Abstract
Purpose
Infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus 2019 disease (COVID-19), poses a serious risk to humanity and represents a huge challenge for healthcare systems worldwide. Since the early days of the COVID-19 pandemic, it has been evident that adequate testing is an essential step in limiting and controlling the spread of SARS-CoV-2. Here, we present an accurate, inexpensive, scalable, portable, and rapid detection kit to directly detect SARS-CoV-2 in biological samples that could even be translated for population testing. We have demonstrated that our method can reliably identify viral load and could be used to reach those fractions of the population with limited access to more sophisticated and expensive tests.
Procedures
The proposed SARS-CoV-2 detection kit is based on the combination of a SARS-CoV-2-targeted antibody (CR3022) that targets spike protein S1 domain on the viral surface. This antibody was radiolabeled with a long-lived isotope (Iodine-125) to allow us to detect bound antibody in samples with SARS-CoV-2. We used a series of in vitro assays to determine sensitivity and specificity and facilitate automation of the testing kit. Bound antibody was extracted from saliva samples via a centrifugation step and a semi-permeable membrane. Our kit was further validated using SARS-CoV-2 virions.
Results
We were able to accomplish radiosynthesis of [125I]I-CR3022 reliably without loss of binding. The SARS-CoV-2-sensing antibody was shown to maintain its spike S1 affinity and to bind to as low as 2.5–5 ng of spike protein. We then used beads-bound spike S1 to develop a separation kit which proved to be both easy to use and inexpensive. The kit made it possible to extract bound antibody from the saliva-like sample. We were able to validate the separation kit using intact SARS-CoV-2 virions and showed that our kit can detect a viral concentration as low as 19,700 PFU/mL (~ 9.22%TBF) and as high as 1,970,000 PFU/mL (45.04%TBF).
Conclusion
Here we report the development and validation of a SARS-CoV-2 detection system based on the combination of a specific radiolabeled antibody and a separation membrane. We demonstrate our system to be comparable to other SARS-CoV-2 detection kits already approved by the FDA and believe this technology could be easily deployed to countries with limited resources for the diagnosis of COVID-19. Furthermore, workflows could be easily adapted to target other antigens and therefore other types of diseases.
Keywords: SARS-CoV-2, COVID-19, Virus, Radiolabeled antibody, Detection kit
Graphical abstract

1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic [1] brought increased attention to a widespread problem threatening humanity since its existence: highly contagious and lethal viral and other pathogenic infections. Methods of conducting and disseminating scientific research quickly shifted, and the scientific community put together an incredible effort to adapt and repurpose their skills and knowledge to address the challenges that persist to today. It became evident that in order to face this healthcare crisis, quick and efficient interventions were needed to follow and slow the viral spread; in other words, it become important to follow the “three Ts”, i.e., Test-Track-Treat [2,3]. So far, the widespread use of mRNA vaccines has been a great scientific success, with more and more people getting vaccinated every day [4]. Nevertheless, the persistent threat of new vaccine-resistant variants or new viruses [5] could resurface the needs that arose at the beginning of the pandemic.
SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as a receptor to enter host cells [6] through the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein [7]. Nasopharyngeal and oropharyngeal swabs can be accurate gold standards for the diagnosis of SARS-CoV-2 using real-time reverse transcription polymerase chain reaction (RT-PCR). Unfortunately, this method of collection is slightly invasive, can cause discomfort, and requires close contact between healthcare workers and patients, which can pose a risk of transmission of the virus that necessitates the use of personal protective equipment (PPE). PPE includes barriers (gowns, gloves, eye shields) and respiratory protection (masks, respirators) to protect mucous membranes, airways, skin, and clothing from contact with infectious agents [8]. Moreover, rRT-PCR requires sterile collection tubes, time (typically in the order of one to three hours), and specialized laboratories with expensive reagents and adequate personnel [9]. For these reasons, saliva, also in the form of droplets and aerosols, was proposed as a valid alternative to nasopharyngeal swabs [10,11]. Saliva has been proven to provide highly concordant results for viral detection [12], though with a much lower sensitivity than RT-PCR technology using nasal swabs [13]. Because the saliva samples can be collected and submitted by patients themselves, PPE requirements are less stringent [14]. Looking at the kinetics of SARS-CoV-2 presence in saliva samples from patients, it has been shown that viral presence peaks in the first week after symptom onset [15]. Of the diagnostic tools for SARS-CoV-2 infection that don't rely on RT-PCR, most are based on the detection of viral effects on the human immune system, i.e. they detect the presence of IgA, IgM, and IgG antibodies that are produced against the virus [16]; other examples of techniques include Raman imaging [17] and machine learning [18].
Based on our early work using an oncology-inspired strategy to radiolabel viruses [8], and our previous experience with radioiodinated agents for nuclear medicine applications [[19], [20], [21], [22]], here we explored a novel, fast, and inexpensive method for the direct detection of SARS-CoV-2 virions in saliva samples. Unlike the other methods already in use [23], our technology directly targets the S1 domain of the spike proteins on the surface of viruses using a radioactive detection output. Preliminary results suggest that this technology provides fast, simple and reliable technology that can be used in a low resource setting.
2. Materials & methods
2.1. General
Chemicals were procured from commercial suppliers and used without further purification. 0.9% Phosphate buffered saline (PBS), Iodogen® and dichloromethane were obtained from Thermo Fisher Scientific (Waltham, MA). Anti-SARS-CoV-2 antibody CR3022 was purchased from Creative Biolabs (Shirley, NY). Recombinant SARS-CoV-2 spike protein - S1 subunit (host cell receptor binding domain - RBD) with N-terminal histidine tag was purchased from Raybiotech (Peachtree Corners, GA, catalog # 230-01102-100). 1-μm diameter magnetic beads functionalized with Ni-NTA (Nickel-Nitrilotriacetic acid; HisPur™ Ni-NTA magnetic beads; Catalog # 88831) used for bead assay were purchased from Thermo Fisher Scientific. Iodogen® (1,3,4,6-tetrachloro-3α,6α-diphenyl-glycoluril, catalog # PI28600) coated glass reaction tubes were prepared by evaporating 50 μL of Iodogen® solution (50 μg, 1 mg/mL) in a borosilicate glass test tube (12 × 75 mm, catalog # 14-961-26). PD MiniTrap G-25 columns (GE Healthcare, catalog # 28918007) were preconditioned with 2 mL of PBS (Catalog # 10-010-023) before using for separating radioiodinated antibody from the free radioiodine.
2.2. Radiosynthesis
Radiosynthesis was performed as previously described [8]. Briefly, 70 μL of PBS was added to an Iodogen (100 μg) precoated culture tube. To the resulting solution, 25 μg of CR3022 mAb (25 μL, 1.0 mg/mL) was added followed by the addition of 9.25 MBq (250 μCi) of Na[125]I (in 17 μL of 0.1 N NaOH). The mixture was allowed to react for 4 min at room temperature. For purification, the crude product was loaded onto a PD MiniTrap G-25 column (GE Healthcare, catalog # 28918007) that had been preconditioned with 2 mL of PBS. The radiolabeled antibody was purified using saline as eluant and fractions were collected and used for the binding studies. The purity of the radiolabeled antibody was measured with SG-ITLC paper using 10% trifluoroacetic acid in water as eluent. The specific activity was about 296–370 MBq/mg (8–10 mCi/mg).
2.3. Spike-ACE2 binding kit assay
To test antibody specificity to spike S1, a commercially available COVID-19 Spike-ACE2 binding in vitro kit was used (RayBiotech, Peachtree Corners, GA; Code: CoV-SACE2-1). Manufacturer instructions were followed for the reagents and sample preparation. 100 μL of each sample were added to each well in triplicate and incubated overnight at 4 °C with shaking. The solution was discarded the following day and washed 4 times in 1× wash solution. 100 μL of 1× horseradish peroxidase (HRP) conjugated IgG was added to each well for 1 h at room temperature with shaking. Samples were washed three times with 1× wash solution. 100 μL of TMB one-step substrate reagent was added to each well, incubated for 30 min at room temperature with shaking in the dark. 50 μL of stop solution were added to each well and the plate was immediately read at 450 nm in a plate reader. As a final step, the content of each well was lysed using 100 μL of 1 M sodium hydroxide and collected into a disposable plastic culture tube (12 × 75 mm). Wells were washed three times with PBS and each wash was added to the corresponding a disposable plastic culture tube (12 × 75 mm). Tubes were then counted on a gamma counter to detect radioactivity.
2.4. Magnetic beads assay
The assay was performed as previously described [24], but modified to meet the hypothesis of this study by changing the concentration of SARS-CoV-2 spike protein S1 subunit to test lower detection limits. Briefly, samples were prepared by aliquoting 20 μL of the magnetic bead slurry into a 1.5 mL lo-bind microcentrifuge tube (13-698-794; Fisher Scientific). The beads were washed by adding 380 μL of PBS-BSA (PBS containing 1% bovine serum albumin) and the tubes were vortexed for 5 s followed by a brief spin in a mini-centrifuge prior to placing the tubes on a magnetic rack (12321D; DynaMag™-2; ThermoFisher Scientific) for 30–45 s to isolate the magnetic beads. The SARS-CoV-2 –S1 antigen was resuspended to achieve a gradient of concentrations of 2.5, 5.0, 50, 1000 ng/mL. The washed beads were resuspended in 390 μL of PBS-BSA and the beads in all tubes except the control arm were incubated with 1 μg (10 μL) of His-tagged or biotinylated antigen for 15 min on an Eppendorf™ Thermomixer at 300 RPM at room temperature. Subsequently, the beads were washed once with 400 μL of % BSA-PBS before adding 3700 Bq (0.1 μCi) of the radiolabeled antibody ([125I]I-CR3022) resuspended in 1% BSA-PBS. [125I]I-CR3022 was incubated with antigen-coated beads for 30 min on a rotating mixer at room temperature. Thereafter, the beads were isolated using a magnet, and the supernatant containing unbound radioligand was aspirated with a pipette and collected in separate tubes. To remove non-specifically-bound radioligand, the beads were washed twice with 400 μL of PBS-BSA. Finally, the beads, supernatant and washes were measured for radioactivity on a gamma counter. The relative binding fractions were determined by dividing the percentage of total activity bound to magnetic beads to the total activity (beads + supernatants + wash).
2.5. Separation kit
For separation, a Vivaspin 500 with 300,000 MWCO PES membrane (Sartorius #VS0152) was used to separate target-bound antibody from unbound antibody by using a tabletop centrifuge (Eppendorff) at 1000 ×g for 30 min. Tubes were primed using 5% BSA-PBS (1000 ×g for 5–10 min) to avoid non-specific binding. The separation kit was used as described in the protocol section with either the magnetic beads or in vitro virions.
2.6. In vitro detection of SARS-CoV-2
All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee-approved BSL3 facilities at Johns Hopkins University School of Medicine using appropriate positive pressure air respirators and protective equipment. SARS-CoV-2/USA-WA1/2020 was obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID) and propagated in Vero E6 TMPRSS cells (ATCC). The virus stocks were stored at −80 °C and titers were determined by tissue culture infectious dose 50 (TCID50) assay. On the day of the experiment, an aliquot of SARS-CoV-2 (1.97 × 106 PFU/mL) was diluted 10× in PBS. Each viral dilution (1 mL) was incubated with 7400 Bq (0.2 μCi) of [125I]I-CR3022 for 30 min at room temperature. Sterile PBS was used as a negative control. Subsequently, the mixture was transferred to a separation unit with a 300 kDa pore size semi-permeable membrane (Vivaspin 500 as described above). The separation unit was centrifuged in a tabletop centrifuge (30 min at 1000 ×g) and the filter was collected for detection the associated radiation using an automated gamma counter (Perkin Elmer).
3. Results
3.1. Specificity
The anti-SARS-CoV-2 spike S1 antibody CR3022 was radiolabeled with a gamma-emitting iodine isotope (either Iodine-131, or iodine-125) at 296–370 kBq/μg (8–10 μCi/μg) specific activity [8]. In order to determine the specificity of the radiolabeled antibody for the spike protein S1 domain, we used a commercially available COVID-19 spike-ACE2 binding kit (RayBiotech, Peachtree Corners, GA; Code: CoV-SACE2-1) with ACE2 protein fixed on the bottom of the 96-well plate (Fig. 1A). the amount of ACE2 and Spike S1 protein was maintained constant in each well, whereas a decreasing gradient of [125I]I-CR3022 antibody was added to inhibit the spike-ACE2 interaction (Fig. 1B). We measured absorbance at 450 nm wavelength and observed an increase determined by the [125I]I-CR3022 decreasing gradient, confirming specificity for the spike S1 of the radiolabeled antibody with an IC50 of 0.24 μCi ≡ 2.4 μg (R2 = 0.88), Fig. 1C. After measuring absorbance, we collected the content of each well and measured the radioactivity (Fig. 1D), confirming the presence of decreasing amounts of [125I]I-CR3022.
Fig. 1.
(A) A binding kit was used to determine specificity of the radiolabeled antibody in a 96-well plate format. (B) The experiment was performed using decreasing amounts of antibody and constant amounts of ACE2 proteins on the bottom of the plate and spike S1 proteins. (C) Decreasing amounts of radiolabeled antibody result in an increasing absorbance signal.
3.2. Sensitivity
In order to evaluate our ability to detect different amounts of spike S1 using the [125I]I-CR3022 antibody, we performed a sensitivity test by modifying a previously published magnetic bead assay [24]. We used HIS-tagged spike S1 proteins bound to magnetic beads and added the radiolabeled anti-spike antibody (Fig. 2A). We used an increasing gradient of spike S1 proteins (0, 2.5, 5, 50, 1000 ng), and a constant amount of [125I]I-CR3022 (3700 Bq (0.1 μCi)/sample, ~0.01 μg), Fig. 2B. The radiolabeled antibody was added to the beads-spike complex and then pulled-down using a magnet. The supernatant was removed. After three washes, tubes were scanned through a gamma counter to calculate the percentage target binding fraction (%TBF) as follows: %TBF = 100 ∗ [CPMbeads] / [CPMbeads + CPMsupernatant+washes], where CPMbeads is the gamma counts per minute of the beads-bound activity, and CPMsupernatant+washes is the gamma counts of the supernatant and the relative washes. Counts were normalized by subtracting the CPM of beads plus no Spike S1 protein (i.e., the non-specific antibody-beads interaction). A Vmax of 2.83 was calculated by fitting the data (R2 = 0.33), with a curve plateau starting at ~5 ng of spike protein, and a normalized %TBF of 1.73 at 2.5 ng (Fig. 2C).
Fig. 2.
(A) A beads assay was used to determine sensitivity of the radiolabeled antibody. (B) The experiment was performed using a gradient of spike proteins. (C) The experiment resulted in a detected sensitivity as low as 2.5 ng of spike protein.
3.3. Automation
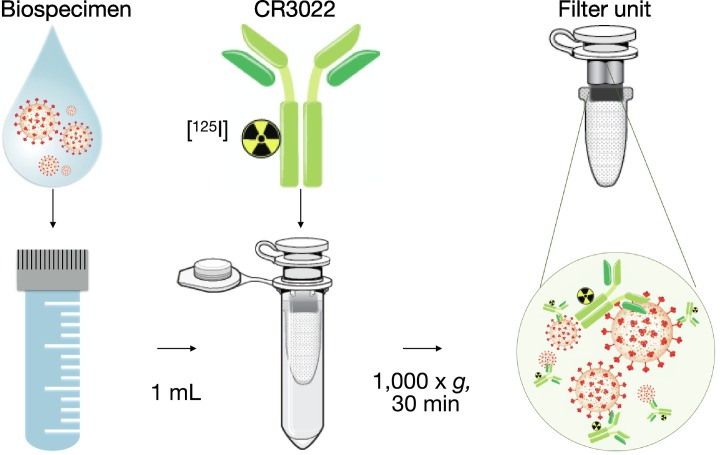
To prepare the radiolabeled antibody [125I]I-CR3022 for more realistic scenarios, we developed a novel detection kit based on the following protocol. We hypothesized that we can isolate SARS-CoV-2-bound antibody based on a size exclusion step and detect it using a gamma counter. The method consists of collecting the human biospecimens in the form of a small volume saliva (~1 mL is sufficient). This sample, which might contain SARS-CoV-2, is diluted in a saline solution (e.g., 1%BSA/PBS) and 500 μL are added to the separation unit, a tabletop centrifuge tube with a separation membrane with a pore size of 300 kDa. The tube must be primed using a 5% BSA/PBS solution prior to use (Fig. 3 ). The radiolabeled antibody [125I]I-CR3022 is then added to the human sample directly in the separation unit. Each separation unit tube is then centrifuged in a tabletop centrifuge (30 min @ 1000 ×g). Measuring the filter and the flow-through in a gamma counter allows us to detect the amount of [125I]I-CR3022 in each fraction and determine the %TBF (Fig. 4 ). The same kit was tested by spiking a saliva sample from a healthy donor to mimic human sample collection and to determine if priming tubes reduce non-specific binding in human biospecimens. We demonstrated reduced non-specific binding (difference in %TBF) in saliva samples between pretreated and untreated tubes, when compared to beads samples.
Fig. 3.
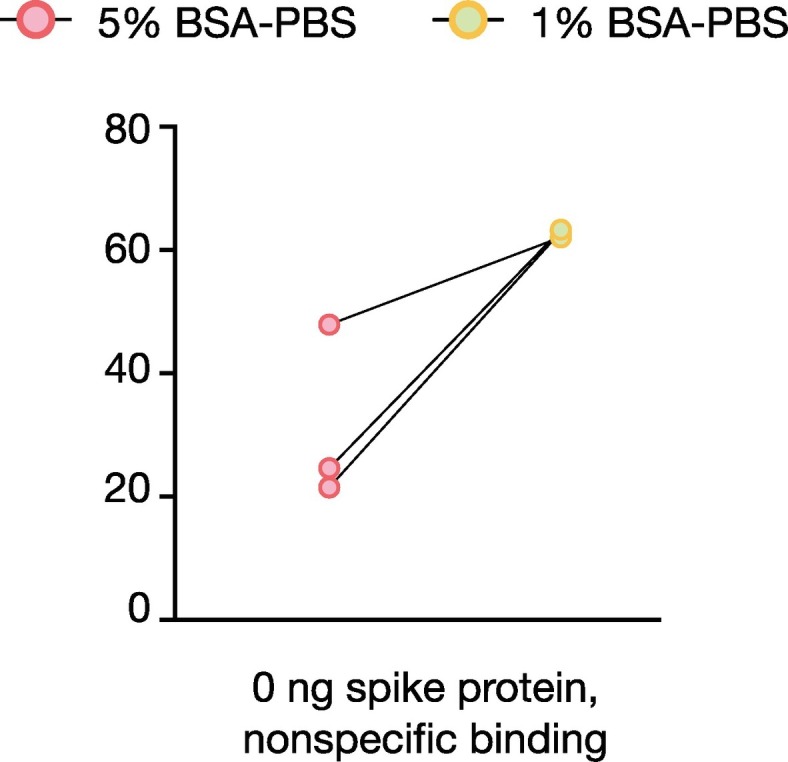
Incubation with 5% BSA in PBS leads to lower non-specific binding as compared to 1% BSA in PBS.
Fig. 4.
Step-by-step graphical explanation of the developed SARS-CoV-2 detection kit.
The SARS-CoV-2 detection kit was tested using spike S1-bound magnetic beads to emulate a SARS-CoV-2 structure (despite the difference in median diameter between SARS-CoV-2 (~0.2–0.05 μm) and the magnetic beads (~ 1 μm). 500 μL of spike-carrying beads in 1% BSA/PBS were added to primed separation unit and the above protocol was followed to trap them into the filter. The %TBF was measured on a gamma counter, as described in the protocol. BSY (spike-carrying beads with [125I]I-CR3022 antibody) sample showed a ~ 100% TBF into the filter unit, as compared to the flow-through (unpaired t-test, ****p-value <0.0001). BY and SY samples (beads with [125I]I-CR3022, and spike S1 with [125I]I-CR3022, respectively) did not show any significant difference between the filter-trapped and the flow-through %TBF. Free Iodine-125 with and without spike S1 (S-FreeI, and FreeI, respectively) presented a significantly higher radioactive fraction flow-through compared to the filter unit (unpaired t-test, ****p-value <0.0001), Fig. 5A.
Fig. 5.
(A) The beads-spike complex was run through the separation kit and the target binding fraction (TBF) was measured. B = beads; S = spikes; Y = radiolabeled antibody, FreeI = unlabeled iodine. (B) The separation kit was tested using increasing concentrations of SARS-CoV-2 and the TBF was measured.
Validation.
In order to validate the results shown in Fig. 5A, we tested whether our separation kit could detect the presence of virulent SARS-CoV-2 in liquid samples. In vitro SARS-CoV-2 virions were diluted at different plaque-forming unit (PFU/mL) concentrations (0.001, 0.0197, 0.197, 1.9700, 19.7000, 197.0000, 1970.0000, 19,700.0000, 197,000.0000, 1,970,000.0000 PFU/mL) in media. Our kit could trap into the filter unit and detect SARS-CoV-2 virions at a concentration as low as 19,700 (~9.22%TBF) and as high as 1,970,000 (45.04%TBF), Fig. 5B, confirming the efficacy of our kit.
4. Discussion
In the current manuscript we present our attempts to validate a simple radioactivity-based assay to measure viral particle load that can be used in low-resource, non-sterile settings and contribute to the development of rapid tests for COVID-19 or the next emerging infection. Based on our experience with radiolabeled antibodies for oncology applications, we hypothesized that the use of a similar strategy could be used to rapidly and reliably detect viral load in patients samples. Previously, as a rapid response to the pandemic, our group published a proof of concept on this use of radiolabeled antibodies [8].
COVID-19, the infectious disease that derives from SARS-CoV-2, has a median incubation period of ~5 days (2 to 14 days), with symptom onset within ~12 days of infection (8 to 16 days) [25]. The fast spread of the virus mainly derives from transmission from pre-symptomatic individuals [26], as even asymptomatic patients contribute substantially to disease transmission [27]. For these reasons, testing large fractions of the population is still a key step in understanding and controlling the spread of the infection. To date, COVID-19 tests can be grouped as nucleic acid, serological, antigen, and ancillary tests, all of which play distinct roles in hospital, point-of-care, or large-scale population testing [28]. Most antigen tests require a nasopharyngeal swab in order to probe for the nucleocapsid (N) or spike (S) proteins of SARS-CoV-2 virus via lateral flow or ELISA, and they typically have the advantage of being fairly fast (~ less than an hour to complete). Ancillary tests comprise a broad category of personal devices (apps and wearable sensors) and hospital laboratory tests.
The ideal test has been described as accurate, economical, scalable, portable, and fast [29]. In the present work we make an effort to show that our approach actually meets most of the above requirements. In addition, we want to emphasize that our method demonstrates reliable results with clean dispensing equipment and collection vials in a clean environment, without the need for sterile equipment, vials or workspace. Accuracy has been proven using a set of laboratory assays to illustrate the specificity of the radiolabeled antibody [125I]I-CR3022 for the spike protein S1 domain target. Furthermore, using a beads assay, we showed detection at spike S1 levels as low as 2.5–5 ng. We integrated our radiolabeled antibody into what we believe to be an inexpensive and easy-to-use detection kit based on size separation of the SARS-CoV-2-bound antibody vs unbound antibody or other agents that could be present in the saliva sample. The radiosynthesis reaction is scalable and the entire kit consists of Eppendorf-sized tubes that can be run in parallel to reach a high throughput where the only limitation would be the size of the tabletop centrifuge. The average viral load of nasal swabs positive for SARS-CoV-2 is around 1.4 × 106 copies/mL [8]. The maximum load seems to be 7.11 × 108 copies/mL [30]. In our assay, 19,700 PFU/mL corresponds to 2.04 × 108 copies/mL, which seems to be the limit of detection. Under stringent laboratory conditions, qRT-PCR for COVID-19 has a limit of detection (LoD) of 500–1000 copies/mL [31]. The currently approved qRT-PCR kits have LoD in range of 1000–6000 copies/mL [32]. The Quidel Sofia2 SARS Antigen FIA kit, an EUA antigen detection assay, has an LoD of approximately 6 million copies/mL in a sample collection [32]. While our method has less sensitivity than approved commercial technologies, we can increase the LoD of our assay by increasing the sample volume (because we are concentrating the sample using centrifugation, volume is not a concern), reducing the non-specific binding using custom manufactured centrifugation filters and further optimizing the buffers. Improvement on these parameters could significantly improve the LoD of our method to match the sensitivity of commercially available antigen detecting kits such as the Quidel Sofia2 SARS Antigen FIA kit.
We demonstrated proof of concept with the long-lived isotope Iodine-125, which has been traditionally used for biological assays and renders the antibody suitable for long storage (125I T1/2 = 59.5 days). The gamma energy emission of Iodine-125 is low-energy (<35 keV), and therefore simple to shield with only a few centimeters of lead. These physical characteristics make Iodine-125 the ideal isotope for shipment and transportation of both the radiolabeled antibody and the filtered biospecimen. The hands-on time for performing the experiment is extremely short (on the order of only a few minutes) and requires only pipetting the saliva sample into the tube, followed by capping the tube. The longest step is the centrifugation step, which requires 30 min. Our kit does not require sophisticated laboratory equipment or intensive training of the laboratory personnel. The small amount of radiation added to each tube (<3700 Bq (0.1 μCi)) makes it safe to handle, requiring just simple protective gloves. The small amount of saliva sample needed is another feature. The ability of the detection kit to work in a real-world scenario was limited only by our access to saliva samples from patients with Covid-19. For this reason, we collaborated with Johns Hopkins University to test our kit in a biosafety level 3 laboratory. The kit was prepped at Memorial Sloan Kettering and shipped the same day to Baltimore with a simple step-by-step guide on how to use it. We were able to obtain the results the same day. Finally, an antibody-based kit such as the one presented here can be easily tweaked to target a different antigen or biomarker and opens the possibility of being used for applications in e.g., liquid biopsies.
5. Conclusion
We demonstrate that we can produce an accurate, secure, and easy-to-use kit for detecting viral loads in small liquid samples containing SARS-CoV-2. This kit could be deployed in difficult-to-reach locales and could significantly improve the way we test for SARS-CoV-2 infection. Because our method is simple to develop and scale, and because target-specific antibodies are increasingly available, we could use a similar — if not nearly identical — strategy for detecting viral or other pathogenic infections in humans using liquid biospecimens.
Declaration of competing interest
T.R. is shareholder and cofounder of Summit Biomedical Imaging, LLC. T.R. is a shareholder and cofounder of Quaero Pharmaceuticals, LLC. T.R. is a coinventor on filed U.S. patent (WO2016164771). T.R., G.P. and N.P. filed patent applications related to the subject matter of this article. T.R. is a paid consultant for Theragnostics, Inc.
Acknowledgements
This work was supported by National Institutes of Health grant P30 CA008748 and a sponsored research agreement from International Isotopes Inc. (Idaho Falls, ID, SK00000014)). The funding sources were not involved in study design, data collection and analysis, writing of the report or the decision to submit this article for publication.
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willem L., Abrams S., Libin P.J.K., Coletti P., Kuylen E., Petrof O., et al. The impact of contact tracing and household bubbles on deconfinement strategies for COVID-19. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munzert S., Selb P., Gohdes A., Stoetzer L.F., Lowe W. Tracking and promoting the usage of a COVID-19 contact tracing app. Nat Hum Behav. 2021;5:247–255. doi: 10.1038/s41562-020-01044-x. [DOI] [PubMed] [Google Scholar]
- 4.Rolla G., Brussino L., Badiu I. Maintaining safety with SARS-CoV-2 vaccines. N Engl J Med. 2021;384 doi: 10.1056/NEJMc2100766. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat Rev Immunol. 2021;21(4):245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillarsetty N., Carter L.M., Lewis J.S., Reiner T. Oncology-inspired treatment options for COVID-19. J Nucl Med. 2020;61:1720–1723. doi: 10.2967/jnumed.120.249748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test To diagnose COVID-19. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azzi L., Carcano G., Gianfagna F., Grossi P., Dalla Gasperina D., Genoni A., et al. Saliva is a reliable tool to detect SARS-CoV-2. J Inf Secur. 2020;81:E45–E50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu R.S., Cui B.M., Duan X.B., Zhang P., Zhou X.D., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020;12 doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To K.K.W., Yip C.C.Y., Lai C.Y.W., Wong C.K.H., Ho D.T.Y., Pang P.K.P., et al. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect. 2019;25:372–378. doi: 10.1016/j.cmi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection (review) Cochrane Database Syst Rev. 2020;8(8) doi: 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor P., Chowdhry A., Kharbanda O.P., Bablani Popli D., Gautam K., Saini V. Exploring salivary diagnostics in COVID-19: a scoping review and research suggestions. BDJ Open. 2021;7 doi: 10.1038/s41405-021-00064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To K.K.W., OTY Tsang, Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., et al. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses-Basel. 2020:12. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlomagno C., Bertazioli D., Gualerzi A., Picciolini S., Banfi P.I., Lax A., et al. COVID-19 salivary Raman fingerprint: innovative approach for the detection of current and past SARS-CoV-2 infections. Sci Rep. 2021;11 doi: 10.1038/s41598-021-84565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts M., Driggs D., Thorpe M., Gilbey J., Yeung M., Ursprung S., et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Machine Intell. 2021;3:199–217. [Google Scholar]
- 19.Wilson T.C., Jannetti S.A., Guru N., Pillarsetty N., Reiner T., Pirovano G. Improved radiosynthesis of (123)I-MAPi, an auger theranostic agent. Int J Radiat Biol. 2020:1–7. doi: 10.1080/09553002.2020.1781283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirovano G., Jannetti S.A., Carter L.M., Sadique A., Kossatz S., Guru N., et al. Targeted brain tumor radiotherapy using an auger emitter. Clin Cancer Res. 2020;26:2871–2881. doi: 10.1158/1078-0432.CCR-19-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jannetti S.A., Carlucci G., Carney B., Kossatz S., Shenker L., Carter L.M., et al. PARP-1-targeted radiotherapy in mouse models of glioblastoma. J Nucl Med. 2018;59:1225–1233. doi: 10.2967/jnumed.117.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangangari K.K., Varadi A., Majumdar S., Larson S.M., Pasternak G.W., Pillarsetty N.K. Imaging Sigma-1 receptor (S1R) expression using Iodine-124-labeled 1-(4-Iodophenyl)-3-(2-adamantyl)guanidine ([(124)I]IPAG) Mol Imaging Biol. 2020;22:358–366. doi: 10.1007/s11307-019-01369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 24.Sharma S.K., Lyashchenko S.K., Park H.A., Pillarsetty N., Roux Y., Wu J., et al. A rapid bead-based radioligand binding assay for the determination of target-binding fraction and quality control of radiopharmaceuticals. Nucl Med Biol. 2019;71:32–38. doi: 10.1016/j.nucmedbio.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 27.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 29.Wu Q., Suo C., Brown T., Wang T., Teichmann S.A., Bassett A.R. INSIGHT: A population-scale COVID-19 testing strategy combining point-of-care diagnosis with centralized high-throughput sequencing. Sci Adv. 2021:7. doi: 10.1126/sciadv.abe5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Dai C., Wang H., Gao Y., Li T., Fang Y., et al. Analysis and validation of a highly sensitive one-step nested quantitative real-time polymerase chain reaction assay for specific detection of severe acute respiratory syndrome coronavirus 2. Virol J. 2020;17 doi: 10.1186/s12985-020-01467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaout R., Lee R.A., Lee G.R., Callahan C., Yen C.F., Smith K.P., et al. SARS-CoV2 testing: the limit of detection matters. bioRxiv. 2020 doi: 10.1101/2020.06.02.131144. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]