Abstract
Pharmacogenetics (PGx) studies the influence of genetic variation on drug response. Clinically actionable associations inform guidelines created by the Clinical Pharmacogenetics Implementation Consortium (CPIC), but the broad impact of genetic variation on entire populations is not well-understood. We analyzed PGx allele and phenotype frequencies for 487,409 participants in the U.K. Biobank, the largest PGx study to date. For fourteen CPIC pharmacogenes known to influence human drug response, we find that 99.5% of individuals may have an atypical response to at least one drug; on average they may have an atypical response to 10.3 drugs. Nearly 24% of participants have been prescribed a drug for which they are predicted to have an atypical response. Non-European populations carry a greater frequency of variants that are predicted to be functionally deleterious; many of these are not captured by current PGx allele definitions. Strategies for detecting and interpreting rare variation will be critical for enabling broad application of pharmacogenetics.
Keywords: Pharmacogenetics, Population Genetics, UK Biobank
Introduction
Drug-based interventions play a primary role in medical treatment; more than 72% of visits to clinics and hospitals in the United States result in drug therapy1. An individual’s genetics can have a profound impact on how they respond to many drugs, with the vast majority of individuals carrying at least one pharmacogenetic variant2–4. Therefore, the field of pharmacogenetics (PGx), is vital to improving modern medicine and prescribing practices5.
The practical value of PGx testing has increased as the field has discovered and characterized high impact haplotypes. These haplotypes are catalogued and named by PharmVar (www.pharmvar.org) using a nomenclature system typically based on “star alleles”6–8. Generally, the relationship between drug response and pharmacogenes is investigated through targeted studies on small groups of human subjects. The findings of these studies are aggregated through curation efforts such as PharmGKB (www.pharmgkb.org)9. The Clinical Pharmacogenetics Implementation Consortium (CPIC; cpicpgx.org) and other organizations assign a clinical function to star alleles based on published experimental research and create peer-reviewed and evidence-based clinical practice guidelines10,11.
PGx testing is not yet capable of robustly handling rare genetic variation. Rare variants can be high impact, but are unlikely to be identified by a genotyping array or included in an established haplotype definition12. Most PGx testing in the US currently uses genotyping arrays. As a result, test results may be based on partial allele definitions or use proxy variants to assign PGx haplotypes, which may not represent the actual haplotype (as would be revealed by full and error-free sequencing) in the subject3,13. Developing more robust methods for assigning function to PGx haplotypes is an active area of research14. The extent to which existing haplotypes definitions capture all important genetic variation within pharmacogenes is not well characterized3,13,15.
We used genotype data from nearly 500,000 participants and exome data from 50,000 participants in UK Biobank to analyze pharmacogenetic variation in fourteen clinically important genes at a population scale. To this end, we developed PGxPOP, a PGx matching engine that is based on PharmCAT16 and uses its PGx allele definitions to characterize PGx allele and phenotype frequencies. PGxPOP extends the capabilities of PharmCAT by generating diplotypes from population scale datasets16. This study represents the largest study of pharmacogenetic allele and phenotype frequencies to date and investigates both the power and limitations of current star allele definitions. Our findings demonstrate the value of characterizing allele frequencies in large populations, the importance of using sequencing platforms that are capable of capturing rare genetic variation, and highlights the need for more PGx research on under-studied populations.
Methods
Data
Participant data from UK Biobank were used in this study. UK Biobank is a prospective study of ~500,000 individuals in the United Kingdom for whom lifestyle, clinical, and genetic data was collected17. We used the genotype data imputed from the Axiom Biobank Array (version 2), and the exome sequencing data from the SPB pipeline (2/12/2020 rerelease)17,18. We removed individuals that were outliers for heterozygosity and missingness rates in the genetic data, as reported by UK Biobank. We excluded any loci with a Hardy-Weinberg equilibrium p-value <1×10−15 or were missing in >10% of individuals using VCFtools19. The imputed data was aligned to hg19 and the exome data was aligned to GRCh38. All data was phased using Eagle v2.4.120.
We created an “integrated call set” by combining coding regions from exome sequencing data and non-coding regions from imputed data. Any region within the exome capture array was taken from the exome data, any region outside, including 20kb upstream and downstream, was taken from the imputed data, discarding the coding region of the imputed data21. We used liftOver to map the imputed data to GRCh3822. The newly merged variants were phased with Eagle v2.4.120.
We assigned genetic ancestry for individuals using principal component analysis (PCA) of their genetic data. We first collapsed individual’s self-reported ethnicity into African, European, East Asian, and South Asian, according to a standardized biogeographical system23. Then, we calculated the mean and standard deviation of the first three principal components from a PCA of the genotype array data for each biogeographical ethnic group, computed by UK Biobank24. Any sample whose principal components did not fall within three standard deviations of the mean of the genetic ancestry for their self-reported ethnicity was referred to as “Other”.
PGxPOP
We developed PGxPOP, a Python program compatible with population scale studies, that rapidly calls PGx star alleles on phased multisample VCFs using matrix operations. PGxPOP uses the PharmCAT allele definition files (https://github.com/PharmGKB/PharmCAT)9,10,16. PGxPOP reports exact matches to the allele definitions based on the provided phased genotype data. If the defining genetic variation for one star allele is a proper subset of those for another star allele, the matching star allele with the greatest number of variants is reported. PGxPOP also reports partial matches or novel combinations of existing pharmacogenetic alleles (i.e. two distinct haplotypes on the same phased chromatid), which would be reported as “not called” by PharmCAT. In cases where there is a complete match to multiple haplotype definitions on the same strand, combinations of non-overlapping haplotypes are reported with “+” notation and overlapping haplotypes are reported with “or” notation. For example, if for CYP2D6 both *2 and *9 alleles were found on the same strand, PGxPOP would report this as a *2+*9 call, since the alleles for these two definitions are mutually exclusive. If instead, variants matching the *35 and *41 alleles, which share two positions, were found on the same strand PGxPOP would report “*35+hg38:chr22.g.42127803C>T or *41+hg38:chr22.g.42130761C>T”, in order to represent all possible combinations of the alleles found at those positions. The identification of combination and overlapping haplotypes is a feature specific to PGxPOP and is not found in PharmCAT. Haplotypes are then mapped to predicted phenotypes based on published guidelines from PharmGKB and CPIC. PGxPOP was validated using the same test cases as PharmCAT, which include 137 genomic samples from Coriell cell lines with genotypes characterized by the GeT-RM Program25.
Cross-platform analysis
We analyzed the ability to call pharmacogenetic haplotypes and phenotypes for fourteen genes (CFTR, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A5, CYP4F2, DPYD, IFNL3, NUDT15, SLCO1B1, TPMT, UGT1A1 and VKORC1) across imputed, exome, and integrated call sets. We limited this analysis to 49,702 samples shared across all three platforms. We calculated diplotype and phenotype concordance between each platform and the integrated call set for each genetic ancestry population and across all populations.
Haplotype and phenotype calling
We generated population specific haplotype, diplotype, and phenotype frequencies for fourteen genes among the ethnic populations reported by UK Biobank. We mapped diplotypes to phenotypes using CPIC guidelines, except VKORC1 and CYP4F2 diplotypes. VKORC1 and CYP4F2 do exist within the CPIC guideline for warfarin, but there is not an explicit phenotype defined by CPIC for these genes, (e.g. Normal Metabolizer). For these two genes we determined the phenotype as related to warfarin dosing. For phenotype prediction, haplotypes were assigned CPIC-associated function or activity values in cases of exact star allele matches. Phenotype was then determined based on the combination of the two alleles in the diplotype or activity score. We assigned a phenotype of “not available” for alleles identified to contain a combination of variants captured within existing star allele definitions. These alleles do not perfectly match any star allele and cannot be mapped to a phenotype.
We use star allele definitions when available for these genes with the following modifications. (1) We made assumptions about the ultimate phenotype of combination alleles and alleles carrying additional variants in order to assess the distribution of likely response phenotypes. In these cases, we assume that if one of these alleles is non-functional, then the new combination of variants will not recover the function26. For example, if we identified a CYP2D6 haplotype combination that includes CYP2D6*4 and CYP2D6*74 on the same strand (CYP2D6*4+*74), the assigned function would be “no function” even though function of CYP2D6*74 is unknown. This logic is extended to alleles with decreased function for SLCO1B1 and UGT1A1.
Additionally, any cystic fibrosis patient carrying a CFTR ivacaftor responsive allele is said to be ivacaftor responsive. (2) We modified the SLCO1B1 allele definitions to exclude synonymous variants. (3) For all INDELs we performed a search for identical INDELs in the sequencing data that may have been aligned differently. This was done by screening 50bp upstream and downstream of each INDEL in the definitions.
Structural variants (SVs) were not called for CYP2D6 or any other gene. Thus, we are not able to call star alleles with whole gene deletions (CYP2D6*5), duplications (e.g. CYP2D6*1×2), CYP2D7–2D6 hybrids (CYP2D6*13) or CYP2D6–2D7 hybrids including CYP2D6*36. This limits the assignment of CYP2D6 function and phenotypes since we are not able to determine CYP2D6 increased function alleles and therefore ultrarapid metabolizers and potentially miss no function alleles.
We calculated the burden of non-typical response phenotypes for each individual by counting the number of diplotypes with predicted non-typical response phenotypes across fourteen genes with phenotypes. Gene phenotypes were determined to have a non-typical response if any CPIC guidance recommended an alternate dosage or drug for that phenotype (Table S1). We determined the CPIC dosage recommendations for each subject for 45 drugs with CPIC guidelines related to genes in this study. For each drug, we determined the percent of the population that has been prescribed the drug by analyzing the general practice prescription data provided by UK Biobank for more than 222,000 subjects. We determined the frequency of prescriptions for each CPIC guidance group. We mapped brand name products to generic drugs using the PharmGKB curated drug list (https://www.pharmgkb.org/downloads)9. We used the intersection of participants with genetic data in the integrated call set and primary care data to determine guidance prescription frequencies (n=28,101).
Deleterious variant analysis
To estimate the burden of deleterious variants in pharmacogenes we identified variants predicted to be deleterious in the exome data. We deemed variants with a high IMPACT rating (e.g. a frameshift INDEL), as deleterious27 and variants predicted to be deleterious based on the ADME-optimized framework for pharmacogenes28. IMPACT classes were determined using VEP29, other annotations were generated using Annovar30. We identified predicted deleterious variants that were not contained within existing star allele definitions and calculated the aggregate deleterious variant allele frequency of all unaccounted-for deleterious variants.
Results
Platform concordance
We evaluated concordance between the imputed, exome, and an integrated call sets derived from the UK Biobank for both diplotype and phenotype calling. We generated diplotype and phenotype calls for all three call sets using PGxPOP (Fig. 1). We used diplotype and phenotype calls for twelve pharmacogenes for concordance analysis, with calls from the integrated set as the basis for comparison (Table 1). Genes with allele definitions consisting of a single non-coding variant were excluded, IFNL3 and VKORC1, as exome data would miss these genes allele definitions entirely. For five genes where the majority of the variants of interest are in exons, we find very high (>96%) correlation between the integrated call set and both the imputed and exome call sets when calling both diplotypes and phenotypes (CFTR, CYP2C9, TPMT, CYP4F2, and DPYD). We observe a variety of concordance patterns for the other seven genes. CYP2C19, which has a common non-coding variant upstream, the exome data is highly discordant with the integrated call set. Several genes have a mix of coding and non-coding variants, have low concordance with the integrated call set for both platforms (UGT1A1, CYP2D6, SLCO1B1). For three genes, the exome data performs well, and the imputed data has lower concordance (CYP2B6, CYP3A5, and NUDT15). The imputed data for NUDT15 has extremely low concordance with the integrated data; a variant that is rare in the population (rs746071566) was imputed for nearly all samples. We provide alluvial diagrams showing the change in haplotypes and phenotypes between the imputed and integrated call sets (Figure S1).
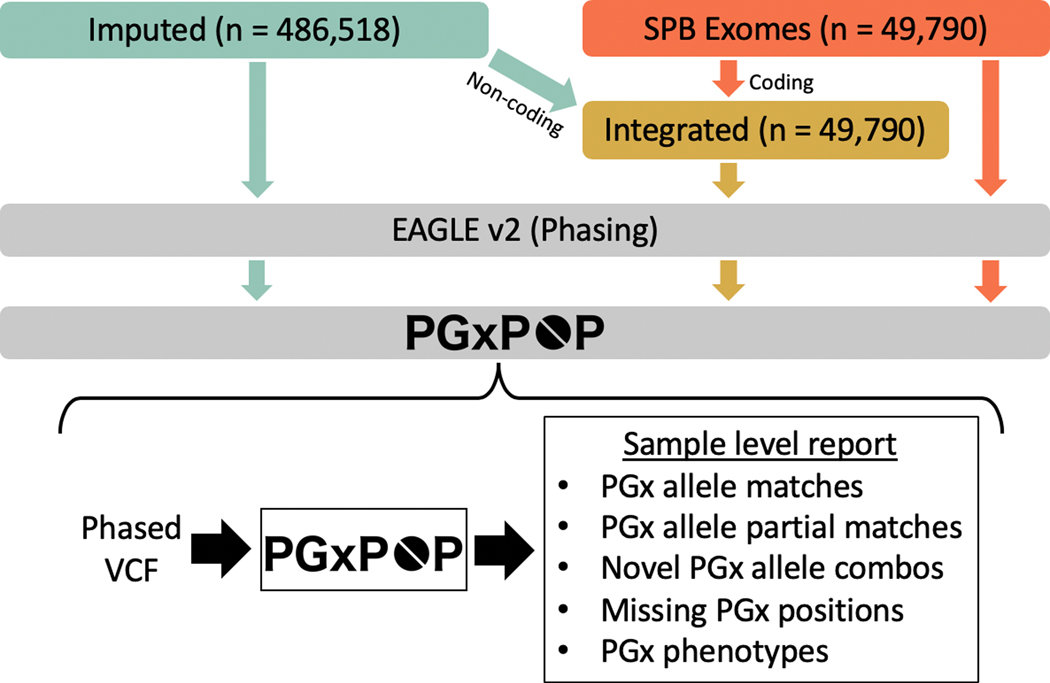
Figure 1.
Analysis workflow. Our analysis comprises three data types, data imputed from genotype arrays, exome sequencing data, and an integrated call set that combines both. We phase all datasets using statistical phasing with Eagle2. We then generate pharmacogenetic alleles for all samples using PGxPOP and generate a report of the matching star allele, the variants contributing to that call, and the resulting phenotype.
Table 1.
Platform concordance with integrated data is variable. We calculated the diplotype and phenotype concordance between the integrated call set and both contributing call sets, exome and imputed. For each gene we show the percent concordance (the percent of diplotypes or phenotypes that exactly match). Haplotypes for IFNL3 and VKORC1 contain only single variants that are in the non-coding regions, so the concordance is not listed for the exome data. SLCO1B1 star alleles are determined excluding synonymous variants.
| Diplotype concordance w/ Integrated | Phenotype concordance w/ Integrated | |||
|---|---|---|---|---|
| Gene | Imputed | Exome | Imputed | Exome |
| NUDT15 | 0.01% | 99.63% | 0.04% | 99.67% |
| UGT1A1 | 9.32% | 29.26% | 77.14% | 48.92% |
| CYP2D6 | 34.23% | 84.50% | 64.86% | 86.64% |
| CYP2B6 | 43.23% | 99.83% | 95.16% | 99.89% |
| SLCO1B1 | 68.41% | 89.73% | 76.64% | 92.64% |
| CYP3A5 | 85.48% | 100.00% | 85.69% | 100.00% |
| CFTR | 96.43% | 99.95% | 96.47% | 99.95% |
| TPMT | 97.76% | 99.93% | 99.17% | 99.93% |
| CYP2C19 | 97.85% | 61.77% | 99.44% | 68.64% |
| CYP2C9 | 98.23% | 99.85% | 98.36% | 99.86% |
| CYP4F2 | 99.44% | 99.91% | 99.49% | 99.91% |
| DPYD | 99.60% | 95.67% | 99.61% | 95.68% |
| IFNL3 | 1.00 | - | 1.00 | - |
| VKORC1 | 1.00 | - | 1.00 | - |
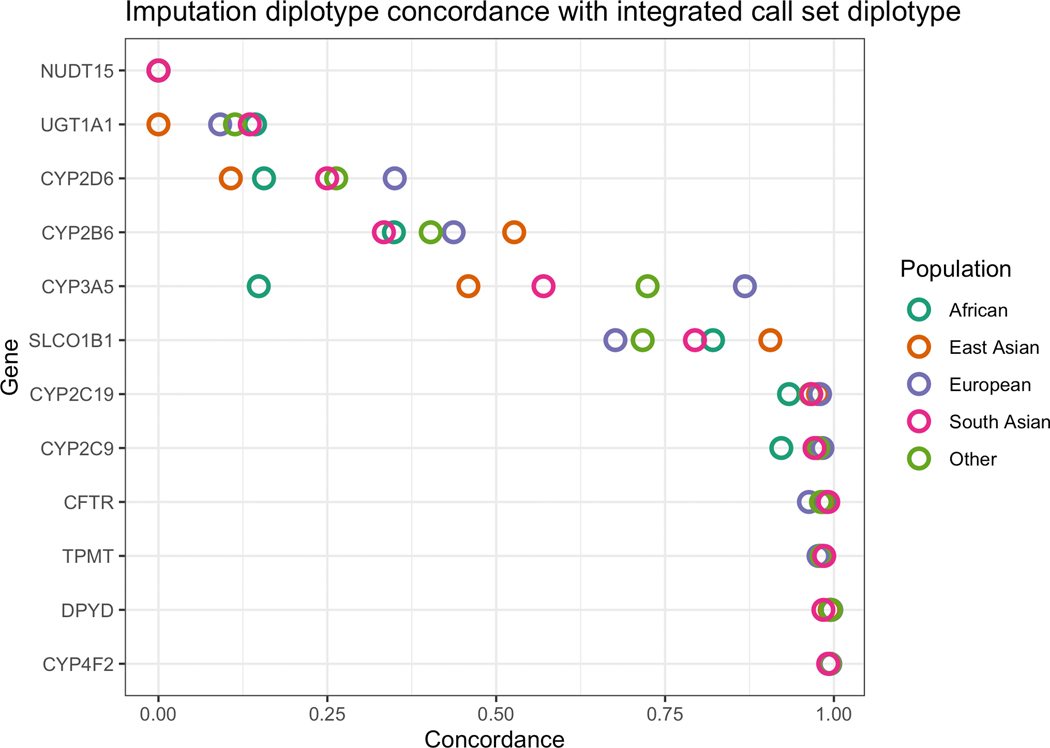
We evaluated population-specific accuracy of imputation by calculating population-aware diplotype concordance between imputed data and integrated data, for the 49,790 individuals who had both exome and imputed data. The method for population assignment is outlined in the Methods (Table S2, Figure S2). In some genetic populations, we found a substantial decrease in imputation accuracy for several genes (Fig. 2). This gap is most extreme in CYP3A5, where subjects with European genetic ancestry have a diplotype concordance of 86.8%, and subjects with African genetic ancestry have a diplotype concordance of 14.7%. In total, four genes have a decrease of 10% diplotype concordance or more.
Figure 2.
Concordance between diplotypes called from imputed data and integrated call sets reveal inefficiencies in data imputed from genotypes. The concordance is the proportion of diplotypes that exactly matched between the two call sets. We calculated population-specific concordance between the imputed data and integrated call sets. This comparison highlights the differences in the coding regions only, as the non-coding regions in the integrated call set are derived from the imputed data. Difference colors represent different global populations.
Haplotype and phenotype calling
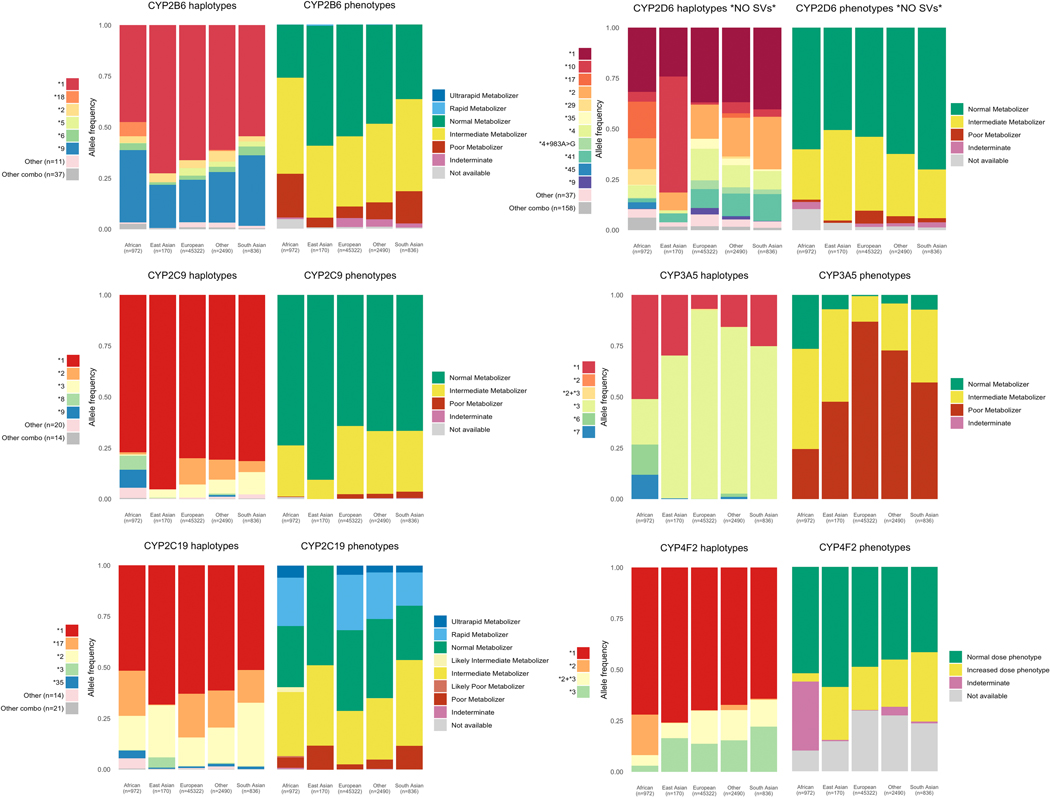
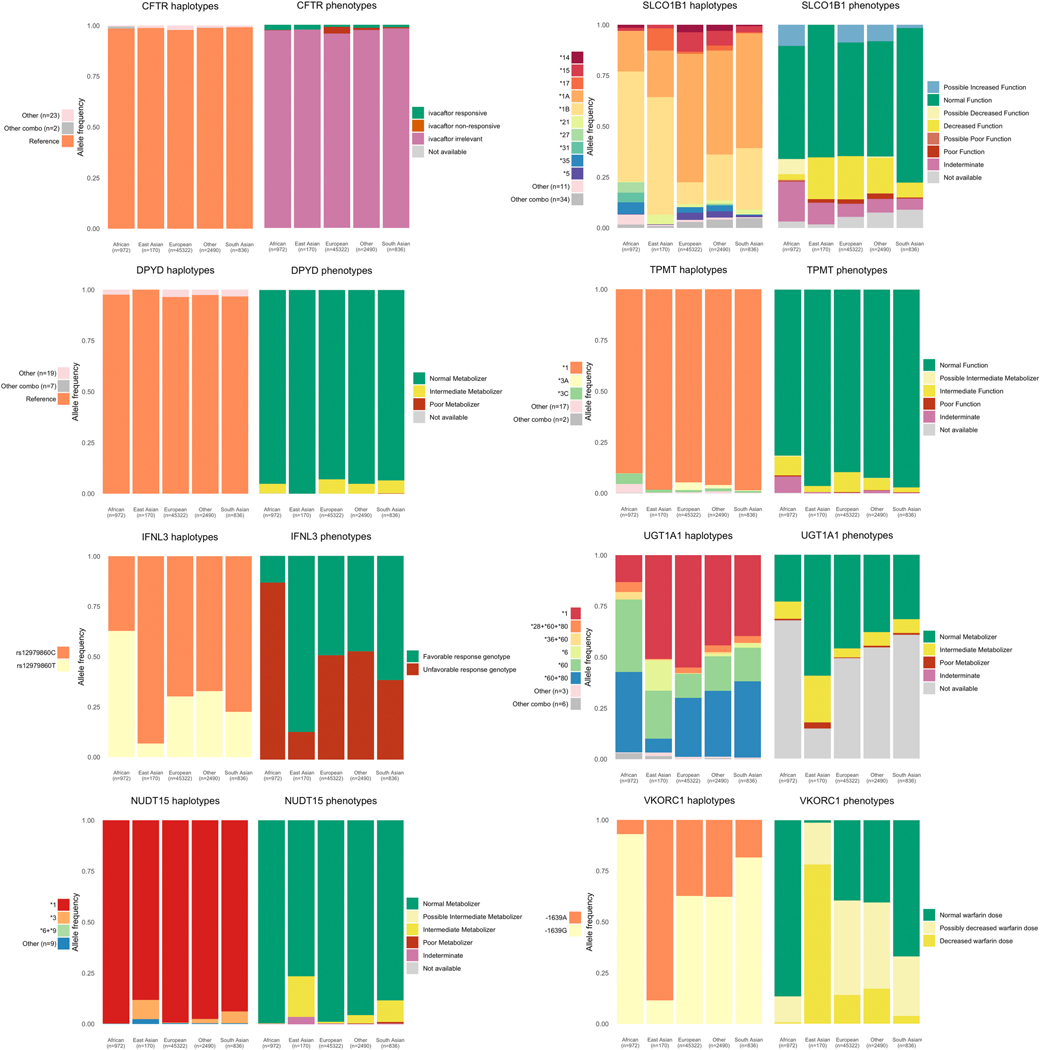
We analyzed haplotype and phenotype allele frequencies in fourteen clinically important pharmacogenes among participants belonging to four global populations in UK Biobank. This included 486,518 participants with imputed data from genotyping arrays, 49,790 with exome sequencing, and 49,790 participants with an integrated call set. Haplotype and phenotype frequencies from the exome and integrated call sets for six cytochrome P450 genes included in our analysis are shown in Figure 3, and eight non-cytochrome genes in Figure 4. We provide a full list of all haplotype, diplotype, and phenotype frequencies for each call set in File S1.
Figure 3.
Star allele and phenotype frequencies for cytochrome P450 genes. Frequencies shown here are generated from the integrated call set which comprises nearly 50,000 subjects. The star allele frequency plots show all star alleles occurring with a frequency of 3% or greater. Any haplotypes with under 3% allele frequency in all populations are grouped into “Other”. Combination alleles, alleles that contain either partial or full matches of more than one star allele on the same strand occurring with less than 3% allele frequency are grouped in “Other combos”. The number of alleles in “Other” and “Other combos” is shown in the legend for each gene. Note that allele and phenotype frequencies for CYP2D6 do not include structural variants.
Figure 4.
Star allele and phenotype frequencies for non-cytochrome P450 genes. Frequencies shown here are generated from the integrated call set which comprises nearly 50,000 subjects. The star allele frequency plots show all star alleles occurring with a frequency of 3% or greater. Any haplotypes with under 3% allele frequency in all populations are grouped into “Other”. Combination alleles, alleles that contain either partial or full matches of more than one star allele on the same strand occurring with less than 3% allele frequency are grouped in “Other combos”. The number of alleles in “Other” and “Other combos” is shown in the legend for each gene. SLCO1B1 star alleles are determined excluding synonymous variants.
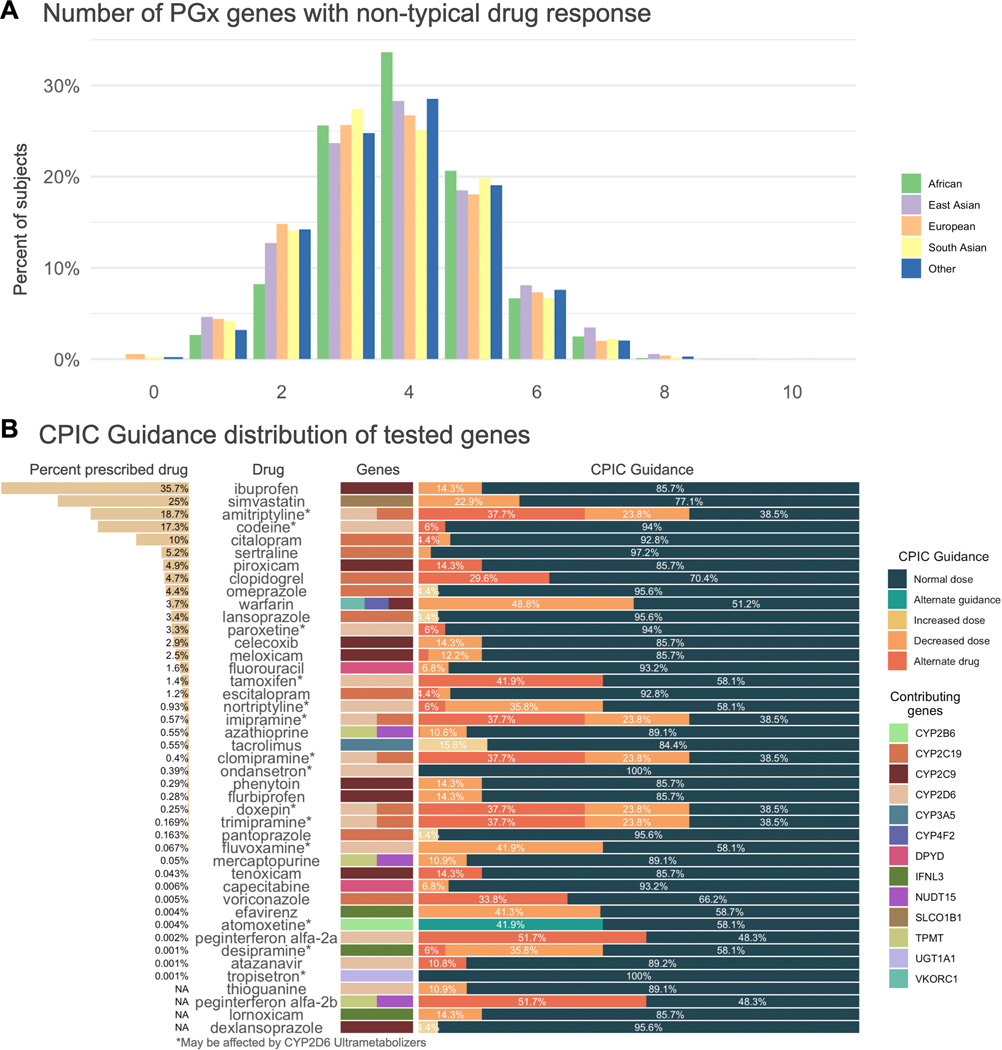
We find that participants carry on average 3.7 non-typical response diplotypes for the fourteen pharmacogenes analyzed in the integrated call set, with 99.5% of participants carrying at least one non-typical drug response diplotype (Fig. 5a). Participants, on average, carry pharmacogene alleles that lead to atypical dosage guidance by CPIC for 10.3 drugs. For several frequently used drugs, we find a high number of people receive atypical dosage guidance (Fig 5b). For example, simvastatin has been prescribed to 25% of the population, and 22.9 percent of all subjects carry either rs4149056 or SLCO1B1 star alleles assigned possible decreased function (*6, *9, *23, *31), which indicates that a lower dose might be recommended due to increased risk of muscle toxicity31. Within the available prescription records, we see that 23.3% of participants who have been prescribed simvastatin are rs4149056 carriers and may need a reduced dose. Overall, 24% of participants have been prescribed a drug for which they may have an atypical response according to CPIC guidelines.
Figure 5.
Frequency of pharmacogenes with a predicted non-typical response across the study population derived from the integrated call set and CPIC guideline recommendations for 45 drugs. a) The distribution of non-typical response alleles across each of the populations included in this study. Frequency of non-typical response pharmacogene alleles per subject range from 0 to 10, with a mean of 3.7. b) CPIC dosage guidance for 45 drugs that include recommendations based on any of the fourteen genes included in this study. We show the percent of the population that has ever been prescribed the drug, the drug name, the genes from this study that contribute to the recommendation, and the distribution of CPIC recommendations.
Star alleles with unknown or uncertain function, leading to an indeterminate phenotype, were found in nine genes. These are diplotypes where both haplotypes exactly match an existing star allele definition, but at least one of those haplotypes has unknown function. We find that 5.0% of subjects carry unknown or uncertain function star alleles in SLCO1B1, 4.2% in CYP2B6, and 1.7% in CYP2D6.
We find that for some genes, many novel combinations of alleles and allelic variants from existing allele definitions occur on a single haplotype in the integrated call set. These allele combinations can be a complete star allele or haplotype definition along with any number of additional variants from another previously defined allele. For example, 29.0% of the study population carries haplotypes that contain both the CYP4F2*2 and CYP4F2*3 variants on a single strand. Large numbers of novel allele combinations are also found in CYP2D6 (159 unique combinations in 6.1% of subjects), SLCO1B1 (34 in 2.9%), and CYP2B6 (37 in 0.9%). At least one such allele combination was identified in twelve genes, the median number of allele combinations was eight, 288 were identified in total. DPYD and CFTR variation are represented by individual variants rather than star alleles, but combinations of variants were identified on a single strand for both genes. Full details of the assumptions we used to make function assignments in cases of combinations are described in the “Haplotype and phenotype calling” section of the Methods. Those assumptions allow us to assign haplotype functions to 102 of the 288 observed variant combinations. The remaining 186 allele combinations cannot reliably be mapped to a function and are designated as ‘not available’ phenotype.
Genes with the most ‘not available’ phenotypes are UGT1A1 (49.9% of subjects) CYP4F2 (30.2%), SLCO1B1 (12.2%), CYP2B6 (5.1%), and CYP2D6 (3.4%). These counts exclude combination alleles for which we estimated function based on the rules defined in the Methods.
We modified the SLCO1B1 star allele definitions to exclude the three synonymous coding variants (chr12.g.21176827G>A, chr12.g.21178665T>C, and chr12.g.21178691C>T). These three variants appear in many combinations with the other core star allele variants and the star alleles that include these variants *18, *19, *20, *21 are assigned uncertain function. Including these three synonymous variants, 315 unique haplotypes were identified. The number of haplotypes decreased to 55 when those variants were removed. We find that when synonymous variants are included in the allele definition 77.9% of SLCO1B1 haplotypes do not perfectly match one of the defined alleles and contain some combination of star allele variants and one or more variants from other definitions. This value drops to 2.9% when synonymous variants are excluded from the SLCO1B1 definitions.
Deleterious variant analysis
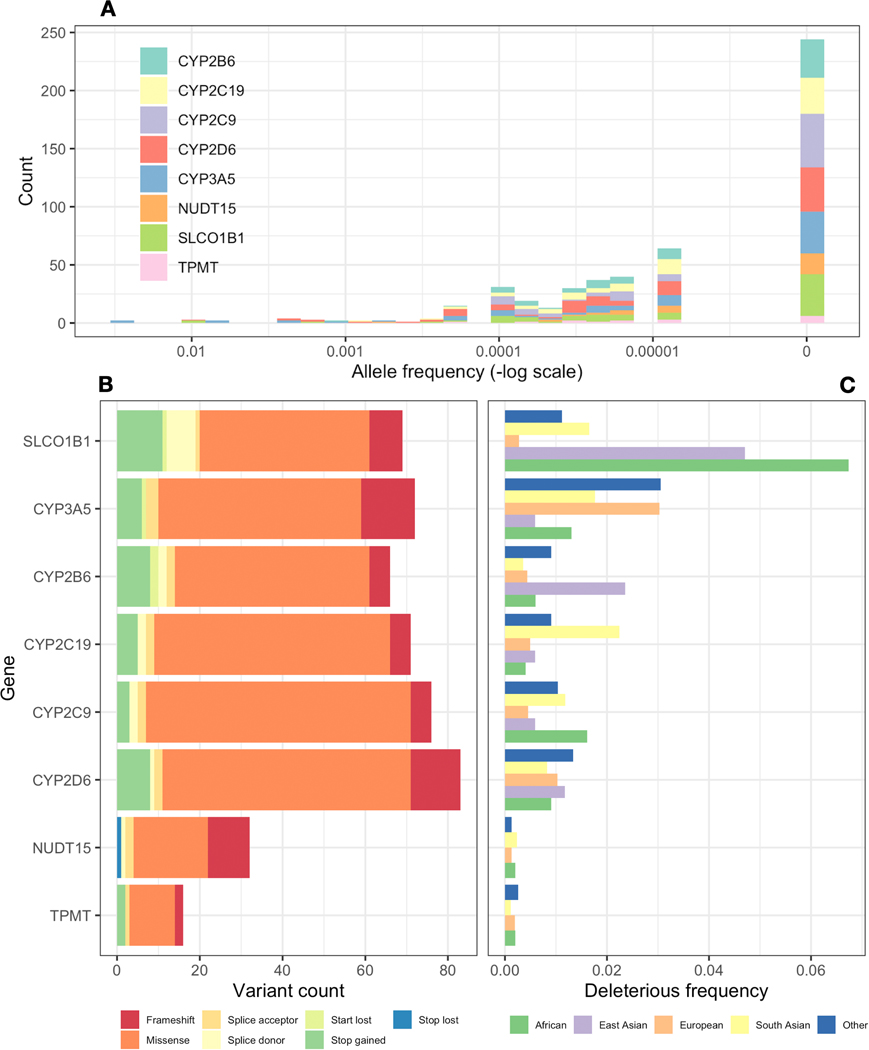
We estimated the burden of deleterious variants that are not currently included in allele definitions for eight of the fourteen genes in our study, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A5, NUDT15, SLCO1B1, and TPMT. We predicted the deleteriousness of each variant found in the exome data and filtered out variants that were included in any existing allele definition, resulting in 478 deleterious variants across all eight genes (Fig. 6). Of the 478 deleterious variants identified, 244 have not been previously observed in gnomAD (Fig. 6c). All identified deleterious variants are rare (minor allele frequency < 1%). However, we find that 6.1% of all subjects carry at least one unaccounted for deleterious variant in one of these eight genes studied. To identify which populations are most underserved by current definitions we calculated the total frequency of all out-of-definitions deleterious variants in a population-specific manner (Fig. 6b). We find that across most genes, non-European populations carry the highest level of out-of-definition deleterious variants. For example, out-of-definition deleterious variants in CYP2B6 have an allele frequency of 0.023 in the East Asian population.
Figure 6.
Analysis of deleterious variants not contained within existing star allele definitions. We identified presumptive deleterious variants in the exome sequencing data for eight genes by identifying probable loss of function variants as well as predicted deleterious missense variants. (a) shows the allele frequency of each probable deleterious variant in gnoMAD. Variants with an allele frequency of 0 were not identified in gnoMAD. (b) shows the number of deleterious variants identified as well as the frequency of each type of variant. (c) shows the total frequency of any deleterious variant in each population in the exome data. Concretely, the frequency represents the sum of allele frequencies for all deleterious variants not found within existing star allele definitions for each population.
Drug Usage Statistics
We determined how many UK Biobank participants have been prescribed drugs for which they are predicted to have a non-typical response using prescription records in UK Biobank. We find that 23.7% of participants have been prescribed a drug for which they may have a non-typical response according to CPIC guidelines; 9.8% of participants have been prescribed a drug where the guidelines suggest an alternate drug (Table S3).
Discussion
We present a pharmacogenetic analysis of 487,409 participants in UK Biobank. Quantifying haplotype and phenotype frequencies at this scale enables a better understanding of the coverage and accuracy of different genetic platforms, limitations of current pharmacogenetic allele definitions, and the potential impact of broader PGx testing.
This analysis includes nearly 50,000 subjects with both genotype array and exome data, providing an opportunity to assess the accuracy of each platform at a large scale. We find that for most genes there is high concordance between imputed genotype data and sequencing data, for both PGx haplotype and phenotype calls. However for some genes we find extremely low concordance. We observe low concordance in highly polymorphic genes such as CYP2D6 where data imputed from genotyping arrays likely does not fully capture the true variation. We show the creation of an integrated call set leads to greater ability to identify haplotypes in genes that have functionally important non-coding variants (e.g. CYP2C19). There are known limitations of exome data for calling PGx alleles32, which are addressed by adding non-coding variation from genotyping arrays.
We find that several very important pharmacogenes are highly discordant between the imputed, exome, and integrated call sets, and some genes have differences in imputation accuracy between populations--highlighting the importance of considering both gene and population of interest when choosing a platform for pharmacogenetic analysis. For genes with splicing and other non-coding variants, exome data may not be sufficient (e.g. CYP2C19). While for highly polymorphic genes, imputed data may not be sufficient (e.g. CYP2D6). This highlights the potential clinical importance of having data from genome sequencing or a targeted capture array that includes coding and non-coding regions, such as PGRNseq33. The addition of genome sequencing data would allow for analysis of SVs, which were not captured by this study. For CYP2D6 analysis, copy number variants (CNVs) and other SVs are common and must be considered to make an accurate assessment of phenotype. Lack of SV analysis is a major limitation of this study’s ability to determine population level phenotype predictions of CYP2D6. However, we believe establishing star allele frequencies for star alleles identified from the variant data is useful.
Across all genes with haplotypes described by a star allele nomenclature, we find that there are haplotypes which are combinations of star allele variants that are currently not found together in any existing star allele definition. We also found combinations of individual variants in DPYD and CFTR on the same chromosome. Using array data can lead to the detection of only one of these alleles or variants, or the assumption that they are on different chromosomes. Either case can lead to mistakes in diplotype and phenotype assignment, potentially resulting in an incorrect prescribing recommendation. We provide the frequency of these star allele and variant combinations in the supplemental material.
Many deleterious variants do not contribute to current allele definitions because they have not yet been submitted to PharmVar6,7, a resource devoted to cataloguing pharmacogene allele variation. None of the variants have a minor allele frequency greater than 1%, but when observations are aggregated among the eight genes analyzed as part of the deleterious variant analysis, 6.1% of the population carries at least one uncatalogued deleterious variant. Deleterious variants within pharmacogenes are likely to have a strong effect on an individual’s PGx phenotype, indicating that 6.1% of the study population could benefit from a PGx guideline, if one were to exist for their variant15. We observed that non-European populations in this cohort carry uncatalogued rare variation at disproportionately high rates. This indicates a need to broaden the diversity of pharmacogenetic research to ensure equitable impact of PGx research.
To date, SLCO1B1 has not been included in PharmVar. Instead, SLCO1B1 alleles *1a-*36 have been defined in 5 publications31,34–37. We find that the 37 star alleles for SLCO1B1 are not commonly found as the only allelic variation for that gene. Only 22.1% SLCO1B1 alleles in the study population exactly match the star allele definitions from these publications. Three synonymous coding variants (chr12.g.21176827G>A, chr12.g.21178665T>C, and chr12.g.21178691C>T) were the most commonly found with other star allele variants and removing them from the star allele definitions increased the allele matches to 97.1%. Studies of the SLCO1B1 haplotypes in other populations would help inform decisions with regards to the inclusion of these three variants in the current star allele definitions.
Our observation of individuals carrying combinations of PGx haplotypes and the observed rate of deleterious variants indicates that PGx allele definitions would benefit from additional population-scale studies. Novel variation could be incorporated into existing or new PGx allele definitions, increasing their coverage. However, our analysis demonstrates the limitations of the current definition based system; to increase the robustness of allele definitions it is important that the community works to identify causal variants, to enable a reduction in the reliance on linkage disequilibrium structure— this is of particular importance for admixed populations, which are a source of vast haplotype diversity. Recent work on the development of data-driven PGx phenotyping methods indicates that given enough data, it might be possible to move away from variant level rule-based systems and towards data-driven machine-learning models capable of robustly handling unobserved genetic variation14,38,39. The challenges posed by rare variation is likely to be a consistent issue for the current PGx system and will likely grow over time as genotyping gives way to genome sequencing and more populations are studied in detail revealing rarer mutations.
One major limitation is that we do not consider the effects of SVs. SVs are relatively frequent and known to be functionally important in CYP2D6. There are tools available for calling CNVs from exomes, but they have not been validated for SVs in CYP2D6 which can include hybridizations with CYP2D7. Without a validated method copy number analysis may not lead to useful results. Other studies have called CNVs in CYP2D6 from genotyping arrays, but the observed frequencies of CNVs from array data are significantly different from those observed in genome sequencing data, calling the accuracy of these methods into question.
Overall, our findings demonstrate the potential impact of pharmacogenetics, with almost all subjects carrying at least one PGx allele that alters drug guidance, reaffirming previous research2,3. We additionally show that on average people carry 3.7 pharmacogenetic variants that may lead to a non-typical drug response, affecting response to 10.3 drugs. Our analysis of individual samples with data from multiple genotyping modalities demonstrates the need for consideration of the variants relevant to the pharmacogene of interest and the ancestral background of the patient when selecting a genotyping modality. Our investigation of the different ancestral groups within UK Biobank shows disproportionate rates of uncatalogued deleterious variants, highlighting the need for large scale PGx studies of diverse populations. We believe the observed rate of uncatalogued pharmacogenetic variation demonstrates a need for further development of the current haplotype definition based approach to PGx.
Supplementary Material
Study Highlights.
What is the current knowledge on the topic?
Most individuals carry actionable pharmacogenetic variants that lead to a change in drug response. Biobanks are collecting data on large numbers of participants enabling population studies.
What question did this study address?
We sought to determine pharmacogenetic allele frequencies in fourteen genes among 500,000 participants in the UK Biobank.
What does this study add to our knowledge?
We find that 99.5% of participants have at least one actionable pharmacogenetic variant, with an average of 3.7 actionable pharmacogenetic variants. Leading to an average of 12.2 drugs that require an alternate drug or dosage according to CPIC guidelines.
How might this change clinical pharmacology or translational science?
These data highlight the widespread nature of actionable pharmacogenetic alleles among commonly used drugs and may motivate increased adoption of clinical pharmacogenetics.
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 33722. We thank all the participants in the UK Biobank study. Most of the computing for this project was performed on the Sherlock cluster. We would like to thank Stanford University, the PharmGKB resource, and the Stanford Research Computing Center for providing the computational resources that contributed to these research results. A.L. is supported by the National Science Foundation Graduate Research Fellowship (DGE – 1656518). G.M. is supported by the Big Data to Knowledge (BD2K) from the National Institutes of Health (T32 LM012409). K.S., M.W.C., T.E.K. and R.B.A are supported by NIH/National Institute of General Medical Sciences PharmGKB resource, (U24HG010615). R.B.A. is also supported by NIH GM102365.
Funding Information
A.L. is supported by the National Science Foundation Graduate Research Fellowship (DGE – 1656518). G.M. is supported by the Big Data to Knowledge (BD2K) from the National Institutes of Health (T32 LM012409). K.S., M.W.C., T.E.K. and R.B.A are supported by NIH/National Institute of General Medical Sciences PharmGKB resource, (U24HG010615). R.B.A. is also supported by NIH GM102365.
Footnotes
Conflict of Interests
R.B.A. is a stockholder in Personalis.com, 23andme.com. All other authors declared no competing interests for this work.
Availability
PGxPOP is freely available and can be downloaded from https://github.com/PharmGKB/PGxPOP. All data used in the study can be obtained by applying to UK Biobank for access.
References
- 1.2016 NAMCS Summary Web Tables. At <https://www.cdc.gov/nchs/data/ahcd/namcs_summary/2016_namcs_web_tables.pdf>
- 2.Van Driest SL et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin. Pharmacol. Ther 95, 423–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reisberg S. et al. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions. Genet. Med (2018).doi: 10.1038/s41436-018-0337-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush WS et al. Genetic variation among 82 pharmacogenes: The PGRNseq data from the eMERGE network. Clin. Pharmacol. Ther 100, 160–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavertu A. et al. Pharmacogenomics and big genomic data: from lab to clinic and back again. Hum. Mol. Genet 27, R72–R78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaedigk A. et al. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaedigk A. et al. The Evolution of PharmVar. Clin. Pharmacol. Ther 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robarge JD, Li L, Desta Z, Nguyen A. & Flockhart DA The star-allele nomenclature: retooling for translational genomics. Clin. Pharmacol. Ther 82, 244–248 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Whirl-Carrillo M. et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relling MV & Klein TE CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther 89, 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swen JJ et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin. Pharmacol. Ther 89, 662–673 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Kozyra M, Ingelman-Sundberg M. & Lauschke VM Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet. Med 19, 20–29 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Caspar SM, Schneider T, Meienberg J. & Matyas G. Added Value of Clinical Sequencing: WGS-Based Profiling of Pharmacogenes. Int. J. Mol. Sci 21, (2020).doi: 10.3390/ijms21072308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauschke VM & Ingelman-Sundberg M. Emerging strategies to bridge the gap between pharmacogenomic research and its clinical implementation. NPJ Genom Med 5, 9 (2020).doi: 10.1038/s41525-020-0119-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingelman-Sundberg M, Mkrtchian S, Zhou Y. & Lauschke VM Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genomics 12, 26 (2018).doi: 10.1186/s40246-018-0157-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sangkuhl K. et al. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin. Pharmacol. Ther 107, 203–210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudlow C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Hout CV et al. Whole exome sequencing and characterization of coding variation in 49,960 individuals in the UK Biobank. bioRxiv 572347 (2019).doi: 10.1101/572347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh P-R, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet 48, 811–816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UKB : Resource 3801. at <http://biobank.ctsu.ox.ac.uk/showcase/refer.cgi?id=3801>. Accessed 22 September 2020.
- 22.Haeussler M. et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 47, D853–D858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huddart R. et al. Standardized Biogeographic Grouping System for Annotating Populations in Pharmacogenetic Research. Clin. Pharmacol. Ther 105, 1256–1262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.UKB : Data-Field 22009. at <https://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=22009>. Accessed 22 September 2020.
- 25.Pratt VM et al. Characterization of 137 Genomic DNA Reference Materials for 28 Pharmacogenetic Genes: A GeT-RM Collaborative Project. J. Mol. Diagn 18, 109–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.PharmVar. at <https://www.pharmvar.org/criteria>. Accessed 22 September 2020.
- 27.Calculated consequences. at <https://m.ensembl.org/info/genome/variation/prediction/predicted_data.html>. Accessed 22 September 2020.
- 28.Zhou Y, Mkrtchian S, Kumondai M, Hiratsuka M. & Lauschke VM An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J. 19, 115–126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsey LB et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 22, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M. van der et al. Repurposing of Diagnostic Whole Exome Sequencing Data of 1,583 Individuals for Clinical Pharmacogenetics. Clin. Pharmacol. Ther 107, 617–627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon AS et al. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet. Genomics 26, 161–168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirona RG, Leake BF, Merino G. & Kim RB Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem 276, 35669–35675 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Nozawa T. et al. Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J. Pharmacol. Exp. Ther 302, 804–813 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Nishizato Y. et al. Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes: consequences for pravastatin pharmacokinetics. Clin. Pharmacol. Ther 73, 554–565 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Niemi M. et al. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1). Pharmacogenetics 14, 429–440 (2004). [DOI] [PubMed] [Google Scholar]
- 38.McInnes G. et al. Transfer learning enables prediction of CYP2D6 haplotype function. bioRxiv 684357 (2020).doi: 10.1101/684357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M. van der et al. A unifying model to predict variable drug response for personalised medicine. bioRxiv 2020.03.02.967554 (2020).doi: 10.1101/2020.03.02.967554 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.