Abstract
Coronavirus disease 2019 (COVID-19) is caused by the SARS-CoV-2 virus and has been affecting the world since the end of 2019. The disease led to significant mortality and morbidity in Turkey, since the first case was reported on March 11th, 2020. Studies suggest a positive association between air pollution and SARS-CoV-2 infection. The aim of the present study was to investigate the role of ambient particulate matters (PM), as potential carriers for SARS-CoV-2. Ambient PM samples in various size ranges were collected from 13 sites including urban and urban-background locations and hospital gardens in 10 cities across Turkey between 13th of May and 14th of June 2020 to investigate the possible presence of SARS-CoV-2 RNA on ambient PM. A total of 203 daily samples (TSP, n = 80; PM2.5, n = 33; PM2.5–10, n = 23; PM10μm, n = 19; and 6 size segregated PM, n = 48) were collected using various samplers. The N1 gene and RdRP gene expressions were analyzed for the presence of SARS-CoV-2, as suggested by the Centers for Disease Control and Prevention (CDC). According to real time (RT)-PCR and three-dimensional (3D) digital (d) PCR analysis, dual RdRP and N1 gene positivity were detected in 20 (9.8%) samples. Ambient PM-bound SARS-CoV-2 was analyzed quantitatively and the air concentrations of the virus ranged from 0.1 copies/m3 to 23 copies/m3. The highest percentages of virus detection on PM samples were from hospital gardens in Tekirdağ, Zonguldak, and Istanbul, especially in PM2.5 mode. Findings of this study have suggested that SARS-CoV-2 may be transported by ambient particles, especially at sites close to the infection hot-spots. However, whether this has an impact on the spread of the virus infection remains to be determined.
Keywords: SARS-CoV-2, Particulate matter, Airborne, 3D-dPCR
Graphical abstract
1. Introduction
Severe acute respiratory syndrome related coronavirus 2 (SARS-CoV-2) disease 19 (COVID-19) started from Wuhan, China, in December 2019 (WHO, 2020; Liu et al., 2020a; Zhu et al., 2020). The spread of the outbreak continues (Maqbool and Khan, 2020), and according to WHO, the COVID-19 pandemic has resulted in 126,888,424 confirmed cases and 2,778,603 related deaths globally as of March 29, 2021 (WHO, 2021). Accordingly, the total number of cases and the number of deaths were 3,208,173 and 31,076, respectively, in Turkey (TMOH, 2021).
Ambient and indoor particulate matter (PM) is a complex matrix that may contain various chemical and biological constituents (bacteria, virus, and fungi etc.) of a great health concern (Gong et al., 2020; Wang et al., 2020; Domingo and Rovira, 2020; Yu et al., 2018; Onat et al., 2016). Studies suggest that these agents can be long-range transported by atmospheric PM, and that this could be a root for transmission of infectious diseases (Chen et al., 2010; Domingo and Rovira, 2020; Wei et al., 2019). Accordingly, aerosol and droplets generated during speaking, sneezing, or coughing by infected people are well-known as the source of short-range transmission pathways for viral infections (Chu et al., 2020). In particular, respiratory viral diseases can spread directly or indirectly through a virus-containing particle among humans (Chu et al., 2020; Buonanno et al., 2020; Liu et al., 2020a). Buonanno et al. (2020) estimated the quanta emission rates of SARS-CoV-2 emitted from contagious subjects, they detected higher emission rates by asymptomatic COVID-19 subjects during exercise as speaking or oral breathing, whereas the symptomatic subjects in resting conditions had low emission rates. Similarly, Liu et al. (2020a) investigated the aerodynamic nature of SARS-CoV-2 in size-segregated indoor aerosol samples from hospital divisions, and the high concentrations were detected in patients' toilets and reported that the size distribution of the SARS-CoV-2 peaked on the smallest particles. Recently, Hadei et al. (2021) reported the presence of SARS-CoV-2 in total suspended particles (TSP) collected from indoor atmosphere of the public places and transportation in Tehran, Iran.
Aerosolized respiratory viruses have great potential for spreading the infections, and studies suggest that ambient air pollutants could be a possible virus carrier (Amoatey et al., 2020; Carraturo et al., 2020; Creager et al., 2017; Groulx et al., 2018; Sedlmaier et al., 2009). Correspondingly, COVID-19 may have a contagion route via airborne transmission on atmospheric PM (Bontempi, 2020; Coccia, 2020; Zhang et al., 2020). In a recent study, Setti et al. (2020) found the marker genes of SARS-CoV-2 in PM10 samples collected in the Bergamo area, Northern Italy suggesting a potential indicator of the transmission of the infection by ambient particles. In contrast, more recent studies from Spain (Linillos-Pradillo et al., 2021) and Italy (Chirizzi et al., 2021) could not demonstrate the presence of SARS-CoV-2 in ambient PM collected from Madrid, and northern (Veneto) and southern (Apulia) Italy.
Unlike the studies investigating the presence of SARS-CoV-2 in indoor environments, atmospheric studies are very scarce. The only study confirming the presence of virus in atmospheric PM has reported qualitative results (negative or positive) (Setti et al., 2020), which does not give any information about the intensity of the virus, where samples are collected. However, the methods such as three-dimensional digital polymerase chain reaction (3D-dPCR) are shown to work with high sensitivity in the detection of SARS-CoV-2 in low viral load specimens (Suo et al., 2020) such as environmentally collected samples. As stated by Maleki et al. (2021), more studies are needed to elucidate the role of atmospheric PM pollution in the SARS-CoV-2 spread. The aim of the present study was to investigate the role of ambient PM, as potential carriers for SARS-CoV-2. We collected ambient PM samples in various size ranges from 13 sites in 10 Turkish cities including urban and suburban locations, as well as hospital gardens from May 13 to June 14, 2020 and demonstrated the presence of SARS-CoV-2 on these particles. To our knowledge, this is the first study reporting quantitative results for SARS-CoV-2 in the atmosphere.
2. Materials and methods
2.1. Sampling locations and methods
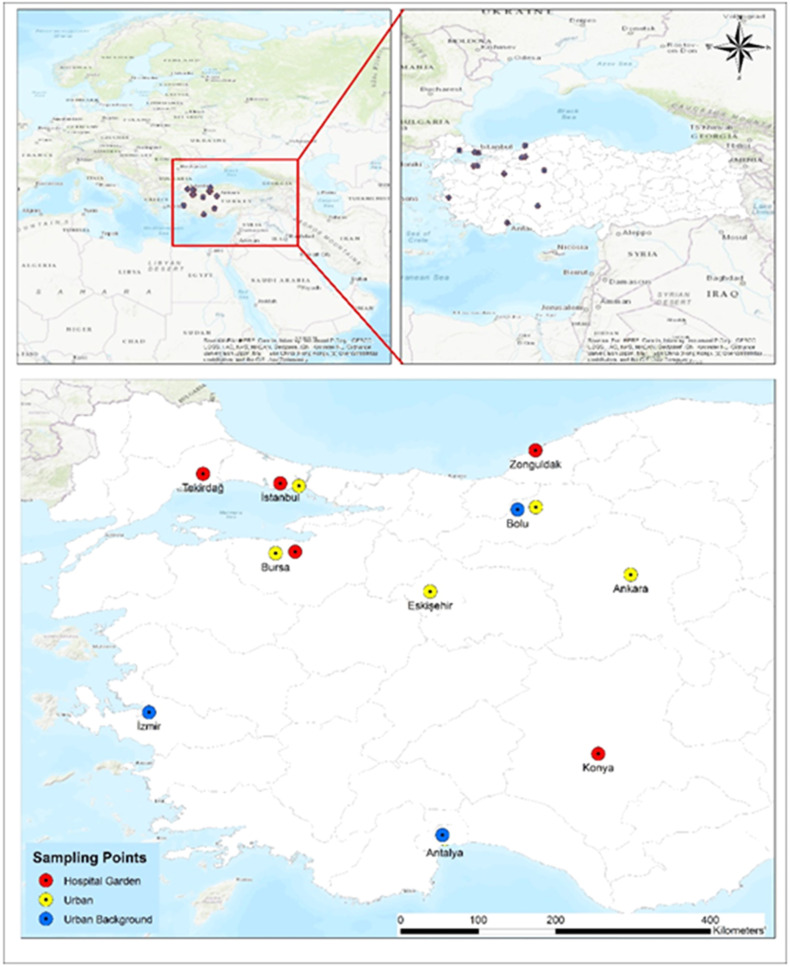
PM samples within various size-ranges were collected from 13 locations within 10 cities in western Turkey between 13th May and 14th June 2020, during the first peak of the outbreak in Turkey. We wanted to include as many cities as possible to adequately represent the whole country, however, it was not possible to cover all regions, because of the local problems and obstacles due to COVID-19 pandemic. Fig. 1 shows the location map of PM sampling sites, and Table 1 summarizes the methodology of PM sampling, cut sizes of collected PM, and the typical features of the sampling locations. A total of 155 samples (TSP, n = 80; <2.5 μm particles-PM2.5, n = 33; 2.5–10 μm particles-PM2.5 – 10, n = 23; >10 μm particles-PM10, n = 19) were collected daily using various PM samplers in each city. One of the sampling stations in Istanbul (Urban station in Esenler) was allocated to collect size segregated PM samples in six size ranges by an impactor sampler. The impactor system consists of 5 stages and a back-up stage for size cutting from 0.49 to >7.2 μm (0.49–0.95; 0.95–1.5; 1.5–3; 3–7.2 and >7.2 μm and a back-up filter <0.49 μm). Therefore, six PM samples of different size ranges per day were collected at this sampling site during 8 days over the study period. Unfortunately, it was not possible to collect samples for eight consecutive days due to COVID-19 restrictions in Turkey, and samples were collected when permitted.
Fig. 1.
Sampling locations in Turkey.
Table 1.
The descriptions of particulate matter sampling methods from 13 sites in 10 cities in Turkey.
| Cities | Site | PM Sampling Equipment | Sampling period | Number of samples | PM Fraction | Filter Types | Sampling volume, m3 day−1 |
|---|---|---|---|---|---|---|---|
| İstanbul | HG | SKC Filter Pack Sampler | 13.05.2020–04.06.2020 | 20 | TSP and PM2.5 | PTFE | 7.2 |
| Tekirdağ | HG | Dichotomous PM Sampler | 17.05.2020–31.05.2020 | 20 | PM2.5–10 and PM2.5 | PTFE | 24 |
| Bursa | HG | High Volume Air Sampler | 21.05.2020–27.05.2020 | 7 | TSP | GF | 278 |
| Zonguldak | HG | Dichotomous PM Sampler | 16.05.2020–28.05.2020 | 26 | PM2.5–10 and PM2.5 | PTFE | 24 |
| Konya | HG | High Volume Air Sampler | 21.05.2020–03.06.2020 | 14 | TSP | GF | 360 |
| Ankara | U | Low Vol Stack Filter Unit | 30.05.2020–14.06.2020 | 10 | TSP | PTFE | 24 |
| Bolu | U | SKC Filter Pack Sampler | 14.05.2020–22.05.2020 | 9 | TSP | PTFE | 7.2 |
| İzmir | U | Zambelli PM Sampler | 14.05.2020–02.06.2020 | 9 | PM10 | PTFE | 24 |
| İstanbul | U | High Vol Cascade Impactor | 13.05.2020–02.06.2020 | 48 | 0.49–7.2 μm (6 fractions) | GF | 1422 |
| Eskişehir | U | SKC Filter Pack Sampler | 16.05.2020–30.05.2020 | 14 | TSP | PTFE | 7.2 |
| Bursa | UB | High Volume Air Sampler | 21.05.2020–27.05.2020 | 7 | TSP | GF | 318 |
| Bolu | UB | SKC Filter Pack Sampler | 14.05.2020–22.05.2020 | 9 | TSP | PTFE | 7.2 |
| Antalya | UB | Low Vol Stack Filter Unit | 18.05.2020–03.06.2020 | 10 | PM10 | NPF | 24 |
U: Urban site, UB: Urban background, HG: Hospital garden, GF: Glass fiber filter, PTFE: Teflon filter, NPF: Nucleopore Polycarbonate filter, TSP: Total Suspended Particulate, PM2.5–10: Particulate Matter between 2.5 and 10 μm, PM10: Particulate Matter ≤10 μm.
Five of the 13 sampling sites were in the hospital gardens. The other five sites were characterized as urban, and the remaining 3 sites were urban background locations. Urban locations, which were located in densely built-up areas, were under the influence of traffic, whereas urban background stations were not affected by nearby traffic and reflected the general exposure of the urban population to air pollutants. Samples were collected on glass fiber filters (GF) and Teflon filters (TF) with different sampling equipments (Table 1). Both low and high-volume samplers were used to collect daily samples. For all sampling locations, sampling was started in the morning and finished at the same time on the following day. All PM samples for each specific location were collected on different days over the study period (Table S1). At the end of 24 h, each sample was placed into sterile petri dish using sterile equipment and stored at −20 °C. PM samples were collected by research teams of several universities of Turkey and transported to biosafety level 2 (BSL 2) microbiology research laboratory at Koç University, School of Medicine for PCR analysis.
At the BSL 2 laboratory, filters were cut into halves with a sterile scalpel, and one half was immediately subjected to PCR analysis. The remaining halves of the filters were kept frozen for further chemical and bacteriological characterization. Since the main purpose of this study was to investigate the presence of SARS-CoV-2, filters were not weighed to avoid any contamination. Field blanks (n = 3) from each site were also analyzed together with samples to detect any contamination.
There are more than 300 air quality monitoring stations in Turkey operated by the Turkish Ministry of Environment and Urbanization (MoEU) according to European and United States standards. To evaluate the air quality and meteorology in the PM sampling cities during the PM sampling time, daily averages of PM2.5, PM10, NO2, SO2, CO, and O3 were acquired from the national air quality monitoring network of the MoEU (MoEU, 2020), and the meteorological parameters were obtained from the National Oceanic and Atmospheric Administration (NOAA), using the locations of PM sampling sites (NOAA, 2021).
2.2. Analytical methods
According to World Health Organization (WHO), real-time (RT)-PCR represents the gold standard for the diagnosis of SARS-CoV-2. Therefore, all samples were first screened by RT-PCR with two different specific markers (nucleocapsid [N]1 and RNA dependent RNA polymerase [RdRP]) genes) (CDC, 2020; Nalla et al., 2020).
Recently, 3D-dPCR has demonstrated the best performance to detect SARS-CoV-2, because it reduces the false negatives (de Almeida et al., 2020; Suo et al., 2020). The samples that were found to be positive for both genes (N1 and RdRP) were further analyzed by 3D-dPCR.
2.2.1. RNA isolation
RNA isolation was performed using the Quick-RNA™ Fecal/Soil Microbe Microprep Kit (ZYMO Research, USA) according to the manufacturer's instructions. Briefly, the half filter was rolled with the upper surface facing inward in a 2-ml polypropylene tube together with the beads provided in the kit. From the initial 1 ml of lysis buffer, ~400 μl of lysate was harvested, and then processed as defined by the instructions resulting in a final eluate of 10 μl. Subsequently, all the eluted RNA was used for SARS-CoV-2 testing.
2.2.2. cDNA synthesis and real-time polymerase chain reaction (RT-PCR)
The cDNA was synthesized from the eluted RNA with Veriti™ Thermal Cycler (Applied Biosystem, USA) using iScript™ cDNA Synthesis Kit (Bio-Rad, USA) according to the instructions of the manufacturer. Real-time (RT)-PCR for SARS-CoV-2 was carried out on a LightCycler 480 RT-PCR (ROCHE, Germany) based on the CDC recommendations. The cycling conditions of Syber Green were as follows; in 5 min at 95 °C, followed by 45 cycles of 10 s at 95 °C, 30 s at 63 °C, and 30 s at 72 °C, and final cooling step at 4 °C. Cycling conditions for Taqman Hybridization was as follows; in 7 min at 95 °C, followed by 45 cycles of 10 s at 95 °C, 1 min at 63 °C, and 30 s at 72 °C followed by maintenance at 4 °C. Gene sets were obtained from the Centers for Disease Control and Prevention (CDC). Primer sequences: N1 Forward: 5’-GACCCCAAAATCAGCGAAAT-3′, N1 Reverse: 5′- TCTGGTTACTGCCAGTTGAATCTG-3′; as an internal control RdRP Forward:5’-AGATTTGGACCTGCGAGCG-3′, RdRP Reverse: 5′- GAGCGGCTGTCTCCACAAGT-3′. Probe: FAM – 5’-TTCTGACCTGAAGGCTCTGCGCG -3′– BHQ1 (CDC, 2020). Positive and negative controls were run in each test. After each Syber Green run, we performed melting curve analysis and gel electrophoresis to verify whether the expected target was amplified as suggested by Ruiz-Villalba et al. (2017). We aimed to detect the viral RNA below 100 copy/ml, therefore we set the threshold cycle to 40. Samples with threshold cycle (Ct) values of 40 and below for the N1 and RdRP genes were identified as positive (Han et al., 2021).
2.2.3. Three-dimensional (3D)-digital (d) PCR
The main difference of 3D-dPCR from RT-PCR is that the reaction volume is split over a high number of small partitions (from 500 up to millions) of a very small volume (currently from 6 nl down to a few picolitres). After the PCR, each partition is scored either as positive or negative (binary or digital read-out). Statistical analysis of the results is then used to determine the absolute quantity of target DNA in a sample (Majumdar et al., 2015; Pavšič et al., 2015). This method shows high sensitivity with the strategy of counting a single molecule, and it also provides a high reliability and repeatability level (Suo et al., 2020). Furthermore, this method has been proven to work with high sensitivity in studies related to SARS-CoV-2 (de Almeida et al., 2020; Suo et al., 2020).
The probe set for N1 gene were obtained from the CDC as described above (CDC, 2020). Primers used in RT-PCR analyses were also used in 3D-dPCR analyses. The probe sequence of N1 gene was FAM-5’-ACCCCGCATTACGTTTGGTGGACC-3’-BHQ1. Cycling conditions were as follows; in 10 min at 96 °C, followed by 39 cycles of 30 s at 63 °C and 2 min at 98 °C, and a final step of 63 °C per 2 min followed by maintenance at 4 °C. The chips were read in the QuantStudio 3D™ reader (Thermo, USA), and the results were interpreted in the dPCR AnalysisSuite™ app in the Thermo Fisher Connect™ Dashboard. For positive control, dilutions of (10−3, 10−4, 10−5 and 10−6) of a clinical isolate of SARS-CoV-2 virus (TCID50 = 106/ml) RNA was used.
The positivity of the SARS-CoV-2 on PM samples was characterized by a multi-parameter decision approach. The 3D-dPCR results (copy number/ μl) were transformed in a threshold of viral particles contained in a single filter (i.e. RNA copies per filter) calculating the volume of RNA and extraction solutions. Later, viral load per filter numbers were normalized using the sampled volume of air to calculate the viral concentration in air per m−3.
3. Results and discussion
3.1. Presence of the virus on ambient atmospheric particulate matter
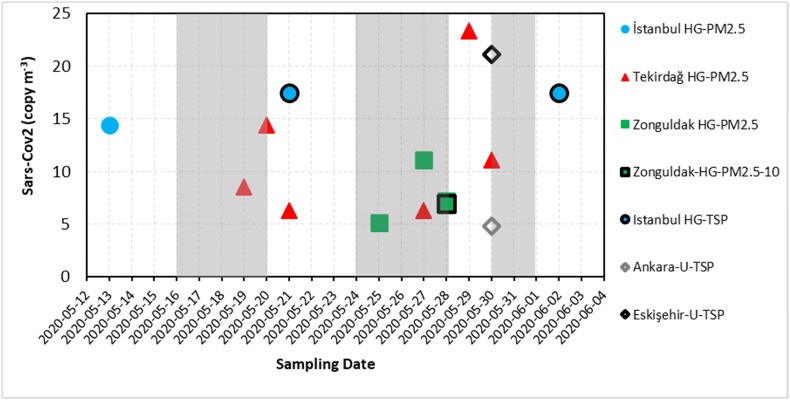
A total of 155 samples including TSP (n = 80), PM2.5 (n = 33), PM2.5 – 10 (n = 23), and PM10 (n = 19) were collected at all locations. Additionally, 48 PM samples segregated in 6 sizes were collected during the 8 days of sampling in urban Istanbul. Fig. 2 and Table S1 show the positive results of the RdRP and N1 genes on filters from various sites inspected within this study. The virus presence along with environmental parameters for all samples collected at each location are given in Table S2 in the supplementary. The positivity of the SARS-CoV-2 on PM samples was characterized by a multi-parameter decision approach. Initially, the N1 gene was studied on all 203 PM filter samples including segregated ones by RT-PCR using Syber Green method. Of these, four samples were positive for the N1 gene with a Ct < 40, whereas 199 samples had a Ct value of ≥40 for the same gene. The latter 199 samples were further analyzed for specific products by checking melting curves, as well as specific products in a 2% agarose gel. Bands of specific products were detected in 52 samples, and these were further analyzed for RdRP genes together with 4 samples positive for the N1 gene by RT-PCR using the Taqman hybridization probe. Of these 56 samples, 4 samples were shown to have a Ct <40 for both N1 and RdRP genes, 35 samples had a Ct of ≥40 for N1 and Ct <40 for RdRP genes, respectively. The remaining 17 samples had a Ct ≥ 40 for both N1 and RdRP genes with no sign of a specific product, and they were treated as negative for SARS-CoV-2. Altogether, 39 samples including 4 samples being positive for N1 gene by RT-PCR and 35 samples with presence of specific products were analyzed by 3D-dPCR for N1 gene, and 20 samples were found to be positive for the N1 gene with an amplified copy number above 10 copies μl−1 (Table S1). The maximum concentrations of viral RNA in all samples were detected as 23 copy/m3 air. The Ct values of all samples were above 35 indicating a low copy number of the virus, and it has been suggested that the culturable virus cannot be isolated from the samples with Ct values >34 (Wolfel et al., 2020). Therefore, it is difficult to predict whether such small copy numbers of the virus will be contagious. The vitality and infectious capacity of the detected virus in our samples could not be evaluated, either.
Fig. 2.
SARS-CoV-2 copy number on TSP, PM10, PM2.5–10 and PM2.5 for sampling sites (excluding size segregated samples). The grey shaded area on the graph shows the lockdown periods in Turkey. The indicator colors of blue, red, green, black and grey represent the samples from İstanbul, Tekirdağ, Zonguldak, Ankara and Eskişehir, respectively. HG, hospital garden; U, urban site; TSP, total suspended particulate; PM, particulate matter. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Airborne transmission of SARS-CoV-2 was previously investigated by Bontempi (2020) in the ambient atmosphere of Northern Italy in relation to the increasing concentrations of PM. After a basic correlation analysis of ambient PM data and confirmed cases obtained from the Italian cities, they reported that it was not possible to conclude that the infection was transmitted through PM10 particles without any additional information. However, Setti et al. (2020) later on reported the presence of the SARS-CoV-2 RNA on ambient aerosol samples collected from the ambient air of Bergamo (Northern Italy), where the incidence of the COVID-19 was extremely high during February 21st and March 13th, 2020. They inspected the marker genes of the virus including E, N, and RdRP, and reported 20 positive results for at least one marker among the 34 RNA extractions. More recently, Hadei et al. (2021) investigated the presence of SARS-CoV-2 in air of public places such as shopping centers, post office, banks, governmental offices, and public transportation facilities including subways, and buses in Tehran, Iran, and they detected SARS-CoV-2 RNA in 64% of 28 TSP samples. These findings are in agreement with the findings of the present study. In contrast, Chirizzi et al. (2021) failed to detect the genetic material of SARS-CoV-2 in PM (0.056 to 18 m) collected from northern and southern Italy during the pandemic in May 2020. Linillos-Pradillo et al. (2021) could not detect SARS-CoV-2 in PM10, PM2.5 and PM1 collected from Madrid, Spain during the same May 2020 period, either. They concluded that the absence of viral genomes could be due to various factors including limited human activities and lower daily PM concentrations or seasonal characteristics.
One of the main differences between our study and those of others is, however, the method used for SARS-CoV-2 analysis. We used a 3D-dPCR method to quantify the copy number of the virus, whereas others used the qualitative detection methods (Setti et al., 2020; Linillos-Pradillo et al., 2021; Hadei et al., 2021). It has been demonstrated that 3D-dPCR provides high reliability and repeatability level (de Almeida et al., 2020; Suo et al., 2020), and gives quantitive information about the intensity of the virus at the location, where samples were collected.
3.2. Impact of particle sampling and location on the presence of the virus
When looked at the locations (Table 1) for sample collection of 20 samples positive for SARS-CoV-2, 13 samples were close to hospitals, and 7 were from urban sites (Table S1). The locations of positive samples included Zonguldak (n = 4, hospital garden), Tekirdağ (n = 6, hospital garden), Istanbul (n = 3, hospital garden), Eskişehir (n = 1, urban site), Istanbul (n = 5, urban site), and Ankara (n = 1, urban site). All analyzed samples were negative for the virus collected from Bolu (urban and urban background), Bursa (urban background and hospital garden), Konya (hospital garden), Antalya (urban background), and Izmir (urban). When urban background samples were analyzed neither the N1 nor the RdRP gene was amplified during the 45 cycles of RT-PCR. Therefore, these samples were negative for the presence of viruses, and no interference was observed from the used filters and equipment. Of 155 PM samples collected from all locations, positive counts of SARS-CoV-2 RNA on ambient particles according to PM sizes were as PM2.5 (n = 10), PM10 (n = 1), and TSP (n = 4). The remaining 48 PM samples were collected by an impactor having different size ranges (0.49 to >7.2 μm [0.49–0.95; 0.95–1.5; 1.5–3; 3–7.2 and >7.2 μm and a back-up filter <0.49 μm]) in urban İstanbul and analyzed for the virus. Among 48 size-segregated filters, the virus was detected only on 5 filters. The PM sizes of positive samples and the number of samples detected in size-segregated samples were PM<0.49 (n = 1), PM0.49 – 0.95 (n = 1), PM0.95 – 1.5 (n = 1), and PM>7.2 (n = 2). The virus was detected in size-segregated PM samples collected on three different days out of 8. On two succeeding days, the virus was detected on particles >7.2 μm and on <0.49 μm and >7.2 μm for 14th and 15th of May, respectively. Since the number of positives in the size segregated samples was low and random, no evaluation could be made regarding the distribution of virus in different sizes of PM. Thus, a detailed description of the size distribution of SARS-CoV-2 in atmospheric particles needs more attention and systematic approaches for further research (Table 1, Table S1 and Table S2).
We demonstrated that SARS-CoV-2 RNA can be present on ambient PM suggesting that the virus may be transported via PM pollution. PM is a complex matrix and consists of a mixture of solid and liquid particles of organic and inorganic substances suspended in the air. It is likely that the liquid like organic layer and surface of particles can provide a medium for the interaction between virus containing particles and PM, and that the virus may also be transferred from a respiratory droplet to PM. It is possible that virus containing particles could be attached to PM by some mechanisms like impaction, interception, and diffusion (Coccia, 2020; Groulx et al., 2018; Bao et al., 2015). Fig. 2 shows the ambient air particle-bound SARS-CoV-2 concentrations for sampling sites excluding the size segregated samples of the urban site of Istanbul in terms of N1 gene copy number (copy/m3). The virus concentrations in the air detected were between 5 and 23 copies/m3. Positive samples for the virus were mostly obtained from the hospital gardens (60%, 50%, and 30% of total samples in Tekirdağ, Istanbul, and Zonguldak, respectively), and the number of positive samples were relatively higher in those collected in PM2.5 mode (Fig. 2 and Table S1). It is possible that the high viral copy numbers were due to increased inhaled droplet numbers exhaled by patients spending time in these areas; however, we do not have any data regarding the number of COVID-19 patients occupying these areas during the period, when the samples were collected.
Interestingly, most of the SARS-CoV-2-positive samples were collected during the lockdown time, which coincided with increased number of daily COVID-19 cases. During the sampling period, there was a lockdown precaution in 9 of the 10 sampling cities due to increasing daily COVID-19 cases. Daily cases and cases per capita were generally high in these cities, particularly in Istanbul, Ankara, Izmir, and Zonguldak exceeding national average. Although the population and daily COVID-19 cases were higher in Izmir and Bursa cities, there were no positive samples detected. Because of the restrictions on disseminating the number of COVID-19 cases in each city of Turkey (Bayram et al., 2020), and only the number of total national cases were shared (RTMH, 2021), we could not evaluate the results comparing with the number of COVID-19 cases. Therefore, a possible association between the presence of SARS-CoV-2 in PM samples from sites studied and the number of patients reported from those areas could not be investigated.
The amount of suspended viral copies in the air is another significant factor affecting the airborne transmission of SARS-CoV-2. Lednicky et al. (2020) reported indoor air virus presence from a hospital room with two COVID-19 patients. They reported 16 to 94 SARS-CoV-2 genome equivalents per L of indoor air in the patients' room after 3 h indoor PM samplings. Liu et al. (2020a) investigated the aerodynamic nature of the virus in two Wuhan hospitals from February to March 2020 by collecting indoor aerosol samples with a cascade sampler and a similar virus detection protocol, as in the present study. They reported indoor air virus concentrations of 0 to 11 copy/m3 on the PM samples collected from public areas, 0 to 42 copy/m3 on medical staff areas, and 0 to 113 copy/m3 in the patient areas including the toilets, workstations, and intensive care units. In a study by Hu et al. (2020), both indoor and ambient air samples were collected and analyzed for the presence of SARS-CoV-2 from selected microenvironments in Wuhan, China. Positive viral RNA percentages were 10% and 20% for the ambient samples collected from a 10 m distance to the inpatient and outpatient building doors, and the air concentrations were 0.89 to 1.65 × 103 copy/m3 (Hu et al., 2020). Those results support the findings of the present study regarding the higher percentage of detection of the virus on PM samples from hospital gardens in Istanbul, Tekirdağ, and Zonguldak. The location of the PM samplers was on close proximities to the hospital main buildings in most of the sampling cities except Bolu (urban and urban background), Antalya (sub-urban), Eskişehir (urban), Istanbul site 1 (urban), Ankara (urban), and Izmir (urban). The sampler in Tekirdağ was a close spot to the exhaust fans of the main ventilation system of the hospital building and showed the highest positive virus incidences. The minimal infectious dose for SARS-CoV-2 is currently not known. However, previous animal studies and modelling of SARS transmission in China estimate that approximately 280 viral particles may cause disease in 50% of population (ID50 dose) (Watanabe et al., 2010). Recently, Karimzadeh et al. (2020) proposed the minimum infective dose of COVID-19 in humans is higher than 100 particles, possibly slightly lower than the 700 particles estimated for H1N1 influenza.
3.3. Effect of air quality and meteorological parameters on the presence of virus
A summary of mean air quality and the meteorological parameters in the PM sampling cities are given in Table S3. Relatively lower pollutant concentrations were obtained due to the decreased anthropogenic activity during the pandemic precautions, stay-at-home advice, and lockdown measures from a nationwide perspective (Brimblecombe and Lai, 2020; Cameletti, 2020; Shrestha et al., 2020; Şahin, 2020). A variable ambient temperature range was observed among 10 cities during PM sampling due to the transitional characteristics of the spring season. Daily average temperatures, wind speeds, relative humidity (RH), as well as precipitation levels all differed substantially, and all were affected by the basic air quality parameters, in particular by PM concentrations. Besides, the synergistic combination of ambient meteorological parameters (temperature and RH), air quality (PM toxicity) may also have a significant effect on vitality and transmission of aerosol biological constituents characterized by the suitability and atmospheric lifetime/travel distance of suspended particles (Domingo and Rovira, 2020). Correspondingly, the RH values observed in Istanbul (76.3%), Zonguldak (75.0%), and Tekirdağ (72.4%), in these cities the virus were detected higher, were all higher than those reported from remaining cities (varying between 51.9% and 67.2%). The data were not sufficient to analyze the impact of humidity or PM levels on the transmission of the virus adsorbed on particles. Both positive and negative non-linear relationships between ambient temperatures and COVID-19 cases were reported in different cities in China by Zhang et al. (2020). More importantly, they reported direct positive correlations between new confirmed cases and air pollution indicators as viral spread increased with air quality index (AQI). Şahin et al. (2020) reported that the wind speed and temperature on the day of the case have high impacts on COVID-19 cases in Turkey. In contrast, Coşkun et al. (2021) showed that winds were effective in spreading, but temperature, humidity, the number of sunny days, and air pollution did not affect the number of cases in Turkey. So, there is not any consensus about the relationship.
Basic air quality parameters like PM, sulfur dioxide (SO2), nitrogen oxides (NOx), ozone (O3), carbon monoxide (CO), volatile organic compounds (VOCs), and polycyclic aromatic hydrocarbons (PAHs) together with meteorological parameters were all inspected to have a potential effect on the transmission of respiratory infections in the literature. Domingo and Rovira (2020) reviewed the literature on ambient air pollution and respiratory diseases. The review supported an obvious relationship between ambient levels of certain pollutants and viruses influencing each other to negatively deteriorate the human respiratory system. Liu et al. (2020b) compiled a daily confirmed case count, ambient temperature, diurnal temperature range, absolute humidity, and migration scale index data from 30 provincial capital cities of China to explain the associations between COVID-19 and meteorological parameters using a non-linear regression analysis. They concluded that the meteorological parameters play an independent functionality in the COVID-19 transmission after controlling for population migration. Local meteorological conditions with low to mild diurnal temperatures together with low humidity likely favored the transmission.
In a similar study, Zhang et al. (2020) inspected ambient meteorology and local air quality on the distribution of the disease in 219 Chinese cities from January 24th to February 29th, 2020. They similarly concluded a negative relationship between ambient temperature and positive correlations among air pollution indicators and confirmed cases. Indeed, both studies were conducted on linear or non-linear statistical relationships with daily confirmed cases and environmental inputs and presented stochastic results among model inputs. The results of the present study could not statistically be related to the ambient air quality or meteorological parameters. Even though the total number of filter samples collected was relatively high, the number of individual samples from each site varied between 7 and 20 and limited the construction of a statistical model for a correlation analysis.
There are several reports on the transmission of influenza viruses by PM (Bao et al., 2015; Chen et al., 2010; Domingo and Rovira, 2020; Sedlmaier et al., 2009; Yu et al., 2018) and they proposed that the long-range transport of influenza viruses is possible by combustion derived particles, which have pores providing a suitable space for the virus settlement. This assumption may also be valid for our case, since a high incidence of daily COVID-19 were observed in Zonguldak and Tekirdağ cities, where higher particulate matter pollutions were reported due to the industries and coal fired power plants, respectively (Akyüz and Çabuk, 2009; Can-Terzi et al., 2021; Tecer et al., 2008), as well as in İstanbul, the biggest metropolitan city of Turkey suffering from traffic and industry-related air pollution (Kuzu and Saral, 2017; Şahin et al., 2020). Thus, more samples need for detailed evaluation of the relationship between air polluted area and the presence of SARS-CoV-2 in atmospheric particles.
4. Conclusions
This study offers a preliminary evaluation of the possible presence of SARS-CoV-2 on ambient PM in characteristic sites of Turkey during one of the most intense periods of COVID-19 pandemic. Most of the SARS-CoV-2 positive samples were taken from hospital gardens, but samples collected from two urban sides were also positive, as demonstrated by the 3D-dPCR method, which has been proven to be highly sensitive and reliable. One of the limitations of our study was that we could not obtain PM samples from all parts of the country due to local problems and the difficulties associated with COVID-19 pandemic. Another constraint was that although various studies from the literature suggested a relationship between SARS-CoV-2 transmission and meteorological parameters and air quality parameters, the data on these parameters were not sufficient to perform such a statistical analysis in the current study. Nevertheless, our findings suggest that SARS-CoV-2 may be transported by ambient particles, whether this has an impact on the spread of the virus remains to be determined. Therefore, the public should use personal protection equipment such as face masks during outdoor activities in “hotspot” areas such as around hospitals. Future studies should focus on the viability and infectivity of the SARS-CoV-2 present on ambient particles.
The following are the supplementary data related to this article.
RdRP and N1 gene positivity of filter samples.
All data collected from 13 stations in 10 cities of Turkey.
Summary of mean air pollutants concentration and meteorological parameters during the sampling periods in each city.
CRediT authorship contribution statement
Özgecan Kayalar: Writing-Original draft preparation, Methodology, Data curation, Investigation, Formal analysis; Akif Arı: Writing Original draft, Resources, Data curation; Gizem Babuççu: Data curation, Methodology, Investigation; Nur Konyalılar: Data curation, Methodology, Investigation; Özlem Doğan: Formal analysis, Methodology, Visualization; Füsun Can: Formal analysis, Methodology, Supervision, Writing - review & editing, Funding acquisition; Ülkü A. Şahin: Data curation, Formal analysis, Methodology, Supervision, Writing - review & editing, Project administration; Eftade O. Gaga: Data curation, Writing - review & editing; S. Levent Kuzu: Data curation, Writing - review & editing; Pelin Ertürk Arı; Data curation, Writing - review & editing; Mustafa Odabası: Data curation, Writing - review & editing; Yücel Taşdemir: Data curation; S.Sıddık Cindoruk: Data curation; Fatma Esen: Data curation; Egemen Sakın: Data curation; Burak Çalışkan: Data curation; Lokman H. Tecer: Data curation, Conceptualization; Merve Fıçıcı: Data curation; Ahmet Altın: Data curation; Burcu Onat: Data curation, Coşkun Ayvaz: Data curation; Burcu Uzun: Data curation; Arslan Saral: Data curation; Tuncay Döğeroğlu; Conceptualization, Project administration, Visualization; Semra Malkoç: Data curation, Özlem Ö. Üzmez: Data curation, Fatma Kunt: Data curation; Senar Aydın: Data curation; Melik Kara: Data curation; Barış Yaman: Data curation; Güray Doğan: Data curation; Bihter Olgun: Data curation; Ebru N. Dokumacı: Data curation; Gülen Güllü: Data curation, Visualization; Elif S. Uzunpınar: Visualization; Hasan Bayram: Conceptualization, Project administration, Funding acquisition; Writing - review & Editing; Visualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Koc University Research Center for Translational Medicine (KUTTAM) for funding the laboratory analysis of SARS-CoV-2 in PM samples. The authors thank to officials of Zonguldak Bülent Ecevit University Science and Technology Research Center, and Health Medical Research and Application Center for device and workforce support during sample collection.
Editor: Jianmin Chen
References
- Akyüz M., Çabuk H. Meteorological variations of PM2.5/PM10 concentrations and particle-associated polycyclic aromatic hydrocarbons in the atmospheric environment of Zonguldak, Turkey. J. Hazard. Mater. 2009;170:13–21. doi: 10.1016/j.jhazmat.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Amoatey P., Omidvarborna H., Baawain M.S., Al-Mamun A. Impact of building ventilation systems and habitual indoor incense burning on SARS-CoV-2 virus transmissions in middle eastern countries. Sci. Total Environ. 2020;733 doi: 10.1016/j.scitotenv.2020.139356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L.M., Zhang G.L., Lei Q.T., Li Y., Li X.L., Hwu Y.K., et al. Microstructure of atmospheric particles revealed by TXM and a new mode of influenza virus transmission. Nucl. Inst. Methods Phys. Res. B. 2015;359:167–172. doi: 10.1016/j.nimb.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram H., Kokturk N., Elbek O., Kilinc O., Sayiner A., Dagli E., et al. Interference in scientific research on COVID-19 in Turkey. Lancet. 2020;396:463–464. doi: 10.1016/S0140-6736(20)31691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontempi E. First data analysis about possible COVID-19 virus airborne diffusion due to air particulate matter (PM): the case of Lombardy (Italy) Environ. Res. 2020;186 doi: 10.1016/j.envres.2020.109639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimblecombe P., Lai Y. Effect of sub-urban scale lockdown on air pollution in Beijing. Urban Clim. 2020;34 doi: 10.1016/j.uclim.2020.100725. [DOI] [Google Scholar]
- Buonanno G., Stabile L., Morawska L. Estimation of airborne viral emission: quanta emission rate of SARS-CoV-2 for infection risk assessment. Environ. Int. 2020;141 doi: 10.1016/j.envint.2020.105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameletti M. The effect of Corona virus lockdown on air pollution: evidence from the city of Brescia in Lombardia region (Italy) Atmos. Environ. 2020;239 doi: 10.1016/j.atmosenv.2020.117794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can-Terzi B., Ficici M., Tecer L.H., Sofuoglu S.C. Fine and coarse particulate matter, trace element content, and associated health risks considering respiratory deposition for Ergene Basin, Thrace. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142026. [DOI] [PubMed] [Google Scholar]
- Carraturo F., Del Giudice C., Morelli M., Cerullo V., Libralato G., Galdiero E., et al. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, 2020. Centers for disease control and prevention. https://www.cdc.gov/coronavirus/2019-ncov/lab.html (Accessed June 06, 2020).
- Chen P.S., Tsai F.T., Lin C.K., Yang C.Y., Chan C.C., Young C.Y., et al. Ambient influenza and avian influenza virus during dust storm days and background days. Environ. Health Perspect. 2010;118:1211–1216. doi: 10.1289/ehp.0901782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirizzi D., Conte M., Feltracco M., Dinoi A., Gregoris E., Barbaro E., et al. SARS-CoV-2 concentrations and virus-laden aerosol size distributions in outdoor air in north and south of Italy. Environ. Int. 2021;146 doi: 10.1016/j.envint.2020.106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schünemann H.J., et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia M. Factors determining the diffusion of COVID-19 and suggested strategy to prevent future accelerated viral infectivity similar to COVID. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coşkun H., Yıldırım N., Gündüz S. The spread of COVID-19 virus through population density and wind in Turkey cities. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creager H.M., Zeng H., Pulit-Penaloza J.A., Maines T.R., Tumpey T.M., Belser J.A. In vitro exposure system for study of aerosolized influenza virus. Virol. 2017;500:62–70. doi: 10.1016/j.virol.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida, P.R., Demoliner, M., Antunes Eisen, A.K., Heldt, F.H., Hansen, A.W., Schallenberger, K., Fleck, J.D., Spilki, F.R., 2020. SARS-CoV2 quantification using RT-dPCR: a faster and safer alternative to assist viral genomic copies assessment using RT-QPCR. bioRxiv. doi: 10.1101/2020.05.01.072728. [DOI]
- Domingo J.L., Rovira J. Effects of air pollutants on the transmission and severity of respiratory viral infections. Environ. Res. 2020;187 doi: 10.1016/j.envres.2020.109650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Qi J.E.B., Yin Y., Gao D. Concentration, viability and size distribution of bacteria in atmospheric bioaerosols under different types of pollution. Environ. Pollut. 2020;257 doi: 10.1016/j.envpol.2019.113485. [DOI] [PubMed] [Google Scholar]
- Groulx N., Urch B., Duchaine C., Mubareka S., Scott J.A. The pollution particulate concentrator (PoPCon): a platform to investigate the effects of particulate air pollutants on viral infectivity. Sci. Total Environ. 2018;628-629:1101–1107. doi: 10.1016/j.scitotenv.2018.02.118. [DOI] [PubMed] [Google Scholar]
- Hadei M., Mohebbi S.R., Hopke P.K., Shahsavani A., Bazzazpour S., Alipour M., et al. Presence of SARS-CoV-2 in the air of public places and transportation. Atmos. Pollut. Res. 2021;12:302–306. doi: 10.1016/j.apr.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Byun J.H., Cho Y., Rim J.H. RT-PCR for SARS-CoV-2: quantitative versus qualitative. Lancet Infect. Dis. 2021;21:165. doi: 10.1016/S1473-3099(20)30424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Lei C., Chen Z., Liu W., Hu X., Pei R., et al. Distribution of airborne SARS-CoV-2 and possible aerosol transmission in Wuhan hospitals, China. Natl. Sci. Rev. 2020;7:1865–1867. doi: 10.1093/nsr/nwaa250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimzadeh, S., Bhopal, R., Nguyen Tien, H., 2020. Review of infective dose, routes of transmission, and outcome of COVID-19 caused by the SARS-CoV-2 virus: comparison with other respiratory viruses. Preprints. 2020070613.
- Kuzu S.L., Saral A. The effect of meteorological conditions on aerosol size distribution in Istanbul. Air Qual. Atmos. Health. 2017;10:1029–1038. doi: 10.1007/s11869-017-0491-y. [DOI] [Google Scholar]
- Lednicky J.A., Lauzardo M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int. J. Infect. Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linillos-Pradillo B., Rancan L., Ramiro E.D., Vara E., Artíñano B., Arias J. Determination of SARS-CoV-2 RNA in different particulate matter size fractions of outdoor air samples in Madrid during the lockdown. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhou J., Yao J., Zhang X., Li L., Xu X., et al. Impact of meteorological factors on the COVID-19 transmission: a multi-city study in China. Sci. Total Environ. 2020;726 doi: 10.1016/j.scitotenv.2020.138513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Majumdar N., Wessel T., Marks J. Digital PCR modeling for maximal sensitivity, dynamic range and measurement precision. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki M., Anvari E., Hopke P.K., Noorimotlagh Z., Mirzaee S.A. An updated systematic review on the association between atmospheric particulate matter pollution and prevalence of SARS-CoV-2. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool A., Khan N.Z. Analyzing barriers for implementation of public health and social measures to prevent the transmission of COVID-19 disease using DEMATEL method. Diabetes Metab. 2020;Syndr.14:887–892. doi: 10.1016/j.dsx.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MoEU, 2020. Turkish Ministry of Environment and Urbanization. https://www.csb.gov.tr/en (Accessed June 06, 2020).
- Nalla A.K., Casto A.M., Huang M.L.W., Perchetti G.A., Sampoleo R., Shrestha L., et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAA, 2021. National Oceanic and Atmospheric Administration. https://www.noaa.gov (Accessed March 31, 2021).
- Onat B., Alver Şahin Ü., Sivri N. The relationship between particle and culturable airborne bacteria concentrations in public transportation. Indoor Built Environ. 2016;26:1420–1428. doi: 10.1177/1420326X16643373. [DOI] [Google Scholar]
- Pavšič J., Žel J., Milavec M. Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Anal. Bioanal. Chem. 2015;408:107–121. doi: 10.1007/s00216-015-9107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RTMH, 2021. Republic of Turkey Ministry of Health, https://covid19.saglik.gov.tr/EN-69532/general-coronavirus-table.html (Accessed March 29, 2021).
- Ruiz-Villalba A., van Pelt-Verkuil E., Gunst Q.D., Ruijter J.M., van den Hoff M.J.B. Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR) Biomol. Detect. Quantif. 2017;14:7–18. doi: 10.1016/j.bdq.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şahin Ü.A. The effects of COVID-19 measures on air pollutant concentrations at urban and traffic sites in Istanbul. Aerosol Air Qual. Res. 2020;20:1874–1885. doi: 10.4209/aaqr.2020.05.0239. [DOI] [Google Scholar]
- Şahin Ü.A., Onat B., Akın Ö., Ayvaz C., Uzun B., Mangır N., et al. Temporal variations of atmospheric black carbon and its relation to other pollutants and meteorological factors at an urban traffic site in Istanbul. Atmos. Pollut. Res. 2020;11:1051–1062. doi: 10.1016/j.apr.2020.03.009. [DOI] [Google Scholar]
- Sedlmaier N., Hoppenheidt K., Krist H., Lehmann S., Lang H., Büttner M. Generation of avian influenza virus (AIV) contaminated fecal fine particulate matter (PM2.5): genome and infectivity detection and calculation of immission. Vet. Microbiol. 2009;139:156–164. doi: 10.1016/j.vetmic.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Setti L., Passarini F., De Gennaro G., Barbieri P., Perrone M.G., Borelli M., et al. SARS-Cov-2RNA found on particulate matter of Bergamo in northern Italy: first evidence. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha, A., Shrestha, U., Sharma, R., Bhattarai, S., Tran, H., Rupakheti, M., 2020. Lockdown caused by COVID-19 pandemic reduces air pollution in cities worldwide. EarthArXiv. 10.31223/osf.io/edt4j.
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes. Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecer L.H., Alagha O., Karaca F., Tuncel G., Eldes N. Particulate matter (PM2.5, PM10-2.5, and PM10) and children’s hospital admissions for asthma and respiratory diseases: a bidirectional case-crossover study. J. Toxicol. Environ. Health, A. 2008;71:512–520. doi: 10.1080/15287390801907459. [DOI] [PubMed] [Google Scholar]
- TMOH, 2021. Turkish Ministry of Health, https://covid19.saglik.gov.tr/ (Accessed March 29, 2021).
- Wang B., Li Y., Xie Z., Du S., Zeng X., Hou J., et al. Characteristics of microbial activity in atmospheric aerosols and its relationship to chemical composition of PM2.5 in Xi'an, China. J. Aerosol Sci. 2020;146 doi: 10.1016/j.jaerosci.2020.105572. [DOI] [Google Scholar]
- Watanabe T., Bartrand T.A., Weir M.H., Omura T., Haas C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010;30:1129–1138. doi: 10.1111/j.1539-6924.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Xu C., Xu X., Zhu C., Li J., Lv G. Size distribution of bioaerosols from biomass burning emissions: characteristics of bacterial and fungal communities in submicron (PM1.0) and fine (PM2.5) particles. Ecotoxicol. Environ. Saf. 2019;171:37–46. doi: 10.1016/j.ecoenv.2018.12.026. [DOI] [PubMed] [Google Scholar]
- WHO, 2020. Rolling updates on coronavirus disease (COVID-19). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen (Accessed July 31, 2020).
- WHO, 2021. Coronavirus (COVID-19) dashboard. https://covid19.who.int/ (Accessed March 29, 2021).
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yu W.L., Liu W.L., Chan K.S., Yang C.C., Tan C.K., Tsai C.L., et al. High-level ambient particulate matter before influenza attack with increased incidence of Aspergillus antigenemia in southern Taiwan, 2016. J. Microbiol. Immunol. Infect. 2018;51:141–147. doi: 10.1016/j.jmii.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Xue T., Jin X. Effects of meteorological conditions and air pollution on COVID-19 transmission: evidence from 219 Chinese cities. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RdRP and N1 gene positivity of filter samples.
All data collected from 13 stations in 10 cities of Turkey.
Summary of mean air pollutants concentration and meteorological parameters during the sampling periods in each city.