To the Editor—Whole-genome sequencing (WGS) analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to important findings related to the origin and evolution of the virus.1-3 The high potential for infectivity of SARS-CoV-2 raises legitimate concerns for person-to-person transmission, particularly in the hospital setting. Evaluation of the viral genome during a pandemic can aid in identifying outbreaks.4
Methods
Viral WGS was performed as previously described5 on all positive SARS-CoV-2 isolates at Children’s Hospital Los Angeles, a quaternary-care, free-standing, pediatric medical center.
To analyze local propagation of the virus, we relied upon direct comparisons of mutations found in viral genomes. We defined the dissimilarity between viral isolates as the size of the symmetric difference between the sets of mutations present relative to the reference genome. The analysis was restricted to the consensus level mutations and SARS-CoV-2 mutations with allele frequency of ≥50%. Only high-quality SARS-CoV-2 genomes, defined as at least 100× coverage (number of reads aligned to a genomic position) across 97% of the genome, were used for cluster analysis.
To analyze emerging groups of comparable isolates, we used hierarchical clustering. The dissimilarity matrix was defined as the sizes of symmetric differences between samples. We used the bottom-up unweighted pair group method with arithmetic mean reference (UPGMA) method with an R function hclust6 to visualize the clustering (Fig. 1). Suspected clusters that warranted investigation were internally defined by the institution’s contact-tracing program as (1) ≥2 SARS-CoV-2–positive cases within the same setting, (2) presence of an epidemiological link between the cases, noting the potential of prolonged close contact within 2 m (6 feet) for 15 minutes or longer, and (3) occurring within 14 days of symptom onset or positive SARS-CoV-2 reverse-transcriptase polymerase chain reaction (RT-PCR) test date if asymptomatic.
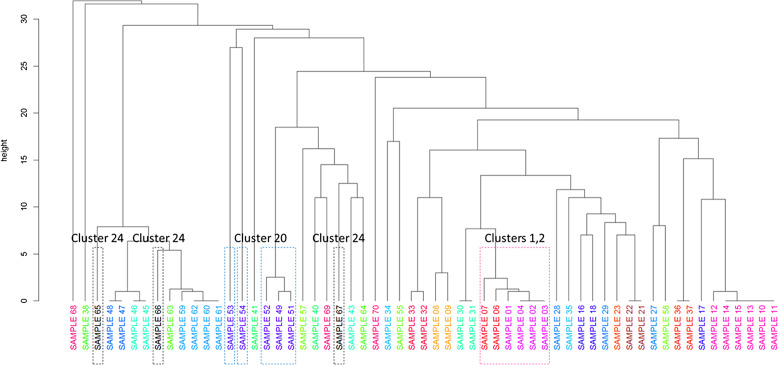
Fig. 1.
Unweighted pair group method with arithmetic mean reference (UPGMA) clustering analysis allows for a global visualization of samples under investigation and a means to assess relatedness across suspected clusters. Relatedness of samples within clusters was based on the symmetric difference calculations: Clusters 1, 2: related (samples 1–4, 6, 7); sample 5 was not included due to low coverage. Cluster 20: related (samples 49, 51, 52); unrelated (samples 53, 54); sample was 50 not included due to low coverage. Cluster 24: unrelated (samples 65–67).
Results
Establishment of protocol
To determine a framework for interpreting dissimilarity between isolates, we compared pairwise differences between repeated samples from a single patient and within-family clusters with epidemiologically unrelated samples. The median pairwise difference between unrelated isolates estimated in the spring and early summer of 2020 was 10 mutations, and as of January 2021, it had increased to 16 mutations. The continued evolution of SARS-CoV-2 genome did not affect our analysis of local transmissions because suspect isolates were subjected to WGS within a relatively short period. Furthermore, the estimate of 0–1 variants between related samples becomes even more robust with increasing divergence between viral isolates.
Consequently, we adopted the following interpretation of pairwise and within-cluster dissimilarities: highly related = 0–1 variant; possibly related = 2–4 variants; probably unrelated = 5–9 variants; unrelated = 10 or more variants. Supplementary Figure 1 and Supplementary Table 1 (online) summarize the sample diversity and pairwise dissimilarities for 3 distinct periods. The 0–1-variant difference is statistically highly unlikely for 2 unrelated viral isolates, with P = .00355, based upon the combined data set.
Institutional cluster analysis
During a 9-month period (April–December 2020), we identified 25 potential clusters, involving a total of 70 SARS-CoV-2–positive individuals, that warranted further exploration by WGS. Of the 25 suspected clusters analyzed, conclusive results were available in 23 cases (92%) (Supplementary Table 2 online). We confirmed some relatedness in 14 clusters (56%) suspected by the contact-tracing team, including an outbreak within a unit. For example, clusters 1 and 2, corresponding to the same unit, were related to each other (Fig. 1 and Supplementary Table 2 online). We encountered cases in which relatedness was confirmed in only a portion of isolates within a suspected cluster. For example, cluster 20 consisted of 3 highly related isolates and 2 distinct isolates that were significantly distant (Fig. 1).
Importantly, WGS allowed us to rule out 9 highly suspicious clusters within the same units (36%), demonstrating that these were not healthcare-associated infections and that, during periods of high community incidence of coronavirus disease 2019 (COVID-19) cases, transmission outside the healthcare setting was a more likely driver of transmission events. The remaining samples were unresolved due to inadequate genome coverage.
Discussion
Similar to other studies,4 WGS analysis of SARS-CoV-2 isolates allowed us to monitor disease spread, to analyze outbreak dynamics, and to identify infection hot spots within our institution. Equally important was the capability provided by the WGS analysis to rule out suspected clusters and to confirm unrelated sources of infection. This capability not only allowed us to implement mitigation efforts with a more targeted approach but also provided us with insights about how our infection prevention and control strategies and use of personal protective equipment effectively prevented disease transmission.
There is currently a lack of standardization and definitions by local public health officials and regulatory bodies regarding the use of genomic epidemiology to identify clusters and hospital-acquired infections. We established a defined cut-off to determine relatedness based on the known mutation rate of SARS-CoV-2, which allowed us to accurately and conservatively interpret genomic data alongside clinical meta-data. We emphasize the need for clinical meta-data as part of the interpretation because 100% identical isolates with absolutely no known association are commonly detected. We do recognize that WGS is not being pursued in many COVID-19 cases tested outside of our facility, and we emphasize the need for more widespread use of WGS given the utility of these data.
In conclusion, genomic analysis during COVID-19 pandemic, as well as other infectious diseases outbreaks, can be highly effective in a clinical setting as a complement to contact-tracing efforts, and WGS will become increasingly important in future pandemics.
Acknowledgments
We would like to acknowledge the Clinical Microbiology and Virology laboratory, Center for Personalized Medicine and the Infection Prevention and Control team at Children’s Hospital Los Angeles.
Financial support
This work was partially funded by The Saban Research Institute at Children’s Hospital Los Angeles intramural support for COVID-19 Directed Research to X.G. and J.D.B.
Conflict of interest
All authors report no conflicts of interest relevant to this article.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/ice.2021.185.
click here to view supplementary material
References
- 1. da Silva Filipe A, Shepherd JG, Williams T, et al. Genomic epidemiology reveals multiple introductions of SARS-CoV-2 from mainland Europe into Scotland. Nat Microbiol 2021;6:112–122. [DOI] [PubMed] [Google Scholar]
- 2. Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science 2020;369:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gudbjartsson DF, Helgason A, Jonsson H, et al. Spread of SARS-CoV-2 in the Icelandic population. N Engl J Med 2020;382:2302–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oude Munnink BB, Nieuwenhuijse DF, Stein M, et al. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med 2020;26:1405–1410. [DOI] [PubMed] [Google Scholar]
- 5. Pandey U, Yee R, Shen L, et al. High prevalence of SARS-CoV-2 genetic variation and D614G mutation in pediatric patients with COVID-19. Open Forum Infect Dis 2020. doi: 10.1093/ofid/ofaa551. [DOI] [PMC free article] [PubMed]
- 6. Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015;31:3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/ice.2021.185.
click here to view supplementary material