Abstract
To reveal the underpinnings of complex biological systems, a variety of approaches have been developed that allow switchable control of protein function. One powerful approach for switchable control is the use of inducible dimerization systems, which can be configured to control activity of a target protein upon induced dimerization triggered by chemicals or light. Individually, many inducible dimerization systems suffer from pre-defined dynamic ranges and overwhelming sensitivity to expression level and cellular context. Such systems often require extensive engineering efforts to overcome issues of background leakiness and restricted dynamic range. To address these limitations, recent tool development efforts have explored overlaying dimerizer systems with a second layer of regulation. Albeit more complex, the resulting layered systems have enhanced functionality, such as tighter control that can improve portability of these tools across platforms.
Keywords: Induced dimerizer, optogenetic, CID, protein control
1. Introduction
Over the past decades, scientists seeking to study a biological process have gained access to powerful approaches for user-directed, inducible control over protein function. One approach, inducible dimerization (Figure 1), utilizes proteins that can be triggered to associate, bringing attached target proteins into proximity. Inducible dimerization systems were initially implemented with chemical inducers (chemical inducers of dimerization or CID systems) [1]. With CID systems, two proteins are induced to associate or dimerize through addition of a bridging chemical ligand (Figure 1A). The proteins that dimerize can be attached to target proteins to bring the targets into proximity with addition of the chemical (Figure 1C). The first developed CID systems include systems for the induced homodimerization of FKBP12 (FK506 binding protein 12) [1], the induced heterodimerization of FKBP12 and cyclosporin A [2], and the induced heterodimerization of FKBP12 and mTor FKBP-rapamycin-binding protein domain (FRB) [3]. These are still used extensively today, in particular the rapamycin-induced FKBP12-FRB system, though due to the cellular effects of rapamycin, second-generation versions have been developed that are activated by orthogonal dimerizers [4,5]. In addition, an expanding number of other orthogonal CID systems are also in use, including a gibberellin-based system that induces interaction of gibberellin insensitive (GAI) and gibberellin insensitive dwarf1 (GID1) [6], an abscisic acid (ABA)-based system that induces pyrabactin resistance like 1 (PYL1) and abscisic acid insensitive 1 (ABI1) interactions [7], an engineered system using the viral protease NS3a and its clinically-used inhibitors [8], versions using SNAP-Tag or Halo-Tag substrates [9–11] as well as several light-sensitive (photocaged or photocleavable) versions [12–15].
Figure 1.
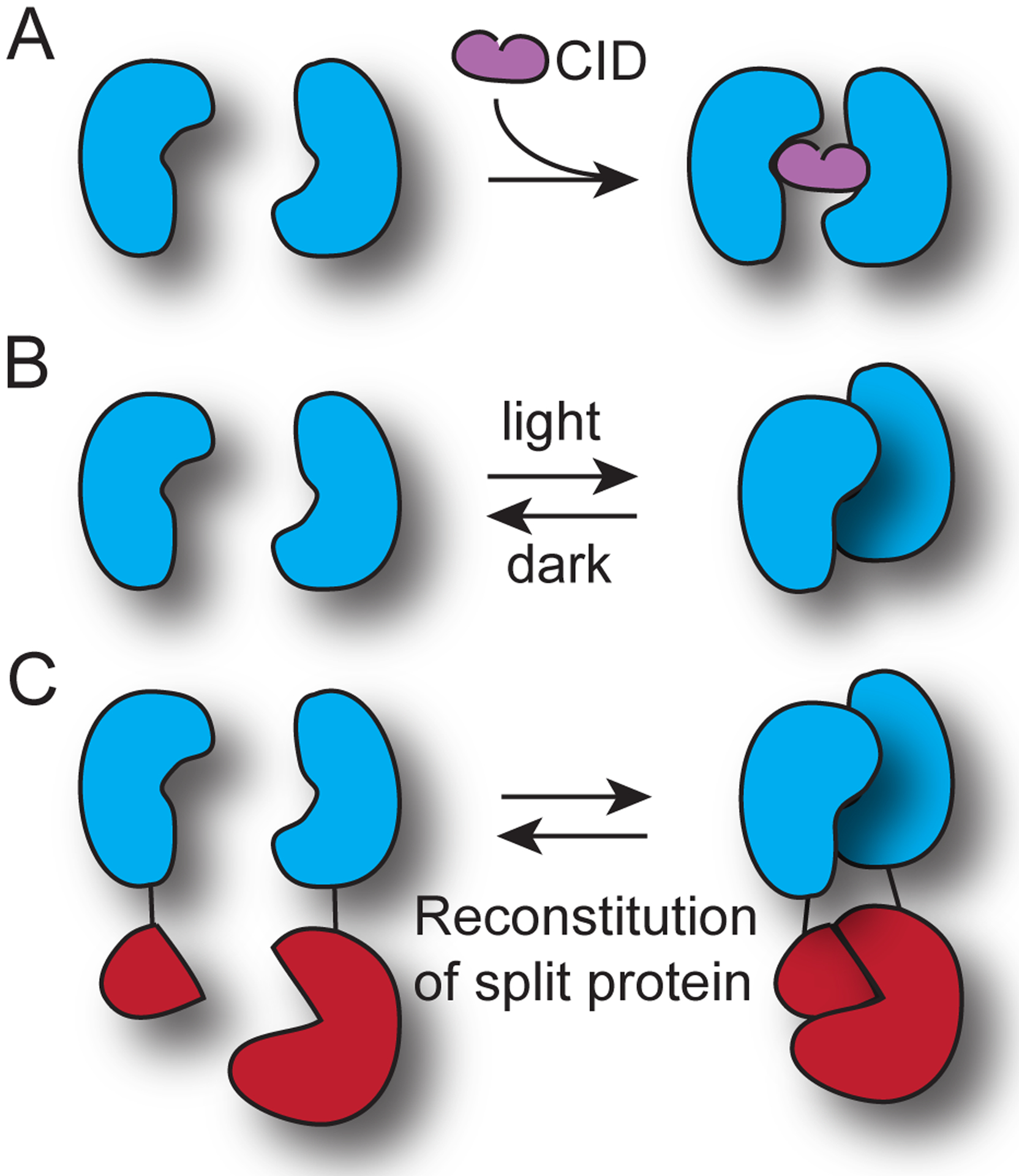
Controlling protein activity with chemical or light-induced association and dissociation. (A) Chemically-induced dimerization system. Addition of a small molecule (CID, chemical inducer of dimerization) that bridges two dimerization domains induces their association. (B) Light induced dimerizer system (photodimerizer system). With illumination, a photosensitive domain transitions to an excited state with altered confirmation, allowing high affinity interaction with a partner. The reaction is inducibly reversed in dark, as the protein recovers the low-affinity, dark state conformation. In some cases the binding affinities can be reversed, such that the partner binds with higher affinity to the dark state conformation. (C) Utilization of a induced dimerization system to reconstitute a split protein, resulting in conditional control of protein activity.
Utilizing a similar mechanism, a number of modular photodimerizer systems triggered by light-regulated protein-protein interactions have been developed [16–23]. These systems are comprised of at least one light-sensitive protein that undergoes a conformational change upon photon absorption, allowing binding to a second protein (Figure 1B). The use of light allows spatial control and enhanced temporal control. Both chemical-triggered and light-triggered inducible dimerizer systems can be configured to regulate target protein activity through a variety of ways, such as altering target protein subcellular localization, reconstituting split protein fragments, induction of oligomerization, and employing steric blocking strategies [24,25]. Biologists have had success applying chemical dimerizers and photodimerizers to regulate protein activity in many contexts without extensive engineering.
Despite their widespread use, dimerization systems can fall short. Traditionally CID systems have suffered from irreversibility, off-target effects of small molecule drugs, and poor spatial resolution owing to drug diffusion, however more recent advances offer improved spatial control with photocaged linkers, and improved reversibility provided by photocleavable linkers or addition of a competing ligand [9–12,15,26]. Induced photodimerization systems offer fast onset kinetics, spatial resolution, and switchable reversibility, but can suffer from limited dynamic ranges and sensitivity to expression level and cellular context. When induced dimerization systems are used to reconstitute split proteins, additional challenges are faced: the affinity of the split proteins must be sufficiently high for the two split protein fragments to assemble when brought into proximity, but low enough that the fragments do not assemble on their own. If expressed at cellular levels that are too high, there can be considerable activity leakiness, while at low levels activity may be insufficient [27,28].
To further probe outstanding questions in biology, systems with improved dynamic range and robustness to cellular noise are needed. The use of dual control mechanisms, whereby a combination of two inducible systems achieves tighter regulation than one alone, offers a solution. Layered control offers the potential for systems with intersectional regulation, increased dynamic range, and more tightly-controlled off-states. Layered or combined approaches that are more tighly regulated should facilitate easier transition between different cellular environments, allowing plug-and-play implementation without optimization in each new context.
To implement dual control with dimerization systems, researchers have taken two basic approaches (Figure 2). In the first approach, the total protein levels of all components within a system remain the same, but a change in localization, activity, or binding is elicited. In the second approach, the level of one or more necessary components of the system is raised or lowered.
Figure 2.
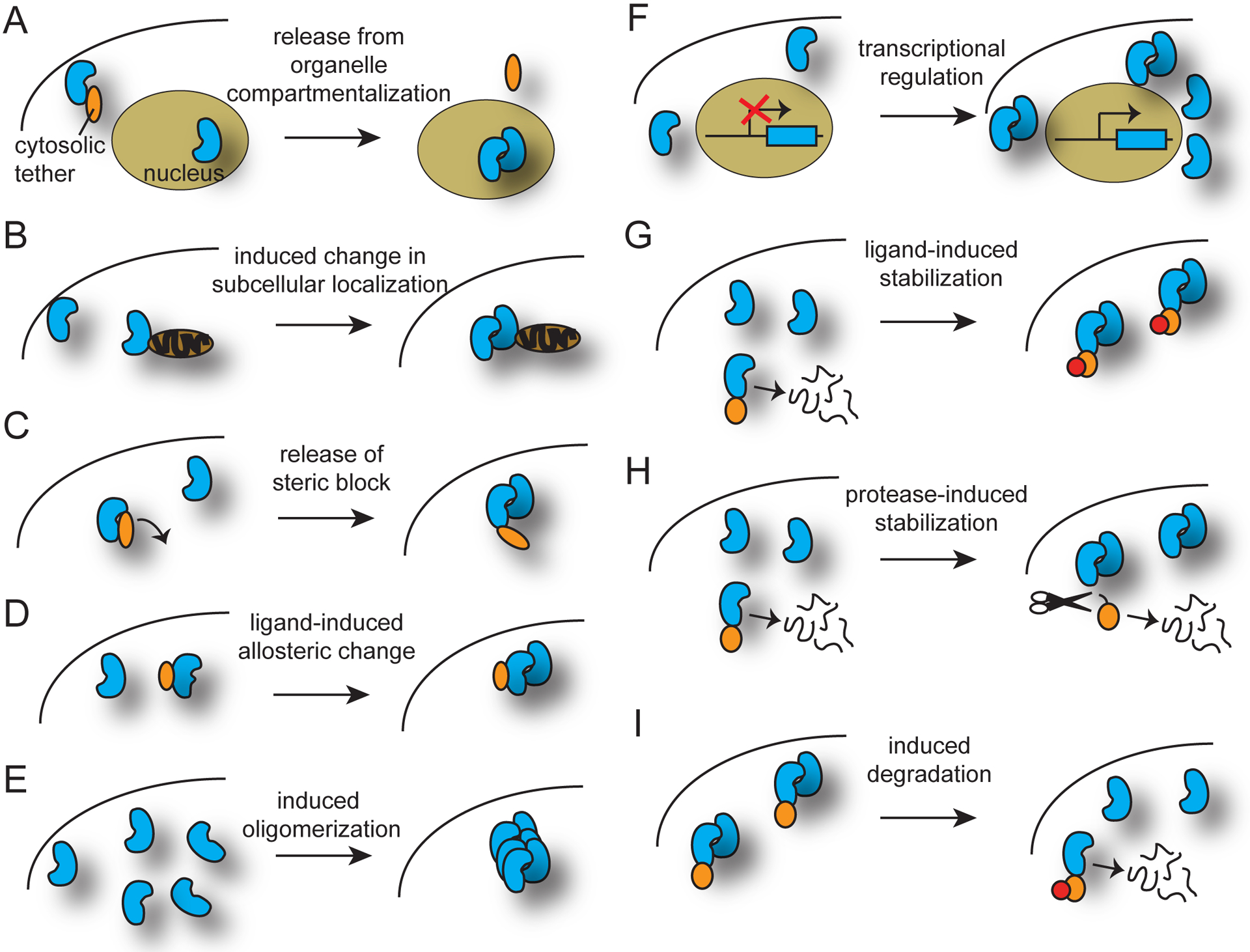
Additional regulation layered upon inducible dimerization systems. Inducible dual system approaches are divided into two categories, regulation of system component properties (protein localization, activity, or binding) without altering actual protein levels and (A-E) and regulation of actual protein abundance (F-I). (A) Release from organelle compartmentalization. In the example shown, a dimerizer component is prevented from entering its functional organelle (the nucleus) through anchoring to a cytosolic tether. The protein can be inducibly released through several mechanisms, including use of light or ligand-activated dimerization systems and inducible protease cleavage. (B) Induced change in subcellular localization. This concept is similar to (A), but does not involve compartmentalization within an organelle. (C) Release of steric block. In the example shown, a light- or ligand-sensitive protein acts to prevent association of a dimerizer-dependent split protein in the uninduced state. With the inducer, the steric block is removed and the protein can associate. (D) Ligand-induced allosteric change. (E) Induced oligomerization. (F) Inducible transcriptional regulation. A light- or ligand-induced transcriptional system is layered onto a dimerization system. (G) Ligand-induced protein stabilization. A destabilized ligand-binding domain is added to one or more components, resulting in protein degradation in the absence of inducer. (H) Protease-induced protein stabilization. Similar to (G), a degron is attached to a component, resulting in constitutive degradation unless a protease is active, removing the degron. The cleavage can be regulated by a dimerized split protease, a photosensitive cleavage site (e.g., PhoCl or AsLOV-caged cleavage site), or a protease inhibitor. (I) Inducible protein degradation. A protein is fused to a domain that can be conditionally targeted to the proteasome (with addition of an inducer).
2. Regulating activity, binding, or localization of a critical system component (system component levels unchanged).
Many effective strategies for cellular regulation involve mechanisms that do not deplete actual levels of a needed component in the cell, but cause changes in local concentrations, temporarily enable or block access to the component, or directly alter a component’s function. For example, recruiting a component to an alternate site where it cannot function reduces its local cellular concentration at the site of action. Central strategies include anchoring at subcellular sites, compartmentalization within organelles, manipulating proximity to itself or other components (through hetero-dimerization, homo-dimerization, or oligomerization), and steric blocking strategies. Such approaches provide simple but powerful means of buffering protein component levels, resulting in tools that are less sensitive to variance in protein expression levels. In addition, these strategies enable fast responses and recovery, as they do not depend on slower-acting protein synthesis or degradation pathways.
2.1. Subcellular manipulation
As most proteins require precise subcellular localization for function, manipulating the location of system components provides an effective means of regulation. For example, one component of a dimerization system can be tethered at a non-functional site (for example, at the golgi, mitochondria, or plasma membrane), then released with an inducer. In another scenario, a system can be functional in the absence of an inducer, then one component is inducibly recruited to a non-functional site, akin to ‘knock-sideways’ approaches that seek to inducibly remove a component from a biological process [29,30].
While subcellular manipulation within the same cellular compartment (for example, moving a protein from the mitochondrial membrane to the plasma membrane) can be effective, additional regulation can be achieved through compartmentalization, where a protein component is sequestered from an entire organelle where it needs to be for function. To conditionally sequester a protein, an inducer promotes relocalization of the protein to or from the functional organelle. Such an approach has been widely used to regulate transcription factors and other nuclear proteins, which are often naturally regulated in the cell through such mechanisms.
Some of the most effective tools for conditionally manipulating subcellular localization have been dimerizers themselves. When combined, dual dimerizers systems can provide additive layers of control, extending the properties of a single system. For example, while the rapamycin-induced FRB-FKBP dimerization system shows fast activation dynamics, the induced dimerization is essentially irreversible due to the high affinity binding of rapamycin [31]. To allow control of the deactivation as well as activation timescales, one group combined an orthogonal gibberellin-based CID system with a rapamycin-dependent CID [6]. Employing a dual and opposing translocation strategy, the researchers demonstrated that Rac1-dependent membrane ruffling can be switched on and off with rapamycin then gibberellin application. The researchers subsequently used the same layered chemical dimerizers to build functional OR and AND logic gates [6,32].
In another design that combines dual CIDs, Chen et al. gained multidirectional control over protein activity, combining a previously developed SLF’-TMP system with a photo-uncagable Nvoc-TMP-Cl (containing a caged TMP fused to HaloTag ligand) [33]. The final system used an opposing translocation strategy to inducibly control more complex subcellular shuttling and trafficking patterns between multiple cell compartments. In one application, the researchers demonstrated the ability to shuttle Rac1 between the cytosol, plasma membrane, and nucleus. While such combined systems often cannot undergo multiple cycles of activation and deactivation without intensive washing steps, they do allow for tighter regulation of off-state activity, as chemical dimerizers can be configured to have essentially no basal off-state activity, and enable more complex spatiotemporal patterns of induced activity.
Another dimerizer tool that has been used in dual systems is LOVTRAP, a reverse dimerization system in which the dimerizing components associate with high affinity in dark and dissociate upon illumination with blue light [34]. The system has two components, AsLOV2, a domain of the plant photoreceptor phototropin 1 that changes conformation upon illumination with blue light, and Zdk (or Zdk2), an engineered interacting partner. Zdk has high affinity for the dark (ground) state of AsLOV2, and much lower affinity for photostimulated AsLOV2, allowing toggling of the interaction with dark and light. An initial demonstration of the promise of LOVTRAP for dual regulation came from a study using the LOVTRAP system to provide additional regulation of a light-gated nuclear localization sequence (NLS) [35]. In this work, LOVTRAP was used to maintain a nuclear protein containing a light-gated NLS sequence in the cytosol in dark. In light, the tethering was released and the NLS exposed, allowing nuclear entry and activity of the nuclear protein. The additional layer of control reduced the background of the NLS system and greatly improved the overall dynamic range. A similar approach as used with the light-caged NLS could provide a second layer of regulation for light-induced dimerization systems that require nuclear entry for function, allowing improved dynamic range and reduced background.
To validate more complex models of dynamic transcriptional activity, the LOVTRAP tool was optimized for use in yeast by swapping the mitochondrial tether for a plasma membrane targeting motif and selecting from more efficient NLS sequences[36]. The resulting controllable light-activated shutting and plasma membrane sequestration (CLASP) system was employed to validate computational models that aim to describe how the cell decodes the dynamics of a naturally pulsatile transcription factor, Crz1. Critical to this analysis was the ability to precisely and reversibly control Crz1 localization repeatedly on the timescale of minutes.
Taking a slightly different approach, several studies used tethering to a cytosolic component to provide tighter regulation of a dimerization-dependent Cre recombinase enzyme [28,37,38]. The studies built on previous work showing that fusion of a steroid hormone ligand binding domain to a protein of interest (POI) can effectively sequester the POI in the cytosol through binding to HSP90 [39]. Addition of hormone triggers release of the ligand binding domain from HSP90, allowing entry of the POI into the nucleus. This concept was applied to more tightly regulate a rapamycin-inducible split CRISPR-Cas9 system [37]. Unique to this system, the high background resulted not from rapamycin-independent dimerization of the split Cas9 fragments, but from the sgRNA itself promoting reconstitution of the split fragments in the absence of the chemical. Compartmentalizing the Cas9 in the cytosol (through fusion to the estrogen ligand binding domain) to keep it away from the sgRNA until after addition of hormone resulted in a system with reduced background and enhanced dynamic range, which could be activated by the combined application of rapamycin and tamoxifen.
In two other examples of a layered inducible system, fusion to the ER ligand binding domain was used to reduce background of split Cre recombinases. Meador et al. applied this strategy to achieve tighter control over a photoactivatable split Cre recombinase using CIBN/CRY2 photodimerizers [28]. Fusing the N-terminal Cre fragment to an estrogen receptor (ER) ligand binding domain sequestered the fragment in the cytosol until estradiol addition, which was added coincident with light to activate the system. This AND gated system resulted in significantly lower background activity. A similar approach was used to regulate a different split Cre under the control of the Magnet photodimerization system, also sequestering the N-terminal Cre fragment (CreN-nMag) in the cytosol until nuclear localization is induced with tamoxifen application [38].
Layering inducible subcellular manipulation with endogenous rather than exogenous dimerization pathways provides a powerful way to gate a system based on cellular context at a user-defined time and space. Several recent approaches to labeling active subsets of neurons at precise times used layered control that combined an endogenous calcium signal (found in active neurons) with a light signal to define the timing of labeling. These approaches, called ‘Cal-Light,’ and ‘FLARE’ use an engineered light-cleavable tether to sequester a transcription factor at the plasma membrane [40,41]. The ‘Cal-Light’ system relies on two combined events: a dimerization event at the plasma membrane that reconstitutes a split tobacco etch virus (TEV) protease, and light-induced uncaging of an AsLOV-caged TEV cleavage site that anchors a transcription factor. An increase in calcium in response to neuronal activity triggers dimerization of a calcium-dependent interaction, bringing together a split TEV protease. If this occurs at the same time or shortly after a light pulse, the light-dependent conformational change in AsLOV exposes the TEV cleavage site, allowing cleavage and release of the anchored transactivator. Taking a similar approach, the FLARE system [41] also uses a transcription factor tethered to the plasma membrane via a light-caged TEV cleavage site. A calcium-dependent interaction induces recruitment of an intact TEV protease to the cleavage site, inducing cleavage and releasing the anchored transcription factor only after uncaging of the cleavage site in the light. With both methods, an increase in intracellular calcium that occurs with neuronal activity acts as one inducer, while light provides a second signal. The released transcription factor localizes to the nucleus and can induce transcription, resulting in marking of active neurons within a window of time defined by light.
2.2. Induced protein-protein interactions (oligomerization and homo-dimerization).
Just as natural systems have been demonstrated to harness dimers, oligomers, and avidity effects to regulate cellular proteins, engineered avidity and oligomerization can provide enhanced control over biological processes. One dimerizer system that also harnesses oligomerization is the CRY2 system, wherein CRY2 interacts in light both with a partner protein CIB1 but also with itself, forming homo-oligomers [19,42–44]. Lee et al. [44] developed a method, light-activated reversible inhibition by assembled trap (LARIAT), that takes advantage of both CRY2-CIB1 and CRY2-CRY2 interactions to sequester a POI from its site of action [44]. The POI is fused to CRY2, and CIB1 fused to a multimeric protein. With light, the POI is pulled into large condensates formed by combining the light-formed CRY2 oligomers and with the multivalent CIB scaffold. The authors also showed the same approach could be used with a GFP nanobody, allowing regulation of GFP-tagged proteins.
In another design that makes use of multivalency, Scheller et al. generated orthogonal phosphoregulated protein switches to layer with and complement existing split transcription factor approaches to regulating protein function [45]. To generate a phosphoregulated orthogonal signal transduction system, the authors re-engineered components of bacterial two-component systems. In their system, a chemical, caffeine, triggers dimerization of a caffeine-binding heavy chain nanobody, which is fused to the kinase domain of the bacterial histidine kinase DcuS. Dimerization activates the kinase, which phosphorylates corresponding response regulator, DcuR, resulting in DcuR dimerization. DcuR is fused to a DNA binding domain and activation domain that becomes activated upon dimerization. Here, one input controls a dimerization relay (one dimerizer controls dimerization of a second, which regulates the POI). The authors show that by layering two dimerization events, they can achieve enhanced dose-response. To allow for further cooperative effects, the authors attached a second caffeine-binding nanobody to each DcuS, allowing the potential for enhanced trans-phosphorylation due to caffeine-induced oligomerization. Changes to the Hill coefficients with the dual system suggested enhanced positive cooperativity compared with the single system.
2.3. Regulation through steric blocking
One strategy that can be easily layered upon dimerization approaches is the use of a conditional steric block. Such an approach is especially useful for split proteins, as a steric block can add a second layer of regulation preventing reconstitution in the uninduced state. To block interaction of two proteins, a steric impediment can be introduced at the interaction interface. For example, Aper et al. used an inducibly cleaved inhibitory peptide to further regulate a 14-3-3 scaffold-based split protein system [46]. In their system, the means of control over protein proximity is two-fold. First, they use a modified caspase-9 containing FGG motifs that can be dimerized by cucurbit[8)uril, allowing inducible activation of caspase. Once activated, the caspase cleaves an inhibitory peptide that blocks access to the 14-3-3 scaffold. Second, addition of fusicoccin allows recruitment of two small protein fragments (C-terminal peptides from the H+-ATPase PMA2: CT52 and CT32), which bind to 14-3-3 proteins in the presence of fusicoccin. The CT52 and CT32 peptides are fused to a split protein (split luciferase fragments), resulting in reconstitution of luciferase activity as the final output in the synthetic signaling cascade.
Another study using steric blocking took a very different approach, using a steric block to prevent association of a split protein in the uninduced state, combined with light-activated dimerizers to reassemble the split protein. To generate a light-regulated chemical labeling system, Photo-SNAP-tag [47], the SNAP-tag enzyme was split into N- and C-terminal fragments and placed under the control of the iLID/SspB photodimerization system. While this approach was effective, the N- and C-terminal fragments retained some ability to associate in dark, leading to background. To gain further control, a photoresponsive AsLOV2 element was fused to the N-terminal SNAP-tag fragment, providing a light-sensitive steric block that reduced the association of the N-terminal fragment with the C-terminal fragment in dark, while in light the steric constraints were released. The combination of two regulatory approaches resulted in improved control and reduced background activity compared with either approach alone.
3. Regulating actual abundance of a critical system component
While not as rapidly responsive, inducible dimerizer systems can also be regulated by modulating the actual cellular abundance of a system component. Because the onset time for inducible regulation of these systems requires either new protein synthesis or protein degradation, these approaches operate at much slower timescales; however, tight control over uninduced state leakiness is gained. Three main strategies that have been used to regulate actual abundance include transcriptional regulation, induced stabilization through the fusion of destabilizing domains or cleaved degrons, and induced degradation.
3.1. Transcriptional regulation
While layered control at the transcriptional level is slow-acting, often on the scale of hours, high background systems with slow temporal dynamics can benefit. Useful inducible transcription systems for layering with optogenetic and chemogenic systems include the Tet-On and Tet-Off gene expression systems [48,49], which allow control of transcription with doxycycline or tetracycline administration or removal. Chen et al. layered the Tet-On and Tet-Off systems with GAVPO, a blue light transcriptional system, to make ‘AND’ and ‘OR’-gated transcriptional systems [50]. In another study, Kempton et al used a doxycycline-inducible transcriptional system to provide further regulation to a split Cas12 enzyme [51]. The doxycycline-induced system was used to regulate the accumulation of the guide RNAs, as their presence resulted in background activity in the split CRISPR-Cas12 system due to promotion of reconstitution. Blocking the accumulation of the guide RNAs until a specific time resulted in substantially reduced leakiness to the system [51].
Layered transcriptional control has also been used to fine-tune other functional components of a system regulated by inducible dimerizers. For example, Kim et al. layered a doxycycline-inducible system with a version of the previously described LARIAT tool used to sequester mRNA (mRNA-LARIAT) [52]. The system consisted of CIB1 fused to a multimeric protein, a GFP nanobody-fused CRY2, a GFP-fused MS2 coat protein, and a mCherry mRNA target (containing MS2 binding sites in the 3’ untranslated region) under control of a tetracycline-responsive promoter element. With light application, it was expected that CRY2 would interact with CIB1-multimeric protein and form clusters, to which the mRNA would be recruited through the nanobody, inhibiting translation. To achieve optimal sequestration resulting in translation inhibition, the authors needed to optimize the level of mRNA transcription. For the resulting system, application of light combined with an optimal doxycycline concentration reduced expression levels by 90%.
Layering of two orthogonal dimerizers can also provide another strategy for fine-tuning transcription, for example providing a means to generate synthetic logic gates. A number of studies layered orthogonal light-induced dimerization systems to achieve more precise multi-chromatic control of experimental processes [53–55]. In a study of dCas9-based transcriptional systems, Gao et al. tested six different inducible dimerizer systems for controlling recruitment of a transcriptional activator or repressor to dCas9, ultimately chosing GA and ABA-based dimerizers for implementation of transcriptional Boolean logic gate operations [56].
3.2. Induced stabilization
One approach for chemical-induced regulatory control involves the fusion to a POI of a destabilizing domain that is stabilized by ligand [57,58]. In its unliganded form, the POI-destabilization domain fusion is targeted for degradation, while addition of a small molecule ligand confers stabilization and protein accumulation. In one study, a split Cre recombinase with activity triggered by photodimerizers was further regulated by addition of a destabilized dihydrofolate reductase (DHFR) to the N-terminal fragment [28]. The DHFR had been engineered to be destabilized in the absence of trimethoprim, a stabilizing ligand [58]. The final dual-regulated system required the addition of trimethoprim prior to light, and did not show activity in dark, light alone, or trimethoprim alone.
While the above study uses regulation by two different inducers, light and trimethoprim, another study developed a CID system in which one of the dimerizer components functioned dually as a destabilization domain [59]. The authors used a destabilized FKBP domain (iFKBP) to provide additional control over split protein systems dimerized by rapamycin. In the absence of rapamycin, the fragment fused to iFKBP is destabilized, reducing spontaneous assembly. The addition of rapamycin served two functions: to stabilize the iFKBP-fused protein half, and to act as CID for the split protein assembly.
3.3. Induced degradation
An opposing approach to induced stabilization is induced degradation, in which an inducer triggers the degradation of a protein target. In one study, a light-caged degron consisting of AsLOV caging a RRRG degron sequence (B-LID, blue light induced degradation domain) [60] was used in a combined approach with a light-induced oligomerizing protein, CRY2, to regulate protein levels with blue light [61]. A transcription factor fused to CRY2 was functional in dark, however light-dependent CRY-CRY oligomerization led to activation of nuclear quality control mechanisms and protein clearance, resulting in loss of activity. Combining the light-disrupted transcriptional system with the light-inducible degron resulted in faster and tighter control of protein expression levels with light onset.
Two other studies also combined light-induced blockage of transcription with a light-activated degron. One approach used EL222, a photoresponsive transcription factor that homodimerizes and binds DNA upon blue light illumination [62]. Baaske et al. showed that when fused to a repressive Krueppel-associated box (KRAB) domain, EL222 can also act as transcriptional repressor [63]. To tightly control the accumulation of proteins in the cell, the authors fused a POI to the same B-LID degron, and placed its expression under control of the KRAB-fused EL222. Upon irradiation with blue light, transcription is repressed by EL222-fused KRAB, while existing protein is depleted through the B-LID degron[60].
Taking a similar approach in yeast, Hasenjager et al. [64] used a different light-sensitive degron consisting of the degron cODC1 caged by AsLOV2 [64]. The authors also generated a new light-disrupted transcriptional system using AsLOV2 and Zdk to control association of a DNA-binding domain and VP16 activation domain (VP16) in dark, and trigger dissociation in light. To VP16, they added the light-induced degron element, to enable degradation of this component with light application. To further enhance control, they placed the induced target protein under light-induced degron control. The combination of the three light-controlled elements resulted in strong expression in dark that could be rapidly switched off with light, quickly downregulating protein levels in yeast.
While dual systems can achieve layered regulation with only one input (i.e. light) inducing two processes, combinatorial approaches can gain functionality as an AND gate when two different inputs are used to induce the system. For example, a system using a cascade of conditional proteases, termed CHOMP, used degrons that could be either removed by protease cleavage (a C-terminal destabilized DHFR) or were generated by protease cleavage (a N-end degron created with cleavage), resulting in presence or loss of a target protein [65]. On top of these systems, they layered ligand regulated proteases (a rapamycin-induced split TEV protease and a protease inhibitor-regulated hepatitis C virus protease), implementing synthetic logic gates through different configurations of these modular degron components. Their system takes advantage of layered control through protease cascades, inducible degradation, protease inhibition, and inducible protease reconstitution to build a variety of chemically-regulated synthetic outputs.
4. Discussion
To develop tools to control protein activity, synthetic biologists have long turned to natural systems for inspiration. Natural systems carry out complex, context-sensitive computation despite noisy environments by spatially organizing biological circuit components and tightly controlling their levels of abundance. Spatial organization can be achieved through mechanisms such as subcellular sequestration, protein scaffolds, and phase separation. In combination, natural systems also engage in complex control over protein abundance, tightly regulating transcription, translation, feedback mechanisms, and protein stability to optimize function. Adopting similar approaches to build synthetic systems, biologists have incorporated artificial tethering systems, scaffolds, and dimerization and oligomerization motifs to spatially sequester or engage system components, and taken advantage of tunable methods for manipulating expression levels. While some basic protein systems can be suitably regulated using only a single component (inducible promoters, for example, for biological processes acting at slower timescales), in many cases the incorporation of multiple layered regulatory approaches can achieve tighter, enhanced control over existing tools.
While layered tools can provide distinct advantages, it is important to note there are drawbacks of such approaches. While layering system A and system B is a simple solution to achieving the properties of both in one system, this approach implies more involved circuit engineering and can result in a higher cellular metabolic load. The introduction of multiple, layered system components into the cell demands increasingly significant resources from the host, and can alter the endogenous biochemical environment [66]. However, by combining inducible systems with different properties, engineers can bypass the extensive protein engineering of a single system that would be needed to match the physiological states of their biological paradigm of interest.
For induced dimerizer systems, in addition to the variety of tools described previously that have been used for layered control, a number of other newly-developed and existing tools offer promise. For example, two very different inducible cleavage systems, PhoCl, a light-cleavable fluorescent protein [67] and TimeSTAMP/StaPLs [68,69], a chemical-regulated protease cleavage system, show high potential for use regulating dimerizer system components. With TimeSTAMP (and StaPLs later), a POI is fused to a protease by means of a protease cleavage site for the same protease, resulting in constitutive cleavage separating the protease from the POI (which can be conditionally regulated by a protease inhibitor). While initial applications explored protein labeling, the approach has since been used to regulate protein degradation, dimerization, and other functions. Using a very different approach, the PhoCl system [67] provides an alternate way to trigger protease cleavage with light using a single light-sensitive engineered fluorescent protein. We envision both PhoCl and StaPLs could be readily applied as layered control for dimerization system components, for regulating subcellular localization or organelle compartmentalization of dimerization system components, for example.
To build enhanced dual systems, another relatively unexplored possibility for layered regulation is allostery. Previous studies have demonstrated that insertion of the photosensitive AsLOV domain within protein loops can confer light-dependent allosteric changes within a variety of protein targets [70–72]. A similar approach could be taken in combination with a split protein half that is inducibly dimerized, preventing background dimerization in the uninduced state. In particular, the combination of two regulatory approaches that use the same inducer, such as blue light, allow the potential for greatly increased regulation with a single signal. Such combined systems can retain the spatiotemporal precision of light-induced protein switches while reducing some of the problematic aspects of each individual dimerizer approach.
In summary, the development of dual systems that provide a second layer of control over inducible dimerizer systems adds complexity to the regulatory approach, but can provide significant benefits including reduced background, increased dynamic range, and greater signal amplification. Dual systems approaches can help facilitate plug-and-play operation, reducing time-consuming optimization between cell types and targets. Systems using two different orthogonal inducers offer the opportunity for integrating multiple signals, such as endogenous and exogenous inducers and light, while systems that use a single inducer for dual regulation (light + light, for example) allow additive regulatory benefits. The modular nature of many regulatory approaches allows easy portability and incorporation into new design approaches, contributing to the armory of tools for dissecting cellular function.
Acknowledgements
This work was supported by funding from the National Institutes of Health (R01 GM100225, R35 GM136367, UF1 NS107710) to C.L.T.
References
- [1].Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR, Science 1993, 262, 1019. [DOI] [PubMed] [Google Scholar]
- [2].Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL, Proc Natl Acad Sci U. S. A 1996, 93, 4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR, Nature 1996, 382, 822. [DOI] [PubMed] [Google Scholar]
- [4].Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR, Chem. Biol 2006, 13, 99. [DOI] [PubMed] [Google Scholar]
- [5].Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffman J, Theiss C, Dehmelt L, Wu Y-W, Angew. Chem. Int. Ed. Engl 2014, 53, 10049. [DOI] [PubMed] [Google Scholar]
- [6].Miyamoto T, DeRose R, Suarez A, Ueno T, Chen M, Sun TP, Wolfgang MJ, Mukherjee C, Meyers DJ, Inoue T, Nat. Chem. Biol 2012, 8, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sen Liang F, Ho WQ, Crabtree GR, Sci. Signal 2011, 4, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Foight GW, Wang Z, Wei CT, Jr Greisen P, Warner KM, Cunningham-Bryant D, Park K, Brunette TJ, Sheffler W, Baker D, Maly DJ, Nat. Biotechnol 2019. 37, 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Feng S, Laketa V, Stein F, Rutkowska A, MacNamara A, Depner S, Klingmuller U, Saez-Rodriguez J, Schultz C, Angew. Chemie Int. Ed 2014, 53, 6720. [DOI] [PubMed] [Google Scholar]
- [10].Zimmermann M, Cal R, Janett E, Hoffmann V, Bochet CG, Constable E, Beaufils F, Wymann MP, Angew. Chem. Int. Ed. Engl 2014, 53, 4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen X, Venkatachalapathy M, Kamps D, Weigel S, Kumar R, Orlich M, Garrecht R, Hirtz M, Niemeyer CM, Wu Y-W, Dehmelt L, Angew. Chemie Int. Ed 2017. 56, 5916. [DOI] [PubMed] [Google Scholar]
- [12].Gutnick A, Banghart MR, West ER, Schwarz TL, Nat. Cell Biol 2019, 21, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Umeda N, Ueno T, Pohlmeyer C, Nagano T, Inoue T, J. Am. Chem. Soc 2011, 133, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen X, Wu Y-W, Angew. Chem. Int. Ed. Engl 2018, 57, 6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ballister ER, Aonbangkhen C, Mayo AM, Lampson MA, Chenoweth DM, Nat. Commun 2014, 5, 5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B, Proc. Natl. Acad. Sci. U. S. A 2015, 112, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kaberniuk A, Shemetov A, Verkhusha V, Nat. Methods 2016, 13, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kawano F, Suzuki H, Furuya A, Sato M, Nat. Commun 2015, 6, 6256. [DOI] [PubMed] [Google Scholar]
- [19].Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL, Nat. Methods 2010, 7, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Levskaya A, Weiner OD, Lim WA, Voigt CA, Nature 2009, 461, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M, Nat. Methods 2012, 9, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen D, Gibson ES, Kennedy MJ, J. Cell Biol 2013, 201, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Crefcoeur RP, Yin R, Ulm R, Halazonetis TD, Nat. Commun 2013, 4, 1779. [DOI] [PubMed] [Google Scholar]
- [24].Spiltoir JI, Tucker CL, Curr. Opin. Struct. Biol 2019, 57, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stanton BZ, Chory EJ, Crabtree GR, Science 2018, 359, eaao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Klewer L, Wu Y-W, Chemistry 2019, 25, 12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Q, Sinnen BL, Boxer EE, Schneider MW, Grybko MJ, Buchta WC, Gibson ES, Wysoczynski CL, Ford CP, Gottschalk A, Aoto J, Tucker CL, Kennedy MJ, Neuron 2019, 101, 863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Meador K, Wysoczynski CL, Norris AJ, Aoto J, Bruchas MR, Tucker CL, Nucleic Acids Res 2019, 47, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB, Cell 2000, 101, 717. [DOI] [PubMed] [Google Scholar]
- [30].Robinson MS, Sahlender DA, Foster SD, Dev. Cell 2010, 18, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lin Y-C, Nihongaki Y, Liu T-Y, Razavi S, Sato M, Inoue T, Angew. Chem. Int. Ed. Engl 2013, 52, 6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Razavi S, Su S, Inoue T, ACS Synth. Biol 2014, 3, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen X, Venkatachalapathy M, Dehmelt L, Wu Y-W, Angew. Chem. Int. Ed. Engl 2018, 57, 11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM, Nat. Methods 2016, 13, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yumerefendi H, Wang H, Dickinson DJ, Lerner AM, Malkus P, Goldstein B, Hahn K, Kuhlman B, ChemBioChem 2018, 19, 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen SY, Osimiri LC, Chevalier M, Bugaj LJ, Nguyen TH, Greenstein RA, Ng AH, Stewart-Ornstein J, Neves LT, El-Samad H, Cell Syst 2020, 11, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nguyen DP, Miyaoka Y, Gilbert LA, Mayerl SJ, Lee BH, Weissman JS, Conklin BR, Wells JA, Nat. Commun 2016, 7, 12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Allen ME, Zhou W, Thangaraj J, Kyriakakis P, Wu Y, Huang Z, Ho P, Pan Y, Limsakul P, Xu X, Wang Y, ACS Synth. Biol 2019, 8, 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Picard D, Trends Cell Biol 1993, 3, 278. [DOI] [PubMed] [Google Scholar]
- [40].Lee D, Creed M, Jung K, Stefanelli T, Wendler DJ, Oh WC, Mignocchi NL, Luscher C, Kwon H-B, Nat. Methods 2017, 14, 495. [DOI] [PubMed] [Google Scholar]
- [41].Wang W, Wildes CP, Pattarabanjird T, Sanchez MI, Glober GF, Matthews GA, Tye KM, Ting AY, Nat. Biotechnol 2017, 35, 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, V Schaffer D, Nat. Methods 2013, 10, 249. [DOI] [PubMed] [Google Scholar]
- [43].Che DL, Duan L, Zhang K, Cui B, ACS Synth. Biol 2015, 4, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Heo WD, Nat. Methods 2014, 11, 633. [DOI] [PubMed] [Google Scholar]
- [45].Scheller L, Schmollack M, Bertschi A, Mansouri M, Saxena P, Fussenegger M, Nat. Commun 2020, 11, 3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aper SJA, den Hamer A, Wouters SFA, Lemmens LJM, Ottmann C, Brunsveld L, Merkx M, ACS Synth. Biol 2018, 7, 2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cleveland JD, Tucker CL, ACS Chem. Biol 2020, 15, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gossen M, Bujard H, Proc. Natl. Acad. Sci. U. S. A 1992, 89, 5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H, Science 1995, 268, 1766. [DOI] [PubMed] [Google Scholar]
- [50].Chen X, Li T, Wang X, Du Z, Liu R, Yang Y, Nucleic Acids Res 2016, 44, 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kempton HR, Goudy LE, Love KS, Qi LS, Mol. Cell 2020, 78, 184. [DOI] [PubMed] [Google Scholar]
- [52].Kim NY, Lee S, Yu J, Kim N, Won SS, Park H, Heo WD, Nat. Cell Biol 2020, 22, 341. [DOI] [PubMed] [Google Scholar]
- [53].Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J, Zurbriggen MD, Weber W, Nucleic Acids Res 2013, 41, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Müller K, Engesser R, Timmer J, Zurbriggen MD, Weber W, ACS Synth. Biol 2014, 3, 796. [DOI] [PubMed] [Google Scholar]
- [55].Redchuk TA, Kaberniuk AA, V Verkhusha V, Nat. Protoc 2018, 13, 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gao Y, Xiong X, Wong S, Charles EJ, Lim WA, Qi LS, Nat. Methods 2016, 13, 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Banaszynski LA, Chen L-C, Maynard-Smith LA, Ooi AGL, Wandless TJ, Cell 2006, 126, 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ, Chem. Biol 2010, 17, 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dagliyan O, Krokhotin A, Ozkan-Dagliyan I, Deiters A, Der CJ, Hahn KM, Dokholyan NV, Nat. Commun 2018, 9, 4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ, ACS Chem. Biol 2014, 9, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pathak GP, Spiltoir JI, Höglund C, Polstein LR, Heine-Koskinen S, Gersbach CA, Rossi J, Tucker CL, Nucleic Acids Res 2017, 45, e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH, Nat. Chem. Biol 2014, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Baaske J, Gonschorek P, Engesser R, Dominguez-Monedero A, Raute K, Fischbach P, Muller K, Cachat E, Schamel WWA, Minguet S, Davies JA, Timmer J, Weber W, Zurbriggen MD, Sci. Rep 2018, 8, 15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hasenjäger S, Trauth J, Hepp S, Goenrich J, Essen L-O, Taxis C, ACS Synth. Biol 2019, 8, 1026. [DOI] [PubMed] [Google Scholar]
- [65].Gao XJ, Chong LS, Kim MS, Elowitz MB, Science 2018, 361, 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Glick BR, Biotechnol. Adv 1995, 13, 247. [DOI] [PubMed] [Google Scholar]
- [67].Zhang W, Lohman AW, Zhuravlova Y, Lu X, Wiens MD, Hoi H, Yaganoglu S, Mohr MA, Kitova EN, Klassen JS, Pantazis P, Thompson RJ, Campbell RE, Nat. Methods 2017, 14, 391. [DOI] [PubMed] [Google Scholar]
- [68].Lin MZ, Glenn JS, Tsien RY, Proc. Natl. Acad. Sci. U. S. A 2008, 105, 7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jacobs CL, Badiee RK, Lin MZ, Nat. Methods 2018, 15, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dagliyan O, Tarnawski M, Chu P-H, Shirvanyants D, Schlichting I, V Dokholyan N, Hahn KM, Science 2016, 354, 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R, Science 2008, 322, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Reynolds KA, McLaughlin RN, Ranganathan R, Cell 2011, 147, 1564. [DOI] [PMC free article] [PubMed] [Google Scholar]


