To the Editor—Infection with severe acute respiratory syndrome virus 2 (SARS-CoV-2) has resulted in >137 million cases globally and >31 million cases in the United States. Whether previous COVID-19 infection is protective against reinfection with original strains or SARS-CoV-2 genomic variants of concern remains unknown. Genomic variants were first reported in South Africa, the United Kingdom, and Brazil.1,2 Variant B.1.1.7 rapidly became the predominant variant in the United Kingdom within 3 months. It is more transmissible, and has caused increased cases and hospitalizations in several European countries.2 Community spread of the B.1.1.7 variant has resulted in >16,000 cases in the United States since first being reported in December 2020 in travelers from the United Kingdom.2−5 Cases of the B.1.1.7 variant are likely underreported in the United States, and the increasing prevalence of this variant in communities adds complexity to the public health and infection control response.5−7 This report illustrates challenges associated with reinfection in hospitalized patients and the importance of genomic sequencing in the evaluation of possible SARS-CoV-2 reinfection.
Methods
Case investigation
In November 2020, a 16-year-old with end-stage renal disease due to focal segmental glomerulosclerosis was electively admitted to our hospital for a trial off hemodialysis. At the time of admission, a nasopharyngeal swab was negative for detection of SARS-CoV-2 by reverse-transcriptase polymerase chain reaction (RT-PCR). On hospital day 2, the patient complained of a sore throat. She had recent exposure to an ill family member, thus repeat testing for SARS-CoV-2 was performed and was positive; cycle threshold (Ct) values for E and S genes were 32.4 and 32.0, respectively. Other symptoms included fatigue, nasal congestion, rhinorrhea, and a nonproductive cough. She remained afebrile and did not require oxygen supplementation. Her symptoms resolved in 8 days and she was discharged.
In January 2021, she was readmitted with leg pain, swelling, and fatigue. Upon readmission, a nasopharyngeal swab was tested by RT-PCR and SARS-CoV-2 was not detected. Diagnostic evaluation revealed Hodgkin’s lymphoma and chemotherapy was initiated on hospital day 9. On hospital day 12, she developed severe mouth and throat pain. On hospital day 16, she developed fever, neutropenia (ANC = 120/µL), and abdominal tenderness. On that day (94 days after her initial COVID-19 infection), she was SARS-CoV-2 positive by RT-PCR (Ct values for E and S genes were 30.6 and 31.0, respectively) and SARS-CoV-2 IgM antibodies were detected though IgG antibodies were not detected.
SARS-CoV-2 testing policy
All patients admitted to our hospital have a nasopharyngeal swab collected for SARS-CoV-2 RNA detection. SARS-CoV-2 molecular testing is repeated if patients develop new symptoms consistent with COVID-19. The infection control and prevention practitioners review all cases with repeat positive SARS-CoV-2 results and follow the CDC common investigation protocol for cases of possible SARS-CoV-2 reinfection, including genomic sequencing.6
SARS-CoV-2 testing
Testing for SARS-CoV-2 RNA was performed using the RealStar SARS-CoV-2 RT-PCR kit (Altona Diagnostics, Hamburg-Altona, Germany), which has US Food and Drug Administration (FDA) emergency use authorization (EUA) and detects the E and S genes of SARS-CoV-2.8
SARS-CoV-2 genomic sequencing
Viral transport media specimens from archived SARS-CoV-2 positive samples were extracted for viral RNA (Allprep PowerViral DNA/RNA kit, Qiagen, Hilden, Germany). Viral genome sequencing (using short-read technology) was performed using the Multiplex PCR CDC protocol with ARTIC Network V3 primers and sequenced on the Illumina MiSeq using the MiSeq Reagent Kit v2, 300 cycles (Illumina, San Diego, CA).
SARS-CoV-2 genome sequence analysis
Viral genomes were assembled using standard protocols outlined by the CDC.9 Briefly, the variant calling and consensus assembly pipeline used cutadapt 2.10 for sequence trimming, bowtie2 v 2.1.0 for aligning reads against the Wuhan-Hu-1 (NC_045512.2) reference genome, SAMtools 1.9 and BCFtools 1.9 for variant calling and file manipulation. Lineage analysis of assembled SARS-CoV-2 genomes was completed using Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN, github.com/cov-lineages/pangolin). SARS-CoV-2 genomes were submitted to GISAID (www.gisaid.org; Accession Numbers EPI_ISL_1482538, EPI_ISL_1482537).
Results
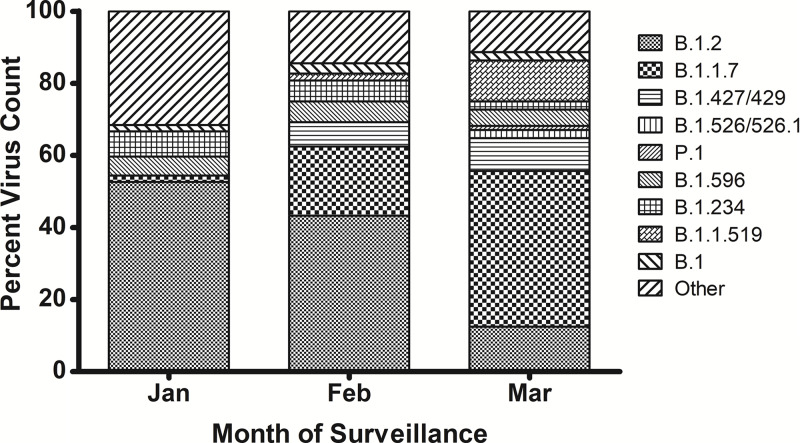
Our patient had clinical and virologic resolution of the first SARS-CoV-2 infection prior to presenting with a second SARS-CoV-2 positive test >90 days later. Reinfection was due to a distinctly different genotype of SARS-CoV-2. Phylogenetic analysis classified the virus from the primary infection as the dominant North American lineage B.1.2 and the second infection as lineage B.1.1.7, a variant of concern that is rapidly becoming the dominant lineage in our pediatric population (Fig. 1). These findings confirm that our case represents a genetically verified SARS-CoV-2 reinfection in an immunocompromised adolescent.
Fig. 1.
Genomic diversity of SARS-CoV-2 circulating in pediatric patients at Texas Children’s Hospital (TCH), January 1–March 23, 2021. Active genomic surveillance of positive SARS-CoV-2 specimens in pediatric patients at TCH reveals a dynamic shift in SARS-CoV-2 genetic diversity over 3 months. Prevalence of variant of concern B.1.1.7 increased from 1.8% in January to 43.2% in March.
Discussion
This is the first report of SARS-CoV-2 reinfection by the B.1.1.7 variant in the United States that occurred >90 days after infection with the dominant North American lineage B.1.2. Neither the patient nor any of her close contacts had traveled, suggesting that this case was community acquired. Reinfection occurred shortly after the B.1.1.7 variant was first reported in Texas.4 Our case emphasizes the importance of SARS-CoV-2 genomic sequence surveillance, not only for public health response but also for hospital infection control and prevention. Although rare, reports of reinfection in both immunocompromised and immunocompetent patients suggest mitigating strategies are important for those who have recovered from COVID-19.6,7 The emergence of variants of concern, such as B.1.1.7, and cases of possible reinfection present unique challenges for patients with recurrent hospitalization. Careful evaluation of possible reinfection and genomic sequencing have implications for infection control measures such as cohorting and duration of isolation.
Acknowledgments
We thank our entire healthcare team for their hard work during the pandemic.
Financial support
No financial support was provided relevant to this article.
Conflicts of interest
All authors report no conflicts of interest relevant to this article.
References
- 1. Mwenda M, Saasa N, Sinyange N, et al. Detection of B.1.351 SARS-CoV-2 variant strain—Zambia, December 2020. Morb Mortal Wkly Rep 2021;70:280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. Morb Mortal Wkly Rep 2021;70:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Firestone MJ, Lorentz AJ, Wang X, et al. First identified cases of SARS-CoV-2 variant B.1.1.7 in Minnesota—December 2020–January 2021. Morb Mortal Wkly Rep 2021;70:278–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ojelade M, Rodriguez A, Gonzalez D, et al. Travel from the United Kingdom to the United States by a symptomatic patient infected with the SARS-CoV-2 B.1.1.7 variant—Texas, January 2021. Morb Mortal Wkly Rep 2021;70:348–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US COVID-19 cases caused by variants. Centers for Disease Control and Prevention website. https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html. Published 2021. Accessed March 28, 2021.
- 6. Babiker A, Marvil CE, Waggoner JJ, Collins MH, Piantadosi A. The importance and challenges of identifying SARS-CoV-2 reinfections. J Clin Microbiol 59:e02769–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis 2021;21:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Visseaux B, Le Hingrat Q, Collin G, et al. Evaluation of the RealStar SARS-CoV-2 RT-PCR kit RUO performances and limit of detection. J Clin Virol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paden CR, Tao Y, Queen K, et al. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020;26:2401–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]