Abstract
A recent study have reported that pre-use of azelastine is associated with a decrease in COVID-19 positive test results among susceptible elderly people. Besides, it has been reported that antihistamine drugs could prevent viruses from entering cells. The purpose of this study is to investigate whether azelastine have antiviral activity against SARS-CoV-2 in vitro and the possible mechanism. Here, we discovered antihistamine azelastine has an affinity to ACE2 by cell membrane chromatography (CMC); Then we determined the equilibrium dissociation constant (KD) of azelastine-ACE2 as (2.58 ± 0.48) × 10−7 M by surface plasmon resonance (SPR). The results of molecular docking showed that azelastine could form an obvious hydrogen bond with Lys353. The pseudovirus infection experiments showed that azelastine effectively inhibited viral entry (EC50 = 3.834 μM). Our work provides a new perspective for the screening method of drug repositioning for COVID-19, and an attractive and promising drug candidate for anti-SARS-CoV-2.
Keywords: Antihistamine, Azelastine, SARS-CoV-2, ACE2, Drug repurposing
Graphical abstract

1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new type of coronavirus reported in 2019, which mainly spreads through respiratory droplets and close contact (Wiersinga et al., 2020). Its infection makes people suffer from coronavirus disease 2019 (COVID-19), accompanied by fever, headache, mild chest pain, loss of smell, and shortness of breath (Yuce et al., 2021). The spike protein encoded by the viral genome has two subunits, of which S1 contains the receptor binding domain (RBD) that enables the virus to bind to its host target (Walls et al., 2020), human angiotensin-converting enzyme 2 (ACE2). ACE2 is a cell surface metalloproteinase, primarily expressed in lung, heart, kidney, and intestine (Gheblawi et al., 2020). Attachment and entry of SARS-CoV2 requires the binding of the spike protein to the target receptor ACE2 (Zhao et al., 2020) on cell surface, which is the same as SARS-CoV infecting the host (Shang et al., 2020). However, the difference is that the binding affinity of the spike protein of SARS-CoV-2 to ACE2 is much stronger than that of SARS-CoV (Gheblawi et al., 2020), so SARS-CoV-2 has an overwhelmingly high transmission rate. It was declared a pandemic by the world health organization (WHO) (Cucinotta and Vanelli, 2020) and has so far caused more than 114 million cases and 2.5 million deaths in 192 countries and regions, according to the Johns Hopkins University COVID-19 dashboard.
The SARS-CoV-2 has triggered a prolonged global health crisis and raised an alarm for the medical community all around the world. Now it has entered the second year of the global fight against the pandemic. However, there is currently no specific medicine to completely treat this infection. Although the advent of vaccines brings hope, it is still an extremely scarce resource. Therefore, for those infected and who cannot get vaccinated, specific medications for treatment are still essential. Unfortunately, in the year-long fight against the epidemic, there is still a lack of reliable specific medicines (Asselah et al., 2021).
Faced with the problem of a long development cycle for de novo during the sudden pandemic, the strategy of drug repurposing can accelerate preclinical and partial clinical evaluations (Riva et al., 2020). It's a feasible and rapid method to identify effective drugs to combat this sudden pandemic (Xu et al., 2020). H1-antihistamines can reduce allergic inflammation and are commonly used to treat allergic diseases (Thangam et al., 2018). In addition to the antihistamine effects, recent studies have described the potential of antihistamine drugs against the Ebola virus, Marburg virus (Schafer et al., 2018), and influenza viruses (Xu et al., 2018). Azelastine is an antihistamine and mast cell stabilizer used as nasal spray for hay fever and eye drops for allergic conjunctivitis (Castillo et al., 2015). A recent study have reported that pre-use of azelastine is associated with a decrease in COVID-19 positive test results among susceptible elderly people (Reznikov et al., 2021). Considering that the SARS-CoV-2 can infect the host through ACE2 expressed in the respiratory tract and eyes (Zhou et al., 2020), and the administration sites of azelastine are nasal cavity and conjunctiva, therefore we speculate that azelastine may act on ACE2 and inhibit the entry of the virus.
In this study, we evaluated the antiviral effect of azelastine and its possible mechanisms. First, we used cell membrane chromatography (CMC) to screen and identify azelastine; its affinity to ACE2 was determined by surface plasmon resonance (SPR); further molecular docking experiments were performed to simulate the interaction site of azelastine and ACE2. Finally, we confirmed the effectiveness of azelastine in inhibiting SARS-CoV-2 through pseudovirus infection experiments. Here we verified the activity of azelastine against SARS-CoV-2. By binding to ACE2, azelastine interferes with the interaction between spike protein and ACE2 and reduces the infection of SARS-CoV-2, which is a promising virus entry inhibitor.
2. Material and methods
2.1. Drugs and reagents
Azelastine was purchased from TargetMol (Boston, USA). Dulbecco's Modification of Eagle's Medium (DMEM) with high glucose (Cat. No. SH30022.01), and fetal bovine serum (FBS) (Cat. No. 16140071) were from HyClone (Logan, UT, USA). Penicillin–streptomycin solution was obtained from Xi'an Hat Biotechnology Co., Ltd (Xi'an, China). Puromycin was purchased from Meilunbio (Dalian, China). Cell Counting Kit was purchased from 7 Sea Biotech (Shanghai, China). SARS-CoV-2 spike pseudovirus (PSC001) was purchased from Sino Biological (Beijing, China). Luciferase assay system was purchased from Proemga Biotech (Madison, USA).
2.2. Cell lines
ACE2-overexpressing HEK293T cell (ACE2h) line was constructed by Genomeditech (Shanghai, China). ACE2h cells were cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 4 μg/mL puromycin, at 37 °C and 5% CO2.
2.3. Western blotting
Total protein in ACE2h cells was extracted on ice for 30 min using RIPA lysis buffer containing 10% protease inhibitor and a phosphatase inhibitor cocktail (Roche Diagnostics). The protein in cell lysates was denatured by boiling the samples with a 5 × loading buffer (Hat Biotechnology, Xi'an, China) for 5 min. Equal amounts of protein were separated on a 10% gel using SDS-PAGE (Hat Biotechnology, Xi'an, China). The separated proteins were transferred onto polyvinylidene fluoride membranes and blocked by constant stirring with 5% non-fat milk in Tris-buffered saline containing Tween-20 for 2 h at room temperature. Then the membranes were incubated overnight at 4 °C with the following primary antibodies: anti-GAPDH (1:2000, a#2118, CST), and anti-ACE2 (1:500, EPR4435, Abcam). The membranes were washed five times with TBST every 10 min followed by incubation with secondary antibodies at a dilution of 1:20,000 in TBST for 1 h, at 37 °C. The membranes were washed five times with TBST and developed using an ECL kit. ChemiDoc MP (Bio-Rad, California, USA) was used to image protein blot and Image-Pro 5.1 software (Media Cybernetics, Inc., Rockville, MD, USA) was used to quantify the protein levels.
2.4. ACE2h/cell membrane chromatography (ACE2h/CMC)
CMC is an affinity chromatography with dual characteristics of biometric and chromatography, which can mimic the binding of drug and receptor in vivo (Han et al., 2018). ACE2h/CMC columns (n = 3) were prepared with ACE2h cells following the published protocol (Hou et al., 2009). The CMC analysis was performed via LC-30A (Shimadzu, Japan). Azelastine was dissolved with methanol and then injected into the ACE2h/CMC column. ACE2h/CMC column was 10.0 mm × 2.0 mm; the flow rate was 0.2 mL/min; column temperature was 37 °C; the mobile phase was 2 mM phosphate-buffer saline (pH 7.4); detection wavelength was 284 nm. Only compounds with ACE2 affinity can be retained on the column, characterized by a long retention time.
2.5. Surface plasmon resonance (SPR) analysis
SPR assay was performed to detect the affinity of azelastine with ACE2 by using Open SPRTM (Nicoya, waterloo, Canada). To prime the sensor surface and charge the nitrilotriacetic acid (NTA) with Ni2+, 200 μL of 200 mM imidazole and 40 mM NiCl2 were injected onto the chip surface sequentially. ACE2 protein with a 6-his tag (20 μg/mL) was loaded onto and captured by the sensor chips coated in NTA (Nicoya, waterloo canada). At a flow rate of 20 μL/min, series concentrations of azelastine were injected into the system after the baseline was stabilized. The one-to-one diffusion-corrected model was fitted to the wavelength shifts corresponding to the series of drug concentrations and the experimental data was processed through TraceDrawer to calculate K D values.
2.6. Molecular docking
Molecular docking were carried out with SYBYL-X 2.0 (Tripos, St. Louis, USA) to investigate the interactions between azelastine and ACE2. The X-ray crystal structure of SARS-CoV-2 spike RBD bound with ACE2 (PDB code: 6LZG) was prepared by removing water, adding hydrogen, and extracting ligand. The force field was AMBER7 FF99 and performed the stage minimization of protein. Azelastine was added to the force field of tripos, assigned charge type of Gasteiger-Huckel, and performed energy minimization.
2.7. Cytotoxicity assay
The cytotoxicity test for azelastine was based on the cell viability after cells were treated with various concentrations of azelastine, and was determined by CCK8 method. 5 × 103 of ACE2h cells were seeded into 96-well microplates and incubated for 24 h under standard conditions (37 °C and 5% CO2). Then the medium was replaced with 100 μL serum-free DMEM or DMEM containing azelastine at concentrations of 0.1, 0.5, 5, 10, 20, 50, 100, and 200 μM. Cells were allowed to grow for an additional 24 h before measurement. 10 μL of CCK8 solution was added in each well and treated for 1.5 h at 37 °C. The optical density of samples at 450 nm (OD450) was measured using a microplate reader (Bio-Rad, USA). At least three independent experiments were performed. The survival rate of ACE2h cells was calculated using the following formula:
2.8. Pseudovirus infection assay
5 × 104 of ACE2h cells seeded into white opaque 96-well microplates were cultured at 37 °C in an incubator containing 5% CO2 until cells were adherent. Then the culture medium was aspirated and 100 μL DMEM containing the corresponding dose of azelastine was added and incubated for 2 h. Next, 5 μL of pseudovirus (104.4 TCID50/mL, 860 ng SARS-CoV-2 spike S1 protein/mL) was added to each well and incubated in a 37 °C incubator for 8 h of infection. The culture medium containing the pseudovirus was aspired and replaced by 200 μL of fresh DMEM and incubated continuously at 37 °C for 48 h. As for the determination, the medium was removed and 20 μL of lysis reagent was added in each well, and then 100 μL of luminescence solution was added. The light produced was detected by FlexStation 3 in a luminescence mode at 560 nm, with an exposure time of 1 s.
2.9. Statistical analysis
The data were analyzed using GraphPad Prism Software 8.0 (GraphPad Software, Inc., San Diego, CA, USA) and presented as the mean ± standard error of the mean (S.D.). Significant differences were determined by one-way ANOVA and Dunnett's test. Two-tailed unpaired Student's t-test was used for two-group comparisons. Differences were deemed statistically significant at P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
3. Results
3.1. ACE2h/cell membrane chromatography
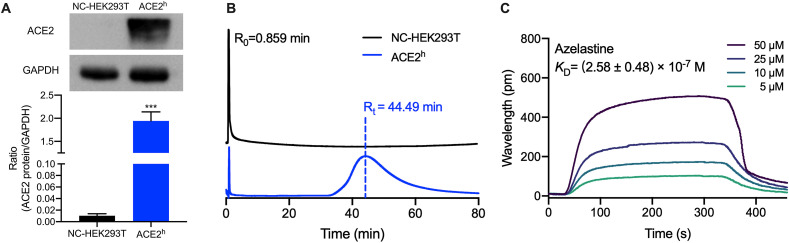
Considering that ACE2 is a transmembrane protein that is located on the cell membrane to form a specific spatial configuration, so it is necessary to investigate the binding of ACE2 in the form of membrane protein with azelastine. Cell chromatography is a technique for studying ligand-membrane protein interaction, which can be used to verify the biological affinity of azelastine to ACE2. The expression level of ACE2 protein in ACE2h cells is significantly higher than that in NC-HEK293T cells (Fig. 1 A), which ensures the specificity of CMC to recognize ligands. As can be seen from Fig. 1B that azelastine was retained on the ACE2h/CMC column, and its retention time is 44.49 min. The retention time of azelastine on the ACE2 column is much longer than the solvent peak's. In the NC-HEK293T group, azelastine passed directly through the column with the solvent, indicating that the non-specific binding of azelastine on the column is slight. This result means azelastine has a strong affinity to ACE2.
Fig. 1.
The binding character of azelastine with ACE2. (A)The protein expression level of NC-HKE293T and ACE2h. (B) The chromatogram of azelastine on the HEK293T/CMC and ACE2h/CMC model. (C) Binding response curves and KD of azelastine to ACE2 protein by SPR. Experiments were repeated three times.
3.2. The binding character of azelastine with ACE2
Now that we know that azelastine has an affinity for ACE2, it is necessary to test its binding force. Thus, SPR with recombined ACE2 protein was performed to verify the actual binding of azelastine to ACE2 protein. Azelastine brought about a concentration-dependent resonance change when flowing through the sensor chip coated with ACE2, indicating the direct binding of azelastine to ACE2 (Fig. 1C). The K D value was calculated by fitting the kinetic data at various concentrations of azelastine and recorded as (2.58 ± 0.48) × 10−7 M, demonstrating a moderately strong binding between azelastine and immobilized ACE2.
3.3. Molecular docking conformation and interaction of azelastine with ACE2
Computer simulation of small molecule-protein binding prediction is feasible since structures of both ACE2 and S protein RBD have been parsed. Therefore, molecular docking was carried out to investigate whether azelastine could interact with the active sites of ACE2 protein. As shown in Fig. 2 A, there were three important residuals in the region where azelastine was binding to the ACE2 protein (within 5 Å): His34, Glu37, and Lys353. Azelastine's carbonyl and Lys353's amino formed a hydrogen bond. Besides, Fig. 2B showed that azelastine's chlorobenzene fits into the active binding pocket of ACE2, which indicates the good interaction between azelastine and ACE2.
Fig. 2.
Molecular docking identifies possible binding pockets for azelastine in ACE2. (A) Structures of azelastine. (B) Docked pose of azelastine at ACE2 protein (inset: conformation of azelastine showing important interaction with the receptor at the active site). Active residues involved in binding are displayed as sticks in purple, with hydrogen bonds shown as yellow dashed lines. (C) Surface representation of the best ranked docking pose of azelastine in ACE2 binding pocket.
3.4. The effect of azelastine on ACE2h cells viability
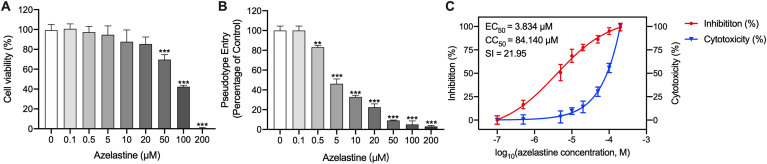
Cytotoxicity assay of azelastine at the concentrations ranged 0–200 μM was performed to determine the maximum concentration of azelastine that was no-toxic to cells. As shown in Fig. 3 A, the cytotoxicity was markedly increased following the up-regulation of azelastine concentration(>50 μM). At concentrations of 50, 100, and 200 μM, the vitality of ACE2h cells was 69.67 ± 2.91%, 42.33 ± 0.89%, 0.67 ± 0.33%. However, azelastine at 20 μM or lower concentrations showed little effect on cell viability. When concentration was 20 μM, cell viability remains 85.33 ± 6.88%.
Fig. 3.
Inhibiton of SARS-CoV-2 by azelastine. (A) Cell viability after incubation of ACE2h cells with azelastine for 24 h. (B) The entance of SARS-CoV-2 spike pseudovirus into ACE2h cells after treated with different concentration azelastine. (C) Inhibition analysis of SARS-CoV-2 spike pseudovirus by different concentration of azelastine. The left and right Y-axis of the graphs represent the mean percentage of inhibition of virus viropexis and cytotoxicity of the ACE2h cells, respectively.
3.5. Azelastine suppress the entry of SARS-cov-2 spike pseudovirus into ACE2h cells
Pseudovirus infection tests were performed to detect the effect of azelastine on the viropexis of SARS-CoV-2. In the control group, ACE2h cells were treated with the only solvent. As can be seen from Fig. 3B, the percentage of cells infected by SARS-CoV-2 spike pseudovirus was reduced significantly under the treatment of azelastine in a concentration-dependent way. There is no effect on viropexis using 0.1 μM azelastine. However, when the concentration raised to 0.5 μM, it began to display a significant difference compared to the control group, reducing the luciferase activity to 83.07 ± 1.16%. The inhibition is concentration-dependent with an EC50 of 3.834 μM of azelastine in the SARS-CoV-2 pseudovirus model (Fig. 3C). However, azelastine has significant cytotoxicity to ACE2h cells at high concentrations (>50 μM), and its CC50 is 84.140 μM (Fig. 3C). The selective index (SI), which was determined as the ratio of CC50 versus EC50 for azelastine is 21.95.
4. Discussion
Given the ongoing pandemic, there is an urgent need for specific drugs to prevent and treat SARS-CoV-2 infection. Although considerable research efforts have been devoted to drug repurposing, such as remdesivir, hydroxychloroquine, and lopinavir, it has been proved that they have little or no effect on hospitalized patients with Covid-19 (Consortium et al., 2021). Therefore, it is necessary to expand the scope of screening and continue to search for effective drugs. Considering that the antiviral effects of histamine receptor antagonists have been reported in recent years, and there is a partial overlap of antihistamine drugs and coronavirus's targets, we speculated that antihistamine drugs may have potential anti-SARS-CoV-2 effects. In this study, CMC with biological recognition characteristics was used to screen histamine H1 receptor antagonists, and we found that azelastine has an affinity for the viral therapeutic target ACE2. Subsequently, SPR and molecular docking were used to determine its binding force and binding site to ACE2. Finally, pseudovirus inhibition tests confirmed that azelastine can effectively inhibit the virus from entering cells.
The completion of the analysis of the crystal structure of the viral protein has facilitated the virtual screening of thousands of FDA-approved drugs for COVID-19. Although the virtual screening based on computer simulation can screen a large number of drugs quickly at a low cost, the actual hit rate of traditional methods is generally low. Cell membrane chromatography uses cell membrane protein receptors as a stationary phase to simulate drug-receptor interactions and can specifically identify target compounds in complex systems. Using this technology, the interaction properties of many receptor agonists and inhibitors have been discovered (Ma et al., 2021). ACE2 has been confirmed as the receptor for SARS-CoV-2 and a promising therapeutic target (Michaud et al., 2020). In our study, cell membrane chromatography was used to identify compounds that directly act on ACE2, exclude non-binding drugs, and improves the efficiency of high through-put screening. The results showed that azelastine has a high affinity with ACE2 (K D = 2.58 × 10−7 M), and forms a hydrogen bond with Lys353 of ACE2. It was worth noting that Lys353 was identified as one of the two virus binding hotspots on ACE2, which hides in a hydrophobic environment and provides a lot of energy for virus-receptor binding interactions (Sehailia and Chemat, 2020). The combination of azelastine weakens the hydrophobic network around the hot spot Lys353 and prevents the binding of Gly496, Gly502, and Tyr505 of the S protein (Rezaei et al., 2020), resulting in its inability to effectively support SARS-CoV-2 infection.
Previous studies have reported that histamine H1 receptor antagonists doxepin and desloratadine can inhibit the entry of viruses by binding to ACE2 Ge et al. (2021); (Hou et al., 2021). Surprisingly, we found that azelastine exhibits a stronger inhibitory effect at low concentrations, and it was statistically significant in reducing virus infection at a concentration of 0.5 μM in vitro. It should be mentioned that recent studies have shown that azelastine can also bind to the main protease of SARS-CoV-2 (Odhar et al., 2020) and the sigma-1 receptor (Reznikov et al., 2021) through molecular docking. Thus it was speculated that azelastine has a potential inhibitory effect on the replication of SARS-CoV-2. On the other hand, the SARS-CoV-2 can activate mast cells (Theoharides, 2020) to cause the early release of histamine and activate the late release of IL-1 from macrophages (Conti et al., 2020). As a H1 receptor antagonist, azelastine is expected to reduce the cytokine storm by inhibiting histamine. Azelastine not only has antiviral activity, but also has anti-inflammatory activity. Therefore, we believe that it is an attractive compound that is conducive to the repurposing of COVID-19 drugs.
In the pseudovirus infection test, the luciferase activity was determined by luciferase assay, and the cells viability during the incubation process is crucial for it. To avoid false-positive results caused by cytotoxicity, we tested the cytotoxicity of azelastine to ACE2h at all experimental concentrations. It was significant cytotoxic to ACE2h cells at high concentrations (>50 μM). This result indicated that the reduction of infection rate mediated by the high concentration of azelastine has the interference of cytotoxicity. However, at low concentrations, azelastine could effectively inhibit virus entry into cells without cytotoxicity, which proved the reliability of the results. Subsequently, the EC50 of azelastine was determined to be 3.834 μM, which was much lower than the CC50 (84.140 μM). The selectivity index of 21.95, indicating that azelastine has a wide safety range.
Azelastine is a third-generation antihistamine, which has been shown to be efficacious with few adverse events including no clinically relevant cytochrome P450 mediated metabolic-based drug-drug interactions or QT interval prolongation/cardiac dysrhythmias (Ten Eick et al., 2001). In acute toxicity study, the median lethal dose of oral azelastine in rats is 580 mg/kg; In the subacute toxicity study, the minimum toxic dose was found to be 30 mg/kg and symptoms of depression of the central nervous system were recognized in the 100 mg/kg group. However, rats administered at oral doses of 10 mg/kg for 5 consecutive weeks were free of particular drug effects, which has proved that azelastine is a substance of, in general, good tolerance (Nakao et al., 1981). The azelastine concentrations used in this study effectively inhibiting viropexis of SARS-CoV-2 spike pseudovirus are far below the minimum toxic dose. The recommended oral dosage of azelastine for prophylaxis and maintenance therapy with bronchial asthma is 4 mg twice daily (McTavish and Sorkin, 1989). Although azelastine has been widely used in clinical practice, it is still unclear whether the conventional dosage of azelastine is suitable for antiviral therapy. Therefore, we still need to cautiously carry out further research and use authentic SARS-CoV-2 and animal models for verification in vivo.
As SARS-CoV-2 is a highly infectious and high-risk virus, its cultivation and animal infection experiments must be carried out in a biosafety level-3 (BSL-3). Although only in vitro tests were performed in the current study, we tried our best to use existing materials and methods to provide insights. In addition, our research team has screened and reported a series of FDA-approved drugs with potential anti-SARS-CoV-2 activity, such as antipsychotic drugs (Lu et al., 2021), antihistamine drugs Ge et al. (2021); (Hou et al., 2021), glucocorticoids (Zhang et al., 2021), and active ingredients of Chinese medicine Gao et al. (2021); Hu et al. (2021); (Lv et al., 2021). Besides.
5. Conclusions
We found that the antihistamine azelastine can inhibit the entry of pseudoviruses into cells in vitro in combination with ACE2. Azelastine is expected to become a valuable dual-target clinical candidate drug for inhibiting the virus and alleviating the inflammation of COVID-19. At the same time, azelastine can also be modified as a lead compound to develop more effective anti-SARS-CoV-2 drugs. We hope our work can provide reasonable help for the drug repurposing for COVID-19 treatment.
Availability of data
All data generated or analyzed during this study are included in this published article.
CRediT authorship contribution statement
Shuai Ge: Conceptualization, Methodology, Investigation, Writing – original draft. Jiayu Lu: Data curation, Investigation. Yajing Hou: Investigation, Software. Yuexin Lv: Formal analysis, Visualization. Cheng Wang: Investigation, Validation. Huaizhen He: Writing – review & editing, Supervision, Resources, All authors read and approved the final manuscript.
Declaration of competing interest
All authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant Number: 81930096, 81903573), the Postdoctoral Research Foundation of China (Grant Number: 2018M643682), and the Natural Science Basic Research Plan in Shaanxi Province of China (Grant Number: 2020JQ-089).
References
- Asselah T., Durantel D., Pasmant E., Lau G., Schinazi R.F. COVID-19: discovery, diagnostics and drug development. J. Hepatol. 2021;74:168–184. doi: 10.1016/j.jhep.2020.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M., Scott N.W., Mustafa M.Z., Mustafa M.S., Azuara-Blanco A. Topical antihistamines and mast cell stabilisers for treating seasonal and perennial allergic conjunctivitis. Cochrane Database Syst. Rev. 2015;(6):1456–1858. doi: 10.1002/14651858.CD009566.pub2. CD009566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium W.H.O.S.T., Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernandez Garcia C., Kieny M.P., Malekzadeh R., Murthy S., Reddy K.S., Roses Periago M., Abi Hanna P., Ader F., Al-Bader A.M., Alhasawi A., Allum E., Alotaibi A., Alvarez-Moreno C.A., Appadoo S., Asiri A., Aukrust P., Barratt-Due A., Bellani S., Branca M., Cappel-Porter H.B.C., Cerrato N., Chow T.S., Como N., Eustace J., Garcia P.J., Godbole S., Gotuzzo E., Griskevicius L., Hamra R., Hassan M., Hassany M., Hutton D., Irmansyah I., Jancoriene L., Kirwan J., Kumar S., Lennon P., Lopardo G., Lydon P., Magrini N., Maguire T., Manevska S., Manuel O., McGinty S., Medina M.T., Mesa Rubio M.L., Miranda-Montoya M.C., Nel J., Nunes E.P., Perola M., Portoles A., Rasmin M.R., Raza A., Rees H., Reges P.P.S., Rogers C.A., Salami K., Salvadori M.I., Sinani N., Sterne J.A.C., Stevanovikj M., Tacconelli E., Tikkinen K.A.O., Trelle S., Zaid H., Rottingen J.A., Swaminathan S. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Ding Y., Wang Y., Liang P., Zhang L., Liu R. Oroxylin A is a severe acute respiratory syndrome coronavirus 2-spiked pseudotyped virus blocker obtained from Radix Scutellariae using angiotensin-converting enzyme II/cell membrane chromatography. Phytother Res. 2021 doi: 10.1002/ptr.7030. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Wang X., Hou Y., Lv Y., Wang C., He H. Repositioning of histamine H1 receptor antagonist: doxepin inhibits viropexis of SARS-CoV-2 Spike pseudovirus by blocking ACE2. Eur. J. Pharmacol. 2021;896:173897. doi: 10.1016/j.ejphar.2021.173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Lv Y., Wei F., Fu J., Hu Q., Wang S. Screening of bioactive components from traditional Chinese medicines using cell membrane chromatography coupled with mass spectrometry. Phytochem. Anal. 2018;29:341–350. doi: 10.1002/pca.2756. [DOI] [PubMed] [Google Scholar]
- Hou X., Zhou M., Jiang Q., Wang S., He L. A vascular smooth muscle/cell membrane chromatography-offline-gas chromatography/mass spectrometry method for recognition, separation and identification of active components from traditional Chinese medicines. J. Chromatogr. A. 2009;1216:7081–7087. doi: 10.1016/j.chroma.2009.08.062. [DOI] [PubMed] [Google Scholar]
- Hou Y., Ge S., Li X., Wang C., He H., He L. Testing of the inhibitory effects of loratadine and desloratadine on SARS-CoV-2 spike pseudotyped virus viropexis. Chem. Biol. Interact. 2021;338:109420. doi: 10.1016/j.cbi.2021.109420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Wang J., Zhang Y., Bai H., Wang C., Wang N., He L. Three salvianolic acids inhibit 2019-nCoV spike pseudovirus viropexis by binding to both its RBD and receptor ACE2. J. Med. Virol. 2021;93(5):3143–3151. doi: 10.1002/jmv.26874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Hou Y., Ge S., Wang X., Wang J., Hu T., Lv Y., He H., Wang C. Screened antipsychotic drugs inhibit SARS-CoV-2 binding with ACE2 in vitro. Life Sci. 2021;266:118889. doi: 10.1016/j.lfs.2020.118889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Wang S., Liang P., Wang Y., Zhang X., Jia Q., Fu J., Han S., He L. Screening and evaluation of anti-SARS-CoV-2 components from Ephedra sinica by ACE2/CMC-HPLC-IT-TOF-MS approach. Anal. Bioanal. Chem. 2021;413(11):2995–3004. doi: 10.1007/s00216-021-03233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Wang C., Liu R., Wang N., Lv Y., Dai B., He L. Advances in cell membrane chromatography. J. Chromatogr. A. 2021;1639:461916. doi: 10.1016/j.chroma.2021.461916. [DOI] [PubMed] [Google Scholar]
- McTavish D., Sorkin E.M. Azelastine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential. Drugs. 1989;38:778–800. doi: 10.2165/00003495-198938050-00005. [DOI] [PubMed] [Google Scholar]
- Michaud V., Deodhar M., Arwood M., Al Rihani S.B., Dow P., Turgeon J. ACE2 as a therapeutic target for COVID-19; its role in infectious processes and regulation by modulators of the RAAS system. J. Clin. Med. 2020;9 doi: 10.3390/jcm9072096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao K., Wakabayashi T., Sugiyama K., Taki T., Sumigama S., Chiba T. Subacute and chronic toxicity studies in rats and dose range finding study in dogs by oral administration of azelastine. Arzneimittelforschung. 1981;31:1220–1225. [PubMed] [Google Scholar]
- Odhar H.A., Ahjel S.W., Albeer A., Hashim A.F., Rayshan A.M., Humadi S.S. Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation. 2020;16:236–244. doi: 10.6026/97320630016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei S., Sefidbakht Y., Uskokovic V. Comparative molecular dynamics study of the receptor-binding domains in SARS-CoV-2 and SARS-CoV and the effects of mutations on the binding affinity. J. Biomol. Struct. Dyn. 2020;1–20 doi: 10.1080/07391102.2020.1860829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikov L.R., Norris M.H., Vashisht R., Bluhm A.P., Li D., Liao Y.J., Brown A., Butte A.J., Ostrov D.A. Identification of antiviral antihistamines for COVID-19 repurposing. Biochem. Biophys. Res. Commun. 2021;538:173–179. doi: 10.1016/j.bbrc.2020.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., Chang M.W., Chan J.F., Cao J., Poon V.K., Herbert K.M., Cheng K., Nguyen T.H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T.P., Sit K.Y., Martinez-Sobrido L., Liu W.C., White K.M., Chapman M.E., Lendy E.K., Glynne R.J., Albrecht R., Ruppin E., Mesecar A.D., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A.I., Garcia-Sastre A., Chatterjee A.K., Yuen K.Y., Chanda S.K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., Cheng H., Xiong R., Soloveva V., Retterer C., Mo F., Bavari S., Thatcher G., Rong L. Repurposing potential of 1st generation H1-specific antihistamines as anti-filovirus therapeutics. Antivir. Res. 2018;157:47–56. doi: 10.1016/j.antiviral.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehailia M., Chemat S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 spike protein than hydroxychloroquine: potential repurposing of artenimol for COVID-19. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Eick A.P., Blumer J.L., Reed M.D. Safety of antihistamines in children. Drug Saf. 2001;24:119–147. doi: 10.2165/00002018-200124020-00003. [DOI] [PubMed] [Google Scholar]
- Thangam E.B., Jemima E.A., Singh H., Baig M.S., Khan M., Mathias C.B., Church M.K., Saluja R. The role of histamine and histamine receptors in mast cell-mediated allergy and inflammation: the hunt for new therapeutic targets. Front. Immunol. 2018;9:1873. doi: 10.3389/fimmu.2018.01873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theoharides T.C. COVID-19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. Biofactors. 2020;46:306–308. doi: 10.1002/biof.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Xu J., Xue Y., Zhou R., Shi P.Y., Li H., Zhou J. Drug repurposing approach to combating coronavirus: potential drugs and drug targets. Med. Res. Rev. 2020;4(13):1375–1426. doi: 10.1002/med.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Xia S., Pu J., Wang Q., Li P., Lu L., Jiang S. The antihistamine drugs carbinoxamine maleate and chlorpheniramine maleate exhibit potent antiviral activity against a broad spectrum of influenza viruses. Front. Microbiol. 2018;9:2643. doi: 10.3389/fmicb.2018.02643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuce M., Filiztekin E., Ozkaya K.G. COVID-19 diagnosis -A review of current methods. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hu S., Wang J., Xue Z., Wang C., Wang N. Dexamethasone inhibits SARS-CoV-2 spike pseudotyped virus viropexis by binding to ACE2. Virology. 2021;554:83–88. doi: 10.1016/j.virol.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., Brindley M.A., Lewis N.E., Tiemeyer M., Chen B., Woods R.J., Wells L. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1016/j.chom.2020.08.004. e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Xu Z., Castiglione G.M., Soiberman U.S., Eberhart C.G., Duh E.J. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul. Surf. 2020;18:537–544. doi: 10.1016/j.jtos.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.