Abstract
Missense and truncating variants in protocadherin 19 (PCDH19) cause PCDH19-related epilepsy. In this study, we aimed to investigate variations in distributional characteristics and the clinical implications of variant type in PCDH19-related epilepsy. We comprehensively collected PCDH19 missense and truncating variants from the literature and by sequencing six exons and intron–exon boundaries of PCDH19 in our cohort. We investigated the distribution of each type of variant using the cumulative distribution function and tested for associations between variant types and phenotypes. The distribution of missense variants in patients was clearly different from that of healthy individuals and was uniform throughout the extracellular cadherin (EC) domain, which consisted of six highly conserved domains. Truncating variants showed two types of distributions: (1) located from EC domain 1 to EC domain 4, and (2) located from EC domain 5 to the cytoplasmic domain. Furthermore, we also found that later onset seizures and milder intellectual disability occurred in patients with truncating variants located from EC domain 5 to the cytoplasmic domain compared with those of patients with other variants. Our findings provide the first evidence of two types of truncating variants in the PCDH19 gene with regard to distribution and the resulting clinical phenotype.
Subject terms: Genetic association study, Epilepsy, Disease genetics
Introduction
Protocadherin 19 (PCDH19)-related epilepsy is an epileptic syndrome with various characteristics. Early-onset seizures (6–36 months), which are the most typical characteristic, can be focal, generalized, tonic–clonic, myoclonic, atonic, or absent and often occur in clusters or as prolonged ictal episodes; their severity and frequency vary among affected females, resembling the seizures observed in patients with Dravet syndrome [1–6]. Furthermore, the clinical features of this condition have expanded to include the presence or absence of intellectual disability, psychiatric features, and behavioral disturbances [7–11].
PCDH19-related epilepsy is caused by pathogenic variants in the PCDH19 gene. This gene is located on chromosome Xq22.1 and encodes subgroup of protocadherins, which are involved in signal transduction at synapses and in the establishment of neuronal connections [12]. PCDH19 is a member of the δ2-protocadherin group and contains six extracellular cadherin (EC) repeats, a transmembrane domain, and a cytoplasmic domain, including two conserved motifs. Crystal structural analysis of zebrafish pcdh19 and solution biophysical resonance analysis of the human δ-protocadherin family have revealed the preferential homophilic trans-interaction of pcdh19 in the region from EC domain 1 (EC1) to EC domain 4 (EC4) [13, 14]. Moreover, the most supported pathogenic mechanism of PCDH19-related epilepsy is mosaicism of two cell populations, expressing either normal or mutant PCDH19 protein [6, 15, 16], which is expected to prevent the normal homophilic adhesive function of PCDH19 proteins.
To date, more than 200 variants have been reported in patients with PCDH19-related epilepsy. Most variants (90%) have been identified in female patients, whereas only 10% of variants have been identified in male patients [17]. Of these variants, missense and truncating variants (nonsense and frameshift variants) account for 58% and 41%, respectively, and are observed mostly in exon 1, which encodes all EC domains. Kolc et al. [18] classified these variants according to variant type, location, inheritance, and clinical features, including seizure onset age and intellectual disability, and examined genotype–phenotype associations. They found that earlier seizure onset (≤12 months) was significantly associated with more severe intellectual disability, whereas cognitive function did not differ between variant types for missense and truncating variants, between variant locations, from EC1 to EC3, and EC4 to the cytoplasmic domain. Currently, no genotype–phenotype correlations have been identified.
In this study, we updated pathogenic variants by collecting data from published and unpublished reports of patients with PCDH19-related epilepsy. We also evaluated differences in variant location and clinical phenotypes among variant types. By analyzing the variant distribution using the cumulative distribution function (CDF), we found that truncating variants in the PCDH19 gene could be separated into two types: (1) located from EC1 to EC4 and (2) located from EC domain 5 to the cytoplasmic domain. Furthermore, a comparison of clinical phenotypes revealed the occurrence of early-onset seizure and milder intellectual disability in patients with the second type of truncating variant compared with that in patients carrying missense variants and the first type of truncating variant.
Materials and methods
Patients and clinical information
Our patients, who were diagnosed with PCDH19-related epilepsy, satisfied the International League Against Epilepsy definition (https://www.epilepsydiagnosis.org/aetiology/gene-abnormalities-overview.html#PCDH19), as follows: (1) seizures starting early (mean age 9 months); (2) tonic–clonic and/or focal seizures; (3) frequent, clustered seizures facilitated by fever; and (4) intellectual development that varied from normal, often with regression, to severe intellectual disability with autistic features and psychiatric disorders.
We collected clinical information using a questionnaire with an organized form, including perinatal history, family history, age at seizure onset, seizure history, psychomotor development, behavioral features, intellectual disability level, neuroimaging findings, electroencephalographic findings, and treatment. After checking clinical information, patients whose parents provided signed informed consent using a protocol approved by the ethics review committee of Fukuoka University underwent genetic analyses.
Identification of PCDH19 pathogenic variants
The presence of PCDH19 gene variants in the above patients was evaluated by Sanger or gene panel sequencing [19–21]. Gene panel sequencing was performed using a customized HaloPlex Target Enrichment System (Agilent, Santa Clara, CA, USA) for 114 genes that are known or suspected to cause epileptic seizures. Samples were sequenced on a MiSeq instrument (Illumina, San Diego, CA, USA). The exon regions of targeted genes were covered with ~100 reads on average. Variants were called from sequencing results using SureCall software (v4.0, Agilent, Santa Clara, CA, USA). Using ANNOVAR software [22], the pathogenicity of variants was estimated based on allele frequencies of 621 East Asian individuals in 1000 Genomes Project data (published in 2015, https://www.internationalgenome.org/), 9977 East Asian individuals in gnomAD (v2.1.1, https://www.internationalgenome.org/; the Genome Aggregation Database [gnomAD], https://gnomad.broadinstitute.org/), and 4456 Japanese individuals in the Human Genetic Variation database (v2.3, Human Genetic Variation Database in the Japanese population, http://www.hgvd.genome.med.kyoto-u.ac.jp/ [23]) and by nucleotide or amino acid conservation and their effects on protein structure predicted by SIFT, PolyPhen2, Mutation Taster, CADD, and PhyloP100way vertebrate. Variants with an allele frequency ≥0.005 in these databases were considered nonpathogenic variants. Furthermore, variants with CADD ≥15 and/or with PhyloP100way vertebrate score ≥3, as well as variants predicted to be deleterious by SIFT, PolyPhen2, and Mutation Taster were preferentially considered pathogenic variants. To investigate large deletions or duplications, multiplex ligation-dependent probe amplification (MLPA) was performed using SALSA MLPA KIT P330-A3 PCDH19 (MRC-Holland, Amsterdam, the Netherlands). When a pathogenic variant was suspected, inheritance was determined by sequencing or by MLPA for available parental DNA.
Collection of PCDH19 variants and clinical information from the literature
For this study, PCDH19 variants were comprehensively gathered from the EpilepsyGene database (accessed on January 6, 2017; http://61.152.91.49/EpilepsyGene/index.php) and from Google Scholar, searched using the words “PCDH19” and “epilepsy” (accessed on May 29, 2020; https://scholar.google.co.jp/). To avoid the risk of bias by overcollection, we adopted the variant and clinical information for one case per family. For collection of intellectual disability information, we adopted only intellectual disability information following the classification of normal, borderline, mild, moderate, severe, or profound, according to the previously established criteria [18].
Statistical analysis
For CDF analysis, we used the coding sequence (CDS) position (transcript ID: NM_001184880.2) of missense and truncating variants (frameshift and nonsense variants) reported in female patients. In CDF analysis, variant distributions were assessed by Anderson–Darling tests, with the hypothesis that the distribution was uniform along the CDS position of PCDH19. The difference in seizure onset age between variant types was assessed by Kruskal–Wallis test and pair-wise Wilcoxon tests with Bonferroni multiple comparison correction, excluding abnormal onset age (more than 36 months). The proportions of patients with each type of clinical feature were compared using pair-wise chi-square tests with Bonferroni multiple comparison correction. The difference in the rate of patient with each intellectual disability level was assessed by hierarchical clustering analysis with Ward’s method and Euclidean distance. These statistical calculations were performed using R software (version 3.6.0), and results with p values of less than 0.05 were considered significant.
Results
Collection of PCDH19 variants
We collected 455 PCDH19 pathogenic variants and clinical features from 13 Japanese patients in our cohort (Supplementary Table 1) and from 442 patients in the literature (Supplemental Table 2). Of these variants, 427 variants (93.8%) were reported in females, and 28 variants (6.15%) were reported in males. The variant types and inheritances were summarized according to sex (Table 1). For both sexes, most commonly reported variants were missense, frameshift, and nonsense variants (198 [46.4%: 198/427], 109 [25.5%: 109/427], and 64 [15.0%: 64/427] in females; 13 [46.4%: 13/28], 3 [10.7%: 3/28], and 9 [32.1%: 9/28] in males). Furthermore, most of these missense, frameshift, and nonsense variants were de novo in both sexes (99 [50.0%: 99/198], 66 [60.6%: 66/109], and 33 [51.6%: 33/64] in females; 8 [61.5%: 8/13], 2 [66.7%: 2/3], 7 [77.8%: 7/9] in males).
Table 1.
Summary of variant types and inheritance by sex
| Female | Male | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| De novo | Paternal | Maternal | Familial | Unknown | Total | De novo | Paternal | Maternal | Familial | Unknown | Total | |
| Missense | 99 | 35 | 32 | 1 | 31 | 198 | 8 | 0 | 0 | 0 | 5 | 13 |
| Compound heterozygous missense | 4 | 1 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Frameshift | 66 | 16 | 4 | 4 | 19 | 109 | 2 | 0 | 1 | 0 | 0 | 3 |
| Nonsense | 33 | 9 | 4 | 6 | 12 | 64 | 7 | 0 | 0 | 0 | 2 | 9 |
| Splicing | 8 | 0 | 1 | 0 | 5 | 14 | 2 | 0 | 0 | 0 | 0 | 2 |
| Inframe-insertion | 1 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Inframe-deletion | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Inframe-insertion/deletion | 1 | 0 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microdeletion | 2 | 0 | 1 | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microduplication | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Whole gene deletion | 16 | 1 | 0 | 0 | 7 | 24 | 1 | 0 | 0 | 0 | 0 | 1 |
| Total | 233 | 62 | 44 | 11 | 77 | 427 | 20 | 0 | 1 | 0 | 7 | 28 |
Of 28 male patients and 62 transmitting fathers of female patients, mosaic and hemizygous males comprised 27 and 12 cases, respectively (Supplementary Tables 1 and 2). Most mosaic males (88.9%: 24/27) showed seizure-related features, whereas, 83.3% (10/12) of hemizygous males were unaffected, in accordance with previously suggested pathomechanisms of PCDH19-related epilepsy [6, 15, 16].
Distribution analysis of PCDH19 variants
Of the reported variants, we used the most commonly reported variants, i.e., missense, frameshift, and nonsense variants, in female patients. In particular, frameshift and nonsense variants were grouped into truncating variants, which generate a premature stop codon, and 198 missense and 173 truncating variants (109 frameshift and 64 nonsense variants) were used for distribution analysis.
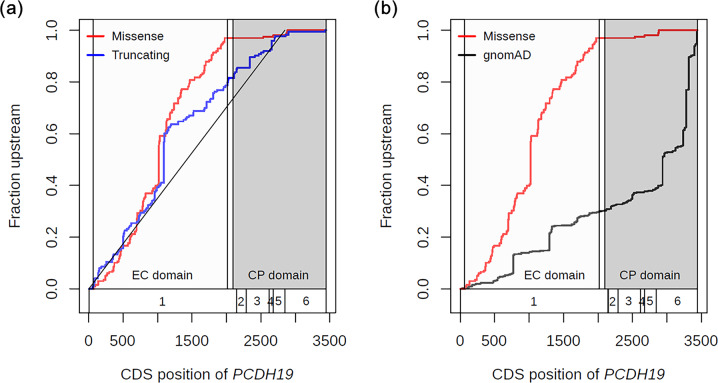
First, we compared the distributions of missense and truncating variants using the CDF (Fig. 1). The CDFs showed extremely high frequencies (more than 20 cases) at CDS positions 1019 and 1091 for missense variant (NM_001184880.2:c.1019A>G [chrX.hg19:g.99662577T>C]) and truncating variants (NM_001184880.2:c.1091dupC [chrX.hg19:g.99662505dupG] and NM_001184880.2:c.1091delC [chrX.hg19:g.99662505delG]; Fig. 1a), respectively. The numbers of patients carrying these missense variant (NM_001184880.2:c.1019A>G) and truncating variants (NM_001184880.2:c.1091dupC and c.1091delC) were 30, 22, and 5, respectively (Supplementary Tables 1 and 2). Furthermore, very low frequencies of missense and truncating variants were observed in the cytoplasmic domain and in the last exon, respectively.
Fig. 1.
CDFs of PCDH19 missense and truncating variants. a CDFs of missense (red line) and truncating variants (blue line). b CDFs of variants from the gnomAD database (black line) and missense variants (red line). The white boxes and numbers at the bottom of the graphs indicate exon regions and exon numbers for the PCDH19 gene. The light and dark gray regions indicate extracellular cadherin (EC) and cytoplasmic (CP) domains, respectively
In order to investigate whether variants located in the cytoplasmic domain region were lethal or noncausal variants of PCDH19-related epilepsy, we compared the CDFs of missense variants with those of variants from healthy individuals in gnomAD database, including various ethnic backgrounds (Fig. 1b). As a result, the distribution of missense variants did not overlap with that of healthy individual variants in any ethnic background, except with seven positions. The maximum allele frequencies of healthy variants at the seven positions were less than 0.005 (0–0.00208). Furthermore, the CDFs of gnomAD database variants showed high frequencies in the cytoplasmic domain and low frequencies in the EC domains, in contrast with the CDFs of missense variants. These results suggested that most missense variants in the cytoplasmic domain were not lethal and did not cause PCDH19-related epilepsy.
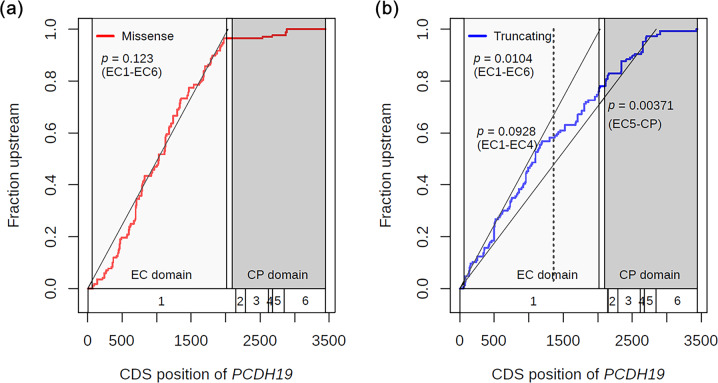
After exclusion of high-frequency variants, we further investigated the uniformity of the distributions of missense and truncating variants using Anderson–Darling tests. The results showed that the distribution of missense variants was uniform throughout the EC domain (p = 0.123, Fig. 2a). In contrast, the distribution of truncating variants was not uniform throughout the EC domain (p = 0.0104, Fig. 2b). As a result of more detailed Anderson–Darling tests, the distribution of truncating variants was found to be uniform from EC1 to EC4 (p = 0.0928, Fig. 2b, Supplementary Table 3). These results indicated that there were at least two types of truncating variants in the PCDH19 gene, i.e., variants located from EC1 to EC4 and variants located from EC5 to the cytoplasmic domain. In addition, truncating variants were not uniform from the start codon to the region upstream of the last exon (p = 0.00371, Fig. 2b).
Fig. 2.
CDFs of PCDH19 missense and truncating variants without hotspots. a CDFs of missense variants without hotspots (red line). b CDFs of truncating variants without hotspots (blue line). The white boxes and numbers at the bottom of the graphs indicate exon regions and exon numbers for the PCDH19 gene. The light and dark gray regions indicate extracellular cadherin (EC) and cytoplasmic (CP) domains, respectively. The dashed line indicates the boundary between EC domains 4 and 5
Patient phenotypes associated with each variant type
CDF analysis revealed that PCDH19 variants could be classified into three types: (1) missense variants, (2) truncating variants located from EC1 to EC4, and (3) truncating variants located from EC5 to the cytoplasmic domain. Next, we assessed the phenotypic differences of patients with PCDH19-related epilepsy, including the age of seizure onset, seizure type, behavioral features, and intellectual disability level, according to variant type using clinical data. In addition, we classified truncating variants as (1) those located from EC1 to EC3 and (2) those located from EC4 to cytoplasmic domain [18] and then assessed phenotypic differences among patients with variant types.
First, we investigated the age at onset of seizures in patients with each variant type (Fig. 3). Of the collected clinical features, seizure onset age under 36 months was observed for 175, 101, and 55 patients carrying missense variants, truncating variants located from EC1 to EC4, and truncating variants located from EC5 to the cytoplasmic domain, respectively. The mean age of patients with missense variants was 10.2 months (standard deviation [SD]: 5.26 months), and those in patients with truncating variants from EC1 to EC4 and from EC5 to the cytoplasmic domain were 10.1 months (SD: 5.37 months) and 12.6 months (SD: 6.66 months), respectively, indicating that the mean ages of patients with truncating variants located from EC5 to the cytoplasmic domain were ~2.44 and 2.54 months older than those of patients with the other types of variants. Furthermore, there were significant differences between truncating variants located from EC5 to the cytoplasmic domain and the other variant types (p = 0.0152 [Kruskal–Wallis test], and p = 0.0173 and 0.0354 [missense versus truncating variants located from EC5 to the cytoplasmic domain and truncating variants located from EC1 to EC4 versus truncating variants located from EC5 to the cytoplasmic domain; pair-wise Wilcoxon tests]). In addition, the mean ages of patients with truncating variants located from EC1 to EC3 and of patients with truncating variants from EC4 to the cytoplasmic domain were 10.9 months (SD: 5.84 months) and 11.0 months (SD: 6.07 months), respectively. There were no significant difference among missense variants and these truncating variant types (p = 0.375 [Kruskal–Wallis test], and p = 0.887, 0.691, and 1.00 [missense versus truncating variants located from EC1 to EC3, missense versus truncating variants from EC4 to cytoplasmic domain, and truncating variants located from EC1 to EC3 versus truncating variants located from EC4 to the cytoplasmic domain, respectively; pair-wise Wilcoxon tests], Supplementary Fig. 1).
Fig. 3.
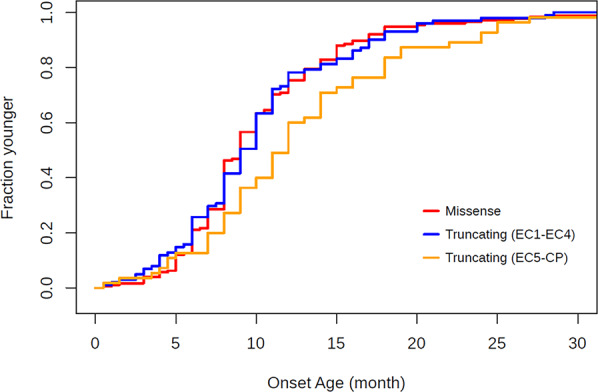
CDFs of seizure onset age. CDFs of the onset age of patients with missense variants, truncating variants located from EC1 to EC4, and truncating variants located from EC5 to the cytoplasmic domain are indicated in red, blue, and orange, respectively
Next, we assessed nine seizure-related features, including “febrile”, “tonic–clonic”, “myoclonic”, “complex partial seizure/absence”, and “status”, as well as behavioral features, including autistic and behavioral disturbances, in patients with PCDH19-related epilepsy (Table 2). Seizure type data were available in 149, 85, and 43 patients with missense variants, truncating variants located from EC1 to EC4, and truncating variants located from EC5 to the cytoplasmic domain, respectively. Behavioral feature data were available for 150, 85, and 43 patients with missense variants, truncating variants located from EC1 to EC4, and truncating variants located from EC5 to the cytoplasmic domain. Of the seizure-related features, high-rate features included “cluster” (65.9–83.7%), “febrile seizures” (55.3–69.8%), “focal seizures” (64.7–67.8%), and “tonic–clonic seizures” (45.9–53.5%), for all variant types. All clinical features showed no significant differences between variant types (p = 0.149–1.00). Similar to seizure-related features, the percentages of patients with autistic features and/or behavioral disturbance were also high (51.2–64.7%) for all variant types and did not differ significantly between variant types (p = 0.594–1.00, Table 2). Furthermore, there were also no significant differences in these clinical features among missense variants, truncating variants located from EC1 to EC3, and truncating variants located from EC4 to the cytoplasmic domain (p = 0.142–1.00, Supplementary Table 4).
Table 2.
Summary of seizure types and behavioral features
| Seizure-related features | Missense (total available data: 149) | Truncating (EC1–EC4) (total available data: 85) | Truncating (EC5–CP) (total available data: 43) | p values (missense vs truncating (EC1–EC4)) | p values (missense vs truncating (EC5–CP)) | p values (truncating (EC1–EC4) vs truncating (EC5–CP)) | |||
|---|---|---|---|---|---|---|---|---|---|
| Patient number | % | Patient number | % | Patient number | % | ||||
| Febrile seizures | 87 | 58.4 | 47 | 55.3 | 30 | 69.8 | 1.00 | 0.726 | 0.495 |
| Tonic-clonic seizures | 72 | 48.3 | 39 | 45.9 | 23 | 53.5 | 1.00 | 1.00 | 1.00 |
| Tonic seizures | 43 | 28.9 | 22 | 25.9 | 13 | 30.2 | 1.00 | 1.00 | 1.00 |
| Hemiclonic/Unilat | 12 | 8.05 | 5 | 5.88 | 2 | 4.65 | 1.00 | 1.00 | 1.00 |
| Myoclonic seizures | 12 | 8.05 | 8 | 9.41 | 2 | 4.65 | 1.00 | 1.00 | 1.00 |
| Complex partial seizure/absence | 26 | 17.4 | 14 | 16.5 | 6 | 14.0 | 1.00 | 1.00 | 1.00 |
| Partial(Focal) seizures | 101 | 67.8 | 55 | 64.7 | 28 | 65.1 | 1.00 | 1.00 | 1.00 |
| Status epilepticus | 29 | 19.5 | 10 | 11.8 | 9 | 20.9 | 0.543 | 1.00 | 0.795 |
| Cluster | 117 | 78.5 | 56 | 65.9 | 36 | 83.7 | 0.149 | 1.00 | 0.168 |
| Behavioral features | Missense (total available data: 150) | Truncating (EC1–EC4) (total available data: 85) | Truncating (EC5–CP) (total available data: 43) | p values (missense vs truncating (EC1–EC4)) | p values (missense vs truncating (EC5–CP)) | p values (truncating (EC1–EC4) vs truncating (EC5–CP)) | |||
|---|---|---|---|---|---|---|---|---|---|
| Patient number | % | Patient number | % | Patient number | % | ||||
| Autistic features/behavioral disturbances | 87 | 58.0 | 55 | 64.7 | 22 | 51.2 | 1.00 | 1.00 | 0.594 |
p values were calculated by pair-wise chi-squared tests and corrected by Bonferroni multiple correction. “EC” and “CP” indicate “extracellular cadherin” and “cytoplasmic”, respectively
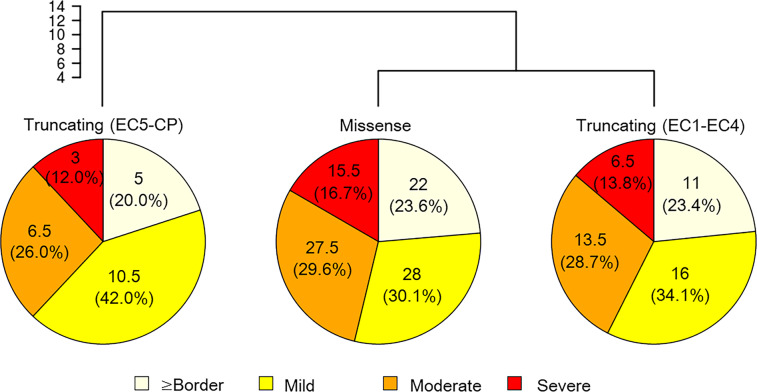
Finally, differences in intellectual disability levels between variant types were assessed using pie charts and clustering analysis based on the numbers and rates of patients with each intellectual disability level (Fig. 4). The intellectual disability levels of patients were available for 93 missense variants, 47 truncating variants located from EC1 to EC4, and 25 truncating variants located from EC5 to the cytoplasmic domain. The most common intellectual disability level was “mild” for all variant types (30.1–42.0%). Furthermore, the total percentage of patients with “≥ border” and “mild” intellectual disability levels was highest in patients with truncating variants located from EC5 to the cytoplasmic domain (62.0%), compared with those in patients with missense variants and truncating variants located from EC1 to EC4 (53.7% and 57.5%, respectively). Although there were no statistically significant differences (p = 1.00), these results suggested that the severity of intellectual disability in patients tends to be milder in patients with truncating variants located from EC5 to the cytoplasmic domain than in patients with other variant types. Furthermore, clustering analysis showed results consistent with those from pie charts. Hierarchical clustering dendrograms demonstrated a higher degree of similarity for smaller distances. The distances between truncating variants located from EC5 to the cytoplasmic domain and the other variant types were 13.8 and 9.26, whereas the distance between missense and truncating variants located from EC1 to EC4 was 4.93, clustering in one group. Taken together, these results suggested that the intellectual disability level of patients with truncating variants located from EC5 to the cytoplasmic domain was separated from those of patients with other variant types. In contrast, although the intellectual disability levels of missense and truncating variants from EC1 to EC3 were classified into one group in the clustering analysis, the distances were high and did not show major differences (7.75–10.5) among missense variants, truncating variants located from EC1 to EC3, and truncating variants located from EC4 to the cytoplasmic domain. These results indicated the differences of intellectual disability levels among patients with these variant types (Supplementary Fig. 2).
Fig. 4.
Comparison of the level of intellectual disability of in patients with PCDH19-related epilepsy. Hierarchical clustering dendrogram (top) showing similarities in the rates of patients with each intellectual disability level. For clustering, Ward’s method and Euclidean distance were utilized. The pie charts (bottom) show the percentages of patients with each level of intellectual disability. Cases with two levels in the literature were counted as 0.5 cases. For example, one case with a “moderate/severe” level was counted as 0.5 cases of moderate level and 0.5 cases of severe level
Discussion
In this study, we investigated the distributional characteristics and clinical implications of each type of PCDH19 pathogenic variant.
Distribution analysis revealed very high frequencies (more than 20 cases) at two CDS positions, 1019 and 1091, for missense and frameshift variants (NM_001184880.2:c.1019A>G, c.1091dupC, and c.1091delC). Of these variants, missense variant (NM_001184880.2:c.1019A>G) and frameshift variant (NM_001184880.2:c.1091dupC) have been called “hotspots” or “recurrent” variants [17, 18]. Focusing on the CDS of PCDH19 mRNA (NM_001184880.2), we found mononucleotide repeats (seven cytosine nucleotide repeats) in the upstream region of the frameshift hotspot variant. Mononucleotide repeats have been founded in gastric and colorectal cancers and are considered targets of frameshift variants [24–29]. Therefore, the seven cytosine nucleotide repeats may be a cause of frameshift hotspot variants. However, there were no mononucleotide repeats or other causal sequences, such as CG-rich sequences, at nearby missense hotspot variants. The occurrence of missense hotspot variants may be due to some external factors, such as the high susceptibility of codons to variants and slow DNA repair rates [30].
After exclusion of hotspot variants, our analysis revealed that although missense variants were distributed uniformly throughout EC domain, truncating variants could be separated into at least two types: (1) located from EC1 to EC4 and (2) located from EC5 to the cytoplasmic domain. Previous our studies have observed that truncating variants distributed uniformly from the start codon to 50 nt upstream region of the last exon in some epileptic genes [20, 21]. Therefore, this non-uniformity of the distribution indicates the specificity of truncating variants in the PCDH19 gene.
In support of this result, we found differences in seizure onset age and intellectual disability levels between the second type of truncating variants and the other variant types. The mean age of seizure onset of patients carrying the second type of truncating variants (about 12 months) was 2 months older than those of patients carrying the other variant types (about 10 months). Furthermore, the intellectual disability of patients carrying the second type of truncating variants was milder than those of patients carrying the other variant types. Notably, these results were concordant with the observations of Kolc et al. [18], who showed that earlier seizure onset (≤12 months) was significantly associated with more severe intellectual disability. Furthermore, they also found that there was no association between patient phenotype and variant location following the classification of variants located from EC1 to EC3, and variants located from EC4 to cytoplasmic domain. Our truncating variant data showed the association between patient phenotypes (seizure onset age and intellectual disability) and variant location (located from EC1 to EC4 and located from EC5 to the cytoplasmic domain), whereas no association was observed when using their classification. Therefore, whether the variant is located upstream of EC5 or not may be crucial for determination of phenotypic severity with regard to seizure onset age and intellectual disability in patients with PCDH19-related epilepsy carrying truncating variants.
Recent crystal structure analysis of zebrafish pcdh19 and surface plasmon resonance data revealed that domains EC1 to EC4 of Pcdh19 encompass the minimal unit of its homophilic adhesive function and that missense mutations in conserved regions of these EC domains abolish the homophilic binding of Pcdh19 [13, 14]. However, the effect of truncating variants on the adhesive function of PCDH19 protein remains unclear. Truncating variants result often contain premature termination codons (PTCs), most of which trigger a process called nonsense-mediated mRNA decay (NMD), thereby preventing the synthesis of truncated and potentially toxic proteins [31–33]. A previous study reported that a PTC-bearing PCDH19 mRNA was downregulated in skin fibroblast cells by NMD [15]. Furthermore, there are no reports on the existence of truncated PCDH19 proteins in human cells. Thus, further studies are needed to elucidate the pathomechanisms underlying the differential phenotypes in patients harboring different types of truncating variants.
In conclusion, we found that variant types in the PCDH19 gene could be separated into three types: missense variants, truncating variants located from EC1 to EC4, and truncating variants located from EC5 to the cytoplasmic domain. In addition, variant distribution, seizure onset age, and severity of intellectual disability in patients were similar among patients with missense and truncating variants located from EC1 to EC4, whereas patients carrying truncating variants located from EC5 to the cytoplasmic domain had later onset seizures and milder intellectual disability. Other epilepsy-causative genes, SCN1A and SCN2A, which encode the voltage-gated sodium channels type 1 and 2, respectively, have shown phenotypic differences based on variant types. As for the SCN1A gene, it has been proposed that compared with those with missense variants, truncating variants result in earlier onset of seizures and more severe phenotypes [34, 35]. Our PCDH19 gene analysis showed similar phenotypes between missense and truncating variants from EC1 to EC4 and showed milder phenotypes with PCDH19-truncating variants from EC5 to the cytoplasmic domain. As for the SCN2A gene, it has shown differential phenotypes within missense variants, which could be classified into two groups, namely gain-of-function and loss-of-function variants. The gain-of-function variants are mostly located at the voltage sensor domain of the channel and cause infantile epileptic encephalopathy and benign infantile seizures. Meanwhile, the loss-of-function variants are mostly located at the pore loop regions and are associated with autism spectrum disorder and/or intellectual disability, resulting in a variety of seizure types [36–38]. PCDH19 variant analysis showed locational and phenotypic differences within truncating variants. To date, no other reports have described similar genotype–phenotype associations with the PCDH19 gene. Our findings could contribute to diagnosis of PCDH19-related epilepsy and could help to elucidate the pathogenic mechanisms of this disease. For more accurate evaluation of patient phenotypes and understanding of pathogenic mechanisms, we will investigate the detailed effects of transcript or protein products generated from each variant type in our future work.
Supplementary information
Acknowledgements
We would like to thank the patients, their families, and the participating doctors for assistance with this study. This work was supported by Grants-in-Aid for Scientific Research (A) (B) from the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant nos. JP15H02548 and JP20H03651 [SH]), Grants-in-Aid for Early Career Scientists from JSPS KAKENHI (grant nos. JP19K17347 [MS] and JP19K17381 [AI]), the Acceleration Program for Intractable Diseases Research utilizing Disease-specific iPS cells (grant nos. 17bm0804014h0001, 18bm0804014h0002, and 19bm0804014h0003 [SH]), a grant for Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development (AMED) (grant nos. 15ek0109038a, 18kk0205014s1803, and 19kk0205014h0004 [SH]), Health Labour Sciences Grants for the Research on Measures for Intractable Diseases (grant nos. H29-Nanji-Ippan-010, H30-Nanji-Ippan-007, H30-Nanji-Ippan-010, and H31-Nanji-Ippan-010 [SH and AI]), Research grants from Kawano Masanori Memorial Public Interest Incorporated Foundation for Promotion of Pediatrics (A.I.), Grants of The Clinical Research Promotion Foundation (AI), the Japan Epilepsy Research Foundation (JERF) Grant (grant no. JERF TENKAN 17007 [MS]), and GlaxoSmithKline (GSK) Japan Research Grant 2016 (MS). This work was also supported in part by Grants from the Science Research Promotion Funds and the Fukuoka University Funds (grant nos. G19001 and G20001 [SH]), a grant from the Central Research Institute of Fukuoka University (grant no. 197007 [AI]). The Research Institute for the Molecular Pathomechanisms of Epilepsy, Fukuoka University is supported by the MEXT-supported Program for the Strategic Research Foundation at Private Universities (grant no. 924).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mami Shibata, Email: shibatamm@fukuoka-u.ac.jp.
Shinichi Hirose, Email: hirose@fukuoka-u.ac.jp.
Supplementary information
The online version of this article (10.1038/s10038-020-00880-z) contains supplementary material, which is available to authorized users.
References
- 1.Scheffer IE, Turner SJ, Dibbens LM, Bayly MA, Friend K, Hodgson B, et al. Epilepsy and mental retardation limited to females: an under-recognized disorder. Brain. 2008;131:918–27.. doi: 10.1093/brain/awm338. [DOI] [PubMed] [Google Scholar]
- 2.Marini C, Mei D, Parmeggiani L, Norci V, Calado E, Ferrari A, et al. Protocadherin 19 mutations in girls with infantile-onset epilepsy. Neurology. 2010;75:646–53.. doi: 10.1212/WNL.0b013e3181ed9e67. [DOI] [PubMed] [Google Scholar]
- 3.Marini C, Darra F, Specchio N, Mei D, Terracciano A, Parmeggiani L, et al. Focal seizures with affective symptoms are a major feature of PCDH19 gene-related epilepsy. Epilepsia. 2012;53:2111–9. doi: 10.1111/j.1528-1167.2012.03649.x. [DOI] [PubMed] [Google Scholar]
- 4.Higurashi N, Nakamura M, Sugai M, Ohfu M, Sakauchi M, Sugawara Y, et al. PCDH19-related female-limited epilepsy: further details regarding early clinical features and therapeutic efficacy. Epilepsy Res. 2013;106:191–9. doi: 10.1016/j.eplepsyres.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Terracciano A, Specchio N, Darra F, Sferra A, Bernardina BD, Vigevano F, et al. Somatic mosaicism of PCDH19 mutation in a family with low-penetrance EFMR. Neurogenetics. 2012;13:341–5. doi: 10.1007/s10048-012-0342-9. [DOI] [PubMed] [Google Scholar]
- 6.Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009;5:e1000381. doi: 10.1371/journal.pgen.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juberg RC, Hellman CD. A new familial form of convulsive disorder and mental retardation limited to females. J Pediatr. 1971;79:726–32.. doi: 10.1016/S0022-3476(71)80382-7. [DOI] [PubMed] [Google Scholar]
- 8.Ryan SG, Chance PF, Zou CH, Spinner NB, Golden JA, Smietana S. Epilepsy and mental retardation limited to females: an X-linked dominant disorder with male sparing. Nat Genet. 1997;17:92–5. doi: 10.1038/ng0997-92. [DOI] [PubMed] [Google Scholar]
- 9.Hynes K, Tarpey P, Dibbens LM, Bayly MA, Berkovic SF, Smith R, et al. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J Med Genet. 2010;47:211–6. doi: 10.1136/jmg.2009.068817. [DOI] [PubMed] [Google Scholar]
- 10.Depienne C, Trouillard O, Bouteiller D, Gourfinkel-An I, Poirier K, Rivier F, et al. Mutations and deletions in PCDH19 account for various familial or isolated epilepsies in females. Hum Mutat. 2011;32:E1959–75. doi: 10.1002/humu.21373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Specchio N, Marini C, Terracciano A, Mei D, Trivisano M, Sicca F, et al. Spectrum of phenotypes in female patients with epilepsy due to protocadherin 19 mutations. Epilepsia. 2011;52:1251–7. doi: 10.1111/j.1528-1167.2011.03063.x. [DOI] [PubMed] [Google Scholar]
- 12.Zipursky SL, Sanes JR. Chemoaffinity revisited: Dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–53.. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Cooper SR, Jontes JD, Sotomayor M. Structural determinants of adhesion by Protocadherin-19 and implications for its role in epilepsy. Elife. 2016;5:e18529. doi: 10.7554/eLife.18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison OJ, Brasch J, Katsamba PS, Ahlsen G, Noble AJ, Dan H, et al. Family-wide structural and biophysical analysis of binding interactions among non-clustered δ-protocadherins. Cell Rep. 2020;30:2655–71.e7. doi: 10.1016/j.celrep.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–81.. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pederick DT, Richards KL, Piltz SG, Kumar R, Mincheva-Tasheva S, Mandelstam SA, et al. Abnormal cell sorting underlies the unique X-linked inheritance of PCDH19 epilepsy. Neuron. 2018;97:59–66.e5. doi: 10.1016/j.neuron.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Niazi R, Fanning EA, Depienne C, Sarmady M, Abou, Tayoun AN. A mutation update for the PCDH19 gene causing early-onset epilepsy in females with an unusual expression pattern. Hum Mutat. 2019;40:243–57.. doi: 10.1002/humu.23701. [DOI] [PubMed] [Google Scholar]
- 18.Kolc KL, Sadleir LG, Scheffer IE, Ivancevic A, Roberts R, Pham DH, et al. A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol Psychiatry. 2019;24:241–51.. doi: 10.1038/s41380-018-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higurashi N, Shi X, Yasumoto S, Oguni H, Sakauchi M, Itomi K, et al. PCDH19 mutation in Japanese females with epilepsy. Epilepsy Res. 2012;99:28–37. doi: 10.1016/j.eplepsyres.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Ishii A, Watkins JC, Chen D, Hirose S, Hammer MF. Clinical implications of SCN1A missense and truncation variants in a large Japanese cohort with Dravet syndrome. Epilepsia. 2017;58:282–90.. doi: 10.1111/epi.13639. [DOI] [PubMed] [Google Scholar]
- 21.Goto A, Ishii A, Shibata M, Ihara Y, Cooper EC, Hirose S. Characteristics of KCNQ2 variants causing either benign neonatal epilepsy or developmental and epileptic encephalopathy. Epilepsia. 2019;60:1870–80.. doi: 10.1111/epi.16314. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higasa K, Miyake N, Yoshimura J, Okamura K, Niihori T, Saitsu H, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet. 2016;61:547–53.. doi: 10.1038/jhg.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alves IT, Cano D, Bottcher R, van der Korput H, Dinjens W, Jenster G, et al. A mononucleotide repeat in PRRT2 is an important, frequent target of mismatch repair deficiency in cancer. Oncotarget. 2017;8:6043–56.. doi: 10.18632/oncotarget.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo YS, Oh HR, Kim MS, Yoo NJ, Lee SH. Frameshift mutations of OGDH, PPAT and PCCA genes in gastric and colorectal cancers. Neoplasma. 2016;63:681–6. doi: 10.4149/neo_2016_504. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz S, Jr, Yamamoto H, Navarro M, Maestro M, Reventos J, Perucho M. Frameshift mutations at mononucleotide repeats in caspase-5 and other target genes in endometrial and gastrointestinal cancer of the microsatellite mutator phenotype. Cancer Res. 1999;59:2995–3002. [PubMed] [Google Scholar]
- 27.Kim NG, Choi YR, Baek MJ, Kim YH, Kang H, Kim NK, et al. Frameshift mutations at coding mononucleotide repeats of the hRAD50 gene in gastrointestinal carcinomas with microsatellite instability. Cancer Res. 2001;61:36–8. doi: 10.1186/bcr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–3. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jo YS, Kim MS, Lee JH, Lee SH, An CH, Yoo NJ. Frequent frameshift mutations in 2 mononucleotide repeats of RNF43 gene and its regional heterogeneity in gastric and colorectal cancers. Hum Pathol. 2015;46:1640–6. doi: 10.1016/j.humpath.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Tornaletti S, Pfeifer GP. Slow repair of pyrimidine dimers at p53 mutation hotspots in skin cancer. Sci (N. Y, NY) 1994;263:1436–8. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]
- 31.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–77.. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 32.Popp MW, Maquat LE. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell. 2016;165:1319–22.. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva AL, Romão L. The mammalian nonsense-mediated mRNA decay pathway: To decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 34.Nabbout R, Gennaro E, Dalla Bernardina B, Dulac O, Madia F, Bertini E, et al. Spectrum of SCN1A mutations in severe myoclonic epilepsy of infancy. Neurology. 2003;60:1961–7. doi: 10.1212/01.WNL.0000069463.41870.2F. [DOI] [PubMed] [Google Scholar]
- 35.Zuberi SM, Brunklaus A, Birch R, Reavey E, Duncan J, Forbes GH. Genotype–phenotype associations in SCN1A-related epilepsies. Neurology. 2011;76:594–600. doi: 10.1212/WNL.0b013e31820c309b. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Shalom R, Keeshen CM, Berrios KN, An JY, Sanders SJ, Bender KJ. Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol Psychiatry. 2017;82:224–32.. doi: 10.1016/j.biopsych.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, Gardella E, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140:1316–36.. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 38.Sanders SJ, Campbell AJ, Cottrell JR, Moller RS, Wagner FF, Auldridge AL, et al. Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci. 2018;41:442–56.. doi: 10.1016/j.tins.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.