Abstract
ID proteins are helix-loop-helix (HLH) transcriptional regulators frequently overexpressed in cancer. ID proteins inhibit basic-HLH transcription factors often blocking differentiation and sustaining proliferation. A small-molecule, AGX51, targets ID proteins for degradation and impairs ocular neovascularization in mouse models. Here we show that AGX51 treatment of cancer cell lines impairs cell growth and viability that results from an increase in reactive oxygen species (ROS) production upon ID degradation. In mouse models, AGX51 treatment suppresses breast cancer colonization in the lung, regresses the growth of paclitaxel-resistant breast tumors when combined with paclitaxel and reduces tumor burden in sporadic colorectal neoplasia. Furthermore, in cells and mice, we fail to observe acquired resistance to AGX51 likely the result of the inability to mutate the binding pocket without loss of ID function and efficient degradation of the ID proteins. Thus, AGX51 is a first-in-class compound that antagonizes ID proteins, shows strong anti-tumor effects and may be further developed for the management of multiple cancers.
Subject terms: Cancer therapy, Metastasis
Introduction
The ID proteins, ID1, ID2, ID3, and ID4, are helix-loop-helix (HLH) transcriptional regulators. They function by binding to and sequestering basic-HLH (bHLH) transcription factors (e.g., the E protein E47). ID proteins are expressed during development and inhibit bHLH transcription factors to block differentiation and maintain self-renewal. ID protein expression is largely silenced in adult tissues but can be reactivated in several disease processes, including cancer1–3. Initial data associating ID1 and ID3 with cancer emerged from studies of xenografts and spontaneous tumors in genetically engineered mouse models, which typically showed decreased tumor growth and impaired angiogenesis in Id1- and/or Id3-deficient backgrounds1,4,5. ID1 and ID3 are highly expressed in virtually all human cancers, in the vasculature and/or the tumor cells, including solid tumors of the breast6–8, pancreas9, bladder6, cervix/uterus10–12, colon13,14, endometrium15,16, stomach17,18, nervous system4,19,20, liver21, ovary22, prostate23–25, esophagus26, lung27–29, and thyroid30,31 as well in hematopoietic tumors such as AML32. More recently, expression of ID1 and ID3 has been associated with anchorage-independent growth of invasive lobular carcinomas of the breast33. In virtually all of these cancer types the presence of ID1 and/or ID3 is associated with an aggressive phenotype and poor clinical outcome. However, mutations in the ID genes or their promoters are rarely found. One notable exception is Burkitt’s lymphoma where ID3 is frequently mutated and appears to act as a tumor suppressor, perhaps due to the growth-promoting properties of E proteins in these cells34.
ID overexpression in cancer is largely due to the convergence of several pro-proliferative and pro-oncogenic signaling cascades on the ID promoters such as MAP kinase35,36, Myc37, BMPs38,39, Src40, FLT332, VEGF41, Tgf-β42, Wnt43, and Notch signaling44. High ID expression is associated with increased metastatic potential. For example, in breast cancer, high levels of ID1, ID3, and ID4 are present in the tumor cells of the highly metastatic, triple-negative subtype45,46. In metastatic tumor cells, ID1 and ID3 facilitate the mesenchymal-to-epithelial conversion required for macro-metastatic disease progression, likely through antagonism of the bHLH protein Twist47.
We recently identified a small-molecule antagonist of ID proteins based on X-ray crystal structure coordinates and an in silico screen that targeted a hydrophobic crevice adjacent to the loop region of the HLH domain48. Approximately 350 molecules with drug-like properties were identified and tested for their ability to perturb ID1 interactions with target bHLH protein E47. The top candidate from this screen, AGX51, caused a change in the secondary structure of purified ID1 and inhibited the ability of ID1 to associate with its target E proteins in cells. This in turn led to the degradation of ID1 via ubiquitin-mediated proteolysis. All 4 members of the ID family were similarly destabilized by AGX51. Consistent with the low-level expression of ID proteins in normal adult tissues, high doses of the drug were well tolerated in mice with no apparent toxicity (see below). Here we report the characterization of AGX51 activity in several cancer models including cell lines, patient-derived xenografts (PDXs), pancreatic ductal adenocarcinoma (PDA) organoid cultures, xenograft and syngeneic mouse models, and a chemically induced murine model of colorectal adenoma. We demonstrate that ID inhibition by AGX51 impairs cell viability, increases reactive oxygen species (ROS) levels, and inhibits tumor growth and cancer cell colonization of the lungs in mice, thus phenocopying the effects seen in genetic animal studies. Importantly, long-term exposure of multiple cancer cell lines to AGX51 fails to demonstrate evidence of acquired resistance likely due to the binding of the drug to a region of the proteins that if mutated renders the protein non-functional.
Results
Effects of AGX51 on ID proteins
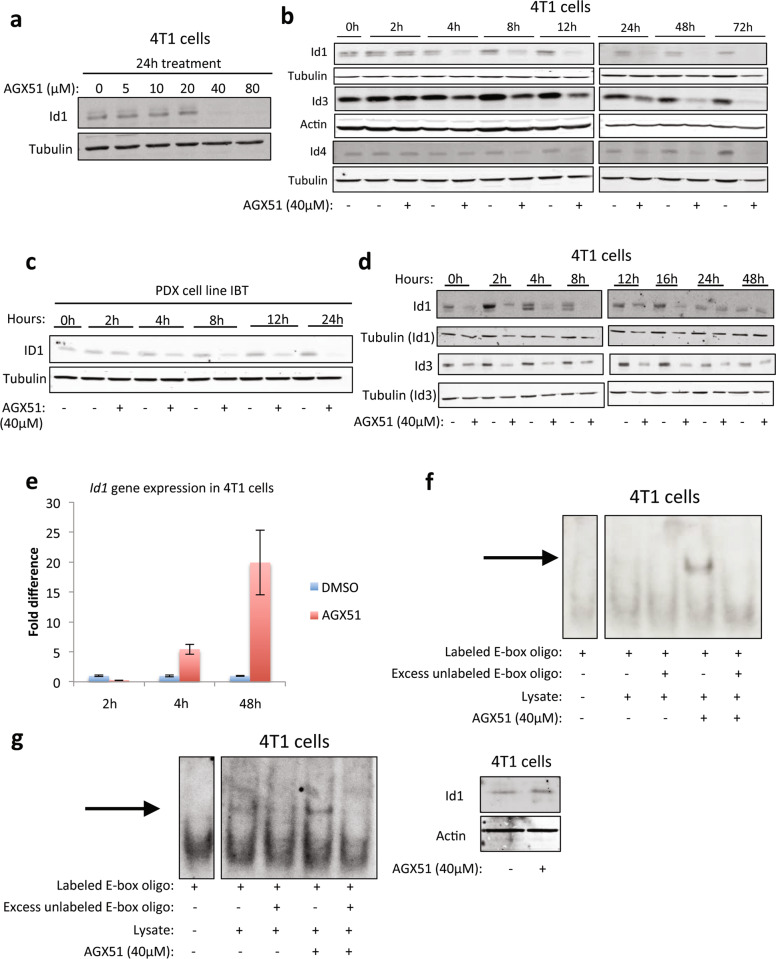
We recently reported that AGX51 treatment of HUVEC and HCT116 cells leads to disruption of the ID–E protein–protein interaction (PPI), resulting in ubiquitination of ID1 and its subsequent degradation48. To further characterize the effects of AGX51 in cancer cells, we treated the 4T1 murine mammary cancer cell line with increasing concentrations of AGX51 (0–80 μM) for 24 h and observed a significant decrease in ID1 protein levels starting at 40 μM (Fig. 1a). 4T1 cells treated with 40 μM AGX51 for 0–72 h showed a decrease in ID1 levels starting at 4 h, with near-complete ID1 loss by 24 h (Fig. 1b and Supplementary Fig. 1a). ID3 and ID4 are also lost albeit with delayed kinetics. ID2 is not expressed in 4T1 cells (data not shown) but we previously showed that ID2 levels decrease in response to AGX51 treatment in HCT116 cells48. Similar to 4T1 cells, HMLE RAS Twist cells (RAS transformed human mammary epithelial cells overexpressing Twist) also showed reduced levels of ID1, ID3, and ID4 when treated with 20 μM AGX51 over similar time courses (Supplementary Fig. 1b). A decrease in ID1 and ID3 protein levels was also observed in a PDX cell line (IBT) generated from a patient with triple-negative breast cancer (TNBC) (Fig. 1c and Supplementary Fig. 1c). The PDX cell line does not express ID2 and ID4 (data not shown). To determine the duration of ID1 and ID3 protein loss, 4T1 cells were treated with 40 μM AGX51 for 24 h, then cultured in AGX51-free media over 48 h. ID1 protein levels increased over time, reaching untreated levels by 24 h (Fig. 1d). ID3 protein levels showed slower recovery (Fig. 1d).
Fig. 1. Effects of AGX51 on ID proteins.
a Western blot for ID1 on whole-cell lysates from 4T1 cells treated with 0–80 µM AGX51 for 24 h. b Western blot for ID1, ID3, and ID4 on whole-cell lysates from 4T1 cells treated with 40 µM AGX51 for 0–72 h. Quantification of western blot bands is shown in Supplementary Fig. 1a. c Western blot for ID1 on whole-cell lysates from the PDX cell line IBT treated with 40 µM AGX51 for 0–24 h. d Western blot for ID1 and ID3 on whole-cell lysates from 4T1 cells treated with 40 µM AGX51 for 24 h, after which compound was washed off and cells were collected for 0–48 h post compound removal. e qRT-PCR analysis for Id1 in 4T1 cells treated with 40 µM AGX51 for 2, 4, or 48 h. Mean fold difference with error bars showing standard error of the mean (SEM) of technical triplicates. The experiment was performed at least three times. f Electrophoretic mobility shift assay (EMSA) on whole-cell lysates from 4T1 cells treated with 40 µM AGX51 for 24 h. Arrow indicates binding to DNA. g EMSA on whole-cell lysates from 4T1 cells treated with 40 µM AGX51 for 1 h, with corresponding western blot for Id1 shown to the right of the EMSA blot. Arrow indicates binding to DNA. Tubulin/actin are used as protein loading controls. See also Supplementary Fig. 1.
To determine if the reduction in ID protein levels was secondary to reduced transcription, we measured Id1 mRNA levels from 4T1 cells treated with AGX51 over a 48 h period. By qRT-PCR, we observed a modest increase of Id1 mRNA at early time points, with a dramatic increase in expression by 48 h (5x and 20x, respectively, Fig. 1e). A similar result was observed for Id3 (data not shown). The increase in Id1 and Id3 mRNA levels suggests that the cell is attempting to compensate for ID1 and ID3 protein loss, likely through the ability of the liberated E proteins to transactivate the Id promoters analogous to what is seen in Drosophila melanogaster49 where daughterless (an E protein orthologue) activates extramachrochaete (an Id orthologue). To address this possibility we obtained T cells from E2A/HEB double knockout mice50,51 and treated them ex vivo with AGX51. These cells express ID3 and ID2 but not ID1. Compared to wild-type T cells, E protein knockout T cells show a reduction in Id3 mRNA levels following treatment with AGX51 (Supplementary Fig. 1d). A similar trend is seen with Id2 mRNA. These findings support the hypothesis that liberation of E proteins contributes to the increase in Id1 mRNA observed after AGX51 treatment of our cancer cells.
We next tested whether loss of ID proteins in response to AGX51 resulted in increased E protein–DNA binding. As expected, treating 4T1 cells with AGX51 for 24 h resulted in an increase in E protein binding (Fig. 1f). Similar electrophoretic mobility shift assay (EMSA) results were obtained using cell lysates from HMLE RAS Twist cells, and the PDX cell line (Supplementary Fig. 1e). To determine if the loss of ID activity precedes ID protein loss, EMSAs were carried out after 1 h of AGX51 treatment. An increase in E protein binding occurred at a time when no ID1 protein loss was observed (Fig. 1g), suggesting that the increase in E protein binding is due to disruption of the ID1–E PPI as opposed to decreased overall ID levels. This conclusion is supported by direct evidence of the disruption of the ID1–E47 interaction observed in HCT116 cells48.
Effects of AGX51 on cell growth
To characterize the effects of AGX51-mediated ID protein loss on cell growth, we determined the IC50 of AGX51 in 4T1 cells and nine other breast cancer cell lines representing the major breast cancer subtypes (ER+, HER2+, and TNBC), and three breast cancer PDX cell lines (Supplementary Table 1 and Supplementary Fig. 1f, g). Due to the observed viability curves of certain cell lines, the IC50 had to be extrapolated to fit the curve (these approximate values are indicated by asterisk in Supplementary Table 1). Of the different breast cancer subtypes, the TNBC cell lines were most sensitive (~25 μM) (Supplementary Table 1). The IC50 of 4T1 cells, a model of TNBC {Aslakson, 1992 #5667; Pulaski, 2001 #5668} was similar to that of the TNBC cell lines (Supplementary Table 1). The effect of AGX51 on 4T1 cells was also determined using Alamar blue viability assays, which show a significant reduction (~3-fold, p = 0.0009) in viability after 24 h of treatment with 40 μM AGX51 (Fig. 2a).
Fig. 2. Effects of AGX51 on cells in culture.
a Alamar blue cell viability assay on 4T1 cells treated with 40 µM AGX51 for 24 h. Mean value of technical triplicates is plotted with error bars representing SEM. * Indicates p < 0.001, as determined by unpaired t-test. The experiment was repeated at least three times. b Cell cycle analysis on 4T1 cells treated with 40 µM AGX51 for 24 and 48 h; the experiment was repeated three times. c BrdU incorporation by 4T1 cells treated with 40 µM AGX51 or vehicle, for 24 h as measured by flow cytometry. d Western blot for phospho-histone H3 on lysates from 4T1 cells treated with 40 µM AGX51 or DMSO for 4 and 24 h. e Annexin V and PI staining of 4T1 cells treated with AGX51 for 4 and 24 h, as analyzed by flow cytometry; the experiment was repeated two times. f Reactive oxygen species accumulation in 4T1 cells induced by AGX51 treatment, as determined by H2DCFDA staining followed by flow cytometry analysis. AGX51-treated cells not stained with H2DCFDA were analyzed as an auto-fluorescence control. The experiment was repeated three times. g Cell viability, as measured by trypan blue exclusion and cell counting of 4T1 cells treated with 40 µM AGX51 or DMSO for 24 h, with or without 1 h pretreatment with vitamin E (50 or 100 µM). Means are plotted with error bars showing standard deviation (SD) of four technical replicates. See also Supplementary Fig. 2.
It has previously been reported that genetic loss of Id1 induces a G0/G1 growth arrest in mouse and human fibroblasts {Barone, 1994 #315; Hara, 1994 #314}. After 24 and 48 h of AGX51 treatment, 4T1 cells showed an accumulation in G0/G1 (Fig. 2b). AGX51 treatment also led to a reduction in BrdU incorporation (Fig. 2c) and phospho-histone H3 levels (Fig. 2d). After 24 h of AGX51 treatment, we saw an increase in the Annexin V+/PI+ fraction (12.7% vs. 40.7%), indicative of late apoptosis and necrosis (Fig. 2e). We observed no overt increase in cleaved caspase 3 or cleaved PARP levels, suggesting that the mechanism of cell death is largely non-apoptotic (data not shown).
A feature of necrotic cell death is the production of ROS. To determine the effect of AGX51 on ROS production in 4T1 cells, we treated cells with 40 μM AGX51 for 24 h, stained with 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate (H2DCFDA) and analyzed the cells by flow cytometry. We observed a significant increase in the fluorescein-positive fraction upon AGX51 treatment, indicative of an increase in ROS production (Fig. 2f). In fact, AGX51 increased ROS production to a greater degree than our positive control, H2O2 (Supplementary Fig. 2a). Similar effects were seen in HCT116 cells (Supplementary Fig. 2b). Furthermore, addition of 50 and 100 μM of the antioxidant vitamin E rescued cell viability, and reduced ROS levels in both cell lines (Fig. 2g and Supplementary Fig. 2c–g). Vitamin E treatment also rescued the AGX51-mediated cell cycle arrest in HCT116 cells (Supplementary Fig. 2h) and 4T1 cells (data not shown), suggesting that the cell cycle arrest is secondary to ROS production. ROS leading to cell cycle arrest has been previously reported52,53. Interestingly, other free radical scavengers such as N-acetyl cysteine (NAC), MnTBAP, MitoQ, MitoTEMPO, Apocynin, and Catalase did not rescue the AGX51-induced phenotypes (Supplementary Fig. 3). This observation raises the possibility that an oxidized lipid may be a key mediator of AGX51-mediated killing activity since vitamin E is an efficient inhibitor of lipid peroxidation54.
Other potential non-apoptotic mechanisms of cell death such as ferroptosis and necroptosis may be at play in mediating AGX51-induced reduced cell viability. To investigate these possibilities we co-treated 4T1 cells with AGX51 and ferrostatin-1 or AGX with necrostatin-1. Neither ferrostatin-1 nor necrostatn-1 were able to rescue the decreases in cell viability seen with AGX51 treatment (Supplementary Fig. 2i, j), suggesting that these cell death mechanisms are not involved in AGX51-mediated effects. Nonetheless, our findings are consistent with a recent report where siRNA-mediated ID1 and ID3 inhibition was associated with increased ROS levels55 and indicate that a main mechanism of AGX51-mediated cell killing is ROS-mediated. We also suggest that the G0/G1 arrest we see prior to cell death may in fact be ROS-mediated as has been observed in other systems52,53.
Enforcement of ID1 expression has previously been reported to play an important role in circumventing Tgf-β’s tumor-suppressive effect in pancreatic cancer. Depletion of the ID proteins by shRNAs has been shown to decrease the ability of pancreatic cancer cells to survive and form orthotopic tumors56. We tested whether AGX51 would have similar effects on pancreatic cancer cells. In line with the results obtained in other cancer models that we tested, AGX51 caused a depletion of the ID1 and ID3 proteins in the pancreatic cancer cell line 806, starting at a dose between 4 and 20 µM (Fig. 3a) with ID3 showing slightly less sensitivity as observed in other cellular and biophysical assays48. Furthermore, AGX51 decreased cell viability in a variety of pancreatic cancer cell and organoid lines, including mouse organoids (Fig. 3b), mouse cell lines 806, NB44 and 4279 (Fig. 3c), and human cell lines Panc1 and A21 (Fig. 3d) with an IC50 of 5.5–19.5 µM. This corresponds to the range of concentrations of AGX51 required for ID protein loss. Of note, when treated with 40 µM of AGX51, well-formed organoid structures collapsed (Fig. 3e).
Fig. 3. Effects of AGX51 on pancreatic ductal adenocarcinoma cells.
a Western blot for ID1 and ID3 on whole-cell lysates from 806 pancreatic ductal adenocarcinoma (PDA) cells treated with 4, 20, and 40 µM AGX51 for 24 h. GAPDH used as loading control. b Dose–response of PDA organoid lines T7 and T8 to AGX51 following 120 h of treatment, with IC50 indicated. c Cell viability as assessed by Cell Titer Glo assay for PDA lines 806, 806 + Smad4, NB44, and 4279 treated with 40 µM AGX51 or DMSO vehicle control for 120 h (three replicates, error bars, standard deviations, SD). d Dose–response of PDA cell lines Panc1 and A21 to 120 h of AGX51 treatment, with IC50 indicated. e Bright field images of T7 and T8 PDA organoid lines treated with 4 and 40 µM AGX51 for 48 h. Scale bars represent 200 µm.
Specificity of AGX51 activity
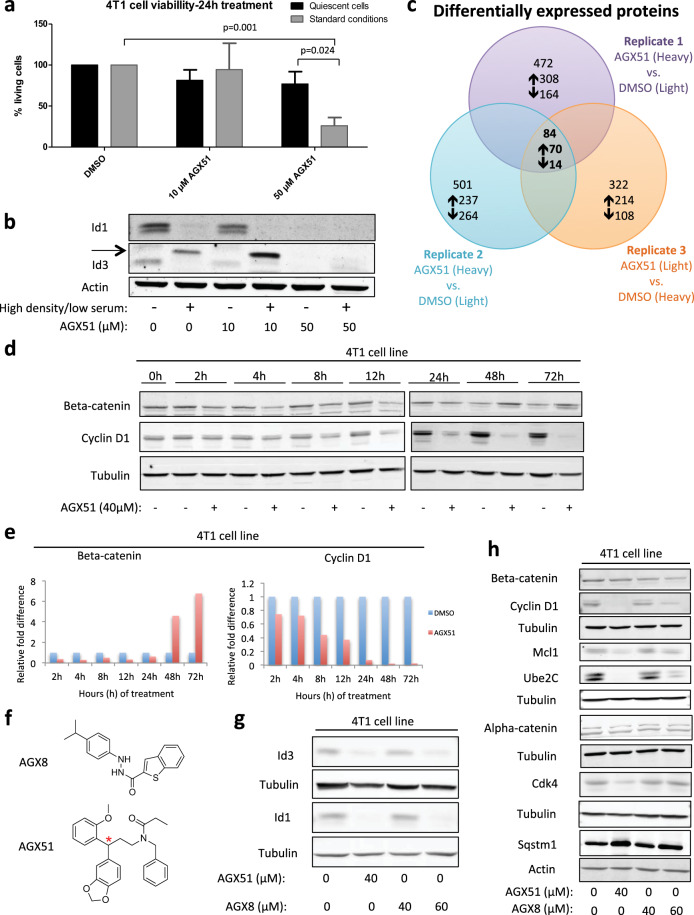
A common concern with any novel small-molecule is specificity. Our results showing loss of ID proteins, an increase in liberated E proteins, and cell phenotypes consistent with genetic models of ID loss indicate that AGX51 is targeting ID proteins. However, AGX51 may also be affecting other, unintended targets. If ID proteins are the primary targets of AGX51 then we reasoned that reducing ID1 or ID3 levels by shRNA should increase the sensitivity of the cells to AGX51 by reducing the concentration of the target. A similar approach was used to show that reduction of Ras genes sensitized cells to a pan-Ras inhibitor57. Similarly, cells able to survive in the absence of ID proteins, such as quiescent cells should be resistant to AGX51. Upon partial knockdown of Id1 or Id3 by shRNAs (Supplementary Fig. 4a) the cells showed increased sensitivity to AGX51 (~3-fold) (Supplementary Table 1). 4T1 cells allowed to reach confluence under low-serum conditions become quiescent and display near-complete loss of ID expression (Fig. 4a, b). Treating these cells with AGX51 led to a dramatic reduction in cell killing relative to cycling control cells. Together these data support the conclusion that the ID proteins are the primary targets in cycling cells.
Fig. 4. Assessing the effect of AGX51 on quiescent cells and the whole proteome.
a 4T1 cells were treated with AGX51 (10 or 50 µM), when they were cultured under standard conditions (standard density, standard serum (10%)) or under quiescence-inducing conditions (high density, low serum (1%)). Following 24 h of AGX51 treatment, cell numbers were determined by cell counting and trypan blue exclusion. Means are plotted with error bars showing SD of four technical replicates. b Western blot for ID1 and ID3 on whole-cell lysates from 4T1 cells presented in (a). Actin serves as the protein loading control. Arrow indicates non-specific band. c Venn diagram of differentially expressed proteins, as identified by mass spectrometry, from three replicates of SILAC-4T1 cells treated with 40 µM AGX51 for 4 h. d Western blot for Beta-catenin and Cyclin D1 on whole-cell lysates from 4T1 cells treated with 40 µM AGX51 for 0–72 h. e Quantification of western blot shown in (d). f Structures of AGX8 and AGX51 (red asterisk indicating chiral center). g Western blot for ID3 and ID1 on whole-cell lysates from 4T1 cells treated with AGX51 and AGX8 at the indicated concentrations. h Western blot for the indicated proteins on whole-cell lysates from 4T1 cells treated with AGX51 and AGX8 at the indicated concentrations. Quantification of western blot bands shown in (g) and (h) are in Supplementary Fig. 7. See also Supplementary Figs. 4–7.
Having established that AGX51 disrupted PPI leading to proteasome-dependent degradation of ID proteins48, we wondered if levels of other proteins would be affected. To determine the full range of consequences of AGX51 exposure, we performed Stable Isotope Labeling of Amino acids in Cell culture (SILAC) on 4T1 cells treated with AGX51 or DMSO for 4 h (at which point we would expect to see a decrease in ID1) and profiled the whole proteome by mass spectrometry. We identified 6869 proteins/protein clusters with an FDR < 1%, where 2735 proteins were found in all three replicate experiments. As expected, ID1 was underexpressed in all three replicates (fold differences: −1.39, −1.71, and −2.07). Using a 1.35-fold cutoff, we identified 13 other underexpressed and 70 overexpressed proteins (Fig. 4c and Supplementary Table 2). ID2 and ID4 were not detected in this analysis and ID3 showed no significant change at 4 h, consistent with the data shown above (see Fig. 1b). The underexpressed proteins included Beta-catenin, Cyclin D1, Cdk4, and Ube2C, which are all involved in cell cycle progression and were validated by western blot (Fig. 4d, e and Supplementary Fig. 4b). The underexpression of these proteins at early time points is modest by SILAC and hence quantitative differences by western blot do not become prominent until later time points, and in some cases are transient (Beta-catenin). An additional three underexpressed proteins were also validated by western blot (Mcl1, Sqstm1, and Alpha-catenin) (Supplementary Fig. 4b) although the Sqstm1 inhibition was also transient. Sqstm1 encodes the protein Sequestosome-1 which targets other proteins to which it binds for degradation by autophagy58. These changes were also observed in the PDX cell line and HMLE RAS Twist cells (Supplementary Fig. 5). We saw no change in Beta-catenin mRNA levels for 24 h of AGX51 treatment of 4T1 cells, and only saw a significant decrease in Cyclin D1 message at later time points (~1-fold at 24 h and ~4-fold at 48 h), suggesting that at early time points the changes in Beta-catenin and Cyclin D1 proteins may be occurring post-transcriptionally (Supplementary Fig. 6a). We tested the gene expression of Cdk4 and Mcl1, and also saw no significant decrease in mRNA over time with AGX51 treatment and saw upregulation at later time points (Supplementary Fig. 6b). Importantly, the loss of these proteins takes place prior to the changes in cell cycle distribution (Supplementary Fig. 6c), arguing against the possibility that these protein changes are secondary to a G0/G1 arrest.
Genetic suppression of Id1–4 could be used to test the possibility that the proteome changes seen after AGX51 treatment are secondary to ID protein loss but a timed, simultaneous reduction of all ID proteins prior to cell cycle arrest has been technically impossible. While overexpression of ID proteins would not necessarily yield the converse result, we nonetheless attempted to assess the proteome changes that occurred in ID1- and ID3-overexpressing 4T1 cells. This approach was also confounding since we found that ID3 overexpression reduced endogenous ID1 levels and that ID1 overexpression reduced ID3 levels so that less-than-expected changes in total ID1/3 levels were observed. Despite this we did see minor increases in Cyclin D1 levels in the overexpressing cell lines as expected (Supplementary Fig. 6d) but many of the other genes were unaffected (data not shown).
Another approach was to examine the effects of a structurally unrelated compound (AGX8) that also scored positive in the initial EMSA screen for perturbation of the ID1–E47 interaction48. AGX51 and AGX8 contain 2 distinct chemical scaffolds with different interaction patterns. While AGX51 has only hydrogen acceptor groups, AGX8 has 2 hydrogen donors and 2 hydrogen acceptor groups. Treating 4T1 cells with AGX8 for 24 h also led to reduced levels of ID1 and ID3 although the concentration needed to see this decrease was greater than that for AGX51 (60 vs. 40 μM) (Fig. 4f, g and Supplementary Fig. 7a). In addition, the cell viability effects of AGX8 were less pronounced than those of AGX51 (Supplementary Table 3). Western blot analyses found that the levels of Cyclin D1, Beta-catenin, Ube2C, and Mcl1 decreased when 4T1 cells were treated with 60 μM AGX8, similar to 40 μM of AGX51 treatment. Very modest decreases were observed for Cdk4 and Alpha-catenin with 40 μM AGX51 but were difficult to discern with 60 μM AGX8 (Fig. 4h and Supplementary Fig. 7b). In addition, there was an increase in Sqstm1 expression after 24 h of AGX8 treatment, similar to that seen with AGX51 treatment (Fig. 4h and Supplementary Fig. 7b). The similarity in the effects of AGX51 and AGX8 on many of the proteins identified by SILAC is consistent with the idea that these changes are secondary to ID loss.
Effects of AGX51 on tumor growth and lung colonization of cancer cells
In spontaneous primary tumors of the breast in the MMTV-Her2/neu model, genetic analyses indicated that loss of Id1 and/or Id3 was insufficient to inhibit tumor growth and needed to be combined with suboptimal doses of chemotherapy to cause regression59. We tested AGX51 in a primary tumor xenograft model using MDA-MB-231 cells with and without paclitaxel. Nude mice were implanted with MDA-MB-231 cells and tumors were allowed to grow to ~100 mm3 before the initiation of the following treatments: vehicle (DMSO); paclitaxel 15 mg/kg qd (quaque die, i.e., once a day) on days 1–5; AGX51 60 mg/kg bid (bis in die, i.e., twice a day) for all 19 days; or paclitaxel 15 mg/kg on days 1–5 plus AGX51 6.7 mg/kg, 20 mg/kg, or 60 mg/kg bid for all 19 days. Also included was a treatment group that received 15 mg/kg paclitaxel qd on days 1–5 and 60 mg/kg of AGX51 for just 7 days. Paclitaxel treatment alone had a modest inhibitory effect on tumor growth while the addition of 6.7 and 20 mg/kg AGX51 to paclitaxel almost completely inhibited tumor growth and the 60 mg/kg dose resulted in tumor regression (Fig. 5a and Supplementary Table 4). Interestingly, just 7 days of AGX51 treatment, in combination with paclitaxel, was sufficient to cause tumor regression, suggesting a long-lived anti-tumor effect that may persist even after treatment has stopped (Fig. 5a and Supplementary Table 4). AGX51 treatment alone did not suppress tumor growth. Thus, similar to the previously conducted genetic loss-of-function experiment59, AGX51 treatment leads to synergistic growth inhibition of primary breast tumors when combined with chemotherapy. This is not due to synergistic killing at the cellular level since we observed no synergistic effect on cell viability when MDA-MB-231 cells in culture are treated with AGX51 and paclitaxel (Supplementary Fig. 8a). While such synergy in vivo may be due to effects of ID loss on neoangiogenesis, in the current in vivo study we observed no obvious changes in angiogenesis or vascularization by gross observation of the tumors, histological evaluation, or CD31 staining by IHC (data not shown, see also ‘Discussion’).
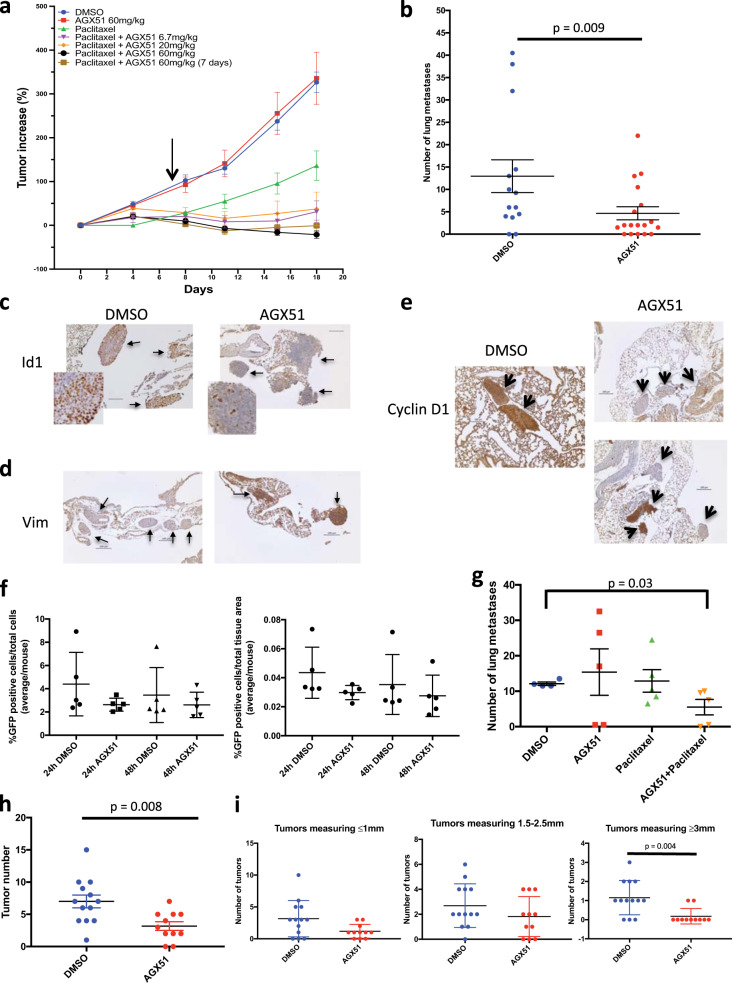
Fig. 5. Effects of AGX51 on tumor growth and metastasis.
a Mean tumor volume increase in athymic nu/nu mice implanted with MDA-MB-231 cells in the mammary fat pad and treated with AGX51, paclitaxel, AGX51 + paclitaxel, or vehicle control (n = 5 mice per treatment group for all groups except paclitaxel alone where n = 4 due to 1 death). Compared to controls, the effect of all AGX51 + paclitaxel treatments was significant by weighted least squares (p ≤ 0.001) and AGX51 + paclitaxel treatments were also significantly different from AGX51 treatment alone (p ≤ 0.001). The mean is plotted with error bars representing the SEM. b Average number of lung metastases in Balb/c mice injected with 5 × 104 4T1 cells via tail vein, treated with 50 mg/kg AGX51 bid or vehicle (n = 18 or n = 14, respectively). Means are plotted with error bars showing SEM. AGX51 treatment had a significant effect on lung tumor development (p < 0.009 by weighted least squares). c Examples of ID1 IHC staining in lung sections from mice described in (b). d Examples of Vimentin IHC staining in lung sections from mice described in (b). e Examples of Cyclin D1 IHC staining in lung sections from mice described in (b), with examples of metastases from AGX51-treated mice showing negative (upper panel) and positive (lower panel) staining for Cyclin D1. Bars are 100 µm in length. Staining in (c–e) was performed on the lesions of 5 animals in each group and representative images are shown. f Quantification of the number of GFP-labeled 4T1 cells in the lungs of Balb/c mice injected with 5 × 104 4T1 cells via tail vein and treated with 50 mg/kg bid or vehicle for 24 and 48 h (n = 5 mice per treatment group). Means are plotted with error bars showing SD. g Average number of lung metastases in Balb/c mice injected with 5 × 104 4T1 cells via tail vein and treated with AGX51, paclitaxel, AGX51 + paclitaxel, or vehicle once lung metastases were established (n = 5 mice per treatment group). Means are plotted with error bars showing SEM. Compared to vehicle, AGX51 + paclitaxel treatment had a significant effect on lung tumor growth (p < 0.03 by weighted linear regression). h Number of colon tumors in AOM colon tumor model in A/J mice that were treated with DMSO (n = 13) or AGX51. Means are plotted with error bars showing SEM. AGX51 treatment resulted in a significant reduction in the number of tumors (p = 0.008). i Size distribution of tumors in (h), grouped by treatment group (AGX51 or DMSO), where AGX51 treatment resulted in a significant reduction in tumors measuring ≥3 mm (p = 0.004). Mean are plotted with error bars showing SD. Colon tumor count data were analyzed using the Wilcoxon rank sum test. For additional data pertaining to the effects of AGX51 on tumor growth and metastases see Supplementary Fig. 9.
Of note, we have previously shown that after 14 days of daily treatment at the highest dose used in any of our studies (60 mg/kg bid), there were no statistically significant differences between the AGX51-treated animals and the vehicle-treated animals with respect to weight, clinical chemistry and hematology plasma values, and histopathological evaluation of major organs at necropsy48. Similarly, when combining AGX51 bid with 15 mg/kg paclitaxel (qd for the first 5 days of a 14-day regimen), a regimen used in the xenograft studies described above, no overt toxicities were observed using any of the above-listed parameters (Supplementary Fig. 8b). Thus, in addition to showing anti-tumor efficacy, AGX51 is well tolerated in mice in this concentration range, alone and in combination with paclitaxel.
Genetic loss of Id1/Id3 prevents the development of breast cancer lung metastasis by blocking the conversion of micro-metastases to macro-metastases47 and by blocking angiogenesis4,60. To determine whether AGX51 phenocopies these results, we used lung colonization assays, a model of experimental metastasis, where luciferase-labeled 4T1 cells, known to be metastatic, were injected into the tail veins of Balb/c mice and after 24 h the mice were treated with 50 mg/kg of AGX51 i.p. bid or vehicle. Colonization of the lungs by 4T1 cells and their growth into tumors (referred to as metastases) was monitored weekly by in vivo bioluminescent imaging. The experiment was stopped at 4 weeks following tail vein injection, due to the deteriorating condition of the control group. Bioluminescent imaging showed significant inhibition of lung metastasis development in the AGX51 group, and this correlated with the metastatic burden observed upon histopathological examination of lung tissues (p = 0.009) (Fig. 5b and Supplementary Fig. 9a). Treatment with AGX51 50 mg/kg once daily also decreased the number of lung metastases, and while still significant (p = 0.0023) the effect was not as dramatic as that seen with the bid regimen (Supplementary Fig. 9b, c). While all macro-metastases showed strong positive IHC staining for ID1, in smaller lung metastases the ID1 staining was decreased in the AGX51 group, relative to metastases of a similar size in the DMSO group (Fig. 5c). Conversely, as predicted from the genetic analyses47, Vimentin staining of small metastatic tumors in the AGX51 group was significantly higher than that in the DMSO group (Fig. 5d), consistent with a reduction in Id1/Id3 levels blocking the mesenchymal-to-epithelial transition during metastatic progression. Interestingly, Vimentin was one of the proteins found overexpressed in our SILAC analysis (see Supplementary Table 2).
To characterize the effects of AGX51 on the expression of EMT-related proteins we assessed Vimentin, E-cadherin, Snail, Twist, and Zeb1 levels in AGX51-treated 4T1 cells. After an 8 h exposure, we observed a decrease in E-cadherin and an increase in Vimentin but by 48 h both proteins were increased. Decreases in Snail and Twist1 were observed throughout the time course, while Zeb1 levels were unchanged (Supplementary Fig. 10). Thus AGX51 modulates the levels of some EMT-related proteins, however, the precise mechanisms leading to these changes and their consequences remain to be elucidated. The changes observed do not appear to adhere to canonical EMT patterns, which may be evidence of AGX51 inducing partial EMT/MET likely secondary to ID protein loss47. The occurrence of partial or intermediate EMT phenotypes in cancer is becoming increasingly recognized61–63. Finally, consistent with the reduced Cyclin D1 levels shown above, IHC staining for Cyclin D1 in the 4T1 lung metastases of the AGX51 group was significantly reduced relative to the DMSO group (p < 0.00001) (Fig. 5e). While all DMSO group metastases stained positive for Cyclin D1, about half of the AGX51 group was negative (Fig. 5e upper panel of AGX51 group).
To determine whether AGX51 inhibits the extravasation and initial seeding at the secondary site or the progression of extravasated cancer cell outgrowth into tumors, we tail vein-injected the mice with GFP-labeled 4T1 cells and treated with AGX51 (50 mg/kg) or DMSO for 24 or 48 h then stained the lungs for GFP (i.e., tumor cells) and quantified the tumor cell number. There was no significant difference in the number of GFP-positive cells between AGX51 and DMSO groups after either 24 or 48 h of treatment (Fig. 5f). These data are consistent with genetic models that found that Id1/Id3 inhibition does not prevent extravasation of metastasizing breast cancer cells, but rather the conversion of micro- to macro-metastatic lesions45,47.
To ascertain whether AGX51 is capable of inhibiting the growth of established lung tumors, we tail vein-injected the mice with 4T1 cells, as described above, and started treatment once lung signal was visible by bioluminescent imaging. Mice were then divided into 4 groups and treated with AGX51, paclitaxel, AGX51 + paclitaxel, or vehicle. Subsequent bioluminescent imaging and histopathological examination of the lungs found that while AGX51 or paclitaxel alone did not have an effect on the lung tumor growth, combination treatment significantly reduced the continued expansion of lung tumors relative to the other groups (p = 0.03) (Fig. 5g). In addition to suppressing the number of lung tumors, the tumors in the treated group were smaller in size (data not show). Thus, consistent with the primary tumor data, established lung tumors can be more effectively treated when AGX51 is combined with paclitaxel.
To determine the effects of AGX51 in an autochronous tumor model, we turned to the azoxymethane (AOM) colon tumor model, a chemically induced autochthonous model of adenoma. Mice were administered AOM once a week for 6 weeks, followed by a 3-week treatment break and then treated with AGX51 (15 mg/kg) or DMSO bid for 3 weeks. AGX51 treatment resulted in a significant decrease in the number of colon tumors (p = 0.008). In addition, the tumors from AGX51 group were smaller than those in the DMSO group, and this difference was significant when tumors measuring >3 mm were compared (p = 0.004) (Fig. 5h, i). Thus AGX51 shows anti-tumor activity in an autochronous cancer setting.
Investigating mechanisms of acquired resistance to AGX51
The efficacy of therapeutics can be hindered by cells developing resistance. Common mechanisms of acquired resistance include mutation of the drug-binding site or overexpression of the target. As described above, mutation of the binding site would likely result in loss of ID1 activity, thereby blocking this mechanism of escape. Overexpression of the ID1 protein could potentially outcompete the drug. However, as described above, endogenous Id1 mRNA levels increase 20-fold in response to AGX51 exposure but the protein is still degraded efficiently making this type of resistance unlikely. Nonetheless, we tested the effects of AGX51 on HMLE RAS Twist cells lines overexpressing exogenous ID1 mRNA (HMLE RAS Twist ID1 cells). Western blot analysis revealed that despite significant ID1 overexpression, AGX51 treatment still dramatically reduced ID1 protein levels (Supplementary Fig. 11a). Similarly, 4T1 cells overexpressing exogenous Id1 mRNA treated with AGX51 also showed a dramatic decrease in ID1 protein levels (Supplementary Fig. 11b). Furthermore, we generated an ID3-overexpressing 4T1 cell line (Supplementary Fig. 6d) and also observed efficient ID3 degradation in response to AGX51, as well as no evidence of measurable cell viability after long-term drug exposure (Supplementary Fig. 11c, d). Together these data suggest that AGX51 is a potent ID degrader, even in the context of Id mRNA overexpression.
To determine whether tumor cells could spontaneously acquire resistance to AGX51, we sought to identify subpopulations of cells capable of continued growth in the presence of AGX51. 4T1 cells were exposed to various AGX51 concentrations, with media replenished every 2–3 days. Above 25 μM no resistant colonies of cells were observed even after 7 weeks of culture (~3 × 107 cells tested). At 20 μM no overt cell death was observed, indicative of an extremely sharp dose–response curve. We then set up new experimental plates, treating the cells with either 24 or 22.5 μM, where ID loss after 24 h is not as pronounced (Fig. 6a). After 6 weeks in culture, we picked 30 seemingly viable clones but 29 eventually died after an average of ~5 passages. One clone survived the 24 μM condition but grew at reduced rates relative to the starting population (Fig. 6b), and eventually died after 11 passages.
Fig. 6. Generation of AGX51-resistant cell lines.
a Western blot for ID1 on whole-cell lysates from 4T1 cells treated with 0–40 µM AGX51 for 24 h. b Cell growth of 4T1 cells and the AGX51 transiently resistant cell line clone 24-13, as determined by cell counting over 7 days. Mean of technical triplicates is shown with error bars representing SEM. c Cells were FACS isolated from the lungs of mice that were injected with 5 × 106 GFP-labeled 4T1 cells via tail vein and treated with 50 mg/kg bid or vehicle for 2 weeks. The isolated cells (from DMSO or AGX51-treated mice) and parental GFP-labeled 4T1 cells were treated with 0, 10, 20, 40, or 60 µM AGX51 for 24 h and whole-cell lysates were prepared. Western blot analysis for ID1 was then carried out. Tubulin serves as a protein loading control. See also Supplementary Figs. 11 and 12.
None of the seven transiently resistant clones tested showed wild-type levels of ID1 although about half did show a possible rebound in ID3 (Supplementary Fig. 12a, b). No ID2 expression was detectable in these cells. In 4/7 transiently resistant cell lines, there was no/low ID1 expression, while 3/7 transiently resistant cell lines maintained ID1 expression at the level of cells treated with AGX51 for 24 h (Supplementary Fig. 12a). We also saw a decrease in Beta-catenin and Cyclin D1 levels in the transiently resistant cell lines (Supplementary Fig. 12c). Targeted sequencing of Id1 and Id3 coding regions in the three clones expressing detectable levels of ID1 and ID3 found no mutations, suggesting that mutation of the predicted compound-binding site, or other regions of the target proteins, is not the mechanism of transient resistance in these clones. While the growth rate of the clones cultured in 22.5 and 24 μM AGX51 was maintained for a time greater than that of the general population, this is not acquired resistance since all clones grew poorly and none remained viable after prolonged passaging. Furthermore, upon AGX51 removal, the cell growth rate and ID protein levels appeared to rebound to near-untreated 4T1 levels, and fell again upon re-treatment with 22.5 or 24 μM AGX51 (not shown). No acquired resistance was similarly observed in four other cell lines tested: HMLE RAS Twist, SW480, HCT15, and HCT116 (data not shown). Thus, despite some cells being capable of propagation in 22.5 or 24 μM AGX51, the cells are under strong growth-inhibitory pressure and fail to acquire genetic or epigenetic changes allowing for normal proliferation and Id expression in the presence of continued AGX51 exposure.
To determine if the small lung tumors that develop after AGX51 treatment are a result of acquired resistance in vivo we injected luciferase and GFP-labeled 4T1 cells into the tail veins of mice and 24 h later started treating the mice with AGX51 (50 mg/kg, bid) or vehicle for 2 weeks. This primary lung metastasis model showed significant anti-tumor efficacy with AGX51 as a single agent, as described above. Consistent with previous results, we observed a significant reduction in lung tumor burden in the AGX51 group. After 2 weeks of treatment, we harvested and dissociated the lungs and collected the GFP-positive cancer cells by FACS. After minimal passaging in cell culture (to obtain sufficient numbers of cells for western blot analysis), we re-challenged cells from both the AGX51- and DMSO-treated animals, as well as the parental cell line, with AGX51. We saw no difference in AGX51 sensitivity between parental (no in vivo exposure), DMSO, and AGX51 in vivo treated cells (Fig. 6c). The IC50 values for AGX51 in the isolated cells were very similar between DMSO- and AGX51-treated mice (23.48 μM and 22.34 μM, respectively). This result demonstrates that the development of small lung tumors after AGX51 treatment is not due to acquired resistance inherent in the cell population, but rather we suspect it is due to incomplete exposure of the tumor cells to AGX51 or the levels achieved in serum (~4 μM, see48) are insufficient to achieve complete regressions.
Discussion
In the present study, we characterized the effects of a small-molecule antagonist of ID proteins, AGX51, in cancer cell lines and mouse models of breast and colonic neoplasia. Importantly, AGX51 leads to a significant reduction in ID protein levels in all cell lines tested, lung tumors, and ocular neovascularization48. ID protein destabilization after perturbation of its ability to antagonize E protein activity is consistent with genetic analyses in which coexpression of E proteins was shown to dramatically increase the stability of ID364,65. AGX51 appears to act as an ID protein antagonist and degrader, thereby accounting for its ability to phenocopy Id genetic loss-of-function phenotypes in multiple settings.
Several approaches were taken to address the question of AGX51 target specificity. SILAC analysis showed that after short exposure to AGX51 when the ID proteins first begin to get degraded, only 14/2735 other proteins were underexpressed. This limited effect on the proteome is unlikely to be a purely off-target effect as AGX8, a molecule structurally unrelated to AGX51 (see Fig. 4f), also reduced ID protein levels and concomitantly the majority of the other proteins affected by AGX51. In addition, partial loss of Id1 and Id3 function reduced the IC50 of AGX51 (see Supplementary Table 1) and quiescent cells in which ID proteins are absent are resistant to AGX51-mediated killing. These data support ID proteins being the primary targets of AGX51. Finally, the ability of AGX51 to recapitulate the effects of Id loss-of-function mutations in various settings also suggests that these proteins are the primary targets in cells: AGX51 induces a G0/G1 arrest; inhibits the conversion of micro-metastases to macro-metastases in the 4T1 breast cancer model of experimental lung metastasis; with paclitaxel leads to regressions of paclitaxel-resistant primary breast tumor xenografts; phenocopies the consequences of Id1/Id3 loss in two models of pathologic neovascularization48; and decreases the viability of PDA organoids, as was seen with shRNAs to the ID proteins56. AGX51 also showed anti-tumor activity in an autochronous model of colorectal neoplasia.
Despite repeated efforts, acquired resistance to AGX51 did not occur in cultured cells indicating that the escape frequency under our culturing conditions must be less than 1 in 107 cells and perhaps significantly lower. These results suggest that despite the observed dramatic upregulation of Id1 mRNA, ID protein levels are not restored efficiently nor are other pathways evoked, to compensate for ID loss. We also observed a lack of resistance in culture in cancer cells isolated from primary metastasis that persisted despite a 2-week AGX51 treatment regimen, suggesting that other factors such as drug exposure or the tumor microenvironment allowed for the survival of these cells. Another mechanism of resistance, mutating the binding site for the compound, is also not observed since this likely results in an inactive protein given the highly conserved nature of this pocket and inability to tolerate directed mutations that maintain activity at these loci in vitro66. Thus, by short-circuiting two major mechanisms of drug resistance (amplification of the ID target or mutation of the drug binding pocket) acquired resistance to AGX51 will be difficult to achieve and will probably serve to enhance the efficacy of AGX51 in a therapeutic setting.
The observed need for combining AGX51 treatment with chemotherapy to see anti-tumor efficacy in established mammary fat pad and lung tumors reflects the situation seen in genetic analyses where Her2/neu breast tumor regression required chemotherapy in Id knockout mice59. The reasons for this are not yet understood but it may be due to there being very few ID-positive resting stem cells, as has been observed in other models35, requiring combination treatment to hit two tumor populations (ID-positive and ID-negative) to see significant effects. Future studies will be required to resolve this question. We note that the use of cannabidiol (CBD), which reduces Id1 transcription, has been reported to inhibit aggressive breast cancer cell behavior as a single agent67. This may in part reflect broad CBD activity against a variety of factors including β-catenin and protein kinase B (AKT) among others68.
ID protein expression in normal adult tissue stem cells is required in response to various stresses: in colonic stem cells, ID1 is required to minimize inflammation after DSS-induced chemical injury in a model of colitis69; in long-term repopulating hematopoietic stem cells they are required for optimal cellular expansion after ablation of the progenitor pool or to maintain long-term serial transplantation70,71; in neural stem cells, ID1 and ID3 are expressed in quiescent stem cell populations but show pronounced phenotypes only when those cells are called into cell cycle upon AraC ablation of progenitor cells72; ID proteins are expressed at very low levels in the liver but are reactivated during regeneration in response to injury73; ID1 is not expressed in normal human epidermis but may be a key regulator of epidermal wound healing74. These observations likely account for the low toxicity of AGX51 in animals but suggest caution in the use under conditions of duress. In cancer, a similar phenomenon may be taking place since in glioma, ID1, and ID3 appear to be in a resting stem-like cell which resists killing of cycling cells by ionizing radiation35. We have recently observed ID1 associated with a resting state in cholangiocarcinoma and TNBC (Robert Benezra 2020, “ID1 expression in resting cancer stem cells”, unpublished). It will be of great interest to determine if combining standard cytotoxic therapies and AGX51 would kill the ID1-negative cycling population and the ID1-positive stem-like cells called into cycle, respectively.
Importantly, other PPIs have been shown to be critical for ID activity in the stem cell character of other tumors: ID2, for example, facilitates stemness in glioma by its association with the VHL complex and antagonism of the ubiquitin-mediated proteolysis of Hif2α75, so determining the effects of AGX51 on this pathway will be of great interest. Given the complete suppression of gliomagenesis upon loss of Id1, Id2, and Id3 in a murine model76 and the ability of AGX51 to antagonize all members of the family, it is likely that sufficient exposure of these brain tumors to AGX51 will provide some therapeutic benefit. It should be noted that in some cancer subtypes ID proteins may play tumor-suppressive roles. As stated above, in Burkitt’s lymphoma ID3 is frequently mutated and appears to act as a tumor suppressor34. ID4, for example, may be a tumor suppressor in ER+ breast cancer77 and ID2 may reduce invasiveness in TNBC78,79. Thus, the lack of specificity of AGX51 for individual ID family member may complicate its use in certain settings.
In conclusion, the first-in-class small-molecule antagonist of the ID protein family, AGX51, dramatically reduces ID protein levels and cancer cell viability, as well as inhibits tumor growth. The ability of AGX51 to phenocopy the consequences of Id genetic loss suggests that in addition to being a useful biologic tool, AGX51 can serve as a lead compound that can be developed to provide clinical benefit for a variety of Id-related cancers.
Methods
Electrophoretic mobility shift assays
EMSAs were carried out on whole-cell lysates from AGX51-treated cells and the EMSA was performed as described previously36.
Cell lines
The 4T1 murine mammary tumor cell line, MDA-MB-157, MDA-MB-436, MDA-MB-231, MDA-MB-453, MDA-MB-361, BT-474, SK-BR-3, MCF-7, T47-D, and HCT116 were purchased from ATCC (Manassas, VA, USA) and grown in RPMI or DMEM (Dulbecco’s Modified Eagle medium) media supplemented with 10% FBS (fetal bovine serum), 1% penicillin–streptomycin and 1% L-glutamine. MDA-MB-231 cells used in xenograft studies were obtained from NCI NIH (Bethesda, MD). Luciferase-labeled 4T1 cells were described previously80, as were luciferase- and GFP-labeled 4T1 cells81. The 4T1 cells’ overexpression Id1 were derived by transducing cells with pBabe-Id1 plasmids as described previously47. HMLE RAS Twist and HMLE RAS Twist ID1 cells were described previously47 and grown in MEBM (Mammary Epithelial Cell Growth Medium) (Lonza), supplemented with BPE (bovine pituitary extract) (70 μg/mL), hEGF (human epidermal growth factor) (5 ng/mL), hydrocortisone (0.5 μg/mL), insulin (5 μg/mL), GA-1000 (Gentamicin 30 μg/mL and Amphotericin 15 ng/mL), hTransferrin (5 μg/mL), and isoproterenol (6 μM). 4T1 cells were authenticated by short tandem repeat analysis and karyotyping. Other cell lines have not been authenticated since being obtained from their source. The cell lines were not tested for mycoplasma contamination recently but routine testing had been negative for years prior and no growth defects associated with such contamination have been observed in any of the cell lines used in the studies presented.
Patient-derived xenograft cell lines BR7, BR11, and IBT
PDXs were established directly from breast cancer bone metastases specimens surgically resected from patients with informed consent (IRB protocol #97-094). BR7 and BR11 specimens are obtained from ER-positive (HER2-negative and PR-negative) metastatic breast cancer patients, while the IBT specimen was obtained from a metastatic, TNBC patient. Fresh tumor tissues were quickly washed with ice-cold PBS and minced into about 1 mm3 pieces in MEM medium (without FBS) using sterile razor blades. A fraction of the minced original tumor tissues was incubated with collagenase/hyaluronidase enzyme mix (1000 Units, Voden Medical) in MEM medium without FBS (5 mL per 250 mg tissue) for 2–4 h. The dissociated tumor tissues were then filtered through a 70 μm nylon filter, cells were concentrated by centrifugation at room temperature and seeded to derive respective primary cell cultures in MEM medium with 3% FBS (Sigma). The primary cell cultures were transduced with the fluorescent td tomato-/EGFP-luciferase fusion protein expressing lentiviral vectors for 18–24 h, the primary cells were then maintained in MEM media supplemented with 3% FBS, 1% penicillin–streptomycin and 1% L-glutamine. Aliquots of the primary cell cultures were also cryopreserved following a minimal number (3–4) of in vitro passages.
E protein knockout cell experiments
DN3/4 thymocytes were isolated from male and female E2Afl/fl HEBfl/fl E8III-cre mice50,51 by bead-based negative selection. Briefly, single-cell suspensions of total thymocytes were stained with biotinylated antibodies against CD4, CD8, CD11b, CD11c, CD16/32, CD19, CD44, c-Kit, B220, Ly6C, GR-1, NK1.1, TER119, and TCRgd. Antibody-labeled cells were then depleted with SPHERO Streptavidin-coated magnetic beads (Spherotech, cat. SVM-40-10) using the Dynabeads biotin binder beads negative isolation protocol (Invitrogen). Purified DN3/4 cells were co-cultured on an OP9-DL1 monolayer at 5.0 × 105 cells/mL in MEM alpha supplemented with 10% FBS, 1× penicillin/streptomycin and 5 ng/mL recombinant IL-7 with 60 μM AGX-51 or DMSO vehicle. After 24 h, DN3/4 cells were harvested and RNA was purified using the Direct-Zol microprep kit (Zymo Research, cat. R2061). cDNA was generated using SuperScript III reverse transcriptase (Invitrogen) with random hexamers according to the manufacturer’s protocol. qPCR was performed using PowerTrack Sybr Green Master Mix (Invitrogen) with an Eppendorf Mastercycler ep realplex thermal cycler. Expression was calculated relative to 0 μM AGX-51 and normalized to actin.
The primers used for qPCR are as follows: Id2-F: CGACCCGATGAGTCTGCTCTA; Id2-R: GACGATAGTGGGATGCGAGTC; Id3-F: GCCTCTTAGCCTCTTGGACG; Id3-R: GTTCCGGAGAGAGCTCAGC; actin-F: AAGGCCAACCGTGAAAAGAT; actin-R: GTGGTACGACCAGAGGCATAC.
Pancreatic ductal adenocarcinoma cells and organoids
The human pancreatic cancer cell line Panc1 was obtained from ATCC in 2012, and A21 was obtained in 2016 from Dr. Christine Iacobuzio-Donahue82. Mouse pancreatic cancer lines 806 (KrasG12D; Ink4a−/−; Smad4−/−) and NB44 (KrasG12D; Ink4a−/−) were obtained from Dr. Nabeel Bardeesy, and 4279 (KrasG12D; Ink4a−/−) was generated in Dr. Joan Massague’s laboratory. Mouse pancreatic organoid cell lines T7 and T8 were obtained from Dr. David Tuveson83. Pancreatic spheroids were grown in Ultra Low Attachment Culture plates (Corning) in DMEM supplemented with Glutamax (2 mM) and heparin (5 µg/mL). Pancreatic organoids were embedded in Matrigel with Advanced DMEM/F12 (Gibco, 12634-028) supplemented with B-27 (Life Technologies, 12587-010), HEPES (10 mM), 50% Wnt/R-spondin/Noggin-conditioned medium (ATCC, CRL-3276), Glutamax (Invitrogen, 2 mM), N-acetyl cysteine (Sigma, 1 mM), nicotinamide (Sigma, 10 mM), epidermal growth factor (Peprotech, 50 ng/mL), gastrin (Sigma, 10 nM), fibroblast growth factor-10 (Peprotech, 100 ng/mL), A83-01 (Tocris, 0.5 µM) as previously described83. All cell lines and organoids were maintained at 37 °C and 5% CO2. Cell viability was measured using Cell Titer-Glo (Promega) according to manufacturer’s instructions.
Generating Id knockdown and overexpression cell lines
To generate 4T1 cells containing inducible short hairpins against Id1 or Id3, short hairpins against Id1 (TGCTGTTGACAGTGAGCGAGCGTGTTTCTGTTTTATTGAATAGTGAAGCCACAGATGTATTCAATAAAACAGAAACACGCGTGCCTACTGCCTCGGA) and Id3 (TGCTGTTGACAGTGAGCGCGCCCTGATTATGAACTCTATATAGTGAAGCCA CAGATGTATATAGAGTTCATAATCAGGGCATGCCTACTGCCTCGGA) were cloned into the tet-regulated all-in-one retroviral miR-E expression vector RT3REVIR84. Retroviral particles were produced by lipofectamine-mediated transfection into 293 GP2 cells. Forty-eight hours after transfection, virus-containing media was collected, filtered and added to 4T1 cells in the presence of 8 μg/mL polybrene. Transduced cells were selected for Venus positivity by FACS. To induce short hairpin expression cell culture media was supplemented with doxycycline (5 μg/mL). RT3REVIR expressing a short hairpin against Renilla was used as a control (shCTL) in experiments.
Human cDNA ID3 (hId3) or ID1 (hId1) was cloned into the lentiviral vector pWPI, obtained from Addgene catalog number: 12254 (addgene.org). A day before transfection, 293T cells were seeded in DMEM media on poly-L-lysine-treated plates to achieve 90% confluency then 18.5 μg of psPax2, 12 μg of VSVG, and 152.4 μL of CaCl2 (2 M) were mixed and brought to a volume of 600 μL with molecular biology-grade water; 600 μL of 2× HBS was added, mixed and incubated for 60 s. Transfection mix was added to 8.8 mL of pre-warmed D10 media (DMEM, 10% FBS, 1% penicillin and streptomycin, 1% L-glutamine, 1% 100 mM sodium pyruvate, 1% sodium bicarbonate) and added to 293T cells. Plates were incubated at 37° C overnight. On day 3, ~12 h post transfection, cells were rinsed with D10 media before incubating in D10 media with 50 μg/mL G418. At ~16 h post transfection, media was replaced with VPM media (Ultraculture media, 1% penicillin and streptomycin, 1% L-glutamine, 1% 100 mM sodium pyruvate, 1% sodium bicarbonate, 5 mM sodium butyrate). On day 4, ~46 h post transfection, viral supernatants were collected into 15 mL tubes, centrifuged at 1100 rpm for 5 min, passed through 0.40 μM filters into clean tubes and aliquoted for storage at −8 °C until transduction.
Infection and western blot analysis
One day before infection, 4T1 cells were plated in DMEM media in 10 cm dishes for 50% confluency the next day. On day 2, 1 mL of viral particles and 8 μL polybrene (10 mg/mL stock) were added to 9 mL of DMEM media, mixed and added to the corresponding dish of 4T1 cells. After overnight incubation, media on infected plates was changed. On day 4 or 5, 4T1 cells transduced with lentiviral particles carrying either pWPI-hId3, pWPI-hId1, or pWPI were subjected to FACS sorting for a high GFP intensity. FACS sorted cells were collected and cultured for western blotting analysis. 4T1-pWPI-hId3 cells, 4T1-pWPI-hId1 cells, or 4T1-pWPI cells were analyzed for the expression of exogenous Id3 protein by western blot. Cells were lysed using the protein extraction buffer containing protease inhibitors (Roche, ref: 04 693 132 001) and 35–40 μg of the total protein lysate was loaded per lane of 15% SDS-PAGE. ID3 protein was detected using anti-ID3 antibodies with the dilution of 1:1000 using the Li-CoR system.
Immunoblotting
For immunoblotting, cells were collected by trypsinization, washed with PBS and lysed in homogenization buffer (0.3 M sucrose, 10 mM Tris (pH 8.0), 400 mM sodium chloride, 3 mM magnesium chloride, 0.5% NP40/IGEPAL, 100 μg/mL Aprotinin+ Protease Inhibitor Cocktail (Roche # 11 836 153 001). Proteins were separated by SDS-PAGE, transferred to a membrane (LI-COR), probed with primary antibodies overnight at 4 °C, and probed with secondary antibodies (LI-COR) for 1–2 h at room temperature. Proteins were visualized using the LI-COR Odyssey Infrared Imaging detection system. The following primary antibodies were used: Id1, Id2, Id3, Id4 (195-14, 9-2-8, 17-3, 82-12, respectively, all from Biocheck, all used at 1:1000), Phospho-Histone H3 (9701, Cell Signaling, 1:500), Cyclin D1 (2978, Cell Signaling, 1:1000), Beta-catenin (9562, Cell Signaling, 1:1000), Cdk4 (sc-260, Santa Cruz, 1:200), Mcl1 (5453, Cell Signaling, 1:500), Ube2C (14234, Cell Signaling, 1:500), Sqstm1 (H00008878-M01, Abnova, 1:1000), Alpha-catenin (C2081, SIGMA-ALDRICH, 1:5000), E-cadherin (3195 Cell Signaling, 1:1000), Vimentin (5741, Cell Signaling, 1:1000), Snail (3879, Cell Signaling, 1:1000), Twist (ab50887, Abcam, 1:200), Zeb1 (NBP1-05987, Novus Biologicals, 1:2000), Actin (A2066, SIGMA, 1:10,000), Tubulin (T4026, SIGMA, 1:10,000). Western blot quantification was carried out using channel 700 and channel 800 intensity data from Odyssey application software version 3.0.30 (LI-COR), subtracting blank values and normalizing to tubulin. All individual blots were derived from the same experiment and processed in parallel.
qRT-PCR
RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA, USA) and cDNA was generated from 1 μg of RNA using SuperScript IV First-Strand Synthesis System (Invitrogen, Grand Island, NY, USA). Quantitative PCR was performed using SYBR Green QuantiTect Primer Assay (Qiagen) according to manufacturer’s instructions in a 7900HT Fast-Real Time PCR System Instrument (Applied Biosystems, Grand Island, NY, USA). Primer pairs for the individual genes were obtained from the bioinformatically validated QuantiTect Library and are as follows: Id1 (QT01743756), Actb (QT00095242), Ctnnb1 (QT00160958), Ccnd1 (QT00154595), Cdk4 (QT00103292), Mcl1 (QT00107436), Idh2 (QT00147301), Idh3b (QT00117341), Nbas (QT00280483), Ifrd1 (QT00093401), Atp5h (QT00175070), C1qbp (QT01057777), Cox5a (QT00164234), Dlst (QT00163275), Glud1 (QT00103411), Vim (QT00159690). The fold changes in gene expression were calculated using the delta-delta CT method.
Cell viability assays
Alamar blue cell viability assay (Invitrogen) was carried out according to manufacturer’s instructions. Briefly, 5000 4T1 cells were seeded in a 96-well plate, the next day they were treated with 40 μM AGX51 for 24 h, after which point a 1:10 dilution of Alamar blue cell viability reagent was added to the cells and absorbance was measured 2, 3, 4, 5, and 6 h after addition of the reagent using a plate reader (Synergy 2, BioTek).
For IC50 determination, cancer cell lines were seeded in a 96-well plate (5000 cells per well). After overnight incubation, cells were treated with AGX51 (0, 5, 10, 20, 40, 60 μM) and incubated for 24, 48, and 72 h, each condition was done in triplicate. At each time point, 40 µL of MTT reagent (5 mg/mL) was added per well and the cells were incubated for 4 h. Following incubation, media was aspirated and 200 µL DMSO was added per well. Absorbance was then measured at 570 nm using a plate reader (Synergy 2, BioTek). Data were analyzed and visualized using GraphPad Prism.
To determine whether vitamin E could rescue AGX51 cell viability effects, cells were pretreated with vitamin E (( ± )-α-Tocopherol, SIGMA) (50 or 100 µM) for 1 h. Following the 1 h treatment, cells were treated with 40 µM AGX51/DMSO for 24 h. Cell viability was then determined by trypan blue exclusion and cell counting.
To measure ferroptosis, 4T1 cells were seeded in 150 μL DMEM media per well of a 96-well plate and treated with vehicle, 1 μM erastin (Millipore Sigma, 329600), 1 μM ferrostatin-1 (Xcessbio, M60042-2), 40 μM AGX51, 1 μM erastin + 1 μM ferrostatin, and 40 μM AGX51 + 1 μM ferrostatin-1. Celltiter-Glo (Promega G8461) was used to determine cell viability using the manufacturer’s instructions. Ferroptosis was observed 12 h after treatment.
To measure necroptosis, 4T1 cells were seeded in 150 μL DMEM media per well of a 96-well plate and treated with vehicle, 40 μM AGX51, 10 ng/mL TNF (R&D Systems, 410-MT-010) + 20 μM z-VAD-FMK (Enzo Life Sciences, ALX-260-020-M001), 30 μM necrostatin-1 (Millipore Sigma, 480065-5MG), 10 ng/mL TNF + 20 μM z-VAD-FMK + 30 μM necrostatin-1, and 40 μM AGX51 + 30 μM necrostatin-1. Celltiter-Glo (Promega G8461) was used to determine cell viability using the manufacturer’s instructions. Necroptosis was observed 12 h after treatment.
Cell proliferation of the AGX51-resistant clone (24-13) was determined by plating 60,000 24-13 or 4T1 cells and collecting and counting the cells on days 1, 3, 5, and 7 after plating using a hemocytometer with the addition of trypan blue to exclude dead cells.
To assess synergistic effect of AGX51 and Paclitaxel (PCT) in vitro, MDA-MB-231 cells were grown in two replicates in DMEM (Dulbecco’s Modified Eagle medium) media supplemented with 10% FBS, 1% L-glutamine and 1% penicillin–streptomycin. When cells reached confluency suitable (40–50%) for drug treatment in 48 h, the cells were treated with DMSO (vehicle), or predetermined optimum PCT concentration (20 nM) alone or various concentrations of AGX51, or combination of various concentrations of AGX51 with PCT for 24 h. The cell numbers were counted using Trypan Blue exclusion to assess the effect of the drugs.
Cell cycle analysis
Cells were treated with AGX51 or DMSO, or vitamin E (as described above), collected by trypsinizaton, washed with 1× PBS, resuspended in 500 μL 1× PBS and then diluted with 6 mL 70% ethanol and stored at −20 °C until analysis. For cell cycle analysis cells were centrifuged 1000 rpm for 5 min, washed with 1× PBS and then resuspended in 0.5 mL PI/RNase staining buffer (550825, BD Biosciences), incubated for 15 min at room temperature and analyzed by flow cytometry (LSR II).
AGX51- or DMSO-treated cells were stained with 10 μM BrdU reagent (550891, BD Biosciences) for 1 h. The cells were then collected, resuspended in 70% ethanol, washed with 1× PBS, incubated with 2 M HCl, washed in 1× PBS and 1× PBS-Tween-BSA, and then incubated with an Alexa Fluor 488 anti-BrdU antibody (558599, BD Biosciences) for 15 min and finally resuspended in RNase/PI buffer (550825, BD Biosciences) and analyzed by flow cytometry.
Annexin/PI staining
To detect apoptosis and necrosis we used the BioVision Annexin V-FITC Apoptosis detection kit according to the manufacturer’s instructions. Briefly, 4T1 cells were treated with 40 μM AGX51, trypsinized and resuspended in 1× Binding Buffer to which 5 μL Annexin V-FITC and 5 μL propidium iodide (PI) (50 μg/mL) were added. The cells were incubated for 5 min protected from light and then analyzed by flow cytometry (LSR II).
Measuring ROS
4T1 cells were treated with 40 μM AGX51 for 24 h, and then 10 μM 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate (H2DCFDA) (Life Technologies) was added to the cells and they were incubated for 1 h. Cells were washed twice with PBS, trypsinized and resuspended in PBS–5% FBS. H2DCFDA is converted to fluorescent DCF upon oxidization, and serves as a readout for ROS generation. Fluorescence was measured using flow cytometry (LSR II). As a positive control for ROS production, cells were treated with 100 μM of H2O2.
Testing the effects of AGX51 on quiescent cells
To induce a senescent state prior to AGX51 treatment, 4T1 cells were plated to reach high density at the time of AGX51 treatment. The day after plating the cells, media was changed to low-serum media (1% FBS). After 24 h in low-serum media, the cells were treated with AGX51. Cells plated at standard density and maintained in 10% FBS served as controls. Following the 24 h of AGX51 treatment, cell numbers were determined by cell counting and trypan blue exclusion and cells collected for whole-cell lysate extraction for western blot analyses.
Generation of SILAC cell lines, and in-gel digestion for mass spectrometry
4T1 cells were grown in DME media supplemented with 10% FBS and penicillin and streptomycin, and either unlabeled L-arginine (Arg0) and L-lysine (Lys0) at 50 mg/L or equimolar amounts of the isotopic variants [U-13C6,15N4]-L-arginine (Arg10) and [U-13C6]-L-lysine (Lys6) (Cambridge Isotope Laboratories). After five cell doublings, cells were >99% labeled with the isotopes, as determined by mass spectrometry. Cell lysates were prepared as described above; 1 mg of total protein from each light and heavy labeled cell line was mixed and the total 2 mg of protein was fractionated using 4–12% gradient SDS-PAGE. Each gel lane cut in equal pieces and gel slices were washed with 1:1 (Acetonitrile: 100 mM ammonium bicarbonate) for 30 min. Gel slices were then dehydrated with acetonitrile for 10 min until gel slices shrunk and excess acetonitrile was removed before slices were dried in speed-vac for 10 min without heat. Gel slices were reduced with 5 mM DTT for 30 min at 56 °C in an air thermostat, then chilled to room temperature, and alkylated with 11 mM IAA for 30 min in the dark. Gel slices were washed with 100 mM ammonium bicarbonate and acetonitrile for 10 min each. Excess acetonitrile was removed and dried in a speed-vac for 10 min without heat and gel slices were rehydrated in a solution of 25 ng/μL trypsin in 50 mM ammonium bicarbonate on ice for 30 min. Digestions were performed overnight at 37 °C in an air thermostat. Digested peptides were collected and further extracted from gel slices in extraction buffer (1:2 v/v) 5% formic acid/50% acetonitrile) at high-speed shaking in an air thermostat. Supernatant from both extractions were combined and dried down in a vacuum centrifuge. Peptides were desalted with C18 resin-packed stage-tips. Desalted peptides were lyophilized and stored at −80 °C until further use.
LC-MS/MS and whole-proteome analysis
Desalted peptides were dissolved in 3% acetonitrile/0.1% formic acid and were injected onto a C18 capillary column on a nano ACQUITY UPLC system (Waters) that was coupled to the Q Exactive plus mass spectrometer (Thermo Scientific). Peptides were eluted with a non-linear 200 m gradient of 2–35% buffer B (0.1% (v/v) formic acid, 100% acetonitrile) at a flow rate of 300 nL/min. After each gradient, the column was washed with 90% buffer B for 5 min and re-equilibrated with 98% buffer A (0.1% formic acid, 100% HPLC-grade water) for 4 min. MS data were acquired with an automatic switch between a full scan and 10 scan data-dependent MS/MS scan (TopN method). Target value for the full scan MS spectra was 3 × 106 charges in the 380–1800 m/z range with a maximum injection time of 30 ms and resolution of 70,000 at 200 m/z in profile mode. Isolation of precursors was performed with 1.5 m/z. Precursors were fragmented by higher energy C-trap dissociation (HCD) with a normalized collision energy of 27 eV. MS/MS scans were acquired at a resolution of 17,500 at 200 m/z with an ion target value of 5 × 104 maximum injection time of 60 ms and dynamic exclusion for 15 s in centroid mode. A total of 6346 proteins (5604 clusters) with an FDR < 1% were identified across the three independent pairs of samples analyzed (4 h treatment of 4T1 cells with AGX51 or DMSO vehicle control). The data were analyzed using Scaffold Q+ version 4.4.5 (Proteome Software, Portland, Oregon, USA). Briefly, fold differences of the fold change ratios were calculated for each pair of samples (AGX51 vs. DMSO) and proteins with fold differences (at least 1.35-fold) in the same direction of change (overexpressed or underexpressed) in each of the three pairs were identified (see Supplementary Table 2).
Animal studies
Animal studies were carried out in accordance with institutional regulations (IACUC protocol 06-10-025) in a non-blinded fashion. Weight measurements and standard blood analyses were carried out on 8–12-week-old male CD1 mice (n = 5 per treatment group) dosed q5d with DMSO, 15 mg/kg qd paclitaxel, 60 mg/kg AGX51 bid, or a combination of paclitaxel and AGX51. Blood samples were collected by retro-orbital puncture on day 6, 12 h after the last dose. Plasma was analyzed for clinical chemistry (albumin, alkaline phosphatase, alanine aminotransferase, amylase, total bilirubin, blood urea nitrogen, calcium, phosphorus, creatine, glucose, sodium, potassium, total protein, and globulin) and hematology (white blood cell, lymphocyte, monocyte, granulocyte, red blood cell, hemoglobin, hematocrit, and platelet) concentrations.
Orthotopic mammary fat pad tumors were generated by injecting 5 × 106 MDA-MB-231 cells (in 1:1 PBS:Matrigel) into the right caudal mammary fat pad of 8–12-week-old, female athymic nu/nu mice (n = 30). Mice were obtained from Simonsen Laboratories (Gilroy, CA). Tumors were allowed to grow to ~100 mm3, at which point they were divided into 6 groups of 5 mice with approximately the same tumor burden and treatment was initiated. Group 1 was vehicle (DMSO) control, group 2 received 5 days of 15 mg/kg paclitaxel, group 3 received 19 days of 60 mg/kg AGX51 bid, group 4 received a combination of paclitaxel and 6.7 mg/kg AGX51 bid, group 5 received a combination of paclitaxel and 20 mg/kg AGX51 bid, group 6 received a combination of paclitaxel and 60 mg/kg bid, and group 7 received a combination of paclitaxel and 60 mg/kg AGX51 but only received AGX51 for 7 days (all other AGX51 groups had 19 days of AGX51 treatment). The AGX51 and paclitaxel were administered i.p. Tumor volumes were determined throughout the study using a digital caliper and the formula: tumor volume = ½ (length × width2) where the greatest longitudinal diameter is the length of the tumor and the greatest transverse diameter is the width. At study termination the mice were sacrificed by cervical dislocation.
Lung metastases were generated by injecting 6–8-week-old, female, Balb/c mice with 50,000 luciferase-labeled 4T1 cells into the tail vein. Twenty-four hours after tail vein injections, mice were treated with DMSO vehicle or 50 mg/kg AGX51 qd (5 mice per treatment group) by i.p. injection. One mouse in the AGX51-treatment group died from the isoflurane anesthesia during the first luciferase imaging session. In another experiment mice were treated bid. The bid experiment was performed three times. In the first experiment n = 5 mice per group, in the second experiment n = 4 vehicle-treated mice and n = 8 AGX51-treated mice, and in the third experiment n = 5 mice per group. No statistically significant differences in the effect was observed across experiments. Development of lung metastases was monitored using the IVIS-200 in vivo imaging system. In the experiment testing the effects of AGX51 on established lung metastases (n = 5 mice per group), once evidence of lung metastases was observable by in vivo imaging, mice were divided into 4 groups of 5 mice with approximately the same tumor burden per treatment group. The groups were: group 1, vehicle; group 2, 50 mg/kg AGX51 bid; group 3, 15 mg/kg paclitaxel qd for 5 days; and group 4, combination of AGX51 + paclitaxel. At the end of the experiments mice were euthanized and tissues were collected for further analyses. Lung tumor burden was quantified in a blinded fashion by a pathologist.
To assess effects of AGX51 on seeding of the lungs, 20 6–8-week-old female Balb/c mice were injected with GFP-labeled 4T1 cells as described above and on the next day treated with AGX51 (50 mg/kg) (n = 10) or DMSO (n = 10). Twenty-four hours later, one set of mice (5 from the AGX51 group and 5 from the DMSO group) was euthanized and their lungs collected for histological analysis. After 48 h of treatment, the remaining 10 mice were euthanized and their lungs collected for histological analysis.
To assess whether in vivo treatment results in acquired resistance to AGX51, 10 6–8-week-old female Balb/c mice were tail vein-injected with GFP-labeled 4T1 cells and treated with AGX51 or DMSO for 2 weeks as described above. After 2 weeks of treatment, the mice were euthanized and the lungs were dissociated according to the kit instructions (Mouse Lung Dissociation Kit, MACS Miltenyi Biotec) and the cells were sorted for the GFP-positive fraction. The cells were allowed to expand to generate enough cells for AGX51 treatment and western blot analysis, at which point the cells were harvested and whole-cell lysates were made.
Spontaneous colon tumors were induced by treating 30, 4-week-old male A/J mice (Jackson Laboratory) with AOM (10 mg/kg; Sigma Aldrich) once a week by i.p. injection for 6 weeks. Mice were maintained on AIN-93G purified diet (Research Diets) for the duration of the experiment. After a 3-week treatment break, mice were treated i.p. with DMSO (n = 13) or AGX51 (n = 11) (15 mg/kg) bid for 3 weeks. Following the last injection, the mice were euthanized and colon tumors were formalin fixed to assess tumor burden. Tumor numbers and size were determined in whole mounts of the tissues following methylene blue staining.
All tumor growth and treatment studies complied with MSKCC Institutional Guidelines and received IACUC approval.
Immunohistochemistry
Slides mounted with sections from formalin-fixed and paraffin-embedded mouse tissues were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E), as well as with the following primary antibodies: Id1 (37-2, Biocheck 1:1000), Vimentin (5741, Cell Signaling 1:500), Cyclin D1 (2978, Cell Signaling 1:500). Detection was performed using the Vectastain Elite ABC kit (Vector), diaminobenzidine was used as the chromogen, and Harris Modified Hematoxylin was used as the nuclear counterstain.
Immunofluorescence
The lungs from mice from the seeding experiment were perfused with 1× PBS, followed by 4% PFA. The lungs were then removed, washed in cold PBS, and fixed in 4% PFA overnight. The lungs were dehydrated in a sucrose solution series (20–30%) and then embedded in OCT and flash frozen. Ten micron cryosections were then boiled for 10 min in citrate buffer, for antigen retrieval, washed with 1× PBS, blocked in PBS, 10% goat serum, 0.05% triton and then incubated with anti-GFP antibody (GFP-1020, Aves labs, Inc.) overnight at 4°. The next day the slides were washed and incubated with Alexa Fluor 555 anti-chicken antibody (A-21437, Thermo Fisher Scientific), washed and counterstained with DAPI followed by mounting and coverslip application. The slides were scanned and ten random fields from each section were analyzed for GFP-positive cell content.
AGX51-resistant clone generation
To generate resistant cell lines, cells were maintained in AGX51-containing media (20–40 μM range). AGX51-containing media was replaced every 2–3 days. Any clones that grew out were expanded, and maintained in AGX51-containing media. Transiently resistant clones expressing ID1/ID3 had their cDNA sequenced for the coding regions of ID1 and ID3 by Sanger sequencing using the primers:
Id1-F:CTTCTTGTTCTCTTCCCACA
Id1-R:GATCAAACCCTCTACCCACT
Id3-F:CACTGTTTGCTGCTTTAGGT
Id3-R:CGTTGAGTTCAGGGTAAGTG
Statistical analyses
Three replicates were generally used for each experimental condition for in vitro experiments and 5 mice per group were typically used in each mouse experiment. The sample sizes were determined based on an expected large effect size. With 3 replicated per condition, an effect size as small as 3 can be detected with 80% power at a two-sided significance level of 0.05 using a two-sample t-test. With 5 mice per group, an effect size as small as 2 can be detected with 80% power at a two-sided significance level of 0.05 using a two-sample t-test. Additional experiments may be performed when larger variation in data was observed and data were pooled for analysis. In general, Welch’s t-test was used to examine differences between two groups. ANOVA was used to examine differences across multiple experimental groups. Data may be transformed to ensure the underlying normality assumptions were met. For example, for tumor number data, square root transformation was used. Weighted linear regression analysis was used when heteroscedasticity was observed and data points in each group were typically weighted by the reciprocal of the standard deviation of data in each group. For data pooled from multiple experiments, the model included both experiments and experiments by treatment group interaction as covariates to account for potential differences in experiments. Significance of linear contrasts of interest was assessed based on estimates obtained from the weighted least squares. Q–Q plot of the residuals was examined to ensure the underlying model assumptions were met. p-value < 0.05 was considered statistically significant. For the colon tumor counts, the data were analyzed using the Wilcoxon rank sum test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors are grateful to members of the Benezra Lab, past and present, for helpful discussions throughout the course of this work. We are also grateful to Andrea Rizzi for additional supportive modeling work of AGX51 interactions. We thank the MSKCC Molecular Cytology Core Facility for assistance in image analyses and the MSKCC Flow Cytometry Core Facility for assistance with flow cytometry. R.B. gratefully acknowledges support from the Sloan Kettering Institute and NIH grant P30 CA008748. R.B. is also supported by grants from the NCI (PO1 CA094060) and The Breast Cancer Research Foundation. A.J.D. is supported by funding from the Breast Cancer Research Foundation and the Botwinick-Wolfensohn foundation in memory of Mr. and Mrs. Benjamin Botwinick. Work was partially supported by 1R43CA150448 01 from NCI/NIH to WAG and National Institute of Health grants to A.I. (R01CA101644 and R01CA131126). P.M.W. is a recipient of a Postdoctoral Training Award from the Fonds de recherche du Quebec – Sante (FRQS). M.G.E. was supported by fellowships issued by the Valencian Government of Spain (GVA) and the European Social Fund (ACIF/2018/004 and BEFPI/2018/029).
Author contributions
Conceptualization, R.B; methodology, P.M.W., R.K.S., J.P., D.C.M., and R.B.; formal analysis, P.M.W., R.K.S., J.P., D.C.M., and X.K.Z.; investigation, P.M.W., M.G.E., B.D., Y.C., R.S., S.X., S.Y., S. Y., Y.Z., J.R., R.K.S., J.P., and D.C.M.; resources, J.H.H. and V.K.; writing – original draft, P.M.W. and R.B.; writing – review & editing, R.B., P.M.W., D.C.M., A.D. A.J.D., and M.G.E.; visualization, P.M.W. and R.B.; supervision, O.O., L.N., N.R., A.I., A.L., and R.B.; funding acquisition, A.I., A.J.D., and R.B.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1422998685. The proteomics data of anti-tumor effects of an ID antagonist with no observed acquired resistance are openly available in the PRIDE Archive via the following accession: https://identifiers.org/pride.project:PXD02459386. The raw data underlying the following figures are openly available as part of the data record: 1E, 2A, 3B, C, D, 4A, E, 5A, B, F, G, 6B, S1A, D, F, G, S2E, I, J, S5A, B, S6, S7A, S8C. A comprehensive list of which file is associated with each figure is also included in the Excel spreadsheet ‘Wojnarowicz_et_al_underlying_data_files_list.xlsx’. All uncropped western blots are in the Supplementary Information section.
Competing interests
R.B. and W.A.G. own shares of Angiogenex, Inc., which holds the patent for the use and development of AGX51. R.B. is the Chair of the Scientific Advisory Board (SAB) of Angiogenex, Inc. and a Board Member, both positions unpaid. O.O. is listed as an inventor and receives royalties from Patents that were filed by MSKCC. O.O. is an unpaid Member of the SAB of Angiogenex, Inc. and owns shares in Angiogenex, Inc.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-021-00266-0.
References
- 1.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 2.Ling F, Kang B, Sun XH. Id proteins: small molecules, mighty regulators. Curr. Top. Dev. Biol. 2014;110:189–216. doi: 10.1016/B978-0-12-405943-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 3.Wang LH, Baker NE. E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev. Cell. 2015;35:269–280. doi: 10.1016/j.devcel.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyden D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 5.Ruzinova MB, et al. Effect of angiogenesis inhibition by Id loss and the contribution of bone-marrow-derived endothelial cells in spontaneous murine tumors. Cancer Cell. 2003;4:277–289. doi: 10.1016/S1535-6108(03)00240-X. [DOI] [PubMed] [Google Scholar]
- 6.Perk J, et al. Reassessment of Id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-Id1 antibody. Cancer Res. 2006;66:10870–10877. doi: 10.1158/0008-5472.CAN-06-2643. [DOI] [PubMed] [Google Scholar]
- 7.Schoppmann SF, et al. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int. J. Cancer. 2003;104:677–682. doi: 10.1002/ijc.11009. [DOI] [PubMed] [Google Scholar]
- 8.Wazir U, Jiang WG, Sharma AK, Newbold RF, Mokbel K. The mRNA expression of inhibitors of DNA binding-1 and -2 is associated with advanced tumour stage and adverse clinical outcome in human breast cancer. Anticancer Res. 2013;33:2179–2183. [PubMed] [Google Scholar]
- 9.Lee KT, et al. Overexpression of Id-1 is significantly associated with tumour angiogenesis in human pancreas cancers. Br. J. Cancer. 2004;90:1198–1203. doi: 10.1038/sj.bjc.6601684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maw MK, Fujimoto J, Tamaya T. Expression of the inhibitor of DNA-binding (ID)-1 protein as an angiogenic mediator in tumour advancement of uterine cervical cancers. Br. J. Cancer. 2008;99:1557–1563. doi: 10.1038/sj.bjc.6604722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindl M, et al. Overexpression of Id-1 protein is a marker for unfavorable prognosis in early-stage cervical cancer. Cancer Res. 2001;61:5703–5706. [PubMed] [Google Scholar]
- 13.Zhang N, et al. Id1 deficiency protects against tumor formation in ApcMin/+ mice but not in a mouse model of colitis-associated colon cancer. Cancer Prev. Res. 2015;8:303–311. doi: 10.1158/1940-6207.CAPR-14-0411. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Zhao Z-R, et al. Overexpression of Id-1 protein is a marker in colorectal cancer progression. Oncol. Rep. 2008;19:419–424. [PubMed] [Google Scholar]
- 15.Takai N, Miyazaki T, Fujisawa K, Nasu K, Miyakawa I. Id1 expression is associated with histological grade and invasive behavior in endometrial carcinoma. Cancer Lett. 2001;165:185–193. doi: 10.1016/S0304-3835(01)00433-5. [DOI] [PubMed] [Google Scholar]
- 16.Takai N, Ueda T, Nishida M, Nasu K, Miyakawa I. The relationship between oncogene expression and clinical outcome in endometrial carcinoma. Curr. Cancer Drug Targets. 2004;4:511–520. doi: 10.2174/1568009043332871. [DOI] [PubMed] [Google Scholar]
- 17.Han S, et al. Expression and significances of Id1 helix-loop-helix protein overexpression in gastric cancer. Cancer Lett. 2004;216:63–71. doi: 10.1016/j.canlet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 18.Yang H-Y, et al. Expression and prognostic values of Id-1 and Id-3 in gastric adenocarcinoma. J. Surgical Res. 2011;167:258–266. doi: 10.1016/j.jss.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Anido J, et al. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Vandeputte DA, et al. Expression and distribution of id helix-loop-helix proteins in human astrocytic tumors. Glia. 2002;38:329–338. doi: 10.1002/glia.10076. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda Y, et al. Overexpressed Id-1 is associated with a high risk of hepatocellular carcinoma development in patients with cirrhosis without transcriptional repression of p16. Cancer. 2005;104:1037–1044. doi: 10.1002/cncr.21259. [DOI] [PubMed] [Google Scholar]
- 22.Schindl M, et al. Level of Id-1 protein expression correlates with poor differentiation, enhanced malignant potential, and more aggressive clinical behavior of epithelial ovarian tumors. Clin. Cancer Res. 2003;9:779–785. [PubMed] [Google Scholar]
- 23.Coppe JP, Itahana Y, Moore DH, Bennington JL, Desprez PY. Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin. Cancer Res. 2004;10:2044–2051. doi: 10.1158/1078-0432.CCR-03-0933. [DOI] [PubMed] [Google Scholar]
- 24.Forootan SS, et al. Increased Id-1 expression is significantly associated with poor survival of patients with prostate cancer. Hum. Pathol. 2007;38:1321–1329. doi: 10.1016/j.humpath.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P, Patel D, Chaudhary J. Id1 and Id3 expression is associated with increasing grade of prostate cancer: Id3 preferentially regulates CDKN1B. Cancer Med. 2012;1:187–197. doi: 10.1002/cam4.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, et al. Expression and prognostic relevance of Id1 in stage III esophageal squamous cell carcinoma. Cancer Biomark. A. 2010;8:67–72. doi: 10.3233/DMA-2011-0831. [DOI] [PubMed] [Google Scholar]
- 27.Castanon E, et al. Id1 and Id3 co-expression correlates with clinical outcome in stage III-N2 non-small cell lung cancer patients treated with definitive chemoradiotherapy. J. Transl. Med. 2013;11:13. doi: 10.1186/1479-5876-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M-S, Park T-I, Lee Y-M, Jo Y-M, Kim S. Expression of Id-1 and VEGF in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2013;6:2102–2111. [PMC free article] [PubMed] [Google Scholar]
- 29.Ponz-Sarvise M, et al. Inhibitor of differentiation-1 as a novel prognostic factor in NSCLC patients with adenocarcinoma histology and its potential contribution to therapy resistance. Clin. Cancer Res. 2011;17:4155–4166. doi: 10.1158/1078-0432.CCR-10-3381. [DOI] [PubMed] [Google Scholar]
- 30.Ciarrocchi A, Piana S, Valcavi R, Gardini G, Casali B. Inhibitor of DNA binding-1 induces mesenchymal features and promotes invasiveness in thyroid tumour cells. Eur. J. Cancer. 2011;47:934–945. doi: 10.1016/j.ejca.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Kebebew E, et al. Id1 gene expression is up-regulated in hyperplastic and neoplastic thyroid tissue and regulates growth and differentiation in thyroid cancer cells. J. Clin. Endocrinol. Metab. 2004;89:6105–6111. doi: 10.1210/jc.2004-1234. [DOI] [PubMed] [Google Scholar]
- 32.Tang R, et al. High Id1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood. 2009;114:2993–3000. doi: 10.1182/blood-2009-05-223115. [DOI] [PubMed] [Google Scholar]
- 33.Tasdemir N, et al. Proteomic and transcriptomic profiling identifies mediators of anchorage-independent growth and roles of inhibitor of differentiation proteins in invasive lobular carcinoma. Sci. Rep. 2020;10:11487. doi: 10.1038/s41598-020-68141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter J, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 2012;44:1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 35.Cook PJ, et al. Cox-2-derived PGE2 induces Id1-dependent radiation resistance and self-renewal in experimental glioblastoma. Neuro Oncol. 2016;18:1379–1389. doi: 10.1093/neuonc/now049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tournay O, Benezra R. Transcription of the dominant-negative helix-loop-helix protein Id1 is regulated by a protein complex containing the immediate-early response gene Egr-1. Mol. Cell Biol. 1996;16:2418–2430. doi: 10.1128/MCB.16.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasorella A, Noseda M, Beyna M, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 38.Ogata T, Wozney JM, Benezra R, Noda M. Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc. Natl Acad. Sci. USA. 1993;90:9219–9222. doi: 10.1073/pnas.90.19.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- 40.Gautschi O, et al. Regulation of Id1 expression by SRC: implications for targeting of the bone morphogenetic protein pathway in cancer. Cancer Res. 2008;68:2250–2258. doi: 10.1158/0008-5472.CAN-07-6403. [DOI] [PubMed] [Google Scholar]
- 41.Ciarrocchi A, et al. Id1 restrains p21 expression to control endothelial progenitor cell formation. PLoS ONE. 2007;2:e1338. doi: 10.1371/journal.pone.0001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003;11:915–926. doi: 10.1016/S1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 43.Rockman SP, et al. Id2 is a target of the beta-catenin/T cell factor pathway in colon carcinoma. J. Biol. Chem. 2001;276:45113–45119. doi: 10.1074/jbc.M107742200. [DOI] [PubMed] [Google Scholar]
- 44.Liu KJ, Harland RM. Cloning and characterization of Xenopus Id4 reveals differing roles for Id genes. Dev. Biol. 2003;264:339–351. doi: 10.1016/j.ydbio.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Gupta GP, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc. Natl Acad. Sci. USA. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen YH, et al. Id4 protein is highly expressed in triple-negative breast carcinomas: possible implications for BRCA1 downregulation. Breast Cancer Res. Treat. 2012;135:93–102. doi: 10.1007/s10549-012-2070-0. [DOI] [PubMed] [Google Scholar]
- 47.Stankic M, et al. TGF-beta-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Rep. 2013;5:1228–1242. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wojnarowicz PM, et al. A small-molecule pan-Id antagonist inhibits pathologic ocular neovascularization. Cell Rep. 2019;29:62–75.e7. doi: 10.1016/j.celrep.2019.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharya A, Baker NE. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell. 2011;147:881–892. doi: 10.1016/j.cell.2011.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dashtsoodol N, et al. Alternative pathway for the development of Valpha14(+) NKT cells directly from CD4(-)CD8(-) thymocytes that bypasses the CD4(+)CD8(+) stage. Nat. Immunol. 2017;18:274–282. doi: 10.1038/ni.3668. [DOI] [PubMed] [Google Scholar]
- 51.Park JH, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, et al. G1 phase cell cycle arrest in NSCLC in response to LZ-106, an analog of enoxacin, is orchestrated through ROS overproduction in a P53-dependent manner. Carcinogenesis. 2019;40:131–144. doi: 10.1093/carcin/bgy124. [DOI] [PubMed] [Google Scholar]
- 53.Zhou M, Shen S, Zhao X, Gong X. Luteoloside induces G0/G1 arrest and pro-death autophagy through the ROS-mediated AKT/mTOR/p70S6K signalling pathway in human non-small cell lung cancer cell lines. Biochem. Biophys. Res. Commun. 2017;494:263–269. doi: 10.1016/j.bbrc.2017.10.042. [DOI] [PubMed] [Google Scholar]
- 54.Tappel AL. Vitamin E and free radical peroxidation of lipids. Ann. NY Acad. Sci. 1972;203:12–28. doi: 10.1111/j.1749-6632.1972.tb27851.x. [DOI] [PubMed] [Google Scholar]
- 55.Bensellam M, Montgomery MK, Luzuriaga J, Chan JY, Laybutt DR. Inhibitor of differentiation proteins protect against oxidative stress by regulating the antioxidant-mitochondrial response in mouse beta cells. Diabetologia. 2015;58:758–770. doi: 10.1007/s00125-015-3503-1. [DOI] [PubMed] [Google Scholar]
- 56.Huang YH, et al. ID1 mediates escape from TGF-beta tumor suppression in pancreatic cancer. Cancer Discov. 2020;10:142–157. doi: 10.1158/2159-8290.CD-19-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welsch ME, et al. Multivalent small-molecule pan-RAS inhibitors. Cell. 2017;168:878–889.e29. doi: 10.1016/j.cell.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc. Natl Acad. Sci. USA. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Candia P, et al. Angiogenesis impairment in Id-deficient mice cooperates with an Hsp90 inhibitor to completely suppress HER2/neu-dependent breast tumors. Proc. Natl Acad. Sci. USA. 2003;100:12337–12342. doi: 10.1073/pnas.2031337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao D, et al. Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science. 2008;319:195–198. doi: 10.1126/science.1150224. [DOI] [PubMed] [Google Scholar]
- 61.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 62.Pastushenko I, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 63.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. doi: 10.1096/fasebj.13.15.2257. [DOI] [PubMed] [Google Scholar]
- 65.Deed RW, Armitage S, Norton JD. Nuclear localization and regulation of Id protein through an E protein-mediated chaperone mechanism. J. Biol. Chem. 1996;271:23603–23606. doi: 10.1074/jbc.271.39.23603. [DOI] [PubMed] [Google Scholar]
- 66.Pesce S, Benezra R. The loop region of the helix-loop-helix protein Id1 is critical for its dominant negative activity. Mol. Cell. Biol. 1993;13:7874–7880. doi: 10.1128/MCB.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McAllister SD, et al. Pathways mediating the effects of cannabidiol on the reduction of breast cancer cell proliferation, invasion, and metastasis. Breast Cancer Res. Treat. 2011;129:37–47. doi: 10.1007/s10549-010-1177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia-Morales L, et al. CBD reverts the mesenchymal invasive phenotype of breast cancer cells induced by the inflammatory cytokine IL-1beta. Int. J. Mol. Sci. 2020;21:2429. doi: 10.3390/ijms21072429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang N, et al. ID1 is a functional marker for intestinal stem and progenitor cells required for normal response to injury. Stem Cell Rep. 2014;3:716–724. doi: 10.1016/j.stemcr.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jankovic V, et al. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perry SS, et al. Id1, but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood. 2007;110:2351–2360. doi: 10.1182/blood-2007-01-069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le Jossic C, et al. Expression of helix-loop-helix factor Id-1 is dependent on the hepatocyte proliferation and differentiation status in rat liver and in primary culture. Cancer Res. 1994;54:6065–6068. [PubMed] [Google Scholar]
- 74.Schaefer BM, Koch J, Wirzbach A, Kramer MD. Expression of the helix-loop-helix protein ID1 in keratinocytes is upregulated by loss of cell-matrix contact. Exp. Cell Res. 2001;266:250–259. doi: 10.1006/excr.2001.5215. [DOI] [PubMed] [Google Scholar]
- 75.Lee SB, et al. An ID2-dependent mechanism for VHL inactivation in cancer. Nature. 2016;529:172–177. doi: 10.1038/nature16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niola F, et al. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat. Cell Biol. 2012;14:477–487. doi: 10.1038/ncb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nasif D, et al. Epigenetic regulation of ID4 in breast cancer: tumor suppressor or oncogene? Clin. Epigenetics. 2018;10:111. doi: 10.1186/s13148-018-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bajikar SS, et al. Tumor-suppressor inactivation of GDF11 occurs by precursor sequestration in triple-negative breast cancer. Dev. Cell. 2017;43:418–435.e13. doi: 10.1016/j.devcel.2017.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stighall M, Manetopoulos C, Axelson H, Landberg G. High ID2 protein expression correlates with a favourable prognosis in patients with primary breast cancer and reduces cellular invasiveness of breast cancer cells. Int. J. Cancer. 2005;115:403–411. doi: 10.1002/ijc.20875. [DOI] [PubMed] [Google Scholar]
- 80.Granot Z, et al. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011;20:300–314. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimel MN, et al. HPMA-copolymer nanocarrier targets tumor-associated macrophages in primary and metastatic breast cancer. Mol. Cancer Ther. 2017;16:2701–2710. doi: 10.1158/1535-7163.MCT-15-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 85.Wojnarowicz, P. M. et al. Metadata record for the manuscript: Anti-tumor effects of an ID antagonist with no observed acquired resistance. figshare. 10.6084/m9.figshare.14229986 (2021). [DOI] [PMC free article] [PubMed]
- 86.PRIDE Archive. https://identifiers.org/pride.project:PXD024593 (2021). Accessed 09 Mar 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1422998685. The proteomics data of anti-tumor effects of an ID antagonist with no observed acquired resistance are openly available in the PRIDE Archive via the following accession: https://identifiers.org/pride.project:PXD02459386. The raw data underlying the following figures are openly available as part of the data record: 1E, 2A, 3B, C, D, 4A, E, 5A, B, F, G, 6B, S1A, D, F, G, S2E, I, J, S5A, B, S6, S7A, S8C. A comprehensive list of which file is associated with each figure is also included in the Excel spreadsheet ‘Wojnarowicz_et_al_underlying_data_files_list.xlsx’. All uncropped western blots are in the Supplementary Information section.