKAGACM | 3 | 17 | 1.1719 | 27 | 100.00% (97/97) | ||
| TMPSLTLACLTKQGQ | 6 | 20 | 1.1134 | 27 | 100.00% (97/97) | ||
| MPSLTLACLTKQGQV | 7 | 21 | 1.0459 | 27 | 100.00% (97/97) | ||
| LCMV | NFSLGAAVKAGAALL | 1 | 15 | 1.2041 | 27 | 84.62% (33/39) | |
| LTDLGLLYTVKYPNL | 8 | 22 | 1.1861 | 27 | 94.87% (37/39) | ||
| ALTDLGLLYTVKYPN | 7 | 21 | 0.9756 | 27 | 94.87% (37/39) | ||
| LNGLDFSEVSNVQRI | 2 | 16 | 0.9237 | 27 | 97.44% (38/39) | ||
| LLNGLDFSEVSNVQR | 1 | 15 | 0.8913 | 27 | 97.44% (38/39) | ||
| VQALTDLGLLYTVKY | 5 | 19 | 0.8413 | 27 | 94.87% (37/39) | ||
| VVQALTDLGLLYTVK | 4 | 18 | 0.8009 | 27 | 94.87% (37/39) | ||
| NGLDFSEVSNVQRIM | 3 | 17 | 0.7633 | 27 | 97.44% (38/39) | ||
| QALTDLGLLYTVKYP | 6 | 20 | 0.6954 | 27 | 94.87% (37/39) | ||
| DVVQALTDLGLLYTV | 3 | 17 | 0.5867 | 27 | 94.87% (37/39) | ||
| Lujo Virus | VWDVKDSSLLNNQFG | 4 | 18 | 1.1337 | 27 | 100.00% (1/1) | |
| WDVKDSSLLNNQFGT | 5 | 19 | 1.1169 | 27 | 100.00% (1/1) | ||
| DVKDSSLLNNQFGTM | 6 | 20 | 0.8891 | 27 | 100.00% (1/1) | ||
| RVWDVKDSSLLNNQF | 3 | 17 | 0.7122 | 27 | 100.00% (1/1) | ||
| VRVWDVKDSSLLNNQ | 2 | 16 | 0.6717 | 27 | 100.00% (1/1) | ||
| VKDSSLLNNQFGTMP | 7 | 21 | 0.6619 | 27 | 100.00% (1/1) | ||
| KDSSLLNNQFGTMPS | 8 | 22 | 0.5533 | 27 | 100.00% (1/1) | ||
| VVRVWDVKDSSLLNN | 1 | 15 | 0.4941 | 27 | 100.00% (1/1) | ||
| SSLLNNQFGTMPSLT | 10 | 24 | 0.4797 | 27 | 100.00% (1/1) | ||
| DSSLLNNQFGTMPSL | 9 | 23 | 0.3935 | 27 | 100.00% (1/1) | ||
| Guanarito Virus | LFDIHGRKDLKLVDV | 11 | 25 | 1.5311 | 27 | 100.00% (22/22) | |
| TSLGLLYTVKYPNLD | 18 | 32 | 1.1723 | 27 | 100.00% (22/22) | ||
| LGLLYTVKYPNLDDL | 20 | 34 | 1.1312 | 27 | 100.00% (22/22) | ||
| SLGLLYTVKYPNLDD | 19 | 33 | 1.1124 | 27 | 100.00% (22/22) | ||
| LTSLGLLYTVKYPNL | 17 | 31 | 1.0659 | 27 | 100.00% (22/22) | ||
| DAKLIADSLDFTQVS | 1 | 15 | 0.9498 | 27 | 100.00% (22/22) | ||
| GLLYTVKYPNLDDLE | 21 | 35 | 0.9362 | 27 | 100.00% (22/22) | ||
| ALTSLGLLYTVKYPN | 16 | 30 | 0.8800 | 27 | 100.00% (22/22) | ||
| FSLSAAVKAGASLID | 1 | 15 | 0.8734 | 27 | 100.00% (22/22) | ||
| LTLEHDCLQIITKDE | 37 | 51 | 0.8608 | 27 | 100.00% (22/22) |
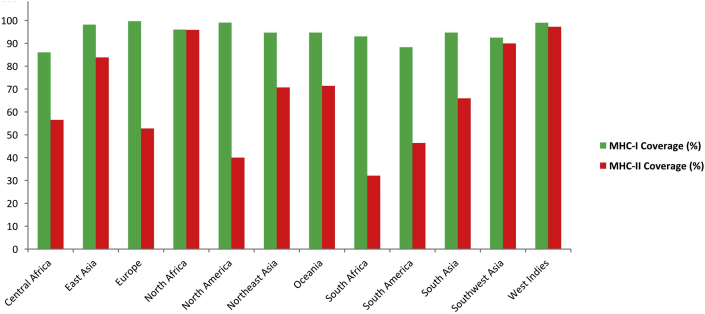
3.4. Population coverage, allergenicity, toxicity and conservancy analysis of T-cell epitopes
Results showed that population from the most geographic areas can be covered by the predicted T-cell epitopes (Figure 2). Non-allergenic epitopes were identified using four distinct allergenicity assessment servers (Supplementary file 5 and Supplementary file 6). Epitopes those were indicated as allergen and toxic for human were removed from the predicted list of epitopes (Table 2). Putative epitopes generated from 4 diverse viral proteins were found to be highly conserved (84%–100%) within different strains (Table 2). Highly conserved epitopes which are expected to confer broad immune response were selected and subjected to molecular docking study (Supplementary file 7).
Figure 2.
Population coverage analysis of predicted T-cell epitopes to MHC-I and MHC-II based antigenicity.
3.5. Designing three-dimensional (3D) epitope structure and molecular docking analysis
A total of 8 T-cell (4 CTL and 4 HTL) epitopes were subjected to PEP-FOLD server for 3D structure conversion and their interactions with HLA molecules were analyzed. The top epitopes were docked against class-I HLA-A∗11:01 and class-II HLA-DRB1∗01:01. Results confirmed that all the predicted epitopes bound in the groove of MHC molecules with a minimal binding energy (Supplementary file 8).
3.6. B-cell epitope prediction and screening
B-cell epitopes were predicted according to the analysis via three different algorithms from IEDB. Top B-cell epitopes for selected four proteins were screened based on their antigenicity scoring and allergenicity pattern, and were used to design the final vaccine construct (Table 3).
Table 3.
Allergenicity assessment and antigenicity analysis of the predicted B-cell epitopes.
| Virus | Proteins | Algorithm | Top CTL epitopes | Allergenicity | Vaxijen score |
|---|---|---|---|---|---|
| LASV | Nucleoprotein | Linear Epitope | PNAKTWMDIEGRPEDPVE | Non-allergen | 0.6123 |
| Surface Accessibility | LMRKERRDDNDLKRLRDL | Non-allergen | 0.9451 | ||
| Antigenicity | SLTLACLTKQ | Non-allergen | 1.5660 | ||
| LCMV | Nucleoprotein | Linear Epitope | SERPQASGV | Non-allergen | 0.9344 |
| Surface Accessibility | IMRKEKRDDKDLQRLR | Non-allergen | 0.9494 | ||
| Antigenicity | PPQVGLSYSQTM | Non-allergen | 1.0820 | ||
| Lujo Virus | Nucleoprotein | Linear Epitope | KVVKDAVSLINGLDFSMV | Non-allergen | 0.5035 |
| Surface Accessibility | PNMDDLDKLKNK | Non-allergen | 0.7144 | ||
| Antigenicity | NLLYKICLSG | Non-allergen | 0.7217 | ||
| Guanarito Virus | Nucleoprotein | Linear Epitope | DERPGNRNPY | Non-allergen | 0.9986 |
| Surface Accessibility | DERPGNRNPY | Non-allergen | 0.9986 | ||
| Antigenicity | PGLLSYVIGLLPQGSVITVQ | Non-allergen | 0.6398 |
3.7. Epitope cluster analysis and vaccine construction
A total 19 clusters were identified among the top epitopes (4 CTL, 4 HTL and 12 B-cell epitopes) as proposed in Supplementary File 7 and Table 3, and were utilized to design the final constructs. Each construct was composed of a protein adjuvant followed by T-cell and B-cell epitopes with their respective linkers. A total 3 vaccine molecules (i.e. V1, V2, V3) of 394, 479 and 508 amino acid residues were designed and further investigated to evaluate their immunogenic potential (Table 4).
Table 4.
Allergenicity, antigenicity and solubility prediction of the designed vaccine constructs.
| Constructs | Composition | Complete Sequence of Vaccine Constructs | Antigenicity (Threshold 0.4) | Allergenicity (Threshold -0.4) | Solubility (Threshold 0.45) |
|---|---|---|---|---|---|
| V1 | Predicted CTL, HTL & B-cell epitopes with β defensin adjuvant and PADRE sequence | EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKAKFVAAWTLKAAAGGGSLDFSEVSNVGGGSIEGRPEDPVGGGSKDSSLLNNQGGGSGWPYIGSRSGPGPGALTDLGLLYTVKYPNGPGPGNNQFGTMPSLTLACLGPGPGVWDVKDSSLLNNQFGGPGPGLTSLGLLYTVKYPNLKKSERPQASGVKKIMRKEKRDDKDLQRLRKKPPQVGLSYSQTMKKPNAKTWMDIEGRPEDPVEKKLMRKERRDDNDLKRLRDLKKSLTLACLTKQKKKVVKDAVSLINGLDFSMVKKPNMDDLDKLKNKKKNLLYKICLSGKKDERPGNRNPYKKPGLLSYVIGLLPQGSVITVQKKAKFVAAWTLKAAAGGGS | 0.703 | -0.45 | 0.65 |
| V2 | Predicted CTL, HTL & B-cell epitopes with L7/L12 ribosomal protein adjuvant & PADRE sequence | EAAAKMAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVKEAAAKAKFVAAWTLKAAAGGGSLDFSEVSNVGGGSIEGRPEDPVGGGSKDSSLLNNQGGGSGWPYIGSRSGPGPGALTDLGLLYTVKYPNGPGPGNNQFGTMPSLTLACLGPGPGVWDVKDSSLLNNQFGGPGPGLTSLGLLYTVKYPNLKKSERPQASGVKKIMRKEKRDDKDLQRLRKKPPQVGLSYSQTMKKPNAKTWMDIEGRPEDPVEKKLMRKERRDDNDLKRLRDLKKSLTLACLTKQKKKVVKDAVSLINGLDFSMVKKPNMDDLDKLKNKKKNLLYKICLSGKKDERPGNRNPYKKPGLLSYVIGLLPQGSVITVQKKAKFVAAWTLKAAAGGGS | 0.630 | 0.010 | 0.67 |
| V3 | Predicted CTL, HTL & B-cell epitopes with HABA adjuvant & PADRE sequence | EAAAKMAENPNIDDLPAPLLAALGAADLALATVNDLIANLRERAEETRAETRTRVEERRARLTKFQEDLPEQFIELRDKFTTEELRKAAEGYLEAATNRYNELVERGEAALQRLRSQTAFEDASARAEGYVDQAVELTQEALGTVASQTRAVGERAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSLDFSEVSNVGGGSIEGRPEDPVGGGSKDSSLLNNQGGGSGWPYIGSRSGPGPGALTDLGLLYTVKYPNGPGPGNNQFGTMPSLTLACLGPGPGVWDVKDSSLLNNQFGGPGPGLTSLGLLYTVKYPNLKKSERPQASGVKKIMRKEKRDDKDLQRLRKKPPQVGLSYSQTMKKPNAKTWMDIEGRPEDPVEKKLMRKERRDDNDLKRLRDLKKSLTLACLTKQKKKVVKDAVSLINGLDFSMVKKPNMDDLDKLKNKKKNLLYKICLSGKKDERPGNRNPYKKPGLLSYVIGLLPQGSVITVQKKAKFVAAWTLKAAAGGGS | 0.650 | -1.004 | 0.63 |
3.8. Allergenicity, antigenicity and solubility prediction of different vaccine constructs
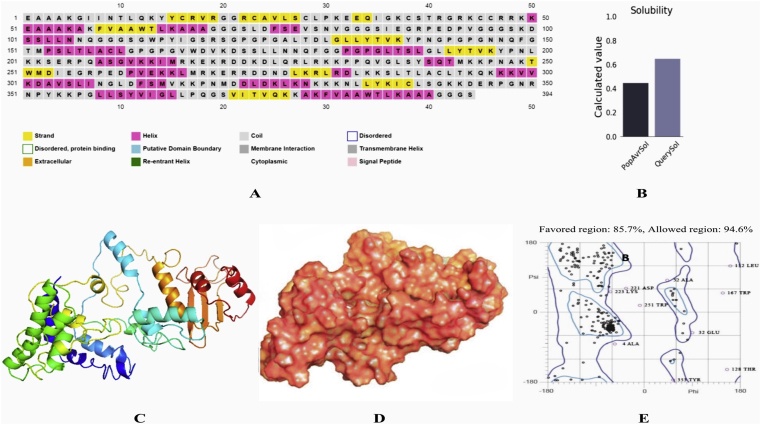
Both vaccine construct V1 and V2 exhibited non-allergic pattern, while V3 showed slightly allergenic behavior (Table 4). Construct V1 was found superior considering the safety assessment results, antigenicity scoring (0.703) and degree of solubility (0.65) (Figure 3B).
Figure 3.
Secondary structure analysis (A) and solubility prediction of vaccine construct V1 (B). Here, QuerySol (scaled solubility value) higher than PopAvrSol (population average for experimental dataset) indicates higher solubility of V1 than average soluble E. coli protein from experimental solubility dataset. Homology modeling of vaccine protein V1 via I-TASSER (C: Cartoon format, D: Surface structure) and validation by Ramachandran plot analysis (E).
3.9. Physicochemical characterization and secondary structure analysis of vaccine protein
Vaccine construct V1 was characterized on the basis of physical and chemical properties. The computed instability index of the protein was 35.87 which classified it as a stable one. The theoretical pI was demonstrated as 9.96, indicating its net negative charge above the specific pI. The extinction coefficient was 43890, considering all the cysteine residues reduced at 0.1%. In mammalian reticulocytes in vitro the vaccine was expected to have a biological half-life of 1 h, and 30 min in yeast in vivo (>10 h in E. coli). The aliphatic index and GRAVY values were 75.25 and -0.625, respectively. Secondary structure of the construct V1 confirmed to have 26.14% alpha helix, 20.56% sheet and 53.30% coil structure (Figure 3A).
3.10. Tertiary structure prediction, refinement, validation and disulfide engineering of the vaccine constructs
5 tertiary structures were generated via I-TASSER for construct V1 using the top 10 templates from LOMETS. TM score and RMSD were estimated based on C score which was minimum for Model 1 (-0.77), thus ensuring its better quality (Figure 3C and D). The refined structure was validated through Ramachandran plot analysis which revealed that 94.6% residues were in the allowed and 5.36% residues in the outlier region (Figure 3E). Modeled tertiary structure of vaccine construct V2 and V3 have been shown in Figure 4. 54 pairs of disulfide bond forming amino acid residues were identified via DbD2. However, after evaluation of the residue pairs in terms of energy, chi3 and B-factor parameter, only 2 pairs (ALA 24-PRO 162 and CYS 38-LYS 44) met the criteria for disulfide bond formation and were replaced with cysteine (Figure 5).
Figure 4.
3D modelled structure of vaccine protein V2 (A) and V3 (B).
Figure 5.
Disulfide engineering of the candidate vaccine V1 (A: Initial form, B: Mutant form).
3.11. Conformational B-cell and IFN-γ inducing epitopes prediction
A total 5 conformational B-cell epitopes were generated from the designed vaccine construct. Epitopes no. 2 (85 residues) and epitope no. 4 (13 residues) were considered as the broadest and smallest conformational B- cell epitopes in the study (Supplementary file 9 and Figure 6). Results also revealed that most of the linear epitopes were included in the conformational B-cell epitopes. 15-mer IFN-γ inducing epitopes were also predicted from the final vaccine constructs. Results confirmed 100 positive IFN-γ inducing epitopes from which 8 had a score ≥1. Regions from 375 to 390 in the vaccine showed highest score of 1.209 (Supplementary file 10).
Figure 6.
Predicted 24 mer (A), 85 mer (B), 75 mer (C), 13 mer (D) and 14 mer (E) conformational epitopes within construct V1 with score ranging from 0.56 up to 0.75.
3.12. Protein-protein docking
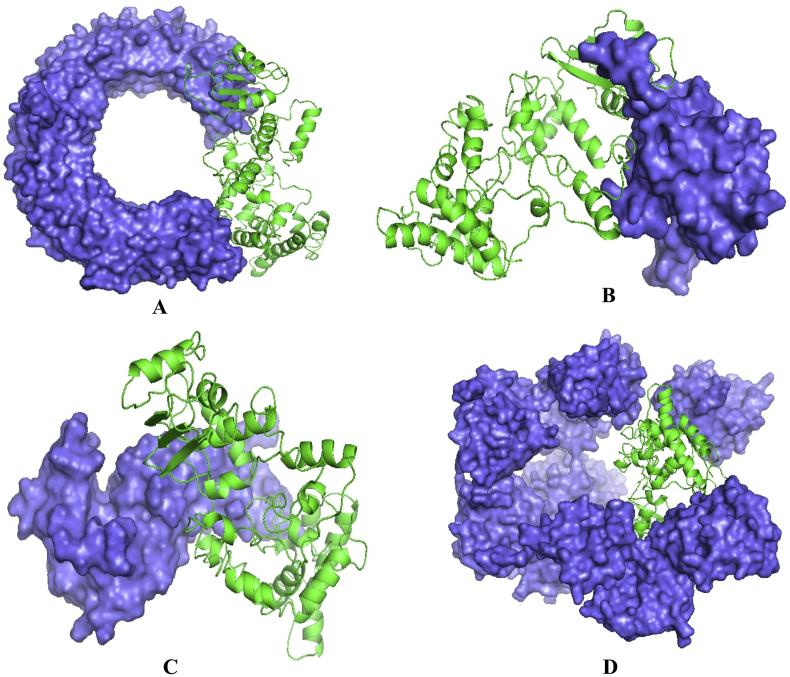
Patchdock server ranked the docked complexes based on Atomic Contact Energy (ACE), complementary score and approximate interface area of the complex. Construct V1 showed lowest binding energy, while compared with other constructs (Table 5). Binding affinity of construct V1 with different human immune receptors was biologically significant as well (Figure 7 and Table 5).
Table 5.
Docking scores of the vaccine constructs with different HLA alleles i.e. HLA-DRB1∗03:01 (1a6a), HLA-DRB5∗01:01 (1h15), HLA-DRB1∗01:01 (2fse), HLA-DRB3∗01:01 (2q6w), HLA-DRB1∗04:01 (2seb) and HLA-DRB3∗02:02 (3c5j).
| Vaccine constructs | PDB IDs of HLAs/receptors | Global Energy |
Hydrogen bond energy | ACE | Score | Area |
|---|---|---|---|---|---|---|
| V1 | 1a6a | -2.65 | -3.20 | 5.18 | 16280 | 2111.50 |
| 1h15 | -6.40 | -2.75 | 7.08 | 17246 | 2552.90 | |
| 2fse | -26.12 | -2.94 | 0.28 | 16062 | 2181.20 | |
| 2q6w | -2.59 | -4.48 | 9.33 | 15968 | 2084.90 | |
| 2seb | -28.54 | -3.71 | 5.51 | 17904 | 2308.00 | |
| 3c5j | -28.29 | -5.12 | 9.40 | 16956 | 2397.50 | |
| V2 | 1a6a | -0.55 | -2.61 | 12.31 | 16394 | 2349.80 |
| 1h15 | -0.55 | -4.21 | 10.79 | 17406 | 2402.80 | |
| 2fse | 12.04 | -5.21 | 12.26 | 18306 | 2646.60 | |
| 2q6w | -0.33 | -3.06 | 6.47 | 18262 | 2940.70 | |
| 2seb | -5.52 | -0.82 | -1.77 | 21390 | 3942.60 | |
| 3c5j | -22.27 | -3.87 | 9.04 | 16846 | 2299.50 | |
| V3 | 1a6a | -0.23 | -4.64 | 10.85 | 17830 | 2324.60 |
| 1h15 | -16.48 | -3.48 | 15.41 | 19260 | 2658.10 | |
| 2fse | -9.38 | -1.68 | 12.37 | 19720 | 2651.90 | |
| 2q6w | -23.12 | -4.09 | 10.44 | 19724 | 2701.10 | |
| 2seb | -19.67 | -0.52 | -3.87 | 18596 | 3097.60 | |
| 3c5j | 1.78 | -5.71 | 12.71 | 18192 | 2430.00 |
Figure 7.
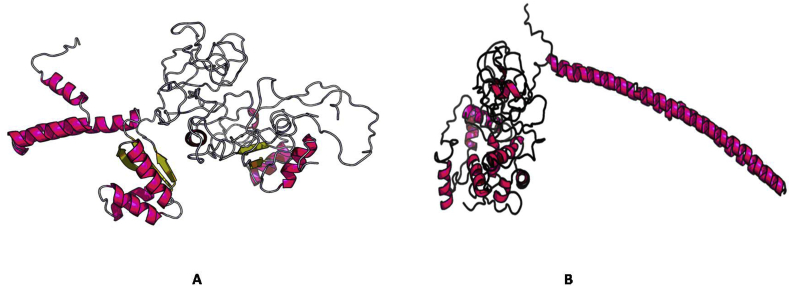
Docked complex of vaccine construct V1 and human TLR3 (A), MDA 5 (B), α dystroglycan (C) and RIG-1 (D) with a binding energy of -10.82, -3.91, -17.17, -0.89 kCal/mole respectively.
3.13. Molecular dynamics and normal mode analysis (NMA)
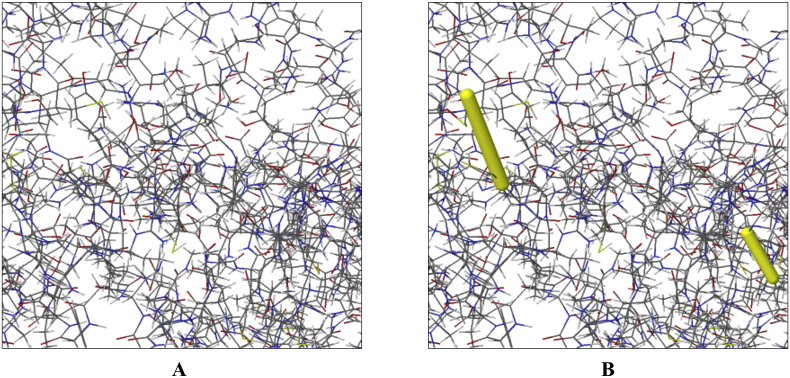
Normal mode analysis was performed to describe the stability of protein complex and large-scale mobility. The location of hinges in the chain (as shown in Figure 8A) was negligible, while an averaged RMS was given by the B-factor column (Figure 8B). The estimated higher eigenvalue (2.2023e−06) indicated low chance of deformation of the protein complex (Figure 8C). Colored bars in Figure 8D show the individual (red) and cumulative (green) variances which were inversely related to eigenvalue. The coupling between residues were revealed by covariance matrix i.e., correlated, uncorrelated and anti-correlated motions indicated by red, white and blue colors respectively (Figure 8E), while the elastic network model (Figure 8F) identified the pairs of atoms connected via springs (the darker the grays, the stiffer the springs).
Figure 8.
Molecular dynamics simulation of vaccine protein V1 and TLR3 complex. Stability of the protein-protein complex was investigated through deformability (A), B-factor (B), eigenvalue (C), variance (D), covariance (E) and elastic network (F) analysis.
3.14. Codon adaptation and in silico cloning
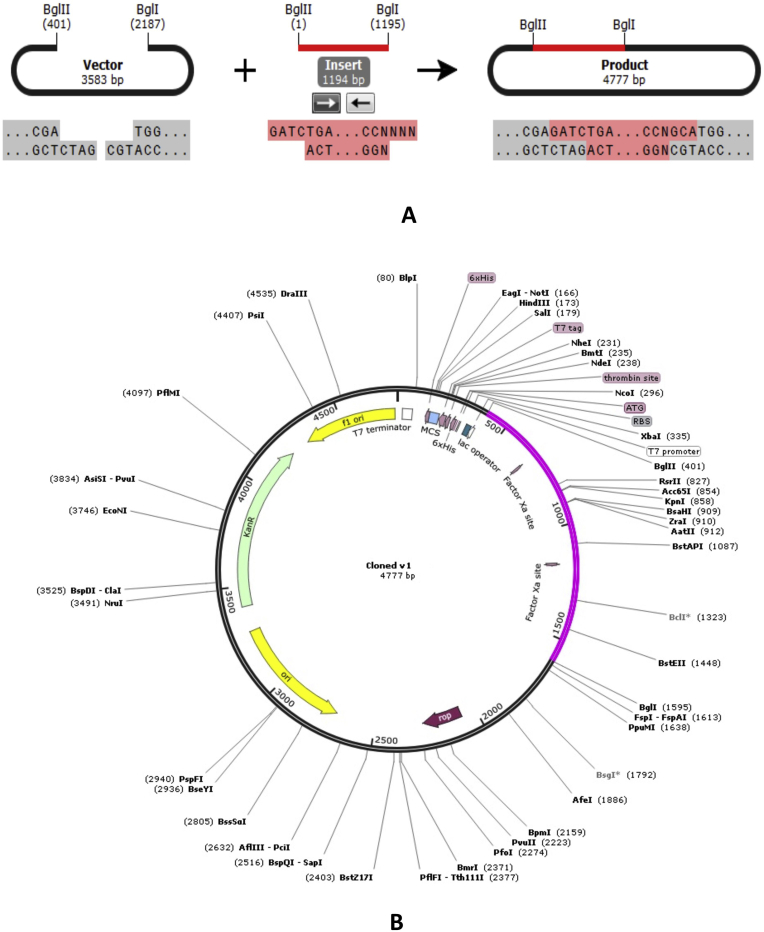
The Codon Adaptation Index (CAI) of construct V1 was 0.969. Results also revealed a significant GC contents (50.93%) for the adapted codons of the vaccine molecule. An insert of 1194 bp was obtained which lacked restriction sites for BglII and BglI, thus ensuring safety for the cloning purpose. The codons were inserted into pET28a(+) vector along with BglII and BglI restriction sites and a clone of 4777 base pair was produced (Figure 9).
Figure 9.
Restriction digestion (A) and in silico cloning (B) of the gene sequence of final construct V1 into pET28a(+) expression vector. Target sequence was inserted between BglII (401) and BGlI (2187) indicated in violate color.
4. Discussion
Designing multivalent vaccine has enormous advantages to prevent infections caused by similar group of viruses [53, 54]. Potential advantages of peptide vaccines such as increased safety and opportunity to rationally engineer the epitopes led the researchers to shift their focus towards the development of subunit vaccines [55, 56]. Hence, designing a cross-reactive vaccine against Arenaviral family has brought our attention owing to the emergence of infection by LASV, LCMV, Lujo and Machupo virus. Due to the fact that these viruses are generally geographically restricted, designing a chimeric construct would save both time and cost to develop a vaccine against arenaviruses and to perform in vivo trials in future. In the present study, attempts were taken to develop a vaccine candidate using the most antigenic viral proteins of Arenaviridae family that could elicit broad spectrum immunity in the host. Kotturi et al. (2009) demonstrated the possibility of designing a universal vaccine against arenaviruses either through the identification of widely conserved epitopes, or by the identification of a collection of epitopes derived from multiple arenavirus species. However, widely conserved CD8+ T cell epitopes among different arenaviruses species were not identified due to significant variation in their amino acid sequences [57]. Consistent with our study, the authors emphasized on multivalent vaccine strategy expressing antigens from different arenavirus species to obtain cross protection. Therefore, combination of epitopes from different arenaviral nucleoproteins as designed in this study could be an effective vaccinomics approach. Sakabe et al. (2020) identified 12 CD8+ T-cell Epitopes from 6 Lassa Fever (LF) survivors in Nigeria and Sierra Leone. However, HLAs were not universally conserved among LF survivors suggesting substantial variability in epitope-specific responses among individuals [58]. The top epitopes in our study do not overlap with the previous one, however showed high population coverage around the world.
Nucleoprotein (NP) specific CD8+ T cells play a major role in virus control and immune stimulation in the host [59]. Barale et al. (2020) explored Lassa virus glycoprotein to identify candidate epitopes in a previous study [60]. However, this study focused on NP protein and the prediction of conserved epitopes in this protein is new. Meulen and coworkers (2000) found that Lassa fever survivors had strong CD4+ T cell responses against LASV Nucleoprotein [61]. In another study, a single inoculation of a plasmid encoding full-length Lassa nucleoprotein induced CD8(+) T cell responses in mice model and protected against LCMV [62]. All these findings suggest that anti-NP response at an early stage effectively controls and contributes to cross-protective immunity against arenavirus infections. The superiority of Nucleoproteins of LASV, LCMV, Lujo and Guanarito virus in terms of antigenicity score were also revealed in this study (Supplementary File 1). Considering the fact, a multivalent vaccine approach was employed to achieve protection against nucleoprotein antigens of different arenaviruses. Protective response against numerous viral strains is possible if the candidate epitopes remain in the highly conserved regions. Thus, only the conserved sequences from each viral protein (Table 1) were used for epitope prediction, which we believe, would generate more acceptable, cross protective and universally effective vaccine constructs [63, 64]. Determination of potent T cell epitopes is a pivotal step during vaccine design, since T-cells play the key role in antibody production through antigen presentation [65]. Therefore, both MHC-I (CTL) and MHC-II (HTL) restricted epitopes were predicted using IEDB T-cell epitope prediction tools. Top candidates were screened based on transmembrane topology screening, antigenicity scoring, allergenicity and toxicity pattern, and conservancy analysis (Table 2). The proposed epitopes showed a high cumulative population coverage in most of the geographic regions, including 88% and 86% MHC-I coverage in South America and Africa respectively (Figure 2). Moreover, the top epitopes were checked for their ability to bind with class-I and class-II MHC alleles and were found to be efficient binders of corresponding alleles (Supplementary File 8). B-cell epitopes boost neutralizing-antibody responses in the host. To achieve protective immunity against arenavirus infection, B-cell epitopes were identified using three different algorithms (Table 3). Results also revealed that among the top epitopes, T cell epitope GWPYIGSRS were conserved in Argentine mammarenavirus (Junin virus) and Brazilian mammarenavirus (Sabia virus), while B cell epitope NLLYKICLSG were conserved in Bolivian mammarenavirus (Machupo virus), indicating the possibility of final vaccine constructs to confer broad range immunity in the host against the arenaviruses.
Specialized spacer sequences enabled researchers to perform rational designing of potent multiepitope vaccine [66]. Therefore, in the present study, previously reported GGGS, GPGPG, and KK linkers were utilized to produce sequences with minimized junctional immunogenicity [54, 67]. Three different constructs were generated using 3 distinct adjuvants, and were investigated in terms of safety and efficacy (Table 4). Vaccine V1 was identified best in terms of immunogenic potential, hydrophobicity, and allergenicity and other physicochemical behavior. Moreover, the predicted instability index 35.87 confirmed that the vaccine protein will be stable upon expression. To strengthen our prediction, all three constructs were subjected to 3D modeling, and interactions between designed vaccine constructs with different HLA molecules were determined. V1 was found superior considering the free binding energy (Table 5). The Ramachandran plot also indicated that the quality of the overall model was satisfactory. IFN-γ is the signature cytokine of both the innate and adaptive immune systems with ability to provoke antiviral immune responses and protection against re-infection. The release of IFN-γ enhances the magnitude of antiviral cytotoxic T lymphocytes (CTLs) responses and aids in production of neutralizing IgG [68]. The proposed vaccine molecule (V1) was also screened in this study to identify IFN-γ producing epitopes (Supplementary File 10). The main hallmark of good vaccines is that they closely mimic the successful natural triggers of dendritic cell (DC) activation. Effective DC-activation signals employ molecularly defined innate immunity receptors such as those belonging to the Toll-like receptor (TLR) family and other adaptive immunity receptors [69, 70]. One effective strategy to design an efficient vaccine is to use Toll-Like Receptor (TLR) ligands as an adjuvant which polarizes CD4+ T helper cells to provoke CTL response [71] and thereby enhancing the efficacy of the multi-epitope subunit vaccine [72]. α-dystroglycan (α-DG), a peripheral membrane protein, acts as an anchor between the sub-membranous cytoskeleton and the extracellular matrix which is widely expressed in host cells [73]. It has been identified as a host receptor for all Old World arenaviruses and certain strains of New World arenaviruses [74, 75]. Therefore, another docking analysis was performed to explore the binding affinity of construct V1 with α-DG and different human immune-receptors (TLR3, MDA 5, RIG-1) to evaluate the efficacy of used adjuvants (Table 5). The stability of V1-TLR3 complex was confirmed by molecular dynamics study. Finally, the designed construct was reverse translated and inserted within pET28a(+) vector for heterologous expression in E. coli srain K12. Due to the encouraging findings, we suggest further wet lab trials involving model animals for the experimental validation of the designed vaccine candidate.
5. Conclusion
In the present study, genome based immunoinformatic study was employed to design a novel cross-reactive vaccine against the deadliest viruses of Arenaviridae family. The results suggest that the proposed vaccine could stimulate both humoral and cell-mediated immune responses, and serve as a potential vaccine against arenaviruses. However, in vitro and in vivo immunological studies are highly recommended to validate the efficacy of the designed construct.
Declarations
Author contribution statement
Kazi Faizul Azim: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Tahera Lasker, Rahima Akter: Performed the experiments; Wrote the paper.
Mantasha Mahmud Hia, Omar Faruk Bhuiyan: Performed the experiments.
Mahmudul Hasana, Md. Nazmul Hossain: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Brisse M.E., Ly H. Hemorrhagic fever-causing arenaviruses: lethal pathogens and potent immune suppressors. Front. Immunol. 2019;10:372–374. doi: 10.3389/fimmu.2019.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salvato M.S., editor. The Arenaviridae. Springer Science & Business Media; 2012. [Google Scholar]
- 3.Shao J., Liang Y., Ly H. Human hemorrhagic fever causing arenaviruses: molecular mechanisms contributing to virus virulence and disease pathogenesis. Pathogens. 2015;4:283–306. doi: 10.3390/pathogens4020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention . 2015. Fact Sheet: Lassa Fever. [Google Scholar]
- 5.Yun N.E., Walker D.H. Pathogenesis of Lassa fever. Viruses. 2012;4:2031–2048. doi: 10.3390/v4102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention . 2014. Fact Sheet: Lymphocytic Choriomeningitis (LCM) [Google Scholar]
- 7.Bonthius D.J., Wright R., Tseng B., Barton L., Marco E., Karacay B., Larsen P.D. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann. Neurol. 2007;62:347–355. doi: 10.1002/ana.21161. [DOI] [PubMed] [Google Scholar]
- 8.Bonthius D.J. Lymphocytic choriomeningitis virus: an under-recognized cause of neurologic disease in the fetus, child, and adult. Semin. Pediatr. Neurol. 2012;19:89–95. doi: 10.1016/j.spen.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendenhall M., Russell A., Juelich T., Messina E.L., Smee D.F., Freiberg A.N., Holbrook M.R. T-705 (favipiravir) inhibition of arenavirus replication in cell culture. Antimicrob. Agents Chemother. 2011;55:782–787. doi: 10.1128/AAC.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sewlall N.H., Paweska J.T. Lujo virus: current concepts. Dovepress. 2017;9:41–47. [Google Scholar]
- 11.Pawesk J.T. In: Lujo Virus Hemorrhagic Fever in Clinical Case Study of Emerging Infectious Diseases. Ergonul O., Can F., Akova M., Madoff L., editors. Elsevier/Academic Press London; 2014. pp. 95–110. [Google Scholar]
- 12.Paweska J. Nosocomial outbreak of novel arenavirus infection: southern Africa. Emerg. Infect. Dis. 2009;15:1598–1602. doi: 10.3201/eid1510.090211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manzione N.D., Salas R.A., Paredes H., Godoy O., Rojas L., Araoz F., Fulhorst C.F. Venezuelan hemorrhagic fever: clinical and epidemiological studies of 165 cases, Clinic. Infect. Dis. 1998;26:308–313. doi: 10.1086/516299. [DOI] [PubMed] [Google Scholar]
- 14.Meulen J.T., Lukashevich I., Sidibe K., Inapogui A., Marx M., Dorlemann A., Yansane M.L., Koulemou K., Chang-Claude J., Schmitz H. Hunting of peridomestic rodents and consumption of their meat as possible risk factors for rodent-to-human transmission of Lassa virus in the Republic of Guinea. Am. J. Trop. Med. Hyg. 1996;55:661–666. doi: 10.4269/ajtmh.1996.55.661. [DOI] [PubMed] [Google Scholar]
- 15.Tesh R.B., Jahrling P.B., Salas R., Shope R.E. Description of Guanarito virus (Arenaviridae: arenavirus), the etiologic agent of Venezuelan hemorrhagic fever. Am. J. Trop. Med. Hyg. 1994;50:452–459. doi: 10.4269/ajtmh.1994.50.452. [DOI] [PubMed] [Google Scholar]
- 16.Shoemaker T., Choi M. 2019. CDC Yellow Book 2020, Health Information for International Travel, Viral Hemorrhagic Fevers, Travel-Related-Infectious-Diseases (Chapter 4)https://wwwnc.cdc.gov/travel/yellowbook/2020/travel-related-infectious-diseases/viral-hemorrhagic-fevers [Google Scholar]
- 17.Ambrosio A., Saavedra M., Mariani M., Gamboa G., Maiza A. Argentine hemorrhagic fever vaccines. Hum. Vaccine. 2011;7:694–700. doi: 10.4161/hv.7.6.15198. [DOI] [PubMed] [Google Scholar]
- 18.Clark L.E., Mahmutovic S., Raymond D.D., Dilanyan T., Koma T., Manning J.T., Shankar S., Levis S.C., Briggiler A.M., Enria D.A., Wucherpfennig K.W. Vaccineelicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-04271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrion R., Jr., Patterson J.L., Johnson C., Gonzales M., Moreira C.R., Ticer A., Brasky K. A ML29 reassortant virus protects Guinea pigs against a distantly related Nigerian strain of Lassa virus and can provide sterilizing immunity. Vaccine. 2007;25:4093–4102. doi: 10.1016/j.vaccine.2007.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ölschläger S., Flatz L. Vaccination strategies against highly pathogenic arenaviruses: the next steps toward clinical trials. PLoS Pathog. 2013;9:e1003212. doi: 10.1371/journal.ppat.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenbaum B.D., Li O.T., Poon L.L., Levine A.J., Rabadan R. Viral reassortment as an information exchange between Viral segments. Proc. Natl. Acad. Sci. 2012;109:3341–3346. doi: 10.1073/pnas.1113300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stratton K., Almario D.A., Wizemann T.M., McCormick M.C. Institute of Medicine (US) Immunization Safety Review Committee, National Academies Press (US); 2003. Immunization Safety Review: Vaccinations and Sudden Unexpected Death in Infancy. [PubMed] [Google Scholar]
- 23.Akhand M.R.N., Azim K.F., Moli M.A., Joy B.D., Afif I.K., Ahmed N., Hasan M. Genome based evolutionary lineage of SARS-CoV-2 towards the development of novel chimeric vaccine. Infect. Genet. Evol. 2020;85:104517. doi: 10.1016/j.meegid.2020.104517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasson S.S., Al-Busaidi J.K.Z., Sallam T.A. The past, current and future trends in DNA vaccine immunisations. Asian Pacific J. Trop. Biomed. 2015;5:344–353. [Google Scholar]
- 25.Kaufmann S.H., McElrath M.J., Lewis D.J.M., Del Giudice G. Challenges and responses in human vaccine development. Curr. Opin. Immunol. 2014;28:18–26. doi: 10.1016/j.coi.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 27.Doytchinova I.A., Flower D.R. Identifying candidate subunit vaccines using an alignment independent method based on principal amino acid properties. Vaccine. 2007;25:856–866. doi: 10.1016/j.vaccine.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 28.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal Omega. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with A hidden markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 30.Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Peters P. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2014;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrov I., Bangov I., Flower D.R., Doytchinova I.A. AllerTOP v.2- A server for in silico prediction of allergens. J. Mol. Model. 2013;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 32.Dimitrov I., Naneva L., Doytchinova I.A., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30:846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 33.Goodman R.E., Ebisawa M., Ferreira F., Sampson H.A., van Ree R., Vieths S., Baumert J.L., Bohle B., Lalithambika S., Wise J., Taylor S.L. AllergenOnline: a peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol. Nutr. Food Res. 2016;60:1183–1198. doi: 10.1002/mnfr.201500769. [DOI] [PubMed] [Google Scholar]
- 34.Fiers M.W., Kleter G.A., Nijland H., Peijnenburg A., Nap J.P., Ham R. Allermatch™, A webtool for the prediction of potential allergenicity according to current FAO/WHO codex alimentarius guidelines. BMC Bioinf. 2004;5:133. doi: 10.1186/1471-2105-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maupetit J., Derreumaux P., Tuffery P. A fast method for large scale de novo peptide and miniprotein structure prediction. J. Comput. Chem. 2010;31:726–738. doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- 36.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emini E.A., Hughes J.V., Perlow D., Boger J. Induction of hepatitis A virus-neutralizing antibody by A virus-specific synthetic peptide. J. Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolaskar A., Tongaonkar P.C. ASemi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 40.Rana A., Akhter Y. A multi-subunit based, thermodynamically stable model vaccine using combined immunoinformatics and protein structure based approach. Immunobiology. 2016;221:544–557. doi: 10.1016/j.imbio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Hebditch M., Carballo-Amador M.A., Charonis S., Curtis R., Warwicker J. Protein–sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33:3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu D., Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011;101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. MOLPROBITY: structure validation and all-atom Contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craig D.B., Dombkowski A.A. Disulfide by design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinf. 2013;14:346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponomarenko J.V., Bui H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a New structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hajighahramani N., Nezafat N., Eslami M., Negahdaripour M., Rahmatabadi S.S., Ghasemi Y. Immunoinformatics analysis and in silico designing of A novel multi-epitope peptide vaccine against Staphylococcus aureus. Infect. Genet. Evol. 2017;48:83–94. doi: 10.1016/j.meegid.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Borrow P., Martinez-Sobrido L., delaTorre J.C. Inhibition of the type I interferon antiviral response during arenavirus infection. Viruses. 2010;2:2443–2480. doi: 10.3390/v2112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ibraghimov-Beskrovnaya O., Milatovich A., Ozcelik T., Yang B., Koepnick K., Francke U., Campbell K.P. Human dystroglycan: skeletal muscle cdna, genomic structure, origin of tissue specific isoforms and chromosomal localization. Hum. Mol. Genet. 1993;2:1651–1657. doi: 10.1093/hmg/2.10.1651. [DOI] [PubMed] [Google Scholar]
- 50.Azim K.F., Ahmed S.R., Banik A., Khan M.M., Deb A., Somana S.R. Screening and druggability analysis of some plant metabolites against SARS-CoV-2: an integrative computational approach. Inf. Med. Unlock. 2020;20:100367. doi: 10.1016/j.imu.2020.100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Blanco, Aliaga J.R., Quintana J.I., Orti E.S., Chacon P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42:271–276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Awan F.M., Obaid A., Ikram A., Janjua H.A. Mutation-structure function relationship based integrated strategy reveals the potential impact of deleterious missense mutations in autophagy related proteins on hepatocellular carcinoma (HCC): a comprehensive informatics approach. Int. J. Mol. Sci. 2017;1:13. doi: 10.3390/ijms18010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell. Mol. Immunol. 2018;15:182–184. doi: 10.1038/cmi.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azim K.F., Hasan M., Hossain M.N., Somana S.R., Hoque S.F., Bappy M.N., Chowdhury A.T., Lasker T. Immunoinformatics approaches for designing a novel multi epitope peptide vaccine against human norovirus (Norwalk virus) Infect. Genet. Evol. 2019;74:103936. doi: 10.1016/j.meegid.2019.103936. [DOI] [PubMed] [Google Scholar]
- 55.Vartak A., Sucheck S.J. Recent advances in subunit vaccine carriers. Vaccines (Basel) 2016;4:12. doi: 10.3390/vaccines4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou W.Y., Shi Y., Wu C., Zhang W.J., Mao X.H., Guo G., Li H.X., Zou Q.M. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27:5013–5019. doi: 10.1016/j.vaccine.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Kotturi M.F., Botten J., Sidney J., Bui H.H., Giancola L., Maybeno M., Babin J., Oseroff C., Pasquetto V., Greenbaum J.A., Peters B. A multivalent and cross-protective vaccine strategy against arenaviruses associated with human disease. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakabe S., Hartnett J.N., Ngo N., Goba A., Momoh M., Sandi J.D., Kanneh L., Cubitt B., Garcia S.D., Ware B.C., Kotliar D. Identification of common CD8+ T cell epitopes from Lassa fever survivors in Nigeria and Sierra Leone. J. Virol. 2020;94 doi: 10.1128/JVI.00153-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schildknecht A., Welti S., Geuking M.B., Hangartner L., van den Broek M. Absence of CTL responses to early viral antigens facilitates viral persistence. J. Immunol. 2008;180:3113–3121. doi: 10.4049/jimmunol.180.5.3113. [DOI] [PubMed] [Google Scholar]
- 60.Baral P., Pavadai E., Gerstman B.S., Chapagain P.P. In-silico identification of the vaccine candidate epitopes against the Lassa virus hemorrhagic fever. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-63640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.ter Meulen J., Badusche M., Kuhnt K., Doetze A., Satoguina J., Marti T., Fleischer B. Characterization of human CD4+ T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J. Virol. 2000;74:2186–2192. doi: 10.1128/jvi.74.5.2186-2192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez Carreno M.P., Nelson M.S., Botten J., Smith Nixon K., Buchmeier M.J., Whitton J.L. Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology. 2005;335:87–98. doi: 10.1016/j.virol.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 63.Toussaint N.C., Maman Y., Kohlbacher O., Louzoun Y. Universal peptide vaccines–optimal peptide vaccine design based on viral sequence conservation. Vaccine. 2011;29:8745–8753. doi: 10.1016/j.vaccine.2011.07.132. [DOI] [PubMed] [Google Scholar]
- 64.Sheikh Q.M., Gatherer D., Reche P.A., Flower D.R. Towards the knowledge-based design of universal influenza epitope ensemble vaccines. Bioinformatics. 2016;32:3233–3239. doi: 10.1093/bioinformatics/btw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amorim K.N., Rampazo E.V., Antonialli R., Yamamoto M.M., Rodrigues M.M., Boscardin S.B. The presence of T cell epitopes is important for induction of antibody responses against antigens directed to DEC205+ dendritic cells. Sci. Rep. 2016;6:39250. doi: 10.1038/srep39250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meza B., Ascencio F., Sierra-Beltrán A.P., Torres J., Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: an in silico approach. Infect. Genet. Evol. 2017;49:309–317. doi: 10.1016/j.meegid.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Khatoon N., Pandey R.K., Prajapati V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017;7:8285. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nosrati M., Behbahani M., Mohabatkar H. Towards the first multi-epitope recombinant vaccine against Crimean-Congo hemorrhagic fever virus: a computer-aided vaccine design approach. J. Biomed. Inf. 2019;93:103160. doi: 10.1016/j.jbi.2019.103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 70.Pichlmair A., Reis e Sous C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 71.van der Burg S.H., Bijker M.S., Welters M.J., Offringa R., Melief C.J. Improved peptide vaccine strategies, creating synthetic artificial infections to maximize immune efficacy. Adv. Drug Deliv. Rev. 2006;58:916–930. doi: 10.1016/j.addr.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Jung I.D., Jeong S.K., Lee C.M., Noh K.T., Heo D.R., Shin Y.K., Yun C.H., Koh W.J., Akira S., Whang J., Kim H.J. Enhanced efficacy of therapeutic cancer vaccines produced by co-treatment with Mycobacterium tuberculosis heparin-binding hemagglutinin, a novel TLR4 agonist. Canc. Res. 2011;71:2858–2870. doi: 10.1158/0008-5472.CAN-10-3487. [DOI] [PubMed] [Google Scholar]
- 73.Durbeej M., Henry M.D., Campbell K.P. Dystroglycan in development and disease. Curr. Opin. Cell Biol. 1998;10:594–601. doi: 10.1016/s0955-0674(98)80034-3. [DOI] [PubMed] [Google Scholar]
- 74.Kunz S., Rojek J.M., Kanagawa M., Spiropoulou C.F., Barresi R., Campbell K.P., Oldstone M.B. Posttranslational modification of alpha-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J. Virol. 2005;79:14282–14296. doi: 10.1128/JVI.79.22.14282-14296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spiropoulou C.F., Kunz S., Rollin P.E., Campbell K.P., Oldstone M.B.A. New World arenaviruses clade C, but not clade A and B viruses, utilizes a-dystroglycan as its major receptor. J. Virol. 2002;76:5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.