Abstract
Chronic myeloid leukemia (CML) accounts for approximately 15% of new adult leukemia cases. The fusion gene BCR-ABL is an important biological basis and target for CML. In the present study, a novel compound, ND-09, was developed and its inhibitory effect and mechanism of action on CML growth were evaluated using RT-PCR and western blot analysis. The results showed that ND-09 demonstrated a high level of inhibitory action toward CML cells overexpressing BCR-ABL and induced K562 cell apoptosis through the mitochondrial pathway. Notably, combined ND-09 and BCR-ABL siRNA treatment could better inhibit cell proliferation and induce apoptosis in K562 cells. Furthermore, this growth effect of BCR-ABL siRNA could be fully rescued by transfection with BCR-ABL. ND-09 exhibited a good fit within BCR-ABL and occupied its ATP-binding pocket, thus altering BCR-ABL kinase activity. Therefore, ND-09 downregulated the phosphorylation of BCR-ABL and ABL, ultimately inhibiting the downstream signaling pathways in K562 cells. These findings suggest that ND-09 induces growth arrest in CML cells by targeting BCR-ABL.
Keywords: ND-09, BCR-ABL, chronic myeloid leukemia, cell apoptosis, cell cycle
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic stem cell disorder that affects the blood and bone marrow and accounts for about 15% of newly diagnosed adult leukemia cases (1). CML is a myeloproliferative disease comprising three stages, including a chronic phase, accelerated phase, and lymphatic/myeloid blast phase, and is characterized by the Philadelphia chromosome. The Ph chromosome is a specific CML diagnostic marker and is present in more than 95% of patients (2,3). This chromosome is the result of a translocation between chromosomes 9 and 22, leading to the generation of a chimeric gene product of the breakpoint cluster region (BCR) and the Abelson murine leukemia (ABL), a fusion gene known as BCR-ABL (4,5). The BCR-ABL fusion gene can form a chimeric protein, resulting in a constitutively active ABL kinase, which can transform hematopoietic stem cells into leukemic stem cells and actuate the overproduction and expansion of leukocytes in the bone marrow (6,7). Thus, the BCR-ABL fusion protein is an important molecular basis for CML pathogenesis.
The oncogenic BCR-ABL fusion protein has persistent kinase activity, resulting in the uncontrolled proliferation of myeloid cells through multiple downstream pathways (2). BCR-ABL phosphorylates several substrates that activate a number of aberrant kinase-dependent pathways, including the Ras-mitogen-activated protein kinase (MAPK) pathway, which results in increased proliferation; the Janus-activated kinase/signal transducer and activator of transcription (JAK/STAT) pathway, which impairs transcriptional activity, and the phosphatidylinositide 3-kinase/protein kinase B (PI3K/AKT) pathway, which leads to enhanced survival (8–10). Thus, BCR-ABL has been recognized as the most important target for CML treatment.
CML therapy has seen impressive advances with the development of tyrosine kinase inhibitors (TKIs) against the BCR-ABL pathway (11), revolutionizing CML management and resulting in a 10-year overall survival rate of more than 90% (12). In terms of CML treatment, nilotinib (a second-generation TKI) has certain advantages over the first-generation TKI imatinib, such as improved response kinetics, a significant progression rate reduction, and deeper molecular responses (13,14). Although existing TKIs are highly effective in CML patients, many patients develop drug resistance. Therefore, the development of novel kinase inhibitors in an effort to overcome drug resistance is crucial (15).
ND-09 (4-methoxyl-3-[[4-(3-pyridyl)-2-pyrimidinyl]amino] benzoic acid pyrimidin-2-ylamino-benzamides) is a novel compound developed by our group (16). The aim of the present study was to investigate whether ND-09 could regulate BCR-ABL signaling to suppress CML growth.
Materials and methods
Chemicals and reagents
ND-09 (Fig. 1A) was developed in the Research and Engineering Center for Natural Medicine, Xi'an Jiaotong University (Shaanxi, China). K562, HUT78 and JURKAT cells were obtained from the Shanghai Institute of Cell Biology in the Chinese Academy of Sciences (Shanghai, China). Iscove's modified Dulbecco's medium (IMDM) and RPMI-1640 medium were purchased from Sigma-Aldrich. 3-(4,5-Dimethylthiazol-2-yl)-2.5-diphenyl-2H-tetrazolium brom-ide (MTT) were purchased from Sigma-Aldrich. Fetal bovine serum (FBS) was obtained from ExCell Bio Co., Ltd.
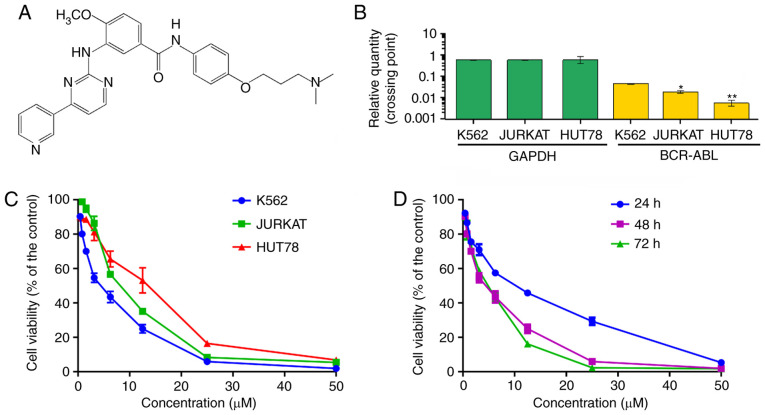
Figure 1.
The effect of ND-09 on hematologic tumor cell proliferation. (A) Chemical structure of ND-09. (B) mRNA level of BCR-ABL in hematologic tumor cells. (C) An MTT assay was used to evaluate the inhibitory effect of ND-09 on K562, JURKAT, and HUT78 cell proliferation. (D) An MTT assay was used to evaluate the inhibitory effect of ND-09 on K562 cell proliferation at different time points. *P<0.05, **P<0.01 compared to the untreated control group.
Thermal Cycle Dice Real time system, PrimeScript RT Master Mix Perfect Real Time kit, and SYBR® Premix Ex Taq™ II were obtained from Takara Biotechnology. Annexin V-FITC cell apoptosis detection kit was obtained from Pioneer Biotechnology Co., Ltd. RNase and propidium iodide (PI) were obtained from Sigma-Aldrich. Opti-MEM medium was purchased from Gibco. Lipofectamine 2000 reagent was obtained from Invitrogen. Phosphatase and protease inhibitors were obtained from Roche Tech. The BCA kit, RNAfast200 kit, and RIPA lysis buffer were obtained from Pioneer Biotechnology Co., Ltd. BCR-ABL and control siRNA were obtained from Shanghai GenePharma Co., Ltd.
The mcl-1 (monoclonal, 66026-1-Ig), Bad (monoclonal, 67830-1-Ig), Bcl-2 (monoclonal, 60178-1-Ig), Bak (polyclonal, 14673-1AP), Bax (monoclonal, 60267-1-Ig), CDC2 (monoclonal, 67575-1-Ig), Cyclin D1 (monoclonal, 60186-1-Ig), Cyclin E (polyclonal, 11554-1-AP), CDK2 (monoclonal, 60312-1-Ig), Cyclin A2 (monoclonal, 66391-1-Ig), and Cyclin B1 (monoclonal, 67686-1-Ig) antibodies were purchased from Protein Technology Group. The phospho-BCR-ABL (polyclonal, Tyr177), BCR-ABL (monoclonal, L99H4), phospho-ABL (monoclonal, 73E5), ABL (polyclonal, 2862), phospho-BCR (polyclonal, Tyr177), and BCR (polyclonal, 3902) antibodies were obtained from Cell Signaling Technology. The JAK2 (monoclonal, D2E12), phospho-JAK2 (monoclonal, D15E2), JAK3 (monoclonal, D1H3), phospho-JAK3 (monoclonal, D44E3), STAT3 (monoclonal, D3Z2G), phospho-STAT3 (monoclonal, D3A7), STAT5 (monoclonal, D3N2B), and phospho-STAT5 (monoclonal, D47E7) antibodies were obtained from Cell Signaling Technology. The p53 (polyclonal, 10442-1-AP), p38 (monoclonal, 66234-1-Ig), and PTEN (monoclonal, 60300-1-Ig) antibodies were obtained from the Protein Technology Group. The phospho-p38 (monoclonal, D3F9), PI3K-p110α (monoclonal, C73F8), PI3K-p110β (monoclonal, C33D4), PI3K-p110γ (monoclonal, D55D5), PI3K-p85 (monoclonal, 19H8), and phospho-PI3K-p85/p55 (monoclonal, E3U1H) antibodies were obtained from Cell Signaling Technology. The AKT (monoclonal, C67E7), phospho-AKT (monoclonal, D9E), PLCγ (monoclonal, D9H10), phospho-PLCγ (monoclonal, D6M9S), Erk1/2 (monoclonal, 137F5), phospho-Erk1/2 (monoclonal, Tyr202/204), MEK1/2 (monoclonal, D1A5), phospho-MEK1/2 (monoclonal, 166F8), mTOR (monoclonal, 7C10), and phospho-mTOR (monoclonal, D9C2) antibodies were obtained from Cell Signaling Technology. Rabbit anti-GAPDH (monoclonal, 60004-1-lg) was obtained from Protein Technology Group. The working solution for the primary antibodies was 1:1000, except for the Bad and Cyclin E antibodies, which was 1:500. HRP-conjugated goat anti-rabbit IgG (secondary antibody, dilution: 1:20,000) was obtained from Pierce Biotechnology. Enhanced Chemiluminescent Plus Reagent was obtained from Biotech Co., Ltd.
Cell culture
K562 and HUT78 cells were cultured in IMDM medium containing 10% FBS, 100 U/ml penicillin, and 100 U/ml streptomycin. JURKAT cells were cultured in RPMI-1640 medium containing 10% FBS, 100 U/ml penicillin, and 100 U/ml streptomycin. All cell lines were grown at 37°C in a 95% humidified atmosphere with 5% CO2.
Cell viability assay
K562, JURKAT and HUT78 cells were seeded in 96-well plates and cultivated in complete medium for 24 h. Then cells were treated with ND-09 (0.39, 0.78, 1.56, 3.12, 6.25, 12.5, 25, and 50 µM) for 48 h. Then, 20 µl MTT solution (5 mg/ml) was added to each well and incubated at 37°C for 4 h. Subsequently, the cells were enriched at the bottom of each well using a plate spinner (4,192.5 × g for 5 min at 25°C) and the medium was slowly removed from each well. Then, 150 µl DMSO was added in each well for 15 min. The absorbance was recorded by a microplate reader (Bio-Rad) at 490 nm.
Flow cytometric analysis of cell apoptosis
Cells were seeded in 6-well plates and treated with ND-09 for 48 h, after which the cells were collected by centrifugation (4,192.5 × g for 5 min at 25°C), washed, and resuspended in PBS. According to the instructions, Annexin V-FITC and PI double staining were used to detect the apoptotic rate. Then, Cell Quest software was used for flow cytometry and data analysis.
Determination of mitochondrial transmembrane potential (Δψm)
Cells were seeded in 6-well plates and treated with ND-09 for 48 h, after which the cells were washed with complete medium, followed by incubation with 1 mM Rhodamine 123 at 37°C in the dark for 30 min. Then BD FACSCalibur Flow cytometer was used to perform flow cytometric analysis and analyze data.
Flow cytometric analysis of cell cycle
Cells were seeded in 6-well plates, treated with ND-09 for 48 h, and then fixed in ice-cold 70% ethanol at 4°C overnight. Then, the cells were washed with cold PBS and stained with RNase and PI for 30 min in the dark. Cell Quest software was used to perform flow cytometric analysis and analyze data.
RNA extraction and RT-PCR assay
Cells were seeded in 6-well plates and treated with ND-09 for 48 h. According to the manufacturer's protocol, total RNA from K562, JURKAT and HUT78 cells was extracted using RNAfast200 kit (Pioneer Biotechnology Co., Ltd). RT-PCR was performed using PrimeScript RT Master Mix Perfect Real Time kit and was performed using SYBR® Premix Ex Taq™ II and Thermal Cycle Dice Real time system. GAPDH was considered the reference gene. The primer sequences used were: GAPDH forward: 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse: 5′-TGGTGAAGACGCCAGTGGA-3′; BCR-ABL forward: 5′-CATTCCGCTGACCATCAATAAG-3′ and reverse: 5′-GATGCTACTGGCCGCTGAAG-3′.
Threshold cycle (Ct) values of BCR-ABL in each sample were normalized with the GAPDH expression. The ratio of BCR-ABL versus the corresponding GAPDH of each sample was determined on the basis of the equation BCR-ABL/GAPDH=2Cq(GAPDH)-Cq(BCR-ABL).
SiRNA transfection
K562 cells were seeded in cell culture dishes at a density of approximately 50%. According to the manufacturer's protocol, Lipofectamine 2000 reagent was used to transfect anti-BCR-ABL siRNA or control siRNA (100 nM) into cells for 24 h. Then, transfected cells were immediately seeded to perform subsequent assays.
Plasmid transfection
K562 cells were seeded into 6-well plates at the density of 2×105 per well. After 24 h, transfection of EphB4 plasmid into BCR-ABL-depleted K562 cells was performed using Lipofectamine 2000 reagent. The ratio of Lipofectamine 2000 (µl) to BCR-ABL plasmid (µg) was 2:1.
Molecular docking (MD)
SYBYL-X 1.1 was used to conduct docking studies to understand the binding mode of ND-09 and BCR-ABL domain (PDB ID: 1IEP). The substrate was constructed by Sybyl/Sketch modul and optimized by Powell's method. Tripos force field and Gasteiger-Hückel charges were used to minimize energy, the convergence criterion was set at 0.005 kcal/(Å mol), and the maximum value was set at 1,000 iterations. To explore intramolecular interactions, the non-bonded cut-off distance was set to 8 Å.
Kinase assay
Firstly, BCR-ABL kinase (2 µl, 5 ng/µl) and substrate (2 µl, 10 µM) were added to 384-well plates, and then drugs (4 µl) were added to the test plate at different concentrations (0.016, 0.008, 0.040, 0.20, 1.02, 5.12, 25.60, 128.00, and 640 nM). ATP solution (2 µl, 1 mM) was added and the system was incubated at 37°C for 30 min. TK-Antibody (5 µl, 1,000 tests, reconstituted with 5 ml of HTRF® detection buffer) and streptavidin-XL665 (5 µl, 125 nM) were then added to the test plate at room temperature for 1 h.
Western blot analysis
Cells treated with ND-09 were seeded in 6-well plates. RIPA buffer with protease and phosphatase inhibitor were used to extract protein from cells. The cell lysate was then concentrated at 12,000 × g for 10 min at 4°C. Protein (40 µg) quantified by using BCA kit was then loaded to 10% SDS-PAGE gel, after which protein was transferred to polyvinylidene difluoride membranes. The membranes with protein were blocked with Tris-buffered saline for 1 h at room temperature. Subsequently, the primary antibody was incubated overnight at 4°C, and the secondary antibody was incubated at room temperature for 1 h. The blot was then exposed to the Enhanced Chemiluminescent substrate. Band intensity was quantified by densitometric analysis using an image quantitative analysis system (Image-Pro Plus 5.1; Media Cybernetics Inc.).
Statistical analysis
Statistical analyses were performed using SPSS 18.0. All the experiments were performed at least three times, and the results are expressed as mean ± SD. Statistical analyses of differences between the groups were performed with ANOVA followed by the Tukey post-hoc test. P<0.05 was considered statistically significant, and P<0.01 was considered extremely significant.
Results
Inhibitory effect of ND-09 on the proliferation of hematologic cancer cells
In order to evaluate the inhibitory effect of ND-09 on hematologic tumor cell growth, CML K562 cells, cutaneous T-cell lymphoma HUT78 cells, and acute T-cell lymphoma JURKAT cells were treated with ND-09. As BCR-ABL is a key factor in the myeloproliferative disorder of CML, BCR-ABL expression was investigated in all three cell lines. The results indicated that the expression of BCR-ABL in K562 cells was higher than that in JURKAT and HUT78 cells (Fig. 1B). As seen in Fig. 1C, K562 cells were more sensitive to ND-09 than JURKAT and HUT78 cells. The IC50 values of ND-09 for K562, JURKAT, and HUT78 cells were 6.08, 7.81, and 14.43 µM, respectively. Furthermore, ND-09 inhibited the proliferation of K562 cells in a dose- and time-dependent manner (Fig. 1D). Thus, the inhibitory effect of ND-09 on K562 cell growth may be related to BCR-ABL.
ND-09 induces cell apoptosis
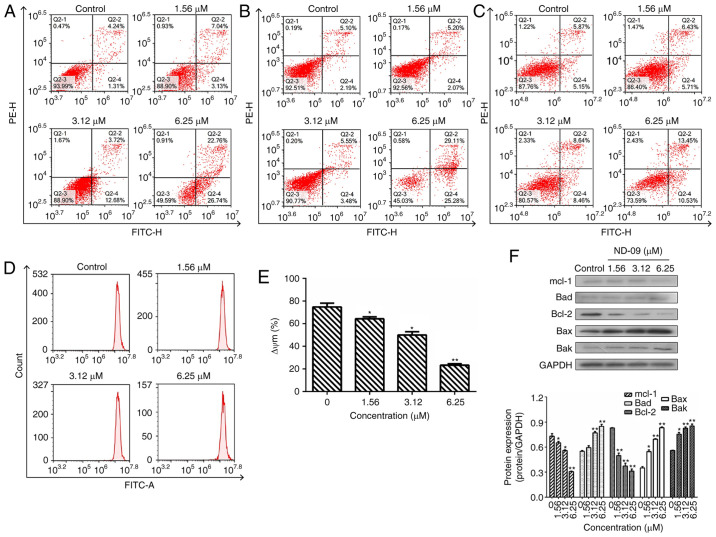
Following ND-09 treatment of 1.56, 3.12, and 6.25 µM, the percentage of apoptotic K562 cells increased to 10.17±1.03, 16.40±1.41, and 49.50±1.25%, respectively, compared to control cells (5.55±0.93%) (Fig. 2A). In addition, ND-09 could induce JURKAT and HUT78 cell apoptosis (Fig. 2B and C). The percentage of apoptotic JURKAT cells increased to 7.27±0.95, 9.03±0.83, and 54.39±1.79%, respectively, compared to control cells (7.29±1.02%). The percentage of apoptotic HUT78 cells increased to 12.14±1.15, 17.10±1.34, and 23.98±1.67%, respectively, compared to control cells (11.02±0.96%). The above data indicated that the apoptosis-inducing effect of ND-09 in the two cell lines was relatively weaker than that in K562 cells.
Figure 2.
Effect of ND-09 treatment on cell apoptosis. The proportion of apoptotic cells was determined by double staining with Annexin V/FITC and PI in (A) K562, (B) JURKAT, and (C) HUT78 cells after treatment with ND-09 (0, 1.56, 3.12, and 6.25 µM). (D) Effect of ND-09 on mitochondrial membrane potential (Δψm). Δψm was assessed through flow cytometry following treatment of K562 cells with ND-09 (0, 1.56, 3.12, and 6.25 µΜ) for 48 h. (E) Quantitative analysis of flow cytometry data. (F) Effects of ND-09 on apoptosis-related protein expression in K562 cells. All results were quantified by densitometric analysis of the bands and were normalized to GAPDH (internal control). Samples were derived from the same experiment, and blots were processed in parallel. Values represent the average of three independent experiments. Data are presented as mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group.
Subsequently, the effect of ND-09 on mitochondrial transmembrane potential was examined. Rhodamine 123 fluorescence intensity of K562 cells exposed to ND-09 decreased significantly from 74.82±2.58% in the control group to 64.26±2.15% (1.56 µΜ), 49.98±1.69% (3.12 µΜ), and 23.25±1.76% (6.25 µΜ) (Fig. 2D and E).
To explore the mechanism of ND-09-induced cell apoptosis, western blot analysis was performed to detect the levels of apoptosis-related proteins. Results indicated that Bak, Bax, and Bad levels significantly increased after ND-09 treatment, while Bcl-2 and Mcl-1 levels decreased in a dose-dependent manner (Fig. 2F).
ND-09 induces G0/G1 phase arrest in K562 cells
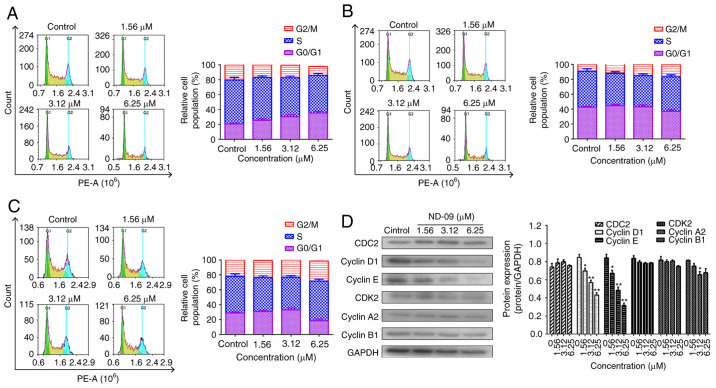
ND-09 treatment led to a significant increase in G0/G1 phase cells (Fig. 3A). At increasing ND-09 concentrations of 1.56, 3.12 and 6.25 µM, the K562 cell population in G0/G1 phase increased by 25.56±1.15, 30.29±1.43, and 35.31±1.37% compared to the untreated cells (20.61±1.26%). A different mode of cell cycle arrest induction was observed in JURKAT and HUT78 cells, as these were arrested in the G2/M phase (Fig. 3B and C).
Figure 3.
Effect of ND-09 treatment on the cell cycle. Representative flow cytometry DNA content histogram of (A) K562, (B) JURKAT, and (C) HUT78 cells after treatment with ND-09 (0, 1.56, 3.12 and 6.25 µM). (D) Effects of ND-09 on cell cycle-related protein expression in K562 cells. All results were quantified by densitometric analysis of the bands and were normalized to GAPDH (internal control). Samples were derived from the same experiment, and blots were processed in parallel. Values represent the average of three independent experiments. Data are presented as mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group.
As ND-09 arrested the K562 cell cycle in the G0/G1 phase, levels of CDK/cyclin proteins were evaluated through western blot analysis. Fig. 3D revealed that cyclin E and cyclin D1 levels decreased in ND-09-treated K562 cells.
Role of BCR-ABL on the effects of ND-09 on cell growth
A siRNA assay was carried out to determine whether BCR-ABL was a key factor for the ND-09-induced inhibition of K562 cell growth. First, BCR-ABL knockdown and BCR-ABL rescue of K562 cells were successfully established (Fig. 4A and B).
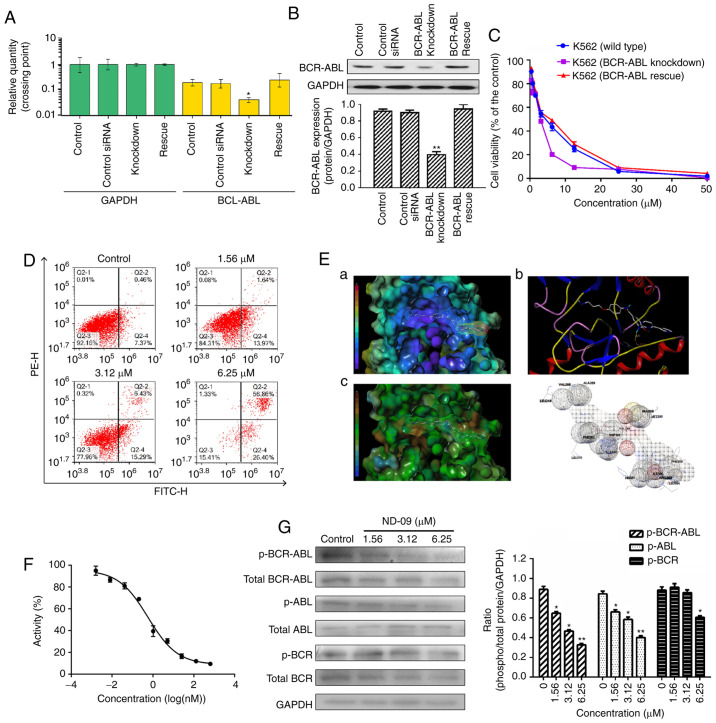
Figure 4.
The role of BCR-ABL in the biological activity of ND-09 treatment. (A) BCR-ABL mRNA expression and (B) protein expression in wild-type, BCR-ABL knockdown, and BCR-ABL rescue K562 cells were determined by RT-PCR analysis and western blot analysis, respectively. Cells transfected with a control siRNA construct served as negative controls. Samples are derived from the same experiment, and blots were processed in parallel. (C) An MTT assay was used to evaluate the effect of ND-09 on cell proliferation in wild-type, BCR-ABL knockdown, and BCR-ABL rescue cells. (D) Effect of ND-09 on apoptosis in BCR-ABL-depleted K562 cells. Values represent the average of three independent experiments. Data are presented as mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group. (E) The binding mode of ND-09 to BCR-ABL (PDB ID: 1IEP). a, ND-09 in the crystal structure of BCR-ABL with electrostatic coloring; b, Hydrogen bonds are depicted by dashed yellow lines; c, ND-09 in the crystal structure of BCR-ABL with hydrophobic coloring. (F) Effect of ND-09 on BCR-ABL kinase activity. Values represent the average of three independent experiments. Values are presented as mean ± SEM (n=3). (G) Levels of BCR-ABL, p-BCR-ABL, ABL, p-ABL, BCR, and p-BCR in K562 cells treated with ND-09 were examined by western blot analysis. Results were quantified by densitometric analysis of the bands and were normalized to GAPDH (internal control). Samples were derived from the same experiment, and blots were processed in parallel. Values represent the average of three independent experiments. Data are presented as the mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group.
Then, to assess the role of BCR-ABL in ND-09-induced cell growth inhibition, wild-type and BCR-ABL-depleted K562 cells were treated with ND-09. ND-09 had a dose-dependent inhibitory effect on the proliferation of the two groups, and combined ND-09 and siRNA treatment resulted in an enhanced inhibitory effect (Fig. 4C). The IC50 values of ND-09 in wild-type and BCR-ABL-depleted K562 cells were 6.08 and 3.61 µM, respectively. Furthermore, this growth effect of BCR-ABL siRNA could be fully rescued by transfection with BCR-ABL. In addition, BCR-ABL knockdown by siRNA led to increased K562 cell apoptosis, and combined ND-09 and siRNA treatment enhanced this effect (Fig. 4D). These observations suggested that BCR-ABL was a key target for ND-09.
Effect of ND-09 on BCR-ABL
An MD assay was performed to evaluate the affinity characteristics of ND-09 binding to the active site of BCR-ABL. The binding energy of ND-09 with BCR-ABL was −7.07. ND-09 bound to the BCR-ABL ATP-binding pocket through an electrostatic interaction (Fig. 4E-a), hydrogen-bond interaction (Fig. 4E-b), and hydrophobic interaction (Fig. 4E-c). Furthermore, ND-09 may interact with VAL256, ALA269, LEU248, GLU288, VET290, VAL299, ASP381, PHE382, ALA380, LEU370, PHE359, ILE360, ARG362, LEU354, and HIS361 amino acid residues of BCR-ABL (Fig. 4E-d). The docking assay results demonstrated that ND-09 fit well within BCR-ABL.
Accordingly, we examined the inhibitory effect of ND-09 on BCR-ABL kinase activity. ND-09 inhibited BCR-ABL kinase activity with an IC50 value of 0.57 nM (Fig. 4F). Furthermore, ND-09 markedly inhibited the phosphorylation of BCR-ABL and ABL in K562 cells (Fig. 4G). These data indicate that ND-09 could inhibit BCR-ABL phosphorylation and kinase activity.
ND-09 regulates the BCR-ABL downstream signaling pathway
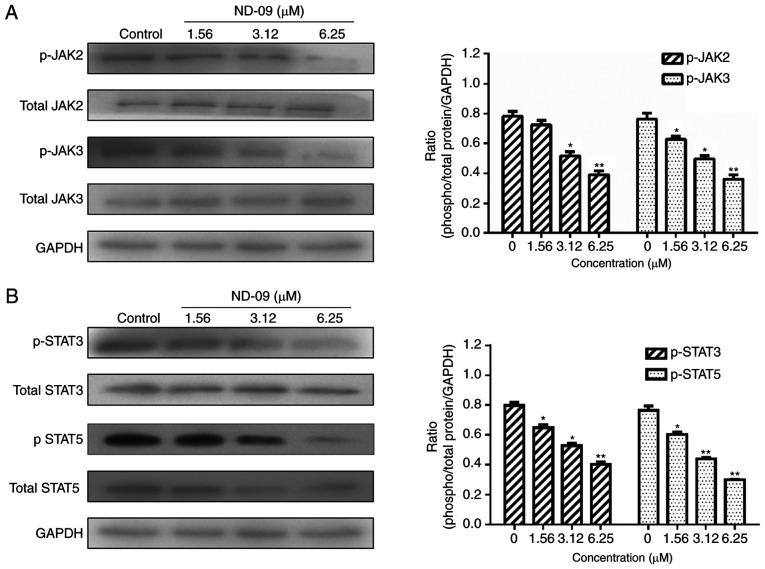
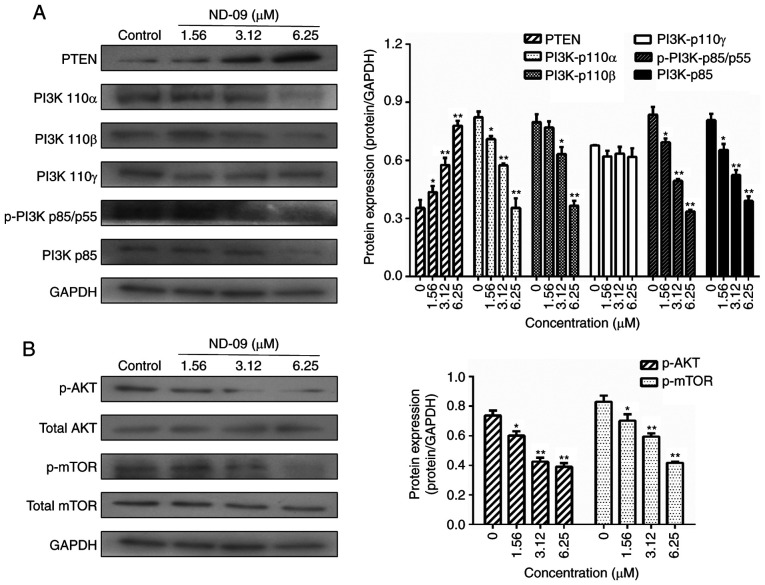
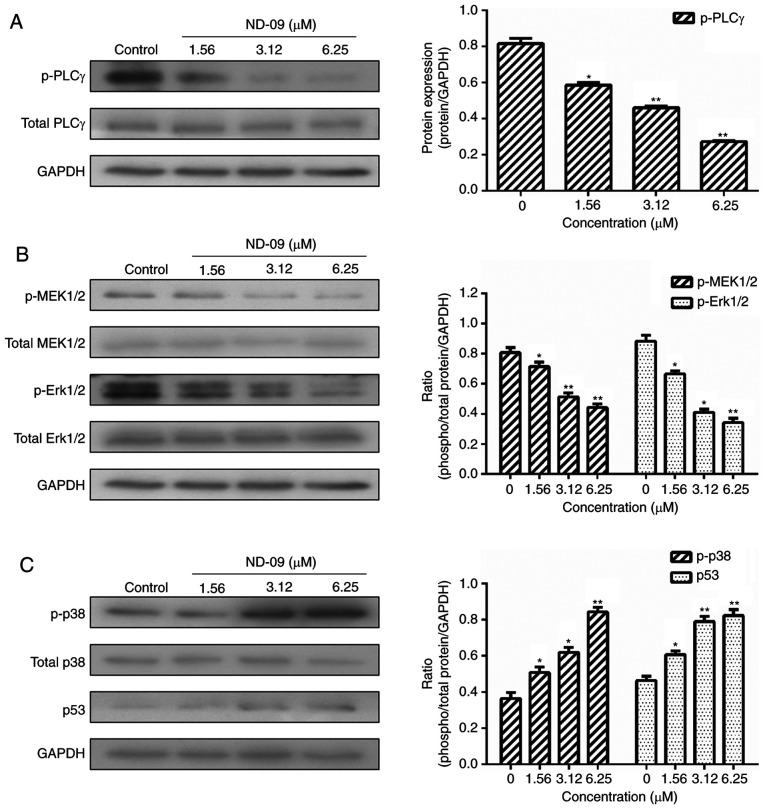
As shown in Fig. 5, ND-09 was found to significantly inhibit the phosphorylation of JAK/STAT signaling pathway members (JAK2, JAK3, STAT3, and STAT5). Furthermore, ND-09 treatment led to an increase in the amount of PTEN and a decrease in the amount of PI3K subunits (110α, 110β, p-p85/p55 and p85), as well as decreased phosphorylation of downstream molecules (AKT and mTOR) in K562 cells (Fig. 6). ND-09 upregulated p-p38 and p53 levels and significantly inhibited the phosphorylation of PLC-γ, which in turn inhibited MEK1/2 and ERK1/2 phosphorylation (Fig. 7).
Figure 5.
Effects of ND-09 on JAK/STAT signaling protein levels. (A) Protein levels of JAK2, p-JAK2, JAK3, and p-JAK3 in K562 cells treated with ND-09 were evaluated by western blot analysis. (B) Protein levels of STAT3, p-STAT3, STAT5, and p-STAT5 in K562 cells treated with ND-09 were evaluated by western blot analysis. Results were quantified by densitometry analysis of the bands and were normalized to GAPDH (internal control). Samples were derived from the same experiment, and blots were processed in parallel. Values represent the average of three independent experiments. Data are presented as mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group.
Figure 6.
Effects of ND-09 on PI3K/AKT signaling protein levels. (A) Protein levels of PTEN, PI3K-p110α, PI3K-p110β, PI3K-p110γ, p-PI3K p85/p55, and PI3K-p85 in K562 cells treated with ND-09 were evaluated by western blot analysis. (B) Protein levels of AKT, p-AKT, mTOR, and p-mTOR in K562 cells treated with ND-09 were evaluated by western blot analysis. Results were quantified by densitometric analysis of the bands and were normalized to GAPDH (internal control). Samples were derived from the same experiment, and blots were processed in parallel. Values represent the average of three independent experiments. Data are presented as mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group.
Figure 7.
Effects of ND-09 on MAPK signaling protein levels. (A) Protein levels of PLCγ and p-PLCγ in K562 cells treated with ND-09 were examined by western blot analysis. (B) Protein levels of MEK1/2, p-MEK1/2, Erk1/2, and p-Erk1/2 in K562 cells treated with ND-09 were examined by western blot analysis. (C) Protein levels of p38, p-p38, and p53 in K562 cells treated with ND-09 were examined by western blot analysis. Results were quantified by densitometric analysis of the bands and were normalized to GAPDH (internal control). Samples were derived from the same experiment, and blots were processed in parallel. The values represent the average of three independent experiments. Data are presented as mean ± SEM (n=3). *P<0.05, **P<0.01 compared to the untreated control group.
Discussion
CML, a myeloproliferative malignancy driven by the constitutively active BCR-ABL1 tyrosine kinase, is currently treated with TKIs that have proven to be clinically effective (17). However, while TKI treatment was initially successful, the emerging development of TKI resistance presents a considerable challenge (18,19). In this study, we developed ND-09, which exhibited potent anticancer activity against CML by targeting BCR-ABL.
K562 cells, which express high levels of BCR-ABL, were most sensitive to ND-09, suggesting that ND-09 potentially inhibits cell proliferation via targeting BCR-ABL (Fig. 1). Furthermore, ND-09 inhibited the growth of K562 cells in a dose- and time-dependent manner. We proceeded to verify whether BCR-ABL is the target of ND-09. The results revealed that combining ND-09 with BCR-ABL siRNA knockdown led to an enhanced inhibitory effect on cell proliferation, and this growth effect of BCR-ABL siRNA could be fully rescued by transfection with BCR-ABL (Fig. 4A-C). Taken together, ND-09 appears to inhibit K562 cell proliferation via targeting BCR-ABL.
As ND-09 targets BCR-ABL to inhibit cell proliferation, MD was used to simulate the binding properties of ND-09 and BCR-ABL. Notably, ND-09 fit well within BCR-ABL and occupied the BCR-ABL ATP-binding pocket (Fig. 4E). As a consequence, ND-09 could alter BCR-ABL kinase activity and inhibit the phosphorylation of BCR-ABL and ABL (Fig. 4F and G). Our data strongly support the hypothesis that ND-09 is a BCR-ABL-specific inhibitor.
Apoptosis is a major mechanism of programmed cell death. Findings have shown that BCR-ABL exerts anti-apoptotic effects that play an important role in the development of CML (20). Furthermore, the intrinsic mitochondrial pathway is one of the two main apoptotic pathways (21). In the current study, ND-09 was found to induce apoptosis in K562 cells, an effect that was relatively higher in K562 cells than in JURKAT and HUT78 cells (Fig. 2A-C). More importantly, combined ND-09 and BCR-ABL siRNA treatment led to an enhanced induction of apoptosis in K562 cells (Fig. 4D), indicating that ND-09 could induce cell apoptosis by targeting BCR-ABL. ND-09 treatment significantly reduced Δψm in K562 cells (Fig. 2D and E). The mitochondrial apoptotic pathway is mainly regulated by the Bcl-2 protein family, which includes the anti-apoptotic members Bad, Bak, and Bax, and pro-apoptotic members Mcl-1 and Bcl-2. Findings have shown that p53 could activate the expression of proapoptotic Bcl-2 proteins, thus triggering apoptosis (12). In the present study, we found that ND-09 could significantly upregulate p53 protein levels (Fig. 7C). Accordingly, ND-09 increased the protein levels of Bad, Bax, and Bak, and downregulated Mcl-1 and Bcl-2 protein levels (Fig. 2F).
Previous findings have shown that the BCR-ABL oncoprotein plays an important role in regulating the cell cycle of cancer cells (22). Furthermore, silencing BCR-ABL causes CML cell cycle arrest in the G0/G1 phase (23,24). Our data revealed that ND-09 treatment arrested K562 cell cycle in the G0/G1 phase (Fig. 3A). As CDK/cyclin proteins play a critical role in cell cycle regulation, we examined whether ND-09 regulated CDK/cyclin protein levels. Results revealed that ND-09 treatment resulted in a decrease in the protein levels of cyclin D1 and cyclin E in K562 cells (Fig. 3D).
As BCR-ABL is a key factor for K562 cell growth, major downstream molecules of the BCR-ABL pathway were evaluated. BCR-ABL activates a number of signaling pathways (JAK/STAT, PI3K/AKT, and MAPK), promotes the proliferation of hematopoietic stem cells, and prevents cell apoptosis (25). Inhibition of JAK/STAT and PI3K/AKT signals can inhibit CML cell growth, indicating JAK/STAT and PI3K/AKT are both involved in BCR-ABL-driven leukemias (26). BCR-ABL reportedly induces Grb2-mediated MAPK activation, which regulates cell proliferation, cell cycle progression, cell survival, differentiation, and induces leukemogenesis (27). Our data revealed that ND-09 could downregulate the phosphorylation of JAK/STAT signaling members, including JAK2, JAK3, STAT3, and STAT5 (Fig. 5). In addition, ND-09 treatment led to an increase in the amount of PTEN. The main PI3K subunit proteins (110α, 110β, p-p85/p55, and p85) and phosphorylated AKT and mTOR were significantly decreased (Fig. 6). Furthermore, ND-09 treatment led to the decreased phosphorylation of PLC-γ and subsequently inhibited the phosphorylation of MEK1/2 and Erk1/2, while upregulating the amount of p-p38 and p53 in K562 cells (Fig. 7). The aforementioned results suggest that ND-09 regulated cell proliferation through modulation of JAK/STAT, PI3K/AKT, and MAPK/ERK signaling.
In conclusion, our study has established ND-09 as a selective inhibitor of BCR-ABL. The findings suggest that BCR-ABL pathway downregulation by ND-09 drives growth arrest in CML.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CML
chronic myeloid leukemia
- BCR
breakpoint cluster region
- ABL
Abelson murine leukemia
- MAPK
mitogen-activated protein kinase
- JAK/STAT
Janus-activated kinase/signal transducer and activator of transcription
- PI3K/AKT
phosphatidylinositide 3-kinase/protein kinase B
- TKIs
tyrosine kinase inhibitors
- IMDM
Iscove's modified Dulbecco's medium
- MTT
3-(4,5-dimethylthiazol-2-yl)-2.5-diphenyl-2H-tetrazolium bromide
- FBS
fetal bovine serum
- PI
propidium iodide
Funding Statement
This study was supported by the Natural Science Basic Research Program of Shaanxi Province (grant no. 2018JQ8019), the Fundamental Research Funds for the Central Universities (grant no. xzy012019078).
Funding
This study was supported by the Natural Science Basic Research Program of Shaanxi Province (grant no. 2018JQ8019), the Fundamental Research Funds for the Central Universities (grant no. xzy012019078).
Availability of data and materials
The authors declare that all the data supporting the results in this study are available within the article.
Authors' contributions
WNM was responsible for the conceptualization, collection of material, and writing and reviewing the manuscript. YHL was responsible for data collection, curation and analysis, as well as writing the original draft. MZ contributed to the curation and interpretation of data. PPL contributed to the design and data analysis, and writing of the manuscript. XYP was responsible for the synthesis of compound ND-09. All authors read and approved the study.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen Y, Wang TT, Du J, Li YC, Wang X, Zhou Y, Yu XX, Fan WM, Zhu QJ, Tong XM, Wang Y. The critical role of PTEN/PI3K/AKT signaling pathway in shikonin-induced apoptosis and proliferation inhibition of chronic myeloid leukemia. Cell Physiol Biochem. 2018;47:981–993. doi: 10.1159/000490142. [DOI] [PubMed] [Google Scholar]
- 2.Kaleem B, Shahab S, Ahmed N, Shamsi TS. Chronic myeloid leukemia-prognostic value of mutations. Asian Pac J Cancer Prev. 2015;16:7415–7423. doi: 10.7314/APJCP.2015.16.17.7415. [DOI] [PubMed] [Google Scholar]
- 3.Yu CJ, Gorantla SP, Müller-Rudorf A, Muller TA, Kreutmair S, Albers C, Jakob L, Lippert LJ, Yue ZY, Engelhardt M, et al. Phosphorylation of BECLIN-1 by BCR-ABL suppresses autophagy in chronic myeloid leukemia. Haematologica. 2020;105:1285–1293. doi: 10.3324/haematol.2018.212027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin XL, Zhou MR, Fu Y, Yang L, Xu M, Sun T, Wang XM, Huang T, Chen CY. Histone demethylase RBP2 mediates the blast crisis of chronic myeloid leukemia through an RBP2/PTEN/BCR-ABL cascade. Cell Signal. 2019;63:109360. doi: 10.1016/j.cellsig.2019.109360. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Serna J, Kucharski C, Chen M, Remant KC, Jiang XY, Brandwein J, Uludag H. siRNA-mediated BCR-ABL silencing in primary chronic myeloid leukemia cells using lipopolymers. J Control Release. 2019;310:141–154. doi: 10.1016/j.jconrel.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Burslem GM, Schultz AR, Bondeson DP, Eide CA, Stevens SL, Druker BJ, Crews CM. Targeting BCR-ABL1 in chronic myeloid leukemia by PROTAC-mediated targeted protein degradation. Cancer Res. 2019;79:4744–4753. doi: 10.1158/0008-5472.CAN-19-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugler J, Kinstrie R, Scott MT, Vetrie D. Epigenetic reprogramming and emerging epigenetic therapies in CML. Front Cell Dev Biol. 2019;7:136. doi: 10.3389/fcell.2019.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rosa V, Monti M, Terlizzi C, Fonti R, Del Vecchio S, Iommelli F. Coordinate modulation of glycolytic enzymes and OXPHOS by imatinib in BCR-ABL driven chronic myelogenous leukemia cells. Int J Mol Sci. 2019;20:3134. doi: 10.3390/ijms20133134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XF, Yang JL, Guo GJ, Feng RY, Chen K, Liao Y, Zhang LF, Sun LP, Huang SL, Chen JL. Novel lncRNA-IUR suppresses Bcr-Abl-induced tumorigenesis through regulation of STAT5-CD71 pathway. Mol Cancer. 2019;18:84. doi: 10.1186/s12943-019-1013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu ZW, Yang C, Wen LL, Liu L, Zuo XB, Zhou FS, Gao JP, Zheng XD, Shi YJ, Zhu CH, et al. Bach2 regulates aberrant activation of B cell in systemic lupus erythematosus and can be negatively regulated by BCR-ABL/PI3K. Exp Cell Res. 2018;365:138–144. doi: 10.1016/j.yexcr.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 11.Kumar H, Chattopadhyay S, Das N, Shree S, Patel D, Mohapatra J, Gurjar A, Kushwaha S, Singh AK, Dubey S, et al. Leprosy drug clofazimine activates peroxisome proliferator-activated receptor-γ and synergizes with imatinib to inhibit chronic myeloid leukemia cells. Haematologica. 2020;105:971–986. doi: 10.3324/haematol.2018.194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter BZ, Mak PY, Mu H, Wang XM, Tao WJ, Mak DH, Dettman EJ, Cardone M, Zernovak O, Seki T, Andreeff M. Combined inhibition of MDM2 and BCR-ABL1 tyrosine kinase targets chronic myeloid leukemia stem/progenitor cells in a murine model. Haematologica. 2020;105:1274–1284. doi: 10.3324/haematol.2019.219261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura M. Therapeutic drug monitoring of imatinib, nilotinib, and dasatinib for patients with chronic myeloid leukemia. Biol Pharm Bull. 2015;38:645–654. doi: 10.1248/bpb.b15-00103. [DOI] [PubMed] [Google Scholar]
- 14.Huguet F, Cayuela JM, Cambier N, Carpentier N, Tindel M, Violet I, Zunic P, Lascaux A, Etienne G, AdheRMC Investigators. Nilotinib efficacy, safety, adherence and impact on quality of life in newly diagnosed patients with chronic myeloid leukaemia in chronic phase: A prospective observational study in daily clinical practice. Br J Haematol. 2019;187:615–626. doi: 10.1111/bjh.16145. [DOI] [PubMed] [Google Scholar]
- 15.Minson AG, Cummins K, Fox L, Costello B, Yeung D, Cleary R, Forsyth C, Tatarczuch M, Burbury K, Motorna O, et al. The natural history of vascular and other complications in patients treated with nilotinib for chronic myeloid leukemia. Blood Adv. 2019;3:1084–1091. doi: 10.1182/bloodadvances.2018028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan X, Wang F, Zhang Y, Gao HP, Hu ZG, Wang SC, Zhang J. Design, synthesis and biological activities of Nilotinib derivates as antitumor agents. Bioorg Med Chem. 2013;21:2527–2534. doi: 10.1016/j.bmc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Braun TP, Eide CA, Druker BJ. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell. 2020;37:530–542. doi: 10.1016/j.ccell.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clapper E, Wang S, Raninga PV, Di Trapani G, Tonissen KF. Cross-talk between Bcr-abl and the thioredoxin system in chronic myeloid leukaemia: Implications for CML treatment. Antioxidants (Basel) 2020;9:207. doi: 10.3390/antiox9030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiti A, Franquiz MJ, Ravandi F, Cortes JE, Jabbour EJ, Sasaki K, Marx K, Daver NG, Kadia TM, Konopleva MY, et al. Venetoclax and BCR-ABL tyrosine kinase inhibitor combinations: Outcome in patients with philadelphia chromosome-positive advanced myeloid leukemias. Acta Haematol. 2020;14:1–7. doi: 10.1159/000506346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H, Kapuria V, Peterson LF, Fang DX, Bornmann WG, Bartholomeusz G, Talpaz M, Donato NJ. Bcr-Abl ubiquitination and Usp9× inhibition block kinase signaling and promote CML cell apoptosis. Blood. 2011;117:3151–3162. doi: 10.1182/blood-2010-03-276477. [DOI] [PubMed] [Google Scholar]
- 21.Zhu M, Yang L, Shi XP, Gong ZY, Yu RZ, Zhang DD, Zhang YM, Ma WN. TPD7 inhibits the growth of cutaneous T cell lymphoma H9 cell through regulating IL-2R signalling pathway. J Cell Mol Med. 2020;24:984–995. doi: 10.1111/jcmm.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 23.Rangatia J, Bonnet D. Transient or long-term silencing of BCR-ABL alone induces cell cycle and proliferation arrest, apoptosis and differentiation. Leukemia. 2006;20:68–76. doi: 10.1038/sj.leu.2403999. [DOI] [PubMed] [Google Scholar]
- 24.Fathi E, Farahzadi R, Valipour B, Sanaat Z. Cytokines secreted from bone marrow derived mesenchymal stem cells promote apoptosis and change cell cycle distribution of K562 cell line as clinical agent in cell transplantation. PLoS One. 2019;14:e0215678. doi: 10.1371/journal.pone.0215678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CW, Lee YL, Liou JP, Liu YH, Liu CW, Chen TY, Huang HM. A novel tubulin polymerization inhibitor, MPT0B206, downregulates Bcr-Abl expression and induces apoptosis in imatinib-sensitive and imatinib-resistant CML cells. Apoptosis. 2016;21:1008–1018. doi: 10.1007/s10495-016-1264-z. [DOI] [PubMed] [Google Scholar]
- 26.Moriyama K, Hori T. BCR-ABL induces tyrosine phosphorylation of YAP leading to expression of Survivin and Cyclin D1 in chronic myeloid leukemia cells. Int J Hematol. 2019;110:591–598. doi: 10.1007/s12185-019-02726-7. [DOI] [PubMed] [Google Scholar]
- 27.Ma L, Xu Z, Wang J, Zhu ZC, Lin GB, Jiang LJ, Lu XZ, Zou C. Matrine inhibits BCR/ABL mediated ERK/MAPK pathway in human leukemia cells. Oncotarget. 2017;8:108880–108889. doi: 10.18632/oncotarget.22353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all the data supporting the results in this study are available within the article.